Abstract
Neuronal nicotinic acetylcholine receptors (nAChRs)1 are pentamers composed of α and β subunits. Different molecular compositions of these subunits constitute various receptor subtypes that are implicated in the pathophysiology and/ or treatment of several disease states, but are difficult to distinguish among pharmacologically. α-Conotoxins are a group of small, structurally defined peptides that may be used to molecularly dissect the nAChR binding site. Heteromeric nAChRs generally contain either a β2 or β4 subunit in addition to an α subunit at the ligand-binding interface. α-Conotoxin BuIA kinetically distinguishes between β2- and β4-containing nAChRs with long off-times for the latter. Mutational studies were used to assess the influence of residues that line the putative acetylcholine- binding pocket, but differ between β2 and β4 subunits. Residues Thr/Lys59, Val/Ile111 and Phe/Gln119 of the respective β2- and β4 subunits are critical to off-rate differences. Among these residues, Thr59 of nAChR β2 may interfere with effective access to the binding site whereas Lys59 may facilitate this binding.
Nicotinic acetylcholine receptors (nAChRs) are involved in numerous physiological CNS functions including learning and memory, reward, analgesia and motor control. These nAChRs are present not only postsynaptically, but also on presynaptic and preterminal sites where stimulation activates the synaptic release of neurotransmitters including dopamine, norepinephrine, serotonin, glutamate and γ-aminobutyric acid (1). The ability to modulate the release of these key neurotransmitters has led to proposals that nicotinic drugs are potential novel therapeutics for the treatment of a broad array of maladies including cognitive dysfunction, addiction, pain and Parkinson’s disease as well as psychotic, mood and anxiety disorders; for review see (2).
nAChRs are pentamers made up of α and β subunits; different combinations of these subunits constitute subtypes of receptors that have discrete anatomical distributions. To date, all neuronal nAChRs that contain an α2, α3, α4, or α6 subunit also contain and require a β2 or β4 subunit in order to function. The presence of either the β2 and/or β4 subunit influences pharmacological properties of the nAChR such as agonist efficacy, desensitization kinetics, and Ca++ permeability (3). Competitive nicotinic ligands generally bind to both the α and β subunit that together form a ligand-binding interface. There is considerable conservation of residues that form the ligand binding sites of nAChR subtypes, contributing to the challenge of designing selective drugs (4). Further complicating matters is the fact that the nAChR fluctuates among different states including (at least) resting, active and desensitized (5). A practical consequence of this is that binding selectivity may not translate into functional selectivity. For instance, the antagonist A-186253 has 200,000-fold binding selectivity for α4β2 vs. α7 nAChRs, yet shows only 20-fold selectivity with respect to functional block of these receptor subtypes (6). Rational design of functionally selective ligands must, therefore, take into account the allosteric nature of nAChR function.
Conotoxins are a large family of peptide ligands from carnivorous mollusks of the genus Conus. Cocktails of these peptides are used to envenomate fish and other prey. Identified macromolecular targets include a broad array of ligand- and voltage-gated ion channels and G-protein coupled receptors (7) (8). Due to their pharmacological specificity some of these peptides are being developed as medications (9, 10). ω-Conotoxin MVIIA (ziconotide) is a potent analgesic that blocks N-type calcium channels and is in current clinical use (11). Contulakin-G (CGX-1160) is a neurotensin type I receptor agonist in phase I human clinical trials for spinal cord injury pain (12). Xen2174, an analog of MrIA, is an allosteric blocker of the norepinephrine transporter in Phase I clinical trials for treatment of cancer pain (13). α-Conotoxin Vc1.1 is a nAChR antagonist in Phase I human clinical trials for neuropathic pain (14, 15). Thus, understanding the mechanism by which conotoxins achieve their target specificity is a priority.
α-Conotoxins are two-disulfide bridge small peptides that target to and are widely used as structural probes of nAChRs (16). The crystal structure of the Lymnae acetylcholine binding protein bound to α-conotoxin PnIA (A10L D14K), a blocker of α7 nAChRs, was solved and indicates that this α-conotoxin binds with high affinity to a structure homologous to the resting state of the nAChR (17). Recently, a novel peptide with unique selectivity features was identified from Conus bullatus (18). In this report, we have investigated nAChR β subunit residues present at the ligand-binding interface that profoundly affect the kinetics of block by this ligand known as α-conotoxin BuIA. Among these, Lys59 may allow more effective access to the high-affinity binding site.
Materials and Methods
Chemical synthesis
α-Conotoxin BuIA was synthesized on an Fmoc amide resin using Fmoc chemistry and standard side protection except on cysteine residues. Cys residues were protected in pairs with either S-trityl on Cys1 and Cys3 (the first and third Cys), or S-acetamidomethyl on Cys2 and Cys4. The peptide was removed from the resin and precipitated. A two-step oxidation protocol was used to selectively fold the peptides as described previously (18).
Mutagenesis of Receptors
The notation used for point mutants is to list the naturally occurring residue, followed by its position, followed by the change made. For example, β2T59G is a β2 subunit with the 59 position threonine replaced by a glycine. Point mutants were constructed using the QuikChange Site-Directed Mutagenesis Kit (Stratagene). The mutant receptors were in either the pGEMHE vector (Liman et al., 1992) or the pSP65 vector (Promega). All PCR mutations were confirmed by sequencing.
Electrophysiology
Oocytes were harvested and injected with cRNA encoding nAChR subunits as described previously (19). All clones were from rat. For off-rate kinetics, a 30 µl cylindrical oocyte recording chamber fabricated from Sylgard was gravity-perfused with ND96A (96.0 mM NaCl, 2.0 mM KCl, 1.8 mM CaCl2, 1.0 mM MgCl2, 1 µM atropine, 5 mM HEPES, pH 7.1–7.5) at a rate of ~2 ml/min. All toxin solutions also contained 0.1 mg/ml bovine serum albumin to reduce nonspecific adsorption of peptide. ACh-gated currents were obtained with a two-electrode voltage clamp amplifier (model OC-725B, Warner Instrument, Hamden, CT), and data captured as previously described (20). The membrane potential of the oocytes was clamped at −70 mV. To apply a pulse of ACh to the oocyte, the perfusion fluid was switched to one containing ACh for 1 sec. This was automatically done at intervals of 1 min. The ACh was diluted in ND96A. For control responses, the ACh pulse was preceded by perfusion with ND96A. The concentration of ACh was 100 µM. Toxin was bath-applied for 5 min, followed by a pulse of ACh. The volume of entering ACh is such that the toxin concentration remains at a level >50% of that originally in the bath until the ACh response has peaked (<2 sec). Thereafter, toxin was washed away and subsequent ACh pulses were given every 1 min, unless otherwise indicated. All ACh pulses contain no toxin, for it was assumed that little if any bound toxin washed away in the brief time (less than 2 s it takes for the responses to peak). In our recording chamber, the bolus of ACh does not project directly at the oocyte but rather enters tangentially, swirls and mixes with the bath solution.
