Abstract
It has recently been suggested that the use of optimal codons limits mistranslation-induced protein misfolding, yet evidence for this remains largely circumstantial. In contrast, molecular chaperones have long been recognized to play crucial roles in misfolding prevention and remedy. We propose that putative error limitation in cis can be elucidated by examining the interaction between codon usage and chaperoning processes. Using Escherichia coli as a model system, we find that codon optimality covaries with dependency on the chaperonin GroEL. Sporadic but not obligate substrates of GroEL exhibit higher average codon adaptation and are conspicuously enriched for optimal codons at structurally sensitive sites. Further, codon optimality of sporadic clients is more conserved in the E. coli clone Shigella dysenteriae. We suggest that highly expressed genes cannot routinely use GroEL for error control so that codon usage has evolved to provide complementary error limitation. These findings provide independent evidence for a role of misfolding in shaping gene evolution and highlight the need to co-characterize adaptations in cis and trans to unravel the workings of integrated molecular systems.
Keywords: codon bias, GroEL, misfolding
Introduction
While mutation is a rare process, errors during all stages of gene expression occur at considerably higher rates (Lynch, 2007). Increasingly, there is interest in the possibility that, because such errors are relatively commonplace, many features of gene and genome anatomy and organization evolve as error-reduction or error-mitigation mechanisms (Maquat and Carmichael, 2001; Willensdorfer et al, 2007; Drummond and Wilke, 2008; Jaillon et al, 2008; Hurst, 2009; Zaher and Green, 2009). An example of the latter has been provided by analysis of intron composition in Paramecium tetraurelia. Across species, a large fraction of alternative splice isoforms probably originates from errors in the splicing process rather than representing functional variation (Melamud and Moult, 2009; Zhang et al, 2009). In line with this notion, short introns in Paramecium are under selection to render transcripts recognizable to the nonsense-mediated decay machinery (they contain premature termination signals) if they fail to be spliced out (Jaillon et al, 2008).
Downstream of splicing, the polypeptide chain that emerges from the ribosome may fail to fold into its native structure. At one extreme, protein function may barely be compromised and the cost of misfolding therefore minimal. At the other extreme, however, some aberrantly folded proteins, exposing hydrophobic residues that would normally be buried, may begin to promiscuously interact with other proteins, become toxic to the cell and thus pose a substantial fitness concern (Gregersen et al, 2006). Unsurprisingly then, echoing the case of splicing, there are signatures of evolved error management.
In trans, molecular chaperones increase the likelihood that proteins reach native state. First, chaperones like the DnaK/DnaJ/GrpE system in Escherichia coli bind to folding intermediates and prevent aggregation in a crowded cellular environment. Second, binding as well as stepwise cycling on and off the polypeptide chain can narrow the folding landscape the nascent protein is allowed to explore, thus channelling the protein towards native state (Hartl and Hayer-Hartl, 2009). Third, some chaperones can unfold misfolded proteins in an energy-dependent process. This allows de novo exploration of alternative folding pathways for proteins that would otherwise be stuck at a local kinetic optimum or ushers the misfolded protein into degradation (Weber-Ban et al, 1999; Wickner et al, 1999; Lin et al, 2008).
In cis, codon usage has recently been attributed adaptive roles both in shaping folding pathways (via local control of the speed of translation) (Komar et al, 1999; Cortazzo et al, 2002; Neafsey and Galagan, 2007) and minimizing folding errors (Drummond and Wilke, 2008). Evidence for the latter is centred on the argument that synonymous codons differ in their propensity to cause mistranslation. Translationally optimal codons (henceforth simply referred to as ‘optimal codons’) are defined as those codons that are relatively enriched in very highly expressed genes. Typically, these codons are represented by more abundant cognate tRNAs (Ikemura, 1981; Akashi, 1994; Duret, 2000) and consequently thought less likely to cause ribosomal stalling and/or incorporation of the wrong amino acid. However, while there is good evidence for some transcriptomes that selection operates to avoid such mistranslation events (‘translational accuracy’) (Stoletzki and Eyre-Walker, 2007; Drummond and Wilke, 2008), whether this actually reflects selection against misfolding remains controversial. Mistranslation may simply be deleterious, even if the protein has folded correctly, because the erroneously inserted amino acid compromises protein function.
Support for the misfolding hypothesis comes from a recent study by Zhou et al (2009). The authors compared solvent-exposed and buried amino-acid residues, mutations at the latter type of site being more likely to disrupt protein structure. Consistent with selection on codon usage to reduce mistranslation-induced misfolding, they found optimal codons in E. coli and other organisms to be moderately enriched at these structurally sensitive sites. However, as the authors acknowledge, the extent to which buried sites represent structurally rather than functionally important sites remains to be established. Consequently, we do not know whether optimal codons might be enriched at these sites, at least in part, to avoid mistranslation-induced malfunctioning. In addition, other studies have failed to detect a link to misfolding. Kudla et al (2009) monitored the expression of >150 constructs all encoding the same green fluorescent protein but with synonymous codon identity randomized across sites. Despite such radically altered codon usage patterns, the authors found no differences in the amount of misfolded protein produced by different constructs, assayed as the ratio of total protein (determined by Coomassie) to functional protein (determined by fluorescence), and no relationship between putative misfolding rates and codon usage bias. This may reflect the fact that misfolding is not related to codon usage or may simply be owing to a lack of power in the experiments to detect small, but evolutionarily significant, misfolding rates.
Here we propose a novel test of the hypothesis that evolution of protein-coding genes is modulated by selection to avoid misfolding. We suggest that the role, if any, of error limitation in cis (for which we examine codon usage) can be revealed by studying its interaction with well-established error management systems in trans (chaperones). If codon usage does indeed play a tangible role in misfolding prevention, we would expect selection on codon identity to vary with the degree to which a protein can rely on other error control mechanisms, namely chaperones.
What direction this covariation should take is not necessarily obvious. Are proteins that are particularly liable to misfolding both regular clients of chaperones and employ a greater number of optimal codons? This could be expected, for example, if substantive energetic costs could be avoided by getting folding right first time around, rather than having to subject substrates to repeated refolding cycles. Alternatively, might selection on codon usage be relaxed, rather than strengthened, in proteins that interact with chaperones to attain native state? This may apply in particular to proteins that are habitually passaged through chaperones, which can therefore serve as a reliable error control.
Support for such a selective relief scenario comes from experiments using E. coli demonstrating that certain deleterious mutations, presumably affecting folding competence, can effectively be buffered by overexpression of the chaperonin GroEL (Fares et al, 2002b; Tokuriki and Tawfik, 2009). Does buffering extend beyond amino-acid substitutions to synonymous codon identity? Such a finding would provide strong support for a role of codons in misfolding prevention. More generally, such a finding would bolster the hypothesis that gene evolution is modulated by selection against misfolding.
