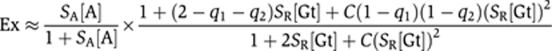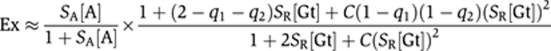Abstract
Systems biology seeks a genomic-level interpretation of transcriptional regulatory information represented by patterns of protein-binding sites. Obtaining this information without direct experimentation is challenging; minor alterations in binding sites can have profound effects on gene expression, and underlie important aspects of disease and evolution. Quantitative modeling offers an alternative path to develop a global understanding of the transcriptional regulatory code. Recent studies have focused on endogenous regulatory sequences; however, distinct enhancers differ in many features, making it difficult to generalize to other cis-regulatory elements. We applied a systematic approach to simpler elements and present here the first quantitative analysis of short-range transcriptional repressors, which have central functions in metazoan development. Our fractional occupancy-based modeling uncovered unexpected features of these proteins’ activity that allow accurate predictions of regulation by the Giant, Knirps, Krüppel, and Snail repressors, including modeling of an endogenous enhancer. This study provides essential elements of a transcriptional regulatory code that will allow extensive analysis of genomic information in Drosophila melanogaster and related organisms.
Keywords: Drosophila, enhancer, modeling, repression, transcription
Introduction
The rapid increase in sequenced genomes has provided an extensive parts list of organisms; however, deeper understanding of the regulatory code of the genome is critical to discerning the dynamic activity of biological systems. (By regulatory code, we mean the relationships reflecting biochemical interactions between transcription factors reflected in the structure of binding sites in cis-regulatory regions.) Subtle changes in regulatory elements are often involved in hereditary diseases, population differences, and the evolution of morphological novelties (Carroll et al, 2001). Comparative studies have shown that regulatory regions can retain function over large evolutionary distances, even though the DNA sequences are divergent and poorly alignable (Ludwig and Kreitman, 1995; Hare et al, 2008). The flexibility in arrangement of binding sites is not unlimited, however. For instance, the effectiveness of short-range transcriptional repressors that have important functions in Drosophila development is strongly influenced by activator–repressor distances (Gray et al, 1994; Arnosti et al, 1996a; Kulkarni and Arnosti, 2005).
The Drosophila blastoderm embryo provides an ideal setting for the analysis of transcriptional enhancers; the cascade of maternally and zygotically supplied transcription factors has been extensively investigated at a molecular level, and many DNA regulatory elements have been identified and functionally dissected. In this system, genes with complex expression patterns are controlled by multiple enhancers, whose modular function depends on the local action of repressor proteins (Small et al, 1993). Although Drosophila features a derived syncytial embryo, it is clear that similar regulatory networks control development in a cellularized environment (Denell, 2008). Similar modular enhancers provide complex developmental signaling in higher metazoans. In light of the similarities between Drosophila and mammalian transcription factors and signal transduction components, it is likely that the fly will provide useful guidelines to enhancer structure and function in metazoans in general. The blastoderm embryo has been used for quantitative analysis of gene expression by reaction diffusion, Boolean, and fractional occupancy modeling (Sánchez and Thieffry, 2001; Jaeger et al, 2004; Segal et al, 2008). Fractional occupancy models draw from simple biophysical principles and statistical physics to predict the overall readout of endogenous enhancers (Bintu et al, 2005a, 2005b). In these models, parameters include the binding affinity of transcription factors to the DNA and cooperativity between proteins. Although these models are based on quantitative modeling of DNA–protein interactions, they are generally joined with a phenomenological description of the gene regulatory process to take into account important, but less accessible, features such as chromatin modifications and RNA polymerase phosphorylation.
Simple prokaryotic systems provide a tractable setting for quantitative studies, and fractional occupancy models have been applied to the lac operon in Escherichia coli and the lysis/lysogeny switch of phage lambda (Von Hippel et al, 1974; Ackers et al, 1982; Shea and Ackers, 1985, Vilar and Leibler, 2003). Use of these models in eukaryotes is more problematic, given the higher degree of enhancer complexity in eukaryotic systems, but Drosophila enhancers have been treated by fractional occupancy models that account for factor spacing and recruitment of co-regulators (Janssens et al, 2006; Zinzen et al, 2006). These models can reproduce the behavior of specific enhancers, but a major limitation of fractional occupancy modeling of endogenous enhancers is that models of a single regulatory region may not generally apply to other elements. In studies of multiple enhancers, the parameter estimation has been difficult, as the different architecture of distinct enhancers, even those regulated by the same proteins, makes it difficult to know which parameters (number of bindings sites, relative arrangements, etc.) are important to determining the particular activity of an enhancer (Segal et al, 2008). As we describe here, a more systematic approach is necessary to parse the contributions of individual physical features to enhancer activity.
One particular area that has been inadequately explored is the important function of the repressor proteins. Giant, Knirps, and Krüppel are regionally deployed short-range repressor proteins that bind to and control the patterning of pair-rule genes such as even skipped. Earlier studies showed that precise positioning of short-range repressors on an enhancer can be used to generate the appropriate expression pattern in a morphogenetic field in which the concentration of these repressors are used to set gene expression thresholds (Hewitt et al, 1999; Clyde et al, 2003). Thus, the flexibility of enhancer architecture incorporating these proteins is constrained by some distance limitations. Our earlier study showed that activator–repressor stoichiometry and arrangement of binding sites also influence the overall readout of developmental enhancers (Kulkarni and Arnosti, 2005). To build tools able to accurately predict the function of novel enhancer sequences, we recognized a need to quantitatively measure the specific contributions of these factors to overall enhancer function. Here, we describe the creation and quantitative assessment of a well-defined set of transcriptional regulatory modules in the Drosophila embryo, in which individual aspects relating to repressor–activator spacing, stoichiometry, and arrangement are systematically explored. Using quantitative data from these genes, we apply a fractional occupancy-based approach to model the interaction of short-range repressors with endogenous transcriptional activators. We show that this approach can correctly decipher the transcriptional regulatory code of endogenous enhancers, pointing the way to a general approach for unlocking the transcriptional regulatory information of genomes.
Results
Gene modules
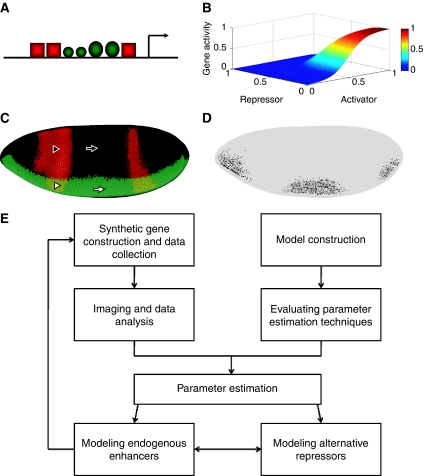
We set out to map regulatory surfaces of genes controlled by short-range repressors; these surfaces show the functional relationship of activator/repressor input and gene expression output (Figure 1). Such regulatory surfaces reflect evolutionary forces that shape gene output, as shown for the lac operon (Setty et al, 2003; Mayo et al, 2006). The design of the enhancers responding to short-range repressors accommodates sensitive distance and binding site parameters within a flexible design framework (Clyde et al, 2003; Kulkarni and Arnosti, 2005). The output of a model of a particular configuration of transcription factor-binding sites should lead to a regulatory surface that allows mapping of known values of regulatory factors, such as Dorsal and Twist activator protein levels, and Giant repressor protein levels, through this surface to produce an expected regulatory outcome (Figure 1).
Figure 1.
Transformation of DNA sequence and protein information by gene modeling. (A) An enhancer with three repressors (red squares) and four activators (green circles) is modeled, to generate the gene expression surface shown in (B). The axes represent normalized activator, repressor, and gene activity levels. (C) A Drosophila embryo with Giant repressor (red stripes) and Dorsal activator (green) staining is shown. Each embryo provides a diversity of potential inputs to the regulatory element: the white arrow points to a region in which activator levels are high and repressor levels are low. The black arrow points to a region in which both activator and repressor levels are low. The white triangle points to a region in which activator and repressor levels are both high, and the black triangle points to a region in which repressor levels are high and activator levels are low. (D) Output of regulatory element shown in (A), which mirrors values from (C) being mapped through surface shown in (B). (E) Formal scheme of data collection, analysis, and modeling.
