Summary
Stem cells exhibit promise in numerous areas of regenerative medicine. Their fate and function are governed by a combination of intrinsic determinants and signals from the local microenvironment, or niche. An understanding of the mechanisms underlying both embryonic and adult stem cell function has been greatly enhanced by the recent development of several high-throughput technologies: microfabricated platforms, including cellular microarrays, to investigate the combinatorial effects of microenvironmental stimuli; and large-scale screens utilizing small molecules and short interfering RNAs to identify critical genetic and signaling elements. Furthermore, the integration of these systems with other versatile platforms, such as microfluidics and lentiviral microarrays, will continue to enable the detailed elucidation of stem cell processes, and thus, greatly contribute to the development of stem cell-based therapies.
Introduction
Stem cells are uniquely positioned at the foundation of potential regenerative medicine therapies due to their distinctive ability to undergo self-renewal combined with the capacity to generate numerous differentiated cell types, including progenitor and effector cell populations. Extensive recent work has begun to delineate the genomic and proteomic signatures underlying the self-renewal and pluripotency of embryonic stem (ES) cells as well as the multipotency of various adult stem cell populations, and the identification within these expression profiles specific interaction and regulatory networks [1,2,3]. Such networks and their associated functional phenotypes are defined and regulated through the complex interplay of intrinsic properties and signals from the stem cell microenvironment (Figure 1). For example, adult or somatic stem cells, which contribute to both tissue formation in development and regeneration in adult life, have been demonstrated to reside within specialized niches which modulate stem cell proliferation, influence symmetric versus asymmetric division, control differentiation, and protect stem cells from physiologic stresses [4•,5]. The components of the stem cell microenvironment that regulate these processes include distinct cell-cell and cell-extracellular matrix (ECM) interactions, localized soluble stimuli and gradients of soluble factors, and the three-dimensional architecture of the niche itself, which shapes and restricts the delivery of these cues. A detailed understanding of the cooperative involvement of these diverse environmental interactions together with knowledge of stem cell genetic programs will be critical for the development of new stem cell-based therapeutic approaches, including transplantation and tissue engineering schemes, stem cell-targeted pharmaceuticals, and gene delivery strategies. Thus, to systematically probe mechanisms of stem cell function, platforms in which stem cells can be evaluated in a high-throughput manner have begun to be developed. Here, we will provide examples of recent efforts utilizing such strategies to identify microenvironmental factors and signaling pathways important in stem cell differentiation as well as highlight some other newly developed systems which should be extremely useful within the context of stem cell studies in the near future.
Figure 1.
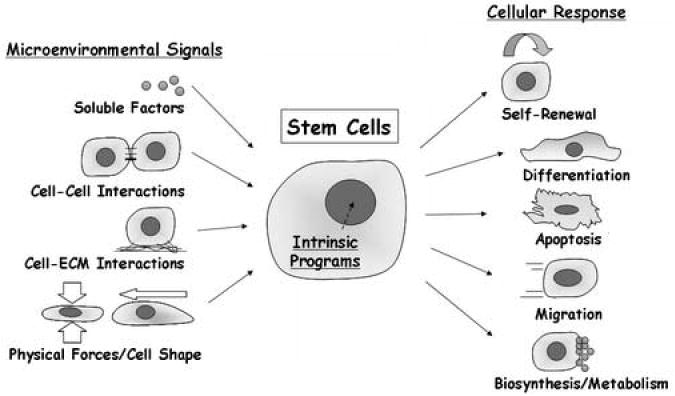
Stem cell fate and function are regulated by a combination of intrinsic programs and signals from the microenvironment. Intrinsic determinants can consist of both genetic and epigenetic components. For example, the molecular mechanisms underlying embryonic stem cell pluripotency have begun to be determined, including transcriptional regulatory networks initiated by the expression of Oct4, Sox2, and Nanog, as well as the expression of Polycomb group proteins and distinct chromatin dynamics [1]. In addition, the importance of environmental signals in stem cell function has been highlighted by the identification of distinct stem cell niches in a wide range of organ systems [4•,5]. Overall, high-throughput analysis of stem cells, utilizing both controlled cellular microenvironments and perturbations of intrinsic elements, can provide substantial insight into the factors governing stem cell biology.
Microfabricated culture platforms for the high-throughput analysis of microenvironmental factors
In order to decouple the complex spatiotemporal cues that cells experience in vivo, microfabrication tools have been applied to in vitro cell culture models, and have found great utility [6]. One approach, termed micropatterning, which has been reviewed extensively elsewhere [7,8], has enabled the generation of patterned heterogeneous surfaces in which cell-cell interactions, cell-matrix interactions, and cell shape can be controlled on the micrometer scale. Studies using this methodology have examined, for instance, the role of homotypic and heterotypic interactions in hepatocyte stabilization within hepatocyte/non-parenchymal cell co-cultures [9], as well as the relative contribution of cell spreading and cell-cell contact in various cellular responses [10,11,12,13]. In particular, the degree of spreading has been demonstrated to modulate endothelial cell proliferation versus apoptosis [10], and the differentiation of human mesenchymal stem cells towards either the osteoblast or adipocyte lineage [11]. In another system, a micropatterned protein surface, patterned on the subcellular length scale, has been shown to influence immunological synapse formation and T cell activation [14]. Collectively, these examples underscore the usefulness of microfabrication approaches for examining biological processes through the highly controlled regulation of environmental signals that these systems afford. Another key feature of microfabrication tools, broadly applicable to numerous cell types including stem cells, is the capacity to miniaturize cell culture platforms for parallel analysis. These high-throughput systems enable the systematic screening of cellular processes on a large scale including an ability to examine the effects of combinations of extracellular signals. One such system recently applied to explicitly investigate microenvironmental regulation of stem cell differentiation is cell microarrays.
