Abstract
The molecular understanding of phenotypes caused by drugs in humans is essential for elucidating mechanisms of action and for developing personalized medicines. Side effects of drugs (also known as adverse drug reactions) are an important source of human phenotypic information, but so far research on this topic has been hampered by insufficient accessibility of data. Consequently, we have developed a public, computer-readable side effect resource (SIDER) that connects 888 drugs to 1450 side effect terms. It contains information on frequency in patients for one-third of the drug–side effect pairs. For 199 drugs, the side effect frequency of placebo administration could also be extracted. We illustrate the potential of SIDER with a number of analyses. The resource is freely available for academic research at http://sideeffects.embl.de.
Keywords: adverse drug reactions, database, drugs, human phenotypes, side effects
Introduction
Side effects are phenotypic responses of the human organism to drug treatment. In recent years, side effects have become an important subject of research in the pharmaceutical industry, which is interested in predicting the possible side effects of drug candidates based on, for example, the binding fingerprint, chemical structure and other properties of the drug candidate (Krejsa et al, 2003; Bender et al, 2007; Fliri et al, 2007). Side effects can also be used to predict novel drug–target interactions and might be utilizable for drug re-purposing (Campillos et al, 2008).
Pharmacological and medical research would greatly benefit from the integration of side effect data with other emerging public resources in chemical biology. For example, the National Institutes of Health Molecular Libraries Roadmap Initiative has led to the creation of the PubChem repository of chemical compounds (Wheeler et al, 2007). Data on cellular phenotypes in response to chemicals are stored in PubChem BioAssay and ChemBank (Seiler et al, 2008). Other databases, such as DrugBank, the PSDP Ki database and BindingDB, contain binding information (Roth et al, 2000; Liu et al, 2007; Wishart et al, 2008). As public databases of protein–chemical interactions are beginning to grow, there is hope that pharmacology may be transformed by the application of large-scale computational methods in the same way that biology has been (Kuhn et al, 2008). However, there is currently no public database of drug side effects that makes these important data readily available for analysis and research.
To ameliorate this situation, we have compiled package inserts from several public sources, in particular, from the US Food and Drug Administration (FDA), in the form of either Structured Product Labeling (SPL) or Portable Document Format (PDF) documents. SPL is a dedicated electronic format for package inserts and is thus more amenable to extracting information. We used text mining to solve the cumbersome task of extracting side effects from the differently formatted, human-readable labels (see Materials and methods). The standardized Coding Symbols for a Thesaurus of Adverse Reaction Terms (COSTART), which are part of the Unified Medical Language System (UMLS) Metathesaurus, were used as the basic lexicon of side effects. To facilitate linking to other databases and reuse for research, we have mapped drug names to PubChem identifiers.
Results and discussion
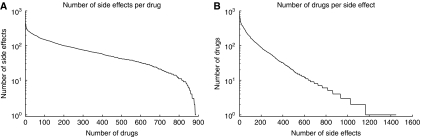
Side effect resource (SIDER) contains 62 269 drug–side effect pairs and covers a total of 888 drugs and 1450 distinct side effects. Altogether, 70% of drugs have between 10 and 100 different side effects (Figure 1A). There is an under-representation of drugs with few side effects, whereas 55% of all side effects occur for <10 drugs (Figure 1B). In all, 33% of all side effects occur for 10–100 drugs; 12% of all side effects occur for >100 drugs. Labels for 79% of the drugs were available in SPL format, and 75% in PDF (i.e., 55% of the drugs are available in both formats). In total, 798 of these drugs are FDA-approved; the remaining 90 drugs have either been previously approved but were since withdrawn from the market (like cerivastatin (Lipobay/Baycol)), or are marketed outside the United States (like gliclazide).
Figure 1.
Statistics of the database. (A) The number of side effects is counted for each drug and the number of drugs is plotted versus the number of side effects per drug. For example, there are about 200 drugs with at least 100 side effects. (B) Similar to A, the number of drugs per side effects is plotted.
