Abstract
Purpose
This study aims to investigate the role of aberrant expression of TKTL1 (Transkelolase-like 1) in Head and Neck Squamous Cell Carcinoma (HNSCC) tumorigenesis and to characterize TKTL1 contribution to HNSCC tumorigenesis via aerobic glycolysis and HIF1α stabilization.
Experimental design
TKTL1 promoter hypomethylation and mRNA/protein aberrant expression were studied in human HNSCC tumor samples and normal mucosas. Oncogenic functions of TKTL1 were examined in HNSCC cell line panels and tumor xenograft models with TKTL1 expression construct. The metabolite levels of fructose-6-phosphate, glyceraldehydes-3-phosphate, pyruvate, lactate, and the levels of HIF1α protein and its downsteam glycolytic targets were compared between the TKTL1-expressing and vehicle-expressing HNSCC cells. Meanwhile, the effects of HIF1α/glycolytic inhibitors were evaluated on the TKTL1 transfectants.
Results
TKTL1 exhibits high frequency of promoter hypomethylation in HNSCC tumors compared with the normal mucosas, correlating with its overexpression in HNSCC. Overexpression of TKTL1 in HNSCC cells promoted cellular proliferation and enhanced tumor growth in vitro and in vivo. Overexpression of TKTL1 increased the production of fructose-6-phosphate and glyceraldehyde-3-phosphate, in turn elevating the production of pyruvate and lactate, resulting in the normoxic stabilization of the malignancy-promoting transcription factor HIF1α and the upregulation of downstream glycolytic enzymes. Notably, reduction of TKTL1 expression decreased HIF1α accumulation and inhibition with HIF1α and/or glycolysis inhibitor could abrogate the growth effects mediated by TKTL1 overexpression.
Conclusion
TKTL1 is a novel candidate oncogene that is epigenetically activated by aberrant hypomethlation and contributes to a malignant phenotype via altered glycolytic metabolism and HIF1α accumulation.
Keywords: TKTL1, hypomethylation, Warburg effect, HIF1α
Introduction
There are more than 40,000 new cases of HNSCC in the United States each year, with a mortality rate of 12,000 annually. HNSCC develops through a prolonged multistage process involving accumulation of genetic and epigenetic alterations. Gene silencing by promoter hypermethylation of CpG islands has been linked to inactivation of tumor suppressor genes, such as DCC, p16, RASSF1 in HNSCC (1, 2). In contrast, less is known about the impact in tumor development or progression of DNA hypomethylation. DNA hypomethylation has recently been proposed to play a role in cancer gene expression activation and genomic instability (3). Rare examples of hypomethylation leading to activation of genes that are important in cancer have resulted in the overexpression of Bcl-2, R-ras, Cyclin D2 (4–6). In HNSCC as in other solid tumor, DNA global hypomethylation is found and is associated with smoking, alcohol consumption and stage (7). Using pharmacologic demethylation in normal oral keratinocyte cells combined with expression analysis in primary tumor tissues as a whole genome discovery approach, we recently identified a set of novel candidate oncogenes that undergo hypomethylation and increased expression in primary human HNSCC cancer, including TKTL1 (8).
As in most solid tumors, HNSCC displays dramatically altered glucose metabolism. Over 75 years ago, Warburg noted that cancer cells typically depend more on increased rates of glycolysis even in the presence of available oxygen, a phenomenon known as aerobic glycolysis or the Warburg effect (9). It is suggested that the Warburg effect confers a selective advantage for survival and proliferation of tumor cells. Recently, the relation between HIF1α and metabolic reprogramming has explored in HNSCC and other solid tumors. HIF1α participates in transcriptional activation of genes that are involved in crucial aspects of cancer biology including angiogenesis, cell survival, glucose metabolism, and invasion (10). It is proposed that HIF1α represents one principal mechanistic axis for regulation of aerobic glycolysis (11). Further, recent studies suggested that several intermediary glucose metabolites could promote HIF1α activation by interacting with the HIF prolyl hydroxylases (PHD 1–3). Expression of a mutant mitochondrial-ND2 mutation contributes to development of a HNSCC malignant phenotype via increased ROS, up-regulation of PDK2, attenuating PDH activity, elevating pyruvate and lactate production, and thereby HIF1α stabilization (12). Moreover, we found that inhibition of PDH activity via enhanced expression of PDK1, is associated with normoxic stabilization of HIF1α by glycolytic metabolites. Out study suggest the buildup of glycolytic metabolites, resulting from high PDK1 expression, may in turn promote HIF1α activation, thus sustaining a feed-forward loop for HNSCC malignant progression (13).
In this study we describe TKTL1, a gene encoding enzyme responsible for the transketolase reaction in pentose phosphate pathway (PPP) (14). The PPP, which facilitates glucose conversion for nucleic acid synthesis, glucose degradation to lactate, and regeneration of redox equivalents, connects with glycolysis via metabolites such as fructose-6-phosphate and glyceraldehyde-3-phosphate (15). In our preliminary study we have found that TKTL1 promoter demethylation and overexpression occurs in HNSCC (8). Here we further explore the functional significance of TKTL1 in HNSCC tumorigenesis, and demonstrate an oncogenic function of TKTL1 in promoting HNSCC tumor growth via enhancement of aerobic glycolysis and increased normoxic HIF1α expression, facilitating HNSCC tumor growth.
Materials and Methods
Tissue samples and cell lines
The tumor samples from patients with HNSCC and normal samples were obtained from patients surgically treated in the Department of Otolaryngology-Head and Neck Surgery at Johns Hopkins Medical Institutions, Baltimore using appropriate informed consent obtained after institutional review board approval. Microdissection of frozen tumor tissue was performed to assure that greater than 80% of tissue contained HNSCC. The normal tissues consisted of tissues obtained from patients that underwent uvulopalatopharyngoplasty from sleep apnea. UM22A, UM22B cells were provided by Ajay Verma. FaDu cells were from ATCC. Human HNSCC cell line JHU-O11, JHU-O22, and JHU-O28, were from the Johns Hopkins University Department of Otolaryngology–Head and Neck Surgery (16, 17).
Bisulfite treatment of DNA and QUMSP
Genomic DNA was extracted using the phenol chloroform, and bisulfate modification of genomic DNA was done as described (16). QUMSP was carried out as described previously (8). In our previous cohort, we have shown that the QUMSP method is highly correlated with bisulfite sequencing method in detection of TKTL1 hypomethylation.
Quantitative RTPCR
Real-time RTPCR was performed on ABI PRISM 7000 Sequence Detection System using the QuantiFast SYBR Green PCR kit (Qiagen). The primers used for real-time RTPCR were available upon request. The β-actin was used for normalization of the expression data of each gene. Each sample was run in triplicate to ensure quantitative accuracy, and the threshold cycle numbers (Ct) were averaged. The results were reported as fold changes and calculated using 2−deltadelta(Ct) method (18, 19).
TKTL1 constructs
A human cDNA clone of TKTL1 was purchased from ATCC. HindIII and XbaI restriction sites flanking the whole open reading frame of TKTL1 were constructed by PCR. The PCR products were digested with HindIII and XbaI restrictases and gel extracted and subcloned between into the corresponding sites of pcDNA3.1 expression vector. The sequence and direction of the insert was confirmed during each step by sequencing.
Cell proliferation and Soft agar assay
The HNSCC cell lines were transfected with TKTL1 expression plasmids at 40–60% confluence by using FuGENE 6 (Roche) as described previously (12). Validated plasmids encoding shRNA for TKTL1 and negative control were introduced into the FaDu cells using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions (8). Cell growth rate was measured by MTT assay (Sigma) (12). HIF1α inhibitor, NSC134754 was obtained from National Cancer Institute through their Developmental Therapeutic Program. HIF1α siRNA was from Ambion. Ascorbate and 3-bromopyruvate was purchased from Sigma. The soft agar assay was carried out as previously described (20).
Tumor Xenograft
NOD/SCID mice (5-week-old, Jackson Laboratories) were injected subcutaneously with 5 × 106 TKTL1- or vector-expressing O11 cells. One week after injection, tumor volume was assessed every 2–3 days using a digital caliper and calculated using the following formula: volume = (a×b2)/2, where a is the widest diameter of the tumor, and b is the diameter perpendicular to a. At 5 weeks after injection, the tumors were dissected and weighed.