In this study, then, we integrate genome-scale sequence, expression, structural and protein interaction data from E. coli to elucidate the interplay between chaperone dependency and codon usage in managing misfolding. In the chaperonin GroEL/GroES (henceforth simply referred to as GroEL) E. coli arguably has the best-characterized chaperone system of any organism. GroEL simultaneously provides a sheltered folding environment (passively preventing aggregation) and guides folding through hydrophilic residues that line the inside of its cylindrical cavity (Sigler et al, 1998). Further, recent evidence suggests that GroEL can partially unfold proteins to allow renewed exploration of the folding landscape (Lin et al, 2008). This is important because it implies that errors introduced during translation, where codon usage reportedly reduces error rates, may be remedied after the event.
Putative in vivo substrates of GroEL have been determined on a genome-wide scale (Kerner et al, 2005; Chapman et al, 2006). Exploiting the fact that substrate release is an energy-dependent process, Kerner et al (2005) could trap substrates by rapid ATP depletion and subsequently co-purify them with GroEL. Based on enrichment in GroEL complexes and validated by in vitro refolding assays, the authors assigned ∼250 proteins to three classes reflecting GroEL dependency.
Class-I proteins, only a small fraction of which (<1%) associates with GroEL, show low propensity to aggregate upon dilution from denaturant, spontaneously regain some enzymatic activity and regain full activity when DnaK (acting as an aggregation inhibitor upstream of GroEL) is added.
Proteins allocated to class-II fail to refold at 37°C, but spontaneous refolding is observed at more permissive temperatures (25°C) and DnaK addition again re-establishes high levels of correct folding. In the following sections, we will sometimes refer to proteins from classes-I and II as sporadic clients.
Finally, class-III proteins are obligate substrates of GroEL, largely failing to refold even under more benign conditions and the DnaK system unable to rescue. Notably, although on average less abundant than class-I/II proteins, class-III proteins occupy ∼80% of GroEL's capacity in vivo (Kerner et al, 2005). Consequently, a higher proportion (∼100% versus ∼20% for class-II and ∼1% for class-I) of these proteins is routinely processed by the GroEL system.
Results
Isolating misfolding propensity as a determinant of codon usage bias
Figure 1A suggests that average codon bias strongly varies by GroEL dependency (Kruskal–Wallis test, P=5.59E−12). However, this observation does not by itself make a case for a functional relationship between codon usage and chaperone dependency as far as the management of misfolding is concerned. Importantly, in as far as codon usage reflects selection, the degree of bias will mirror the strength of selection, which is proportional to the overall cost of misfolding. Yet, for any individual gene, cost is the product not only of error propensity, but also of the deleterious effects of any one individual error (e.g., toxicity), and of the absolute frequency at which the error occurs. That the latter is a key determinant of cost is manifest in the observation that expression level is the strongest known predictor of codon bias in E. coli (Stoletzki and Eyre-Walker, 2007).
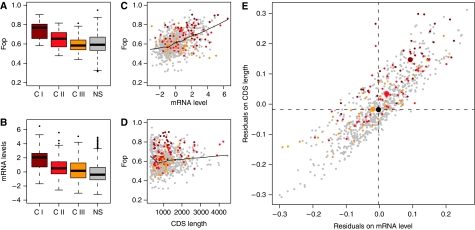
Figure 1.
Obtaining codon bias residuals in E. coli. (A) The frequency of optimal codons (Fop) varies as a function of GroEL dependency. (B) Fop also varies as a function of expression level. (C) LOWESS regression fitted for Fop on the natural logarithm of mRNA levels (from Bernstein et al (2002); LB medium) and (D) CDS length. (E) Residuals extracted from both regressions plotted against each other. The median residual value by substrate class on both axes is indicated by a larger dot of the corresponding colour. GroEL substrate classes are coloured gray (non-substrates; ‘NS’; n=706), dark red (class-I; ‘C I’; n=33), red (class-II; ‘C II’; n=49) and orange (class-III; ‘C III’; n=37).
Comparison of Figure 1A and B deepens the suspicion that elevated codon bias in class-I/II genes is first and foremost owing to higher expression of these genes. As our objective is to isolate misfolding propensity as a determinant of codon usage differentials between GroEL dependency classes, we thus need to control for expression. We can do this by studying residuals from a regression of codon usage bias (measured as the frequency of optimal codons, Fop) on expression levels. The regression line provides us with an approximate expectation of cost (and therefore selection on codon usage) for a given expression level. Subsequently, we can ask whether there are systematic deviations for certain groups of genes (here GroEL dependency classes) to exhibit codon usage above or below what we would expect given their expression.
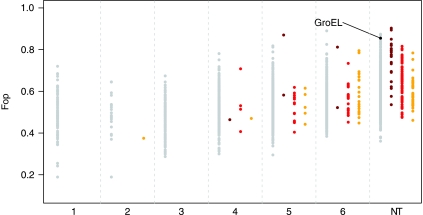
In the absence of comprehensive translation rate or protein abundance measures for E. coli, we use microarray data to approximate translation frequencies. Microarray measurements are typically noisy and agreement across studies and platforms is often unsatisfactory (dos Reis et al, 2003). This may be cause for concern when analysing the relationship between expression and codon usage. In particular, dos Reis et al (2003) reported a curious U-shaped relationship between codon usage bias and mRNA levels in the largest data set they analysed. We considered four genome-wide expression data sets and recovered the same apparently inconsistent relationship in data from the ASAP database (Figure 2C, top row) when we fitted non-parametric LOWESS regressions. However, we note the following: first, regressing mRNA level on codon usage inverts the likely underlying biological relationship (also pointed out by Wu et al, 2007). Recent evidence strongly supports the view that optimal codons are unlikely to be employed to boost protein levels. More likely, codon usage adapts to a given expression level in order to reduce errors (more costly in more frequently translated transcripts) and/or minimize ribosomal dwelling time (Andersson and Kurland, 1990; Kudla et al, 2009). Regressing codon usage on expression level yields not a mirror image (as LOWESS regressions are locally fitted) but a consistent picture across data sets (Figure 2, centre row). More importantly, even the inverse regressions harmonize across studies when we exclude genes with evidence for lateral gene transfer (LGT) (Figure 2, bottom row). Rather than merely conveniently censoring the offending data set, exclusion of LGT candidate genes is a meaningful constraint to impose on our data. We are principally interested in the interaction between chaperone dependency and codon usage at equilibrium. Yet genes that enter the E. coli genome via LGT are rarely codon-adapted, evident in the amelioration of codon usage towards the host genome over time (Figure 3). Moreover, GroEL interactors tend to be long-term residents of the E. coli genome (clustering of substrates towards the right-hand side in Figure 3), further suggesting that interactions between this chaperone system and codon usage might differ systematically for LGT genes. Thus, our analyses below focus on genes not derived from suspected LGT events (see Materials and methods). All results reported below hold across all expression data sets.