We group the 888 drugs in SIDER by their drug class, that is, the main anatomical group of their indication area as derived from first-level Anatomical Therapeutic Chemical Classification System (ATC) code, and analyze how specific the side effects are to drug classes. It becomes apparent that most side effects can occur for more than one drug class (Figure 2A). In fact, even when excluding drugs that belong to more than one drug class, only 347 out of 1344 side effects (25.8%) occur in only one drug class. This overlap of side effects between drugs from different anatomical classes points to common molecular mechanisms for drugs of different classes or multiple anatomical effects of the same drug. That is, on one hand, drugs can have off-targets in other tissues and, on the other hand, the main targets themselves can be expressed in different tissues or have effects in other tissues (Liebler and Guengerich, 2005). At the level of individual drugs, side effect similarity has been found to be predictive of common drug targets. For example, the proton pump inhibitor rabeprazole (indicated against stomach ulcers) and the dopamine receptor agonist pergolide (previously used to treat Parkinson's disease) share many side effects (Supplementary Figure S1). Indeed, rabeprazole has been shown to bind to dopamine receptors (Campillos et al, 2008).
Figure 2.
Analysis of side effects. (A) Overview of the different categories of drugs. Drugs are grouped by their drug class. The number of shared and unique drugs and side effects is shown for each class. (Some drugs, e.g., aspirin, have more than one anatomical class assigned to them.) (B) Using Fisher's exact test, over-represented side effects were determined for each category. A few over-represented medical concepts might describe indications for the drugs (e.g., ‘cancer') that were not caught by our filtering mechanisms (see Materials and methods). These concepts are marked with an asterisk. (C) Associations between drug classes and anatomical classes of side effects are shown (see Supplementary Information). Positive values indicate an association between a drug and an anatomical class. Negative values represent an under-representation, for example, drugs indicated for disorders of the blood system cause few sensory side effects.
At the level of drug classes, common side effects reveal important information as they can point, for example, to shared underlying mechanisms of action. We therefore calculated which side effects were most over-represented per drug class using Fisher's exact test for a non-redundant set of drugs (at a q-value cutoff of 0.05, see Supplementary Information). For 12 out of 14 drug classes, we find significantly over-represented side effects (Figure 2B, see Supplementary Table 3 for a full list of over-represented side effects and their q-values). To investigate which side effects are related to the primary indication area of the drug and which ones occur in other areas of the body, we take into account anatomical classes as defined in the COSTART ontology and assign anatomical classes to drug classes (see Supplementary Table 2). We find that 28% of all over-represented side effects are directly related to the anatomical region of the drug class, while 43% are related to a different anatomical class, and thus represent the phenotypic expression of off-target and off-tissue effects. (For the remaining 29%, either drugs or side effects have no corresponding anatomical class, see Supplementary Table 3.) Of the ten drug classes that have a corresponding anatomical class, eight have one or more significantly over-represented side effects outside their anatomical classes. In all cases in which over-represented side effects have been found, this is highly significant, with P-values below 0.01 when the assignments between drugs and drug classes are randomized (controlling for multiple testing by Bonferroni correction). For example, cardiovascular drugs also cause dizziness, impotence (erectile dysfunction) and weakness. Dizziness and weakness can be probably attributed to a decreased blood pressure caused by medications like β-blockers. Impotence has been found to be a side effect of several kinds of cardiovascular drugs such as calcium channel inhibitors, angiotensin II antagonists, non-selective β-blockers and diuretics (Shiri et al, 2007). The occurrence of erectile dysfunction for non-selective β-blockers has been explained with their action on α-adrenergic receptors in penile tissue (Lue, 2000), thus providing an example of closely related off-targets in different tissues. In conclusion, almost all drug classes have significantly over-represented side effects, some of which even point to a shared action of drug of the same therapeutic area on targets outside the intended indication area.
To get a global overview of the anatomical distribution of the side effects of drug classes, we calculate association scores (see Supplementary Information) between drug classes and anatomical classes. We find that drugs from all drugs classes cause side effects also in anatomical classifications that do not correspond to the respective indication area (Figure 2C); however, the extent to which this occurs is not the same for all drug classes. Some drug classes are very specific, especially those applied topically like dermatologicals (ATC code D) and sensory organ drugs (S), whereas others are more spread, such as antineoplastic and immunomodulating drugs (L).