Cell microarrays consist of printed spots of biomolecules onto which cells are seeded [15,16]. These spots normally include adhesive factors to retain the seeded cells, as well as other elements for influencing cellular function or detection of specific cellular processes. However, in addition to simply serving a capture role, it is becoming increasingly clear that certain adhesive factors, such as ECM molecules, can play an important part in cellular function through the binding of integrin receptors [17]. Thus, specifically investigating the effect of combinations of these factors in an array context is of substantial interest. Notably, the ECM components of stem cell niches have been suggested to be involved in retaining stem cells within the niche and regulating stem cell signaling and proliferation [18,19,20,21,22]. As a means to analyze cellular interactions with combinatorial mixtures of ECM molecules, an ECM microarray platform was developed [23••]. Utilizing this system, the differentiation of ES cells containing a β-galactosidase reporter for the fetal liver-specific gene Ankrd17 was assessed in the presence of 32 different combinations of collagen I, collagen III, collagen IV, laminin, and fibronectin (Figure 2). An approximately 140-fold difference in β-galactosidase signal was observed between the least and most efficient conditions, suggesting that environmental matrix composition can influence early hepatic lineage specification. In addition, since soluble factors are also important components of the stem cell microenvironment and soluble factor/ECM crosstalk has been suggested in many settings [24,25], our group has recently extended this matrix array platform into a multiwell format with a 96-well plate footprint to simultaneously investigate stem cell differentiation in 1200 parallel experiments representing 240 unique soluble factor/ECM environments (CJ Flaim et al., unpublished).
Figure 2.
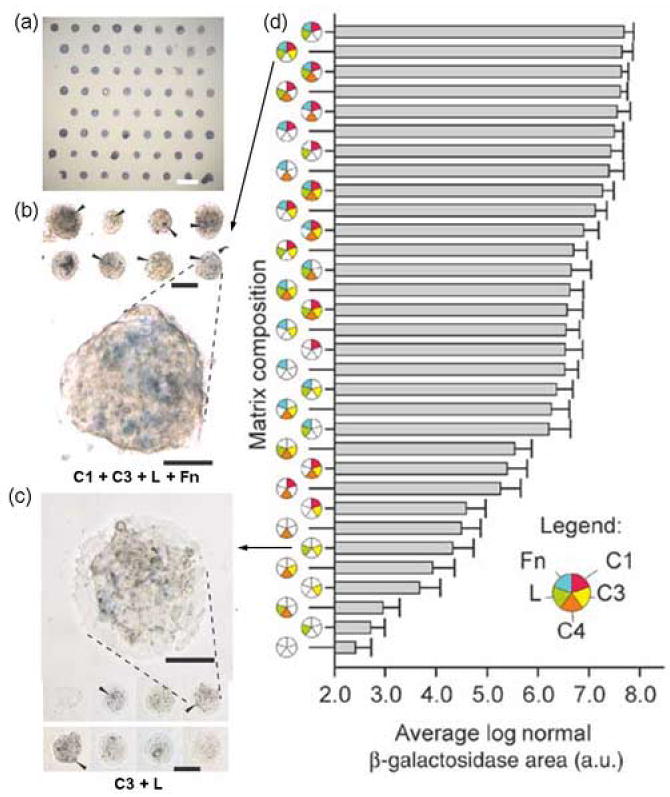
Extracellular matrix (ECM) microarray utilized to investigate embryonic stem cell differentiation towards an early hepatic lineage. (a) Alkaline phosphatase staining of day 1 ES cultures on ECM microarrays (scale bar, 1 mm). (b,c) Bright-field micrograph of selected X-gal–stained conditions after 3 d of culture in retinoic acid. Collagen I (C1) + collagen III (C3) + laminin (L) + fibronectin (Fn) (b) induced higher reporter activity (arrowheads) for Ankrd17, a fetal liver-specific gene, than was seen in cells cultured on C3 + L (c). Scale bars, 250 μm. Magnified views of reporter activity: scale bars, 50 μm. (d) Hierarchical depiction of ‘blue’ image area (pooled data from four microarrays) for each of the matrix mixtures. Error bars, s.e.m. (n = 32). The C1 + C3 + L + Fn culture condition induced –27-fold more reporter positive image area than the C3 + L cultures. (Figure adapted from [23••] with permission).
Another approach for exploring stem cell differentiation with arrays of signaling molecules was demonstrated by Soen et al. [26••], and utilized printed combinations of ECM molecules, growth factors, and other signaling proteins. In this study, the proliferation and differentiation of human neural precursor cells towards a neuronal or glial fate was examined by quantitative image analysis in response to these various exogenous stimuli (Figure 3). Some of the notable findings from this work include a dose responsive role of the Notch ligand, Jagged-1, in shifting differentiation towards the glial fate, and the observation that co-stimulation of the Notch and Wnt signaling pathways resulted in the proliferation of cells exhibiting undifferentiated characteristics. Also, the presence of bone morphogenetic protein 4 induced the acquisition of a hybrid phenotype, with cells expressing markers of both neurons and glial cells. One important benefit of the wealth of data obtained from high-throughput cellular arrays, which was particularly highlighted in this work, is the ability to begin to dissect mechanistically the responses of cells to complex environmental stimuli, including conflicting signals. For example, the presence of Jagged-1 appeared to exhibit dominance over other factors in determining differentiation direction but not in the response to mitogenic stimuli.
Figure 3.