For determination of toxin on-rate and when longer than 5 min of toxin application was needed to reach maximum block, toxin was applied by continuous perfusion to the oocytes as previously described (21). When the on-rate was rapid, ACh was applied every one min, but 15 sec time intervals were determined by staggering the start time of these one min intervals in 15 sec increments and then combining the data. The average peak amplitude of three control responses just preceding exposure to toxin was used to normalize the amplitude of each test response to obtain a “% response” or “% block.” Each data point of a dose-response curve represents the average value ± SE of measurements from at least three oocytes.
Results
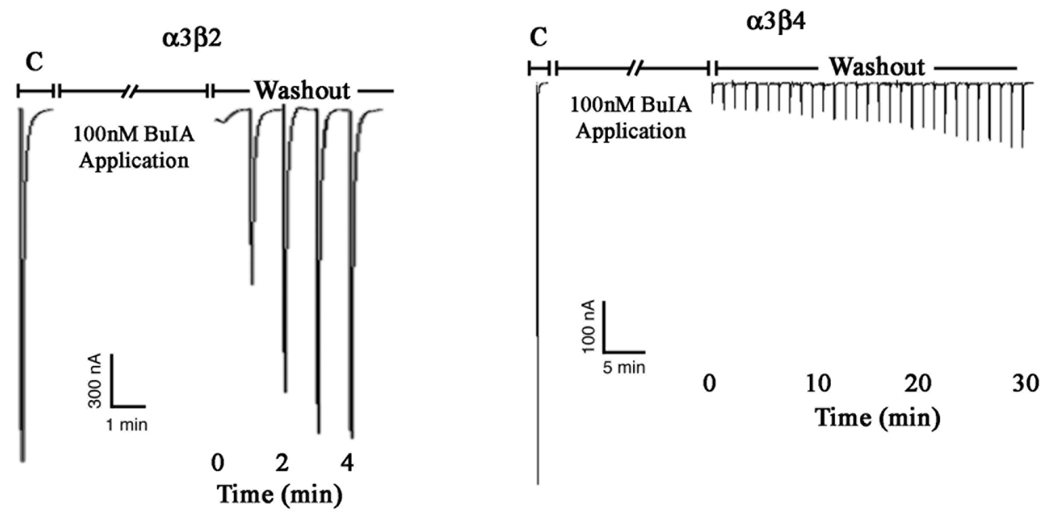
α-Conotoxin BuIA blocks both β2*1 and β4* nAChRs. However, the off-rate from α×β4* nAChRs is much slower than that of α×β2 nAChRs (18). An example of this difference for rat α3β2 vs. α3β4 nAChRs is shown in Fig. 1.
Fig. 1.
α-conotoxin BuIA has slow washout kinetics for α3β4 vs. α3β2 nAChRs. Toxin (100 nM) was applied to oocytes expressing α3β2 nAChRs (panel A) and α3β4 nAChRs (Panel B). After a 5 min. incubation, toxin was washed out and responses to a 1 sec pulse of ACh were measured every 1 min. C, control response to ACh prior to the addition of toxin.
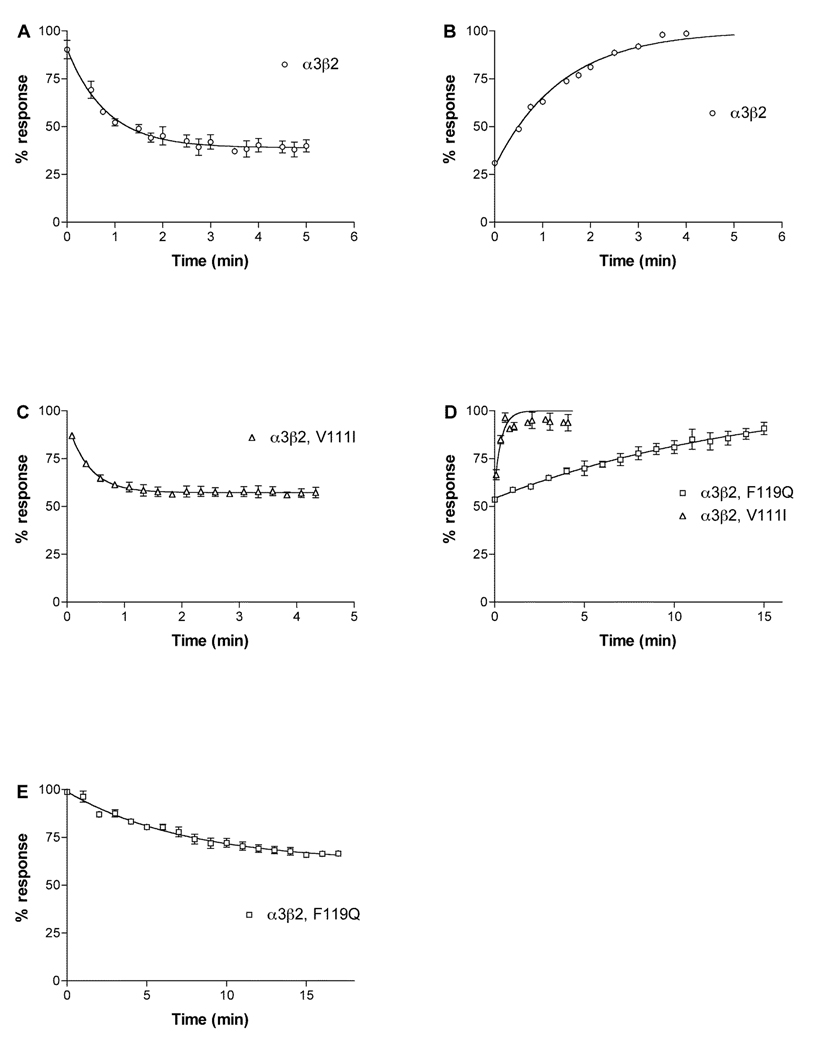
Data from X-ray crystallography, receptor labeling, mutagenesis and receptor modeling based on the molluscan acetylcholine binding protein indicate that the acetylcholine binding pocket is constituted by a hydrophobic cage of conserved aromatic residues from both the α and β subunits in proximity to the two disulfide linked vicinal cysteines in loop C of the α subunit (3, 5). The conservation of these residues among nAChR subtypes restricts the ability of ligands to discriminate among these subtypes. However, non-conserved residues appear to line the binding pocket and could allow for ligand subtype specificity (17, 22). Three residues in β2, Val111 Phe119 and Leu121 form a hydrophobic partial rim around the binding pocket. Val111 is replace by Ile and Phe119 is replaced by Gln in β4. We therefore tested these residues to assess their role in the kinetics of block by α-conotoxin BuIA. Substitution of Val with Ile (β2 Val111Ile) led to an increase in koff (Fig. 2 and Table 1). Due to the fast kinetics, measuring kobs and koff was difficult. To ensure that we had obtained reliable measurements we used a second method to determine kinetic constants. kobs was plotted vs. toxin concentration according to the equation kobs = kon (F) + koff, where F is the free toxin concentration, the slope is kon and the y-intercept is koff . Using this method, we found kon to be 5.995 × 107 min−1M−1 and koff to be 2.446 min−1 (data not shown), giving a ki of 4.08 × 10−8 M in good agreement with values determined by the methods shown in Fig. 2 and detailed in Table 1. This ki value is 4.5-fold higher than the IC50, which may represent the error associated with calculating kon when kobs is approximately equal to koff. In contrast to the results withVal111Ile, substitution of the hydrophilic Gln for Phe (β2 Phe119Gln) led to a 7-fold decrease in koff, partially explaining the longer off-time of α-conotoxin BuIA for α3β2 vs. α3β4 nAChRs. The kon for β2 Phe119Gln was not significantly different than wildtype β2. Results for both mutations are shown in Table 1 and Fig. 2.
