Abstract
Recently, it has emerged that palindrome-mediated genomic instability contributes to a diverse group of genomic rearrangements including translocations, deletions, and amplifications. One of the best studied examples is the recurrent t(11;22) constitutional translocation in humans that has been well documented to be mediated by palindromic AT-rich repeats (PATRRs) on chromosomes 11q23 and 22q11. De novo examples of the translocation are detected at a high frequency in sperm samples from normal healthy males, but not in lymphoblasts or fibroblasts. Cloned breakpoint sequences preferentially form a cruciform configuration in vitro. Analysis of the junction fragments implicates frequent double-strand-breaks (DSBs) at the center of both palindromic regions, followed by repair through the non-homologous end joining (NHEJ) pathway. We propose that the PATRR adopts a cruciform structure in male meiotic cells, creating genomic instability that leads to the recurrent translocation.
Keywords: Translocation, Palindrome, Cruciform
1. Identification of the PATRRs at the breakpoints of the recurrent t(11;22) constitutional translocation
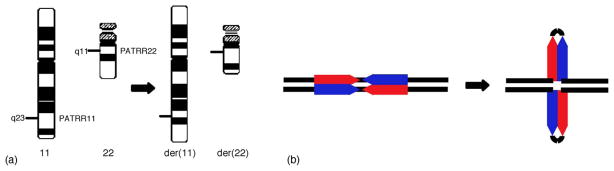
The constitutional t(11;22)(q23;q11) is the only known recurrent, non-Robertsonian translocation in humans (Fig. 1(a)) [1]. Whereas balanced carriers of the translocation usually manifest no clinical symptoms, they often have problems in reproduction, such as male infertility, recurrent abortion, and the birth of offspring with chromosomal imbalance. Severely affected offspring demonstrate the supernumerary-der(22) t(11;22) syndrome, as a result of 3:1 meiotic malsegregation of the der(22) [2]. The syndrome is characterized by severe mental retardation, preauricular tag or sinus, ear anomalies, cleft or high-arched palate, micrognathia, microcephaly, kidney abnormalities, heart defects, and genital abnormalities in males [1]. This clinical entity was recently named after the investigator who has been studying this disorder since 1976 as Emanuel syndrome (MIM#609029).
Fig. 1.
Palindrome-mediated translocation in humans. (a) Schematic representation of the t(11;22)(q23;q11). The PATRR11 and the PATRR22 are located at the breakpoints on 11q23 and 22q11, respectively. (b) Palindromic sequence possesses the potential for forming a cruciform configuration. Short palindromic sequences have the potential to form such double-stranded cruciform structures by intra-strand base pairing in the single-stranded DNA. DNA sequences indicated by blue arrows are the complement for those depicted by red arrows.
Its recurrent nature implicates a specific genomic structure at the t(11;22) breakpoints, which prompted us to examine the translocation breakpoint in detail. Fluorescence in situ hybridization (FISH) localized the breakpoints of all independent t(11;22) cases within the same small intervals on 11q23 and 22q11 [2–5]. The translocation breakpoint at 22q11 was mapped within one of the low-copy-repeats (LCRs) on 22q that produce another human chromosomal disorder, the 22q11 deletion syndrome. Each of the LCRs on 22q11 extends more than several 100 kb, and shares greater than 95% homology. Thus, the translocation breakpoint on 22q11 is not amenable to standard cloning strategies. Instead, we successfully identified a BAC encompassing the breakpoint region on 11q23, which allowed us to analyze the breakpoint sequence in detail. Scanning the BAC sequence for transcripts revealed that the translocation does not disrupt any functional genes at either of the breakpoint regions, which is in agreement with the lack of clinical symptoms in the balanced carriers. However, 11q23 breakpoint sequence was partially deleted from the BAC sequenced by the Human Genome Project. We eventually amplified and cloned the breakpoint region by PCR of normal genomic DNA. The breakpoint region deleted from the BAC is ~450bp in length, with an AT content of 93%. It comprises a nearly perfect palindromic structure, with 98% identity between the proximal and distal arm (Fig. 1(b), Table 1). We designated it as a palindromic AT-rich repeat on 11q23 (PATRR11) [6,7].
Table 1.