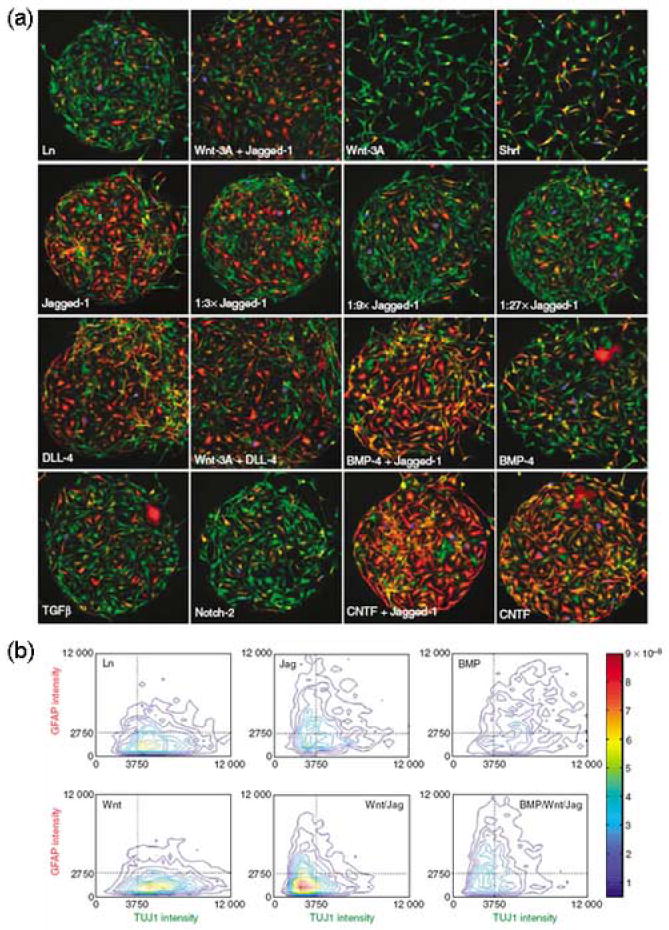
Differentiation of neural precursor cells on a spotted array of microenvironmental signals. (a) Following 70 h culture on an array containing laminin (Ln) alone or Ln in combination with various other indicated factors, cells were stained for markers of proliferation (BrdU, blue), glial differentiation (GFAP, red), and neuronal differentiation (TUJ1, green). (b) High-resolution imaging of multiple parameters enabled the quantification of the differentiation status of single cells. Contour plots of the probability density of cells in response to a selection of stimuli are shown. BMP-4 or BMP (bone morphogenetic protein 4), CNTF (ciliary neurotrophic factor), DLL-4 (delta-like protein 4), Jag (Jagged-1), Shh (sonic hedgehog), TGFβ (transforming growth factor β), Wnt (Wnt-3a). (Figure adapted from [26••] with permission).
In contrast to the ECM array developed by Flaim et al. [23••], which employed a polyacrylamide hydrogel surface, the array system described by Soen et al. [26••] utilized aldehyde-derivatized glass substrates for biomolecule retention. In another study, an array of growth factor and ECM molecule combinations for neural stem cell culture was generated using a technique based on patterned chemically active self-assembled monolayers [27]. Due to the fact that mode of presentation can modulate the function of some ligands [28,29], and mechanical cues have been implicated in stem cell differentiation [11,30], systems that incorporate surface chemistry or material modifications to systematically explore these additional issues will likely be key extensions of cellular array platforms in the future. Precedence for such material-based systems has been demonstrated by Anderson et al. [31], which described a synthetic polymer array consisting of 1700 cell-biomaterial interactions that was utilized to identify biomaterial compositions influencing human ES cell attachment, growth, and differentiation.
Additionally included among the important components of stem cell niches in numerous organ systems are the distinct cell-cell interactions that can exist within these specialized environments [32,33,34]. Similar to the methodologies described for purified proteins, approaches that would enable a high-throughput analysis of cell-cell signaling could provide important clues towards a more thorough understanding of microenvironmental regulation of cellular function. For example, regarding in vitro ES cell differentiation, the significance of cell-cell contacts is demonstrated by the important role of heterogeneous aggregates termed embryoid bodies in the efficient differentiation of these cells towards certain lineages [35]. In order to better dictate cell-cell interactions in ES cell culture, schemes have utilized the fabrication of microwell substrates, both for the generation of controlled size embryoid bodies for differentiation [36], and controlled size aggregates of undifferentiated human ES cells in expansion cultures [37,38]. Scale-up of these platforms and integration with systems to control additional environmental stimuli should provide substantial information regarding ES cell functions.
One approach that is clearly amenable to high-throughput development is microfluidics [39]. Microfluidic channels are normally formed through the casting of polydimethylsiloxane (PDMS), a biocompatible silicone rubber, on a micropatterned photoresist (a photosensitive polymer). These microscale channels can be utilized for cell culture and enable a large degree of control, both spatially and temporally, over the delivery of nutrients and other soluble mediators. For example, a recent study utilized a microfluidic device to analyze in parallel ES cell culture in channels with a logarithmic range of flow rates and demonstrated that flow rate can significantly influence colony formation [40]. Independent of flow rate, microfluidic systems have also been used to generate culture models exhibiting a range of oxygen concentrations [41], as well as growth factor and chemotactic gradients [42,43].
In addition, incorporation of hydrodynamic traps within microfluidic channels has been utilized as a means to create an ordered individual cell array [44]. Recently, there has been a growing interest in examining the functional characteristics of single cells as opposed to often heterogeneous bulk populations. This is particularly true for stem and progenitor cells for which clonal analysis is the most rigorous assessment of cellular potential [45]. Specifically, experiments examining the fate of single cells have proven to be critical in examining stem cell self-renewal capacity, lineage restriction, and in the identification of factors influencing proliferation and differentiation [46,47,48,49]. However, the analysis of individual stem cells within standard (96- or 384-well) multiwell formats can be inefficient and time consuming, and may not provide the necessary number of data points required in some studies. In order to examine clonal stem cell function in a high-throughput manner, microfabrication tools were used to create an array of approximately 10,000 microwells (Figure 4) [50]. The well dimensions were configurable, ranging from 10-500 μm in height and 20 to > 500 μm in diameter. This system was exploited to analyze the proliferation dynamics of adult neural progenitor cells, and confirmed the heterogeneous nature of this proliferation response [51]. Collectively, platforms that enable high-throughput analysis of individual stem and progenitor cells will provide key insights into the differentiation potential of prospectively isolated subpopulations, possible stochastic variations, and the further examination of microenvironmental regulation of stem cell functions.
Figure 4.
