Abstract
Defining the protein profiles of tissues and organs is critical to understanding the unique characteristics of the various cell types in the human body. In this study, we report on an anatomically comprehensive analysis of 4842 protein profiles in 48 human tissues and 45 human cell lines. A detailed analysis of over 2 million manually annotated, high-resolution, immunohistochemistry-based images showed a high fraction (>65%) of expressed proteins in most cells and tissues, with very few proteins (<2%) detected in any single cell type. Similarly, confocal microscopy in three human cell lines detected expression of more than 70% of the analyzed proteins. Despite this ubiquitous expression, hierarchical clustering analysis, based on global protein expression patterns, shows that the analyzed cells can be still subdivided into groups according to the current concepts of histology and cellular differentiation. This study suggests that tissue specificity is achieved by precise regulation of protein levels in space and time, and that different tissues in the body acquire their unique characteristics by controlling not which proteins are expressed but how much of each is produced.
Keywords: antibody-based analysis, bioimaging, global protein expression, immunofluorescence, immunohistochemistry
Introduction
Recently, a detailed study of 1% of the human genome showed that chromosomes are pervasively transcribed and that the majority of all bases are included in primary transcripts (Birney et al, 2007). This has been recently confirmed by extensive parallel sequencing of transcripts, which has shown that a large fraction (70%) of the predicted 20 400 (Clamp et al, 2007) protein-encoded genes can be detected in a single human cell line (Sultan et al, 2008). In addition, a vast number of alternative splicing events have been identified, adding to the complexity and ubiquitous expression of the human transcriptome (Tress et al, 2007; Wang et al, 2008). This plasticity at the RNA level is even further accentuated by the presence of an immense numbers of inhibitory RNAs (Yelin et al, 2003; Katayama et al, 2005) and the recent discovery that tens of thousands of binding sites are present across the genome, as shown by genome-wide profiles of the DNA binding of mammalian transcription factors (Robertson et al, 2007). The question arises whether this ubiquitous RNA expression is also translated to the protein level and how this relates to central biological questions regarding the link between protein expression profiles and cellular phenotypes, and the divergence of protein levels in differentiated cells from normal and disease tissues.
We have previously described the high-throughput generation of antibodies and subsequent creation of a Human Protein Atlas (http://www.proteinatlas.org) based on tissue microarrays (TMAs), immunohistochemistry, and immunofluorescence (Berglund et al, 2008). In this study, we describe, for the first time, a systematic analysis of global protein expression patterns generated from this public resource. We have examined the spatial distribution and the relative abundance of proteins in the different cell populations of various tissues in all major human tissues and organs, including the brain, liver, kidney, lymphoid tissues, heart, lung, skin, GI tract, pancreas, endocrine tissues, and the reproductive organs. Thus, it has been possible to assess the functional associations between phenotypically different cells and to study the relationship between protein expression profiles and developmental origin.
Results and discussion
Global protein profiling in 65 normal human cell types
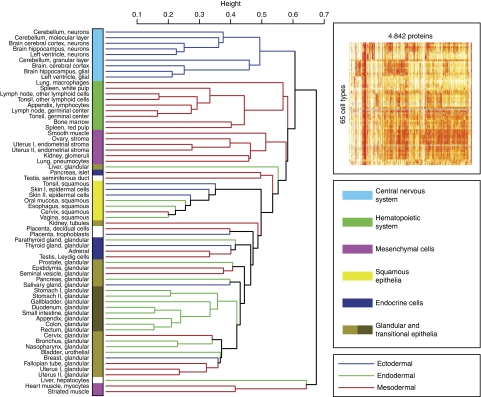
An unsupervised cluster analysis (Eisen et al, 1998) was carried out based on the protein levels in 65 normal cell types. The variability introduced by the individual experimental staining protocol, including the choice of antibody dilution and antigen retrieval methods, was addressed by the use of TMAs (Kononen et al, 1998), thus allowing parallel determination of the relative levels of a particular protein target, within its dynamic range, across hundreds of biosamples (Warford et al, 2004; Taylor and Levenson, 2006). Annotations of more than 2 million images were performed by certified pathologists, and the relative expression level of a particular protein was translated into a four-color code ranging from strong (red), moderate (orange), weak (yellow), and no (white) protein expression (Kampf et al, 2004; Bjorling et al, 2008). It is important to point out that this color code represents the relative expression levels of a particular protein across tissues and organs, but the absolute levels of each protein have not been determined and could vary by many orders of magnitude. Although the level at which it is appropriate to divide cell types into categories is arbitrary, the resulting heat map (Figure 1) shows that the cells cluster into groups that could be expected on the basis of traditional embryology, histology, and anatomy, with most of the cells divided into six major groups: (i) cells of the central nervous system (CNS); (ii) hematopoietic cells; (iii) mesenchymal cells; (iv) cells with squamous differentiation; (v) endocrine cells; and (vi) glandular and transitional epithelial cells. Further subdivision is also evident, as exemplified by (i) separate subgroups containing neuronal and glial cells in the CNS cluster; (ii) subdivision of cells from the male and female genital tracts; and (iii) a distinct subcluster of glandular cells from the GI tract. The liver hepatocytes, together with striated and heart muscle cells (myocytes), have the most divergent protein profiles.