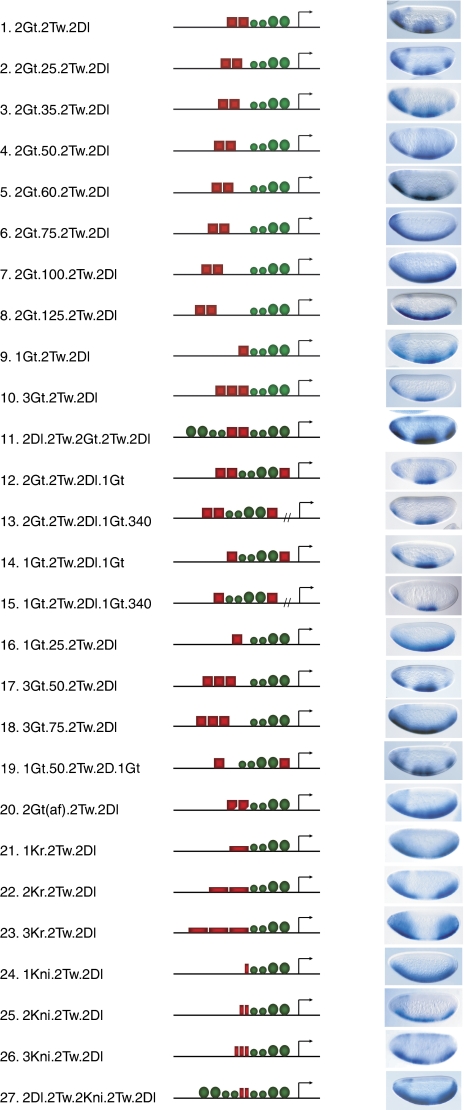
To carry out this scheme on a practical level, we created a series of genes to test in a systematic manner the effect of parameters affecting repression. The quantitative measurement of these genes was used to create a database suitable for quantitative modeling, identification of parameters related to repressor activity, and analysis of endogenous regulatory elements (Figure 1E). We used endogenous activators and repressors that are active in the blastoderm embryo. A convenient juxtaposition of anterior-posteriorly expressed repressor proteins Giant, Krüppel, or Knirps are superimposed on the patterns derived from activators working on the dorsal–ventral axis to generate readouts as shown in Figure 1. This design permits the simultaneous monitoring of repressed and unrepressed states in a single embryo. Twenty-seven P-element-based genes were inserted into the Drosophila germline to produce stably integrated lacZ reporters. We tested multiple lines for each; position effects had some effect on overall expression levels, but not on relative repression effectiveness. As described below, activator signals are normalized before parameter estimation and modeling, removing this potential source of variability. On the basis of the earlier studies, we knew that spacing between activators and repressors would be a critical element to model, thus a series of genes (1–8, Figure 2) tested variable distances between Giant repressor-binding sites and the nearest Twist activator sites. As revealed by conventional in situ staining, repression effectiveness was markedly attenuated by this increase in spacing. Genes for which the most proximal-binding site for Giant was located at least 81 bp from the nearest Twist site failed to show any repression (genes 6–8, Figure 2). A gene containing a single Giant-binding site adjacent to the Twist activators was weakly repressed, consistent with earlier reports (Hewitt et al, 1999), and this repression was also found to be distance-dependent (genes 9, 16). Increasing the number of binding sites to three (genes 10, 17, 18) seemed to generate an especially effective repression context, one that was similarly susceptible to distance effects; at this level of resolution, it was not clear whether the distance function is appreciably different with different numbers of repressors. We also tested the effect of arranging the repressors in a distinct pattern, so that some sites were located 3′ of the activator cluster, adjacent to the Dorsal activator sites. In this way, we were able to test whether overall stoichiometry of repressors to activators was the sole determinant of repression effectiveness when binding sites are close to the activators. We noted that different distributions of two or three sites seemed to yield similar results, whether all sites were located 5′ of the activator cluster, proximal to the Twist activator sites, or with some of the Giant repressor sites located 3′ of the activator cluster, adjacent to Dorsal (genes 12, 14, 19). Insertion of a 340 bp neutral spacer sequence between the transcription factor cluster and the basal promoter did not change the pattern of gene expression, suggesting that the repressor is not acting directly on the basal promoter in this context (genes 12 versus 13; 14 versus 15). Most blastoderm enhancers characterized for these regulatory proteins are located some distance from transcriptional start sites; thus, the distance independence of these modules mimics the activity of endogenous enhancers. We furthermore tested the effect of increasing the number of activators located in the vicinity of the repressors (genes 11, 27) and found that repression effectiveness was little compromised in the case of Giant, but seemed to be attenuated in the case of the weaker Knirps repression. Weaker-binding sites for Giant produced attenuated repression, as expected (gene 20). Finally, a series of genes with increasing numbers of binding sites for Knirps and Krüppel allowed for direct comparison of repressor effectiveness and effects of stoichiometry (genes 21–26); as noted for Giant, more sites were generally more effective, but overall repression effectiveness of Knirps was lower. This difference may be attributed to weaker-binding sites, lower absolute levels of the protein, or protein activity, as discussed below. The quantitative analysis of these genes was followed by quantitative measurements described below.
Figure 2.
Structures of genes assayed to determine context dependence of short-range repressor activity, and representative in situ images showing lacZ activity. Mid blastoderm embryos are oriented dorsal up, anterior to the left. Genes 1–8 test activator–repressor spacing, 9–10 and 16–18 activator–repressor stoichiometry and spacing, 12–15 and 19 arrangement and promoter proximity, 11 and 20 activator number and affinity, and 21–27 alternative short-range repressors.
Image processing and data analysis
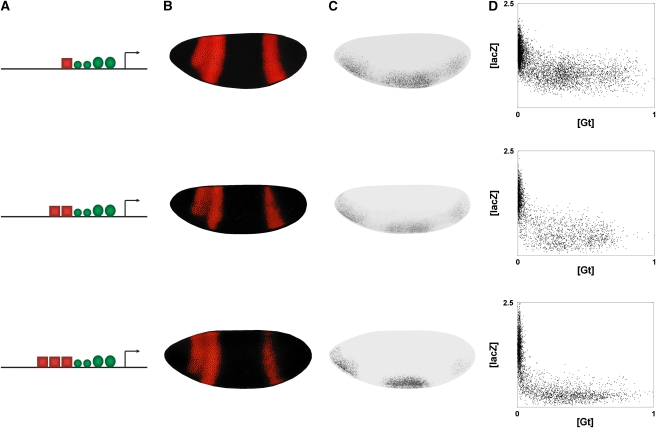
To simplify modeling, we initially restricted our measurements to the regions of the embryos containing peak levels of the Dorsal and Twist activators, which were identified as ventral regions expressing >60% of peak lacZ levels. To identify gene responses to varying repressor levels, we generated correlated Giant protein/lacZ mRNA plots (Figure 3). This step involved a series of image-processing procedures, as described in Ay et al (2008). The relative levels of gene expression as a function of repressor protein were plotted for individual images and compiled into composite plots (Figure 3) (Ay et al, 2008). These plots were used to infer cis-regulatory rules by fractional occupancy models as described below. Further information is provided in Materials and methods.
Figure 3.
Representative [lacZ] versus [Gt] plots. (A) Structures of three genes assayed (1, 9, and 10). (B, C) Representative embryos imaged for Giant protein and lacZ reporter gene activity. (D) The data from multiple confocal embryo images was processed and compiled to provide normalized reporter gene [lacZ] versus normalized repressor [Gt].
Fractional occupancy modeling
Fractional occupancy models of transcriptional regulatory regions enumerate all possible states of an enhancer based on potential transcription factor–DNA interactions, and then calculate the probability of a gene firing as the fraction of the successful states, that is those with activators bound, and without excessive interference by repressors (Bintu et al, 2005a; Janssens et al, 2006; Zinzen et al, 2006; Segal et al, 2008). To capture the important function of short-range repressors on activator elements, we used a modified fractional site occupancy model that explicitly accounts for distances between activators and short-range repressors, as well as cooperativity and binding affinity of short-range repressors. We allow for change in repression with distance, but make no a priori assumptions about how the repression efficiency changes.