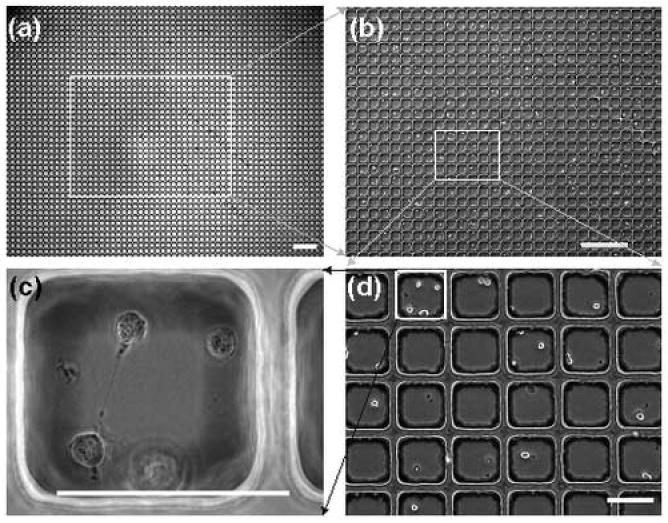
Microwell platform for examining stem cell fates. (a) A low magnification image to illustrate the scale of the system. (b) A higher magnification image of (a) in which distinct cells can be seen. (c) A high numerical aperture (NA) image of a single well. This image was taken with an oil immersion 100x objective to demonstrate the compatibility with high NA objectives. (d) Higher magnification image of an area outlined in white in (b). Scale bars, a-b (500 μm); c-d (100 μm). (Figure adapted from [50] with permission).
Notwithstanding the critical role of environmental cues in stem cell function, these complex extracellular signals interact with and are interpreted by cell intrinsic networks that can significantly influence responses. In the next section, we discuss high-throughput approaches for identifying and modulating intrinsic cellular properties utilizing technologies such as small molecules, RNA interference (RNAi)-mediated gene silencing, and other genetic strategies.
High-throughput manipulation of intrinsic cellular programs
Small molecule screens have proven to be important in cell manipulation, signaling pathway analysis, and have also formed the basis for the development of many novel clinical agents [52,53]. For stem cells, various synthetic and natural small molecules have been shown to influence the processes of self-renewal and differentiation [54]. In a recent study, multiwell high-throughput analysis was performed utilizing a library of 50,000 compounds to find compounds that promoted the self-renewal of mouse ES cells in the absence of supportive feeder cells and without exogenously added leukemia inhibitory factor (LIF) [55•]. From this screen, a class of compounds, 3,4-dihydropyrimido[4,5-d]pyrimidines, was initially discovered that maintained the undifferentiated state of these cells, which subsequently led to the identification of an analog, termed SC1, that exhibited both higher activity and lower toxicity. Experiments aimed at elucidating the underlying mechanism suggested that SC1 acts through the inhibition of RasGAP and ERK1 signaling pathways.
Analogous to the protein-based systems described above, small molecule screening assays have also recently been adapted to a microarray format. In one approach, compounds were localized to arrayed regions on a surface prior to cell seeding through their encapsulation and slow release from microscale biodegradable poly-(D),(L)-lactide/glycolide (PLGA) scaffolds [56]. In another design, an agarose gel sheet containing cells was overlaid onto an array of small molecules to identify factors that bind to a dopamine receptor and increase intracellular calcium levels [57]. Although these types of systems remain primarily proof-of-concept demonstrations at this stage, their potential utility towards the future identification of molecules impacting the function of a wide range of primary cells, including stem cells, is extensive.
Concomitant with the development of small molecule platforms, high-throughput RNAi screens are similarly emerging as important tools in many cell and developmental biology contexts [58]. For instance, in a recent study examining ES cell function, a subtractive library approach identified multiple genes involved in the regulation of Oct-4 expression and self-renewal [59]. In addition to more conventional multiwell approaches, several recently developed methodologies may offer advantages in screening efficiency. One method, referred to as a siRNA bar-code screen, employs a short hairpin RNA (shRNA) library with each vector carrying a unique 19-mer oligonucleotide. After transfection and exposure to a selectable stimulus (e.g. drug exposure, differentiation, migration, etc.), the bar-code sequences are recovered by polymerase chain reaction (PCR) and hybridized to an oligonucleotide array to determine relative abundance of each sequence. This approach has been used to identify anticancer drug targets [60•], tumor suppressors [61], and components of the p53 pathway [62]. In addition, as a means to enhance throughput and minimize reagent requirements of RNAi screens, miniaturization using microarray strategies has also been explored. Various permutations of these systems have utilized arrayed double stranded RNAs [63], small interfering RNAs (siRNAs) [64,65], or vectors encoding shRNAs [66]. To extend the application of these systems to mammalian cell types for which transfection can be difficult, such as nondividing primary cells, Bailey et al. [67••] recently described the fabrication of lentiviral microarrays (Figure 5). This system was compatible with a range of primary and transformed cell types, and was also shown to be useful for so-called high-content screening, or the detection of subcellular changes such as protein localization. Furthermore, both the delivery of siRNAs and cDNAs for overexpression could be performed in parallel.
Figure 5.
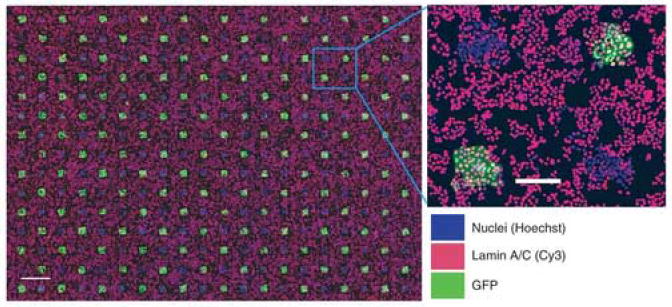
Lentiviral microarrays for high-throughput screening of gene function. Illustrated is a microarray printed with an alternating pattern of lentiviruses expressing either GFP or short hairpin RNA specific for lamin A/C and subsequently seeded with HeLa cells. Hoechst staining of nuclei (blue), anti-lamin A/C immunofluorescence (red), and GFP fluorescence (green) are displayed as indicated. Right panel, higher magnification image of a selected region of the array (boxed). Scale bars, left (1 mm); right (200 μm). (Figure adapted from [67••] with permission).
