Abstract
The purpose of this study was to characterize myosin light chain kinase (MLCK) expression in cardiac and skeletal muscle. The only classic MLCK detected in cardiac tissue, purified cardiac myocytes, and in a cardiac myocyte cell line (AT1) was identical to the 130-kDa smooth muscle MLCK (smMLCK). A complex pattern of MLCK expression was observed during differentiation of skeletal muscle in which the 220-kDa-long or “nonmuscle” form of MLCK is expressed in undifferentiated myoblasts. Subsequently, during myoblast differentiation, expression of the 220-kDa MLCK declines and expression of this form is replaced by the 130-kDa smMLCK and a skeletal muscle-specific isoform, skMLCK in adult skeletal muscle. These results demonstrate that the skMLCK is the only tissue-specific MLCK, being expressed in adult skeletal muscle but not in cardiac, smooth, or nonmuscle tissues. In contrast, the 130-kDa smMLCK is ubiquitous in all adult tissues, including skeletal and cardiac muscle, demonstrating that, although the 130-kDa smMLCK is expressed at highest levels in smooth muscle tissues, it is not a smooth muscle-specific protein.
Keywords: AT1 cells, C2C12 cells, differentiation
Myosin light chain kinases (MLCK) are Ca2+/calmodulin-regulated enzymes that catalyze the phosphorylation of the 20-kDa regulatory light chains of myosin. In smooth muscle, phosphorylation of the myosin regulatory light chain is an obligatory step involved in the initiation of contraction (17). In striated muscle, myosin light chain phosphorylation increases the isometric tension generated at submaximally activating Ca2+ concentrations (22), by decreasing the apparent Michaelis constant (Km) of actin for myosin. At maximally activating Ca2+ concentrations, light chain phosphorylation has no effect on force. In adult striated muscles, myosin light chain phosphorylation thus plays an important regulatory role, although it is not obligatory for muscle contraction. Recent studies have also suggested that phosphorylation of cardiac myosin light chains is important for maintaining normal cardiac function during development. Transgenic mice in which the normal cardiac myosin light was replaced by a form that could not be phosphorylated displayed atrial hypertrophy and chamber enlargement (29).
The molecular identity of the MLCK responsible for light chain phosphorylation in cardiac muscle is unclear. On the basis of the similar molecular masses characterized for purified skeletal and cardiac MLCKs (36, 37), and the amino acid sequence similarity between skeletal and cardiac myosin regulatory light chains, it has been assumed that the cardiac MLCK is similar or identical to the skeletal muscle enzyme. However, Northern blot and RNase protection analysis have failed to detect any of the 3.3-kb rat skeletal muscle MLCK (skMLCK) mRNA in rat cardiac muscle (14, 28). There are two known families of classic myosin light chain kinases, one includes the skeletal muscle isoforms and one includes the smooth muscle/nonmuscle isoforms (reviewed in Ref. 8). skMLCKs from chicken, rat, and rabbit vary greatly in apparent molecular mass determined by SDS-PAGE from 82 kDa in rat to 150 kDa in chicken. However, all have a similar domain structure. An elongated amino-terminal tail region displays very little sequence homology between species and largely accounts for the differences in molecular mass of the skMLCKs (20). A highly conserved catalytic core is located in the center of the molecule, followed by a regulatory region and the calmodulin-binding domain at the carboxy terminus. Within a given species there is a single skMLCK that is expressed in both fast- and slow-twitch muscles (14). Undifferentiated skeletal muscle myoblasts express nonmuscle myosin light chains and have been proposed to contain a unique Ca2+/calmodulin-independent form of MLCK that is distinct from the adult skMLCK (30). The 130-kDa smooth muscle MLCK (smMLCK) is the product of a distinct gene. Overall it has a domain organization similar to that of the skMLCK, except for an extended carboxy-terminal region, which is also expressed as the independent protein, telokin (6). A 210- to 220-kDa-long form of the classic smMLCK has been cloned from chicken embryo fibroblasts and human endothelial cells (10, 19, 31). These long forms of MLCK are identical in sequence to the corresponding smMLCKs but contain additional residues extending the amino terminus of the proteins. Western blotting of adult smooth and nonmuscle tissues demonstrated that the major form of MLCK expressed in these tissues has the same mass as the 130-kDa smMLCK (7). The tissue expression pattern or potential function of the 220-kDa MLCK has not been well characterized. Preliminary evidence in chicken suggest that this may represent an embryonic MLCK isoform (5), although in humans it has been reported to be expressed in adult tissues and endothelial cells (10, 19).
To resolve the identity of the MLCKs expressed in cardiac muscle and in skeletal muscle during development, expression of MLCK was examined in cardiac myocytes and during in vitro differentiation of skeletal muscle using immunological and Northern blotting techniques. A MLCK expressed in heart was cloned from an AT2 cardiac myocyte library. The identity of the MLCK expressed in AT1 cardiac muscle cells and adult cardiac muscle tissue was then confirmed by RT-PCR cloning and sequencing. Results demonstrate that the only classical MLCK detectable in cardiac myocytes is indistinguishable from the 130-kDa smMLCK. No skeletal muscle-like MLCK was detected in heart by Western blotting, while both the 130-kDa smMLCK and skMLCK are readily detectable in adult skeletal muscle. Western blotting experiments revealed that undifferentiated skeletal muscle myoblasts express the 220-kDa-long form of MLCK; however, expression of this 220-kDa MLCK was undetectable in adult skeletal muscle tissues.
METHODS
Western blot analysis
Extracts were prepared from cells or tissues by homogenization in a lysis buffer containing 1% Nonidet P-40, 20 mM MOPS, pH 7, 2 mM ethylene bis(oxy-ethylenenitrilo) tetra-acetic acid, 50 mM MgCl2, 300 mM NaCl, 50 mM dithiothreitol, 20 μg/ml leupeptin, 40 μg/ml aprotinin, 60 μg/ml tosyl-phenylalanyl chloromethyl ketone, 60 μg/ml N-p-tosyl-L-lysine chloromethyl ketone, and 100 μg/ml phenylmethylsulfonyl fluoride. Extracts were clarified by brief centrifugation, divided into aliquots, and stored at −70°C until analysis. For analysis of skMLCK, extracts were separated by SDS-PAGE on a 7.5% polyacrylamide gel and transferred to nitrocellulose in 25 mM Tris, 192 mM glycine, 0.5% SDS at 60 mA for 14–16 h. For analysis of smMLCK and myosin, extracts were separated on 5% polyacrylamide gels and transferred to nitrocellulose at 80 mA for 16 h. Blots were blocked in 10 mM Tris, pH 7.5, 150 mM NaCl, 0.05% Tween 20 (TBST) containing 2% gelatin for 2 h, and incubated with the antibodies indicated diluted in 2% gelatin-TBST for a further 2 h. Blots were washed three times for 5 min each in TBST and then incubated with the appropriate second step antibody conjugated to horseradish peroxidase for 45 min. After extensive washing (total 30 min), blots were developed using chemiluminescence according to the manufacturers directions (ECL; Amersham, Arlington Heights, IL).