Characterization of the PATRRs identified to date
| PATRR | Location | Size | AT content (%) | Homology (%)a | Acc. Nob | References |
|---|---|---|---|---|---|---|
| PATRR11 | 11q23 | 450 | 93 | 98 | AF391129 | [7] |
| PATRR22 | 22q11 | 590 | 74c | 99c | Unclonable | [7] |
| PATRR17 | 17q11 | 197c | 80 | 97 | AB195812 | [16,17] |
Homology between the proximal and distal arms of the PATRR.
Accession number for GenBank database.
Inferred from isolated junction fragments without cloned normal sequence.
Next, we attempted to clone junction fragments from one of the balanced carriers of the t(11;22) rearrangement to identify the unknown sequence at the 22q11 breakpoint. By means of cloning rearranged fragments detected by Southern hybridization, we successfully isolated junction fragments both on the der(11) and the der(22). Thus, the putative structure of the breakpoint region on 22q11 could be inferred from sequence information derived from the junction fragments of the two derivative chromosomes by comparison with that from the normal PATRR11 [6]. Surprisingly, a similar PATRR-like sequence was reconstituted in the region surrounding the breakpoint on 22q11 (PATRR22). The size of the putative PATRR22 is ~590 bp, comparable to the size of the PATRR11. No substantial homology was observed between the PATRR11 and PATRR22 (identities = 58% between PATRR11 and PATRR22).
Using the sequence data of the junction fragments, we established a t(11;22)-specific PCR assay. PATRR-flanking primers were designed on 11q23 and 22q11 to amplify the junction fragments of the der(11) and the der(22). This system allowed us to analyze the translocation breakpoints of more than forty independent t(11;22) cases, and we localized virtually all of the breakpoints within the same PATRR11 and PATRR22 [8]. Other groups analyzed different subsets of translocation carriers, and demonstrated similar results (Table 2) [9,10]. Thus, the recurrent constitutional t(11;22) has been shown to take place within two different PATRRs located at 11q23 and 22q11. This observation provided a turning point in the understanding of the mechanism of this recurrent chromosomal translocation.
Table 2.
Reported examples of translocations mediated by palindromic sequence
| Karyotype | Case no. | Palindromic partner | References | |
|---|---|---|---|---|
| 40 | PATRR22 | 445 bp (PATRR11) | [8] | |
| t(11;22)(q23;q11) | 13 | PATRR22 | >230bpa | [9] |
| 5 | PATRR22 | >166bpa | [10] | |
| t(17;22)(q11;q11) | 2 | PATRR22 | 197 bp (PATRR17) | [12,16] |
| t(4;22)(q35;q11) | 1 | PATRR22 | 554 bp (547 bp spacer) | [18] |
| t(1;22)(p21;q11) | 1 | PATRR22 | 278 bp | [19] |
| t(X;22)b | 1 | PATRR22 | Not palindromic | [14] |
The sizes were estimated from junction fragments without normal sequence analyzed.
This is the only reported PATRR22-related translocation with a partner chromosome that does not appear to involve palindromic sequence.
2. PATRR-mediated chromosomal translocations
Although many efforts have been directed toward obtaining complete sequence information for the original PATRR22, the entire PATRR22 has not yet been cloned. The PATRR22 is located within one of the unclonable gaps remaining from the human genome project. It is reasonable to assume that the PATRR22 is highly unstable in cloning hosts like bacteria or yeast, similar to the observed deletion of the PATRR11 from the sequenced BAC clone. Although sequence of the junction fragments of the translocation allow for reconstitution of PATRR22-like sequence, indicating a size as short as ~590 bp, genomic PCR fails to bridge this gap.
Indeed, the PATRR22 appears to be highly unstable in the human genome. 22q11 has been deemed to be a hotspot for translocation breakpoints, since cytogenetic studies demonstrated that a large number of translocation breakpoints cluster at 22q11. FISH mapping indicates that the breakpoints of a variety of translocations involving 22q11 cluster within the unclonable LCR region that encompasses the PATRR22 [11–15]. These data implicate involvement of the PATRR22 in the etiology of these 22q11-related translocations.