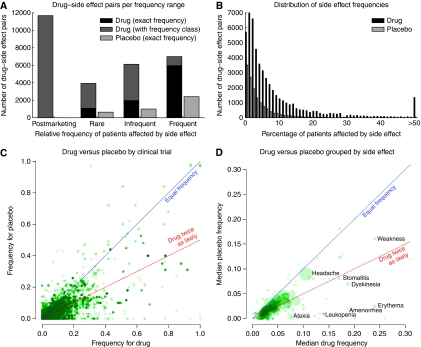
By automatically interpreting the text and tables on package inserts, we were able to obtain information on the frequencies of the side effects for more than half of the drugs (500 of 888), either as a general frequency range (e.g., ‘frequent') or as an exact frequency (e.g., ‘3.1%', see Materials and methods). We determined by manually inspection of 20 randomly chosen examples that for half of the remaining drugs, the available labels do not contain any frequency information and thus even a human expert could not derive such information. Nonetheless, frequency information could be deduced for 23 631 drug–side effect pairs (38% of all pairs, Figure 3A). Exact frequencies could be extracted from tables in SPL documents for 6448 (27%) of all pairs with frequency information by analyzing the content of the table caption and rows to deduce the format of the table (e.g., percentage or number of cases). The median frequency in patients of ‘frequent' side effects (occurring in >1% of patients) is 5%. Thus, it is reassuring that even frequent side effects mostly occur in a minority of the patients (Figure 3B).
Figure 3.
(A) For the different side effect frequency classes, the number of drug–side effect pairs is shown. For a fraction of the side effects an exact frequency (e.g., ‘3%') is known, whereas for others only the general frequency class (e.g., ‘frequent') is given in package inserts. When an exact frequency is given on the labels, the corresponding frequency of the side effect in the control (placebo) group is often also available. (B) The number of drug–side effect pairs with specific frequencies is counted. Exact frequency ranges are in many cases only given for side effects occurring in at least 1% of the patients, therefore the first bin (<1%) of the histogram has a lower abundance than the second bin (1–2%). (C) Frequencies obtained from the administration of drug and placebo during clinical trials are compared. In panels C and D, the blue line stands for equal frequency and the red line for side effects that are twice as frequent after drug treatment than in the control group. (D) For each side effect, the median frequency in drug and placebo administration is computed. The area of the circles corresponds to the number of drugs with the side effect and available placebo information. (See Supplementary Figure S2 for adapted versions of C and D that resolve the cloud of points near the origin.).
The SPL documents often also contain information about the frequency of the side effects when a placebo had been administered during clinical trials. Frequency information is available for 3426 placebo–side effect pairs (53% of the pairs with exact frequency information and 14% of all drug–side effect pairs). To illustrate the utilization of placebo information, we compared the frequencies reported during clinical trials for drug and placebo administration (Figure 3C). The low fraction (14%) of side effects that are more frequent for placebo than drug administration reflects that side effects are generally only reported if the patients receiving the drug have a higher frequency than those receiving placebo. The main purpose of this limitation is to filter out the symptoms (or phenotypes) that occur in all patients with a given medical condition, regardless of actual treatment. If a symptom occurs at the expected rate for the population or if it is related to the pre-existing condition and not to drug treatment, then the frequency will be the same in both groups. However, it would be very valuable for research to also have access to phenotypes that decrease in frequency on drug treatment. If a drug strongly reduces the frequency of certain phenotypes compared with the background frequency observed in the control (placebo) group, this might give hints towards novel indications for this drug. For example, it had been noticed that selective serotonin reuptake inhibitors (SSRIs) often have the side effect of delayed ejaculation, or conversely, a reduced rate of premature ejaculation compared with the background frequency. Dapoxetine, a short-acting SSRI, is therefore now used to treat premature ejaculation (Ashburn and Thor, 2004). A large-scale analysis of all drugs might be able to uncover further novel indication areas. Many side effects occur almost equally often in placebo as in drug treatment, for example, weakness and headache. Those side effects are either associated with the underlying disease, or are a result of the nocebo effect: patients who expect certain side effects are more likely to experience these effects (Barsky et al, 2002). In contrast, a few side effects have a median frequency for drug administration that is much larger than placebo (Figure 3D), for example, erythema, amenorrhea and leukopenia. Further investigation on the nature of these side effects that cannot be induced by placebo treatment could yield insights into the etiology of side effects occurring during placebo treatment.
To facilitate research on drugs and their side effects, we have created a website (http://sideeffects.embl.de) where users can download the whole database and examine the side effects of individual drugs of interest. In particular, researchers can explore the package inserts through the concept of ‘augmented browsing', in which the side effect terms are highlighted and the user can click on the highlighted terms to retrieve additional information about proteins, chemicals and side effects (Pafilis et al, 2009). In this way, even scientists who do not have a background in pharmacology can easily work with this diverse set of entities.