Northern blot analysis
Northern blots were performed as described previously (7). Northern blots were probed with a 556-bp mouse MLCK cRNA corresponding to nucleotides 3018–3574 of the rabbit uterine MLCK cDNA (7). This fragment of mouse smMLCK was generated by reverse transcription coupled to PCR using mouse uterine mRNA and oligonucleotide primers complementary to nucleotides 3018–3042 and 3551–3587 of the rabbit cDNA. Final wash conditions were 1.25 mM sodium phosphate, pH 7.4, 30 mM NaCl, 0.2 mM EDTA (0.1× SSPE) and 0.1% SDS at 68°C for 10 min. Blots were exposed for 2 days at −70°C.
RNase protection analysis
An antisense RNA probe was transcribed from the 335-bp EcoR I-BamH I fragment of the rat skMLCK cDNA (14) subcloned into pGEM 3Z (Promega, Madison, WI), using SP6 polymerase on an EcoR I linearized template. The probe generated contains an additional 38 nucleotides from the pGEM polylinker. RNase protection assays were performed as described previously (14). RNase digestion was performed using 6 μg/ml RNase A and 0.03 μg/ml RNase T1 at 37°C for 1 h.
Cloning of mouse cardiac muscle MLCK
A cDNA probe encoding the mouse smMLCK/telokin region (15) was utilized to screen a mouse AT2 cell cardiac myocyte gt11 cDNA library, generously provided by Dr. Loren Field (Indiana University, IN), and positive isolates were subcloned and sequenced. A full-length cDNA encoding the 1,031 amino acids of the mouse 130-kDa MLCK was identified by sequence comparison to the sequence of clones encoding MLCK from several other species.
RT-PCR analysis
Total RNA was prepared from adult mouse heart, skeletal and bladder tissues, and AT1 cardiac myocyte cells. Five micrograms of RNA from each source were reverse-transcribed using oligo(dT) and random primers. PCR amplification was performed on an aliquot of the RT mixture for 35 cycles at 94°C for 1 min, 50°C for 1 min, and 72°C for 1 min, using Vent DNA polymerase (New England Biolabs). Oligonucleotide primers corresponding to three overlapping regions spanning the mouse smMLCK cDNA described in this report were employed to amplify the MLCK expressed in these tissues and cells. Negative control PCR reactions were run with mock reverse-transcribed RNA that was not treated with RT. Positive control reactions utilized the full-length mouse smMLCK cDNA as a template as well as RT-PCR from mouse bladder, a smooth muscle tissue. The PCR products from selected reactions were subcloned and sequenced. The three sets of oligonucleotide primers used were: PCR-1F, ATGGATTTCCGCGCC and PCR-1R, CTGCCACATGGACATCCT; PCR-2F, AAGTCTGCTGAGACCTCGAAAC and PCR-2R, GCCGCTCTCGCCATTC; PCR-3F, TGCAAGTCGATCAAGTTC and PCR-3R, ATCATCAGCGAGCATTGCC.
Cell culture
C2C12 myoblasts, obtained form Dr. W. Wright (University of Texas Southwestern, TX), were grown in DMEM/199 (4:1) containing 10% FCS, 10% newborn calf serum, and 50 μg/ml genticin. Myoblasts were induced to differentiate by switching to media containing 2% horse serum for 3–4 days. AT1 and AT2, SV40 large T-antigen transformed cardiac myocytes, were kindly provided by Dr. Loren Field (Indiana University, IN) and grown in DMEM containing 10% FCS. Adult rat cardiac myocytes were a generous gift from Dr. X.-D. Huang (Indiana University, IN), and neonatal day 1 purified rat cardiac myocyte and fibroblast fractions were kindly provided by Dr. M. LaCorbiere and Dr. K. Chien (University of California, San Diego, CA).
Immunofluorescence
For immunofluorescence analysis, AT1 cells were grown on glass coverslips coated with fibronectin under the same conditions as described above. Cells were fixed in 3.7% formaldehyde in PBS for 5 min, washed in 50 mM Tris, pH 7.6, 150 mM NaCl, 0.01% NaN3 (TBS) for 5 min, permeabilized in 0.2% Triton X-100 in TBS for 5 min, washed for 5 min in TBS, and then incubated with the antibodies indicated in the legend for Fig. 3 for 1 h at 37°C. After incubation, the cells were washed in TBS and incubated with either rhodamine or fluorescein-conjugated second antibodies for 1 h at 37°C. Washed cover chips were mounted in gelvatol and stored at 4°C.
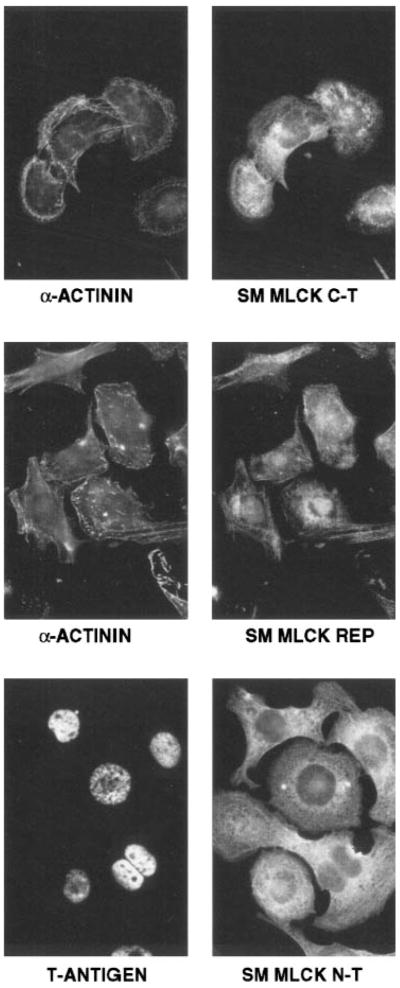
Fig. 3.