We analyzed a case of neurofibromatosis type 1 (NF1) with a constitutional t(17;22)(q11;q11) [16], together with a previously reported NF1 case with t(17;22) [12]. Both translocations disrupt the NF1 gene on 17q11, which produces the phenotype in the patients. As was expected, FISH analysis localized the 22q11 breakpoints within the same LCR, where the breakpoint of t(11;22) resides. Further analysis of the chromosome 17 breakpoints within the NF1 gene revealed the presence of a ~200 bp PATRR within intron 31 of the gene (PATRR17) (Table 1) [17]. Subsequent molecular cloning of other translocation breakpoints has demonstrated similar palindromic sequences on partner chromosomes, such as 4q35.1, and 1p21.2 (Table 2) [18,19]. Hence, palindrome-mediated chromosomal translocation appears to be one of the universal pathways of human genomic rearrangements. This subset of translocations appears to occur in a non-random fashion possibly mediated by the genomic instability of palindromic DNA.
3. PATRR forms cruciform structure in vitro
Recently, it has emerged that palindrome-mediated genomic instability contributes to a diversity of genome rearrangements including not only constitutional translocations, but also large germ line deletions [20]. Cancer-related gross chromosomal rearrangement and genomic amplification are also reported to be associated with palindromic DNA [21,22]. The breakpoints usually constitute large palindromes, ranging in sizes from 40 kb to ~1 Mb. Homologous recombination might cause the large deletions, while “breakage-fusion-bridge” cycles are proposed as a plausible model for genomic amplifications.
Small palindromes (<1 kb) might behave differently from such large palindromes. Small palindromic sequences have the potential to form stem-loop structures by intrastrand base pairing in single-stranded DNA (Fig. 1(b)). As a consequence, they form specific tertiary structures, a single-stranded hairpin or a double-stranded cruciform.
We analyzed the tertiary structure of the cloned PATRR11 in vitro [23]. A plasmid containing the PATRR11 undergoes a conformational change, causing temperature-dependent retardation of electrophoretic mobility in standard agarose gels. Analysis using two-dimensional gel electrophoresis indicates that the mobility shift results from cruciform formation. S1 nuclease cuts the PATRR11-containing plasmid at the center of the palindromic structure that is a tip of putative cruciform resulting in a linear configuration. In contrast, T7 endonuclease cleaves the plasmid at the base of the cruciform. Further, anti-cruciform DNA antibody reduces the electrophoretic mobility of the PATRR11-containing fragment. Finally, using atomic force microscopy, we visualized cruciform extrusion of the PATRR11 plasmid directly.
To date, three translocation-related PATRRs have been identified (Table 1). They share common features; (1) several hundred base pairs with a very AT-rich palindrome; and (2) relatively non-AT-rich regions at both ends of the palindrome. Both of these features facilitate formation of a potential secondary structure. The AT-rich nature of the sequence promotes the initiation of strand unpairing, while the non-AT-rich regions at both ends contribute to the stability of the cruciform conformation. Taken together, the process that leads to the recurrent translocation has been proposed to involve the formation of cruciform structure as the putative mutagenic intermediates [6].
Whether the palindromic DNA adopts a cruciform conformation in vivo has remained enigmatic for years and is still somewhat controversial. In Escherichia coli, a substantial fraction of plasmids harboring palindromic sequence extrude a cruciform, as evidenced by in vivo crosslinking experiments by means of psoralen treatment and ultraviolet exposure [24]. Recently, one of the most common cancer-related translocations, the t(14;18), has been shown to be caused by the instability of non-B DNA structure [25,26]. The breakpoint region of the translocation manifests potential single-stranded DNA or triplex DNA. This observation implicates the presence of a non-B DNA structure in vivo. Further, this circumstantial evidence lends support to our proposal that such unusual DNA structures give rise to genomic instability leading to recurrent genomic rearrangements in somatic as well as meiotic cells.
4. PATRR is a hotspot of double-strand-breaks (DSBs)—studies in model organisms
Genomic dynamism of palindromic regions has been examined extensively for the past 20 years. As a consequence, instability of palindromic regions has been consistently demonstrated in many experimental organisms such as E. coli, Saccharomyces cerevisiae, and mouse.