For a general description of our model, we use three parameter types: SR, a repressor-scaling factor, indicating the potency of the repressor, C, representing cooperativity between repressor proteins binding to sites that are close together, and q, representing the distance-dependent ‘quenching’ efficiency of the short-range repressors. In genes assayed here, the activator-binding sites do not vary; therefore, additional parameters representing activator potency or binding cooperativity are not required. A more sophisticated general model incorporating these features is described below for endogenous sequences.
To apply this model to one of our genes, 2Gt.2Tw.2Dl (gene 1), we express normalized activator and Giant repressor concentrations, respectively, as [A] and [Gt], activator and Giant repressor-scaling factors as SA and SR (which represent binding affinity and concentration scaling combined into one scaling factor) (1⩽SA, SR⩽100), and cooperativity between Giant repressor proteins for binding to DNA as C (0.1⩽C⩽100). Quenching, the distance-dependent repression efficiency, is represented by q1 and q2 for the two Giant repressors in this gene (0⩽q1, q2⩽1). As derived in Materials and methods, the expression of this gene when fully bound by activators and repressors will be:
Comparable expressions are generated for each of the genes (Supplementary Table I).
Parameter estimation
Parameter estimation is a critical step in implementation of modeling. We used evolutionary strategy a global parameter estimation approach described by Runarsson and Yao (2005), which was shown in a recent study to work well in biological modeling (Fomekong-Nanfack et al, 2007).
Testing/implementing nine forms of the model
To analyze the quantitative data obtained from the embryos, we built nine forms of the model featuring increasing complexity in terms of number of parameters used; the models differ in their treatment of cooperativity and quenching distance. In the simpler case, a single parameter represents cooperativity between adjacent Giant repressor-binding sites, as well as the interaction of all three sites involved in genes 10, 17, and 18. Alternatively, we also used a more complex treatment in which adjacent sites are fit to C1 and sites separated by intervening Giant sites are fit to C2. Similarly, quenching efficiency parameters of repressors can be defined either as unique parameters for each distance or as parameters for a range of distances, as described in Materials and methods.
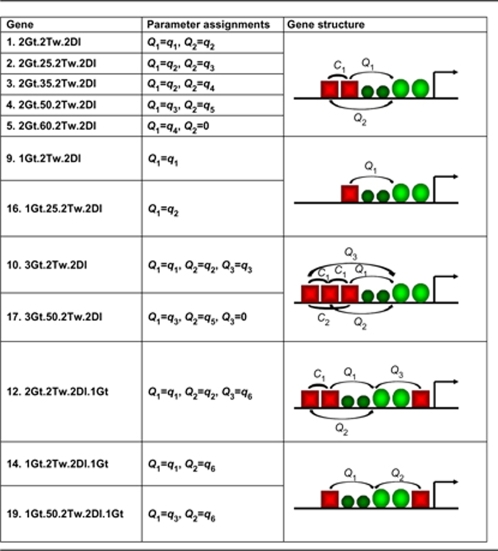
We show a pictorial description of the parameter assignments for scheme 2, a simpler form, in Table I. Supplementary Table II provides a pictorial description of the parameter assignments for all schemes.
Table 1. Parameter descriptions for scheme 2.
| In the first column, 12 synthetic enhancers used for parameter estimation in this study are listed. In the second column, parameter selections are shown. In the third column structure of the synthetic enhancers are depicted. | ||
|---|---|---|
|
We compared the nine schemes as explained in model validation section below. As judged by the error comparison, schemes 1–4, 8, and 9 work better than schemes 5–7 in this data set, probably because of the smaller number of parameters (Supplementary Figure 6). Here, for further analysis we showed the results of scheme 2. The results of the schemes 1, 3–6, 8, and 9 were comparable, suggesting that conclusions drawn from scheme 2 are representative (Supplementary Figure 5).
Model predictions
Earlier identified qualitative relationships about quenching and cooperativity/activity provide the backdrop for this work; the quantitative relationships presented here constitute the heart of this study, obtained after modeling our quantitative data set. It was striking that certain qualitative and quantitative insights became apparent only after analysis of the complete data set; these were not relationships that would necessarily be evident by inspection of individually stained embryos in Figure 2. First, our model predicts rather modest levels of Giant–Giant cooperativity, greater than simply additive, but lower than earlier estimates (Figure 4A) (Segal et al, 2008).
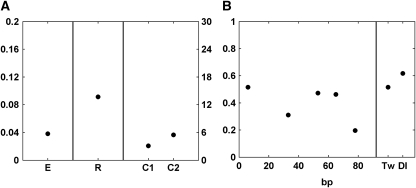
Figure 4.
Parameters found by the ES parameter estimation technique for scheme 2 of the model. (A) Root mean square error, E, is shown on the left, with corresponding scale shown on the left axis. Repressor-scaling factor R (referred to as SR in fractional occupancy model in Materials and methods) and cooperativity C are shown in the central and right portions, respectively, with scale shown on the right axis. (B) Quenching efficiency parameters are shown for increasing distances of repressors located 5′ of the activators on the left. Quenching efficiency levels relative to Twist proximal (T) sites and Dorsal proximal (D) sites are shown in the right panel. A non-monotonic decrease in quenching efficiency for increasing distances is observed.
Second, earlier qualitative observations show that the effect of short-range repressors decreases with distance, and is lost around 100–150 bp. To our knowledge, our study is the first that analyzes distance dependency of the short-range repressors systematically. Short-range repressor-quenching efficiency is represented by several parameters in the model as described earlier. We noted a general decrease in quenching efficiency with distance, consistent with earlier qualitative observations, but at (52–55) bp, relative efficiency is predicted to increase, before dropping off with greater distance (Figure 4B). This trend was evident for multiple formulations of the model (Supplementary Figure 5), and persisted when we carried out parameter estimation with subsets of the data (see below), indicating that the non-monotonic behavior reflects a real biochemical property of the Giant repressor. The change in this monotonic behavior may be a reflection of specific phasing effects, perhaps relating to nucleosomal structure. The non-monotonic decline in repression effectiveness was an unexpected result of our modeling and contrary to the simple step functions or linear functions used in earlier modeling efforts (Janssens et al, 2006; Zinzen et al, 2006). Note that the reduction in repression efficiency at ∼30 bp does not imply that gene 3 (2Gt.35.2Tw.2Dl) should have weak repression, because this gene has an additional more distal-binding site that also contributes to activity through quenching and cooperativity.
Third, the repressor-quenching efficiency parameters are similar whether the repressor was located adjacent to the Twist or to the Dorsal activator site, which suggests that short-range repressors have similar effects on different activators (Figure 4B). The short-range repression mechanism seems to involve chromatin modification, which may allow for more promiscuous action on many types of transcription factors, rather than a mechanism based on specific contacts between repressor and activator (Li Li (Arnosti Lab), unpublished data). This activator insensitivity is consistent with the action of short-range repressors on a range of enhancers that bind diverse transcriptional enhancers (Gray et al, 1994; Kulkarni and Arnosti, 2005). Parameters identified in this study are, therefore, likely to be generally applicable to diverse settings.
We tested whether the non-linear quenching is critical to obtaining reasonable parameters by repeating our procedure with a constraint that required a monotonic decrease for quenching efficiency. As shown in Figure 5, this constraint produced parameter sets that predicted repressor-quenching efficiency would remain almost constant between 6 and 77 bp, which is not supported by this or earlier studies. For example, the 35 bp increase in spacing between gene 2 and gene 5 has a measurable effect. Therefore, the non-monotonic decrease in quenching efficiency is likely to indicate some actual biological property of the repressors and should be validated experimentally.
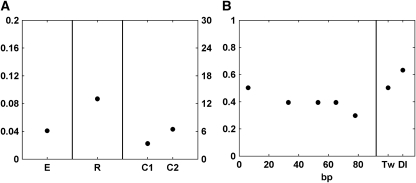
Figure 5.
Parameters for scheme 2 with the constraint that quenching efficiency parameters decrease monotonically. (A) Root mean square error E, repressor-scaling factor R, and cooperativity C labeled as in Figure 4. (B) Quenching efficiency parameters and relative quenching of Dorsal and Twist sites. Under this constraint, the level of quenching efficiency changes very little from 28 to 66 bp, in contrast to observed trends (Figure 2).