Liquid Chromotography-Mass Spectrometry/Mass Spectrometry (LC-MS/MS) analysis of metabolites
Metabolic extracts were prepared and measured by LC-MS/MS as described previously (13, 21). Prior to extraction, the frozen cell samples were suspended in 2 mL of 100% MeOH and mixed vigorously. The cells were sonicated before the resulting suspension was centrifuged at 9500× g for 10 min. The supernatant was then dried under a nitrogen flow and reconstituted in 500 µL of methanol/water (10:90, v/v). Chromatographic analysis was performed using an Agilent 1100 Series HPLC. The column effluent was monitored using an API 3000 triple-quadruple mass-spectometric detector (Applied Biosystems). The spectrometer was programmed to monitor the ion of glyceraldehyde-3-p at m/z 168.7, ribose-5-p at m/z 228.7 and that of fructose-6-phosphate at m/z 258.7. For each measurement, at least triplicates were performed.
Measurement of pyruvate, lactate, glucose and ATP
Pyruvate and Lactate production was measured by the enzymetic method using a commercially available fluorescence-based assay kit or using the CMA 600 Analyzer as described previously (13, 22). Glucose in the medium were quantitated using the amplex red glucose/glucose oxidase kit (Invitrogen) (23). ATP levels were assessed using an ATP bioluminescence assay kit (Roche) (22, 24). Cell number was determined using a Coulter particle analyser.
Western blotting
~30 µg of whole protein extract were electrophoresed and transferred to a PVDF membrane, probed overnight at 4°C with the antibody against TKTL1 (R-Biopharm, kindly provided by Dr. Coy), HIF1α (BD Biosciences), actin (Sigma). Quantification of band intensity was performed using Image J software (NIH).
Statistical analysis
Differences between experimental variables were estimated using Student's t test or Mann-Whitney tests as appropriate. A probability level of 0.05 was chosen for statistical significance. All computations were done in R 2.8.1 (http://cran.r-project.org/).
Results
1. TKTL1 is hypomethylated and overexpressed in HNSCC tumors
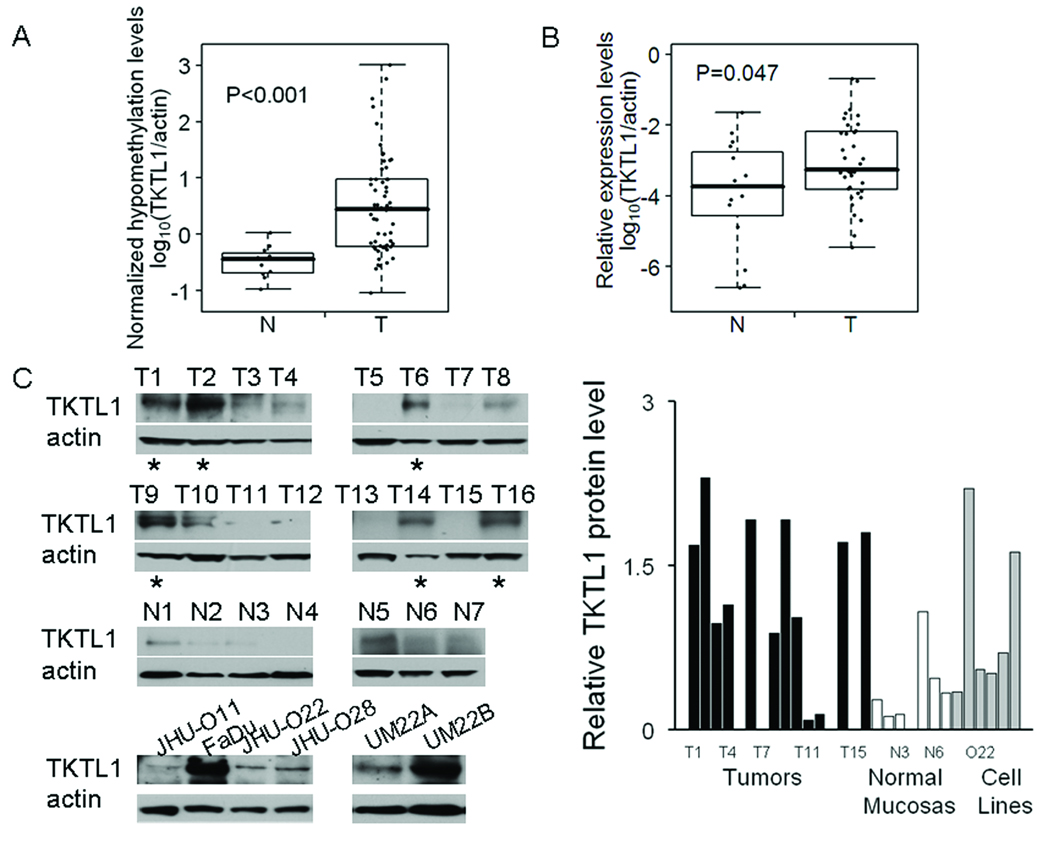
Our laboratory has previously described coordinated activation of candidate proto-oncogenes via promote hypomethylation in HNSCC and lung cancer (8). We found that promoter hypomethylation and overexpression of TKTL1, a candidate oncogene, occurred in HNSCC. To examine the frequency of TKTL1 promoter hypomethylation in HNSCC, we further tested tissues from 58 HNSCC cancers and 11 normal mucosas in non-cancer control subjects, using a method we devised previously termed TKTL1 Quantitative Unmethylation-specific PCR (QUMSP), which is a rapid, quantitative assay for specifically measuring non-methylated TKTL1 promoter. We found that TKTL1 promoter was significantly demethylated in primary HNSCC (log10 median: 0.45, log10 range: −1.04–3.01) compared with that in normal mucosa (log10 median: −0.44, log10 range: −0.97–0.02) (P < 0.001), confirming that TKTL1 is hypomethylated in HNSCC (Figure 1A). To validate the overexpression of TKTL1 at the transcriptional level in HNSCC, we further compared TKTL1 expression patterns in 36 HNSCC tumor tissues to those seen in 16 normal mucosas using quantitative RTPCR. Our analysis revealed the mean expression levels for TKTL1 in HNSCC tumor tissues was roughly 7-fold higher than that in the normal mucosa (Figure 1B, P=0.047). Further, western blot was performed to confirm TKTL1 overexpression in HNSCC tumor samples and different HNSCC cell lines. Of the 16 HNSCC tumor samples we examined, 6 were found to have increased expression of TKTL1 protein compared with the normal mucosas. Meanwhile, among the HNSCC cell lines blotted, we found that TKTL1 was relatively overexpessed in FaDu and UM22B cell lines in comparison to levels in nomal mucosal samples (Figure 1C).
Figure 1. TKTL1 is hypomethylated and overexpressed in HNSCC tumors.
A, Hypomethylation of TKTL1 in HNSCC tumor tissues. QUMSP percentage of TKTL1 was conducted in a cohort of HNSCC cancer patients using 58 tumors and 11 mucosal samples to assay promoter demethylation. Each dot represents the relative mean levels of TKTL1 hypomethylation from triplicate experiments in the sample normalized by actin. N, normal mucosas, T, tumors. P<0.001. B, Quantitative RTPCR analysis of TKTL1 on 36 HNSCC tumors and 16 normal samples. N denoted normal mucosa, T, tumor. Each dot represented the relative mean expression of TKTL1 from triplicate experiments in the sample. P=0.047. C, left, Immunoblot of TKTL1 in 16 HNSCC tumor tissues and 7 normal mucosal samples and 6 cancer cell lines using a mouse monoclonal anti-TKTL1 antibody. Blots were probes with mouse monoclonal anti-β-actin antibody as a control for loading and transfer. Right, relative TKTL1 protein levels in all samples examined by western blot. Quantification of bands was performed using NIH ImageJ software. Asterisk, tumor samples with TKTL1 protein overexpression. Overexpression of TKTL1 were found in tumor sample T1, T2, T6, T9, T14 and T16, as well as FaDu and UM 22B HNSCC cells in comparison to that in the normal mucosal samples we examined. T denotes tumor sample, and N normal mucosa.