Fig. 2.
Kinetics of block of α-conotoxin BuIA on α3β2 and α3 with point mutations in β2. Note that β2 Val111Ile speeds recovery, whereas Phe119Gln slows recovery from block by α-conotoxin BuIA . A, 5 nM α-conotoxin BuIA applied to α3β2. B, Washout of α-conotoxin BuIA from α3β2. C, 6 nM α-conotoxin BuIA applied to α3β2Val111Ile. D, washout of α-conotoxin BuIA from α3β2Val111Ile and α3β2Phe119Gln. E, 500 pM α-conotoxin BuIA applied to α3β2Phe119Gln. Toxin was applied by perfusion to oocytes expressing the indicated nAChRs as described in Materials and Methods. Response to a 1 sec pulse of ACh was measured. Results are summarized in Table 1.
Table 1.
α-conotoxin BuIA kinetic constants for block of α3β2 nAChRs
| kon | koff | ki | IC50 | |
|---|---|---|---|---|
| min−1 M−1 | min−1 | M | M | |
| α3β2 | 1.12 (0.19–2.05) × 108 | 0.656 (0.483–0.829) | 5.85 × 10−9 | a5.72 × 10−9 |
| aα3β4 | 4.68 (3.06–6.30) × 107 | 0.0111 | 2.38 × 10−8 | 2.77 × 10−8 |
| α3β2,V111I | 5.21 (0–18.2) × 107 | 2.614 (1.58–3.64) | 4.91 × 10−8 | 8.98 (7.6–10.6) × 10−9 |
| α3β2,F119Q | 1.05 (0.864–1.24) × 108 | 0.0936 (0.0813–0.106) | 8.91 × 10−10 | 7.35 (5.92–9.13) × 10−10 |
| α3β2,T59D | 6.72 (2.19–11.3) × 107 | 0.0448 (0.0274–0.0622) | 6.65 × 10−10 | 5.66 (5.00–6.40) × 10−10 |
| α3β2,T59G | 1.17 (0.26–2.08) × 108 | 0.0581 (0.0489–0.0660) | 5.01 × 10−10 | 3.62 (2.15–6.08) × 10−10 |
| α3β2,T59K | 6.03 (5.34–6.72) × 107 | 0.0178 (0.0164–0.0192) | 2.95 × 10−10 | 2.43 (1.86–3.19) × 10−10 |
| α3β2,T59S | 2.83 (1.71–3.94) × 107 | 0.0878 (0.0636–0.112) | 3.11 × 10−10 | 4.93 (4.30–5.68) × 10−10 |
| α3β2,T59V | 1.61 (1.08–2.14) × 108 | 0.301 (0.259–0.343) | 1.87 × 10−9 | 1.08 (0.993–1.18) × 10−9 |
Numbers in parentheses are 95% confidence intervals
Values from (18).
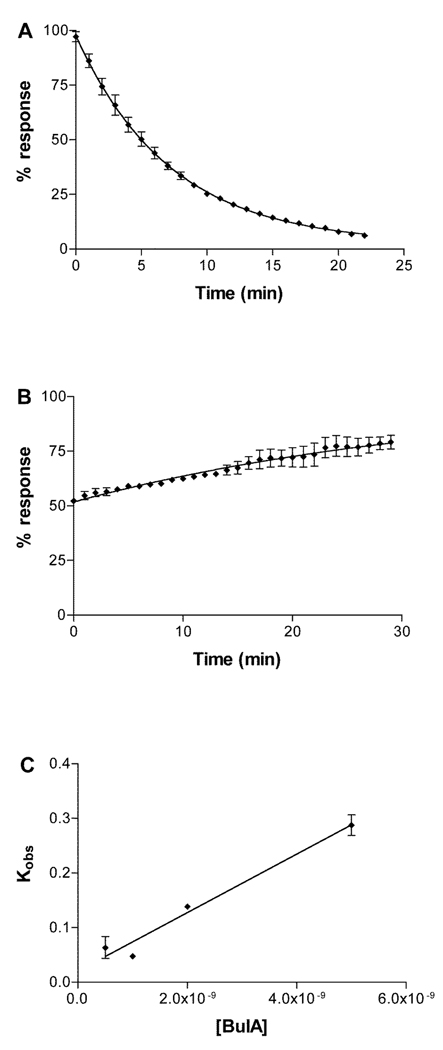
β2 Thr59 is located at the opposite edge of the binding pocket rim. This residue is replaced by a positively charged Lys in β4. Kinetics of unblock for this mutation were difficult to quantitate due to the very long on-and off times (Fig. 3) combined with limitations of oocyte recording duration and a tendency for the ACh response to drift over time. Kinetics were therefore also assessed by determining the kobs at 4 different toxin concentrations (Fig. 3C). In this instance, accuracy is hindered by the uncertainty of the y intercept where the error includes the ordinate. Despite these technical limitations, the ki values calculated by the two methods are in reasonable agreement, 2.95 × 10−10 vs. 3.81 × 10−10 M, respectively. The mutation β2 Thr59Lys had a large effect on decreasing the koff of α-conotoxin BuIA (36.9-fold) compared to a less than 2-fold decrease in kon.