Together with RNAi-mediated gene silencing approaches, additional high-throughput strategies aimed at genome-wide analysis are becoming increasingly utilized, in particular in stem cell biology. For example, large scale gene trapping techniques have been used to generate ES cell lines and subsequent mouse models with a wide range of single gene mutations [68,69]. Also, a genome-scale gain of function screen coupled with gene expression profiling identified a host of genetic elements that may be important for ES cell self-renewal [70]. Overall, as the genomic and proteomic signatures of various stem cell populations continue to be identified, high-throughput approaches for the manipulation of intrinsic genetic and signaling programs will continue to be important in the interpretation of the mechanisms of stem cell function.
Conclusions
A more thorough understanding of the pathways governing both embryonic and adult stem cell functions has been facilitated by the development and application of several high-throughput platforms. These include miniaturized cell-based assays (e.g. cell microarrays), which have provided insight into the important roles of microenvironmental signals such as extracellular matrix, growth factors, and other signaling proteins in stem cell differentiation. Notably, one of the major benefits of these systems is the ability to efficiently analyze the effects of combinations of extracellular signals. Consequently, the convergence of immobilized protein-based platforms with tools to control the soluble milieu, biomechanical influences, and cell-cell interactions would enable an unprecedented control over the design of ex vivo stem cell microenvironments and greatly aid investigations of stem cell function. Moreover, together with factors derived from the analysis of microenviromental influences, the identification of factors that can modulate the intrinsic regulatory networks of stem cells (e.g. small molecules, siRNAs) could be equally important in providing fundamental insights and serving as the foundation for stem cell therapies. In the future, such therapies could be broadly applicable to a wide range of degenerative diseases as well as the potential selective targeting of cancer stem cells in various malignancies.
Acknowledgments
Funding was generously provided by NSF CAREER, NIH NIDDK, and the David and Lucile Packard Foundation. G.H.U. was supported by a Ruth L. Kirschstein National Research Service Award.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Boyer LA, Mathur D, Jaenisch R. Molecular control of pluripotency. Curr Opin Genet Dev. 2006;16:455–462. doi: 10.1016/j.gde.2006.08.009. [DOI] [PubMed] [Google Scholar]
- 2.Van Hoof D, Passier R, Ward-Van Oostwaard D, Pinkse MW, Heck AJ, Mummery CL, Krijgsveld J. A quest for human and mouse embryonic stem cell-specific proteins. Mol Cell Proteomics. 2006;5:1261–1273. doi: 10.1074/mcp.M500405-MCP200. [DOI] [PubMed] [Google Scholar]
- 3.Eckfeldt CE, Mendenhall EM, Verfaillie CM. The molecular repertoire of the ‘almighty’ stem cell. Nat Rev Mol Cell Biol. 2005;6:726–737. doi: 10.1038/nrm1713. [DOI] [PubMed] [Google Scholar]
- 4•.Scadden DT. The stem-cell niche as an entity of action. Nature. 2006;441:1075–1079. doi: 10.1038/nature04957.. Insightful review of recent advances identifying stem cell niches in multiple organ systems. Important components of stem cell niches are highlighted including paracrine signals, extracellular matrix composition, as well as metabolic elements such as reactive oxygen species and calcium concentrations. The dynamic nature of stem cell microenvironments and the potential for targeting normal or cancer stem cell niches for therapeutic purposes is also discussed.
- 5.Fuchs E, Tumbar T, Guasch G. Socializing with the neighbors: stem cells and their niche. Cell. 2004;116:769–778. doi: 10.1016/s0092-8674(04)00255-7. [DOI] [PubMed] [Google Scholar]
- 6.Folch A, Toner M. Microengineering of cellular interactions. Annu Rev Biomed Eng. 2000;2:227–256. doi: 10.1146/annurev.bioeng.2.1.227. [DOI] [PubMed] [Google Scholar]
- 7.Pirone DM, Chen CS. Strategies for engineering the adhesive microenvironment. J Mammary Gland Biol Neoplasia. 2004;9:405–417. doi: 10.1007/s10911-004-1410-z. [DOI] [PubMed] [Google Scholar]
- 8.Voldman J, Gray ML, Schmidt MA. Microfabrication in biology and medicine. Annu Rev Biomed Eng. 1999;1:401–425. doi: 10.1146/annurev.bioeng.1.1.401. [DOI] [PubMed] [Google Scholar]
- 9.Bhatia SN, Balis UJ, Yarmush ML, Toner M. Effect of cell-cell interactions in preservation of cellular phenotype: cocultivation of hepatocytes and nonparenchymal cells. Faseb J. 1999;13:1883–1900. doi: 10.1096/fasebj.13.14.1883. [DOI] [PubMed] [Google Scholar]
- 10.Chen CS, Mrksich M, Huang S, Whitesides GM, Ingber DE. Geometric control of cell life and death. Science. 1997;276:1425–1428. doi: 10.1126/science.276.5317.1425. [DOI] [PubMed] [Google Scholar]
- 11.McBeath R, Pirone DM, Nelson CM, Bhadriraju K, Chen CS. Cell shape, cytoskeletal tension, and RhoA regulate stem cell lineage commitment. Dev Cell. 2004;6:483–495. doi: 10.1016/s1534-5807(04)00075-9. [DOI] [PubMed] [Google Scholar]