2. TKTL1 promotes HNSCC cell growth in vitro
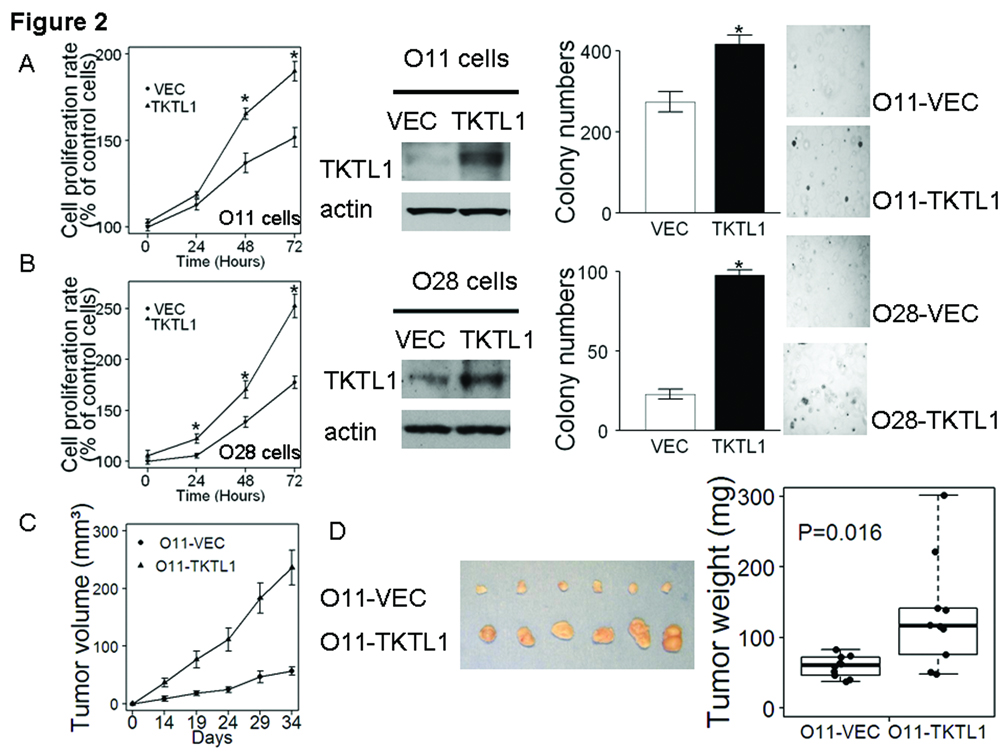
To understand the function of TKTL1 in HNSCC, we first explored the effect of ectopic TKTL1 expression on cellular growth in vitro in JHU-O11 cell line, which expresses relatively lower endogenous levels of TKTL1. As shown in Figure 2A, left, following transfection with a TKTL1 cDNA expression construct, JHU-O11 cells demonstrated significant increases in cellular proliferation (P<0.05). To further functionally characterize the oncogenic property of TKTL1, we established stable cell lines overexpressing TKTL1 in O11 cells and performed anchorage independent growth assays in cells stably expressing TKTL1. Forced expression of TKTL1 in the stable clone was verified by immunobloting with the monoclonal anti-mouse TKTL1 antibody (Figure 2A, middle). Two weeks after seeding on soft agar, we noted a dramatic increase in size and colonies in TKTL1 expressing cells compared with the vehicle-expressing cells (417 ± 22 vs 274 ± 25, n=3, P<0.05, Figure 2A, right). Similarly, the JHU-O28 cells transiently transfected with the TKTL1 expression construct grew faster than that transfected with vector (Figure 2B). Further, stable expression of TKTL1 in O28 cells exhibited markedly increased colony formation relative to vector controls (n=3, P<0.05, Figure 2B). Taken together, these data suggest that overexpression of TKTL1 provides a growth advantage for HNSCC tumor cells in vitro.
Figure 2. TKTL1 promoted HNSCC tumor growth in vitro and in vivo.
A, TKTL1 promoted cell growth in JHU-O11 cells in vitro. Shown are the MTT proliferation assay in TKTL1-transiently transfected JHU-O11 cells (*P<0.05), western blot displaying the overexpression of TKTL1 protein levels in TKTL1–expressing O11 cells and soft agar assay showing increased anchorage-independent growth in TKTL1-expressing O11 cells compared with the vector-expressing cells (*P<0.05). Representative pictures of soft-agar colonies in TKTL1- and vector-expressing O11 were shown. B, TKTL1 promoted cell growth in JHU-O28 cells in vitro. Shown are the MTT proliferation assay in TKTL1-transiently transfected JHU-O28 cells (*P<0.05), western blot displaying the overexpression of TKTL1 protein levels in TKTL1–expressing O28 cells and soft agar assay showing increased anchorage-independent growth in TKTL1-expressing O28 cells compared with the vector-expressing cells (*P<0.05). Representative pictures of soft-agar colonies in TKTL1- and vector-expressing O28 cells were shown. C, Plot of mean tumor-volume trajectories over time for the mice inoculated with TKTL1- and vector-expressing O11 stable cells. 5 mice per group. Error bars represent the SEMs at each time point. D, left, dissected tumors from the NOD/SCID mice. The representative tumors derived from the O11 cells stably expressing TKTL1 are shown (bottom row), and these tumors were larger than the tumors from the O11 cells stably transfected with vector (upper row). Right, mass of the dissected tumors. Each dot represented the tumor mass from the mice. P=0.016.
3. TKTL1 promotes HNSCC tumor growth in vivo
To examine the effects of TKTL1 overexpression on HNSCC tumor growth in vivo, we subcutaneously injected the TKTL1- or vehicle-expressing O11 cells into NOD/SCID mice. The tumor growth was monitored over a 5-week period. As shown in Figure 2C, mice injected with the TKTL1-expressing cells showed a significant increase in tumor development as compared with those injected with the vector control cells (P<0.01). The tumors developed from the TKTL1-expressing cells were larger in size compared with that from the vehicle expressing cells (Figure 2D, left). The TKTL1-expresssing cells gave rise to larger tumors than the vector-expressing cells (TKTL1-expressing cells vs vector-expressing cells, 132 mg (68.4—180.1 mg) vs 59.1 mg (49.8—84.2 mg ), mean and 95% CI, P=0.016, Figure 2D, right). These data support the role of TKTL1 in promoting HNSCC tumor growth.