Fig. 3.
Kinetics of block of α-conotoxin BuIA on α3β2Thr59Lys. A, 2 nM α-conotoxin BuIA perfusion applied to α3β2Thr59Lys. The response to ACh was measured every one min. B, Washout of α-conotoxin BuIA. C, Plot of the observed on-rate vs. toxin concentration. Toxin (500 pM, 1 nM, 2 nM and 5 nM) was applied by perfusion to oocytes expressing α3β2Thr59Lys as described in Materials and Methods. Response to a 1 sec pulse of ACh was measured. Kinetics constants were calculated according to the equation kobs = kon (F) + koff where F is the free toxin concentration and the y-intercept = koff. Using this equation, kon = 5.36 ± 0.67 × 107 min−1 M−1 and koff = 0.0204 ± 0.018 min−1.
α-Conotoxin MII is a peptide with an α 4/7 structure whose nAChR selectivity profile is functionally distinct from that of α-conotoxin BuIA (23). It was previously reported that the mutation β2 Thr59Lys decreased the affinity of α-conotoxin MII. We therefore examined what effects the Thr59Lys mutation had on the kinetics of binding of α-conotoxin MII. In contrast to α-conotoxin BuIA, the Thr59Lys affected kon, but had no effect on koff (Fig. 4 and Table 2).
Fig. 4.
Kinetics of block of α-conotoxin MII on α3β2Thr59Lys. Note that the Thr59Lys mutation affects kon, but not koff. A, 10 nM α-conotoxin MII applied to α3β2. B, Washout of α-conotoxin MII from α3β2 nAChRs. C, 10 nM α-conotoxin MII applied to α3β2Thr59Lys. D, Washout of α-conotoxin MII from α3β2Thr59Lys. Toxin was applied by perfusion to oocytes expressing α3β2Thr59Lys as described in Materials and Methods. Response to a 1 sec pulse of ACh was measured. Results are summarized in Table 2.
Table 2.
α-conotoxin MII kinetic constants for block of α3β2 nAChRs
| kon | koff | ki | |
|---|---|---|---|
| min−1 M−1 | min−1 | M | |
| α3β2 | 0.460 (0.378–0.542) × 107 | 0.106 (0.0565–0.156) | 2.32 × 10−9 |
| α3β2,T59K | 0.180 (0.130–0.232) × 107 | 0.102 (0.0847–0.119) | 5.65 × 10−9 |
Numbers in parentheses are 95% confidence intervals.
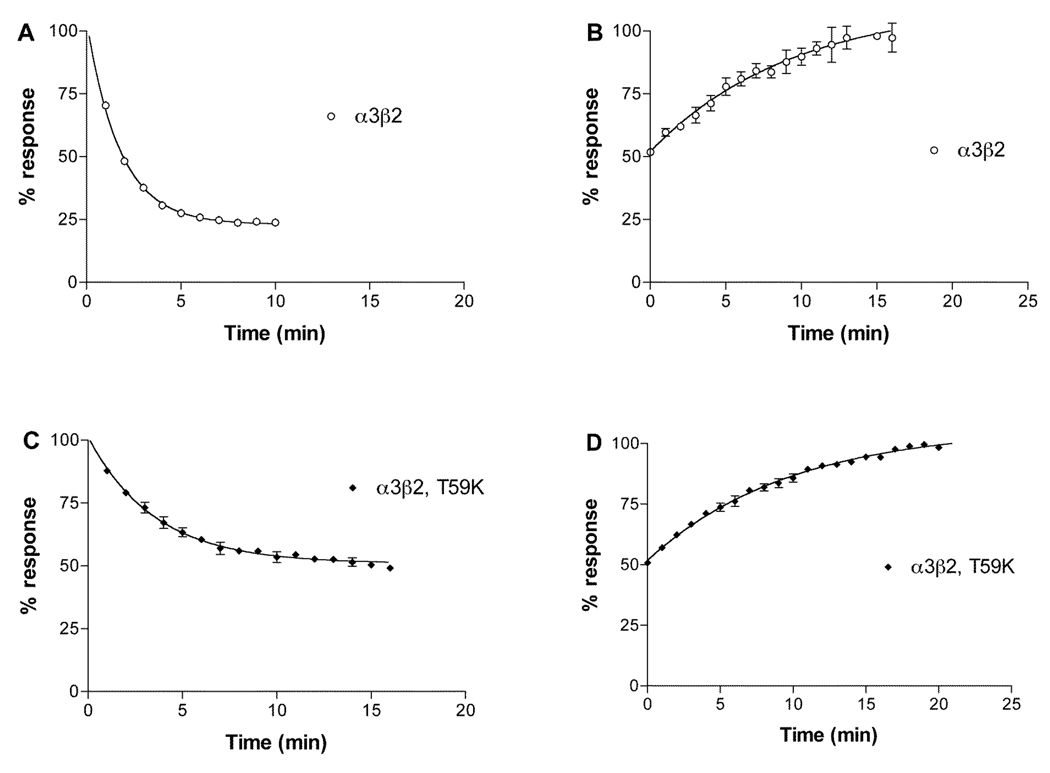
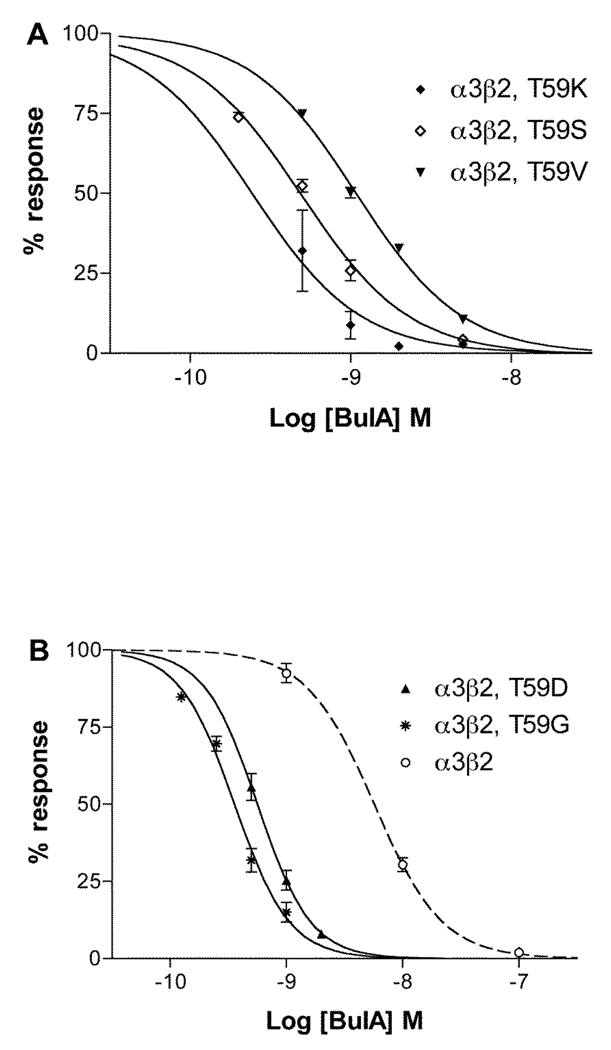
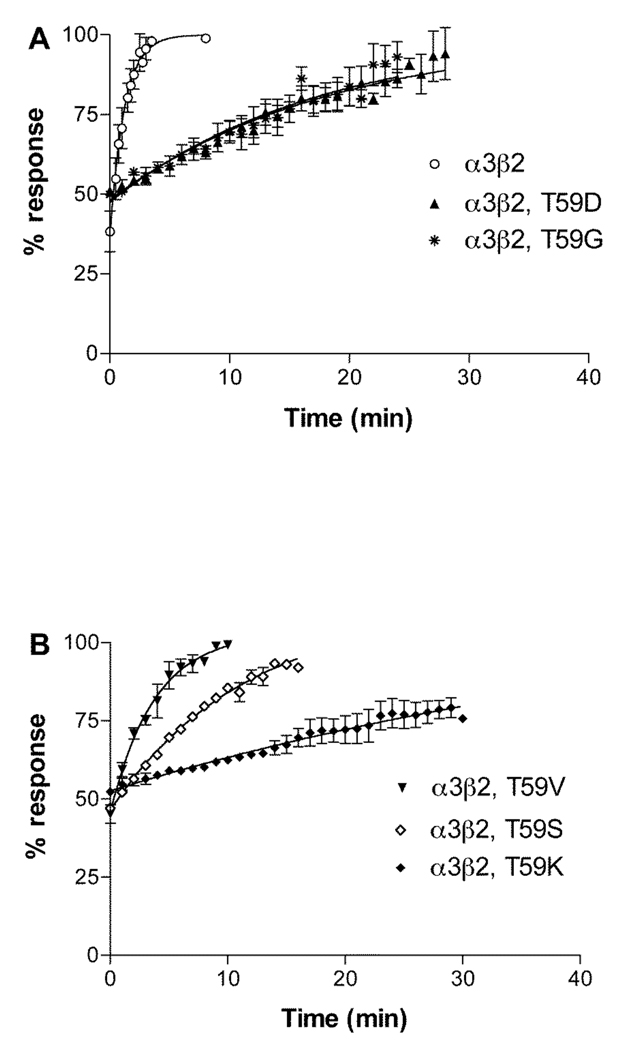