- 12.Nelson CM, Pirone DM, Tan JL, Chen CS. Vascular endothelial-cadherin regulates cytoskeletal tension, cell spreading, and focal adhesions by stimulating RhoA. Mol Biol Cell. 2004;15:2943–2953. doi: 10.1091/mbc.E03-10-0745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu WF, Nelson CM, Pirone DM, Chen CS. E-cadherin engagement stimulates proliferation via Rac1. J Cell Biol. 2006;173:431–441. doi: 10.1083/jcb.200510087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Doh J, Irvine DJ. Immunological synapse arrays: patterned protein surfaces that modulate immunological synapse structure formation in T cells. Proc Natl Acad Sci U S A. 2006;103:5700–5705. doi: 10.1073/pnas.0509404103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Castel D, Pitaval A, Debily MA, Gidrol X. Cell microarrays in drug discovery. Drug Discov Today. 2006;11:616–622. doi: 10.1016/j.drudis.2006.05.015. [DOI] [PubMed] [Google Scholar]
- 16.Chen DS, Davis MM. Molecular and functional analysis using live cell microarrays. Curr Opin Chem Biol. 2006;10:28–34. doi: 10.1016/j.cbpa.2006.01.001. [DOI] [PubMed] [Google Scholar]
- 17.Miranti CK, Brugge JS. Sensing the environment: a historical perspective on integrin signal transduction. Nat Cell Biol. 2002;4:E83–90. doi: 10.1038/ncb0402-e83. [DOI] [PubMed] [Google Scholar]
- 18.Jones PH, Watt FM. Separation of human epidermal stem cells from transit amplifying cells on the basis of differences in integrin function and expression. Cell. 1993;73:713–724. doi: 10.1016/0092-8674(93)90251-k. [DOI] [PubMed] [Google Scholar]
- 19.Campos LS, Leone DP, Relvas JB, Brakebusch C, Fassler R, Suter U, ffrench-Constant C. Beta1 integrins activate a MAPK signalling pathway in neural stem cells that contributes to their maintenance. Development. 2004;131:3433–3444. doi: 10.1242/dev.01199. [DOI] [PubMed] [Google Scholar]
- 20.Garcion E, Halilagic A, Faissner A, ffrench-Constant C. Generation of an environmental niche for neural stem cell development by the extracellular matrix molecule tenascin C. Development. 2004;131:3423–3432. doi: 10.1242/dev.01202. [DOI] [PubMed] [Google Scholar]
- 21.Stier S, Ko Y, Forkert R, Lutz C, Neuhaus T, Grunewald E, Cheng T, Dombkowski D, Calvi LM, Rittling SR, et al. Osteopontin is a hematopoietic stem cell niche component that negatively regulates stem cell pool size. J Exp Med. 2005;201:1781–1791. doi: 10.1084/jem.20041992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nilsson SK, Johnston HM, Whitty GA, Williams B, Webb RJ, Denhardt DT, Bertoncello I, Bendall LJ, Simmons PJ, Haylock DN. Osteopontin, a key component of the hematopoietic stem cell niche and regulator of primitive hematopoietic progenitor cells. Blood. 2005;106:1232–1239. doi: 10.1182/blood-2004-11-4422. [DOI] [PubMed] [Google Scholar]
- ••23.Flaim CJ, Chien S, Bhatia SN. An extracellular matrix microarray for probing cellular differentiation. Nat Methods. 2005;2:119–125. doi: 10.1038/nmeth736.. The authors developed an extracellular matrix microarray platform in order to examine the combinatorial impact of extracellular matrix mixtures on cellular function. In one application, extracellular matrix components that influenced the differentiation of embryonic stem cells towards an early hepatic fate were identified.
- 24.Sastry SK, Horwitz AF. Adhesion-growth factor interactions during differentiation: an integrated biological response. Dev Biol. 1996;180:455–467. doi: 10.1006/dbio.1996.0319. [DOI] [PubMed] [Google Scholar]
- 25.Comoglio PM, Boccaccio C, Trusolino L. Interactions between growth factor receptors and adhesion molecules: breaking the rules. Curr Opin Cell Biol. 2003;15:565–571. doi: 10.1016/s0955-0674(03)00096-6. [DOI] [PubMed] [Google Scholar]
- 26••.Soen Y, Mori A, Palmer TD, Brown PO. Exploring the regulation of human neural precursor cell differentiation using arrays of signaling microenvironments. Mol Syst Biol. 2006;2:37. doi: 10.1038/msb4100076.. Utilizing a microarray platform consisting of spotted combinations of extracellular matrix, growth factors, and other signaling proteins the authors investigated, in a high-throughput manner, microenvironmental regulation of neural precursor differentiation. This system represents an important example of the utility of microarray approaches for the examination of stem cell responses within complex, multi-factorial environments.
- 27.Nakajima M, Ishimuro T, Kato K, Ko IK, Hirata I, Arima Y, Iwata H. Combinatorial protein display for the cell-based screening of biomaterials that direct neural stem cell differentiation. Biomaterials. 2007;28:1048–1060. doi: 10.1016/j.biomaterials.2006.10.004. [DOI] [PubMed] [Google Scholar]
- 28.Varnum-Finney B, Wu L, Yu M, Brashem-Stein C, Staats S, Flowers D, Griffin JD, Bernstein ID. Immobilization of Notch ligand, Delta-1, is required for induction of notch signaling. J Cell Sci. 2000;113(Pt 23):4313–4318. doi: 10.1242/jcs.113.23.4313. [DOI] [PubMed] [Google Scholar]
- 29.Hicks C, Ladi E, Lindsell C, Hsieh JJ, Hayward SD, Collazo A, Weinmaster G. A secreted Delta1-Fc fusion protein functions both as an activator and inhibitor of Notch1 signaling. J Neurosci Res. 2002;68:655–667. doi: 10.1002/jnr.10263. [DOI] [PubMed] [Google Scholar]
- 30.Engler AJ, Sen S, Sweeney HL, Discher DE. Matrix elasticity directs stem cell lineage specification. Cell. 2006;126:677–689. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- 31.Anderson DG, Levenberg S, Langer R. Nanoliter-scale synthesis of arrayed biomaterials and application to human embryonic stem cells. Nat Biotechnol. 2004;22:863–866. doi: 10.1038/nbt981. [DOI] [PubMed] [Google Scholar]
- 32.Song X, Zhu CH, Doan C, Xie T. Germline stem cells anchored by adherens junctions in the Drosophila ovary niches. Science. 2002;296:1855–1857. doi: 10.1126/science.1069871. [DOI] [PubMed] [Google Scholar]