4. TKTL1 enhances the production of fructose-6-phosphate and glyceraldehydes-3-phosphate
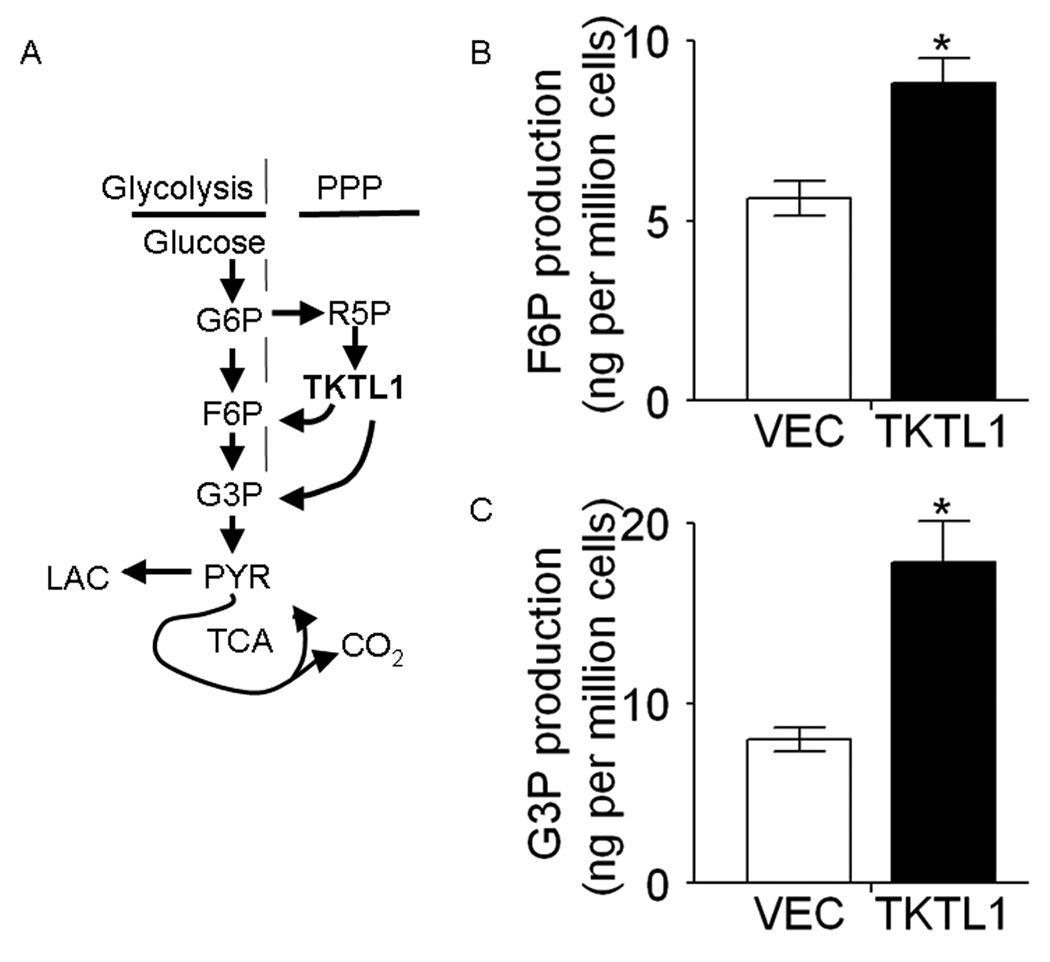
We sought to address the means by which overexpression of TKTL1 promotes HNSCC tumor growth. TKTL1 encodes a member of PPP, which has an important role in controlling the nonoxidative PPP (25). As shown in Figure 3A, the PPP is directly connected to glycolysis, as fructose-6-phosphate and glyceraldehyde-3-phosphate are the intermediates in both pathways. We hypothesized that TKTL1 could enhance increase the production of fructose-6-phosphate and glyceraldehydes-3-phosphate, increasing aerobic glycolysis. The fructose-6-phosphate and glyceraldehydes-3-phosphate levels in TKTL1-expressing O11 cells were determined by LC-MS/MS-based analysis of metabolites. As shown in Figure 3B and 3C, we found an increase in fructose-6-phosphate (8.8 ± 0.7 vs 5.6 ± 0.5, n=3, P = 0.019) and glyceraldehydes-3-phosphate production (17.8 ± 2.3 vs 8.0 ± 0.7, n=3, P=0.039) in TKTL1-expressing O11 cells compared with vehicle-expressing cells. This suggests that TKTL1 may promote the aerobic glycolysis via enhanced metabolite production including fructose-6-phosphate and glyceraldehydes-3-phosphate.
Figure 3. TKTL1 enhanced the production of fructose-6-phosphate and glyceraldehyde-3-phosphate.
A, simplified overview of glycolysis, TCA, and PPP. G6P, glucose-6-phosphate; F6P, fructose-6-phosphate; G3P, glyceraldehyde-3-phosphate; PYR, pyruvate; LAC, lactate; R5P, ribose-5-phosphate. B and C, metabolite levels in the O11 cells stably expressing TKTL1 and vector, as determined by LC-MS/MS. Shown are the quantitative changes of F6P and G3P in TKTL1-expressing cells compared with that in vector expressing cells. Error bars denote the SEMs (n = 3). *P<0.05.
5. TKTL1 promotes increased aerobic glycolysis in HNSCC
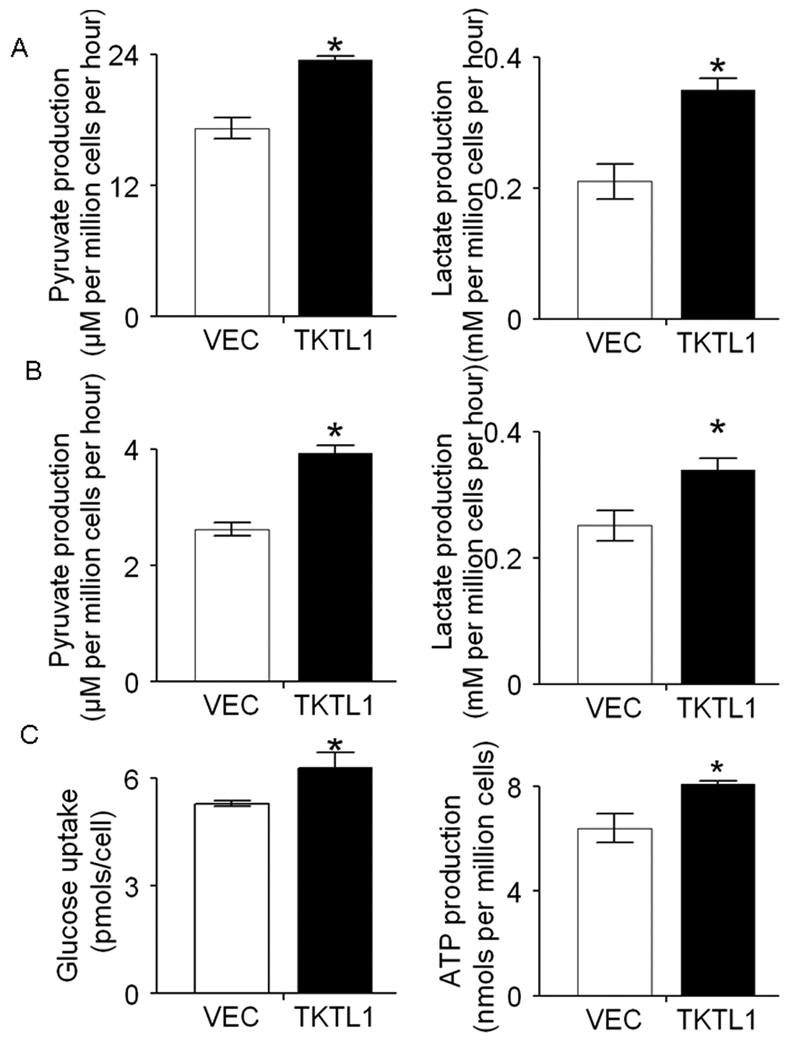
Given the enhanced fructose-6-phosphate and glyceraldehydes-3-phosphate we observed, we were interested in examining the effects of TKTL1 overexpression on aerobic glycolysis. It is known that pyruvate and lactate, the end products of aerobic glycolysis, are elevated in cancer cells even in the presence of oxygen (26). We proposed that TKTL1 enhances generation of fructose-6-phosphate and of glyceraldehyde-3-phosphate, resulting in elevated production of pyruvate and lactate. We found that significantly increased pyruvate and lactate production in TKTL1-expressing O11 cells compared with the corresponding vector controls (Pyruvate, 23.4 ± 0.4 vs 17.2 ± 1.0, P<0.01, Figure 4A; Lactate, 0.35 ± 0.03 vs 0.21 ± 0.03, P<0.01, Figure 4A). Similar results were obtained in O28 cells ectopically expressing TKTL1 (P<0.05, Figure 4B). As the Warburg effect described, tumor cells rapidly use glucose and convert the majority of it to lactate. To determine if the increased pyruvate and lactate production are accompanied by the increased glucose consumption, we monitored the glucose uptake in the TKTL1 expressing O11 cells. As shown in Figure 4C, glucose uptake was significantly higher in TKTL1-expressing O11 cells than that in the vector control cells (6.3 ± 0.4 vs 5.3 ± 0.1, P=0.012). Further, to evaluate the impact of ectopic expression of TKTL1 on the cellular energy balance, ATP levels were measured in the TKTL1-expressing O11 cells and its corresponding vector controls. In consistent, we found that TKTL1-expressing O11 cells displayed increased ATP levels in comparison to the vehicle-expressing cells (8.1 ± 0.2 vs 6.4 ± 0.5, P=0.037, Figure 4D). Together, these data supported that TKTL1 increased aerobic glycolysis in HNSCC tumorigenesis.
Figure 4. TKTL1 increased aerobic glycolysis in HNSCC.