Since the mutation β2 Thr59Lys had the most profound effect on α-conotoxin BuIA off-rates, we further investigated the role of position 59 in leading to the substantial difference in koff for α-conotoxin BuIA by creating additional position 59 point mutations in the β2 subunit. Concentration response analysis of α-conotoxin BuIA on the various point mutants in shown in Fig. 5 and Table 3 and data on toxin kinetics is shown in Fig. 6 and Table 3. Thr has an aliphatic hydroxyl side chain. Non-conservative substitution of Thr59 by Gly, which has only a hydrogen atom side chain, surprisingly also led to a large decrease in the α-conotoxin BuIA koff (11.3-fold) and 15.1-fold decrease in Ki. Similarly, substitution of β2 Thr59 with a negatively charged residue, Asp, also led to a large decrease (14.6-fold) in α-conotoxin BuIA koff as well as an 11.4-fold decrease in Ki. Thus, non-conservative substitution of Thr by Lys, Gly, or Asp leads to an increase in affinity in each instance. These results suggest that the primary effect of Thr59 in the β2 subunit is to interfere with α-conotoxin BuIA binding. Such a result might be explained by steric hindrance by Thr but not Lys, Gly or Asp. Thr is the only residue in this set that has a side chain that is branched at the β carbon. We reasoned this branching could be a source of steric hindrance to toxin binding and if so, that replacement of Thr with Val (an aliphatic hydrophobic residue, branched at the β carbon) would, like Thr, also hinder binding. Indeed this is the case; substitution of β2 Thr59 by Val caused only a relatively modest change (2.18-fold decrease) in koff compared to Thr. In contrast, Ser normally acts as a conservative substitution for Thr. Both Ser and Thr have polar –OH groups in their side chain and differ by only a single methyl group. However, unlike Thr, Ser is not branched at the β-carbon. The results indicate that replacement of Thr with Ser, leads to a larger change (7.46-fold decrease) in koff. It will be of future interest to compare receptor position 59 Leu vs. Ile to further examine the effects of branching at the β-carbon.
Fig. 5.
Concentration response analysis of α-conotoxin BuIA on α3β2 nAChRs with point mutations in position 59 of β2. Toxin was applied by perfusion to oocytes expressing nAChRs as described in Materials and Methods. Results are summarized in Table 1. Dashed line shows wildtype α3β2 data from (18).
Table 3.
Effect of β2 Thr59Lys
Fig. 6.
Recovery from block by α-conotoxin BuIA of α3β2 nAChRs with point mutations in position 59 of β2. Response to a 1 sec pulse of ACh was measured was measured every one min after toxin washout. Results are summarized in Table 1.