Immunolocalization of smMLCK expression in AT1 cardiac myocytes. AT1 cardiac myocytes were prepared for analysis as described in METHODS. A monoclonal antibody to sarcomeric α-actinin was obtained from Sigma (clone EA-53), and polyclonal antisera to T-antigen was a generous gift from Dr. Loren Field. Top right: a polyclonal antibody (C-T) that binds to the carboxy-terminal “telokin” region of the smMLCK (6) was used. Middle: an anti-peptide antibody (REP) directed against the 15-amino acid repeated motif in the amino terminus of the smMLCK (7) was used. Bottom: a monoclonal antibody (N-T) that binds to the amino terminus of smMLCK (Sigma, clone K56) (9) was used. Monoclonal antibodies were visualized using rhodamine-conjugated anti-mouse IgG and polyclonal antibodies using fluorescein-conjugated anti-rabbit IgG. Cells are shown at ×1,000 original magnification.
RESULTS
Expression of smMLCK in skeletal and cardiac muscle tissues
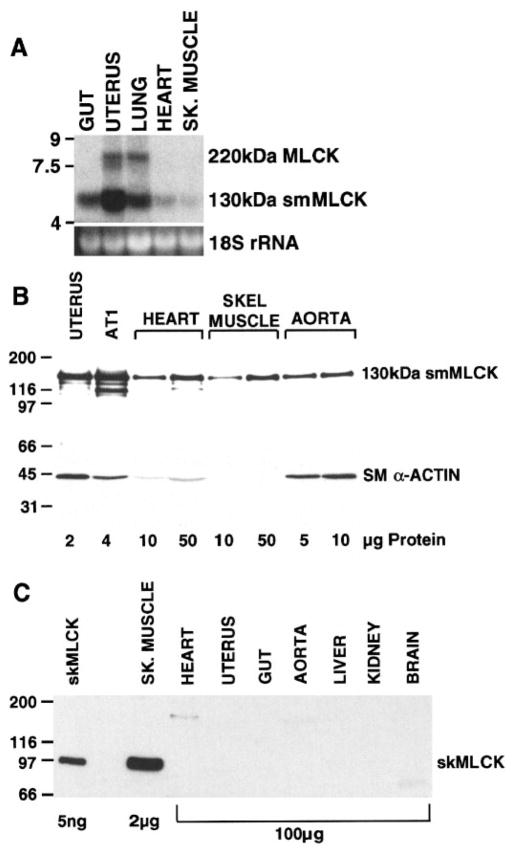
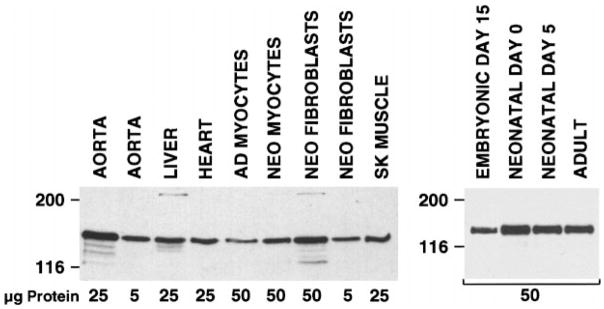
Northern blot analysis of RNA isolated from mouse skeletal and cardiac muscle demonstrated the presence of low levels of the 5.8-kb smMLCK mRNA (Fig. 1A). To examine the pattern of MLCK protein expression, Western blot analysis of mouse skeletal, cardiac, and smooth muscle tissues was performed using a monoclonal antibody that binds to the amino terminus of the 130-kDa smMLCK (9) (Figs. 1B and 2) and a polyclonal antibody to skMLCK (25) (Fig. 1C). The 130-kDa smMLCK was readily detectable in cardiac and skeletal muscle tissues in addition to smooth muscle tissues. Densitometric analysis of the immunoblot shown in Fig. 1B revealed that the 130-kDa smMLCK was expressed at approximately three- and fourfold lower levels in mouse cardiac and skeletal muscle extracts, respectively, compared with mouse aortic extracts. The 130-kDa smMLCK was detected in embryonic, neonatal, and adult cardiac tissue (Fig. 2). The 220-kDa MLCK was expressed at low levels in adult uterus, lung, and liver and in neonatal cardiac fibroblasts (Figs. 1 and 2). No expression of the 220-kDa MLCK isoform was detected in embryonic, neonatal, or adult cardiac muscle or in adult skeletal muscle (Figs. 1 and 2). In contrast to the 130- and 220-kDa smMLCKs, the 97-kDa skMLCK was only detected in skeletal muscle tissue and could not be detected in cardiac muscle, smooth muscle or nonmuscle tissues, even when 50-fold greater amounts of these extracts were analyzed (Fig. 1C). Similar results were obtained using a monoclonal antibody (clone 12a; Ref. 24) to skMLCK that we have previously shown binds to a region of the skMLCK that is conserved across species (13) (data not shown).
Fig. 1.
Northern and Western blot analysis of myosin light chain kinase (MLCK) expression in mouse tissues. A: Northern blot in which 15 μg of total RNA, isolated from the indicated mouse tissues, were separated on a 1.4% agarose gel. A 5.8-kb mRNA corresponding to the 130-kDa smooth muscle MLCK (smMLCK) and an 8-kb mRNA corresponding to the 220-kDa MLCK were detected using an anti-sense cRNA probe to smMLCK as described in METHODS (top). Bottom: ethidium bromide stain of 18S rRNA. B: Western blot analysis of extracts from various mouse tissues and AT1 cardiac myocytes, reacted with a monoclonal antibody to smMLCK (top) or to smooth muscle α-actin (bottom). The amount of each extract analyzed is indicated in μg below the blots. C: Western blot analysis of mouse tissue extracts reacted with a polyclonal antibody specific for skeletal muscle MLCK (skMLCK). Lane 1, 5 ng of purified rabbit skMLCK. The amount of each extract analyzed is indicated in μg below the blot. Note that 50-fold less extract from skeletal muscle was analyzed compared with the other tissues. The cross-reacting band at 180–200 kDa in heart extract likely represents nonspecific binding to cardiac myosin.
Fig. 2.
smMLCK expression in cardiac myocytes. Immunoblot analysis of various rat tissue and cell extracts (left) and mouse heart extracts (right) as indicated. Amounts of extract analyzed are indicated under the blot. Adult heart cells were isolated by retrograde perfusion of collagenase through an adult rat heart, and unfractionated cells were analyzed. Neonatal myocytes and fibroblasts were prepared by digestion of hearts from newborn rats. Myocytes and fibroblasts were then separated on a Percoll gradient as described previously (16). This procedure results in a myocyte fraction that is >95% cardiac myocytes. Mouse hearts were harvested from embryonic, neonatal, and adult mice at the developmental stages indicated.
