Abstract
Respiring Saccharomyces cerevisiae cells respond to a sudden increase in glucose concentration by a pronounced drop of their adenine nucleotide content ([ATP]+[ADP]+[AMP]=[AXP]). The unknown fate of ‘lost’ AXP nucleotides represented a long-standing problem for the understanding of the yeast's physiological response to changing growth conditions. Transient accumulation of the purine salvage pathway intermediate, inosine, accounted for the apparent loss of adenine nucleotides. Conversion of AXPs into inosine was facilitated by AMP deaminase, Amd1, and IMP-specific 5′-nucleotidase, Isn1. Inosine recycling into the AXP pool was facilitated by purine nucleoside phosphorylase, Pnp1, and joint action of the phosphoribosyltransferases, Hpt1 and Xpt1. Analysis of changes in 24 intracellular metabolite pools during the respiro-fermentative growth transition in wild-type, amd1, isn1, and pnp1 strains revealed that only the amd1 mutant exhibited significant deviations from the wild-type behavior. Moreover, mutants that were blocked in inosine production exhibited delayed growth acceleration after glucose addition. It is proposed that interconversion of adenine nucleotides and inosine facilitates rapid and energy-cost efficient adaptation of the AXP pool size to changing environmental conditions.
Keywords: ATP homeostasis, metabolic regulation, purine nucleotide metabolism, respiro-fermentative transition, yeast
Introduction
Yeast cells are exposed to strongly fluctuating nutrient concentrations in their environment, which requires rapid and efficient adaptation through rearrangements on all levels of their metabolism. These rearrangements serve to ensure energy supply from different carbon sources, they enable tight coordination of metabolic processes that provide and consume energy, and ultimately facilitate growth or survival. The quantitative understanding of these adaptation processes represents the basis for a directed optimization of the microorganism to suit the needs of biochemical production processes that often impose non-uniform or harsh cultivation conditions (Lara et al, 2006), or require a redirection of metabolic fluxes to improve productivity (Bailey, 1991).
In this context, controlled perturbation experiments represent a valuable tool, as they allow studying the transition from one defined physiological state to another under well-characterized conditions. Examples for investigations of such type are experiments in which yeast cultures are exposed to sudden changes in nutrient concentration (Visser et al, 2004), oxygen supply (van den Brink et al, 2008), the administration of electron acceptors other than oxygen (Mashego et al, 2006), or metabolic uncouplers (Kresnowati et al, 2008), respectively, to investigate the metabolic response. Measurements of metabolite pool dynamics and enzymatic activities in response to different perturbations enable the quantitative mathematical analysis of glycolytic dynamics, which is ultimately meant to discriminate the impact of allosteric regulation and changes in the enzymatic make-up of the cell on the overall metabolic response (Rizzi et al, 1997; Teusink et al, 2000; Daran-Lapujade et al, 2007; van den Brink et al, 2008).
Owing to its economic role as the most important ethanol producer, the transition from respiratory to fermentative growth of Saccharomyces cerevisiae received particularly strong attention. After the pioneering work of Theobald et al (1993) and the optimization of sampling and analytical techniques in recent years (Visser et al, 2002; Mashego et al, 2006; Loret et al, 2007; Canelas et al, 2008), it is now well established that the immediate response of the cell's metabolism implies strong changes in glycolytic metabolite and cofactor levels, which manifest within few seconds after glucose addition. The presence of glucose and the changes in intracellular metabolite levels activate signaling cascades (Thevelein et al, 2005), among which the cAMP protein kinase A (PKA) pathway has the predominant role in controlling the respiro-fermentative transition (Mbonyi et al, 1988; Rolland et al, 2002; Wang et al, 2004; Zaman et al, 2009). The activation of PKA triggers posttranslational modification of enzymes involved in gluconeogenesis (Mazon et al, 1982), glycolysis (Francois et al, 1984) and storage of carbohydrates (Francois and Parrou, 2001), and eventually controls the transcriptional downregulation of respiratory and gluconeogenetic pathways (Thevelein et al, 2005; Zaman et al, 2009).
A long standing problem in the context of these studies is the apparent loss of adenine nucleotides, which immediately follows the relief from glucose limitation (Theobald et al, 1997; Kresnowati et al, 2006). Given the fact that adenine nucleotides are important regulators of several glycolytic enzymes, the unknown fate of AXPs must compromise the mathematical investigation of dynamic metabolic interactions (Teusink et al, 2000; Bosch et al, 2008; Nikerel et al, 2009). In addition, metabolic rearrangements on changing environmental conditions serve to ensure the equilibrium between energy supply and demand to prevent the catastrophic loss of energy that may cause substrate accelerated death (Thevelein and Hohmann, 1995; Teusink et al, 1998; Gancedo and Flores, 2004). Thus, the fact that the fate of apparently lost adenine nucleotides is still unclear points to a significant lack in our understanding of mechanisms that rule the respiro-fermentative transition in yeast. Consequently, the search for the destination of AXP nucleotides, and the functional role and regulation of their removal has attained strong attention in recent studies (Visser et al, 2004; Kresnowati et al, 2006), without, however, identifying the pathway(s) implied in AXP homeostasis.
In this study, we showed that the transient accumulation of the purine salvage pathway (PSP) metabolites, IMP and inosine, accounted for the apparent loss of AXP nucleotides. We identified the pathway for inosine accumulation and recycling, and demonstrated that inosine formation is a specific response to perturbations of energy homeostasis under fermentative conditions. In the absence of fermentable sugars, perturbation of energy homeostasis rather resulted in the formation of AMP and no inosine was formed. The impact of defective AXP cycling on the global metabolic response to glucose addition was tested under conditions in which respiration was inhibited by antimycin A: in the amd1 mutant, in which adenine nucleotide cycling was completely blocked, adenylic energy charge exhibited a strong drop after glucose addition, and recovered much slower than in wild-type cells. Furthermore, deletion of AMD1 resulted in strong accumulation of AMP and pronounced changes in the dynamics of several metabolite pools after glucose addition. Although PSP mutants did not exhibit any growth phenotype under constant cultivation conditions, the amd1 and isn1 mutant strains showed delayed growth acceleration after glucose addition. Differences in growth dynamics were not accompanied by changes in the formation of the fermentative end products: ethanol and glycerol.
Results
Shake flask cultivation on trehalose mimics glucose-limited growth and can be used for metabolic analyses after glucose upshift
Analysis of the respiro-fermentative transition is classically carried out with yeast cells cultivated in chemostat at low dilution rate, resulting in a constant growth rate and fully respiratory growth. However, this cultivation mode requires comparatively sophisticated instrumentation and is, thus, less suitable for high throughput studies using a large collection of mutants. In a recent study, we showed that the yeast S. cerevisiae cultivated on trehalose grew at a constant growth rate of 0.045 h−1, which is well below 0.1 h−1 commonly set in chemostat experiments. This constant growth rate resulted from the rate-limiting cleavage of the disaccharide trehalose into two glucose molecules by the periplasmic acid trehalase encoded by ATH1 (Jules et al, 2004). Consequently, the metabolic regimen was strictly respiratory, with no fermentative end products, ethanol and glycerol, detected. In this study, we used the fact that glucose-limited growth can be conveniently mimicked by shake flask cultivations using trehalose as the limiting carbon source (Jules et al, 2005).
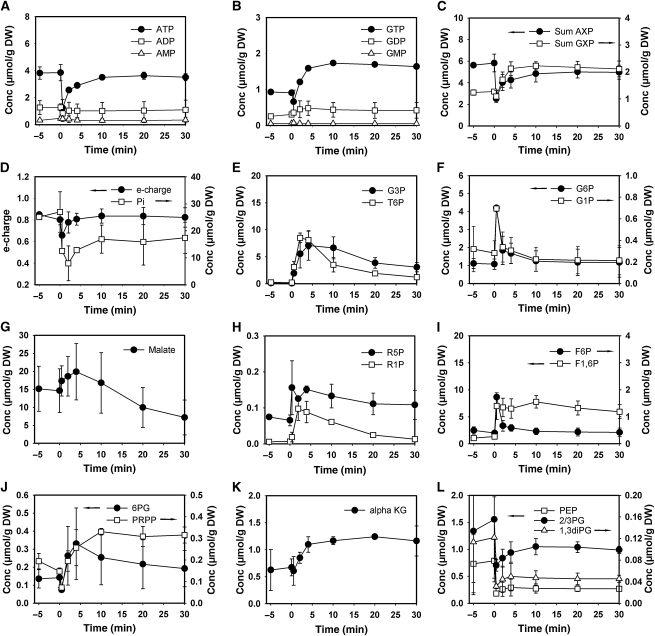
Figure 1 shows typical profiles of intracellular metabolite pools in response to the addition of glucose to wild-type cells growing on trehalose. Immediately after the relief from glucose limitation, the phosphorylated sugars: glucose-6-phosphate (G6P), trehalose-6-phosphate (T6P), glucose-1-phosphate (G1P), and fructose-1,6-bisphosphate (F1,6P) exhibited a strong increase (Figure 1E, F and I). High F1,6P levels were maintained in the presence of excess glucose, which is consistent with its positive effect on the yeast's fermentation capacity brought about by the activation of pyruvate kinase. In contrast, G6P, F6P, G1P accumulated only transiently and decreased to their initial levels within few minutes. Similar to what was first observed by Hohmann et al (1996), T6P concentration passed through a maximum of ∼9 μmol/g DW, which was 30 times higher than before glucose administration, and stabilized at a concentration of ∼0.5 μmol/g DW after 30 min, which was only 2–3 times higher than under fully respiratory conditions. Metabolites in the lower part of glycolysis (Figure 1L), that is, 1,3-phosphoglycerate (1,3-diPglyc), 2-phosphoglycerate and 3-phosphoglycerate (measured as a single pool denoted 2/3-Pglyc), as well as phosphoenolpyruvate (PEP), exhibited the characteristic decrease reported earlier (Groussac et al, 2000; Visser et al, 2004). In agreement with previous studies (Theobald et al, 1997; Kresnowati et al, 2006), a pronounced transient drop of adenine (AXP) nucleotide concentrations was observed (Figure 1C), which was caused by the decrease of ATP (Figure 1A). Changes in the AXP balance were accompanied by a transient decrease in the adenylic energy charge (e-charge=([ATP]+0.5[ADP])/([ATP]+[ADP]+[AMP]) (Chapman and Atkinson, 1977)). Similarly, GXP nucleotide concentration exhibited a slight decrease immediately after glucose addition (Figure 1C). However, although the AXP concentration quickly recovered to about 80% of its initial level, the GXP concentration reached a new steady state that was significantly higher than that before glucose addition. Finally, we report an increase in the concentrations of the pentose phosphate pathway (PPP) metabolites: 6-phosphogluconate (6PG), ribose-1-phosphate (R1P), and the ribose-5-phosphate/ribulose-5-phosphate pool (measured as a single pool denoted R5P), which was accompanied by a significant augmentation in the concentration of the purine nucleotide precursor PRPP (Figure 1H and J). Altogether, the intracellular metabolite pool dynamics in trehalose-grown cells exposed to glucose addition were in good agreement with earlier studies that were exclusively carried out in chemostat cultivations (Theobald et al, 1997; Vaseghi et al, 1999; Visser et al, 2004; Kresnowati et al, 2006). Therefore, we conclude that transitory phenomena observed during the shift from glucose limitation to glucose excess can be conveniently studied in our comparatively simple experimental set-up.