- 33.Shen Q, Goderie SK, Jin L, Karanth N, Sun Y, Abramova N, Vincent P, Pumiglia K, Temple S. Endothelial cells stimulate self-renewal and expand neurogenesis of neural stem cells. Science. 2004;304:1338–1340. doi: 10.1126/science.1095505. [DOI] [PubMed] [Google Scholar]
- 34.Adams GB, Scadden DT. The hematopoietic stem cell in its place. Nat Immunol. 2006;7:333–337. doi: 10.1038/ni1331. [DOI] [PubMed] [Google Scholar]
- 35.Keller GM. In vitro differentiation of embryonic stem cells. Curr Opin Cell Biol. 1995;7:862–869. doi: 10.1016/0955-0674(95)80071-9. [DOI] [PubMed] [Google Scholar]
- 36.Khademhosseini A, Eng G, Yeh J, Fukuda J, Blumling J, 3rd, Langer R, Burdick JA. Micromolding of photocrosslinkable hyaluronic acid for cell encapsulation and entrapment. J Biomed Mater Res A. 2006;79:522–532. doi: 10.1002/jbm.a.30821. [DOI] [PubMed] [Google Scholar]
- 37.Khademhosseini A, Ferreira L, Blumling J, 3rd, Yeh J, Karp JM, Fukuda J, Langer R. Co-culture of human embryonic stem cells with murine embryonic fibroblasts on microwell-patterned substrates. Biomaterials. 2006;27:5968–5977. doi: 10.1016/j.biomaterials.2006.06.035. [DOI] [PubMed] [Google Scholar]
- 38.Mohr JC, de Pablo JJ, Palecek SP. 3-D microwell culture of human embryonic stem cells. Biomaterials. 2006;27:6032–6042. doi: 10.1016/j.biomaterials.2006.07.012. [DOI] [PubMed] [Google Scholar]
- 39.Whitesides GM. The origins and the future of microfluidics. Nature. 2006;442:368–373. doi: 10.1038/nature05058. [DOI] [PubMed] [Google Scholar]
- 40.Kim L, Vahey MD, Lee HY, Voldman J. Microfluidic arrays for logarithmically perfused embryonic stem cell culture. Lab Chip. 2006;6:394–406. doi: 10.1039/b511718f. [DOI] [PubMed] [Google Scholar]
- 41.Vollmer AP, Probstein RF, Gilbert R, Thorsen T. Development of an integrated microfluidic platform for dynamic oxygen sensing and delivery in a flowing medium. Lab Chip. 2005;5:1059–1066. doi: 10.1039/b508097e. [DOI] [PubMed] [Google Scholar]
- 42.Li Jeon N, Baskaran H, Dertinger SK, Whitesides GM, Van de Water L, Toner M. Neutrophil chemotaxis in linear and complex gradients of interleukin-8 formed in a microfabricated device. Nat Biotechnol. 2002;20:826–830. doi: 10.1038/nbt712. [DOI] [PubMed] [Google Scholar]
- 43.Chung BG, Flanagan LA, Rhee SW, Schwartz PH, Lee AP, Monuki ES, Jeon NL. Human neural stem cell growth and differentiation in a gradient-generating microfluidic device. Lab Chip. 2005;5:401–406. doi: 10.1039/b417651k. [DOI] [PubMed] [Google Scholar]
- 44.Di Carlo D, Wu LY, Lee LP. Dynamic single cell culture array. Lab Chip. 2006;6:1445–1449. doi: 10.1039/b605937f. [DOI] [PubMed] [Google Scholar]
- 45.Wagers AJ, Weissman IL. Plasticity of adult stem cells. Cell. 2004;116:639–648. doi: 10.1016/s0092-8674(04)00208-9. [DOI] [PubMed] [Google Scholar]
- 46.Lemischka I. Searching for stem cell regulatory molecules. Some general thoughts and possible approaches. Ann N Y Acad Sci. 1999;872:274–287. doi: 10.1111/j.1749-6632.1999.tb08472.x. discussion 287-278. [DOI] [PubMed] [Google Scholar]
- 47.Wagers AJ, Sherwood RI, Christensen JL, Weissman IL. Little evidence for developmental plasticity of adult hematopoietic stem cells. Science. 2002;297:2256–2259. doi: 10.1126/science.1074807. [DOI] [PubMed] [Google Scholar]
- 48.Ema H, Sudo K, Seita J, Matsubara A, Morita Y, Osawa M, Takatsu K, Takaki S, Nakauchi H. Quantification of self-renewal capacity in single hematopoietic stem cells from normal and Lnk-deficient mice. Dev Cell. 2005;8:907–914. doi: 10.1016/j.devcel.2005.03.019. [DOI] [PubMed] [Google Scholar]
- 49.Moretti A, Caron L, Nakano A, Lam JT, Bernshausen A, Chen Y, Qyang Y, Bu L, Sasaki M, Martin-Puig S, et al. Multipotent embryonic isl1+ progenitor cells lead to cardiac, smooth muscle, and endothelial cell diversification. Cell. 2006;127:1151–1165. doi: 10.1016/j.cell.2006.10.029. [DOI] [PubMed] [Google Scholar]
- 50.Chin VI, Taupin P, Sanga S, Scheel J, Gage FH, Bhatia SN. Microfabricated platform for studying stem cell fates. Biotechnol Bioeng. 2004;88:399–415. doi: 10.1002/bit.20254. [DOI] [PubMed] [Google Scholar]
- 51.Palmer TD, Markakis EA, Willhoite AR, Safar F, Gage FH. Fibroblast growth factor-2 activates a latent neurogenic program in neural stem cells from diverse regions of the adult CNS. J Neurosci. 1999;19:8487–8497. doi: 10.1523/JNEUROSCI.19-19-08487.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Seethala R, Fernandes PB. Handbook of drug screening. New York: Marcel Dekker; 2001. [Google Scholar]
- 53.Luesch H. Towards high-throughput characterization of small molecule mechanisms of action. Mol Biosyst. 2006;2:609–620. doi: 10.1039/b609384a. [DOI] [PubMed] [Google Scholar]
- 54.Ding S, Schultz PG. A role for chemistry in stem cell biology. Nat Biotechnol. 2004;22:833–840. doi: 10.1038/nbt987. [DOI] [PubMed] [Google Scholar]
- 55•.Chen S, Do JT, Zhang Q, Yao S, Yan F, Peters EC, Scholer HR, Schultz PG, Ding S. Self-renewal of embryonic stem cells by a small molecule. Proc Natl Acad Sci U S A. 2006;103:17266–17271. doi: 10.1073/pnas.0608156103.. In this study, the authors used a high-throughput iterative screening approach to identify a small molecule which promoted the self-renewal of mouse embyronic stem cells in the absence of LIF and feeder cells. The mechanism of action for this molecule, termed SC1, was determined to be dual inhibition of ERK1 and RasGAP signaling pathways.