Figure 1.
Global protein profiling in 65 normal human cell types. A dendrogram showing the relationships, based on global expression profiling, between the various cell types. The dendrogram was constructed using a hierarchical clustering model; the inset shows the original heat map. The underlying data are based on manual annotation of protein expression patterns in 65 normal cell types using 5934 antibodies corresponding to 4842 proteins. The dendrogram bars are labeled according to the proposed origin within the embryonic germ cell layers: ectoderm (blue), mesoderm (red), and endoderm (green). The cell types have been classified into six categories according to the color code to the right. A list of all the cell types can be found in Supplementary Table S2. The dendrograms for proteins encoded on single chromosomes are shown in Supplementary Figures S1–S3 and for random sets of 200 antibodies in Supplementary Figures S4–S6.
Sensitivity analyses were also carried out using proteins encoded from single human chromosomes to obtain a random stratification of a substantially smaller subset of the proteome. Similar dendrograms were obtained for the chromosome specificity (Supplementary Figures S1–S3), as well as random groups of 200 antibodies (Supplementary Figures S4–S6), suggesting that the phenotype of the cells is generated by a large fraction of human proteins, as a random sampling of only ∼1% of the protein-encoded genes (200 proteins) are sufficient to group the cells in a nearly identical pattern compared with the whole data set. The dendrogram shows that cells with similar cellular functions exhibit similar protein profiles, as exemplified within the hematopoietic cell cluster, in which germinal center cells and peri-follicular lymphoid cells have a more closely related expression profile than the more distant hematopoietic cells in the bone marrow. Similarly, the myocytes in cardiac and striated muscle have similar expression profiles, and these are distinctly different from smooth muscle cells and other stroma cells in the mesenchymal cell cluster.
Protein profiles and developmental origin of the cells
The similarity in protein profiles often coincides with the putative developmental origin (endoderm, ectoderm, or mesoderm) of cell types, as shown by different color codes for branches of the dendrogram (Figure 1). This can be exemplified by glandular cells in the GI tract, which are derived from the endoderm, cells in the CNS originating from the neuro-ectoderm, and hematopoietic/mesenchymal cells derived from the mesoderm. For certain cell types, morphological differentiation supersedes developmental origin, as exemplified in the cluster of cells with squamous differentiation, in which cells from all three germ layers are represented: surface epithelia of the esophagus (endoderm), epidermal cells from the skin (ectoderm), and surface epithelia from intra-vaginal elements of the cervix (mesoderm). These patterns show that global protein profiles in differentiated normal cells reflect the pluripotent origin of the corresponding stem cells in different germ layers, but that functional convergence also exists resulting in similar expression profiles that are independent of developmental origin.
The tissue-specific protein expression in 65 cell types corresponding to 48 tissues and organs
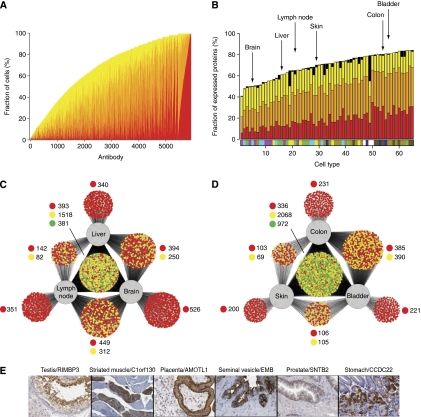
Expression analysis can be used to estimate the relative level of different protein expressions in each cell type. An analysis of the fraction of cell types containing each protein is shown in Figure 2A, with the cells classified into groups exhibiting strong (red), medium (orange), or weak (yellow) expression. The analysis indicates that a large proportion of the proteome is expressed across many of the 65 cell types: 20% (949) of the proteins are found at detectable levels in ⩾60 cell types, whereas only 3% (150) are detected in less than six cell types. A similar analysis was conducted to study the fraction of analyzed proteins (4842) that are detected in each cell type (Figure 2B). This showed that a large fraction of protein-encoding genes are present in any given cell type, with an average of 68% (range 40–84%) of all proteins expressed. The supportive cells are the most specialized, for example, glial cells in the CNS and stroma cells in the endometrium and ovary. In contrast, the study showed that several glandular cells have as many as 80% of the analyzed proteins present at detectable levels, and this raises the question how much the result is influenced by background staining due to nonspecific binding or cross-reactivity to homologous proteins. To explore this issue, a sensitivity analysis was carried out, using various subfractions of the antibodies (see Supplementary Table S1) with the selection based on paired antibodies with high confidence or documented supportive western blots. The analysis showed essentially the same results as when all antibodies were used (Supplementary Figures S7, S8 and Table S2), with an average of 61–71% of the proteins detected across the 65 cell types.
Figure 2.