Figure 2.
Codon usage–expression correlations harmonize across data sets upon removal of LGT candidates. Panels in the top row show the relationship between codon usage and mRNA levels for (A) Bernstein et al (2002) (LB medium); (B) Bernstein et al (2002) (M9 medium); (C) the ASAP database and (D) Covert et al (2004), with LOWESS regressions fitted using a range of smoothing parameters f. Inverse regressions of the same data sets are depicted in the centre row. The bottom row corresponds to the top row with LGT candidates removed (see Materials and methods). Expression values (hybridization intensities) have been transformed with the natural logarithm.
Figure 3.
Non-equilibrium codon usage in laterally transferred genes. Genes are ordered based on the time of transfer into the E. coli genome: ‘1’ comprising the most recent transfer events and ‘NT’ signifying no evidence for transfer within the γ-proteobacteria (see Materials and methods). Genes transferred into the E. coli genome exhibit below-average codon usage bias, but appear to ameliorate over time to resemble the codon usage of long-term residents. GroEL substrates are typically long-term residents in the E. coli genome. GroEL substrate classes are coloured gray (non-substrates), dark red (class-I), red (class-II) and orange (class-III).
Expression level is the main factor that needs to be taken into account in order to establish to what extent codon usage might reflect misfolding propensity. Ideally, however, we also want to control for the varying severity of individual errors. Severity is difficult to predict on a transcriptome-wide basis, certainly for costs like toxicity, which will be strongly dependent on the context in which the error occurs. We can, however, approximate one particular aspect of severity: from a perspective of energy expenditure, errors during the synthesis of longer proteins should be more costly, if only because more energy is wasted in futile peptide synthesis and degradation. As a result, longer proteins in E. coli exhibit greater codon bias (Stoletzki and Eyre-Walker, 2007). We thus also control for protein length in attempting to isolate misfolding propensity as the causative agent of codon usage variation between GroEL substrate classes.
Residual analysis
We fitted LOWESS regressions of Fop on mRNA levels and protein length (see Materials and methods; Figure 1C and D) to obtain residual codon usage values. Figure 1E demonstrates that proteins occupy typical ranges in the Fop residual space according to their dependency on GroEL. Class-I proteins in particular have higher residual codon usage than non-substrates (Mann–Whitney U-test, P(expression)=1.29E−05, P(protein length)=7.61E−12), while class-III proteins are indistinguishable from non-substrates (P(expression)=0.28, P(protein length)=0.87). These effects are evident for all expression data sets analysed (Supplementary Figure 1). To confirm these differences using an alternative approach, we also matched each substrate (class-I–III) gene with a non-substrate gene expressed at a similar level. To be conservative we required the non-substrate gene to be at least as highly expressed and at least as long as the class-I/II/III gene, so that, if anything, we biased expected codon optimality towards non-substrates. Despite the reduced power of this approach, we recovered above-expectation codon optimality for class-I proteins (Mann–Whitney U-test, P=0.033) and no difference for class-III proteins (P=0.56).
We suggest the following explanation: As mentioned above, class-III substrates are defined not only by GroEL being critical for proper folding, but also by occupying most of GroEL's capacity (∼80%). With a high proportion of class-III protein passaged through the GroEL system, mistranslation errors in these proteins weigh less severely as GroEL can remedy at least some misfolding that ensues. In contrast, class-I and II genes are more highly expressed and cannot routinely rely on GroEL to rectify folding errors. Yet class-I/II proteins are clearly liable to misfold as testified by their sporadic association with GroEL. We hypothesize that augmenting GroEL's capacity to address the misfolding propensity of these genes would be prohibitively costly to the organism and that, as an alternative strategy, these genes employ optimal codons to reduce the rate of misfolding error.
Interestingly, we find 100% of genes in our sample annotated for involvement in unfolded protein binding (GO:0051082), that is, chaperones themselves, to have positive residuals (adjusted P=0.019, see Materials and methods). This might simply highlight that the strength of selection on these genes (as far as translational frequency is concerned) is poorly estimated under standard conditions—after all, many chaperones are much more highly expressed in response to stress. However, it is equally consistent with a model where codon usage complements chaperone activity to achieve adequate folding—chaperones, especially at times of cellular trauma, should be prime targets of selection to reduce error rates to an absolute minimum.
Sporadic GroEL clients exhibit strong enrichment of optimal codons at structurally sensitive sites
A recent study by Zhou et al (2009) exploited information on three-dimensional protein structures to compare codon usage patterns at sites of different solvent accessibility. The authors found that amino-acid residues with restricted solvent accessibility (<25%, ‘buried sites’), considered structurally sensitive, were more likely to be encoded by optimal codons than exposed sites, consistent with the hypothesis that codon choice can function to limit the frequency of folding errors during translation.
The above model of complementarity between codon usage and chaperone activity predicts that sporadic GroEL clients (class-I/II) should exhibit a stronger tendency for optimal codons to be associated with buried sites than class-III genes, which presumably experience relaxed selection in the presence of regular error correction provided by GroEL. Replicating the analysis of Zhou et al (see Materials and methods), but distinguishing by substrate class, we find strong support for our model. Odds ratios for finding optimal codons more frequently employed at buried versus exposed sites are dramatically higher for class-I/II genes (Table I). In contrast, class-III genes exhibit no significant enrichment and, in fact, have a smaller odds ratio than non-substrates. Trends are enhanced when considering only amino acids with odds ratios significantly >1 (L/N/Q/S/T) across all proteins analysed by Zhou et al. These findings are consistent with the hypothesis that habitual association with GroEL mediates release from selective constraint. They also support the notion that buried sites chiefly represent structurally rather than functionally sensitive sites, as for the latter we would not have expected any systematic covariance with GroEL dependency.