To examine the possibility that the 130-kDa smMLCK detected in the cardiac and skeletal tissues was contributed from vascular smooth muscle contamination of the tissue samples, the relative levels of vascular smooth muscle in the samples were evaluated. To do this, the relative levels of smooth muscle α-actin, a well-characterized marker of vascular smooth muscle cells, were determined. Densitometric analysis of the immunoblot shown in Fig. 1B revealed that the ratios of 130-kDa smMLCK to smooth muscle α-actin in each sample were higher in skeletal and cardiac muscle compared with vascular smooth muscle. For example, there was more 130-kDa MLCK detected in 50 μg of skeletal muscle extract than there was detected in 5 μg of aortic extract. In contrast, smooth muscle α-actin was readily detectable in 5 μg of aortic extract, although smooth muscle α-actin was undetectable in 50 μg of skeletal muscle extract. Similarly, although the 130-kDa MLCK was expressed at 3-fold lower levels in heart compared with aorta, smooth muscle α-actin was expressed at 28-fold lower levels. These data suggest that there is a greater amount of 130-kDa smMLCK expressed in cardiac and skeletal muscle than can be accounted for by vascular smooth muscle contamination.
Smooth muscle MLCK expression in cardiac muscle cells
To further confirm that smMLCK was expressed in cardiac muscle cells, smMLCK expression was examined in isolated adult cardiac myocytes, purified neonatal cardiac myocytes and neonatal cardiac fibroblasts (Fig. 2), and AT1 cardiac myocytes (Fig. 1B). In extracts prepared from each of these cells, 130-kDa smMLCK was readily detectable. No smooth muscle myosin could be detected in the purified neonatal cardiac myocytes or the isolated adult myocytes, indicating that these cells are free from contaminating smooth muscle cells (data not shown). The fibroblast fraction from the neonatal cardiac cell preparation also contained vascular smooth muscle cells, as determined by positive staining for smooth muscle myosin (data not shown). It is likely that the presence of these contaminating cells accounts for the relatively high levels of smMLCK found in the purified fibroblast cell fraction.
The expression of the 130-kDa smMLCK in AT1 cardiac myocytes was further analyzed by indirect immunofluorescence using three antibodies that bind to different regions of smMLCK. A monoclonal antibody (N-T, Fig. 3) that binds to the amino terminus of the kinase, an anti-peptide polyclonal antibody (REP, Fig. 3) directed against the repeated region of amino acids near the amino terminus of the mammalian kinases, and a polyclonal antibody (C-T, Fig. 3) directed against the carboxy terminus of the kinase all stain AT1 cardiac cells. The staining pattern observed for each antibody was identical and is a distinct filamentous pattern consistent with the MLCK being localized to cytoskeletal elements (Fig. 3). The cardiac origin of the AT1 cells was confirmed by costaining with antibodies to sarcomeric α-actinin or to T-antigen.
Cloning of the cardiac MLCK
To determine whether the 130-kDa MLCK expressed in cardiac muscle tissue was identical to the form expressed in smooth muscle cells, a probe derived from the 3′ end of mouse smMLCK cDNA (15) was used to screen a cDNA library prepared from mouse cardiac AT2 cells. Both AT1 and AT2 cells are derived from atrial myocytes that have been transformed with SV40 large T-antigen. AT1 cells beat spontaneously in culture, and both AT1 and AT2 express cardiac myosin isoforms (Ref. 4 and unpublished observations). Twenty-two overlapping clones were isolated and completely sequenced. All of the clones encoded a protein with high homology to the other characterized mammalian smMLCKs. The level of amino acid identity between the mouse, rabbit, bovine, and human MLCKs is 96% in the catalytic domain, 100% in the calmodulin binding domain, 92% in the carboxy-terminal telokin region, and 80% in the amino-terminal region (excluding the variable repeat domain located between residues 87 and 172 of the mouse MLCK). Two overlapping clones were joined to result in a full-length cDNA that, when expressed in COS cells, encodes a 130-kDa protein that is catalytically active and Ca2+/calmodulin dependent (data not shown). Alignment of the predicted sequence of mouse cardiac MLCK revealed a high degree of identity between this sequence and the sequences of the other mammalian smMLCKs (Fig. 4).
Fig. 4.
Alignment of the protein sequence of the mouse cardiac 130-kDa MLCK to mammalian smMLCKs. The deduced amino acid sequence of the mouse 130-kDa smMLCK, obtained from AT2 cardiac myocytes, is shown aligned to the sequences reported for the bovine (18), human (27), and rabbit (7) smMLCKs. The symbols below the alignment refer to residues that are identical (★) in all 4 of the sequences. The positions of the 3 sets of oligonucleotide primer pairs used for PCR are shown above the sequence, and the position of the catalytic core is bracketed. Sequences of all fragments isolated by RT-PCR from mouse bladder and AT1 cells were identical to the sequence of the cDNA clone obtained from AT2 cells.
Molecular identity of the 130-kDa MLCK expressed in adult skeletal and cardiac muscle and AT1 cardiac myocytes
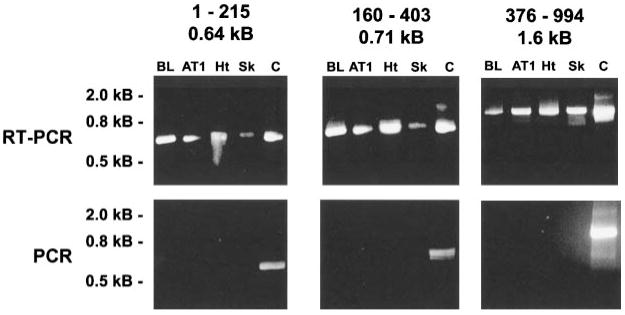
To determine whether the 130-kDa MLCK expressed in skeletal and cardiac muscle tissue and cardiac AT1 cells was identical to the form cloned from cardiac AT2 cells, RT-PCR analysis was performed. Three pairs of primers were designed to span the mouse smooth/cardiac muscle MLCK cDNA as shown in Fig. 4. Reverse transcription of total RNA isolated from adult mouse heart, skeletal muscle, bladder tissue, and AT1 cardiac cells was used to generate cDNA. The cDNA was then used to PCR amplify each of three fragments that span 994 of 1,030 amino acids comprising the 130-kDa MLCK from mouse heart. The sizes of the amplified PCR products were directly compared using agarose gel electrophoresis. This analysis revealed that identical sized fragments were generated from each of the tissues (Fig. 5). To further confirm the identity of the amplified products, all of the fragments amplified from bladder and AT1 cells, together with select fragments from skeletal and cardiac tissue, were subcloned and sequenced. The sequence of the fragments obtained from each tissue were found to be identical to the sequence of the cDNA encoding the mouse 130-kDa MLCK isolated from AT2 cardiac myocytes, shown in Fig. 4. Control PCR reactions performed using primer pairs and total RNA without reverse transcription were negative, suggesting that the products obtained by RT-PCR did not result from genomic DNA contamination (Fig. 5).