The tissue-specific protein expression in 65 cell types corresponding to 48 tissues and organs. (A) The fraction (%) of cells in which a particular protein was detected, including the fraction of cells with the relative expression levels strong (red), moderate (orange), and weak (yellow). A total of 5934 antibodies against 4842 proteins are arranged according to abundance of the corresponding protein target with cell-type-specific proteins to the left and ‘housekeeping proteins' to the right. The results for the various subfractions of antibodies are presented in Supplementary Figure S7. (B) The fraction (%) of the analyzed proteins detected in a specific cell type. Cells are arranged according to the fraction of proteins detected. A bar displaying the different color codes representing the six major categories of normal cell types (defined in Figure 1) is shown for each cell type. The name of each cell type is shown in Supplementary Table S2 and the results for the various subfractions of antibodies are presented in Supplementary Figure S8. Black represents missing data, i.e., where there was no representative cell type for a given immunostaining. The six cell types analyzed in C and D are pointed out by arrows. (C) A Cytoscape network plot (Shannon et al, 2003) showing the distribution of the analyzed proteins detected in at least one of the three cell types analyzed; liver hepatocytes, neurons of the cerebral cortex of the brain, and lymphoid cells from the germinal center of the lymph node. Each antibody/protein is represented by a small circle that is connected by a line/lines to the cells it was detected in. The color of the circle indicates the variability of staining intensity between the different cell lines; green indicates that all cell lines belonged to the same staining intensity category, yellow indicates that two cell lines belonged to the same staining intensity category, and red indicates that the staining intensity category was different for all three cell lines or only detected in a single cell. (D) A similar network plot based on the analysis of protein expression in glandular cells in the colon, epidermal cells from the skin, and urothelial cells from the bladder. (E) Six examples of proteins with essentially unknown functions that exhibit cell-type-specific expression. Testis—maturing spermatocytes and spermatids in the testicular seminiferous duct show strong partly membranous positivity with an antibody generated toward the uncharacterized protein RIMS-binding protein 3A. Muscle—striated skeletal muscle is shown with a fiber-type-specific sarcoplasmic positivity with an antibody directed toward an unknown protein encoded by C1orf130. Placenta—the expression of angiomotin-like protein 1 in placental tissue (immature) shows strong membranous positivity in basal cytotrophoblasts with moderate cytoplasmic positivity in syncytiotrophoblasts and exhibits distinct expression in the brush-border membrane. Seminal vesicle—glandular cells in the seminal vesicle were the only cells found to express embigin, a previously unknown protein. Prostate—in the prostate, moderate positivity was found with a membranous expression pattern for beta-2-syntrophin protein (SNTB2), a protein with unknown functions that has been shown to co-purify, with dystrophin, the protein product of the Duchenne muscular dystrophy locus. Stomach—in the stomach mucosa the previously unknown coiled-coil domain-containing protein 22 (CCDC22) was expressed in the parietal cells, producers of hydrochloric acid in response to histamine, acetylcholine, and gastrin.
Differential expression in selected human cell types
A study was performed to explore the difference in protein expression in three cells with distinctly different phenotypes, namely hepatocytes from the liver, neurons from the cerebral cortex of the brain, and lymphoid cells from the germinal center of the lymph nodes. A network analysis (Shannon et al, 2003) showed (Figure 2C) that as much as 90% of the antibodies (n=5138) detect proteins that are expressed in at least one of the three cells and few proteins are detected exclusively in one of the cells, as exemplified by the brain (9%). However, the cells still display a highly differentiated global expression pattern as shown by the fact that only 6% of the proteins are expressed at the same level in all three cell types. We extended this study to explore the protein profiles in three more closely related cell types: glandular cells in the colon, epidermal cells from the skin, and urothelial cells from the bladder. In this case, a smaller fraction (59%) of the antibodies (n=3376) detect proteins in all three cells and a larger fraction (17%) of the proteins were scored with the same expression level in all three cells (Figure 2D). However, considering the fact that these cells share a common epithelial phenotype, it is interesting that a large fraction (74%) of the proteins still have differential expression across the three cell lines. As expected, cells with more similar functions show less differences in global protein expression patterns compared with those with widely different functions. Despite the high fraction of overall expression of proteins, the given examples show that only 6% of expressed proteins are expressed at the same level in widely different cell types compared with 17% in more closely related cell types. The network analysis provides a further insight into the dynamics of cellular phenotypes and functions as a consequence of differences in protein signatures and allows for a novel angle to determine which cell types are most similar (and most dissimilar) irrespective morphological traits and functions.