Figure 1.
Intracellular concentrations of glycolytic, TCA cycle, and pentose phosphate pathway metabolites after exposure of trehalose-grown wild-type cells to sudden increase of glucose concentration to 110 mmol/l. (A) ATP, ADP, and AMP; (B) GTP, GDP, and GMP; (C) sum of adenine and guanine nucleotides; (D) energy charge, inorganic phosphate (Pi); (E) glycerol-3-phosphate (G3P); trehalose-6-phosphate (T6P); (F) glucose-6-phosphate (G6P); glucose-1-phosphate (G1P); (G) malate; (H) ribose-5-phosphate (R5P); ribose-1-phosphate (R1P); (I) fructose-6-phosphate (F6P); fructose-1,6-bisphosphate (F1,6P); (J) 6-phosphogluconate (6PG); phosphoribosyl pyrophosphate (PRPP); (K) alpha-ketoglutarate (alphaKG); (L) phosphoenolpyruvate (PEP); pooled 2 and 3-phosphoglycerate (2/3PG); 1,3-phosphodiglycerate (1,3diPG). Data shown represent the average of at least two independent experiments. Metabolite concentrations are provided in tabular form in Supplementary information II.
IMP and inosine are the major sinks for adenine nucleotides
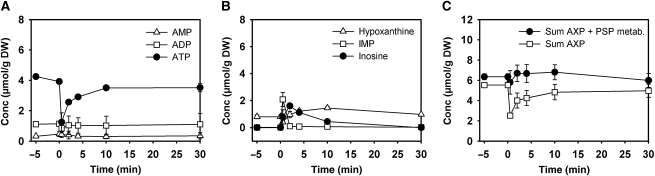
The fact that the decrease of the AXP concentration was not counterbalanced by an increase in the other purine and pyrimidine nucleotide pools ((Kresnowati et al, 2006), also see Figure 1C) led us to investigate the behavior of PSP metabolites after glucose addition (Loret et al, 2007). Our analyses revealed that only IMP and inosine responded significantly to glucose addition. As can be seen in Figure 2, IMP and inosine concentration increased transiently after glucose administration, with IMP being detected only within the first ∼2 min, and inosine being detectable in the cells for at least 10 min. The sum of AXP, IMP, inosine, and hypoxanthine did not decrease after the glucose pulse, therefore, indicating that accumulation of IMP and inosine fully account for the apparent loss of adenine nucleotides commonly observed under these conditions.
Figure 2.
Intracellular concentrations of adenine nucleotides and selected purine salvage pathway metabolites after exposure of trehalose-grown wild-type cells to sudden increase of glucose concentration to 110 mmol/l. (A) ATP, ADP, and AMP; (B) hypoxanthine, inosine, IMP; (C) sum of AXP nucleotides, sum of (AXP+IMP+inosine+hypoxanthine). Data represent the average of at least two independent experiments.
Inosine formation is a specific response to perturbations of the AXP balance that requires the presence of a fermentable carbon source or a sugar derivative that undergoes phosphorylation by hexokinase
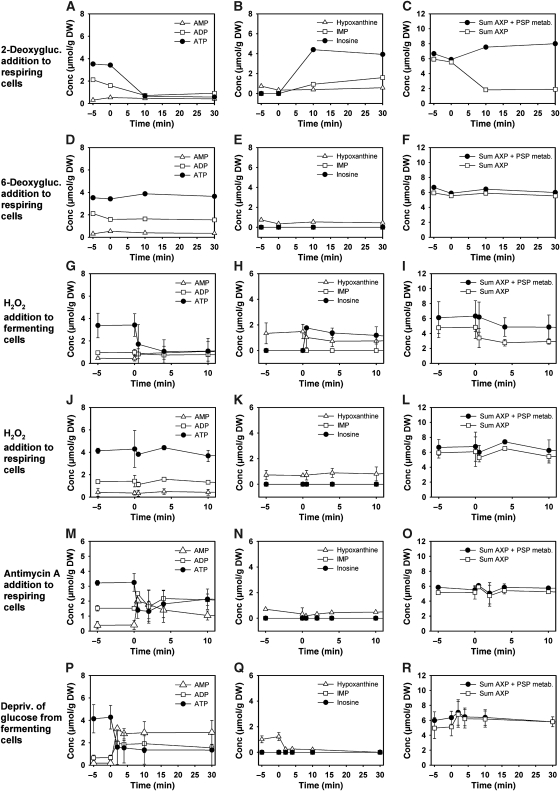
We next set out to investigate which factors cause the accumulation of inosine and exposed respiring yeast cells to different carbon sources. Table I summarizes the results of these experiments. Fermentable sugars, addition of which provoked a drop in ATP level similar to glucose, induced formation of inosine. Furthermore, the administration of 2-deoxyglucose, a sugar derivative that can only undergo the first glycolytic phosphorylation step by hexokinase, caused a drop in ATP level and formation of inosine (Figure 3A–C). Administration of 6-deoxyglucose that is not phosphorylated and does not provoke changes in the AXP balance did not suffice to induce accumulation of inosine (Figure 3D–E). Similarly, when glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was rapidly inactivated by the addition of hydrogen peroxide (Grant et al, 1999) to the cultivation medium of fermenting cells, inosine accumulated in response to the resulting drop in ATP concentration (Figure 3G–I). When GAPDH was inactivated in respiring cells, AXP levels remained constant and no inosine was formed (Figure 3J–L; inactivation of GAPDH under both fermentative and respiratory conditions was verified experimentally; data not shown). When intracellular ATP concentrations were caused to drop in the absence of a fermentable carbon source, for example, by addition of the mitochondrial inhibitor, antimycin A, to respiring cells (Figure 3M–O), or by rapidly depriving fermenting cells from glucose (Figure 3P–R), inosine was not formed but AMP accumulated instead. Therefore, we conclude that inosine formation is a specific response to perturbations of the AXP balance that requires the presence of a fermentable carbon source or a sugar derivative that undergoes rapid phosphorylation by hexokinase.
Table I. Accumulation of inosine in response to exposure of trehalose-grown wild-type cells to different nutrients and sugar derivatives.
| Condition | Concentration [μmol/g DW] |
|||
|---|---|---|---|---|
| ATP | Inosine | Sum AXP | Sum (AXP+inosine) | |
| Trehalose-grown wild-type cells were exposed to addition of different carbon sources and sugar derivatives. | ||||
| aIntracellular metabolite concentrations before substrate addition are shown. Data represent the average of at least two independent experiments. | ||||
| bIntracellular metabolite concentrations 3 min after substrate addition are shown. Data represent the average of at least two independent experiments. | ||||
| Trehalose mediuma | 3.7±0.50 | 0 | 5.7±0.56 | 5.7±0.56 |
| 20 g/l Glucoseb | 2.4±0.26 | 1.8±0.4 | 3.9±0.56 | 5.7±0.30 |
| 20 g/l Fructoseb | 1.8±0.31 | 2.5±0.4 | 3.5±0.26 | 5.9±0.30 |
| 20 g/l Mannoseb | 1.8±0.33 | 2.4±0.4 | 3.1±0.34 | 5.5±0.30 |
| 100 g/l Ethanolb | 3.5±0.25 | 0 | 5.6±0.36 | 5.6±0.36 |
| 20 g/l 6-Deoxyglucoseb | 3.9±0.18 | 0 | 5.5±0.20 | 5.5±0.40 |
| 20 g/l 2-Deoxyglucoseb | 0.6±0.15 | 4.1±0.33 | 1.8±0.10 | 5.9±0.26 |
Figure 3.
Intracellular concentrations of adenine nucleotides and selected purine salvage pathway metabolites in response to addition of sugar derivatives and different perturbations of energy homeostasis under fermentative and respiratory conditions. Addition of (A–C) 6-deoxyglucose, (D–F) 2-deoxyglucose (each at a final concentration of 110 mmol/l) to trehalose-grown cells. Addition of H2O2 (final concentration 2 mM) to (G–I) glucose-grown or (J–L) trehalose-grown cells. (M–O) Addition of antimycin A (final concentration 2 μg/ml) to trehalose-grown cells. (P–R) Transfer of glucose-grown cells to medium containing 110 mmol/l trehalose as the only carbon source. Cells were cultivated on glucose until mid-exponential phase, spun down by centrifugation, and resuspended in trehalose medium having the same composition except for the carbon source. (Cells that were resuspended in glucose medium as a control did not exhibit any change in AXP concentration; data not shown.) All experiments were carried out using wild-type cells. Left panel of figures: ATP, ADP, and AMP. Middle panel of figures: hypoxanthine, inosine, and IMP. Right panel of figures: Sum of AXP nucleotides, sum of (AXP+IMP+inosine+hypoxanthine). Data represent the average of at least two independent experiments.