A, the production of pyruvate (left) and lactate (right) in the culture buffer was measured in TKTL1- and vector-expressing JHU-O11 cells. B, the production of pyruvate (left) and lactate (right) in the culture buffer was measured in TKTL1- and vector-expressing JHU-O28 cells. The cells were allowed to grow in Krebs media for 6 h, and then media were collected and assayed for concentration of pyruvate and lactate. Data represent mean ± SEM. of three individual experiments. *P < 0.05. C, The glucose uptake of TKTL1 expressing O11 cells was significantly increased compared with that of the control cells after 48 hour culture in RPMI with 11 mM glucose. Triplicate samples were used in each group. *P< 0.05. D, The ATP level in TKTL1-expressing O11 cells was significantly increased compared with that of the vector-expressing cells. The cells were cultured with normal medium for 48 hours and harvested for intracellular ATP content (nmols per million cells ± SEM) measurement by luciferin-luciferase assay. *P<0.05.
6. TKTL1 contributes to HIF1α accumulation in HNSCC
We next explored possible downstream mediators of TKTL1’s growth promoting effect. Recently the cancer-specific aerobic glycolytic metabolism reported by Warburg has been linked to HIF1α degradation induced by pyruvate and lactate (27). We predicted that TKTL1 overexpression results in HIF1α accumulation through the increased pyruvate and lactate. We examined the expression level of HIF1α in TKTL1-expressing O11 cells. As shown in Figure 5A, enhanced HIF1α accumulation was observed in the TKTL1-expressing O11 cells compared with the vehicle-expressing cells. Further, we found that, two HIF1α downstream target genes, including GLUT1 and LOX, which are considered to be under the regulation of hypoxia, were upregulated in TKTL1-expresing O11 cells (Figure 5A). Of note, the HIF1α accumulation and the upregulation of GLUT1, were also found in the tumor xenografts originated from the TKTL1-expressing O11 cells (Figure 5B). Conversely, TKTL1 shRNA-mediated silencing of endogenous TKTL1 expression significantly decreased the HIF1α levels in FaDu cells (Figure 5C). To address the contribution of HIF1α to growth, a HIF1α small molecule inhibitor, NSC134754, was applied to the TKTL1-expressing O11 cells (12, 28). Our results found that treatment with 0.5 µM of NSC134754 significantly inhibited the TKTL1-expressing O11 cell growth (Figure 5D). We found that, upon treatment with ascorbate, an agent which selectively reverses the HIF1α accumulation induced by pyruvate and lactate, the growth-promoting effect mediated by TKTL1 in O11 cells was abolished (Supplementary Figure 1A and B) (27). Furthermore, we observed that introduction of HIF1α siRNA resulted in growth inhibition that reversed the TKTL1-mediated growth stimulation in O11 cells (Supplementary Figure 1C). Overall, these data indicate that TKTL1’s growth promoting effects are significantly dependent on HIF1α stabilization.
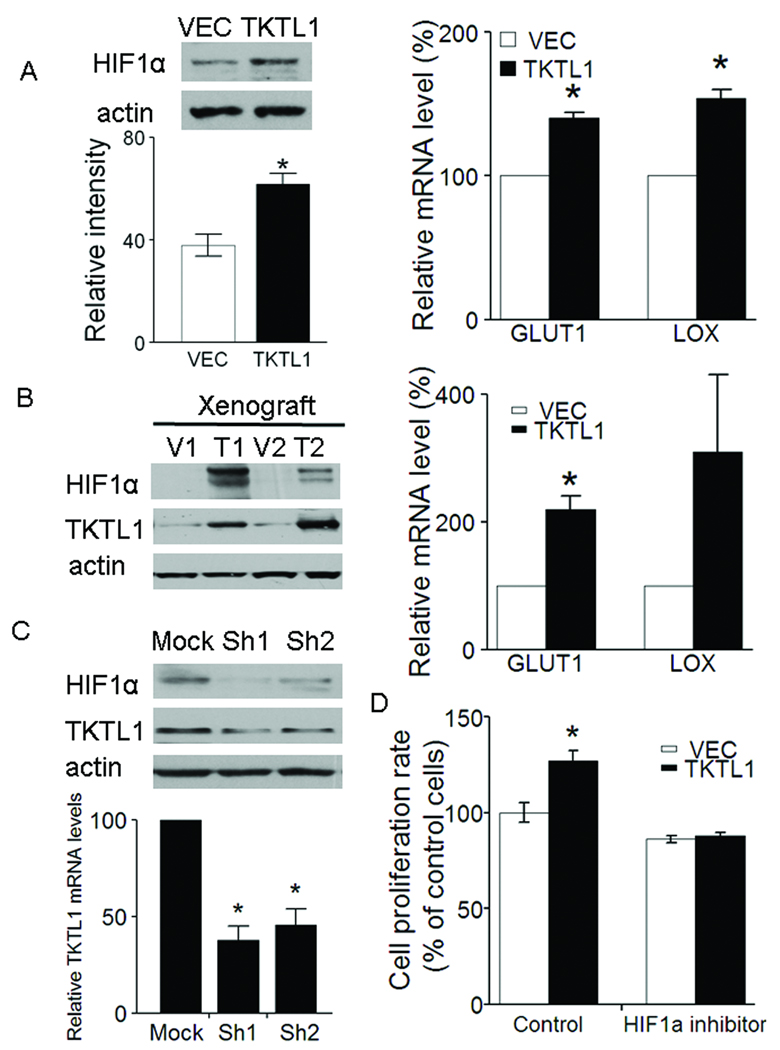
Figure 5. TKTL1 contributes to HIF1α accumulation in HNSCC.
A, left, the protein levels of HIF1α were determined 4 h following switching from culture medium to Krebs-Henseleit buffer in O11 cells stably expressing TKTL1. HIF1α was overexpressed by 1.5-fold in the O11 cells stably expressing TKTL1 versus its corresponding vehicle cells (n = 3, P < 0.05). Right, quantitative RTPCR analysis for two HIF-regulated downstream genes, GLUT1 and LOX in O11 cells expressing TKTL1. Shown are mean values of triplicate assays ± SEM. *P<0.05. B, left, the protein levels of HIF1α and TKTL1 in tumor xenografts originated from TKTL1-expressing O11 cells compared with those from vector-expressing cells. Representative images from three independent experiments were shown. Right, quantitative RTPCR analysis for GLUT1 and LOX in tumor xenografts originated from TKTL1-expressing O11 cells. Shown are mean values of triplicate assays ± SEM. *P<0.05. C, Western blot analysis on HIF1α and TKTL1 in from FaDu cells after transient transfection of TKTL1 shRNAs. After 48 h of transfection, cells were incubated with glucose-free Krebs buffer for 4 h. The cells then were harvested for Western blotting. Results are representative of three independent experiments. Bar graph shows the inhibition of TKTL1 expression in Fadu cells after TKTL1 shRNA transfection, as quantitated by quantitative real-time RTPCR. *P<0.05. D, the growth of O11 cells stably expressing TKTL1, was inhibited after treatment with NSC134754, a HIF1α small inhibitor. Cell growth was estimated by using MTT assay, after exposure for 48 h to 0.5 µM of NSC134754. Data are mean ± SEM. values from three independent experiments. *P<0.05.
7. HIF1α provides a feed-forward loop in TKTL1-mediated HNSCC tumorigenesis by upregulation of glycolytic enzymes
We hypothesized that enhancement of HIF1α constitutes a feed-forward mechanism via upregulation of glycolytic enzymes contributing to the TKTL1-mediated HNSCC tumorigenesis. To validate the involvement of HIF1α on glycolysis during TKTL1-mediated HNSCC tumorigenesis, we next performed quantitative RTPCR of mRNAs corresponding to four well-established HIF1α target genes implicated in glycolysis, including HK2, ALDOC, PGK1, and PGM1 in the TKTL1-expressing O11 cells. As shown in Figure 6A, elevated expression of HK2, ALDOC, PGK1 and PGM1 were found in TKTL1-expressing O11 cells in comparison with vector-expressing cells. We found that HK2, ALDOC and PGK1 were also upregulated in the tumor xenograft originated from TKTL1-expressing O11 cells (Supplementary Figure 1D). Conversely, we found that treatment with 1 µM of HIF1α small molecule inhibitor for 48 h, the upregulation of HK2, ALDOC and PGK1 was eliminated (Figure 6A). To examine whether this increased glycolytic phenotype is essential for the TKTL1-mediated tumor growth, 3-bromopyruvate, an agent which inhibits glycolysis through inhibition of hexokinase, was then applied to the TKTL1-expressing O11 cells (29). We found that treatment of 3-bromopyruvate significantly inhibited the TKTL1-expressing O11 cell growth (Figure 6B). It is previously reported that the PPP including two distinctive branches, oxidative branch and nonoxidative branch, that fulfill the requirement for ribose-5-phosphate for the synthesis of nucleotides and nucleic acids (15). It is thought that the nonoxidative branch of PPP is the main source for ribose-5-phosphate synthesis in the tumor cells (30, 31). To determine whether the production of ribose-5-phosphate is coordinated with the enhanced production of fructose-6-phosphate and glyceraldehydes-3-phosphate, we also measured the generation of ribose-5-phosphate in TKTL1-expressing O11 cells. Intriguingly, we noted that ribose-5-phosphate is significantly increased in the TKTL1-expressing O11 cells in comparison to the vehicle-expressing cells (Figure 6C, 15.2 ± 1.5 vs 8.6 ± 1.5, P=0.035). This observation suggests that HIF1α promotes a feed-forward loop in TKTL1-mediated HNSCC tumorigenesis by upregulating glycolytic target genes, as well as indirectly provides increasing substrate for TKTL1 mediated supply of glycolytic metabolites (Figure 6D).