The recent fractional occupancy modeling of 44 endogenous Drosophila enhancers identified potential cooperativity values that were somewhat greater than those found here. We ran our parameter estimation algorithm with fixed Giant cooperativity values found in Segal et al (2008) and estimated the remaining parameter values in our model. We observed that although the main conclusions of our study did not change, the overall fitting was slightly worse (Figure 6). We extended this analysis by running our parameter estimation algorithm with eight more choices of Giant cooperativity values. Although we tested Giant cooperativity values ranging from 0 to 30, the root mean square errors between predicted and observed values did not change drastically, with minimum at cooperativity value 3. We note that cooperativity may reflect DNA-binding or post-DNA-binding effects; explicit measurements of in vivo protein occupancy may help differentiate these two.
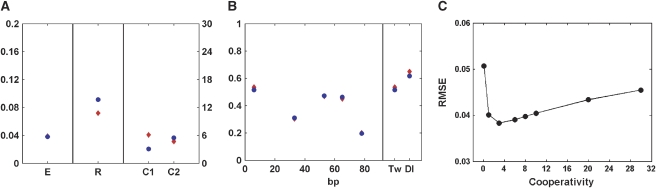
Figure 6.
Parameters for scheme 2 with cooperativity parameters set to different levels. (A, B) Parameters found in our study (circles) and parameters found by constraint of cooperativity parameters to those from Segal et al (2008) (diamonds). The increased cooperativity value is compensated by a decreased repressor-scaling factor R. (C) Root mean square errors (RMSE) for cooperativity parameters (constrained to values between 0 and 30). Estimated cooperativity values from our model lie near the lowest point in this curve.
Model validation
The analysis described above involved identifying parameters using all data available. An important question is whether such values are overfit, and whether the model and parameter estimation technique are robust, that is relatively insensitive to contributions of individual portions of the data set. Robustness of the parameter estimation technique is described in the supplementary material; here, we assess the model's effectiveness at predicting subsets of the data. We tested whether parameter estimation was markedly affected by removal of individual genes from the data set (leave-one-out analysis) (Figure 7A). We used nine different forms of the model to evaluate the effects including different assumptions of cooperativity and quenching. We calculated the average of 12 leave-one-out prediction root mean square errors for each scheme, and used these error values for comparison of schemes (Pizarro et al, 2000). As judged by the error comparison, schemes 1–4, 8, and 9 work better than schemes 5–7 in this data set, probably because of the smaller number of parameters (Supplementary Figure 6). Leave-one-out analysis was extended by excluding nine separate, specific groups of genes that share structural properties (Figure 7B). The sets used for this analysis are described in Table II. The results of excluding individual genes or sets of related genes suggest that genes that depend on fewer parameters, such as 1Gt.2Tw.2Dl (gene 9), which has no contribution by repressor–repressor cooperativity, may not be predicted well in our analysis. Thus, the contributions of certain classes of gene can be great. Parameters found by leave-one-out analysis did not change much, but the parameters found by leaving out specific sets of genes changed depending on the genes chosen (Figure 7A and B). The predictions for genes 1, 10, and 12 by the parameters estimated from set 8, which excludes genes 1, 10, and 12, are shown in Figure 7C. We conclude that the set of gene modules tested here adequately sample enhancer design to identify critical elements for repressor activity in a robust manner.
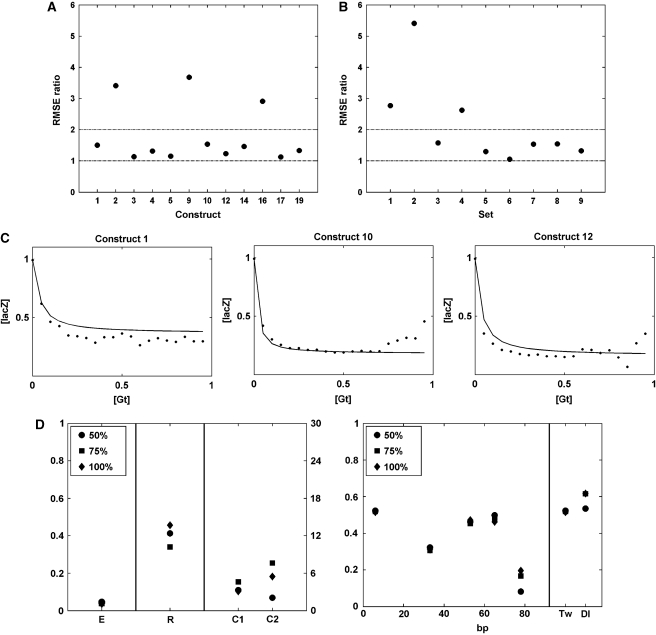
Figure 7.
Validation of modeling by prediction of subsets of the data from parameters derived from the remainder of the data. (A) Leave-one-out analysis. Root mean square errors are calculated using parameters found by 11 genes excepting the genes indicated, and all the genes. Relative RMSE ratios, indicating greater errors for prediction of genes 2, 9 and, 16, indicating their greater contribution to the parameter constraints. (B) Leave-sets-out analysis for nine distinct sets of genes defined by their shared properties (Table II). Root mean square errors are calculated using parameters found from the reduced set and the entire set. Relative RMSE ratios, indicating greater errors for prediction of sets 1, 2, and 4, indicating their greater contribution to the parameter constraints. (C) Predictions for leaving out set 8. Genes 1, 10, and 12 are predicted by using parameters found from other 9 genes. Points represent average values for [lacZ] versus [Gt] data, which was divided into 20 bins. (D) Parameter estimation results are shown for different amounts of data 50, 75, and 100%. The data is cut randomly from each gene at the same percentage.
Table 2. Functionally grouped sets of gene constructs used for leave-sets-out analysis shown in Figure 7B.
| Set# | Excluded genes |
|---|---|
| 1 | Genes with one or three Giant-binding sites (9, 10, 12, 16, and 17) |
| 2 | Stoichiometry genes (1, 9, and 10) |
| 3 | Genes with adjacent Giant-binding sites in both 5′ and 3′ end of activators (12, 14, and 19) |
| 4 | Genes with only one Giant-binding site (9 and 16) |
| 5 | Genes with exactly three Giant-binding sites (10, 12, and 17) |
| 6 | Genes with one Giant-binding site at 5′ end of activators (9, 14, 16, and 19) |
| 7 | Genes with one Giant-binding site immediately adjacent to the 5′ end of activators (9 and 14) |
| 8 | Genes with at least two Giant-binding sites immediately adjacent to the 5′ end of activators (1, 10, and 12) |
| 9 | Genes with three Giant-binding sites adjacent to the 5′ end of activators (10 and 17) |
Each embryo, with its thousands of imaged nuclei representing different levels of transcription factors, provides a matrix of input and output values that should in theory suffice to describe the response of a gene construct. However, variations in embryo age, staining, and orientation necessitate multiple images for each gene. We obtained between 30 and 53 good quality images for each gene used in our parameter estimation. To test whether this data set is sufficient, or additional individual images would significantly change the conclusions reached, we sampled randomly 50 or 75% of the images from each reporter gene construct, and repeated the parameter estimation. Reducing the data set by one quarter or even one half does not change the value of estimated parameters drastically or the main conclusions of the paper (Figure 7D). This result suggests that our data set is sufficiently complete, allowing us to draw significant conclusions. In contrast, as shown above, decreasing the number of genes rather than just the number of images obtained for each gene, can affect our results drastically (Figure 7A and B).