Analysis of tissue-specific proteins
To identify tissue-specific proteins, we analyzed how many proteins could be detected exclusively in a single cell and this query resulted in a list of 74 proteins (Supplementary Table S4). The list included several previously well-known cell-type-specific proteins, such as insulin, glucagon, IAPP (Langerhans islets), troponins (muscle), PSA, ACPP (prostate), and several CD markers (hematopoietic cells). The analysis also identified a subset of cell-type-specific proteins for which there is no or little information, including proteins exclusively expressed in the testis, skeletal muscle, placenta, seminal vesicle, prostate, and stomach (Figure 2E). Several of these proteins showed a remarkable specificity, with expression in only a subset of the entire annotated cell population, for example, expression in parietal cells of the stomach mucosa and fiber-type-specific expression myocytes. Expanding the query to include similar cell types at different locations, for example, the nine annotated cell populations in the brain, also showed a surprisingly low number (n=30) of proteins exclusively expressed in the brain (data not shown). These results are somewhat surprising considering the numerous reports describing genes expressed in a tissue-specific manner (Saito-Hisaminato et al, 2002), in particular examples of genes exclusively expressed in the brain, such as the KIAA genes (Ishikawa et al, 1997) In summary, our analysis shows a surprisingly low fraction (<2%) of proteins expressed in a single or only few distinct types of cells. The few cell- or tissue-specific proteins that were found are, of course, interesting starting points for further studies and this is facilitated by the fact that all the annotation results and the underlying original images are available as a public resource from the Human Protein Atlas portal.
Global protein expression in 45 human cell lines
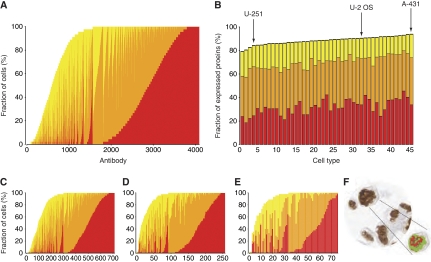
As all the immunohistochemical images from the TMAs were manually annotated by pathologists involving subjective scoring, we decided to carry out the same analysis on 45 human cell lines in which an automated image analysis algorithm have been used (Stromberg et al, 2007; Lundberg et al, 2008). The data from 5349 antibodies corresponding to 4349 genes were analyzed, involving more than 450 000 additional images, and the results are shown in Figure 3. A pattern of protein expression similar to that for tissues and organs was recorded for the in vitro cultured cells, with most proteins expressed in the majority of the 45 cell lines (Figure 3A) and nearly 80% of the proteins expressed across all the analyzed human cell lines (Figure 3B and Supplementary Table S3). A sensitivity analysis using antibodies with supportive western blots (Figure 3C) or paired antibodies with highly correlated expression patterns (Figure 3D and E, and Supplementary Figure S9) produced similar results. An example of the automated image analysis algorithm can be seen in Figure 3F.
Figure 3.
Global protein expression in 45 human cell lines. (A) The fraction (%) of the 45 cell lines in which a particular protein was detected, including the fraction of the three relative expression levels: strong (red), moderate (orange), and weak (yellow). Each bar represents one of the 4096 antibodies with no missing data, i.e., where all cell lines were represented. (B) The fraction (%) of a total number of 5349 antibodies against 4349 proteins detected in a specific cell line, and with the cell lines ordered according to the fraction of proteins detected. The corresponding name and number of each cell line is shown in Supplementary Table S3 and the results for the various subfractions of antibodies are presented in Supplementary Figure S9. The same three staining categories were used and the black (top) part of the bar represents antibodies with missing data for the particular cell line. Arrows point out the three cell lines used in immunofluorescence analysis. (C) The fraction of cell lines in which each protein from a data set of 714 antibodies with supportive results from western blot analysis was detected. (D) A plot similar to C, with each bar representing one of the 257 antibodies remaining from a data set of paired HPA-antibodies, i.e., toward the same target protein, with no missing data for any of the cell lines, and a correlation coefficient of ⩾0.5 when cell line expression profiles were analyzed. (E) Same as D, but displaying only the results from the 75 antibodies with a correlation coefficient of ⩾0.8 and no missing data. (F) An example of cells, visualizing the interpretation of immunostaining by an automated image analysis software.
Global protein expression in cell lines using confocal microscopy
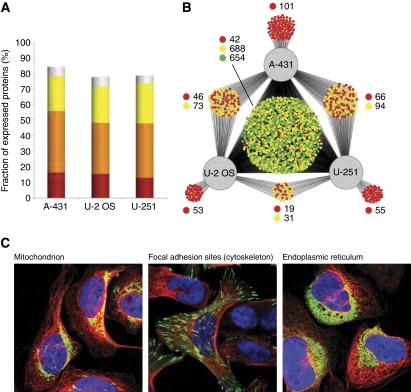
The immunohistochemical analysis, based on an enzyme amplification method, is semiquantitative and we therefore decided to extend the study using immunofluorescence analysis with confocal microscopy. An analysis of three selected human cell lines (Barbe et al, 2008) of epithelial (A-431), glial (U251-MG), and mesenchymal (U-2 OS) origin was carried out for 2,064 proteins (see Figure 4C). More than 70% of the proteins were detected in each of the three cell lines (Figure 4A), even when proteins without a defined subcellular localization (i.e., with weak, granular, and cytoplasmic staining) were excluded. Only 14% of the proteins could not be detected in any of the three cell lines. A plot was generated to show the number of proteins expressed in one, two, or all cell lines (Figure 4B), and the results show that the majority (72%) of the antibodies (n=1384) detect proteins in all three cell lines and that only 11% of the proteins are detected in just one out of the three cell lines. As to the tissue analysis (Figure 2C and D), the majority (66%) of the proteins were expressed at a different level in at least one of the three cell lines.