Table 1. Enrichment of optimal codons at structurally sensitive sites in E. coli varies by GroEL dependency.
| Amino acids | Zhou et al (odds ratio) | P | Non-substrates (odds ratio) | P | Class-III (odds ratio) | P | Class-II (odds ratio) | P | Class-I (odds ratio) | P |
|---|---|---|---|---|---|---|---|---|---|---|
| All | 1.06 | <0.001 | 1.09 | 1.16E−05 | 1.02 | 0.83 | 1.24 | 0.001 | 1.25 | 0.002 |
| L/N/Q/S/T | 1.25 | 5.29E−32 | 1.29 | 8.72E−16 | 1.08 | 0.51 | 1.59 | 8.36E−05 | 1.79 | 5.31E−05 |
Conservation of codon optimality varies across substrate classes
The evidence presented above coherently points towards codon choice at structurally sensitive sites being of greater selective significance for highly expressed sporadic clients of GroEL. To further corroborate this hypothesis we examined the extent to which codon optimality at buried sites has been retained in orthologous codons of Shigella dysenteriae. Shigella ‘species’ are clones of E. coli that have adopted an intracellular parasitic lifestyle (Lan et al, 2004). Concomitant with this lifestyle, these clones have experienced reductions in effective population size. This is a useful feature for our analysis: In small populations, the fate of slightly deleterious mutations is principally determined by genetic drift. Synonymous codon choice is generally considered to be a weakly selected trait so that mutations to non-optimal codons should typically fall into the ‘slightly deleterious’ category. Consistent with this notion, severely reduced codon optimality has been detected in species with long-term bottlenecking populations such as the aphid endosymbiont Buchnera (Wernegreen and Moran, 1999; Moya et al, 2002; Rispe et al, 2004). Thus, in S. dysenteriae we find a system where codon usage is under reduced purifying selection. At the same time, the S. dysenteriae lineage diverged relatively recently from the E. coli K12 strain (Balbi et al, 2009) so that chaperoning dynamics should be virtually identical. In contrast, one outstanding feature of Buchnera and other longstanding intracellular species is dramatically increased GroEL output, quite possibly under selective pressure to handle the increasing number of proteins that have accumulated destabilizing mutations (Moran, 1996; Fares et al, 2002a).
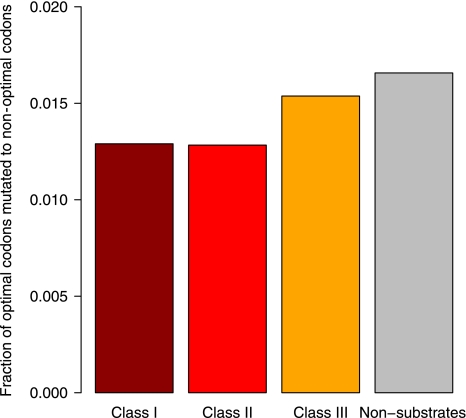
We followed the fate of ancestrally optimal codons at buried sites in the S. dysenteriae and E. coli K12 genomes, considering only sites at which codon optimality has been retained in E. coli (thus enriching for structurally sensitive sites, see Materials and methods). Our complementarity hypothesis predicts that the decay in optimality should be less prominent for class-I/II substrates. This can be tested by comparing substrate classes with regard to the fraction of buried sites that lose optimal codons in S. dysenteriae. Class-I/II substrates should experience proportionally fewer changes from optimal to non-optimal codons. Figure 4 suggests that this is indeed the case. However, as differences in expression between substrate classes are still evident in this subset of genes (Kruskal–Wallis test statistic=12.7909, P<0.006), might this simply be an artefact of stronger selection owing to higher expression? To rule out this possibility, we pooled sporadic GroEL clients (class-I/II) in order to boost sample size (and thus power) and paired each gene with a non-substrate expressed at a similar level as described above. Despite the relatively small sample size (see legend to Figure 4), we find that class-I/II genes do indeed show fewer changes than expected (Fisher test odds ratio 0.65, P=0.034), whereas class-III genes do not (Fisher test odds ratio: 1.10, P=0.79).
Figure 4.
The fraction (by substrate class) of codon-optimal buried sites in E. coli where an ancestrally optimal codon has mutated to a non-optimal codon in S. dysenteriae. The absolute numbers of optimal to non-optimal transitions observed across all amino acids are 454, 35, 28, 29 for non-substrates, class-I, II and III genes, respectively.
Discussion
While the majority of E. coli proteins (∼70%) reach their native folding conformation spontaneously, a persistent minority require assistance via one or more chaperone systems (Hartl and Hayer-Hartl, 2009). Based on the analysis of experimentally identified GroEL substrates in E. coli, we have uncovered an interaction between optimal codon usage and chaperone dependency, providing strong independent support that molecular evolution at the codon level is partially driven by misfolding concerns. Prior limited analysis (Noivirt-Brik et al, 2007) suggested that above-average codon adaptation was a general hallmark of GroEL substrates. This is not so; rather, sporadic GroEL clients experience selection to use optimal codons, whereas obligate substrates do not. We argue that this can be understood in a simple framework of energy economics: although occasional errors in highly expressed proteins can be handled by GroEL, these proteins cannot be quality-controlled in bulk by chaperones so that selection in cis complements the error control capacity of chaperones by reducing the incidence of misfolding errors.
The combined burden of non-synonymous and synonymous mutations
Our findings also suggest that the capacity of GroEL to buffer deleterious mutations (Fares et al, 2002b; Tokuriki and Tawfik, 2009) extends to synonymous changes. This has strong implications for assessing the mutational load that has to be shouldered by chaperones, particularly in the context of Buchnera and other intracellular bacteria. Computational analyses suggest that Buchnera proteins have low intrinsic folding efficiency owing to unfavourable amino-acid content (van Ham et al, 2003; Bastolla et al, 2004). Observations that purifying selection on Buchnera GroEL is atypically high (Wernegreen and Moran, 1999; Fares et al, 2002a), that positive selection has occurred in its apical domain, possibly to broaden substrate specificity (Fares et al, 2002a) and that GroEL levels have been substantially increased, are all consistent with the notion that GroEL has adopted a critical role in buffering the deleterious effects of such unfavourable mutations (Moran, 1996). Yet the burden on GroEL to provide an error correction facility might go well beyond what has been anticipated, for Buchnera also exhibits severely compromised codon adaptation (Wernegreen and Moran, 1999; Moya et al, 2002; Rispe et al, 2004).
Cis–trans complementarity in misfolding management beyond E. coli
Codon usage patterns consistent with selection against mistranslation-induced misfolding have been reported not only for E. coli but also several eukaryotes, notably yeast, Drosophila and mammals (Drummond and Wilke, 2008; Zhou et al, 2009).
In addition, comprehensively handling highly expressed yet folding-sensitive proteins through energy-dependent chaperones should be challenging, if not unfeasible, for any cellular system, regardless of differences in chaperone repertoire and action between prokaryotes and eukaryotes (Young et al, 2004). Can we then find a similar interaction between chaperone dependency and codon usage in molecular systems other than that of E. coli?
Protein interaction data for the eukaryotic chaperonin CCT/TRiC provides us with an opportunity to address this question in a comparative manner. Initially believed to be specifically required only for the folding of cytoskeletal proteins actin and tubulin (Leroux and Hartl, 2000), a small number of other obligate interactors have since been identified (e.g., Siegers et al, 2003). As our focus is as much on sporadic as it is on obligate chaperonin clients, we aimed to cast our net widely and analysed the data of Dekker et al (2008) who identified interactors of CCT/TRiC (including putative substrates) by a combination of physical and genetic interaction screens.