Extension of the model to other repressors and endogenous regulatory elements
Our modeling focused on repression mediated by Giant, which possesses quenching properties similar to those of Snail, Krüppel, and Knirps (Gray et al, 1994; Hewitt et al, 1999; Kulkarni and Arnosti, 2005). To extend these findings to other short-range repressors, Krüppel and Knirps were tested in parallel genes containing one, two, or three binding sites (genes 21–26). As was evident from qualitative staining, both proteins mediated repression, but Krüppel seemed to be a more effective repressor in terms of completeness of reduction of lacZ activity. We measured Knirps or Krüppel protein levels with antibodies as was carried out with Giant, and created [lacZ] versus [repressor] plots for parameter fitting. The limited number of genes tested for these factors did not exhaustively explore possible architectural features, thus making it difficult to differentiate effects of spacing, cooperativity, and relative activity. We judged distance parameters most likely to be conserved between these different factors, based on earlier tested genes; therefore, the modeling was carried out using quenching parameters from Giant (Gray et al, 1994; Arnosti et al, 1996a). Modeling was performed to identify likely scaling factors and cooperativity constants. Using the same form of the model used for Figure 4, we found that cooperativity parameters were low (e.g. Krüppel=2; Knirps=0.67), similar to those observed for Giant (Supplementary Figure 5; Supplementary Table V). The major difference between Krüppel and Knirps was the repressor-scaling factor, which was low in the case of Knirps (∼1.4), and more robust for Krüppel (∼30), similar to that of Giant (∼14). Differences in repression efficiency may be attributed to distinct levels of cooperativity, but the model suggests that such homotypic interactions are of minor importance. This prediction suggests that the higher effectiveness of Krüppel is likely because of greater potency of this protein on a molar basis, a more complete occupancy of the binding sites because of their higher affinity, or higher concentrations of the repressor. Further analysis will be required to separate these effects.
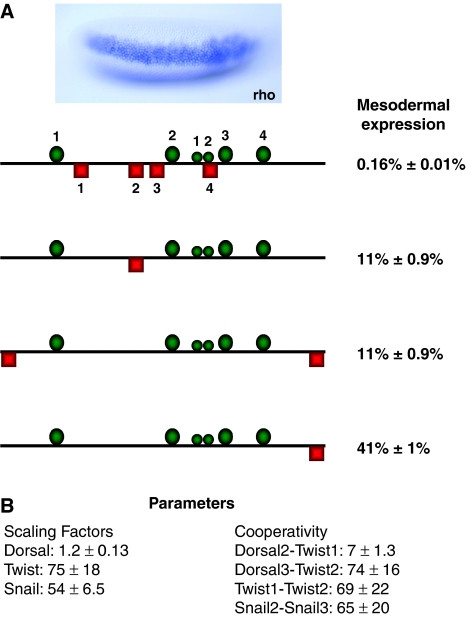
Dorsal and Twist activators were studied earlier in the context of the rhomboid (rho) neuroectodermal enhancer (NEE), in which their activity was used to identify properties of short-range repressors, including Snail. This protein is required to block expression of rho in the mesoderm, resulting in two lateral stripes of expression in the presumptive neuroectoderm of the blastoderm embryo (Figure 8A). Four Snail-binding sites are located within the 330 bp minimal NEE enhancer, and loss of these sites strongly attenuates repression, permitting expression in the mesoderm (Gray et al, 1994). A single Snail site (#2) is sufficient to mediate repression, and similar repression is effected by ectopic Snail, Krüppel, or Knirps sites introduced 5′ and 3′ of the Dorsal 1 and 4 sites, respectively, or even a single Snail site 3′ of the Dorsal 4 site (Figure 8A) (Gray et al, 1994; Arnosti et al, 1996a).
Figure 8.
Extension of the model to endogenous regulatory elements. (A) The rhomboid gene is expressed in the blastoderm embryo in two lateral stripes (one shown in focal plane), under control of the Dorsal and Twist activators. Ventral expression is inhibited by the Snail short-range repressor, which is expressed in the presumptive mesoderm. The cis-regulatory modules used for analysis are shown. Different forms of rhomboid NEE enhancer are depicted, with varying number and arrangements of Snail short-range repressor-binding sites. Dorsal and Twist activators are shown by large and small green circles, respectively, and Snail repressors are shown by red squares. On the right are the predicted repression levels caused by Snail-binding sites shown in each module based on parameter estimation using this group of enhancers. (B) Predicted parameters for scaling factors for each transcription factor and cooperativity. Average and standard deviation for 20 estimation runs are shown.
As an extension of our analysis, we tested quenching parameters produced from our model on this element, and carried out parameter estimation to determine values of cooperativity and scaling factors. This modeling is more complex than that used for genes in Figure 2, because we now consider scaling factors for each transcription factor, not just for the repressor, and binding sites of different qualities are considered. Position weight matrix (PWM) information was used to score Dorsal, Twist, and Snail sites within the rho NEE. In addition, we consider cooperativity not just between repressor sites, but also between activator sites, both of heterotypic (Dorsal–Twist) and homotypic (Twist–Twist) nature. A further consideration is that information about these rho NEE variants is qualitative; a single Snail site can repress, but may not be as effective as four Snail-binding sites.
Simultaneous parameter estimation was carried out using the forms of the rho NEE shown in Figure 8A. We estimated levels of mesodermal repression to be >90% for the endogenous gene, 70–90% for genes carrying one Snail #2-binding site or two ectopic-binding sites located 5′ and 3′ of the element, and 50–70% for one Snail site located 3′ of Dorsal #4. Evolutionary strategy parameter estimation was performed multiple times to identify parameters for cooperativity and scaling factors, as well as the predicted effect on expression within ranges specified above. Several striking outcomes were evident from this exercise; first, to find optimal values, the model consistently predicts that the wild-type rho NEE, containing four Snail sites, will have output at the lowest end of the allowed range, close to zero, whereas the internal Snail site #2, or the two ectopic flanking Snail sites, generates values close to the bottom of the allowed range, at about 10% residual activity (Figure 8A). The single ectopic Snail site 3′ of Dorsal #4 is predicted to mediate repression in the middle of the allowable range, about 40% residual activity, consistent with published images (Gray et al, 1994). The scaling factor for Dorsal (i.e. its overall activity) is considerably lower than that predicted for Twist, whereas the scaling factor for Snail is similar to those of Krüppel and Giant (Figure 8B). Dorsal–Twist cooperativity values vary considerably, with Dorsal2–Twist1 cooperativity predicted to be lower than Twist2–Dorsal3, consistent with the closer spacing of the latter two factors (Crocker et al, 2008). Twist–Twist cooperativity is also predicted to be high. These relative differences in activator-scaling factors and cooperativity values support known features of the rho NEE; the low-scaling factors for Dorsal sites are consistent with the inability of individual Dorsal sites to mediate robust activation (Ip et al, 1992); but in combination with Twist sites they add considerably to the output of the enhancer. A single repressor-binding site that is not close to most of the activator sites would in this model still be able to impair enhancer function by initiating a chain-reaction collapse of cooperative interactions. The native rho NEE does not seem to rely solely on this mechanism, as most of the identified activator sites lie within a short distance of one of the four Snail sites, suggesting a redundant approach to repression. It will be interesting to survey the entire set of enhancers targeted by short-range repressors to determine whether this feature is consistently observed in most elements.
Discussion
In the past 20 years, essential features of the complex biochemistry of gene regulation have come into focus, but we still lack a comprehensive picture of how a transcriptional enhancer operates. Quantitative models, based on aspects of the system that are readily quantifiable, such as DNA sequence of a regulatory region, quantities of regulatory proteins, and transcript levels, offer an alternative route to learn about important features of regulatory systems. When combined with biochemical and genomic information, such models may provide the bridge that will allow deeper understanding of the function and evolution of cis-regulatory elements, which are the nexus of many biological processes.
In this study, by using a reductionist analysis of short-range repression, we explored a relatively untouched, yet central aspect of gene regulation in Drosophila. Earlier qualitative studies highlighted the extreme distance dependence of short-range repressors, and comparative analysis has shown many instances of evolutionary plasticity of regulatory regions controlled by these proteins (Gray et al, 1994; Ludwig and Kreitman, 1995; Hewitt et al, 1999; Hare et al, 2008). Knowing that transcription factors influence each other in a local manner permitted the identification of novel enhancers, based on the clustering of binding sites (Berman et al, 2002; Schroeder et al, 2004). Yet, clustering studies alone do not provide the basis for predicting evolutionary changes that reshape transcriptional output, or predicting activity of coregulated enhancers. For example, the original hypothesis that the affinity and or number of Bicoid-binding sites dictates the output of regulated genes has been replaced by an understanding that other, as-yet unknown features, seem to have more decisive functions (Driever et al, 1989; Gao et al, 1996; Ochoa-Espinosa et al, 2009).