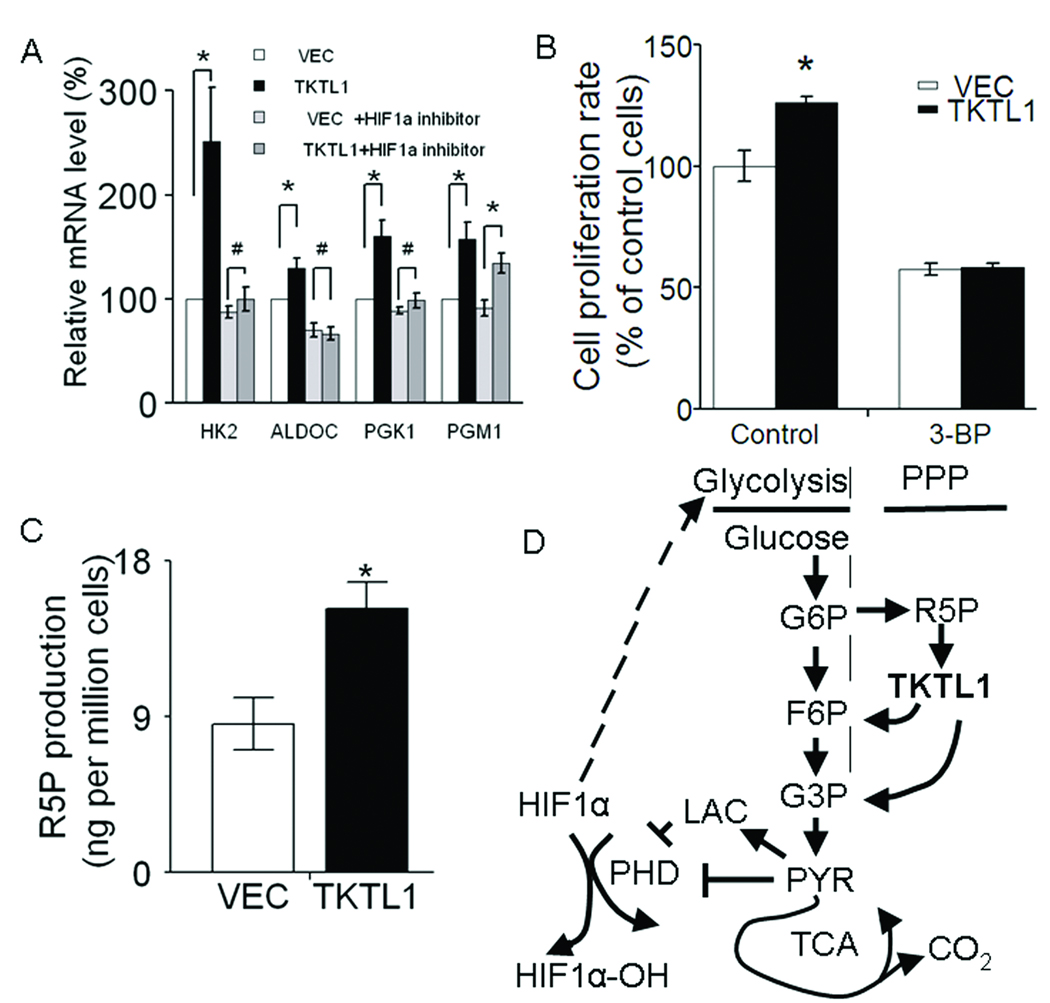
Figure 6. HIF1α provides a feed-forward loop in TKTL1-mediated HNSCC tumorigenesis by regulation of glycolytic enzymes.
A, quantitative RTPCR analysis for four HIF1α -regulated downstream genes implicated in glycolysis (HK2, ALDOC, PGK1, and PGM1) in O11 cells stably expressing TKTL1. Student’s t test showed significance for expression of HK2, ALDOC, PGK1, and PGM1 between TKTL1- and VEC-expressing O11 stable cells, whereas there was no significance for expression of HK2, ALDOC, and PGK1 after 48 h treatment with 1µM of NSC134754 (the HIF1α inhibitor). Actin gene served as controls. Shown are mean values of triplicate assays ± SEM. *P<0.05, #P>0.05. B. the growth of O11 cells stably expressing TKTL1, was inhibited after treatment with 3-bromopyruvate, a glycolysis inhibitor through inhibition of hexokinase. Cell growth was estimated by using MTT assay, after exposure for 48 h to 30 µM of 3-bromopyruvate. Data are mean ± SEM. values from three independent experiments. *P<0.05. C, Ribose-5-phosphate (R5P) levels in TKTL1-expressing O11 cells. Cells were cultured with normal RPMI1640 medium for 48 h and harvested for R5P assay using LC-MS/MS as described in Methods. *P<0.05. D, the proposed mechanism of TKTL1-mediated HNSCC tumorigenesis.
Discussion
TKTL1 has been reported to be overexpressed in a number of different cancer types, including lung, cervical, nasopharyngeal, breast, colon, gastric cancers. Recent studies have correlated TKTL1 expression with poor survival in laryngeal carcinoma, colon and urothelial cancers, as well as distant metastasis in ovarian carcinoma (25, 32). In our current study QUMSP analysis revealed a dramatic increase of TKTL1 hypomethylation in the HNSCC tumors compared with the normal control mucosa. By evaluation of TKTL1 overexpression at transcriptional level and protein level, we observed that TKTL1 is significantly overexpressed in HNSCC cancers in comparison to that in normal tissues. It is likely that hypomethylation could be attributable to the overexpression of TKTL1. In support of this, we previously identified the HNSCC primary tissues showing both hypomethylation and overexpression of TKTL1. In the TKTL1 methylated human oral keratinocyte cell line OKF6, we reported transcriptional upregulation of TKTL1 after 5-aza/TSA treatment, supporting the concordance between methylation and expression (8).
In exploring the oncogenic function of TKTL1, we found that HNSCC cells (O11 and O28 cells, as well as 22A cells, unpublished data) with exogenous TKTL1 overexpression grew at an accelerated rate and gained the ability to undergo anchorage-independent growth. Further, we firstly showed that TKTL1-overexpressing HNSCC cells grow faster and larger tumor xenografts compared with the vector-expressing cells. Conversely, we also reported that knockdown of TKTL1 in HNSCC cells (FaDu and UM22B) decrease the cellular proliferation, anchorage-dependent and independent tumor growth (8). Taken together, our studies provide convincing evidence indicating that TKTL1 is a candidate oncogene.