The database will be updated periodically with the incorporation of new drug labels. It is available under Creative Commons Attribution-Noncommercial-Share Alike 3.0 License with separate licensing for commercial entities. A snapshot of the database has been deposited as Supplementary Information. The download files also contain information extracted from package inserts that could not be mapped to individual drugs, either because the particular brand name is not in our dictionary of drugs, or because the package insert describes a combination of drugs.
The present resource for the first time makes a set of side effects for drugs from all indications freely available for academic research. Although we have calculated only a few global statistics to illustrate the potential of this resource, future studies might investigate the relations between side effects and chemical structure, pharmacophores, gene expression or target sharing on a molecular level. In particular, the data about side effect frequencies should be a valuable resource to determine the correlation between drug targets, plasma concentrations and side effect incidence, and to understand from a molecular point of view the observed variability in the population to drug response and their side effects. This knowledge will be very helpful in designing personalized medicines in which drug treatment will be ideally adjusted according to particular genomic, proteomic or environmental personal features. All these venues of research would furthermore greatly benefit from a greater availability of data generated during clinical trials. The SPL format is a considerable step towards making this information accessible. Nonetheless, all relevant data that have to be shown by law on the drug labels should be directly deposited into computer-readable repositories to avoid loss of information.
Materials and methods
The extraction procedure is based on the method used in Campillos et al (2008). We only include labels from public sources (which are partly more difficult to parse because of a greater variability in formats) and extend the set of FDA-approved drugs from 746 used in the earlier study to 798. Although new information could be found for 97 drugs, labels for 45 drugs were not publicly available and hence could not be included in this public resource. For example, this is the case for hydrocodone, which is exclusively used in combination with other drugs. Adding information for 90 non-FDA-approved drugs yields a total of 888 drugs in the database.
Drug labels are provided by the FDA and the other sources in two kinds of files: PDF and SPL documents. Among the five public sources from which we compiled information (Supplementary Table 1), Facts@FDA is the only source providing SPL documents. Analyzing the content of PDF files is hampered by the fact that the format only describes the coordinates of individual pieces of text, but not the logical structure of the document, for example, paragraphs or table rows. Besides, PDF files had to be converted to text files to be amenable for text mining. In contrast, SPL documents are Extensible Markup Language (XML) documents that provide information in a structured and machine-readable way. Both types of labels were analyzed by text-mining tools using a dictionary of side effects derived from the UMLS Metathesaurus. COSTART was used as seed dictionary, to which synonyms from equivalent terms of the UMLS were added. The sections describing the drugs' indication areas and side effects were used to extract terms corresponding to side effects. Terms that occur as an indication for the drug were subsequently excluded from the drug's side effects to filter terms used in the adverse effect section that actually describe the drug's indication area (e.g., in the description of clinical trials).
For 52% of all drugs, the sections describing adverse reactions on the labels contain information about the frequencies of side effects. The FDA SPL labels were amenable to a thorough analysis and exact frequency information could be extracted from the tables detailing the side effect frequencies by analyzing the contents of the table captions and cells (e.g., caption in the first row: ‘Percentages of Patients Reporting Event', caption in the first column: ‘anemia', cell: ‘12': 12% of the patients report anemia). Although it was not possible to deduce exact frequency information for PDF labels, standardized general frequency ranges (‘rare', ‘infrequent' and ‘frequent') could be extracted for both label types from sections that listed the frequency and a number of side effects (e.g., ‘frequent: headache, dizziness, …') Furthermore, side effects that occurred in the post-marketing phase were also extracted from all labels. For any given drug–side effect pair, multiple frequencies might be available, for example, from clinical trials in different indication areas. On the website, we display all reported frequencies to the user.
Supplementary Material
Supplementary Methods, Supplementary Figures S1-2, Supplementary Tables S1-2
List of over-represented side effects
A snapshot of the database version released on November 6, 2009. Unless otherwise noted, all files in this archive are under a Creative Commons Attribution-Noncommercial 3.0 License (CC-BY-NC)
Acknowledgments
We thank A Rzhetsky and L Yao for helpful discussions. This work was partially funded by the National Institute on Drug Abuse (contract number HHSN271200900215P).
Footnotes
In addition to his employment at the European Molecular Biology Laboratory (EMBL), IL is also the Chief Executive Officer of biobyte solutions GmbH, which handles commercial licensing of SIDER for EMBL.