Earlier modeling studies focused on endogenous enhancers, which have complex arrangements of transcription factor-binding sites. Our studies focused on detecting quantitative differences resulting from subtle differences in binding sites, allowing modeling with a tractable number of parameters. We used a common block of Dorsal and Twist activator sites, allowing us to focus on changes made in the number and arrangement of repressor sites; clearly, differences in affinity, number, and arrangement of activator sites also have decisive functions in dictating transcriptional output; thus, future modeling efforts will need to integrate these elements as well. The tight focus on short-range repressors with the analysis of a relatively small number of reporter genes provided sufficient data for robust estimation of important parameters (Figure 7). From our comparison of repression by other short-range repressors, it is likely that the analysis of Giant can guide studies of other similarly acting repressors, including Krüppel, Knirps, and Snail (Figure 8).
Relating to transcriptional regulatory code, our study uncovered specific quantitative features that seem to apply to short-range repressors in a general context. We identified a complex non-linear quenching relationship that suggests that within the range of activity, Giant, and probably other short-range repressors, have an optimum distance of action that may reflect steric constraints (Figure 4). Multiple formulations of the model generated very similar predictions, suggesting that this non-linear distance function is a real feature of the system (Supplementary Figure 5). Consistent with this notion, an earlier study of transcription factor-binding sites in Drosophila enhancers discovered an overall preference of Krüppel sites to be found 17 bp from Bicoid activator sites, which may be an indication that other short-range repressors also have preferred distances for optimal activity (Makeev et al, 2003).
The similar quenching efficiencies for repressors acting adjacent to Dorsal or Twist activator sites were an additional significant finding (Figure 4). The similar effect on disparate activator proteins indicates that the effects of short-range repression are general, and are likely to be translatable to distinct contexts. Earlier empirical tests had already pointed in this direction; for example, insertion of ectopic-binding sites for Knirps and Krüppel into rho NEE sequences is sufficient to induce repression, although these proteins do not usually cross-regulate (Gray et al, 1994; Arnosti et al, 1996a). In addition, short-range repressors can counteract a variety of transcriptional activation domains with similar efficiency, suggesting that specific protein–protein contacts are not essential (Arnosti et al, 1996b; Kulkarni and Arnosti, 2005). In one area we found quantitative differences between parameters derived from the synthetic gene modules and the endogenous regulatory regions. The importance of homotypic cooperativity predicted for Snail sites in the context of the rho NEE was overall much higher than that found for Giant, Krüppel, and Knirps sites acting on the synthetic gene constructs; this might be an example in which the individual proteins do exhibit different context dependencies perhaps because the proteins differ in level of stickiness. Alternatively, the distance between the Snail sites in question, 23 bp, might facilitate cooperative interactions much more than the closely apposed spacing used in our genes, in which steric interference may have an opposing function.
In modeling mutant forms of the endogenous rho NEE, we uncovered several important features of the architecture of this regulatory region. This enhancer seems to use redundancy in use of Snail to mediate repression; based on earlier experiments, it seems that even a single Snail site is sufficient to mediate repression (Gray et al, 1994). Such redundancy may provide the correct dynamical response, with a swift repression of rho at an early enough time in which Snail levels are still low, or it may ensure that gene output is robust to environmental and genetic noise.
The rho NEE modeling also highlighted features of transcriptional activators. Activator-scaling factors for Dorsal were reproducibly lower than those of Twist, and this was apparent for several different assumptions of expression level (Figure 8 and data not shown). The relative differences in contribution to activation can be explained by examination of the structure of the enhancer; contribution by the low intrinsic values of Dorsal is amplified by strong cooperativity with Twist, setting up a chain of interacting weak sites that together are highly active. Experimental evidence bears out these conclusions: isolated Dorsal sites tested on reporter genes mediate relatively weak activation, and a rho NEE lacking Twist sites, but containing four Dorsal sites, is similarly compromised (Ip et al, 1992; Szymanski and Levine, 1995).
Our earlier studies suggested that many developmental enhancers, including those regulated by short-range repressors, may possess a flexible ‘billboard’ design, in which individual factors or small groups of proteins would independently communicate with the promoter region, so that the net output of an enhancer would reflect the cumulative set of contacts over a short time period (Kulkarni and Arnosti, 2003). Such a view of enhancers would account for the evolutionary plasticity observed in regulatory sequences. No DNA-scaffolded superstructure, reflecting the formation of a unique three-dimensional complex, would be necessary in this scenario. Yet, our modeling suggests that the rho NEE might involve communication between relatively distant-binding sites, through sets of cooperative interactions. In this case, it is possible that such distant interactions might be compatible with a flexible structure, if many distinct configurations of binding sites provide such a cooperative network. Current studies have indeed highlighted potential frameworks involving Dorsal and interacting factors on same classes of enhancer (Erives and Levine, 2004; Papatsenko and Levine, 2007). Application of a transcriptional regulatory code integrating activities of activators and repressors is a critical next step to illuminate enhancer design and evolution.
Materials and methods
Reporter genes
The binding motifs for the Giant, Krüppel, and Knirps short-range repressors and the Twist and Dorsal activators used in this study were characterized in earlier studies (Szymanski and Levine, 1995; Hewitt et al, 1999; Kulkarni and Arnosti, 2005).
Regulatory modules were constructed in pBluescript KS(+) using the EcoRI, BamHI, XbaI, and SacII restriction sites, amplified by PCR using T3 and T7 primers, and amplicons were digested with EcoRI and SacII and subcloned into the compatible sites of C4PLZ (Wharton and Crews, 1993). Gene 1 contains two Giant-binding sites inserted between EcoRI and BamHI, two Twist sites inserted between BamHI and XbaI, and two Dorsal-binding sites between XbaI and SacII.
Gene 2 includes a 25 bp spacer inserted between Giant and Twist sites using BamHI. For genes 4, 6, 7, and 8, the same 25 bp spacer was concatemerized and inserted at BamHI. For genes 3 and 5, a 35 or 60 bp spacer was inserted at BamHI, between the Giant- and Twist-binding sites. Spacer DNAs were analyzed for putative-binding sites to known blastoderm regulatory proteins. Gene 9 contains a single Giant-binding site inserted between EcoRI and BamHI. Gene 10 was constructed by digestion of the parent gene 1 pBluescript plasmid with EcoRI and insertion of the single Giant-binding site, preserving a single 5′ EcoRI site. For genes 12, 13, 14, 15, and 19, the same strategy was used to insert an extra Giant-binding site 3′ of the Dorsal sites using SacII. For genes 13 and 15, in which the binding sites are moved away from the basal promoter of lacZ reporter gene, a 340 bp spacer was amplified from the coding region of knirps gene and inserted into the SpeI of C4PLZ plasmid (Kulkarni and Arnosti, 2005). A weaker Giant site was tested in gene 20. The sequences for all oligos used are shown in Supplementary Table III. All gene cassettes were confirmed by sequencing, and at least five transgenic lines of each gene were analyzed by in situ hybridization for lacZ expression pattern. Lines showing enhancer trapping were not included in the analysis. Fixed embryos from two to three transgenic lines of each gene were used for confocal laser scanning microscopy.
Image processing
A five-step procedure was applied to all embryo images as described in Ay et al (2008), involving binary image generation, rotation, outlier removal, background subtraction, and normalization. We first identified and subtracted non-specific signals (outliers) observed in the Giant channel, then identified and subtracted background from each embryo. Background intensities for the lacZ and Giant channels were subtracted using average values from regions lacking activators and repressors. Next, we normalized the Giant channel for similarly aged and oriented embryos. The lacZ channel was normalized using the average signal in a region defined by 50–60% egg length of the embryo (anterior-posterior) and peak to 60% of the peak (dorsal–ventral). Our data set comprises expression data from 20 lacZ reporter genes regulated by Giant, 3 lacZ reporter genes regulated by Krüppel, and 4 lacZ reporter gene constructs regulated by Knirps. Over 900 blastoderm embryos were quantified to aid in parameterization of repressor and lacZ expression. Images were processed as described above and [lacZ] versus [repressor] (Giant, Krüppel, or Knirps) plots were created. Further details are provided in Supplementary information.
Data set
A total of 769 embryos bearing lacZ reporter gene constructs regulated by the Giant repressor protein were analyzed and an additional 45 and 88 were analyzed for genes regulated by Krüppel and Knirps, respectively. Genes 6, 7, 8, and 18 were not used in the quantitative modeling because no Giant repression was ever observed, and an ectopic modulation of the reporter gene by unknown factors in a fraction of the imaged embryos made this data especially noisy. All primary data are available on our server at: http://www.arnosti-lab.bmb.msu.edu/moreArnostipubs.html.