It has long been considered that aerobic glycolysis associated with the Warburg effect is an intrinsic component of a malignant phenotype. TKTL1 encodes transketolase of the PPP, which catalyzed the production of metabolites including fructose-6-phosphate and glyceraldehydes-3-phosphate. Interestingly, glyceraldehydes-3-phosphate has been shown significantly increased on disease progression from benign to clinically localized prostate cancer to metastatic prostate cancer (33). Here we propose that TKTL1 increases the production of fructose-6-phosphate and glyceraldehydes-3-phosphate, resulting in an increased aerobic glycolysis. Several lines of evidence support this. First, enhanced production of fructose-6-phosphate and glyceraldehydes-3-phosphate was observed in TKTL1-expressing cells compared with that in the vector control cells. Second, production of pyruvate and lactate were significantly elevated in TKTL1-expressing cells in comparison with their vector control cells (in O11 cells and O28 cells, as well as 22A cells, unpublished data). Third, increased glucose consumption, accompanied by increased ATP yield was shown in the TKTL1-expressing cells. In agreement to our study, Xu et al also reported that inhibition of TKTL1 resulted in a significantly slowed glucose consumption and lactate production (34).
Lu and colleagues recently reported that the metabolites of Warburg effect, pyruvate and lactate, could dramatically increase HIF1α accumulation in cancer cells by inhibiting the prolyl hydroxylase enzyme activity, which could result in degradation of HIF1α protein through hydroxylation (27). Given the increased production of pyruvate and lactate we observed in TKTL1-expressing HNSCC cells, we further investigate the effect of TKTL1 overexpression on HIF1α stabilization. We found the HIF1α accumulation, together with the upregulation of two HIF1α downstream target genes, GLUT1 and LOX, in TKTL1-expression O11 cells and/or tumor xenografts. In addition, use of a HIF1α small molecule inhibitor, as well as HIF1α siRNA blocked TKTL1-mediated cancer cell growth. Conversely, shRNA knockdown of TKTL1 expression in FaDu cells decreased HIF1α accumulation, which is consistent with the decreased cellular growth after TKTL1 shRNA transfection as we reported previously. Of note, we also found that overexpression of TKTL1 in human immortal oral keratinocyte OKF6 resulted in HIF1α accumulation accompanied with increased lactate production (unpublished data). These suggest that TKTL1 may induce HNSCC development through constitutive activation of HIF1α.
We also propose that the enhanced HIF1α accumulation represents a feed-forward mechanism for the TKTL1-mediated tumorigenesis. In this respect, our study showed that four glycolytic genes directly targeted by HIF1α, including HK2, ALDOC, PGK1 were upregulated in TKTL1-expressing O11 cells, as well as in tumor xenografts. The upregulation of HK2, ALDOC, PGK1 in TKTL1-expressing O11 cells could be abolished after treatment with HIF1α small molecule inhibitor, suggesting the specificity of pathway via HIF1α mediated action. Meanwhile, treatment of 3-bromopyruvate, a glycolytic inhibitor through inhibition of hexokinase abolished TKTL1-mediated O11 tumor cell growth. These data indicate that the induction of HIF1α, coupled in turn with the HIF1α-promoted expression of glycolytic enzymes, could be an essential feed-forward loop for sustaining the glycolytic phenotype and malignant progression mediated by TKTL1.
The data in the present study support the following model for the mechanism by which TKTL1 functions to promote carcinogenesis in HNSCC cancer cells. As shown in Figure 6D, in HNSCC cancer cells expressing high levels of TKTL1, TKTL1 enhances the production of fructose-6-phosphate and glycerealdehyde-3-phosphate (two essential metabolites required for glycolysis), thereby facilitating an increased production of pyruvate and lactate, and subsequent HIF1α accumulation and activation of its downstream target genes. Altogether, these studies define an oncogenic role of TKTL1 in human tumorigenesis and define a novel mechanism for increasing aerobic glycolysis and HIF1α accumulation in cancer. Although these data do not support TKTL1 as sufficient to induce carcinogenesis in an HNSCC model, they do support the role of TKTL1 as a significant contributor to a malignant phenotype.
Translational relevance
The "Warburg effect”, an elevation in aerobic glycolysis, is a fundamental property of cancer cells. Aberrant expression of TKTL1 (Transketolase-like 1), occurs in HNSCC and other epithelial tumors, but the functional significance of TKTL1 aberrant expression in cancer and its association with aerobic glycolysis has not been defined. Here we showed that TKTL1 is a novel candidate oncogene that is activated by promoter hypomethylation, accelerates aerobic glycolysis and HIF1α mediated tumor growth. Our data demonstrate a mechanism of tumorigenesis that bridges the pentose phosphate pathway and the glycolytic pathway, resulting in an amplified feed forward pathway that results in HIF1α mediated tumor growth. These studies provide evidence for an important role of TKTL1 in HNSCC tumorigenesis, and may contribute to effective therapeutic targeting of metabolic pathways in cancer.
Supplementary Material
A, HIF1α expression in TKTL1-expressing O11 cells was abolished after treatment with 100 µM ascorbate for 24 h, as demonstrated by Western Blot. B, the growth of TKTL1-expressing O11 cells was inhibited after treatment with 200 µM ascorbate for 48 h. Columns, mean from three independent experiment, bars, SEM. *, P<0.05. Student’s t ttest showed significance between TKTL1- and Vector-expressing O11 cells in the absence of ascorbate, whereas there was no significance in the presence of ascorbate. C, the growth of TKTL1-expressing O11 cells was significantly inhibited by transient transfection of HIF1α siRNA for 36 h, as demonstrated by MTT assay. Columns, mean from three independent experiment, bars, SE. *, P<0.05. Student’s t test showed significance between TKTL1- and Vector-expressing O11 cells in the transfection of scramble siRNA, whereas there was no significance in the transfection of HIF1α siRNA. D, quantitative RTPCR analysis for three HIF1α-regulated downstream genes (HK2, ALDOC, and PGK1) in tumor xenografts originated from TKTL1-expressing O11 cells. Actin gene served as controls. Shown are mean values of triplicate assays ± SEM.
Acknowledgements
We thank M. Ochs for statistical advice. This work was supported by Specialized Program of Research Excellence grant P50 CA96784 and a Clinical Innovator Award from Flight Attendant Medical Research Institute.
References
- 1.Carvalho AL, Jeronimo C, Kim MM, et al. Evaluation of promoter hypermethylation detection in body fluids as a screening/diagnosis tool for head and neck squamous cell carcinoma. Clin Cancer Res. 2008;14:97–107. doi: 10.1158/1078-0432.CCR-07-0722. [DOI] [PubMed] [Google Scholar]
- 2.Hoque MO, Kim MS, Ostrow KL, et al. Genome-wide promoter analysis uncovers portions of the cancer methylome. Cancer Res. 2008;68:2661–2670. doi: 10.1158/0008-5472.CAN-07-5913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Feinberg AP, Tycko B. The history of cancer epigenetics. Nat Rev Cancer. 2004;4:143–153. doi: 10.1038/nrc1279. [DOI] [PubMed] [Google Scholar]
- 4.Hanada M, Delia D, Aiello A, Stadtmauer E, Reed JC. bcl-2 gene hypomethylation and high-level expression in B-cell chronic lymphocytic leukemia. Blood. 1993;82:1820–1828. [PubMed] [Google Scholar]
- 5.Nishigaki M, Aoyagi K, Danjoh I, et al. Discovery of aberrant expression of R-RAS by cancer-linked DNA hypomethylation in gastric cancer using microarrays. Cancer Res. 2005;65:2115–2124. doi: 10.1158/0008-5472.CAN-04-3340. [DOI] [PubMed] [Google Scholar]
- 6.Oshimo Y, Nakayama H, Ito R, et al. Promoter methylation of cyclin D2 gene in gastric carcinoma. Int J Oncol. 2003;23:1663–1670. [PubMed] [Google Scholar]