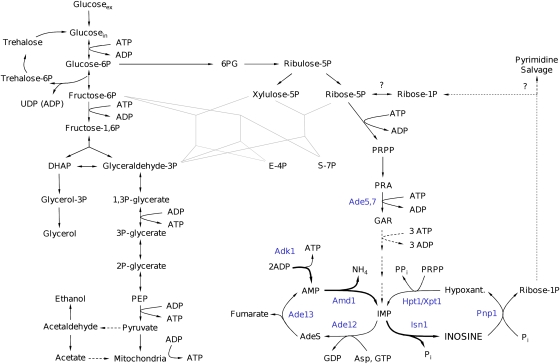
Identification of the pathway for inosine accumulation and recycling
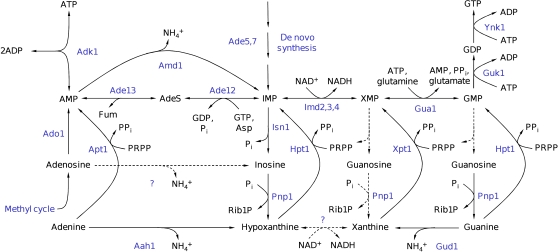
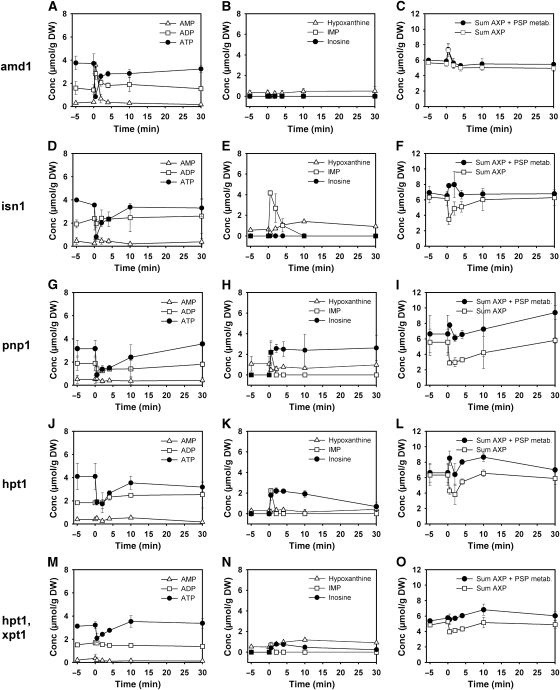
In subsequent experiments, the metabolic pathway for inosine formation and recycling was identified. Figure 4 depicts a schematic representation of the purine salvage pathway to show metabolic reactions that are potentially involved in this process. Not all PSP reactions that were identified according to the KEGG database (Kanehisa et al, 2008) have a known representation in S. cerevisiae. Figure 4 accounts for these uncertainties by plotting potentially relevant (but in yeast still unknown) reactions as dashed lines. To identify the reactions responsible for AXP cycling, different PSP mutants were cultivated on trehalose medium, exposed to an increase in glucose concentration, and sampled for intracellular metabolites. Results of these experiments are summarized in Figure 5. The first step of the AXP-to-inosine conversion is catalyzed by AMP deaminase, encoded by AMD1 (Meyer et al, 1989), which transforms AMP into IMP (Figure 4). Glucose addition to the amd1 mutant caused a transient drop in ATP level, which was accompanied by an equimolar increase in AMP, whereas no detectable amounts of IMP or inosine were formed (Figure 5A–C). The sum of AXP nucleotides even exhibited a transient increase contrary to the drop observed in wild-type cells. Whether or not this transient increase was caused by a rapid response of the AXP de novo synthesis pathway or by transiently draining a neighboring metabolite pool into AXP could not be answered by our analyses. The deletion of AMD1 was the only mutation that resulted in the accumulation of AMP after glucose addition (Figure 5). The IMP-specific 5′-nucleotidase, encoded by ISN1 (Itoh et al, 2003), is the only enzyme that catalyses the conversion of IMP into inosine, as proven by the complete block of inosine formation in the isn1 mutant and the pronounced accumulation of IMP (Figure 5D–F). Recycling of inosine is facilitated by the purine nucleoside phosphorylase, encoded by PNP1 (Lecoq et al, 2001b), which converts inosine into hypoxanthine. In the pnp1 strain, inosine levels remained constantly elevated for more than 30 min after glucose addition (Figure 5G–I). Under these conditions, replenishment of the AXP pool and recovery of the initial ATP concentration took considerably longer than in the wild-type strain. The conversion of hypoxanthine to IMP is facilitated by the phosphoribosyltransferase, encoded by HPT1 (Woods et al, 1983). Unexpectedly, hypoxanthine did not accumulate in the hpt1 mutant, but inosine recycling was significantly slowed down (Figure 5J–L). The deletion of another phosphoribosyltransferase, encoded by XPT1, which is known to catalyze the conversion of xanthine into XMP and to rescue hpt1 phenotypes when overexpressed (Guetsova et al, 1999), did not have any effect on the rate of inosine recycling (data not shown). In an attempt to completely abolish inosine recycling, an hpt1 xpt1 double mutant was produced and exposed to sudden glucose addition. Curiously, the double mutant hpt1 xpt1 exhibited a smaller drop in ATP level after glucose addition, and consequently accumulated much less inosine (Figure 5M–O). Contrary to the single phosphoribosyltransferase mutants, inosine was detectable even 60 min after the glucose pulse (60 min time point not shown). However, both single and double phosphoribosyltransferase mutants could not completely block the disappearance of inosine from the cells, which indicates the presence of yet another metabolic route for inosine degradation.
Figure 4.
Schematic representation of purine salvage pathway reactions. Metabolites are depicted in black and enzymes in blue. Dashed lines with question marks indicate reactions that exist according to the KEGG database but which were not yet identified in Saccharomyces cerevisiae. (Metabolites: AdeS, adenylosuccinate; Asp, aspartate; Fum, fumarate; PRPP, phosphoribosyl pyrophosphate; PPi, pyrophosphate; Pi, phosphate. Enzymes: Aah1, adenine deaminase; Ade5,7, aminoimidazole ribotide synthetase and glycinamide ribotide synthetase; Ade13, adenylosuccinate lyase; Ade12, adenylosuccinate synthase; Adk1, adenylate kinase; Ado1, adenosine kinase; Amd1, AMP deaminase; Apt1, adenine phosphoribosyltransferase; Gua1, GMP synthase; Gud1, guanine deaminase; Guk1, guanylate kinase; Hpt1, hypoxanthine–guanine phosphoribosyltransferase; Imd2,3,4, IMP dehydrogenase; Isn1, IMP-specific 5′-nucleotidase; Pnp1, purine nucleoside phosphorylase; Xpt1, xanthine–guanine phosphoribosyltransferase; Ynk1, nucleoside diphosphate kinase; adapted from Lecoq et al (2001a), the KEGG database (Kanehisa et al, 2008), and personal communication from Drs B Daignan-Fornier and B Pinson.
Figure 5.
Intracellular concentrations of adenine nucleotides and selected purine salvage pathway metabolites after exposure of trehalose-grown purine salvage pathway mutants to sudden increase of glucose concentration to 110 mmol/l. Data corresponding to each mutant are arranged row wise. Left panel of figures: ATP, ADP, and AMP. Middle panel of figures: hypoxanthine, inosine, and IMP. Right panel of figures: Sum of AXP nucleotides, sum of (AXP+IMP+inosine+hypoxanthine). Data represent the average of at least two independent experiments. Metabolite concentrations are provided in tabular form in Supplementary information I.
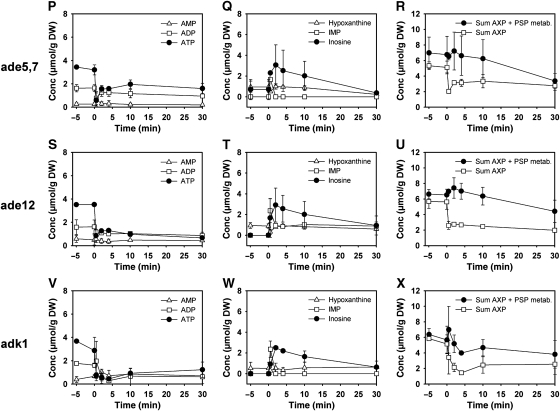
The contribution of the purine nucleotide de novo synthesis to the kinetics of AXP recovery was investigated using an ade5,7 mutant. As this strain was purine base auxotroph, cells were grown in the presence of 0.1 g/l adenine, washed and resuspended in the standard cultivation medium without adenine 1 h before the relief from glucose limitation. We found that inosine formation was not affected in the ade5,7 mutant. However, inosine degradation was slowed down and resulted only in a partial (but significant) recovery of AXP levels (Figure 5P–R).
Figure 5.
Continued.
Transformation of IMP into AMP requires the subsequent action of adenylosuccinate synthase and adenylosuccinate lyase, encoded by ADE12 and ADE13, respectively (Figure 4). The impact of mutations that prevent the conversion of IMP into AMP on AXP and purine nucleotide levels after glucose addition was tested by using an ade12 strain using the same experimental procedures as used for the ade5,7 mutant. The decrease in AXP nucleotides after glucose addition could not be compensated in ade12 (Figure 5S–U), proving that inosine recycling and/or purine nucleotide de novo synthesis are the only pathways that significantly contribute to the regulation of the AXP pool size. Inosine was detectable in the cells over the whole observed time span of 60 min, but continuously decreased without causing an increase in neither hypoxanthine, IMP, nor GXP (data not shown) concentration.
Finally, the contribution of adenylate kinase, encoded by ADK1 (Tomasselli et al, 1986), to AXP homeostasis was tested. When cultivated on trehalose, adk1 cells exhibited the same AXP concentrations as the wild-type cells, indicating that the UMP kinase, encoded by URA6 (Schricker et al, 1992), can compensate for the lack of Adk1 under these conditions. However, after glucose addition initial ATP concentrations were not reestablished and inosine concentration remained elevated for up to 60 min (60 min time point not shown). Similar to the situation in the ade5,7, hpt1 xpt1, and ade12 mutants, the decrease in inosine concentration did not result in the replenishment of the AXP pool. In contrast, an overall drop in purine nucleotide and nucleoside concentrations was observed. Furthermore, the adk1 mutant was the only tested strain incapable to reestablish its adenylic energy charge (e-charge) to the level before glucose addition (Supplementary Figure S1). In the course of our investigation, we additionally tested strains with deletions in ADO1, YNK1, and AAH1 and found that these mutations did not have any significant impact on the kinetics of inosine formation or recycling (data not shown).