Schemes
Distinct forms of our model were implemented in which the quenching parameters were grouped in different ‘bins’ of distances. For distances >81 bp, quenching efficiencies of the repressors are taken as 0, motivated by our genes 6–8, which shows no repression.
Schemes 1, 2, and 8: q1 (6 bp), q2 (28–41 bp), q3 (50–56 bp), q4 (63–66 bp), q5 (78 bp), q6 (6 bp from 3′ end of activators).
Schemes 3, 4, and 9: q1 (6 bp), q2 (28–41 bp), q3 (50–53 bp), q4 (56–66 bp), q5 (78 bp), q6 (6 bp from 3′ end of activators).
Schemes 5: q1 (6 bp), q2 (28–31 bp), q3 (41–50 bp), q4 (53–56 bp), q5 (63–66 bp), q6 (78 bp), q7 (6 bp from 3′ end of activators).
Schemes 6: q1 (6 bp), q2 (28–31 bp), q3 (41 bp), q4 (50–56 bp), q5 (63–66 bp), q6 (78 bp), q7 (6 bp from 3′ end of activators).
Schemes 7: q1 (6 bp), q2 (28 bp), q3 (31 bp), q4 (41 bp), q5 (50 bp), q6 (53 bp), q7 (56 bp), q8 (63 bp), q9 (66 bp), q10 (78 bp), q11 (6 bp from 3′ end of activators).
We also tried different expressions of cooperativity. In schemes 1, 3, 5, 6, and 7, only one parameter is used for Giant–Giant cooperativity. In schemes 2, 4, 8, and 9, two parameters are used for Giant–Giant cooperativity, in which the first parameter describes cooperativity of Giant proteins with 10 bp distance and the second describes cooperativity for 32 bp distance. In schemes 2 and 4, we described the cooperativity of observing all three Giant proteins on the DNA as the summation of the two cooperativity parameters, and in schemes 8 and 9 as the multiplication of the two cooperativity parameters.
Derivation of the model for gene 1
We express efficiency of the activator group bound and not bound, respectively, as EA and EN, and efficiency of the Giant repressor as EGt. We represent the efficiency vector that represents efficiency for each state of activator set and Giant repressors as E, the state vector of activator set and Giant repressors as F, the regulatory function that transforms each efficiency vector input to transcription level as T, and total steady state transcription level as Ex. The probability of each state of those proteins on the DNA can be calculated. As the activator-binding sites do not vary within the genes tested here, the Dorsal and Twist activators are not parameterized, and are considered as one group. We set SA[A] equal to 100 with the assumption that in the absence of Giant repressor protein, the activators are fully functional. A set of eight equations describes all possible states of this gene. For simplification, we use the following formulas: Z=(1+SA[A])(1+2SR[Gt]+C(SR[Gt])2).
No activator and repressor bound: FN=1/Z.
Activator set is bound: FA=(SA[A])/Z.
Proximal Giant to the activator set is bound: FGt1=SR[Gt]/Z
Distal Giant to the activator set is bound: FGt2=SR[Gt]/Z
Activator set and proximal Giant to the activator set is bound: FAGt1=SA[A]SR[Gt]/Z
Activator set and distal Giant to the activator set is bound: FAGt2=SA[A]SR[Gt]/Z
Both Giant repressors are bound: 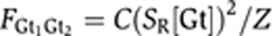
Activator set and both Giant repressors are bound: 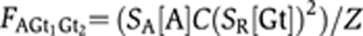
Then the states vector of one activator set and two repressors can be written as 
In the above expressions, we stated all the Boltzmann states of an enhancer with one activator set and two repressor-binding sites. We claim that the binding of the repressors modulates the probability of states with activators and repressors simultaneously bound by a quenching factor. For example, binding of the activator A and repressor R1 simultaneously is reduced by a factor of (1−q1). We can modify the probability of states, in the following way: 
Next, we calculate the total efficiency of the enhancer when factors are bound on the DNA. We model cooperativity between Giant sites, which might be, for example, because of cooperative cofactor recruitment, with an additive function, so the total efficiency of Giant repressors bound to the DNA at the same time is 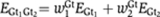 where
where 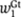 and
and 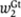 are the cooperativity terms after binding. The efficiency of one activator set and two repressors are expressed in the following way, each term representing one state of the all possible states:
are the cooperativity terms after binding. The efficiency of one activator set and two repressors are expressed in the following way, each term representing one state of the all possible states: 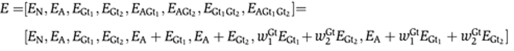
Expression contributions from each state are added to obtain the total expression: 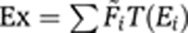
If we set the following simple assumptions EN=0, EA=10, EGt1=0, EGt2=0 and 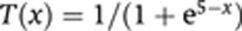
, the total expression of the enhancer with 1 activator set and 2 repressor-binding sites can be written as:
Expression functions (Ex) for all cases are shown in Supplementary Table I. Further details about the model are explained in the Supplementary information.
Modeling endogenous enhancer sequences
We made the following assumptions to simplify the parameter estimation for modeling of the rho NEE: (1) we model activity of the NEE in the mesoderm, in which Dorsal and Twist levels are high, and Snail is present at uniform levels. We used values for expression contribution of Dorsal and Twist as +5 each, and for Snail, −5. We also carried out parameter estimation with values of +3 or +7 for activators and −3 or −7 for Snail, and obtained essentially equivalent results. (2) We set quenching parameters to those obtained from our modeling, as shown in Figure 4, on the reasonable assumption that these are functionally equivalent among short-range repressors. (3) To reduce the number of possible parameters, we only included cooperative interactions between factors that are nearest neighbors, and are located within 25 bp of each other. (4) We allow that the relative effectiveness of repression with four Snail sites might be higher than that seen with one or two, and stipulate ranges of repression in which parameter space is investigated (Figure 8). (5) We set ranges for cooperativity and scaling factors from 1 to 100. (6) For each transcription factor, we took the score of the strongest site among all those that bind that transcription factor as a free parameter and constrain the other values by treating the PWM score as a free energy of binding (Stormo, 2000). We used PWMs created from FlyReg database by Daniel A. Pollard, which are available at: http://www.flyreg.org/. As an example, the two Twist sites differ considerably in terms of their match to a consensus PWM, with Twist site #2 predicted to have a 47-fold lower score than Twist site #1, although it still has a considerably higher score than background sequences.
Supplementary Material
Supplementary information, Supplementary figures S1–6, Supplementary tables SI–V
dataset
Acknowledgments
We thank Hin Siong Chong and Jennifer Chen for their contributions to initial stages of this project, Xiaozhou Liu and Chike Anadumaka for technical assistance, Li Li for photography, Melinda Frame for assistance with confocal microscopy, and Chichia Chiu (Department of Mathematics) and members of the Arnosti laboratory for thoughtful discussions. WDF created and analyzed the activities of transgenes with help from RS and ED. Data analysis and modeling was conducted by AA, with help from ED and JD. The manuscript was written by AA and DNA with input from RS and WDF, and overall project conceived and guided by DNA. Special thanks to Bill Wedemeyer and Ian Dworkin for comments on the manuscript. This projected was supported by a Special Projects Grant from the MSU Foundation and NIH GM 56976 to DNA and fellowships from the MSU Quantitative Biology Initiative to AA and JD.
Footnotes
The authors declare that they have no conflict of interest.