- 7.Smith IM, Mydlarz WK, Mithani SK, Califano JA. DNA global hypomethylation in squamous cell head and neck cancer associated with smoking, alcohol consumption and stage. Int J Cancer. 2007;121:1724–1728. doi: 10.1002/ijc.22889. [DOI] [PubMed] [Google Scholar]
- 8.Smith IM, Glazer CA, Mithani SK, et al. Coordinated activation of candidate proto-oncogenes and cancer testes antigens via promoter demethylation in head and neck cancer and lung cancer. PLoS ONE. 2009;4:e4961. doi: 10.1371/journal.pone.0004961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hsu PP, Sabatini DM. Cancer cell metabolism: Warburg and beyond. Cell. 2008;134:703–707. doi: 10.1016/j.cell.2008.08.021. [DOI] [PubMed] [Google Scholar]
- 10.Kaelin WG, Jr, Ratcliffe PJ. Oxygen sensing by metazoans: the central role of the HIF hydroxylase pathway. Mol Cell. 2008;30:393–402. doi: 10.1016/j.molcel.2008.04.009. [DOI] [PubMed] [Google Scholar]
- 11.Kroemer G, Pouyssegur J. Tumor cell metabolism: cancer's Achilles' heel. Cancer Cell. 2008;13:472–482. doi: 10.1016/j.ccr.2008.05.005. [DOI] [PubMed] [Google Scholar]
- 12.Sun W, Zhou S, Chang SS, McFate T, Verma A, Califano JA. Mitochondrial mutations contribute to HIF1alpha accumulation via increased reactive oxygen species and up-regulated pyruvate dehydrogenease kinase 2 in head and neck squamous cell carcinoma. Clin Cancer Res. 2009;15:476–484. doi: 10.1158/1078-0432.CCR-08-0930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McFate T, Mohyeldin A, Lu H, et al. Pyruvate dehydrogenase complex activity controls metabolic and malignant phenotype in cancer cells. J Biol Chem. 2008;283:22700–22708. doi: 10.1074/jbc.M801765200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Coy JF, Dressler D, Wilde J, Schubert P. Mutations in the transketolase-like gene TKTL1: clinical implications for neurodegenerative diseases, diabetes and cancer. Clin Lab. 2005;51:257–273. [PubMed] [Google Scholar]
- 15.Horecker BL. The pentose phosphate pathway. J Biol Chem. 2002;277:47965–47971. doi: 10.1074/jbc.X200007200. [DOI] [PubMed] [Google Scholar]
- 16.Carvalho AL, Chuang A, Jiang WW, et al. Deleted in colorectal cancer is a putative conditional tumor-suppressor gene inactivated by promoter hypermethylation in head and neck squamous cell carcinoma. Cancer Res. 2006;66:9401–9407. doi: 10.1158/0008-5472.CAN-06-1073. [DOI] [PubMed] [Google Scholar]
- 17.Tokumaru Y, Yamashita K, Osada M, et al. Inverse correlation between cyclin A1 hypermethylation and p53 mutation in head and neck cancer identified by reversal of epigenetic silencing. Cancer Res. 2004;64:5982–5987. doi: 10.1158/0008-5472.CAN-04-0993. [DOI] [PubMed] [Google Scholar]
- 18.Liu Y, Sun W, Zhang K, et al. Identification of genes differentially expressed in human primary lung squamous cell carcinoma. Lung Cancer. 2007;56:307–317. doi: 10.1016/j.lungcan.2007.01.016. [DOI] [PubMed] [Google Scholar]
- 19.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 20.Zhou S, Kachhap S, Sun W, et al. Frequency and phenotypic implications of mitochondrial DNA mutations in human squamous cell cancers of the head and neck. Proc Natl Acad Sci U S A. 2007;104:7540–7545. doi: 10.1073/pnas.0610818104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Luo B, Groenke K, Takors R, Wandrey C, Oldiges M. Simultaneous determination of multiple intracellular metabolites in glycolysis, pentose phosphate pathway and tricarboxylic acid cycle by liquid chromatography-mass spectrometry. J Chromatogr A. 2007;1147:153–164. doi: 10.1016/j.chroma.2007.02.034. [DOI] [PubMed] [Google Scholar]
- 22.Christofk HR, Vander Heiden MG, Harris MH, et al. The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumour growth. Nature. 2008;452:230–233. doi: 10.1038/nature06734. [DOI] [PubMed] [Google Scholar]
- 23.Samudio I, Fiegl M, McQueen T, Clise-Dwyer K, Andreeff M. The warburg effect in leukemia-stroma cocultures is mediated by mitochondrial uncoupling associated with uncoupling protein 2 activation. Cancer Res. 2008;68:5198–5205. doi: 10.1158/0008-5472.CAN-08-0555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fantin VR, St-Pierre J, Leder P. Attenuation of LDH-A expression uncovers a link between glycolysis, mitochondrial physiology, and tumor maintenance. Cancer Cell. 2006;9:425–434. doi: 10.1016/j.ccr.2006.04.023. [DOI] [PubMed] [Google Scholar]
- 25.Langbein S, Zerilli M, Zur Hausen A, et al. Expression of transketolase TKTL1 predicts colon and urothelial cancer patient survival: Warburg effect reinterpreted. Br J Cancer. 2006;94:578–585. doi: 10.1038/sj.bjc.6602962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lu H, Forbes RA, Verma A. Hypoxia-inducible factor 1 activation by aerobic glycolysis implicates the Warburg effect in carcinogenesis. J Biol Chem. 2002;277:23111–23115. doi: 10.1074/jbc.M202487200. [DOI] [PubMed] [Google Scholar]
- 27.Lu H, Dalgard CL, Mohyeldin A, McFate T, Tait AS, Verma A. Reversible inactivation of HIF-1 prolyl hydroxylases allows cell metabolism to control basal HIF-1. J Biol Chem. 2005;280:41928–41939. doi: 10.1074/jbc.M508718200. [DOI] [PubMed] [Google Scholar]
- 28.Chau NM, Rogers P, Aherne W, et al. Identification of novel small molecule inhibitors of hypoxia-inducible factor-1 that differentially block hypoxia-inducible factor-1 activity and hypoxia-inducible factor-1alpha induction in response to hypoxic stress and growth factors. Cancer Res. 2005;65:4918–4928. doi: 10.1158/0008-5472.CAN-04-4453. [DOI] [PubMed] [Google Scholar]
- 29.Yun J, Rago C, Cheong I, et al. Glucose Deprivation Contributes to the Development of KRAS Pathway Mutations in Tumor Cells. Science. 2009 doi: 10.1126/science.1174229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Boros LG, Torday JS, Lim S, Bassilian S, Cascante M, Lee WN. Transforming growth factor beta2 promotes glucose carbon incorporation into nucleic acid ribose through the nonoxidative pentose cycle in lung epithelial carcinoma cells. Cancer Res. 2000;60:1183–1185. [PubMed] [Google Scholar]
- 31.Tong X, Zhao F, Thompson CB. The molecular determinants of de novo nucleotide biosynthesis in cancer cells. Curr Opin Genet Dev. 2009;19:32–37. doi: 10.1016/j.gde.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Volker HU, Scheich M, Schmausser B, Kammerer U, Eck M. Overexpression of transketolase TKTL1 is associated with shorter survival in laryngeal squamous cell carcinomas. Eur Arch Otorhinolaryngol. 2007;264:1431–1436. doi: 10.1007/s00405-007-0394-x. [DOI] [PubMed] [Google Scholar]
- 33.Sreekumar A, Poisson LM, Rajendiran TM, et al. Metabolomic profiles delineate potential role for sarcosine in prostate cancer progression. Nature. 2009;457:910–914. doi: 10.1038/nature07762. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 34.Xu X, Zur Hausen A, Coy JF, Lochelt M. Transketolase-like protein 1 (TKTL1) is required for rapid cell growth and full viability of human tumor cells. Int J Cancer. 2009;124:1330–1337. doi: 10.1002/ijc.24078. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A, HIF1α expression in TKTL1-expressing O11 cells was abolished after treatment with 100 µM ascorbate for 24 h, as demonstrated by Western Blot. B, the growth of TKTL1-expressing O11 cells was inhibited after treatment with 200 µM ascorbate for 48 h. Columns, mean from three independent experiment, bars, SEM. *, P<0.05. Student’s t ttest showed significance between TKTL1- and Vector-expressing O11 cells in the absence of ascorbate, whereas there was no significance in the presence of ascorbate. C, the growth of TKTL1-expressing O11 cells was significantly inhibited by transient transfection of HIF1α siRNA for 36 h, as demonstrated by MTT assay. Columns, mean from three independent experiment, bars, SE. *, P<0.05. Student’s t test showed significance between TKTL1- and Vector-expressing O11 cells in the transfection of scramble siRNA, whereas there was no significance in the transfection of HIF1α siRNA. D, quantitative RTPCR analysis for three HIF1α-regulated downstream genes (HK2, ALDOC, and PGK1) in tumor xenografts originated from TKTL1-expressing O11 cells. Actin gene served as controls. Shown are mean values of triplicate assays ± SEM.