Inhibition of respiration affected recovery of AXP nucleotides and altered glycolytic metabolite pool dynamics in the amd1 mutant on glucose addition
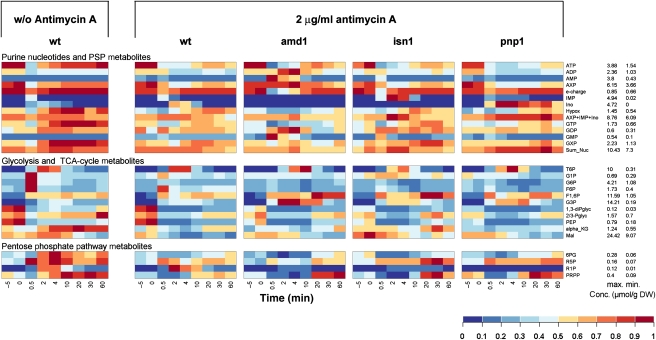
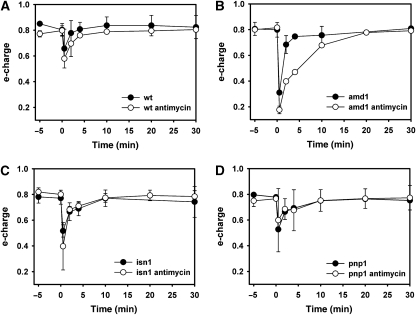
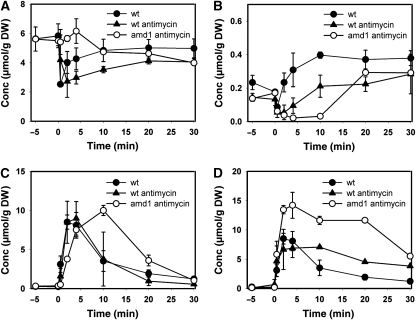
As adenine nucleotides are potent regulators of several glycolytic enzymes (e.g. Zimmermann (1997); Teusink et al (2000)), we asked whether abolishment of inosine formation or recycling, respectively, causes significant differences in the intracellular metabolite pool kinetics of glycolysis, PPP, and PSP during the respiro-fermentative transition. When respiring yeast cells undergo transition to fermentative growth, both glycolytic substrate level phosphorylation and mitochondrial ATP production contribute to the regeneration of ATP levels after glucose addition (Pronk et al, 1996). Thus, a potential deregulation of glycolysis due to impaired conversion of adenine nucleotides into inosine might be partially compensated by mitochondrial activity. Therefore, we investigated the behavior of PSP mutants during relief from glucose limitation when respiration was blocked by the presence of antimycin A (in control experiments it was verified that incubation with antimycin A alone did not cause inosine formation (compare to Figure 3M–O)). We measured 24 intracellular metabolites in the wild-type, amd1, isn1, and pnp1 strains in response to glucose addition in the presence of antimycin A. A color-coded representation was chosen to highlight changes between different mutants and conditions (Figure 6). In addition, graphs showing the metabolite concentrations individually are provided in Supplementary Figures S2–S6. Inosine recycling and ATP regeneration were significantly slowed down in the wild-type strain (compare Figures 6, 2A, B and Supplementary Figure S2), indicating that mitochondrial function contributed significantly to the comparatively fast recovery of ATP levels when antimycin A was absent. Similarly, elevated AMP and IMP concentrations were significantly longer detectable in the amd1 and isn1 mutant strains, respectively (Figures 5, 6 and Supplementary Figure S2). In the absence of antimycin A, ATP concentration in all strains recovered to almost the same level as before glucose addition, whereas presence of this respiratory inhibitor caused the ATP concentration to reach only ∼65% of its initial value (Figures 5, 6 and Supplementary Figure S2). As shown in Figure 7, e-charge dropped to lower levels and recovered much slower in the amd1 mutant than in the wild-type strain and the other tested PSP mutant strains.
Figure 6.
Intracellular metabolite concentrations in trehalose-grown wild-type, amd1, isn1, and pnp1 mutant strains after a sudden increase in glucose concentration to 110 mmol/l in the presence of 2 μg/ml antimycin A. Antimycin A was added 1.5 min before glucose. The left panel shows intracellular concentrations for the WT strain in the absence of antimycin A as a reference. Metabolite concentrations were normalized to the highest concentration measured in all mutants over all time points. Actual concentrations corresponding to maximum and minimum values are shown on the right of the figure. Data represent the average of at least two independent experiments. Metabolite concentrations are provided in tabular form in Supplementary information II.
Figure 7.
Time course of energy charge in trehalose-grown (A) wild-type, (B) amd1, (C) isn1, and (D) pnp1 mutant strains after sudden increase in glucose concentration to 110 mmol/l in the presence or absence of 2 μg/ml antimycin A. Antimycin A was added 1.5 min before glucose. Data represent the average of at least two independent experiments.
The kinetics of intracellular metabolite concentrations in response to glucose were significantly altered in the wild-type when mitochondrial function was impeded by the presence of the inhibitor antimycin A (Figure 6). When cells were treated with antimycin A before glucose addition, the phosphorylated sugars: G1P, G6P and F6P exhibited a less pronounced peak. Furthermore, individual guanosine nucleotide (GTP, GDP, and GMP) concentrations and their sum (GXP) did not show the significant increase after glucose addition, as observed in the absence of antimycin A. Similarly, 6PG, R5P, and PRPP concentrations exhibited a less pronounced increase on glucose addition when respiration was inhibited. Antimycin-treated wild-type, isn1, and pnp1 cells had a very similar metabolite profile in response to glucose addition, whereas that of the amd1 mutant was significantly different. The increase in PRPP concentration was strongly delayed in the amd1 mutant (Figures 6, 8B, and Supplementary Figures S3–S6). Furthermore, deletion of AMD1 resulted in significant changes in the dynamics of glycolytic metabolite concentrations. In particular, G3P concentration increased two-fold higher than in the wild-type strain, and T6P levels decreased significantly slowly than in all other tested strains (Figures 6, 8C, and Supplementary Figures S3–S6). Finally, we observed that R1P only accumulated to significant concentrations when Pnp1 was functional, and inosine was converted into hypoxanthine (Figure 6).
Figure 8.
Intracellular metabolite concentrations in trehalose-grown wild-type and amd1 mutant strain after a sudden increase of glucose concentration to 110 mmol/l in the presence and absence of 2 μg/ml antimycin A. Antimycin A was added 1.5 min before glucose. (A) adenine nucleotides (AXP); (B) phosphoribosyl pyrophosphate (PRPP); (C) trehalose-6-phosphate (T6P); (D) glycerol-3-phosphate (G3P). Data represent the average of at least two independent experiments.
Phenotypic characterization of purine salvage pathway mutants
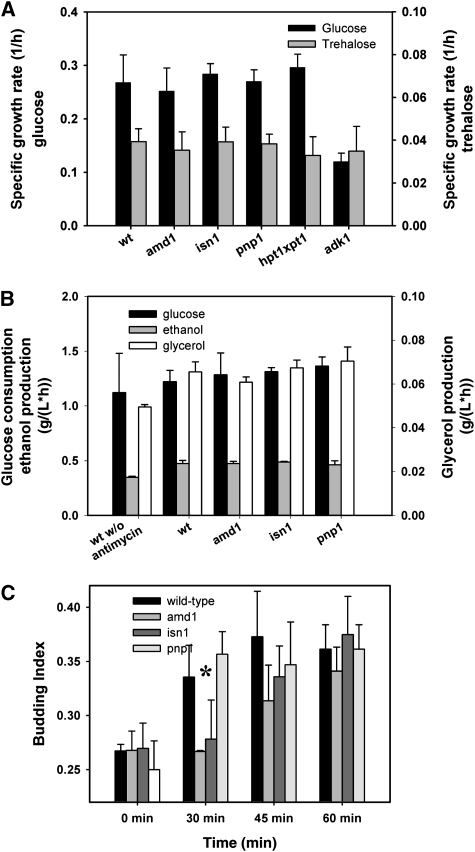
In search for potential functions of adenine nucleotide cycling, we asked whether mutants defective in this process exhibit differences to the wild-type behavior under constant or transient cultivation conditions. To answer this question, strains carrying mutations in the PSP were cultivated on minimal medium using trehalose as a non-fermentable, and glucose as a fermentable carbon source. As shown in Figure 9A, the investigated mutants neither exhibited a growth phenotype under respiratory nor under fermentative growth conditions. Only the adk1 mutant had a strongly decreased growth rate on glucose. This behavior has already been reported previously (Konrad, 1988), and is consistent with the inability of this mutant to restore its ATP pool when exposed to glucose (Figure 5V). Furthermore, growth of the wild-type and amd1 strains was compared under anaerobic conditions, but no differences could be observed (data not shown). Thus, we conclude that mutants with impaired adenine nucleotide cycling do not exhibit a growth phenotype under the constant cultivation conditions that we have tested.
Figure 9.
(A) Growth rates of mutants implicated in purine nucleotide cycling on glucose or trehalose as the limiting carbon source. Growth rates were measured in shake flask experiments at 30°C with initial concentrations of 20 g/l carbon source. Data represent the average of at least 3 independent experiments. (B) Glucose consumption, ethanol and glycerol production rates, respectively, in wild-type amd1, isn1, and pnp1 mutant strains after a sudden increase of glucose concentration to 22 mmol/l. Antimycin A was added at 2 μg/ml unless otherwise stated. (C) Budding index of trehalose-grown wild-type, amd1, isn1, and pnp1 mutant strains after a sudden increase of glucose concentration to 110 mmol/l in the presence of 2 μg/ml antimycin A. Antimycin A was added 1.5 min before glucose. Data represent the average of at least two independent experiments from which at least 600 cells were counted for each time point. The differences in budding index of the different strains were statistically analyzed for the time point at 30 min after glucose addition. (*) The budding index of wild-type and pnp1 mutant was significantly higher than for the amd1 and isn1 mutants with a confidence level of P=0.013. Measured values are provided in tabular form in Supplementary information III.