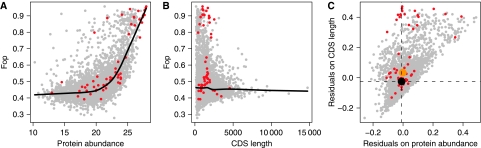
Following the same recipe as for E. coli, we fitted LOWESS regressions predicting Fop on expression level (here protein abundance, see Materials and methods) and CDS length (Figure 5A and B). Note that the relationship between Fop and CDS length in Saccharomyces cerevisiae is negative, chiefly owing to highly expressed genes being short. As a consequence, residual deviations on length or mRNA level partially subsume each other. As Dekker et al did not discriminate between substrate classes, we would expect the set of ∼60 interactors they describe, as far as these do in fact represent bona fide substrates, to comprise a mixture of sporadic and obligate clients. Hence, the overall trend should go towards higher residual deviation of interactors. The data suggest that this is indeed the case. We find a significant tendency towards higher residuals on CDS length (MWU, P=3.5E−10) but not expression level (MWU, P=0.14) for CCT/TRiC interactors (Figure 5C), a finding echoed when we apply the gene-matching approach described above (matching by length: P=1.54E−10; matching by expression: P=0.87). This trend is, to a certain extent, also evident for individual protein components of the CTT/TRiC complex (Supplementary Figure 2), whose interaction with other proteins was recently investigated using TAP-tagged proteins (Gong et al, 2009).
Figure 5.
Obtaining codon bias residuals in S. cerevisiae. (A) LOWESS regressions fitted for Fop on the natural logarithm of protein abundance measured by de Godoy et al (2008) and (B) CDS length. (C) The residuals extracted from both regressions are plotted against each other. The median residual value by substrate class on both axes is indicated by a larger dot. CCT/TRiC substrates are coloured red/orange.
For the same reason as above, we expect odds ratios to be generally higher at buried sites in putative substrates versus non-substrates. Analysing residues from 303 protein structures annotated by Zhou et al, results across all amino acids defy expectations: the combined odds ratio of substrates is lower than that of non-substrates (Table II). Note, however, that only 17 substrate proteins are present in the set of 303 protein structures so that estimates are likely to be noisy. If we focus on amino acids (G/I/R/S) with evidence for enrichment of optimal codons at structurally sensitive sites across all 303 proteins, we do observe the expected higher odds for CCT/TRiC substrates (Table II).
Table 2. Enrichment of optimal codons at structurally sensitive sites in S. cerevisiae varies by CCT/TRiC interaction status.
| Amino acids | Zhou et al, 2009 (odds ratio) | P | Non-interactors (odds ratio) | P | Interactors (odds ratio) | P |
|---|---|---|---|---|---|---|
| All | 1.10 | <0.001 | 1.12 | 1.62E−13 | 1.005 | 0.96 |
| G | 1.42 | <0.001 | 1.48 | 3.24E−12 | 2.28 | 0.27 |
| I | 1.17 | <0.05 | 1.08 | 0.59 | 1.44 | 0.89 |
| R | 1.19 | <0.05 | 1.24 | 0.04 | 5.45 | 0.38 |
| S | 1.25 | <0.001 | 1.19 | 0.005 | 1.85 | 0.38 |
| G/I/R/S | 1.28 | 4.77E−17 | 1.28 | 5.66E−15 | 2.15 | 0.003 |
Substrate residuals appear to fall into two clusters along the CDS length-derived residual dimension (Figure 5C). Given that residual size was partly predictive of sporadic and obligate clients in E. coli, it is tempting to speculate that one might be able to exploit this relationship to predict degrees of dependency on CCT/TRiC. We heuristically chose a cut-off of 0.2, which separates the two apparent clusters (Figure 5C), and examined a small number of known obligate substrates of CCT/TRiC. We find some support for our prediction, with substrates such as tubulin and myosin proteins exhibiting residuals <0.2 (Supplementary Table I). However, actin, a well-known obligate client of CCT/TRiC, constitutes a notable exception (residual=0.35).
We did not examine higher eukaryotes in this study, principally because (a) data quality on chaperonin dynamics does not approach that of E. coli or yeast and (b) contributory factors to misfolding costs are harder to tease apart because the expression-codon bias correlation is distorted by regional biases in nucleotide content (‘isochore effects’) (Eyre-Walker and Hurst, 2001). We note, however, that Zhou et al (2009) reported enrichment of optimal codons at structurally sensitive sites not only for E. coli and yeast, but also for mouse and Drosophila, suggesting that misfolding-related selection on codon usage might be phylogenetically widespread. In fact, mammals might provide a particularly interesting system to analyse how codons function in protein folding, not only in relation to error reduction but also concerning the active regulation of folding. With regard to the latter, it has been suggested that non-optimal codons can be adaptively placed to regulate translation speed and thus allow stepwise folding at the ribosome (Oresic and Shalloway, 1998; Komar et al, 1999; Cortazzo et al, 2002; Neafsey and Galagan, 2007). This mode of folding control should be particularly pertinent to organisms such as mammals with many multidomain proteins because (a) slowdown could facilitate stepwise folding of individual domains (Thanaraj and Argos, 1996; Oresic et al, 2003) and (b) multidomain proteins would be typically unable to exploit the full power of the chaperonin cage owing to size constraints. Current experimental support for codon-mediated folding control remains limited to a small number of experimentally confirmed examples and genome-scale analysis does not necessarily suggest such a mechanism to be common (e.g., Neafsey and Galagan, 2007). At first it seems paradoxical that both optimal and non-optimal codons should be simultaneously involved in facilitating adequate folding. The apparent contradiction, however, can be resolved if we assume that optimal codons populate structurally sensitive sites, whereas non-optimal codons are employed to achieve slowdown in between domains, where occasional mistranslation would not ordinarily be detrimental.
What this highlights, above all, is that codon usage can be adaptive for more than one mechanistic reason, and that these mechanisms need by no means be mutually exclusive. In fact, codon usage represents the integrated product of a diverse collection of mutational and selective forces. In this regard, the results of Kudla et al (2009), which failed to implicate protein folding, but suggested codon choice to be important for generating adequate mRNA secondary structure, do not invalidate protein folding as a prominent mediator of codon choice, but add another facet to an already complex kaleidoscope of determinants. Future research will doubtlessly find yet more hidden functionality in codon choice. The key task, then, will be to disentangle and quantify the contribution of individual mechanisms to shaping diversity in codon usage within and across genomes.
Materials and methods
Sequence data acquisition and analysis
The E. coli K12 genome (NC_000913) was downloaded from NCBI and protein-coding sequences extracted using custom scripts. To control for known biases in codon composition at the CDS termini, we followed the of protocol of Eyre-Walker (1996) and determined codon usage patterns with the first 50 and the last 20 codons of the CDS removed. Only genes longer than 30 codons after trimming were considered for further analysis. As we are interested in codon usage differentials across the proteome, we did not confine analysis to cytosolic proteins.