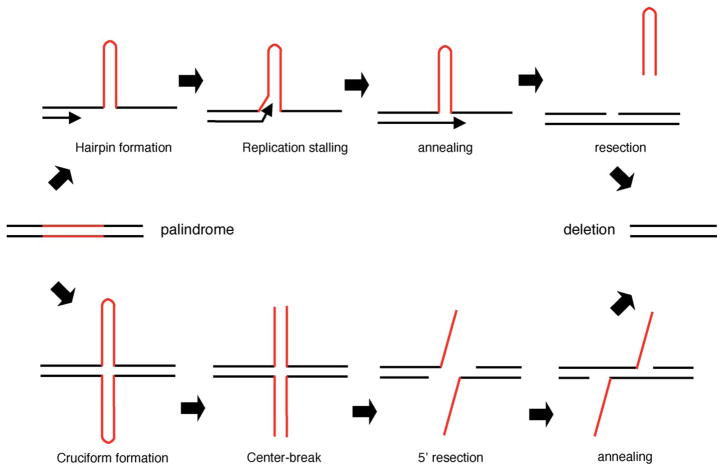
In E. coli, a palindromic region is either partially or completely deleted, whether it is transfected as a plasmid [27–29] or introduced into the bacterial genome [30]. It is easy to imagine that replication of the palindromic region is slowed or even stalled because of intra-strand base pairing, which might lead to formation of a hairpin structure. During synthesis, deletion is mainly mediated by slippage of the lagging strand because of the relative single-strandedness of the lagging strand template during replication. Formation of a free end followed by homology-dependent repair is the major pathway for producing a deletion (Fig. 2). Another important pathway is endonuclease cleavage of DNA secondary structure [29]. The key component of the pathway might be the SbcCD complex, which functions as an ATP-dependent double-strand DNA exonuclease and an ATP-independent single-strand DNA endonuclease. This enzyme complex, the orthologue of the mammalian Rad50/Mrell nuclease complex, is also known to cleave hairpin DNA. Indeed, the SbcCD-deficient strains are well documented as useful for the cloning of DNA fragments that are difficult to isolate because of their secondary structure [31].
Fig. 2.
Palindrome-mediated deletion. Upper panel shows replication-dependent pathway of the deletion. A hairpin structure is formed in the template strand because of intra-strand base pairing of the palindrome. Replication machinery may stall just before the palindrome, or may enter the palindrome and stall after progressing some distance. Since the deletion often utilizes short direct repeats within or flanking the palindrome, the palindromic region is partially or completely deleted. Lower panel depicts an alternative pathway, cruciform-mediated resection of the palindromic DNA. Red lines indicate palindromic regions.
In yeast, palindrome-induced recombination was first observed in frequent post-meiotic segregation in S. cerevisiae [32]. Since then, many researchers have investigated the role of palindromic sequences in chromosomal recombination using yeast. Now it has been generally acknowledged that palindromic sequences induce meiotic recombination by creating a hotspot for double-strand-breaks [33,34]. Similarly, there is accumulating evidence that palindromic sequences are also hotspots for mitotic recombination in S. cerevisiae. In vegetative yeast, the presence of a palindrome most often induces intrachromosomal recombination between direct repeats in the vicinity of the palindrome. The consequence is usually a deletion, suggestive of a mechanism similar to that seen in E. coli [35–37]. Likewise, frequent interchromosomal recombination has also been reported in meiotic and mitotic diploids in Schizosaccharomyces pombe [38,39].
In mammals, the nature of palindromic DNA has also been extensively studied using transgenic mice and/or their derivative cell lines. Several copies of a transgene that are accidentally integrated in a “head-to-head” or “tail-to-tail” orientation provide a good model for the analysis of palindromic DNA. Analysis of transgenic mice has demonstrated frequent deletions or insertions at such artificially created palindromic regions, indicative of substantial meiotic and mitotic instability [40,41]. More detailed studies using cell lines derived from such mice show that the palindromic region is primarily cleaved at its center [42]. This “center-break mechanism” has been proposed, as will be discussed in detail later.
Thus, the majority of experimental data that demonstrates palindromic instability is based on the presence of deletion. Indeed, several lines of evidence in humans also suggest that palindromic regions are susceptible to deletion. Alu-Alu inverted repeats are underrepresented relative to the number of direct repeats in the human genome database [43,44]. Further, PATRRs often manifest size polymorphisms in humans. We identified a rare short 205 bp PATRR11 in addition to the typical 445 bp PATRR11. Sequence analysis demonstrated that the short variant is likely a deleted version derived from the typical PATRR11 [7]. The PATRR17 also exhibits size polymorphism, with the shorter versions appearing to be derived from the longer PATRR17 by central deletion [17]. These deletions always include the center of the PATRR, converting a symmetric palindrome into an asymmetric variant. Symmetric palindromes more readily form a hairpin/cruciform structure, which might be more predisposed to deletion to adopt a stable asymmetric form.