We next asked whether inhibition of respiration and absence of AXP cycling in amd1, isn1, and pnp1 mutant strains had an impact on the formation of fermentative end products and sugar consumption during the respiro-fermentative transition. During the first 20 min after glucose addition, wild-type cells produced ∼25% less glycerol and ethanol when antimycin A was absent (Figure 9B). Thereafter, cultures without antimycin A produced ethanol and glycerol at higher rates identical to cultures exposed to antimycin A (data not shown). No differences with respect to glucose consumption, as well as ethanol and glycerol production could be detected between wild-type and the tested PSP mutants (Figure 9B).
Furthermore, we monitored growth acceleration of the PSP mutants after glucose addition by assessing the budding index of the cell populations. As shown in Figure 9C, the proportion of budded cells in the wild-type and pnp1 cultures increased significantly already within the first 30 min after the pulse and remained constant after 45 min. In contrast, the increase of the budding index for the amd1 and isn1 strains exhibited a strong delay. During the first 30 min after glucose addition almost no new buds were formed by those strains, and wild-type levels of budded cells were only reached at around 60 min after glucose addition.
Discussion
The purine salvage pathway is implicated in the control of AXP concentration during the respiro-fermentative growth transition
The purine salvage pathway facilitates the salvage and redistribution of purine bases and nucleotides, respectively, which are released on RNA degradation, or which are taken up from extracellular sources. In human, deficiency of enzymes of this pathway has been associated to pathologies, such as mental retardation and severe immunodeficiency (Seegmiller, 1967; Dissing and Knudsen, 1972). In this study, we extend this view showing that the PSP, at least in yeast, contributes to AXP homeostasis by facilitating the transient drainage of AXP nucleotides into IMP and inosine. The pathway for inosine formation and recycling, as well as the interplay between different pathways during the respiro-fermentative transition are schematically summarized in Figure 10. The presence of a quickly metabolizable sugar causes accumulation of the phosphorylated metabolites F1,6P, T6P and G3P, which results in a transient imbalance of ATP-consuming and ATP-regenerating reactions, and provokes a drop in both ATP and intracellular phosphate (Pi) concentration. The joint action of fast ATP consumption and the Adk1 reaction results in the net-production of AMP. The accumulation of AMP, however, is prevented by Amd1 and Isn1, which readily convert AMP via IMP into inosine. Recycling of inosine into IMP is facilitated by Pnp1 and the concomitant action of Hpt1 and Xpt1, with Hpt1 having the predominant role.
Figure 10.
Schematic representation of pathways implicated in purine nucleotide cycling. Metabolites are depicted in black and enzymes in blue.
Purine nucleotide (i.e. IMP) de novo synthesis seems to contribute significantly to the re-establishment of initial AXP levels, as witnessed by the incomplete recovery of adenine nucleotide levels in the ade5,7 mutant. However, in this context it is important to note that the ade5,7 strain was cultivated in the presence of adenine to complement its purine base auxotrophy. The presence of adenine is known to strongly repress the expression of genes implicated in purine nucleotide de novo synthesis (Guetsova et al, 1997), and may thus have also affected activities of enzymes involved in inosine recycling. Therefore, we consider it likely that wild-type cells are able to reconvert a larger fraction (if not all) of the inosine into AMP without implying de novo nucleotide synthesis. Our results also showed that the conversion of inosine into hypoxanthine by the Pnp1 reaction was the only significant source of R1P, as concentrations of this metabolite in the amd1 and isn1 mutants were almost undetectable. Whether the R1P released during the conversion of inosine to hypoxanthine is recycled into the pentose phosphate pathway, or is used in pyrimidine salvage reactions, as proposed by Tozzi et al (2006) remains an open question.
In contrast to the behavior observed in the presence of fermentable carbon sources, under respiratory conditions neither IMP nor inosine were formed in response to perturbations of the cellular energy balance. Instead, the drop in ATP level caused a concomitant increase in AMP concentration. Therefore, we conclude that the conversion of AXP nucleotides into inosine represents a specific short-term metabolic response to the perturbation of cellular energy homeostasis, which is controlled by the presence of a fermentable carbon source (or a sugar derivative that can undergo rapid phosphorylation). The discrimination between AMP accumulation and AMP-to-inosine conversion seems to be controlled by Amd1 activity. At the present stage of investigation, we cannot conclude whether regulation of Amd1 is brought about by allosteric control or by posttranslational modification. However, given that inorganic phosphate is a potent inhibitor of Amd1 (Merkler et al, 1989) and intracellular phosphate concentration transiently drops when cells become exposed to fermentable carbon sources (our data, and e.g. Hohmann et al (1996)), phosphate availability is likely to have a pivotal role in the regulation of AXP pool size and inosine formation.
Under constant respiratory and fermentative cultivation conditions, the tested PSP mutant strains did not exhibit any growth phenotype. Furthermore, no altered accumulation of inosine or hypoxanthine was observed under these conditions in PSP mutant cells. We conclude that AXP cycling through inosine is restricted to transitory growth conditions in the presence of a fermentable carbon source, and is negligible during the tested constant respiratory or fermentative cultivation conditions. This idea is supported by a whole-genome metabolic network model of S. cerevisiae, which accounts for all PSP reactions found to be implicated in the control of AXP levels during the respiro-fermentative transition (Forster et al, 2003), and that predicts no flux across the reactions catalyzed by Amd1, Isn1, Pnp1 and Hpt1 during constant respiratory or fermentative growth (Famili et al, 2003).
Inhibition of respiration alters AXP level recovery
Our study revealed a strong impact of mitochondrial function on both rate and final level of ATP recovery. In the absence of the mitochondrial inhibitor, antimycin A, ATP concentration recovered to almost the same level as before glucose addition, whereas inhibition of respiration caused the ATP concentration to reach only ∼65% of its initial value. When mitochondrial ATP production is inhibited, glycolytic flux, that is, the rate of substrate level phosphorylation, has to be augmented to compensate for the lacking energy supply coming from respiration. As ATP is a potent inhibitor of several glycolytic enzymes (Teusink et al, 2000), the decrease in ATP level derepresses glycolysis, thereby facilitating a higher fermentation rate to supply energy. Indeed, during the first 20 min after glucose addition we estimated ethanol and glycerol production rates that were ∼25% lower in the absence of the mitochondrial inhibitor antimycin A. This finding is in line with earlier studies that reported the coincidence of decreasing ATP concentrations on increased glycolytic flux (Larsson et al, 1997, 2000; Koebmann et al, 2002). We provide a refined description of this phenomenon by showing that the decrease of ATP under completely fermentative conditions does not cause augmentation of ADP nor AMP, but rather results in a decrease of the sum of all AXP nucleotides to ∼65% of its concentration under respiratory conditions.
Adenylic energy charge can serve as a measure of the fitness of a cell (Chapman and Atkinson, 1977). The response of this measure to glucose addition with and without antimycin A was compared for different mutants. Our data showed that only in the amd1 mutant kinetics of e-charge restoration depended on functional respiration. These results demonstrated that, in accordance with previous in vitro data (Yoshino and Murakami, 1985), mutants defective in AMP deaminase were significantly more sensitive to inhibition of respiration than the wild-type and other PSP mutants.
Absence of AXP nucleotide cycling interferes with metabolic regulation
The AXP-to-inosine conversion prevents unscheduled AMP accumulation during the respiro-fermentative growth transition. Adenosine monophosphate is a potent activator of phosphofructokinase (Banuelos et al, 1977; Przybylski et al, 1985). Furthermore, accumulation of AMP displaces the equilibrium of adenylate kinase toward higher ADP and lower ATP concentrations, respectively (see Figures 5A and 6, Supplementary Figure S2D). The ATP/ADP ratio in turn controls activities of the glycolytic enzymes: hexokinase, phosphofructokinase, pyruvate kinase and phosphoglycerate kinase (Teusink et al, 2000). Hence, accumulation of AMP can be expected to interfere with the regulation of the glycolytic pathway. Indeed, our results showed that the increase in AMP concentration in the amd1 mutant provoked trans-acting effects in glycolytic metabolite levels, such as the hyper-accumulation of G3P, and delayed degradation of T6P after glucose addition. Changes in metabolite concentration indicate altered flux through adjacent metabolic reactions. However, they do not allow us to conclude whether these changes were caused by alterations in the production or consumption of the corresponding metabolite, nor do they permit to make conclusions on the duration of these alterations. However, variations in metabolite concentrations allow identifying cross-talk between different pathways, thereby guiding further research to potential hotspots of metabolic regulation. In the case of T6P, the concentration of the metabolite exerts a regulatory effect on glycolysis by inhibiting hexokinase and, thus, sugar influx (Thevelein and Hohmann, 1995). The persistence of a higher T6P concentration in the amd1 mutant was not caused by alterations of the G6P level that was indistinguishable from the wild-type cells in the relevant time span. Therefore, a delayed T6P degradation in the amd1 mutant points to the possibility that the accumulation of this glycolytic regulator is controlled by AMP and/or the ATP/ADP ratio.
Despite pronounced changes in metabolite pool dynamics caused by the deletion of AMD1, no alterations in the production of the fermentative end-products ethanol and glycerol, nor in the consumption of glucose were observed. We suggest that rearrangements of glycolytic metabolite levels in the amd1 mutant, for example, the prolonged accumulation of the hexokinase inhibitor, T6P, may have compensated the effect of altered AXP ratios on glycolytic enzymes. This interpretation is supported by the fact that altering the activity of one or more glycolytic enzymes does not result in the change of overall flux across this pathway, but rather in changing glycolytic metabolite concentrations (Müller et al, 1997). Moreover, the observation of unaltered fermentation capacity in amd1 points to the possibility that regulation of glycolysis is not the major target of AMP. Indeed, our study revealed a delayed increase in PRPP concentration in the amd1 strain after glucose addition. PRPP has a central role as a precursor for purine nucleotide de novo synthesis and in the synthesis of amino acids (Vavassori et al, 2005). Hence, limitation of this important precursor may negatively affect growth. In bacteria and humans, PRPP synthase is inhibited by ADP and AMP but not by ATP (Fox and Kelley, 1972; Arnvig et al, 1990; Willemoes et al, 2000). Our results indicated that AMP and/or ADP nucleotides interfere with PRPP-producing or -consuming reactions even in yeast, thereby supporting the idea of a regulatory link between purine nucleotide synthesis and energy homeostasis (Atkinson and Fall, 1967).