S. cerevisiae CDSs were downloaded from the Saccharomyces Genome Database (SGD) (www.yeastgenome.org).
Fop was calculated using codonW, with default parameter settings for E. coli and S. cerevisiae.
Expression data acquisition and analysis
We obtained microarray hybridization intensities as follows: (a) For E. coli grown on rich (LB) and basic (M9) medium from Supplementary Table 6 in Bernstein et al (2002); (b) for wild-type E. coli under aerobic conditions from Supplementary Table 7 in Covert et al (2004), where we only considered genes with at least two present and no absent calls across three replicates and (c) for E. coli grown on LB medium from the ASAP database (https://asap.ahabs.wisc.edu/asap/experiment_data.php), where we averaged intensities from two calibrated data sets (PALSP49 and PALSP50).
Data on protein abundance for log-phase yeast was obtained from Supplementary Table 4 of de Godoy et al (2008) as the summed extracted ion chromatograms of all isotopic patterns detected for the respective peptide.
Chaperonin substrates
Identity and classification of GroEL substrates was taken from Supplementary Table 3 in Kerner et al (2005). Proteins assigned to overlapping classes (n=4, class-I/II) were allocated to class-I. All eligible (see above), expressed CDSs in the current genome annotation that were not considered as GroEL substrates by Kerner et al were allocated to the ‘non-substrate’ class. Proteins were matched to genes, in the first instance, by their SwissProt ID, which is provided both in Kerner et al (2005) and in the NCBI GenBank file. Where the ID given by Kerner et al could not be found in the current genome annotation, matches were made by querying GenProtEC (http://genprotec.mbl.edu/) for all available gene synonyms. Two hypothetical proteins in the work of Kerner et al (ypt1 and ypt2) could not be related to the current genome build and were excluded.
For yeast, proteins interacting with the CCT/TRiC complex were derived from Supplementary Tables 2 and 6 in Dekker et al (2008). Further, Gong et al (2009) used TAP-tagged proteins to define interaction partners (including but not limited to bona fide substrates) for 64 proteins with chaperone activity. We analysed the components of the CCT/TRiC complex (Supplementary Figure 2). Gene identities were matched across studies using the gene registry file from SGD.
Lateral gene transfer
Data on inferred timings of LGT events were obtained from Martin Lercher (personal communication). Briefly, based on the presence/absence of orthologous genes across a phylogeny of >20 proteobacteria, the most parsimonious LGT scenarios were reconstructed in PAUP* using generalized parsimony (see Lercher and Pal, 2008 for details). The authors approximated the age of individual transfers into the E. coli genome by the number of nodes between extant E. coli K12 and the branch on which LGT occurred. The number of nodes ranges from 0 (no evidence for LGT), over 1 (recent LGT on the terminal E. coli branch) to 6 (ancient LGT). Genes where no explicit inference was available were considered anciently resident in the E. coli genome and allocated to node=0. We obtain qualitatively identical results when we exclude these genes. Further, we compared timings inferred using ACCTRAN and DELTRAN algorithms as well as different LGT/gene loss penalties, and found the results to be very similar (data not shown). Following Lercher and Pal (2008), we present results obtained using the DELTRAN algorithm with an LGT:gene loss penalty ratio of 2:1.
LOWESS regressions
Non-parametric LOWESS regressions were fitted and all other statistical analyses were conducted in R (R Development Core Team, 2009). We explored a range of smoothing parameters f (0.2–0.8) and found parameter choice within this range to yield almost identical results (also see Figure 2). We present analysis for f=0.3. We confirmed that residuals for any given regression model and expression data set did not correlate with fitted values or along the predictor variable (expression level or CDS length).
GO analysis
GO terms for E. coli were obtained from EBI (ftp://ftp.ebi.ac.uk/pub/databases/GO/goa/proteomes/18.E_coli_K12.goa).
To investigate whether genes with high positive or negative residuals are more likely to belong to certain functional classes, we applied the FatiScan tool (Al-Shahrour et al, 2006) on a list of genes ranked by their Fop residual. FatiScan tests whether biological labels (here GO terms) are asymmetrically distributed in a ranked list of genes. As mRNA- and CDS length-derived residuals are highly linearly correlated (Figure 1E), we present data for mRNA-derived residuals only. A full list of significant terms derived using default parameter settings can be found in Supplementary Table II.
Analysis of structurally sensitive sites
Annotation of buried (<25% solvent accessibility; see Zhou et al, 2009) and exposed sites for E. coli and S. cerevisiae proteins was kindly provided by Claus Wilke (downloaded from http://openwetware.org/index.php?title=Wilke:Data_Sets). Following the approach of Zhou et al, we confined analysis to genes with sequence identity >40% to those in Protein Data Bank to obtain 822 and 303 eligible genes for E. coli and S. cerevisiae, respectively. To ensure comparability, we checked that optimal codons used in this analysis corresponded to those in Supplementary Table 2 of Zhou et al.
Contingency tables by individual amino acid and gene were constructed as by Zhou et al, excluding tables with a total frequency count <2. Tables were then combined across (selected) amino acids and genes by substrate status (non-substrate, class-I and so on) using the Mantel–Haenszel procedure, a detailed description of which can be found in Zhou et al (2009). P-values were adjusted for multiple testing using the method of Benjamini and Hochberg (1995).
We investigated the effects of sample size on odds ratios and P-values by sub-sampling the same number of contingency tables available in the smallest data set (class-III) from other substrate classes. We find sub-samples from class-I and II genes to show consistently higher average odds ratios than sub-samples taken from non-substrates (Supplementary Figure 3), with class-I and II sub-samples odds ratios significantly >1 in 890 and 1000 out of 1000 repeat random samples, respectively. For other expression data sets, odds ratios are consistently significant for >98% of sub-samples both for class-I and II substrates.
S. dysenteriae
We obtained the alignments of nine E. coli/Shigella strains from Eduardo Rocha through Edward Feil (personal communication; see Rocha (2003) for details on orthologue identification and alignment). These data have been used in previous published research (Rocha, 2003; Balbi et al, 2009). The phylogenetic relationships between strains was taken from Balbi et al (2009). We chose to focus on S. dysenteriae in favour of Shigella strains more recently diverged from E. coli K12 because the latter are likely to contain a higher proportion of sites where selection has not yet had the chance to purge slightly deleterious mutations. We analysed buried sites from all non-LGT genes among the 822 genes for which structural annotation was available (see above). We only considered sites (a) that encoded the same degenerate amino acid in E. coli K12 and S. dysenteriae, (b) where the ancestrally used codon was optimal (inferred by parsimony from strains EAEC 042, CFT073 and UT189) and (c) where the site had retained codon optimality in E. coli K12. We then simply determined, by substrate class, the proportion of sites where a transition to a non-optimal codon had occurred in S. dysenteriae.