Figure 4.
Global protein expression in three human cell lines using immunofluorescence-based confocal microscopy. (A) The fraction (%) of proteins detected in the three analyzed cell lines. The stainings are classified into the categories strong (red), moderate (orange), and weak (yellow) based on their measured intensity. Stainings annotated as weak with a granular cytoplasmic subcellular distribution (i.e., not a distinct cytoplasmic organelle or component) are considered less reliable (shown in gray). (B) A Cytoscape network plot (Shannon et al, 2003) showing the distribution of the analyzed proteins detected in at least one of the three cell lines. Each antibody/protein is represented by a small and the color of the circle indicates the variability of staining intensity between the different cell lines; green indicates that all cell lines belonged to the same staining intensity category, yellow indicates that two cell lines belonged to the same staining intensity category, and red indicates that the staining intensity category was different for all three cell lines. (C) Three example images of immunofluorescently stained U-2 OS showing proteins from the different categories (green, yellow, and red) in panel B and with different subcellular localizations. The protein of interest is shown in green, microtubules in red, and nuclei in blue. The first image (left) shows the 60-kDa heat shock protein (HSPD1) to be localized in the mitochondria and give a strong staining intensity in all three cell lines (green category) as detected by the antibody HPA001523. The second image (middle) shows the four and a half LIM domains protein 2 (FHL2) to be localized at focal adhesion sites in the cytoskeleton and give a strong staining intensity in U-2 OS and a moderate staining intensity in A-431 and U-251 MG (yellow category) as detected by the antibody HPA006028. The third image (right) shows the uncharacterized protein KIAA1467 to be localized in the endoplasmic reticulum and give a strong staining intensity in U-2 OS, moderate in A-431, and weak in U-251 MG (red category) as detected by the antibody HPA010803.
Conclusions
Our findings suggest that few proteins are expressed in a cell-type-specific manner, and that the phenotype and function of a cell is determined by localization and fluctuations in concentration of a large portion of the proteome, as opposed to a binary ‘on/off' expression pattern. Although antibody-based assays are sensitive, it is possible that even more sensitive assays, such as the use of sandwich-based analysis (Larsson et al, 2004), could allow the detection of lower levels of proteins, thus showing an even more ubiquitous expression. It is important to point out that the regulation of proteins is also mediated through protein modification, in which certain fractions of the proteins are activated by chemical modification (Olsen et al, 2006) or proteolysis (Yen et al, 2008). The separate functions of a particular cell are, thus, a consequence of local concentrations and modifications of proteins that are carefully regulated to ensure proper functionality in each cell type. For example, the concentrations of a majority of our proteins in a human kidney cell provide an interaction network appropriate for kidney functions (filtration), whereas the protein interaction network in a specific neuron in the brain is targeted toward neurological functions. These data therefore suggest that the phenotype of a particular cell is a consequence of the local concentration of a large portion of the human proteome. This underlines the importance of systems biology approaches (Kislinger et al, 2006) based on quantitative measurements of protein levels (Cox and Mann, 2008) and network predictions of protein interactions (Shlomi et al, 2008) to study mammalian biology. In this context, it would be interesting to add quantitative expression data from complementary technology platforms, such as mass spectrometry (Mann and Kelleher, 2008), to further explore the protein space in individual cells. The analysis could also be integrated with numerous RNA-based expression studies (Kilpinen et al, 2008) to gain further in-depth understanding of the relationship between transcript and protein profiles. To facilitate such bioinformatics comparisons, the expression data used in this analysis are available for downloads at the public Human Protein Atlas portal (http://www.proteinatlas.org/download.php). In conclusion, this study suggests that tissue specificity is achieved by precise regulation of protein levels in space and time, and the results emphasis the need for quantitative systems biology approaches to understand the molecular mechanisms of human biology and diseases.
Materials and methods
Data collection and extraction
To determine the level of protein expression of each protein in this study, antibodies were used to immunohistochemically stain human tissues assembled in TMA blocks (Kononen et al, 1998). Tissue cores with 1 mm diameter, sampled from 144 individuals, corresponding to 48 different normal human tissues types, were included in the study. In addition, a microarray containing human cell lines was assembled (CMA) (Kampf et al, 2004). In addition, the protein levels were estimated for three human cell lines (A-431, U-2OS, and U-251 MG) using immunofluorescence-based confocal microscopy (Barbe et al, 2008). Immunohistochemically stained sections from TMA/CMA blocks were scanned in high-resolution scanners and separated to individual spot images representing each core. For TMAs, all images were evaluated by certified pathologists in a web-based annotation system to collect parameters regarding distribution, the extent and level of protein expression (P Oksvold and E Björling, unpublished results). Parameters from the annotation included staining intensity, fraction of stained cells in a defined cell population, and subcellular localization of staining. The annotation was performed for selected cell types for each tissue, as most tissue types include several defined cell phenotypes, e.g., neurons and glial cells in brain tissue and glomeruli and tubules in the kidney (Bjorling et al, 2008). CMAs were evaluated using automated image analysis (Stromberg et al, 2007) where five output parameters were combined to calculate a score for the protein expression level. For immunofluorescence images, subcellular localizations were annotated and the relative expression levels were classified as strong, moderate, weak, or negative based on the employed laser power and detector gain settings.