Discussion
In this study we have investigated the kinetics of functional block of nAChRs by α-conotoxin BuIA. These kinetics are dependent on the β nAChR subunit; the β4 subunit but not the β2 subunit is associated with slow recovery from toxin block. Residues 59 and 119 of the rat β subunit are shown to be key determinants of this aspect of toxin action.
Two hydrophobic residues that line the β2 subunit-binding cleft were initially examined. Phe119 in β2 is Gln in β4 and Val111in β2 is Ile in β4. The effects of these two residue changes are opposite. When Phe119 of β2 is replaced by Gln, there is a decrease in α-conotoxin BuIA koff leading to a 6.6-fold increase in toxin affinity. When β2 V111 is replaced by Ile, there is both an increase in koff as well as a decrease in kon leading to an 8.3-fold decrease in toxin affinity. The largest change occurs when β2 Thr59 is substituted with Lys that is in the homologous position of β4. There is a 37-fold decrease in koff partially offset by a 1.9-fold decrease in kon producing a 19.8-fold increase in affinity.
Thr59 has been documented as a positive determinant of high affinity binding for certain competitive antagonists of nAChRs. Changing β2Thr59 to Lys as occurs in β4 caused a 9-fold decrease in sensitivity to dihydro-β-erythroidine and a 71-fold decrease in sensitivity to k-bungarotoxin (24) (see Table 3). This change in the nAChR also lead to a small (2- to 4- fold) decrease in sensitivity of block by the 4/7 α-conotoxin MII (25) and this report. In the present work we show that this decrease for α-conotoxin MII is due to a decrease in the binding on-rate of MII. Significantly, the situation for BuIA is opposite. That is, a change from Thr59 to Lys59 leads to a 20-fold increase in sensitivity to toxin block, due to a substantial decrease in koff.
Thus, the decrease in koff of α-conotoxin BuIA for α3β4 compared to α3β2 nAChRs can be largely explained by the decrease in koff seen in the β2 Thr59Lys and Phe119Gln mutations, partially offset by the increase in koff observed in the Val111Ile mutation. However, given that residue differences can cause either increases or decreases in the α-conotoxin koff, the present data do not exclude the contribution of other subunit residues to the long α-conotoxin off-times observed for β4-containing nAChRs. The slow recovery from α-conotoxin BuIA block has been demonstrated for α3β4 vs. α3β2 nAChR subtypes in different species including rat, mouse and human (18). Position 59 and 111 differences between β2 and β4 subunits are conserved among rat, mouse and human. In position 119, the β2 residues are conserved, whereas β4 is Gln in rat and mouse but is Leu in human (Fig. 7).
Fig. 7.
Alignment of the N-terminal extracellular domains (preceding transmembrane I) of β2 and β4 nAChR subunits. Residue positions (59, 111 and 119) tested in this study are indicated with dots. Bars indicate the agonist binding domain loops D, E and F. The locations of the loops are taken from (5).
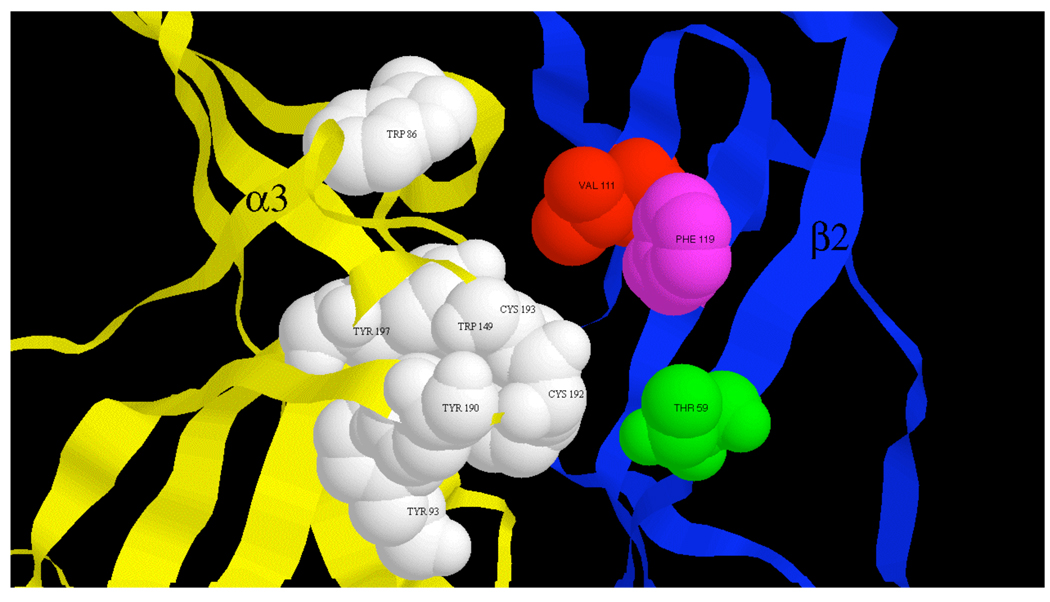
Cryoelectron microscopy images of the Torpedo nAChR have led to near atomic scale resolution of this muscle nAChR (26, 27) and the molluscan acetylcholine binding protein has provided a valuable crystal structure for homology modeling of neuronal nAChRs (28). Modeling of the α3β2 nAChR based on this structure (29) shows β2 Thr59 at the edge of a cleft believed to be the acetylcholine binding pocket (Fig. 8). The present results are consistent with α-conotoxin BuIA acting as an antagonist by preventing access of acetylcholine to this binding pocket. The results further suggest, that Lys59, (present in the homologous position of the β4 subunit) may allow better access of the toxin to the binding cavity. Such increased access may lead to more favorable interaction with receptor residues. Although more favorable interaction could come by either increased kon and/or decreased koff, in the case of α-conotoxin BuIA binding, the improvement comes almost entirely from the latter. Thus, residue 59 appears to be situated at a critical location on the β-portion of the α/β nAChR subunit interface. Previously studied toxins, including alkaloids (dihydro-β-erythroidine from the seeds of the trees an shrubs of the genus Erythrina), proteins (κ-bungarotoxin from the Tawainese banded krait, Bungarus multicinctus) and peptides (α-conotoxin MII) from the marine snail Conus may bind to β2 Thr59 and replacement of Thr59 with Lys decreases affinity for these ligands. In contrast, replacement of Thr59 with residues with widely varying physicochemical properties in each instance leads to an increase in affinity for α-conotoxin BuIA.