What is the function of AXP nucleotide cycling through the purine salvage pathway?
Unscheduled AMP accumulation interferes with metabolic regulation during the respiro-fermentative growth transition. To prevent AMP-induced metabolic rearrangements, S. cerevisiae uses a system to convert this nucleotide into IMP and further into inosine. To the best of our knowledge, IMP and inosine do not interfere with glycolytic regulation. This idea is supported by the fact that no differences in metabolite pool dynamics after glucose addition were detected in the isn1 and pnp1 mutant strains when compared with the wild-type cells. We therefore suggest that inosine formation serves to prevent unscheduled AMP accumulation, and to store AXP nucleotides in a metabolically ‘neutral’ form until re-equilibration of glycolysis eventually allows the recovery of ATP levels. Recycling of inosine through IMP represents an energy-saving way to replenish the AXP pool, as de novo synthesis of IMP starting from PRPP requires 4 ATP molecules, whereas IMP production from inosine and PRPP is an ATP-neutral process (Figure 10).
However, on following this explanation, the role of Isn1, that is, the function of the conversion of IMP into inosine, remains somewhat enigmatic. Considering that high IMP concentrations that accumulate in the isn1 strain will provide IMP dehydrogenase with elevated substrate levels, one might speculate that IMP-to-inosine conversion prevents a potentially catastrophic overflow of GXP nucleotides similar to what was found by Breton et al (2008). However, neither GMP nor other GXP nucleotides were found to be increased after glucose addition in the isn1 mutant. As the isn1 strain exhibited no deviations from the wild-type behavior with respect to the monitored 24 metabolite concentrations, one may thus ask why the yeast uses this additional step. A potential explanation might lie in the regulation of the Pnp1 reaction that controls the recycling of inosine: the thermodynamic equilibrium of this reaction actually favors the production of inosine from hypoxanthine (Bzowska et al, 2000). Only high concentrations of phosphate, which are normally present in live cells displace the equilibrium toward the production of hypoxanthine. This subtle regulation makes Pnp1 the ideal candidate to control the recycling of inosine into AXP nucleotides once increasing phosphate concentrations indicate re-equilibration of glycolysis. Indeed, inosine recycling coincided with the increase in intracellular phosphate concentration (compare Figures 1D and 2B).
Besides the discussed direct effects on metabolic regulation, unscheduled AMP accumulation may also have an important impact on signaling pathways. The cAMP/PKA signaling pathway has the predominant role in the control of the respiro-fermentative growth transition (Thevelein et al, 2005; Zaman et al, 2009). The formation of cAMP by adenylate cyclase, Cyr1, is ATP dependent (Kataoka et al, 1985) and may be affected by changes in the AXP balance. However, to the best of our knowledge, no data exist that demonstrate a direct impact of AMP concentration or the ATP/ADP ratio on cAMP formation and/or PKA activation. In mammals, the AMP-activated protein kinase (AMPK) controls a large number of metabolic pathways and serves to ensure energy homeostasis of the cells (for review, see Hardie et al (2003)). The S. cerevisiae AMPK homolog, Snf1, was shown to be indispensable for the transcription of glucose-repressed genes (Celenza and Carlson, 1984), and unscheduled activation of this kinase by elevated AMP levels might negatively affect the transition to fermentative growth. However, in contrast to the mammalian AMPK, the yeast homolog, Snf1, could not be shown to be directly activated by AMP, although its instantaneous activation was demonstrated under conditions in which AMP levels increased (Wilson et al, 1996). Thus, the assumption of a signaling role of AMP through the cAMP/PKA cascade or Snf1 in yeast remains speculative, although intriguing, when asking for the actual function of AXP-to-inosine conversion during the respiro-fermentative transition.
Concluding remarks
On the basis of this study, we put forward the implication of adenine nucleotide cycling through the PSP in energy homeostasis in yeast. Although it was earlier proposed that Amd1 facilitates regulation of AXP levels in response to changes in the adenylic energy charge (Chapman and Atkinson, 1973; Yoshino and Murakami, 1981), our results extend this view by demonstrating that, at least in yeast, the IMP formed by the action of Amd1 is further converted into inosine. Answering the question whether this pathway is also active in humans could potentially contribute to a better understanding of metabolic processes that control the metabolic transition from respiratory to respiro-fermentative energy supply in muscle on heavy exercise. In particular, it may help to understand conflicting results on the impaired physical performance of individuals carrying an AMP deaminase dysfunction under these conditions (Tarnopolsky et al, 2001; Fischer et al, 2007).
Materials and methods
Strains and cultivation conditions
All strains used in this study were derived from the strain BY4741 Mat a ura3 met1 his3 leu2 background (referred to as wild type). Single deletion mutants were taken from the EUROSCARF collection (Brachmann et al, 1998). The BY hpt1–xpt1 double mutant was obtained by crossing and sporulating the corresponding single deletion mutants, and identifying double knockouts in the segregant population by PCR.
Glucose-limited growth was mimicked by growing the strains in a medium containing 10 g/l trehalose (Sigma), 5 g/l (NH4)2SO4, 1.76 g/l YNB (yeast nitrogen base without amino acids and ammonium sulfate; Difco), as well as amino-acid supplements at a final concentration of 100 mg/l to complement the auxotrophies of the strains. The medium was buffered at pH 4.8 by adding 14.6 g/l succinic acid and 6 g/l NaOH (Jules et al, 2005). Cells were cultivated in Erlenmeyer flasks at 30°C, and shaken on a rotary shaker (Infors) at 200 r.p.m. When yeast cultures reached a biomass concentration of 1 g/l, glucose was added at a final concentration of 20 g/l from a concentrated stock solution of 400 g/l. To block mitochondrial function, antimycin A (0.4 mg/ml dissolved in ethanol) was added at a final concentration of 2 μg/ml 1.5 min before glucose. Block of respiratory activity by the adjusted antimycin A concentration was confirmed by the inability of the cells to grow on the non-fermentable carbon sources: trehalose and glycerol (data not shown).
Metabolite sampling and analysis
Sampling for intracellular metabolites was carried out by filtering 5 ml of culture medium on a polyamide membrane (pore size 0.45 μm, Sartorius) and quenching the metabolism of cells in 80°C hot buffered ethanol (HEPES 10 mM, 75% ethanol, pH 7.1). Processing of the samples and metabolite quantification by high-performance ionic chromatography was done as described previously (Gonzalez et al, 1997; Loret et al, 2007).
Samples of glucose pulses on wild-type (with and without antimycin A), amd1, isn1, and pnp1 mutant strains (all in the presence of antimycin A) were additionally analyzed by LC-MS spectrometry according to the following procedures. High performance liquid anion exchange chromatography: liquid anion exchange chromatography was performed on an ICS 2000 system from Dionex (Sunnyvale, CA, USA) equipped with an eluent generator system (EG 50, Dionex) for automatic base generation (KOH). Analytes were separated by anion exchange chromatography on an IonPac AS11 (250 × 2 mm, Dionex) column equipped with an AG11 (50 × 4 mm, Dionex) pre-column at a flow rate of 0.35 ml/min. Column temperature was 22°C. Metabolite separation was based on a previously described method (Groussac et al, 2000). Eluent was a gradient of KOH carried out as follows: 0 min 0.5; 13 min, 4.1; 20 min, 4.1; 30 min, 10; 45 min, 20; 60 min: 40, 65 min, 40; 65 min, 0.5; and 70 min, 0.5. Gradients were always linear. In all cases the injected sample volume was 15 μl. For background reduction, an AMMS 2-mm chemical suppressor from Dionex was used. The reagent flow (25 mM H2SO4) was set to 3 ml/min. Mass spectrometry: tandem mass analyses were carried out with a 4000 Q Trap triple mass spectrometer (Applied Biosystems, Foster City, CA, USA) with Turbo V source MDS Sciex (Toronto, Canada) for electro spray ionization. Fragmentation was done by collision-activated dissociation using nitrogen as collision gas at moderate pressure. The ion chromatograph was coupled to the mass spectrometer without splitting. The nebulizer gas pressure was 40 psi, the desolvation gas pressure was 50 psi, the desolvation gas temperature was 650°C and the capillary voltage was −3.3 kV. The MS/MS analysis has been performed as described previously (Kiefer et al, 2007).
Glucose consumption and glycerol and ethanol productions were followed over 60 min after glucose addition. Glucose consumption was estimated by a glucose peroxidase-based enzymatic assay (Sigma). Ethanol and glycerol productions were estimated using enzymatic assays (Boehringer Mannheim).
Analysis of budding index
To monitor changes in the budding index of the populations, cells were fixed in 80% ethanol and kept at 4°C until further analysis. To prepare cells for counting, they were washed once with distilled water, resuspended in 1 ml water containing 5 μl of Tween80, and sonicated (Bioblock Scientific, Vibra Cell 72412) for 10 s at 20% intensity. At least 600 cells from two independent experiments were counted per time point.
Supplementary Material
supplementary data I
supplementary data II
supplementary data III
Content of supplementary Excel spreadsheets
Supplementary figures S1–6
Acknowledgments
We thank Drs B Daignan-Fornier and B Pinson (IBGC-CNRS, University of Bordeaux II) for sharing their expertise on the purine base metabolism, and for providing a large number of strains used in this study. We also thank S Durand for providing technical expertise and assistance for LC-MS analyses. This study was partially supported by ANR Blanc no. 05-2-42128 and by Genopole (to JMF). TW was supported by a postdoctoral DFG fellowship (grant WA2163). The procurement of LC-MS equipment used in this study was financed by the Région Midi-Pyrénées and the European Regional Development Fund (ERDF), which are gratefully acknowledged.
Footnotes
The authors declare that they have no conflict of interest.