- 56.Bailey SN, Sabatini DM, Stockwell BR. Microarrays of small molecules embedded in biodegradable polymers for use in mammalian cell-based screens. Proc Natl Acad Sci U S A. 2004;101:16144–16149. doi: 10.1073/pnas.0404425101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gopalakrishnan SM, Moreland RB, Kofron JL, Helfrich RJ, Gubbins E, McGowen J, Masters JN, Donnelly-Roberts D, Brioni JD, Burns DJ, et al. A cell-based microarrayed compound screening format for identifying agonists of G-protein-coupled receptors. Anal Biochem. 2003;321:192–201. doi: 10.1016/s0003-2697(03)00425-1. [DOI] [PubMed] [Google Scholar]
- 58•.Perrimon N, Mathey-Prevot B. Applications of high-throughput RNA interference screens to problems in cell and developmental biology. Genetics. 2007;175:7–16. doi: 10.1534/genetics.106.069963.. Important review on the utility of high-throughput RNA interference technologies, focusing on current work in Drosophila. The authors discuss both strengths and weaknesses of these approaches and applications towards systems biology and gene discovery.
- 59.Zhang JZ, Gao W, Yang HB, Zhang B, Zhu ZY, Xue YF. Screening for genes essential for mouse embryonic stem cell self-renewal using a subtractive RNA interference library. Stem Cells. 2006;24:2661–2668. doi: 10.1634/stemcells.2006-0017. [DOI] [PubMed] [Google Scholar]
- •60.Brummelkamp TR, Fabius AW, Mullenders J, Madiredjo M, Velds A, Kerkhoven RM, Bernards R, Beijersbergen RL. An shRNA barcode screen provides insight into cancer cell vulnerability to MDM2 inhibitors. Nat Chem Biol. 2006;2:202–206. doi: 10.1038/nchembio774.. The authors utilized a RNAi barcode screening technique to elucidate the cytotoxicity mechanisms of the anticancer drug, nutlin-3, in breast cancer cells. Included among the several factors identified to be involved in this process was 53BP1, a component of the DNA damage signaling pathway.
- 61.Westbrook TF, Martin ES, Schlabach MR, Leng Y, Liang AC, Feng B, Zhao JJ, Roberts TM, Mandel G, Hannon GJ, et al. A genetic screen for candidate tumor suppressors identifies REST. Cell. 2005;121:837–848. doi: 10.1016/j.cell.2005.03.033. [DOI] [PubMed] [Google Scholar]
- 62.Berns K, Hijmans EM, Mullenders J, Brummelkamp TR, Velds A, Heimerikx M, Kerkhoven RM, Madiredjo M, Nijkamp W, Weigelt B, et al. A large-scale RNAi screen in human cells identifies new components of the p53 pathway. Nature. 2004;428:431–437. doi: 10.1038/nature02371. [DOI] [PubMed] [Google Scholar]
- 63.Wheeler DB, Bailey SN, Guertin DA, Carpenter AE, Higgins CO, Sabatini DM. RNAi living-cell microarrays for loss-of-function screens in Drosophila melanogaster cells. Nat Methods. 2004;1:127–132. doi: 10.1038/nmeth711. [DOI] [PubMed] [Google Scholar]
- 64.Mousses S, Caplen NJ, Cornelison R, Weaver D, Basik M, Hautaniemi S, Elkahloun AG, Lotufo RA, Choudary A, Dougherty ER, et al. RNAi microarray analysis in cultured mammalian cells. Genome Res. 2003;13:2341–2347. doi: 10.1101/gr.1478703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yoshikawa T, Uchimura E, Kishi M, Funeriu DP, Miyake M, Miyake J. Transfection microarray of human mesenchymal stem cells and on-chip siRNA gene knockdown. J Control Release. 2004;96:227–232. doi: 10.1016/j.jconrel.2004.01.024. [DOI] [PubMed] [Google Scholar]
- 66.Silva JM, Mizuno H, Brady A, Lucito R, Hannon GJ. RNA interference microarrays: high-throughput loss-of-function genetics in mammalian cells. Proc Natl Acad Sci U S A. 2004;101:6548–6552. doi: 10.1073/pnas.0400165101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67••.Bailey SN, Ali SM, Carpenter AE, Higgins CO, Sabatini DM. Microarrays of lentiviruses for gene function screens in immortalized and primary cells. Nat Methods. 2006;3:117–122. doi: 10.1038/nmeth848.. The authors demonstrate an extension on previous ’reverse transfection’-based gene function screening platforms through the development of lentivirus-infected cell microarrays. This system would enable high-throughput analysis of the effects of both gene silencing and overexpression in a wider range of cell types, including various nondividing primary cell populations.
- 68.Stanford WL, Cohn JB, Cordes SP. Gene-trap mutagenesis: past, present and beyond. Nat Rev Genet. 2001;2:756–768. doi: 10.1038/35093548. [DOI] [PubMed] [Google Scholar]
- 69.Schnutgen F, De-Zolt S, Van Sloun P, Hollatz M, Floss T, Hansen J, Altschmied J, Seisenberger C, Ghyselinck NB, Ruiz P, et al. Genomewide production of multipurpose alleles for the functional analysis of the mouse genome. Proc Natl Acad Sci U S A. 2005;102:7221–7226. doi: 10.1073/pnas.0502273102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pritsker M, Ford NR, Jenq HT, Lemischka IR. Genomewide gain-of-function genetic screen identifies functionally active genes in mouse embryonic stem cells. Proc Natl Acad Sci U S A. 2006;103:6946–6951. doi: 10.1073/pnas.0509861103. [DOI] [PMC free article] [PubMed] [Google Scholar]