References
- Ashburn TT, Thor KB (2004) Drug repositioning: identifying and developing new uses for existing drugs. Nat Rev Drug Discov 3: 673–683 [DOI] [PubMed] [Google Scholar]
- Barsky AJ, Saintfort R, Rogers MP, Borus JF (2002) Nonspecific medication side effects and the nocebo phenomenon. JAMA 287: 622–627 [DOI] [PubMed] [Google Scholar]
- Bender A, Scheiber J, Glick M, Davies JW, Azzaoui K, Hamon J, Urban L, Whitebread S, Jenkins JL (2007) Analysis of pharmacology data and the prediction of adverse drug reactions and off-target effects from chemical structure. Chem Med Chem 2: 861–873 [DOI] [PubMed] [Google Scholar]
- Campillos M, Kuhn M, Gavin AC, Jensen LJ, Bork P (2008) Drug target identification using side-effect similarity. Science 321: 263–266 [DOI] [PubMed] [Google Scholar]
- Fliri AF, Loging WT, Volkmann RA (2007) Analysis of system structure-function relationships. Chem Med Chem 2: 1774–1782 [DOI] [PubMed] [Google Scholar]
- Krejsa CM, Horvath D, Rogalski SL, Penzotti JE, Mao B, Barbosa F, Migeon JC (2003) Predicting ADME properties and side effects: the BioPrint approach. Curr Opin Drug Discov Devel 6: 470–480 [PubMed] [Google Scholar]
- Kuhn M, Campillos M, González P, Jensen LJ, Bork P (2008) Large-scale prediction of drug-target relationships. FEBS Lett 582: 1283–1290 [DOI] [PubMed] [Google Scholar]
- Liebler DC, Guengerich FP (2005) Elucidating mechanisms of drug-induced toxicity. Nat Rev Drug Discov 4: 410–420 [DOI] [PubMed] [Google Scholar]
- Liu T, Lin Y, Wen X, Jorissen RN, Gilson MK (2007) BindingDB: a web-accessible database of experimentally determined protein-ligand binding affinities. Nucleic Acids Res 35: D198–D201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lue TF (2000) Erectile dysfunction. N Engl J Med 342: 1802–1813 [DOI] [PubMed] [Google Scholar]
- Pafilis E, O'Donoghue SI, Jensen LJ, Horn H, Kuhn M, Brown NP, Schneider R (2009) Reflect: augmented browsing for the life scientist. Nat Biotechnol 27: 508–510 [DOI] [PubMed] [Google Scholar]
- Roth B, Kroeze W, Patel S, Lopez E (2000) The multiplicity of serotonin receptors: uselessly diverse molecules or an embarrassment of riches? Neuroscientist 6: 252–262 [Google Scholar]
- Seiler KP, George GA, Happ MP, Bodycombe NE, Carrinski HA, Norton S, Brudz S, Sullivan JP, Muhlich J, Serrano M, Ferraiolo P, Tolliday NJ, Schreiber SL, Clemons PA (2008) ChemBank: a small-molecule screening and cheminformatics resource database. Nucleic Acids Res 36: D351–D359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiri R, Koskimaki J, Hakkinen J, Auvinen A, Tammela TL, Hakama M (2007) Cardiovascular drug use and the incidence of erectile dysfunction. Int J Impot Res 19: 208–212 [DOI] [PubMed] [Google Scholar]
- Wheeler DL, Barrett T, Benson DA, Bryant SH, Canese K, Chetvernin V, Church DM, DiCuccio M, Edgar R, Federhen S, Geer LY, Kapustin Y, Khovayko O, Landsman D, Lipman DJ, Madden TL, Maglott DR, Ostell J, Miller V, Pruitt KD et al. (2007) Database resources of the National Center for Biotechnology Information. Nucleic Acids Res 35: D5–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wishart DS, Knox C, Guo AC, Cheng D, Shrivastava S, Tzur D, Gautam B, Hassanali M (2008) DrugBank: a knowledgebase for drugs, drug actions and drug targets. Nucleic Acids Res 36: D901–D906 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Methods, Supplementary Figures S1-2, Supplementary Tables S1-2
List of over-represented side effects
A snapshot of the database version released on November 6, 2009. Unless otherwise noted, all files in this archive are under a Creative Commons Attribution-Noncommercial 3.0 License (CC-BY-NC)