Fig. 5.
RT-PCR analysis of MLCK expression. Top: cDNA was generated by reverse transcription from RNA isolated from the indicated tissues and cell lines. MLCK cDNAs were amplified using oligonucleotide primers based on the sequence of the 130-kDa mouse smMLCK. Three sets of primers were used to generate three fragments spanning the coding region. The fragments from these reactions were compared by agarose gel electrophoresis and compared with the size of a positive control that used PCR and each primer set to amplify the corresponding fragment from the AT2-derived cDNA (Fig. 4). Bottom: negative control reactions incubated in the absence of RT. The numbers above each gel indicate the first and last amino acid residues of the mouse smMLCK that correspond to each fragment and the size of the PCR fragment. Molecular mass markers are indicated at left. PCR fragments from bladder and AT1 cells were subcloned and sequenced. BL, bladder; AT1, AT1 mouse cardiac myocytes; Ht, heart; Sk, skeletal muscle; C, positive control MLCK plasmid.
MLCK expression during skeletal muscle development in vitro
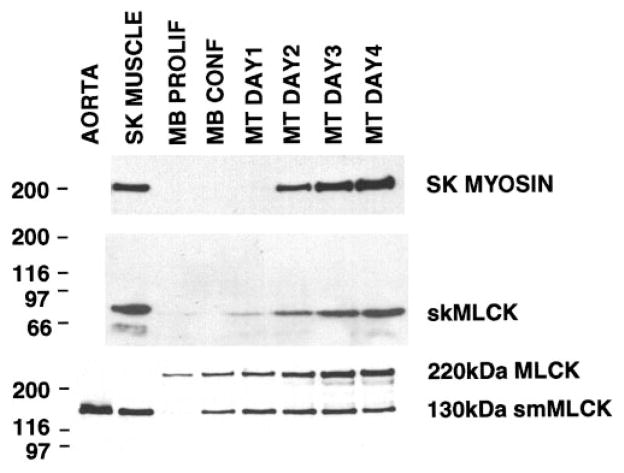
To further analyze the developmental expression of MLCK in skeletal muscle, MLCK expression was examined during the in vitro differentiation of mouse C2C12 skeletal muscle myoblasts. Differentiation was induced by switching confluent cultures of myoblasts from media containing 20% serum to media containing 2% horse serum. skMLCK protein and mRNA expression were first detectable 1–2 days following the induction of differentiation and continued to increase up to 4 days (Figs. 6 and 7). Skeletal muscle myosin was induced over a similar time period. In contrast, the 130-kDa smMLCK was first detected in confluent myoblast cultures before serum deprivation and continued to be expressed at similar levels in differentiated myotubes (Fig. 6). The predominant classic MLCK seen in C2C12 myoblasts was the 220-kDa long form of smMLCK. Expression of the 220-kDa MLCK was further induced during C2C12 cell differentiation, although it was not detectable in adult skeletal muscle tissue (Figs. 1A, 2, and 6).
Fig. 6.
MLCK expression during C2C12 cell differentiation. Western blot analysis of skeletal, 130-kDa smooth, and 220-kDa MLCK and skeletal muscle myosin expression during in vitro differentiation of C2C12 skeletal muscle cells. Extracts were prepared from adult mouse aorta and skeletal (SK) muscle, from C2C12 myoblasts grown to low density (MB Prolif) or high density (MB Conf) in the presence of 20% FCS, or from confluent myoblasts that were switched to media containing 2% horse serum for 1–4 days as indicated. Twenty-five micrograms of cell extracts were analyzed in each lane with the exception of the skeletal muscle extract (top), and the aorta extract (bottom), which represent only 5 μg of extract. The following antibodies were used for the analysis; polyclonal anti-rabbit skMLCK (middle), monoclonal anti-fast skeletal myosin (Sigma clone MY32; top), monoclonal anti-smMLCK (Sigma clone K57, bottom). The positions of molecular mass markers (in kDa) are indicated at left.
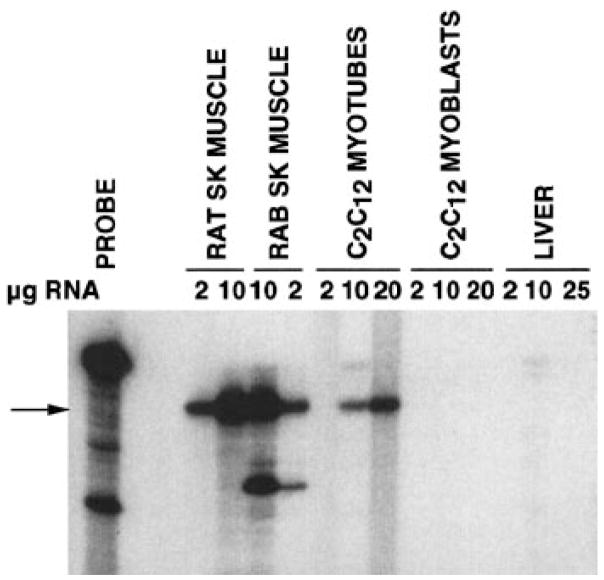
Fig. 7.
RNase protection analysis of skMLCK mRNA in C2C12 cells. A 32P-labeled 335-bp antisense RNA probe derived from the catalytic core of the rat skMLCK cDNA (14) was hybridized in solution to the indicated amounts of total RNA isolated from adult rat and rabbit (RAB) skeletal muscle, C2C12 myoblasts or C2C12 myotubes (differentiated for 4 days) as described previously (14). After hybridization (42°C, 80% formamide), unprotected probe was digested with RNase as described in METHODS. Protected fragments were separated by electrophoresis on denaturing polyacrylamide gels and visualized by autoradiography. Undigested probe is shown in the left lane. Arrow, position of the 335-bp fragment fully protected from digestion by the skMLCK mRNA. The smaller fragment seen in the rabbit lanes corresponds to a partially protected fragment that results from species differences in the nucleotide sequence between rat and rabbit skMLCK.