In total, 5934 antibodies against 4842 proteins with 298 annotations were assembled from TMA measurements and 5349 antibodies against 4349 proteins with 45 annotations from CMAs. The annotation parameters for intensity and quantity (fraction of positively stained cells) were combined into a four-grade scale represented by the colors white (negative), yellow (weak), orange (moderate), and red (strong) level of protein expression. All data are presented in this format on the protein atlas (http://www.proteinatlas.org). For statistical analysis regarding protein expression, the color codes representing the staining levels were converted to numerical values using a red to 4, orange to 3, yellow to 2 and white to 1 transformation. In cases where the protein expression value could not be derived, because of low image quality, a not available (NA) value was introduced. These data were ordered into a matrix with m (number of antibodies) × n (number of tissues (n=65) or cell lines (n=45)) dimensions. The number of tissues is a combined tissue and cell type parameter, where the number of tissues and cell types give rise to × the number of tissues cell type parameters. In all, two matrices were constructed; one matrix contains protein expression data from human normal tissues and a second matrix contains protein expression data from human cell lines. The number of antibodies in the two matrices and all subsets used in the different figures is presented in Supplementary Table 1.
Hierarchical clustering
For the normal tissue data set, two correlation matrices based on Spearman's ρ were calculated for two dimensions (m × m and n × n, respectively) (Spearman, 1987). The correlation matrices were converted to a distance metric using a 1−correlation value transformation. These data were clustered using unsupervised top-down hierarchical clustering (Eisen et al, 1998; Golub et al, 1999), where at each stage the distances between clusters are recomputed by the Lance–Williams dissimilarity update formula according to average linkage. The algorithm consistently sorted the tighter cluster in each division to the left in the resulting dendrogram representing the hierarchical cluster output. The antibodies with no defined correlation due to constant expression across all tissues or cell lines were removed in the clustering procedure.
Statistical analysis
To estimate protein expression values for each protein across all tissues and cell lines, the different intensity categories (weak, moderate, and strong) were added as separate units into a marginal distribution, which constitutes of 4 × the number antibodies values. The marginal distribution can be seen as a proxy for the total expression level for the respective protein across the 65 tissues and cell types used in this study. A similar procedure was conducted for the protein expression for each tissue and cell line across all antibodies, resulting in a marginal distribution of 4 × the number of tissues or cell lines values.
Validity estimation of dendrograms and marginal distributions
In order to investigate the quality and the conclusions made from data, different subsets were constructed and used in the same analysis approaches as the full data set. For the hierarchical clustering, three additional data sets were constructed, where the subsets were selected based on chromosomal appurtenance using 215, 203, and 206 antibodies representing proteins from chromosome 9, 10, and 22 respectively. The similarity between the dendrograms generated with the three subsets and the dendrogram using all antibodies were investigated using cophenetic correlation coefficients (Sokal and Rohlf, 1962). To estimate the reliability of the marginal distributions, different subsets of antibodies were chosen (Supplementary Table S1). One of the subsets was chosen based on western blot data, where the expected size of the protein matches the correct band from a western blot gel image. Two additional subsets were built based on correlation analysis of paired antibodies, where the two antibodies in a pair are generated to different parts of the same protein. For each antibody pair, a correlation coefficient was calculated using Spearman's ρ. A cutoff value of 0.5 was applied to construct subsets used for the tissue and cell type data set and for the human cell lines. A cutoff of 0.8 was implemented to construct an additional data set for the human cell lines. To estimate the similarities of the sub sets and the full data set, a χ2 test statistic was used on the marginal distributions.
Network analysis
In order to visualize the protein expression overlap between different tissues or cell lines, Cytoscape (Shannon et al, 2003), a software package for analyzing biomolecular interaction networks, was used. The resulting images contain schemes that indicate the overlap of different proteins across different sets of cell types. The first combination was hepatocytes from the liver, neurons from the cerebral cortex of the brain, and lymphoid cells from the germinal center of the lymph nodes, where all three cell types have distinctly different phenotypes. The second combination consisted of three more closely related cell types, namely glandular cells in the colon, epidermal cells from the skin, and urothelial cells from the bladder. The third group of cells consisted of the three cell lines used for immunofluorescence analysis, namely A-431, U-2OS, and U-251 MG. Antibodies with missing protein expression data for any of these cell types were removed from the analysis. This resulted in three different sets of 5710, 5700, and 2250 antibodies, respectively. In each of the resulting networks, a node represents an antibody with a specific protein expression profile, and the edges connect the node to the cell(s) where certain protein is expressed, generating nodes with a degree (number of edges) of 1, 2, or 3. Thus, only antibodies corresponding to proteins expressed in the analyzed cell types are parts of the network. The total number of nodes in the three networks was 5138, 5186, and 1921. Each node was colored according to the variability of staining intensity between the analyzed cell types. Red nodes indicate different staining categories for the three cell types, yellow nodes indicate two cell types in the same staining category, and green nodes indicate that all three cell types belong to the same staining category and are therefore only found for nodes with a degree of three.