Taken together, palindromic DNA sequences are susceptible to strand breakage in many organisms. However, palindrome-mediated recurrent chromosomal translocation seems to occur exclusively in humans. Perhaps substantial differences reside in the cleavage or repair processes in the different organisms, resulting in different consequences; translocation or deletion. It is possible that different mechanisms surrounding palindrome stability operate between prokaryotes and higher organisms, between mitosis and meiosis, and between integrated chromatin and episomal DNA.
5. High frequency of de novo translocations in sperm from normal healthy males
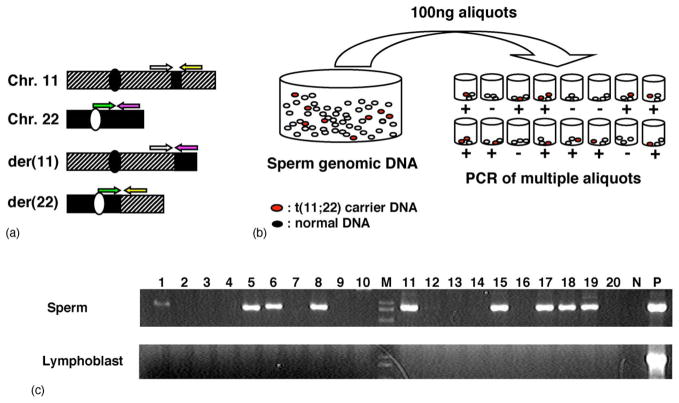
Getting back to the t(11;22) translocation, sequence derived from junction fragments of the der(11) and the der(22) allowed us to establish a t(11;22)-specific PCR assay system (Fig. 3(a)). Using translocation-specific PCR, we examined the frequency of de novo translocations in sperm DNA samples obtained from normal, healthy male volunteers [45]. PCR was performed with conditions that would allow for the detection of a single molecule of target DNA. We amplified multiple aliquots from each sperm sample (Fig. 3(b)). When we amplified multiple individual aliquots of sperm DNA each containing 33,000 haploids (100 ng), we found translocation-specific PCR products in a substantial number of reactions. The presence of both positive and negative PCR reactions clearly indicates the de novo origin of the translocation (Fig. 3(c)). Translocation-specific PCR products were never found in DNA from blood or cheek swabs from the volunteers, which excludes the possibility that positive PCR reactions in sperm were caused by contamination or PCR artifacts.
Fig. 3.
High frequency of de novo t(11;22) in sperm from normal males. (a) Translocation-specific PCR. Hatched boxes indicate chromosome 11, whereas dotted boxes depict chromosome 22. Centromeres are depicted as circles, while the PATRRs are indicated as filled boxes. The four PCR primers used for translocation-specific PCR (arrows) can be distinguished by the different colors. (b) Strategy for estimation of translocation frequency by PCR. Genomic DNA was extracted from sperm samples. Translocation-specific PCR was performed using multiple batches of template DNA. The translocation frequency was calculated using the equation, q = 1 − (1 − p)1/n; n = number of haploid genomes per aliquot, p = the probability that an aliquot contains a translocation product, and q = the probability that one randomly selected haploid genome in a given aliquot sustained a translocation. (c) The results of PCR. Upper panel shows the results from sperm DNA, whereas the lower panel indicates those from lymphoblast DNA. Lane M, size marker; lane N, negative control; lane P, genomic DNA from a t(11;22) carrier serving as a positive control.
The frequency of de novo translocation events was calculated based on the presence of positive PCR reactions. We counted the number of positive PCR reactions per total reactions. The frequency was calculated on the basis that the probability of observing a positive PCR reaction corresponds to the total sum of a binomial series of the translocation frequency calculated as previously described [45]. The estimated frequency was ~10−5, which is unexpectedly high. The frequency of the der(11) and the der(22) was approximately equal, suggesting that the de novo translocation occurs as a reciprocal rearrangement. This observation agrees with the fact that majority of patients with supernumerary-der(22) syndrome arise as offspring of a balanced translocation carrier, not as a de novo event [1,2].
De novo occurrence of the translocation was never detected in cultured somatic cells like lymphoblasts, fibroblasts nor in diverse human cell lines, but only in sperm.