DISCUSSION
This study shows that the only classic MLCK detectable in adult cardiac muscle and in AT1 cardiac myocytes is indistinguishable from the previously described 130-kDa smMLCKs (reviewed in Ref. 8). Additionally, a complex pattern of MLCK expression was observed during differentiation of skeletal muscle in which the 220-kDa-long form of smMLCK is expressed in undifferentiated myoblasts. Subsequently, during myoblast differentiation, expression of the 220-kDa MLCK declines, and expression of this form is replaced by the 130-kDa smMLCK and skMLCK in adult skeletal muscle.
Western blot analysis in this study, together with previously reported RNase protection experiments (14), have failed to detect a skeletal muscle isoform of MLCK in cardiac muscle even when excessive amounts of cardiac tissue are analyzed. Previously, by Northern blotting, we detected a 2.2-kb mRNA present in cardiac muscle that weakly hybridized to a probe derived from rat skeletal muscle MLCK (14). This cross-hybridizing mRNA was only detected when 50 μg of cardiac poly(A)+ mRNA were analyzed. However, when the same probe was used for RNase protection analysis, no signal was detected in 60 μg of cardiac poly(A)+ mRNA. In contrast, in the current study we were able to detect an mRNA, that hybridized to a probe derived from the mouse 130-kDa MLCK, in as little as 15 μg of total RNA from heart (Fig. 1A). These results suggest that the 2.2-kb mRNA reported previously is either present at very low levels or that it represented non-specific hybridization resulting from the large amount of mRNA analyzed.
As a result of the highly vascular nature of cardiac muscle, it is possible that the smMLCK detected in adult heart is derived from vascular smooth muscle cells. However, several experimental results suggest that this possibility is unlikely. These results include the finding that antibodies to the 130-kDa smMLCK detect a corresponding protein in highly purified neonatal rat cardiomyocytes and in adult cardiomyocytes (Fig. 2). Second, the level of 130-kDa MLCK expression in cardiac tissue cannot be accounted for by vascular smooth muscle as judged by a comparison of the relative amounts of the kinase and smooth muscle α-actin (Fig. 1). Finally, the sequence of the MLCK expressed in AT1 cardiac cells is identical to the nucleotide sequence of RT-PCR clones obtained from mouse bladder smooth muscle and to cDNA clones representing the 130-kDa smMLCK, obtained from AT2 cardiac cells (Figs. 4 and 5). In support of these results, immunolocalization experiments also confirm that the 130-kDa smMLCK as well as cardiac muscle markers are both coexpressed in beating AT1 cardiac myocytes (Fig. 3). Although smMLCK is the major, classic MLCK that is detectable in cardiac muscle, our results do not exclude the possibility that other nonclassic MLCKs such as Rho kinase or PAK-2 (p21 activated kinase) may also contribute to cardiac myosin regulatory light chain phosphorylation (1, 3). However, the low levels of 130-kDa MLCK detected in heart, together with its poor kinetics using cardiac light chains as a substrate (33), are consistent with the low rates of light chain phosphorylation observed in vivo, in cardiac muscle (12).
The 130-kDa smMLCK is also expressed in adult skeletal muscle tissue and in differentiated skeletal muscle cells (Figs. 1, 2, and 6). However, in contrast to cardiac muscle, skeletal muscle tissue and differentiated C2C12 cells also express the tissue-specific skM-LCK. During differentiation of skeletal muscle, expression of 130-kDa smMLCK was induced before the expression of the skMLCK or skeletal muscle myosin; however, the 130-kDa smMLCK isoform continues to be expressed at low levels in adult skeletal muscle tissue. The high levels of expression of the skMLCK, as opposed to the 130-kDa smMLCK, in adult skeletal muscle and its lower Km and higher maximum velocity for skeletal myosin light chains suggest that it is likely that the skMLCK mediates the leftward shift in the force-pCa relationship and posttetanic twitch potentiation in skeletal muscle (34). Although the functional significance of the expression of the 130-kDa smMLCK in adult skeletal muscle is not yet clear, it is possible that 130-kDa smMLCK may be required to regulate cytoplasmic nonmuscle actomyosin interactions in skeletal muscle cells. Consistent with this proposal, studies have demonstrated that nonmuscle myosin heavy chain II-B is expressed in both fetal, neonatal, and to a lesser extent adult skeletal muscle tissues (23, 32, 35). The finding that the 130-kDa smMLCK is induced during C2C12 cell differentiation before the expression of skeletal muscle specific proteins may explain the previous observation of coexpression of the smMLCK and skeletal muscle differentiation markers in the developing mouse esophagus (26). The presence of cells expressing the 130-kDa smMLCK and skeletal muscle differentiation markers has been used to suggest that the striated muscle cells of the esophagus transdifferentiate from smooth muscle cells. Although our results do not preclude this possibility, they do demonstrate that the 130-kDa smMLCK is not an exclusive marker of differentiated smooth muscle cells and that it is normally induced during the differentiation of skeletal muscle.
The 220-kDa MLCK was the most abundant of the classic MLCKs detected in undifferentiated skeletal muscle myoblasts (Fig. 6); however, consistent with expression in human tissues (19), this isoform was not detected in adult mouse or rat skeletal muscle (Figs. 1 and 2). The functional significance of expression of the 220-kDa MLCK isoform compared with the 130-kDa smMLCK in myoblasts is uncertain. Both enzymes have identical catalytic domains and are regulated by Ca2+/calmodulin. It is likely that the intracellular localization of the enzymes differ as the amino-terminal region of the kinases appear to play a role in the cytoskeletal localization of the proteins (2, 9, 11, 21). Presumably, the 220-kDa MLCK plays a specific role in myoblasts, perhaps to phosphorylate either fetal skeletal myosin or nonmuscle myosin and regulate its activity during cell division and migration in a manner that cannot be mimicked by the 130-kDa smMLCK.