References
- Ackers GK, Johnson AD, Shea MA (1982) Quantitative model for gene regulation by λ phage repressor. Proc Natl Acad Sci USA 79: 1129–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnosti DN, Gray S, Barolo S, Zhou J, Levine M (1996a) The gap protein knirps mediates both quenching and direct repression in the Drosophila embryo. EMBO J 15: 3659–3666 [PMC free article] [PubMed] [Google Scholar]
- Arnosti DN, Barolo S, Levine M, Small S (1996b) The eve stripe 2 enhancer employs multiple modes of transcriptional synergy. Development 122: 205–214 [DOI] [PubMed] [Google Scholar]
- Ay A, Fakhouri WD, Chiu C, Arnosti DN (2008) Image processing and analysis for quantifying gene expression from early Drosophila embryos. Tissue Eng Part A 14: 1517–1526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman BP, Nibu Y, Pfeiffer BD, Tomancak P, Celniker SE, Levine M, Rubin GM, Eisen MB (2002) Exploiting transcription factor binding site clustering to identify cis-regulatory modules involved in pattern formation in the Drosophila genome. Proc Natl Acad Sci USA 2: 757–762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bintu L, Buchler NE, Garcia HG, Gerland U, Hwa T, Kondev J, Phillips R (2005a) Transcriptional regulation by the numbers: models. Curr Opin Genet Dev 15: 116–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bintu L, Buchler NE, Garcia HG, Gerland U, Hwa T, Kondev J, Kuhlman T, Phillips R (2005b) Transcriptional regulation by the numbers: applications. Curr Opin Genet Dev 15: 125–135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll SB, Grenier JK, Weatherbee SD (2001) From DNA to Diversity. Malden, Massachusetts, USA: Blackwell Science [Google Scholar]
- Clyde DE, Corado MS, Wu X, Paré A, Papatsenko D, Small S (2003) A self-organizing system of repressor gradients establishes segmental complexity in Drosophila. Nature 426: 849–853 [DOI] [PubMed] [Google Scholar]
- Crocker J, Tamori Y, Erives A (2008) Evolution acts on enhancer organization to fine-tune gradient threshold readouts. PLoS Biol 6: e263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denell R (2008) Establishment of tribolium as a genetic model system and its early contributions to evo-devo. Genetics 180: 1779–1786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driever W, Thoma G, Nüsslein-Volhard C (1989) Determination of spatial domains of zygotic gene expression in the Drosophila embryo by the affinity of binding sites for the bicoid morphogen. Nature 340: 363–367 [DOI] [PubMed] [Google Scholar]
- Erives A, Levine M (2004) Coordinate enhancers share common organizational features in the Drosophila genome. Proc Natl Acad Sci USA 101: 3851–3856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fomekong-Nanfack Y, Kaandorp JA, Blom J (2007) Efficient parameter estimation for spatio-temporal models of pattern formation: case study of Drosophila melanogaster. Bioinformatics 23: 3356–3363 [DOI] [PubMed] [Google Scholar]
- Gao Q, Wang Y, Finkelstein R (1996) Orthodenticle regulation during embryonic head development in Drosophila. Mech Dev 56: 3–15 [DOI] [PubMed] [Google Scholar]
- Gray S, Szymanski P, Levine M (1994) Short-range repression permits multiple enhancers to function autonomously within a complex promoter. Genes Dev 8: 1829–1838 [DOI] [PubMed] [Google Scholar]
- Hare EE, Peterson BK, Iyer VN, Meier R, Eisen MB (2008) Sepsid even-skipped enhancers are functionally conserved in Drosophila despite lack of sequence conservation. PLoS Genet 4: e1000106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewitt GF, Strunk BS, Margulies C, Priputin T, Wang XD, Amey R, Pabst BA, Kosman D, Reinitz J, Arnosti DN (1999) Transcriptional repression by the Drosophila giant protein: cis element positioning provides an alternative means of interpreting an effector gradient. Development 126: 1201–1210 [DOI] [PubMed] [Google Scholar]
- Ip YT, Park RE, Kosman D, Bier E, Levine M (1992) The dorsal gradient morphogen regulates stripes of rhomboid expression in the presumptive neuroectoderm of the Drosophila embryo. Genes Dev 6: 1728–1739 [DOI] [PubMed] [Google Scholar]
- Jaeger J, Surkova S, Blagov M, Janssens H, Kosman D, Kozlov KN, Manu, Myasnikova E, Vanario-Alonso CE, Samsonova M, Sharp DH, Reinitz J (2004) Dynamic control of positional information in the early Drosophila blastoderm. Nature 430: 368–371 [DOI] [PubMed] [Google Scholar]
- Janssens H, Hou S, Jaeger J, Kim AR, Myasnikova E, Sharp D, Reinitz J (2006) Quantitative and predictive model of transcriptional control of the Drosophila melanogaster even-skipped gene. Nat Genet 38: 1159–1165 [DOI] [PubMed] [Google Scholar]
- Kulkarni MM, Arnosti DN (2003) Information display by transcriptional enhancers. Development 130: 6569–6575 [DOI] [PubMed] [Google Scholar]
- Kulkarni MM, Arnosti DN (2005) Cis-regulatory logic of short-range transcriptional repression in Drosophila. Mol Cell Biol 9: 3411–3420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig MZ, Kreitman M (1995) Evolutionary dynamics of the enhancer region of even-skipped in Drosophila. Mol Biol Evol 12: 1002–1011 [DOI] [PubMed] [Google Scholar]
- Makeev VJ, Lifanov AP, Nazina AG, Papatsenko DA (2003) Distance preferences in the arrangement of binding motifs and hierarchical levels in organization of transcription regulatory information. Nucleic Acids Res 31: 6016–6026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayo AE, Setty Y, Shavit S, Zaslaver A, Alon U (2006) Plasticity of the cis-regulatory input function of a gene. PLoS Biol 4: e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochoa-Espinosa A, Yu D, Tsirigos A, Struffi P, Small S (2009) Anterior-posterior positional information in the absence of a strong Bicoid gradient. Proc Natl Acad Sci USA 106: 3823–3828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papatsenko D, Levine M (2007) A rationale for the enhanceosome and other evolutionarily constrained enhancers. Curr Biol 17: 955–957 [DOI] [PubMed] [Google Scholar]
- Pizarro J, Guerrero E, Galindo PL (2000) A statistical model selection strategy applied to neural networks. Proc ESANN 2000: 55–60 [Google Scholar]
- Runarsson TP, Yao X (2005) Search biases in constrained evolutionary optimization. IEEE Trans Syst Man Cybern C 35: 233–243 [Google Scholar]
- Sánchez L, Thieffry D (2001) A logical analysis of the Drosophila gap-gene system. J Theor Biol 211: 115–141 [DOI] [PubMed] [Google Scholar]
- Schroeder MD, Pearce M, Fak J, Fan H, Unnerstall U, Emberly E, Rajewsky N, Siggia ED, Gaul U (2004) Transcriptional control in the segmentation gene network of Drosophila. PLoS Biol 2: e271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal E, Raveh-Sadka T, Schroeder M, Unnerstall U, Gaul U (2008) Predicting expression patterns from regulatory sequence in Drosophila segmentation. Nature 451: 535–540 [DOI] [PubMed] [Google Scholar]
- Setty Y, Mayo AE, Surette MG, Alon U (2003) Detailed map of a cis-regulatory input function. Proc Natl Acad Sci USA 100: 7702–7707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shea MA, Ackers GK (1985) The OR control system of bacteriophage lambda. A physical-chemical model for gene regulation. J Mol Biol 181: 211–230 [DOI] [PubMed] [Google Scholar]
- Small S, Arnosti DN, Levine M (1993) Spacing ensures autonomous expression of different stripe enhancers in the even-skipped promoter. Development 119: 762–772 [PubMed] [Google Scholar]
- Stormo GD (2000) DNA binding sites: representation and discovery. Bioinformatics 16: 16–23 [DOI] [PubMed] [Google Scholar]
- Szymanski P, Levine M (1995) Multiple modes of dorsal-bHLH transcriptional synergy in the Drosophila embryo. EMBO J 14: 2229–2238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilar JM, Leibler S (2003) DNA looping and physical constraints on transcription regulation. J Mol Biol 331: 981–989 [DOI] [PubMed] [Google Scholar]
- Von Hippel PH, Revzin A, Gross CA, Wang AC (1974) Non-specific DNA binding of genome regulating proteins as a biological control mechanism: 1. The lac operon: equilibrium aspects. Proc Natl Acad Sci USA 71: 4808–4812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wharton KA Jr, Crews ST (1993) CNS midline enhancers of the Drosophila slit and Toll genes. Mech Dev 40: 141–154 [DOI] [PubMed] [Google Scholar]
- Zinzen RP, Senger K, Levine M, Papatsenko D (2006) Computational models for neurogenic gene expression in the Drosophila embryo. Curr Biol 16: 1358–1365 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary information, Supplementary figures S1–6, Supplementary tables SI–V
dataset