This finding highlights the novel properties of the PATRR in translocation generation. It is not unreasonable to imagine that palindrome-mediated deletions arise in somatic cells, since the proposed mechanism involves the formation of secondary structure during DNA replication. But sperm-specificity implicates a replication-independent mechanism for the translocation. Recently, a yeast strain with low DNA polymerase activity has been shown to cause chromosomal rearrangements, whose breakpoints include a palindromic region [46]. However, this does not provide a good model for the t(11;22) because of the mitotic origin of the yeast translocation. Thus, it could be hypothesized; (1) that the secondary structure required for the t(11;22) is specifically formed in germ cells; or (2) that a protein that cleaves the secondary structure is expressed specifically in germ cells.
The translocation likely depends on the ability of the PATRRs to fold into unusual secondary structures when their DNA strands dissociate. Therefore, DNA breakages that occur normally during or after meiosis are likely to provide the initiating events in strand dissociation and formation of secondary structure. In mammalian spermatogenesis, there are two physiological DNA breakages. (1) A substantial number of DSBs occur as an initiating step for meiotic recombination. Spo1l, a DSB catalytic enzyme, might mainly functions in this step. (2) During meiotic recombination, Holliday junctions are formed as intermediates of homologous recombination. When a Holliday junction finally resolves, DNA cleavage should take place to generate a crossover or non-crossover. Further, the four-way junction of a cruciform DNA structure is analogous to a Holliday junction. Either of these steps provides a good “candidate process” for the generation of the DSBs leading to the translocation.
It is still not known for certain whether the translocation is meiosis-specific or male germ cell specific. Despite the observed high frequency of de novo translocations in sperm, we cannot analyze female germ cell using a similar strategy. Indeed, segregation analysis in our patient population demonstrated only one de novo balanced t(11;22) carrier. This event was of paternal origin (H.K. and B.S.E., unpublished data). The DNA breakage might occur again during sperm elongation as DNA is packaged into chromatin. Topological constraints applied to the DNA during chromatin packaging is likely to partially unwind the DNA duplex and exacerbate secondary structure within the PATRR, resulting in strand breaks. For example, triplet-repeat expansion has been demonstrated to be of post-meiotic origin [47]. A plausible explanation is that chromatin packaging may have a role in facilitating strand breaks within the secondary structure of the PATRR. Alternatively, the kinetic analysis of the PATRR-containing plasmid indicated that the PATRR favors to extrusion of the cruciform at a slightly lower temperature than body temperature [23]. Such temperature-dependent cruciform formation of the PATRR might imply another hypothesis that the relatively low temperature of the testis might contribute to the sperm-specific origin of the translocation. Thus, there are still numerous factors surrounding the mechanism of translocation formation that remain to be elucidated.
6. Proposed mechanism
The recurrent t(11;22) represents a good model for studying translocations in humans. To gain a better understanding of DSB-repair mechanisms specific for palindromes is one of the most important goals of t(11;22) research. We have assumed the mechanism based on the sequence data of breakpoint derived from analysis of the junction fragments. Previous studies indicate that the breakpoints for the human t(11;22) appear to be located at the center of the palindrome when deduced by examination of junction fragment sequence in comparison with normal PATRR11 sequence [7,9,10]. Therefore, as far as DNA breakage is concerned, a DSB is likely to occur at the center of the palindrome. In terms of the repair, simple homologous recombination does not seem to contribute to the translocation, since PATRR11 and PATRR22 do not share substantial homology except for their AT-richness. We still do not know exactly what takes place since we do not have complete sequence information for the normal PATRR22. However, the sequence inferred from the two junction fragments indicates that the repair requires few or no nucleotide number of microhomology (H.K., unpublished data). The presence of a small deletion at the junction is reminiscent of the non-homologous end joining (NHEJ) pathway [7].
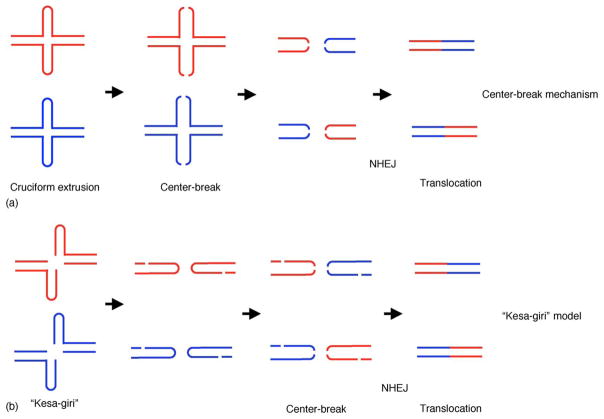
Despite the fact that whether there is a cruciform structure at the PATRR in vivo is still the subject of much discussion/debate, it is likely that the translocation is frequently preceded by a DSB specific for the PATRR, followed by repair through the NHEJ pathway. Although there is no mammalian model for palindrome-mediated translocation, the study of transgenic mice and cell lines derived from them demonstrates that deletion often resolves the palindromic structure by a “center-break mechanism” (Fig. 4(a)) [41,42]. Sequence data of the junction fragments also lend support to a “center-break mechanism” for the translocation. Indeed, polymorphism of human PATRRs has been shown to occur by deletion at the central region, suggesting that the center of the PATRR is somewhat fragile [7,17].