In Supplementary Table III we provide a list of all genes analysed in this study along with the corresponding substrate classifications, expression parameters, and so on, that should allow easy replication of our results.
Supplementary Material
includes Supplementary Figures 1–3 and Supplementary Table 1
FatiScan results
E. coli data
Acknowledgments
We thank Claus Wilke and Tong Zhou for making their annotation of protein structures available promptly upon request. Similarly, we are grateful to Eduardo Rocha and Edward Feil for provision of and help with the E. coli/Shigella alignments. We also thank Allan Drummond for valuable comments during the early phase of this research and Catherine Pink for critical reading of the paper. TW is funded by the Medical Research Council, UK. LDH is a Royal Society-Wolfson Research Merit Award Holder.
Footnotes
The authors declare that they have no conflict of interest.
References
- Akashi H (1994) Synonymous codon usage in Drosophila melanogaster: natural selection and translational accuracy. Genetics 136: 927–935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Shahrour F, Minguez P, Tarraga J, Montaner D, Alloza E, Vaquerizas JM, Conde L, Blaschke C, Vera J, Dopazo J (2006) BABELOMICS: a systems biology perspective in the functional annotation of genome-scale experiments. Nucleic Acids Res 34: W472–W476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson SG, Kurland CG (1990) Codon preferences in free-living microorganisms. Microbiol Rev 54: 198–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balbi KJ, Rocha EP, Feil EJ (2009) The temporal dynamics of slightly deleterious mutations in Escherichia coli and Shigella spp. Mol Biol Evol 26: 345–355 [DOI] [PubMed] [Google Scholar]
- Bastolla U, Moya A, Viguera E, van Ham RC (2004) Genomic determinants of protein folding thermodynamics in prokaryotic organisms. J Mol Biol 343: 1451–1466 [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B 57: 289–300 [Google Scholar]
- Bernstein JA, Khodursky AB, Lin PH, Lin-Chao S, Cohen SN (2002) Global analysis of mRNA decay and abundance in Escherichia coli at single-gene resolution using two-color fluorescent DNA microarrays. Proc Natl Acad Sci USA 99: 9697–9702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman E, Farr GW, Usaite R, Furtak K, Fenton WA, Chaudhuri TK, Hondorp ER, Matthews RG, Wolf SG, Yates JR, Pypaert M, Horwich AL (2006) Global aggregation of newly translated proteins in an Escherichia coli strain deficient of the chaperonin GroEL. Proc Natl Acad Sci USA 103: 15800–15805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortazzo P, Cervenansky C, Marin M, Reiss C, Ehrlich R, Deana A (2002) Silent mutations affect in vivo protein folding in Escherichia coli. Biochem Biophys Res Commun 293: 537–541 [DOI] [PubMed] [Google Scholar]
- Covert MW, Knight EM, Reed JL, Herrgard MJ, Palsson BO (2004) Integrating high-throughput and computational data elucidates bacterial networks. Nature 429: 92–96 [DOI] [PubMed] [Google Scholar]
- de Godoy LM, Olsen JV, Cox J, Nielsen ML, Hubner NC, Frohlich F, Walther TC, Mann M (2008) Comprehensive mass-spectrometry-based proteome quantification of haploid versus diploid yeast. Nature 455: 1251–1254 [DOI] [PubMed] [Google Scholar]
- Dekker C, Stirling PC, McCormack EA, Filmore H, Paul A, Brost RL, Costanzo M, Boone C, Leroux MR, Willison KR (2008) The interaction network of the chaperonin CCT. EMBO J 27: 1827–1839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- dos Reis M, Wernisch L, Savva R (2003) Unexpected correlations between gene expression and codon usage bias from microarray data for the whole Escherichia coli K-12 genome. Nucleic Acids Res 31: 6976–6985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond DA, Wilke CO (2008) Mistranslation-induced protein misfolding as a dominant constraint on coding-sequence evolution. Cell 134: 341–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duret L (2000) tRNA gene number and codon usage in the C. elegans genome are co-adapted for optimal translation of highly expressed genes. Trends Genet 16: 287–289 [DOI] [PubMed] [Google Scholar]
- Eyre-Walker A (1996) Synonymous codon bias is related to gene length in Escherichia coli: selection for translational accuracy? Mol Biol Evol 13: 864–872 [DOI] [PubMed] [Google Scholar]
- Eyre-Walker A, Hurst LD (2001) The evolution of isochores. Nat Rev Genet 2: 549–555 [DOI] [PubMed] [Google Scholar]
- Fares MA, Barrio E, Sabater-Munoz B, Moya A (2002a) The evolution of the heat-shock protein GroEL from Buchnera, the primary endosymbiont of aphids, is governed by positive selection. Mol Biol Evol 19: 1162–1170 [DOI] [PubMed] [Google Scholar]
- Fares MA, Ruiz-Gonzalez MX, Moya A, Elena SF, Barrio E (2002b) Endosymbiotic bacteria: groEL buffers against deleterious mutations. Nature 417: 398. [DOI] [PubMed] [Google Scholar]
- Gong Y, Kakihara Y, Krogan N, Greenblatt J, Emili A, Zhang Z, Houry WA (2009) An atlas of chaperone-protein interactions in Saccharomyces cerevisiae: implications to protein folding pathways in the cell. Mol Syst Biol 5: 275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregersen N, Bross P, Vang S, Christensen JH (2006) Protein misfolding and human disease. Annu Rev Genomics Hum Genet 7: 103–124 [DOI] [PubMed] [Google Scholar]
- Hartl FU, Hayer-Hartl M (2009) Converging concepts of protein folding in vitro and in vivo. Nat Struct Mol Biol 16: 574–581 [DOI] [PubMed] [Google Scholar]
- Hurst LD (2009) Evolutionary genomics and the reach of selection. J Biol 8: 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikemura T (1981) Correlation between the abundance of Escherichia coli transfer RNAs and the occurrence of the respective codons in its protein genes: a proposal for a synonymous codon choice that is optimal for the E. coli translational system. J Mol Biol 151: 389–409 [DOI] [PubMed] [Google Scholar]
- Jaillon O, Bouhouche K, Gout JF, Aury JM, Noel B, Saudemont B, Nowacki M, Serrano V, Porcel BM, Segurens B, Le Mouel A, Lepere G, Schachter V, Betermier M, Cohen J, Wincker P, Sperling L, Duret L, Meyer E (2008) Translational control of intron splicing in eukaryotes. Nature 451: 359–362 [DOI] [PubMed] [Google Scholar]