In summary, Western and Northern blotting, RNase protection, and cDNA cloning and RT-PCR analysis of classic MLCK expression in various mouse tissues demonstrate that only the skMLCK isoform is expressed in a tissue-specific manner. The 130-kDa smMLCK is ubiquitous in all adult tissues, including skeletal and cardiac muscle, and represents the major classic MLCK detectable in cardiac muscle. Together these results demonstrate that smMLCK is not a tissue-specific MLCK, and, although it is expressed at highest levels in smooth muscle tissues, it is not a smooth muscle-specific protein. Expression of the 130-kDa smMLCK in striated muscle tissues likely regulates the motor activity of nonmuscle myosin II expressed in these tissues in addition to regulating the activity of cardiac myosin. These data imply that the smMLCK is likely to play a critical role in regulating myosin motor activities during heart development.
Acknowledgments
This work was supported in part by Grant-in-Aid GA4 from the American Heart Association, Indiana Affiliate (to B. P. Herring), and National Heart, Lung, and Blood Institute Grants HL-58571 (to B. P. Herring) and HL-54118 (to P. J. Gallagher).
Footnotes
We are indebted to the following people for supplying cells used in this study: Dr. W. Wright (University of Texas Southwestern Medical Center at Dallas, TX) for C2C12 cells, Dr. L. Field (Indiana University, IN) for AT1 and AT2 cells, X-D. Huang (Krannert Institute of Cardiology, Indianapolis, IN) for adult rat cardiac myocytes, and Drs. M. La Corbiere and K. Chien (University of California, San Diego, CA) for the purified cardiac myocytes and fibroblasts. We thank Dr. Loren Field for the AT2 cDNA library and Dr. Aiping Smith for expert technical assistance.
References
- 1.Amano M, Ito M, Kimura K, Fukata Y, Chihara K, Nakano T, Matsuura Y, Kaibuchi K. Phosphorylation and activation of myosin by Rho-associated kinase (Rho-kinase) J Biol Chem. 1996;271:20246–20249. doi: 10.1074/jbc.271.34.20246. [DOI] [PubMed] [Google Scholar]
- 2.Cavadore JC, Molla A, Harricane MC, Gabrion J, Benyamin Y, Demaille JG. Subcellular localization of myosin light chain kinase in skeletal, cardiac, and smooth muscles. Proc Natl Acad Sci USA. 1982;79:3475–3479. doi: 10.1073/pnas.79.11.3475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chew TL, Masaracchia RA, Goeckeler ZM, Wysolmerski RB. Phosphorylation of nonmuscle myosin II regulatory light chain by p21-activated kinase (gamma-PAK) J Muscle Res Cell Motil. 1998;19:839–854. doi: 10.1023/a:1005417926585. [DOI] [PubMed] [Google Scholar]
- 4.Field LJ. Atrial natriuretic factor-SV40 T antigen transgenes produce cardiac tumors and cardiac arrhythmias in mice. Science. 1988;239:1029–1033. doi: 10.1126/science.2964082. [DOI] [PubMed] [Google Scholar]
- 5.Fisher SA, Ikebe M. Developmental and tissue distribution of expression of nonmuscle and smooth muscle isoforms of myosin light chain kinase. Biochem Biophys Res Commun. 1995;217:696–703. doi: 10.1006/bbrc.1995.2829. [DOI] [PubMed] [Google Scholar]
- 6.Gallagher PJ, Herring BP. The carboxyl terminus of the smooth muscle myosin light chain kinase is expressed as an independent protein, telokin. J Biol Chem. 1991;266:23945–23952. [PMC free article] [PubMed] [Google Scholar]
- 7.Gallagher PJ, Herring BP, Griffin SA, Stull JT. Molecular characterization of a mammalian smooth muscle myosin light chain kinase. J Biol Chem. 1991;266:23936–23944. [PMC free article] [PubMed] [Google Scholar]
- 8.Gallagher PJ, Herring BP, Stull JT. Myosin light chain kinases. J Muscle Res Cell Motil. 1997;18:1–16. doi: 10.1023/a:1018616814417. [DOI] [PubMed] [Google Scholar]
- 9.Gallagher PJ, Stull JT. Localization of an actin binding domain in smooth muscle myosin light chain kinase. Mol Cell Biochem. 1997;173:51–57. doi: 10.1023/a:1006876318155. [DOI] [PubMed] [Google Scholar]
- 10.Garcia JG, Lazar V, Gilbert-McClain LI, Gallagher PJ, Verin AD. Myosin light chain kinase in endothelium: molecular cloning and regulation. Am J Respir Cell Mol Biol. 1997;16:489–494. doi: 10.1165/ajrcmb.16.5.9160829. [DOI] [PubMed] [Google Scholar]
- 11.Guerriero V, Jr, Rowley DR, Means AR. Production and characterization of an antibody to myosin light chain kinase and intracellular localization of the enzyme. Cell. 1981;27:449–458. doi: 10.1016/0092-8674(81)90386-x. [DOI] [PubMed] [Google Scholar]
- 12.Herring BP, England PJ. The turnover of phosphate bound to myosin light chain-2 in perfused rat heart. Biochem J. 1986;240:205–214. doi: 10.1042/bj2400205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Herring BP, Fitzsimons DP, Stull JT, Gallagher PJ. Acidic residues comprise part of the myosin light chain-binding site on skeletal muscle myosin light chain kinase. J Biol Chem. 1990;265:16588–16591. [PMC free article] [PubMed] [Google Scholar]
- 14.Herring BP, Nunnally MH, Gallagher PJ, Stull JT. Molecular characterization of rat skeletal muscle myosin light chain kinase. Am J Physiol Cell Physiol. 1989;256:C399–C404. doi: 10.1152/ajpcell.1989.256.2.C399. [DOI] [PubMed] [Google Scholar]
- 15.Herring BP, Smith AF. Telokin expression is mediated by a smooth muscle cell-specific promoter. Am J Physiol Cell Physiol. 1996;270:C1656–C1665. doi: 10.1152/ajpcell.1996.270.6.C1656. [DOI] [PubMed] [Google Scholar]