Fig. 8.
α3β2 nAChR binding site. The ACh binding site at an α3/β2 interface is shown within a model of the α3β2 extracellular domain. Proposed α3 subunit agonist binding residues (33) shown in white space filling. Residues affecting α-conotoxin BuIA binding kinetic differences between β2 and β4 subunits shown in color space filling.
The α-conotoxins are two disulfide loop peptides that may be structurally grouped according to loop size (30). Docking simulations of the 4/7 family of α-conotoxins (4 residues in loop 1 and 7 residues in loop 2) suggest that these toxins bind at a cleft located at one entrance to the acetylcholine binding site and above the β9/β10 hairpin (22, 31). The recent crystal structure of a 4/7 α-conotoxin analog bound to the Lymnae acetylcholine binding protein is consistent with this binding mode (17). Residues Thr59, Val111, Phe119 and Leu121 line this cleft (Fig. 8). Only residue, Leu121, is conserved between the β2 and β4 nAChR subunits; Leu121 is also found in homologous position in δ, γ, and ε subunits as well as the (-) face of the α7 subunit. Thus, Thr59, Val111, Phe119, but not Leu121, represent potential residues that may be exploited to achieve discrimination between β2-containing and β4-containing nAChRs.
Acknowledgments
We thank Greg Bulaj for helpful discussions.
Footnotes
This work was supported by the National Institutes of Health Grants MH53631 (JMM) and DA08102 (CWL). D.S. and RR also received partial support from the University of Utah Bioscience Undergraduate Research Program.
Abbreviations: nAChR, nicotinic acetylcholine receptor; ACh, acetylcholine, CI, 95% confidence interval.
References
- 1.Wonnacott S. Presynaptic nicotinic ACh receptors. Trends Neurosci. 1997;20:92–98. doi: 10.1016/s0166-2236(96)10073-4. [DOI] [PubMed] [Google Scholar]
- 2.Gotti C, Clementi F. Neuronal nicotinic receptors: from structure to pathology. Prog. Neurobiol. 2004;74:363–396. doi: 10.1016/j.pneurobio.2004.09.006. [DOI] [PubMed] [Google Scholar]
- 3.Jensen AA, Frolund B, Liljefors T, Krogsgaard-Larsen P. Neuronal nicotinic acetylcholine receptors: structural revelations, target identifications, and therapeutic inspirations. J Med Chem. 2005;48:4705–4745. doi: 10.1021/jm040219e. [DOI] [PubMed] [Google Scholar]
- 4.Gotti C, Riganti L, Vailati S, Clementi F. Brain neuronal nicotinic receptors as new targets for drug discovery. Curr Pharm Des. 2006;12:407–428. doi: 10.2174/138161206775474486. [DOI] [PubMed] [Google Scholar]
- 5.Corringer PJ, Le Novere N, Changeux J-P. Nicotinic receptors at the amino acid level. Annu. Rev. Pharmacol. Toxicol. 2000;40:431–458. doi: 10.1146/annurev.pharmtox.40.1.431. [DOI] [PubMed] [Google Scholar]
- 6.Itier V, Schonbachler R, Tribollet E, Honer M, Prinz K, Marguerat A, Bertrand S, Bunnelle WH, Schubiger PA, Meyer MD, Sullivan JP, Bertrand D, Westera G. A-186253, a specific antagonist of the alpha 4 beta 2 nAChRs: its properties and potential to study brain nicotinic acetylcholine receptors. Neuropharmacology. 2004;47:538–557. doi: 10.1016/j.neuropharm.2004.05.006. [DOI] [PubMed] [Google Scholar]
- 7.Terlau H, Olivera BM. Conus venoms: a rich source of novel ion channel-targeted peptides. Physiol. Rev. 2004;84:41–68. doi: 10.1152/physrev.00020.2003. [DOI] [PubMed] [Google Scholar]
- 8.Lewis RJ. Conotoxins as selective inhibitors of neuronal ion channels, receptors and transporters. IUBMB Life. 2004;56:89–93. doi: 10.1080/15216540410001668055. [DOI] [PubMed] [Google Scholar]
- 9.Livett BG, Gayler KR, Khalil Z. Drugs from the sea: conopeptides as potential therapeutics. Curr. Med. Chem. 2004;11:1715–1723. doi: 10.2174/0929867043364928. [DOI] [PubMed] [Google Scholar]
- 10.Layer RT, McIntosh JM. Conotoxins: Therapeutic Potential and Application. Marine Drugs. 2006;4:119–142. [Google Scholar]
- 11.Miljanich GP. Ziconotide: neuronal calcium channel blocker for treating severe chronic pain. Curr. Med. Chem. 2004;11:3029–3040. doi: 10.2174/0929867043363884. [DOI] [PubMed] [Google Scholar]
- 12.Craig AG, Norberg T, Griffin D, Hoeger C, Akhtar M, Schmidt K, Low W, Dykert J, Richelson E, Navarro V, Macella J, Watkins M, Hillyard D, Imperial J, Cruz LJ, Olivera BM. Contulakin-G, an O-glycosylated invertebrate neurotensin. J. Biol. Chem. 1999;274:13752–13759. doi: 10.1074/jbc.274.20.13752. [DOI] [PubMed] [Google Scholar]
- 13.Sharpe IA, Palant E, Schroeder CI, Kaye DM, Adams DJ, Alewood PF, Lewis RJ. Inhibition of the norepinephrine transporter by the venom peptide chi-MrIA. Site of action, Na+ dependence, and structure-activity relationship. J Biol Chem. 2003;278:40317–40323. doi: 10.1074/jbc.M213030200. [DOI] [PubMed] [Google Scholar]
- 14.Lang PM, Burgstahler R, Haberberger RV, Sippel W, Grafe P. A conus peptide blocks nicotinic receptors of unmyelinated axons in human nerves. Neuroreport. 2005;16:479–483. doi: 10.1097/00001756-200504040-00012. [DOI] [PubMed] [Google Scholar]