Supplementary Material
Supplementary figures S1-9, Supplementary table S1 and Supplementary information regarding tables S2-S4
Supplementary Table S2
Supplementary Table S3
Supplementary Table S4
Acknowledgments
This work was supported by grants from the Knut and Alice Wallenberg Foundation, the Swedish Research Council (VR), and the EU 7th Framework Program Prospects. We are grateful to Claes Wilhelmsson, Kenneth Nilsson, and Per-Åke Nygren for useful comments and advice.
Footnotes
The authors declare that they have no conflict of interest.
References
- Barbe L, Lundberg E, Oksvold P, Stenius A, Lewin E, Bjorling E, Asplund A, Ponten F, Brismar H, Uhlen M, Andersson-Svahn H (2008) Toward a confocal subcellular atlas of the human proteome. Mol Cell Proteomics 7: 499–508 [DOI] [PubMed] [Google Scholar]
- Berglund L, Bjorling E, Oksvold P, Fagerberg L, Asplund A, Szigyarto CA, Persson A, Ottosson J, Wernerus H, Nilsson P, Lundberg E, Sivertsson A, Navani S, Wester K, Kampf C, Hober S, Ponten F, Uhlen M (2008) A genecentric Human Protein Atlas for expression profiles based on antibodies. Mol Cell Proteomics 7: 2019–2027 [DOI] [PubMed] [Google Scholar]
- Birney E, Stamatoyannopoulos JA, Dutta A, Guigo R, Gingeras TR, Margulies EH, Weng Z, Snyder M, Dermitzakis ET, Thurman RE, Kuehn MS, Taylor CM, Neph S, Koch CM, Asthana S, Malhotra A, Adzhubei I, Greenbaum JA, Andrews RM, Flicek P et al. (2007) Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature 447: 799–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjorling E, Lindskog C, Oksvold P, Linne J, Kampf C, Hober S, Uhlen M, Ponten F (2008) A web-based tool for in silico biomarker discovery based on tissue-specific protein profiles in normal and cancer tissues. Mol Cell Proteomics 7: 825–844 [DOI] [PubMed] [Google Scholar]
- Clamp M, Fry B, Kamal M, Xie X, Cuff J, Lin MF, Kellis M, Lindblad-Toh K, Lander ES (2007) Distinguishing protein-coding and noncoding genes in the human genome. Proc Natl Acad Sci USA 104: 19428–19433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J, Mann M (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol 26: 1367–1372 [DOI] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D (1998) Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA 95: 14863–14868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golub TR, Slonim DK, Tamayo P, Huard C, Gaasenbeek M, Mesirov JP, Coller H, Loh ML, Downing JR, Caligiuri MA, Bloomfield CD, Lander ES (1999) Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science 286: 531–537 [DOI] [PubMed] [Google Scholar]
- Ishikawa K, Nagase T, Nakajima D, Seki N, Ohira M, Miyajima N, Tanaka A, Kotani H, Nomura N, Ohara O (1997) Prediction of the coding sequences of unidentified human genes. VIII. 78 new cDNA clones from brain which code for large proteins in vitro. DNA Res 4: 307–313 [DOI] [PubMed] [Google Scholar]
- Kampf C, Andersson AC, Wester K, Bjorling E, Uhlen M, Ponten F (2004) Antibody-based tissue profiling as a tool for clinical proteomics. Clin Proteomics 1: 285–299 [Google Scholar]
- Katayama S, Tomaru Y, Kasukawa T, Waki K, Nakanishi M, Nakamura M, Nishida H, Yap CC, Suzuki M, Kawai J, Suzuki H, Carninci P, Hayashizaki Y, Wells C, Frith M, Ravasi T, Pang KC, Hallinan J, Mattick J, Hume DA et al. (2005) Antisense transcription in the mammalian transcriptome. Science 309: 1564–1566 [DOI] [PubMed] [Google Scholar]
- Kilpinen S, Autio R, Ojala K, Iljin K, Bucher E, Sara H, Pisto T, Saarela M, Skotheim RI, Bjorkman M, Mpindi JP, Haapa-Paananen S, Vainio P, Edgren H, Wolf M, Astola J, Nees M, Hautaniemi S, Kallioniemi O (2008) Systematic bioinformatic analysis of expression levels of 17,330 human genes across 9,783 samples from 175 types of healthy and pathological tissues. Genome Biol 9: R139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kislinger T, Cox B, Kannan A, Chung C, Hu P, Ignatchenko A, Scott MS, Gramolini AO, Morris Q, Hallett MT, Rossant J, Hughes TR, Frey B, Emili A (2006) Global survey of organ and organelle protein expression in mouse: combined proteomic and transcriptomic profiling. Cell 125: 173–186 [DOI] [PubMed] [Google Scholar]
- Kononen J, Bubendorf L, Kallioniemi A, Barlund M, Schraml P, Leighton S, Torhorst J, Mihatsch MJ, Sauter G, Kallioniemi OP (1998) Tissue microarrays for high-throughput molecular profiling of tumor specimens. Nat Med 4: 844–847 [DOI] [PubMed] [Google Scholar]