- Kerner MJ, Naylor DJ, Ishihama Y, Maier T, Chang HC, Stines AP, Georgopoulos C, Frishman D, Hayer-Hartl M, Mann M, Hartl FU (2005) Proteome-wide analysis of chaperonin-dependent protein folding in Escherichia coli. Cell 122: 209–220 [DOI] [PubMed] [Google Scholar]
- Komar AA, Lesnik T, Reiss C (1999) Synonymous codon substitutions affect ribosome traffic and protein folding during in vitro translation. FEBS Lett 462: 387–391 [DOI] [PubMed] [Google Scholar]
- Kudla G, Murray AW, Tollervey D, Plotkin JB (2009) Coding-sequence determinants of gene expression in Escherichia coli. Science 324: 255–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan R, Alles MC, Donohoe K, Martinez MB, Reeves PR (2004) Molecular evolutionary relationships of enteroinvasive Escherichia coli and Shigella spp. Infect Immun 72: 5080–5088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lercher MJ, Pal C (2008) Integration of horizontally transferred genes into regulatory interaction networks takes many million years. Mol Biol Evol 25: 559–567 [DOI] [PubMed] [Google Scholar]
- Leroux MR, Hartl FU (2000) Protein folding: versatility of the cytosolic chaperonin TRiC/CCT. Curr Biol 10: R260–R264 [DOI] [PubMed] [Google Scholar]
- Lin Z, Madan D, Rye HS (2008) GroEL stimulates protein folding through forced unfolding. Nat Struct Mol Biol 15: 303–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M (2007) The Origins of Genome Architecture. Sunderland, Massachusetts: Sinauer Associates [Google Scholar]
- Maquat LE, Carmichael GG (2001) Quality control of mRNA function. Cell 104: 173–176 [DOI] [PubMed] [Google Scholar]
- Melamud E, Moult J (2009) Stochastic noise in splicing machinery. Nucleic Acids Res 37: 4873–4886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran NA (1996) Accelerated evolution and Muller's rachet in endosymbiotic bacteria. Proc Natl Acad Sci USA 93: 2873–2878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moya A, Latorre A, Sabater-Munoz B, Silva FJ (2002) Comparative molecular evolution of primary (Buchnera) and secondary symbionts of aphids based on two protein-coding genes. J Mol Evol 55: 127–137 [DOI] [PubMed] [Google Scholar]
- Neafsey DE, Galagan JE (2007) Positive selection for unpreferred codon usage in eukaryotic genomes. BMC Evol Biol 7: 119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noivirt-Brik O, Unger R, Horovitz A (2007) Low folding propensity and high translation efficiency distinguish in vivo substrates of GroEL from other Escherichia coli proteins. Bioinformatics 23: 3276–3279 [DOI] [PubMed] [Google Scholar]
- Oresic M, Dehn M, Korenblum D, Shalloway D (2003) Tracing specific synonymous codon-secondary structure correlations through evolution. J Mol Evol 56: 473–484 [DOI] [PubMed] [Google Scholar]
- Oresic M, Shalloway D (1998) Specific correlations between relative synonymous codon usage and protein secondary structure. J Mol Biol 281: 31–48 [DOI] [PubMed] [Google Scholar]
- R Development Core Team (2009) R: a Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing [Google Scholar]
- Rispe C, Delmotte F, van Ham RC, Moya A (2004) Mutational and selective pressures on codon and amino acid usage in Buchnera, endosymbiotic bacteria of aphids. Genome Res 14: 44–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocha EP (2003) DNA repeats lead to the accelerated loss of gene order in bacteria. Trends Genet 19: 600–603 [DOI] [PubMed] [Google Scholar]
- Siegers K, Bolter B, Schwarz JP, Bottcher UM, Guha S, Hartl FU (2003) TRiC/CCT cooperates with different upstream chaperones in the folding of distinct protein classes. EMBO J 22: 5230–5240 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Sigler PB, Xu Z, Rye HS, Burston SG, Fenton WA, Horwich AL (1998) Structure and function in GroEL-mediated protein folding. Annu Rev Biochem 67: 581–608 [DOI] [PubMed] [Google Scholar]
- Stoletzki N, Eyre-Walker A (2007) Synonymous codon usage in Escherichia coli: selection for translational accuracy. Mol Biol Evol 24: 374–381 [DOI] [PubMed] [Google Scholar]
- Thanaraj TA, Argos P (1996) Ribosome-mediated translational pause and protein domain organization. Protein Sci 5: 1594–1612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokuriki N, Tawfik DS (2009) Chaperonin overexpression promotes genetic variation and enzyme evolution. Nature 459: 668–673 [DOI] [PubMed] [Google Scholar]
- van Ham RC, Kamerbeek J, Palacios C, Rausell C, Abascal F, Bastolla U, Fernandez JM, Jimenez L, Postigo M, Silva FJ, Tamames J, Viguera E, Latorre A, Valencia A, Moran F, Moya A (2003) Reductive genome evolution in Buchnera aphidicola. Proc Natl Acad Sci USA 100: 581–586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber-Ban EU, Reid BG, Miranker AD, Horwich AL (1999) Global unfolding of a substrate protein by the Hsp100 chaperone ClpA. Nature 401: 90–93 [DOI] [PubMed] [Google Scholar]
- Wernegreen JJ, Moran NA (1999) Evidence for genetic drift in endosymbionts (Buchnera): analyses of protein-coding genes. Mol Biol Evol 16: 83–97 [DOI] [PubMed] [Google Scholar]
- Wickner S, Maurizi MR, Gottesman S (1999) Posttranslational quality control: folding, refolding, and degrading proteins. Science 286: 1888–1893 [DOI] [PubMed] [Google Scholar]
- Willensdorfer M, Burger R, Nowak MA (2007) Phenotypic mutation rates and the abundance of abnormal proteins in yeast. PLoS Comput Biol 3: e203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, Nie L, Freeland SJ (2007) The effects of differential gene expression on coding sequence features: analysis by one-way ANOVA. Biochem Biophys Res Commun 358: 1108–1113 [DOI] [PubMed] [Google Scholar]
- Young JC, Agashe VR, Siegers K, Hartl FU (2004) Pathways of chaperone-mediated protein folding in the cytosol. Nat Rev Mol Cell Biol 5: 781–791 [DOI] [PubMed] [Google Scholar]
- Zaher HS, Green R (2009) Fidelity at the molecular level: lessons from protein synthesis. Cell 136: 746–762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Xin D, Wang P, Zhou L, Hu L, Kong X, Hurst LD (2009) Noisy splicing, more than expression regulation, explains why some exons are subject to nonsense-mediated mRNA decay. BMC Biol 7: 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou T, Weems M, Wilke CO (2009) Translationally optimal codons associate with structurally sensitive sites in proteins. Mol Biol Evol 26: 1571–1580 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
includes Supplementary Figures 1–3 and Supplementary Table 1
FatiScan results
E. coli data