References
- Arnvig K, Hove-Jensen B, Switzer RL (1990) Purification and properties of phosphoribosyl-diphosphate synthetase from Bacillus subtilis. Eur J Biochem 192: 195–200 [DOI] [PubMed] [Google Scholar]
- Atkinson DE, Fall L (1967) Adenosine triphosphate conservation in biosynthetic regulation. Escherichia coli phosphoribosylpyrophosphate synthase. J Biol Chem 242: 3241–3242 [PubMed] [Google Scholar]
- Bailey JE (1991) Toward a science of metabolic engineering. Science 252: 1668–1675 [DOI] [PubMed] [Google Scholar]
- Banuelos M, Gancedo C, Gancedo JM (1977) Activation by phosphate of yeast phosphofructokinase. J Biol Chem 252: 6394–6398 [PubMed] [Google Scholar]
- Bosch D, Johansson M, Ferndahl C, Franzen CJ, Larsson C, Gustafsson L (2008) Characterization of glucose transport mutants of Saccharomyces cerevisiae during a nutritional upshift reveals a correlation between metabolite levels and glycolytic flux. FEMS Yeast Res 8: 10–25 [DOI] [PubMed] [Google Scholar]
- Brachmann CB, Davies A, Cost GJ, Caputo E, Li J, Hieter P, Boeke JD (1998) Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14: 115–132 [DOI] [PubMed] [Google Scholar]
- Breton A, Pinson B, Coulpier F, Giraud MF, Dautant A, Daignan-Fornier B (2008) Lethal accumulation of guanylic nucleotides in Saccharomyces cerevisiae HPT1-deregulated mutants. Genetics 178: 815–824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bzowska A, Kulikowska E, Shugar D (2000) Purine nucleoside phosphorylases: properties, functions, and clinical aspects. Pharmacol Ther 88: 349–425 [DOI] [PubMed] [Google Scholar]
- Canelas AB, van Gulik WM, Heijnen JJ (2008) Determination of the cytosolic free NAD/NADH ratio in Saccharomyces cerevisiae under steady-state and highly dynamic conditions. Biotechnol Bioeng 100: 734–743 [DOI] [PubMed] [Google Scholar]
- Celenza JL, Carlson M (1984) Cloning and genetic mapping of SNF1, a gene required for expression of glucose-repressible genes in Saccharomyces cerevisiae. Mol Cell Biol 4: 49–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman AG, Atkinson DE (1973) Stabilization of adenylate energy charge by the adenylate deaminase reaction. J Biol Chem 248: 8309–8312 [PubMed] [Google Scholar]
- Chapman AG, Atkinson DE (1977) Adenine nucleotide concentrations and turnover rates. Their correlation with biological activity in bacteria and yeast. Adv Microb Physiol 15: 253–306 [DOI] [PubMed] [Google Scholar]
- Daran-Lapujade P, Rossell S, van Gulik WM, Luttik MA, de Groot MJ, Slijper M, Heck AJ, Daran JM, de Winde JH, Westerhoff HV, Pronk JT, Bakker BM (2007) The fluxes through glycolytic enzymes in Saccharomyces cerevisiae are predominantly regulated at posttranscriptional levels. Proc Natl Acad Sci USA 104: 15753–15758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dissing J, Knudsen B (1972) Adenosine-deaminase deficiency and combined immunodeficiency syndrome. Lancet 2: 1316. [DOI] [PubMed] [Google Scholar]
- Famili I, Forster J, Nielsen J, Palsson BO (2003) Saccharomyces cerevisiae phenotypes can be predicted by using constraint-based analysis of a genome-scale reconstructed metabolic network. Proc Natl Acad Sci USA 100: 13134–13139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer H, Esbjornsson M, Sabina RL, Stromberg A, Peyrard-Janvid M, Norman B (2007) AMP deaminase deficiency is associated with lower sprint cycling performance in healthy subjects. J Appl Physiol 103: 315–322 [DOI] [PubMed] [Google Scholar]
- Forster J, Famili I, Fu P, Palsson BO, Nielsen J (2003) Genome-scale reconstruction of the Saccharomyces cerevisiae metabolic network. Genome Res 13: 244–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox IH, Kelley WN (1972) Human phosphoribosylpyrophosphate synthetase. Kinetic mechanism and end product inhibition. J Biol Chem 247: 2126–2131 [PubMed] [Google Scholar]
- Francois J, Parrou JL (2001) Reserve carbohydrates metabolism in the yeast Saccharomyces cerevisiae. FEMS Microbiol Rev 25: 125–145 [DOI] [PubMed] [Google Scholar]
- Francois J, Van Schaftingen E, Hers HG (1984) The mechanism by which glucose increases fructose 2,6-bisphosphate concentration in Saccharomyces cerevisiae. A cyclic-AMP-dependent activation of phosphofructokinase 2. Eur J Biochem 145: 187–193 [DOI] [PubMed] [Google Scholar]
- Gancedo C, Flores CL (2004) The importance of a functional trehalose biosynthetic pathway for the life of yeasts and fungi. FEMS Yeast Res 4: 351–359 [DOI] [PubMed] [Google Scholar]
- Gonzalez B, Francois J, Renaud M (1997) A rapid and reliable method for metabolite extraction in yeast using boiling buffered ethanol. Yeast 13: 1347–1355 [DOI] [PubMed] [Google Scholar]
- Grant CM, Quinn KA, Dawes IW (1999) Differential protein S-thiolation of glyceraldehyde-3-phosphate dehydrogenase isoenzymes influences sensitivity to oxidative stress. Mol Cell Biol 19: 2650–2656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groussac E, Ortiz M, Francois J (2000) Improved protocols for quantitative determination of metabolites from biological samples using high performance ionic-exchange chromatography with conductimetric and pulsed amperometric detection. Enzyme Microb Technol 26: 715–723 [DOI] [PubMed] [Google Scholar]
- Guetsova ML, Crother TR, Taylor MW, Daignan-Fornier B (1999) Isolation and characterization of the Saccharomyces cerevisiae XPT1 gene encoding xanthine phosphoribosyl transferase. J Bacteriol 181: 2984–2986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guetsova ML, Lecoq K, Daignan-Fornier B (1997) The isolation and characterization of Saccharomyces cerevisiae mutants that constitutively express purine biosynthetic genes. Genetics 147: 383–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardie DG, Scott JW, Pan DA, Hudson ER (2003) Management of cellular energy by the AMP-activated protein kinase system. FEBS Lett 546: 113–120 [DOI] [PubMed] [Google Scholar]
- Hohmann S, Bell W, Neves MJ, Valckx D, Thevelein JM (1996) Evidence for trehalose-6-phosphate-dependent and -independent mechanisms in the control of sugar influx into yeast glycolysis. Mol Microbiol 20: 981–991 [DOI] [PubMed] [Google Scholar]
- Itoh R, Saint-Marc C, Chaignepain S, Katahira R, Schmitter JM, Daignan-Fornier B (2003) The yeast ISN1 (YOR155c) gene encodes a new type of IMP-specific 5′-nucleotidase. BMC Biochem 4: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jules M, Francois J, Parrou JL (2005) Autonomous oscillations in Saccharomyces cerevisiae during batch cultures on trehalose. FEBS J 272: 1490–1500 [DOI] [PubMed] [Google Scholar]
- Jules M, Guillou V, Francois J, Parrou JL (2004) Two distinct pathways for trehalose assimilation in the yeast Saccharomyces cerevisiae. Appl Environ Microbiol 70: 2771–2778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Araki M, Goto S, Hattori M, Hirakawa M, Itoh M, Katayama T, Kawashima S, Okuda S, Tokimatsu T, Yamanishi Y (2008) KEGG for linking genomes to life and the environment. Nucleic Acids Res 36: D480–D484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kataoka T, Broek D, Wigler M (1985) DNA sequence and characterization of the S. cerevisiae gene encoding adenylate cyclase. Cell 43: 493–505 [DOI] [PubMed] [Google Scholar]
- Kiefer P, Nicolas C, Letisse F, Portais JC (2007) Determination of carbon labeling distribution of intracellular metabolites from single fragment ions by ion chromatography tandem mass spectrometry. Anal Biochem 360: 182–188 [DOI] [PubMed] [Google Scholar]
- Koebmann BJ, Westerhoff HV, Snoep JL, Nilsson D, Jensen PR (2002) The glycolytic flux in Escherichia coli is controlled by the demand for ATP. J Bacteriol 184: 3909–3916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konrad M (1988) Analysis and in vivo disruption of the gene coding for adenylate kinase (ADK1) in the yeast Saccharomyces cerevisiae. J Biol Chem 263: 19468–19474 [PubMed] [Google Scholar]
- Kresnowati MT, van Winden WA, Almering MJ, ten Pierick A, Ras C, Knijnenburg TA, Daran-Lapujade P, Pronk JT, Heijnen JJ, Daran JM (2006) When transcriptome meets metabolome: fast cellular responses of yeast to sudden relief of glucose limitation. Mol Syst Biol 2: 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kresnowati MT, van Winden WA, van Gulik WM, Heijnen JJ (2008) Energetic and metabolic transient response of Saccharomyces cerevisiae to benzoic acid. FEBS J 275: 5527–5541 [DOI] [PubMed] [Google Scholar]
- Lara AR, Galindo E, Ramirez OT, Palomares LA (2006) Living with heterogeneities in bioreactors: understanding the effects of environmental gradients on cells. Mol Biotechnol 34: 355–381 [DOI] [PubMed] [Google Scholar]
- Larsson C, Nilsson A, Blomberg A, Gustafsson L (1997) Glycolytic flux is conditionally correlated with ATP concentration in Saccharomyces cerevisiae: a chemostat study under carbon- or nitrogen-limiting conditions. J Bacteriol 179: 7243–7250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson C, Pahlman IL, Gustafsson L (2000) The importance of ATP as a regulator of glycolytic flux in Saccharomyces cerevisiae. Yeast 16: 797–809 [DOI] [PubMed] [Google Scholar]