- 16.Iwaki K, Sukhatme VP, Shubeita HE, Chien KR. Alpha- and beta-adrenergic stimulation induces distinct patterns of immediate early gene expression in neonatal rat myocardial cells: fos/jun expression is associated with sarcomere assembly; Egr-1 induction is primarily an alpha 1-mediated response. J Biol Chem. 1990;265:13809–13817. [PubMed] [Google Scholar]
- 17.Kamm KE, Stull JT. The function of myosin and myosin light chain kinase phosphorylation in smooth muscle. Annu Rev Pharmacol Toxicol. 1985;25:593–620. doi: 10.1146/annurev.pa.25.040185.003113. [DOI] [PubMed] [Google Scholar]
- 18.Kobayashi H, Inoue A, Mikawa T, Kuwayama H, Hotta Y, Masaka T, Ebashi S. Isolation of cDNA for bovine stomach 155kDa protein exhibiting myosin light chain kinase activity. J Biochem (Tokyo) 1992;112:786–791. doi: 10.1093/oxfordjournals.jbchem.a123976. [DOI] [PubMed] [Google Scholar]
- 19.Lazar V, Garcia JG. A single human myosin light chain kinase gene (MLCK; MYLK) Genomics. 1999;57:256–267. doi: 10.1006/geno.1999.5774. [DOI] [PubMed] [Google Scholar]
- 20.Leachman SA, Gallagher PJ, Herring BP, McPhaul MJ, Stull JT. Biochemical properties of chimeric skeletal and smooth muscle myosin light chain kinases. J Biol Chem. 1992;267:4930–4938. [PMC free article] [PubMed] [Google Scholar]
- 21.Lin P, Luby-Phelps K, Stull JT. Binding of myosin light chain kinase to cellular actin-myosin filaments. J Biol Chem. 1997;272:7412–7420. doi: 10.1074/jbc.272.11.7412. [DOI] [PubMed] [Google Scholar]
- 22.Morano I, Hofmann F, Zimmer M, Ruegg JC. The influence of P-light chain phosphorylation by myosin light chain kinase on the calcium sensitivity of chemically skinned heart fibres. FEBS Lett. 1985;189:221–224. doi: 10.1016/0014-5793(85)81027-9. [DOI] [PubMed] [Google Scholar]
- 23.Murakami N, Trenkner E, Elzinga M. Changes in expression of nonmuscle myosin heavy chain isoforms during muscle and nonmuscle tissue development. Dev Biol. 1993;157:19–27. doi: 10.1006/dbio.1993.1108. [DOI] [PubMed] [Google Scholar]
- 24.Nunnally MH, Hsu LC, Mumby MC, Stull JT. Structural studies of rabbit skeletal muscle myosin light chain kinase with monoclonal antibodies. J Biol Chem. 1987;262:3833–3838. [PubMed] [Google Scholar]
- 25.Nunnally MH, Stull JT. Mammalian skeletal muscle myosin light chain kinases: a comparison by antiserum cross-reactivity. J Biol Chem. 1984;259:1776–1780. [PubMed] [Google Scholar]
- 26.Patapoutian A, Wold BJ, Wagner RA. Evidence for developmentally programmed transdifferentiation in mouse esophageal muscle. Science. 1995;270:1818–1821. doi: 10.1126/science.270.5243.1818. [DOI] [PubMed] [Google Scholar]
- 27.Potier MC, Chelot E, Pekarsky Y, Gardiner K, Rossier J, Turnell WG. The human myosin light chain kinase (MLCK) from hippocampus: cloning, sequencing, expression, and localization to 3qcen-q21. Genomics. 1995;29:562–570. doi: 10.1006/geno.1995.9965. [DOI] [PubMed] [Google Scholar]
- 28.Roush CL, Kennelly PJ, Glaccum MB, Helfman DM, Scott JD, Krebs EG. Isolation of the cDNA encoding rat skeletal muscle myosin light chain kinase: sequence and tissue distribution. J Biol Chem. 1988;263:10510–10516. [PubMed] [Google Scholar]
- 29.Sanbe A, Fewell JG, Gulick J, Osinska H, Lorenz J, Hall DG, Murray LA, Kimball TR, Witt SA, Robbins J. Abnormal cardiac structure and function in mice expressing nonphosphorylatable cardiac regulatory myosin light chain 2. J Biol Chem. 1999;274:21085–21094. doi: 10.1074/jbc.274.30.21085. [DOI] [PubMed] [Google Scholar]
- 30.Scordilis SP, Uhlendorf BW, Scarpa S, Cantoni GL, Miles JM, Adelstein RS. Changes in myosin and myosin light chain kinase during myogenesis. Biochemistry. 1981;20:3511–3516. doi: 10.1021/bi00515a032. [DOI] [PubMed] [Google Scholar]
- 31.Shoemaker MO, Lau W, Shattuck RL, Kwiatkowski AP, Matrisian PE, Guerra-Santos L, Wilson E, Lukas TJ, Van Eldik LJ, Watterson DM. Use of DNA sequence and mutant analyses and antisense oligodeoxynucleotides to examine the molecular basis of nonmuscle myosin light chain kinase autoinhibition, calmodulin recognition, and activity. J Cell Biol. 1990;111:1107–1125. doi: 10.1083/jcb.111.3.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Simons M, Wang M, McBride OW, Kawamoto S, Yamakawa K, Gdula D, Adelstein RS, Weir L. Human nonmuscle myosin heavy chains are encoded by two genes located on different chromosomes. Circ Res. 1991;69:530–539. doi: 10.1161/01.res.69.2.530. [DOI] [PubMed] [Google Scholar]
- 33.Stull JT, Blumenthal DK, Miller JR, DiSalvo J. Regulation of myosin phosphorylation. J Mol Cell Cardiol. 1982;14(Suppl 3):105–110. doi: 10.1016/0022-2828(82)90137-7. [DOI] [PubMed] [Google Scholar]
- 34.Sweeney HL, Bowman BF, Stull JT. Myosin light chain phosphorylation in vertebrate striated muscle: regulation and function. Am J Physiol Cell Physiol. 1993;264:C1085–C1095. doi: 10.1152/ajpcell.1993.264.5.C1085. [DOI] [PubMed] [Google Scholar]
- 35.Takeda K, Yu ZX, Qian S, Chin TK, Adelstein RL, Ferrans VJ. Nonmuscle myogin II localizes to the Z-lines and intercalated discs of cardiac muscle and to the Z-lines of skeletal muscle. Cell Motil Cytoskeleton. 2000;46:54–68. doi: 10.1002/(SICI)1097-0169(200005)46:1<59::AID-CM6>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 36.Walsh MP, Vallet B, Autric F, Demaille JG. Purification and characterization of bovine cardiac calmodulin-dependent myosin light chain kinase. J Biol Chem. 1979;254:12136–12144. [PubMed] [Google Scholar]
- 37.Wolf H, Hofmann F. Purification of myosin light chain kinase from bovine cardiac muscle. Proc Natl Acad Sci USA. 1980;77:5852–5855. doi: 10.1073/pnas.77.10.5852. [DOI] [PMC free article] [PubMed] [Google Scholar]