- Larsson C, Koch J, Nygren A, Janssen G, Raap AK, Landegren U, Nilsson M (2004) In situ genotyping individual DNA molecules by target-primed rolling-circle amplification of padlock probes. Nat Methods 1: 227–232 [DOI] [PubMed] [Google Scholar]
- Lundberg E, Gry M, Oksvold P, Kononen J, Andersson-Svahn H, Ponten F, Uhlen M, Asplund A (2008) The correlation between cellular size and protein expression levels—normalization for global protein profiling. J Proteomics 71: 448–460 [DOI] [PubMed] [Google Scholar]
- Mann M, Kelleher NL (2008) Precision proteomics: the case for high resolution and high mass accuracy. Proc Natl Acad Sci USA 105: 18132–18138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen JV, Blagoev B, Gnad F, Macek B, Kumar C, Mortensen P, Mann M (2006) Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell 127: 635–648 [DOI] [PubMed] [Google Scholar]
- Robertson G, Hirst M, Bainbridge M, Bilenky M, Zhao Y, Zeng T, Euskirchen G, Bernier B, Varhol R, Delaney A, Thiessen N, Griffith OL, He A, Marra M, Snyder M, Jones S (2007) Genome-wide profiles of STAT1 DNA association using chromatin immunoprecipitation and massively parallel sequencing. Nat Methods 4: 651–657 [DOI] [PubMed] [Google Scholar]
- Saito-Hisaminato A, Katagiri T, Kakiuchi S, Nakamura T, Tsunoda T, Nakamura Y (2002) Genome-wide profiling of gene expression in 29 normal human tissues with a cDNA microarray. DNA Res 9: 35–45 [DOI] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13: 2498–2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shlomi T, Cabili MN, Herrgard MJ, Palsson BO, Ruppin E (2008) Network-based prediction of human tissue-specific metabolism. Nat Biotechnol 26: 1003–1010 [DOI] [PubMed] [Google Scholar]
- Sokal R, Rohlf J (1962) The comparison of dendrograms by objective methods. Taxon 11: 33–40 [Google Scholar]
- Spearman C (1987) The proof and measurement of association between two things. By C. Spearman, 1904. Am J Psychol 100: 441–471 [PubMed] [Google Scholar]
- Stromberg S, Bjorklund MG, Asplund C, Skollermo A, Persson A, Wester K, Kampf C, Nilsson P, Andersson AC, Uhlen M, Kononen J, Ponten F, Asplund A (2007) A high-throughput strategy for protein profiling in cell microarrays using automated image analysis. Proteomics 7: 2142–2150 [DOI] [PubMed] [Google Scholar]
- Sultan M, Schulz MH, Richard H, Magen A, Klingenhoff A, Scherf M, Seifert M, Borodina T, Soldatov A, Parkhomchuk D, Schmidt D, O′Keeffe S, Haas S, Vingron M, Lehrach H, Yaspo ML (2008) A global view of gene activity and alternative splicing by deep sequencing of the human transcriptome. Science 321: 956–960 [DOI] [PubMed] [Google Scholar]
- Taylor CR, Levenson RM (2006) Quantification of immunohistochemistry—issues concerning methods, utility and semiquantitative assessment II. Histopathology 49: 411–424 [DOI] [PubMed] [Google Scholar]
- Tress ML, Martelli PL, Frankish A, Reeves GA, Wesselink JJ, Yeats C, Olason PL, Albrecht M, Hegyi H, Giorgetti A, Raimondo D, Lagarde J, Laskowski RA, Lopez G, Sadowski MI, Watson JD, Fariselli P, Rossi I, Nagy A, Kai W et al. (2007) The implications of alternative splicing in the ENCODE protein complement. Proc Natl Acad Sci USA 104: 5495–5500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB (2008) Alternative isoform regulation in human tissue transcriptomes. Nature 456: 470–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warford A, Howat W, McCafferty J (2004) Expression profiling by high-throughput immunohistochemistry. J Immunol Methods 290: 81–92 [DOI] [PubMed] [Google Scholar]
- Yelin R, Dahary D, Sorek R, Levanon EY, Goldstein O, Shoshan A, Diber A, Biton S, Tamir Y, Khosravi R, Nemzer S, Pinner E, Walach S, Bernstein J, Savitsky K, Rotman G (2003) Widespread occurrence of antisense transcription in the human genome. Nat Biotechnol 21: 379–386 [DOI] [PubMed] [Google Scholar]
- Yen HC, Xu Q, Chou DM, Zhao Z, Elledge SJ (2008) Global protein stability profiling in mammalian cells. Science 322: 918–923 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figures S1-9, Supplementary table S1 and Supplementary information regarding tables S2-S4
Supplementary Table S2
Supplementary Table S3
Supplementary Table S4