- Lecoq K, Belloc I, Desgranges C, Daignan-Fornier B (2001a) Role of adenosine kinase in Saccharomyces cerevisiae: identification of the ADO1 gene and study of the mutant phenotypes. Yeast 18: 335–342 [DOI] [PubMed] [Google Scholar]
- Lecoq K, Belloc I, Desgranges C, Konrad M, Daignan-Fornier B (2001b) YLR209c encodes Saccharomyces cerevisiae purine nucleoside phosphorylase. J Bacteriol 183: 4910–4913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loret MO, Pedersen L, Francois J (2007) Revised procedures for yeast metabolites extraction: application to a glucose pulse to carbon-limited yeast cultures, which reveals a transient activation of the purine salvage pathway. Yeast 24: 47–60 [DOI] [PubMed] [Google Scholar]
- Mashego MR, van Gulik WM, Vinke JL, Visser D, Heijnen JJ (2006) In vivo kinetics with rapid perturbation experiments in Saccharomyces cerevisiae using a second-generation BioScope. Metab Eng 8: 370–383 [DOI] [PubMed] [Google Scholar]
- Mazon MJ, Gancedo JM, Gancedo C (1982) Inactivation of yeast fructose-1,6-bisphosphatase. In vivo phosphorylation of the enzyme. J Biol Chem 257: 1128–1130 [PubMed] [Google Scholar]
- Mbonyi K, Beullens M, Detremerie K, Geerts L, Thevelein JM (1988) Requirement of one functional RAS gene and inability of an oncogenic ras variant to mediate the glucose-induced cyclic AMP signal in the yeast Saccharomyces cerevisiae. Mol Cell Biol 8: 3051–3057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkler DJ, Wali AS, Taylor J, Schramm VL (1989) AMP deaminase from yeast. Role in AMP degradation, large scale purification, and properties of the native and proteolyzed enzyme. J Biol Chem 264: 21422–21430 [PubMed] [Google Scholar]
- Meyer SL, Kvalnes-Krick KL, Schramm VL (1989) Characterization of AMD, the AMP deaminase gene in yeast. Production of amd strain, cloning, nucleotide sequence, and properties of the protein. Biochemistry 28: 8734–8743 [DOI] [PubMed] [Google Scholar]
- Müller S, Zimmermann FK, Boles E (1997) Mutant studies of phosphofructo-2-kinase do not reveal an essential role of fructose-2,6-biphosphate in the regulation of carbon fluxes in yeast cells. Microbiology 143: 3055–3061 [DOI] [PubMed] [Google Scholar]
- Nikerel IE, van Winden WA, Verheijen PJ, Heijnen JJ (2009) Model reduction and a priori kinetic parameter identifiability analysis using metabolome time series for metabolic reaction networks with linlog kinetics. Metab Eng 11: 20–30 [DOI] [PubMed] [Google Scholar]
- Pronk JT, Yde Steensma H, Van Dijken JP (1996) Pyruvate metabolism in Saccharomyces cerevisiae. Yeast 12: 1607–1633 [DOI] [PubMed] [Google Scholar]
- Przybylski F, Nissler K, Schellenberger W, Hofmann E (1985) Inorganic phosphate amplifies the effects of AMP and fructose-2,6-bisphosphate on yeast phosphofructokinase. Biomed Biochim Acta 44: 1559–1565 [PubMed] [Google Scholar]
- Rizzi M, Baltes M, Theobald U, Reuss M (1997) In vivo analysis of metabolic dynamics in Saccharomyces cerevisiae. Mathematical model. Biotechnol Bioeng 55: 592–608 [DOI] [PubMed] [Google Scholar]
- Rolland F, Winderickx J, Thevelein JM (2002) Glucose-sensing and -signalling mechanisms in yeast. FEMS Yeast Res 2: 183–201 [DOI] [PubMed] [Google Scholar]
- Schricker R, Magdolen V, Kaniak A, Wolf K, Bandlow W (1992) The adenylate kinase family in yeast: identification of URA6 as a multicopy suppressor of deficiency in major AMP kinase. Gene 122: 111–118 [DOI] [PubMed] [Google Scholar]
- Seegmiller JE (1967) Genetic and molecular basis of human hereditary diseases. Clin Chem 13: 554–564 [PubMed] [Google Scholar]
- Tarnopolsky MA, Parise G, Gibala MJ, Graham TE, Rush JW (2001) Myoadenylate deaminase deficiency does not affect muscle anaplerosis during exhaustive exercise in humans. J Physiol 533: 881–889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teusink B, Passarge J, Reijenga CA, Esgalhado E, van der Weijden CC, Schepper M, Walsh MC, Bakker BM, van Dam K, Westerhoff HV, Snoep JL (2000) Can yeast glycolysis be understood in terms of in vitro kinetics of the constituent enzymes? Testing biochemistry. Eur J Biochem 267: 5313–5329 [DOI] [PubMed] [Google Scholar]
- Teusink B, Walsh MC, van Dam K, Westerhoff HV (1998) The danger of metabolic pathways with turbo design. Trends Biochem Sci 23: 162–169 [DOI] [PubMed] [Google Scholar]
- Theobald U, Mailinger W, Baltes M, Rizzi M, Reuss M (1997) In vivo analysis of metabolic dynamics in Saccharomyces cerevisiae: I. Experimental observations. Biotechnol Bioeng 55: 305–316 [DOI] [PubMed] [Google Scholar]
- Theobald U, Mailinger W, Reuss M, Rizzi M (1993) In vivo analysis of glucose-induced fast changes in yeast adenine nucleotide pool applying a rapid sampling technique. Anal Biochem 214: 31–37 [DOI] [PubMed] [Google Scholar]
- Thevelein JM, Gelade R, Holsbeeks I, Lagatie O, Popova Y, Rolland F, Stolz F, Van de Velde S, Van Dijck P, Vandormael P, Van Nuland A, Van Roey K, Van Zeebroeck G, Yan B (2005) Nutrient sensing systems for rapid activation of the protein kinase A pathway in yeast. Biochem Soc Trans 33: 253–256 [DOI] [PubMed] [Google Scholar]
- Thevelein JM, Hohmann S (1995) Trehalose synthase: guard to the gate of glycolysis in yeast? Trends Biochem Sci 20: 3–10 [DOI] [PubMed] [Google Scholar]
- Tomasselli AG, Mast E, Janes W, Schiltz E (1986) The complete amino acid sequence of adenylate kinase from baker's yeast. Eur J Biochem 155: 111–119 [DOI] [PubMed] [Google Scholar]
- Tozzi MG, Camici M, Mascia L, Sgarrella F, Ipata PL (2006) Pentose phosphates in nucleoside interconversion and catabolism. FEBS J 273: 1089–1101 [DOI] [PubMed] [Google Scholar]
- van den Brink J, Canelas AB, van Gulik WM, Pronk JT, Heijnen JJ, de Winde JH, Daran-Lapujade P (2008) Dynamics of glycolytic regulation during adaptation of Saccharomyces cerevisiae to fermentative metabolism. Appl Environ Microbiol 74: 5710–5723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaseghi S, Baumeister A, Rizzi M, Reuss M (1999) In vivo dynamics of the pentose phosphate pathway in Saccharomyces cerevisiae. Metab Eng 1: 128–140 [DOI] [PubMed] [Google Scholar]
- Vavassori S, Wang K, Schweizer LM, Schweizer M (2005) Ramifications of impaired PRPP synthesis in Saccharomyces cerevisiae. Biochem Soc Trans 33: 1418–1420 [DOI] [PubMed] [Google Scholar]
- Visser D, van Zuylen GA, van Dam JC, Eman MR, Proll A, Ras C, Wu L, van Gulik WM, Heijnen JJ (2004) Analysis of in vivo kinetics of glycolysis in aerobic Saccharomyces cerevisiae by application of glucose and ethanol pulses. Biotechnol Bioeng 88: 157–167 [DOI] [PubMed] [Google Scholar]
- Visser D, van Zuylen GA, van Dam JC, Oudshoorn A, Eman MR, Ras C, van Gulik WM, Frank J, van Dedem GW, Heijnen JJ (2002) Rapid sampling for analysis of in vivo kinetics using the BioScope: a system for continuous-pulse experiments. Biotechnol Bioeng 79: 674–681 [DOI] [PubMed] [Google Scholar]
- Wang Y, Pierce M, Schneper L, Güldal C, Zhang X, Tavazoie S, Broach J (2004) Ras and Gpa2 mediate one branch of a redundant glucose signalling pathway in yeast. PloS Biol 2: 128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willemoes M, Hove-Jensen B, Larsen S (2000) Steady state kinetic model for the binding of substrates and allosteric effectors to Escherichia coli phosphoribosyl-diphosphate synthase. J Biol Chem 275: 35408–35412 [DOI] [PubMed] [Google Scholar]
- Wilson WA, Hawley SA, Hardie DG (1996) Glucose repression/derepression in budding yeast: SNF1 protein kinase is activated by phosphorylation under derepressing conditions, and this correlates with a high AMP:ATP ratio. Curr Biol 6: 1426–1434 [DOI] [PubMed] [Google Scholar]
- Woods RA, Roberts DG, Friedman T, Jolly D, Filpula D (1983) Hypoxanthine: guanine phosphoribosyltransferase mutants in Saccharomyces cerevisiae. Mol Gen Genet 191: 407–412 [DOI] [PubMed] [Google Scholar]
- Yoshino M, Murakami K (1981) In situ studies on AMP deaminase as a control system of the adenylate energy charge in yeasts. Biochim Biophys Acta 672: 16–20 [DOI] [PubMed] [Google Scholar]
- Yoshino M, Murakami K (1985) AMP deaminase reaction as a control system of glycolysis in yeast. Role of ammonium ion in the interaction of phosphofructokinase and pyruvate kinase activity with the adenylate energy charge. J Biol Chem 260: 4729–4732 [PubMed] [Google Scholar]
- Zaman S, Lippmann SI, Schneper L, Broach JR (2009) Glucose regulates transcription in yeast through a network of signalling pathways. Mol Syst Biol 5: 245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann FK (1997) Yeast Sugar Metabolism. Biochemistry, Genetics, Biotechnology and Applications. Technomic Publishing Co, Lancaster, PA, USA [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
supplementary data I
supplementary data II
supplementary data III
Content of supplementary Excel spreadsheets
Supplementary figures S1–6