- 15.Sandall DW, Satkunanathan N, Keays DA, Polidano MA, Liping X, Pham V, Down JG, Khalil Z, Livett BG, Gayler KR. A novel alpha-conotoxin identified by gene sequencing is active in suppressing the vascular response to selective stimulation of sensory nerves in vivo. Biochemistry. 2003;42:6904–6911. doi: 10.1021/bi034043e. [DOI] [PubMed] [Google Scholar]
- 16.Nicke A, Wonnacott S, Lewis RJ. Alpha-conotoxins as tools for the elucidation of structure and function of neuronal nicotinic acetylcholine receptor subtypes. Eur J Biochem. 2004;271:2305–2319. doi: 10.1111/j.1432-1033.2004.04145.x. [DOI] [PubMed] [Google Scholar]
- 17.Celie PH, Kasheverov IE, Mordvintsev DY, Hogg RC, van Nierop P, van Elk R, van Rossum-Fikkert SE, Zhmak MN, Bertrand D, Tsetlin V, Sixma TK, Smit AB. Crystal structure of nicotinic acetylcholine receptor homolog AChBP in complex with an alpha-conotoxin PnIA variant. Nat Struct Mol Biol. 2005;12:582–588. doi: 10.1038/nsmb951. [DOI] [PubMed] [Google Scholar]
- 18.Azam L, Dowell C, Watkins M, Stitzel JA, Olivera BM, McIntosh JM. Alpha-conotoxin BuIA, a novel peptide from Conus bullatus, distinguishes among neuronal nicotinic acetylcholine receptors. J Biol Chem. 2005;280:80–87. doi: 10.1074/jbc.M406281200. [DOI] [PubMed] [Google Scholar]
- 19.Cartier GE, Yoshikami D, Gray WR, Luo S, Olivera BM, McIntosh JM. A new α-conotoxin which targets α3β2 nicotinic acetylcholine receptors. J. Biol. Chem. 1996;271:7522–7528. doi: 10.1074/jbc.271.13.7522. [DOI] [PubMed] [Google Scholar]
- 20.Luo S, Kulak JM, Cartier GE, Jacobsen RB, Yoshikami D, Olivera BM, McIntosh JM. α-Conotoxin AuIB selectively blocks α3β4 nicotinic acetylcholine receptors and nicotine-evoked norepinephrine release. J. Neurosci. 1998;18:8571–8579. doi: 10.1523/JNEUROSCI.18-21-08571.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Luo S, Nguyen TA, Cartier GE, Olivera BM, Yoshikami D, McIntosh JM. Single-residue alteration in α-conotoxin PnIA switches its nAChR subtype selectivity. Biochemistry. 1999;38:14542–14548. doi: 10.1021/bi991252j. [DOI] [PubMed] [Google Scholar]
- 22.Dutertre S, Lewis RJ. Computational approaches to understand alpha-conotoxin interactions at neuronal nicotinic receptors. Eur J Biochem. 2004;271:2327–2334. doi: 10.1111/j.1432-1033.2004.04147.x. [DOI] [PubMed] [Google Scholar]
- 23.McIntosh JM, Azam L, Staheli S, Dowell C, Lindstrom JM, Kuryatov A, Garrett JE, Marks MJ, Whiteaker P. Analogs of α-conotoxin MII are selective for α6-containing nicotinic acetylcholine receptors. Mol Pharmacol. 2004;65:944–952. doi: 10.1124/mol.65.4.944. [DOI] [PubMed] [Google Scholar]
- 24.Harvey SC, Maddox FN, Luetje CW. Multiple determinants of dihydro-b-erythroidine sensitivity on rat neuronal nicotinic receptor a subunits. J. Neurochem. 1996;67:1953–1959. doi: 10.1046/j.1471-4159.1996.67051953.x. [DOI] [PubMed] [Google Scholar]
- 25.Harvey SC, McIntosh JM, Cartier GE, Maddox FN, Luetje CW. Determinants of specificity for α-conotoxin MII on α3β2 neuronal nicotinic receptors. Mol. Pharmacol. 1997;51:336–342. doi: 10.1124/mol.51.2.336. [DOI] [PubMed] [Google Scholar]
- 26.Miyazawa A, Fujiyoshi Y, Stowell M, Unwin N. Nicotinic acetylcholine receptor at 4.6 A resolution: transverse tunnels in the channel wall. J Mol Biol. 1999;288:765–786. doi: 10.1006/jmbi.1999.2721. [DOI] [PubMed] [Google Scholar]
- 27.Unwin N, Miyazawa A, Li J, Fujiyoshi Y. Activation of the nicotinic acetylcholine receptor involves a switch in conformation of the alpha subunits. J Mol Biol. 2002;319:1165–1176. doi: 10.1016/S0022-2836(02)00381-9. [DOI] [PubMed] [Google Scholar]
- 28.Brejc K, van Dijk WJ, Klaassen RV, Schuurmans M, van Der Oost J, Smit AB, Sixma TK. Crystal structure of an ACh-binding protein reveals the ligand-binding domain of nicotinic receptors. Nature. 2001;411:269–276. doi: 10.1038/35077011. [DOI] [PubMed] [Google Scholar]
- 29.Everhart D, Reiller E, Mirzoian A, McIntosh JM, Malhotra A, Luetje CW. Identification of residues that confer α-conotoxin PIA sensitivity on the α3 subunit of neuronal nicotnic acetylcholine receptors. J. Pharmacol. Exp. Ther. 2003;306:665–670. doi: 10.1124/jpet.103.051656. [DOI] [PubMed] [Google Scholar]
- 30.McIntosh JM. Toxin antagonists of the neuronal nicotinic acetylcholine receptor. In: Clementi F, Fornasari D, Gotti C, editors. Handbook of Experimental Pharmacology. Volume 144. Neuronal Nicotinic Receptors. Berlin, Germany: Springer-Verlag; 2000. pp. 455–476. [Google Scholar]
- 31.Dutertre S, Nicke A, Lewis RJ. Beta2 subunit contribution to 4/7 alpha-conotoxin binding to the nicotinic acetylcholine receptor. J Biol Chem. 2005;280:30460–30468. doi: 10.1074/jbc.M504229200. [DOI] [PubMed] [Google Scholar]
- 32.Harvey SC, Luetje CW. Determinants of competitive antagonist sensitivity on neuronal nicotinic receptor β subunits. J. Neurosci. 1996;16:3798–3806. doi: 10.1523/JNEUROSCI.16-12-03798.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Costa V, Nistri A, Cavalli A, Carloni P. A structural model of agonist binding to the alpha3beta4 neuronal nicotinic receptor. Br J Pharmacol. 2003;140:921–931. doi: 10.1038/sj.bjp.0705498. [DOI] [PMC free article] [PubMed] [Google Scholar]