Fig. 4.
Model for the mechanism of PATRR-mediated translocation. (a) The PATRR might be cleaved at the tip of the hairpin structure, which corresponds to the center of the palindrome (“center-break mechanism”). (b) Alternatively, the PATRR might be cleaved diagonally at the base of the cruciform (“kesa-giri” refers to the way a Japanese samurai kills an enemy with his sword). Two PATRRs, located on different chromosomes, are indicated by red and blue lines, respectively.
In contrast, breakage at the palindromic region could take place by the diagonal cleavage of a cruciform configuration (Fig. 4(b)). An extruded cruciform structure resembles the Holliday junction that forms during homologous recombination and potentially could be a target for Holliday junction resolvase or another four-way junction-cutting enzyme. Although a Holliday junction resolvase of T7 phage, T7 endonuclease, cleaves cruciform DNA in vitro [23], such an enzyme has not yet been identified in higher organisms like yeast or mammals [48]. However, cleavage of a palindromic DNA in this manner was demonstrated by the presence of a hairpin-capped DSB for Alu inverted repeats experimentally introduced in yeast [49].
Exactly what happens as a result of a DSB at the PATRR is also still a subject of debate. Products produced by a “center-break mechanism” could directly provide substrate for the NHEJ pathway. On the other hand, a diagonal cleavage hypothesis appears to conflict with sequence data derived from the junction fragments, but it cannot be totally excluded. An integrative hypothesis for the disparity might be that the second break at the palindromic center occurs after diagonal cleavage of the putative cruciform structure (Fig. 4(b)). Such diagonal cleavage might produce a structure similar to the coding ends generated by somatic rearrangement of immunoglobulin and T-cell receptor genes, which might be a target for NHEJ at the two hairpin tips [50]. The resulting junction fragments would appear to be the product of a “center-break mechanism”. Additional studies will be required to shed light on the precise mechanism of palindrome-mediated translocations in humans.
7. Future direction
Another question that still remains unsolved is why the partner of a PATRR-mediated translocation is preferentially a PATRR located on another chromosome. Two hypotheses have been entertained; (1) that DSBs occur at strikingly high frequency at the PATRR regions; and (2) that an as yet unknown mechanism dictates the PATRR-specific translocation. The first possibility is based on the previous finding that presence of two DSBs on different chromosomes is sufficient to create a translocation [51]. Based on this hypothesis, the DSBs arise most frequently at the PATRR22, and at the PATRR11 as the second most frequent site. One reported case with a t(X;22) demonstrates that the non-palindromic breakpoint may represent a good example of an accidental DSB that was captured by the PATRR22 [14]. On the other hand, the concept of specific chromosomal territories in the interphase nucleus might provide a potential inducer for the site-specific recurrent translocation. The proximity of chromosomes 11 and 22 has been reported in oogenesis [52]. The latter interesting hypothesis that there is a palindrome-specific mechanism that dictates the translocation requires an explanation for how two DSBs proceed all the way to joining. One conceivable explanation is that a pathway like NHEJ during immunoglobulin gene rearrangement might rejoin the two hairpin tips specifically, as was mentioned above [50].
To answer all of these questions, it will be necessary to establish a model system for t(11;22) research. A yeast model system can provide a means to examine the meiotic behavior of the PATRR and provide an outline for the translocation pathway. Similar experiments using mammalian cell lines might lend additional support to elucidation of the mechanism for human palindrome-mediated translocations. Since the mouse genome does not possess PATRR-like sequences, there is little doubt in postulating that introduction of the two copies of the PATRRs will give rise to palindrome-mediated translocation in mouse.
However, establishment of PATRR-transgenic mice or knock-in mice still remains a significant challenge, since it is evident that the PATRR22 plays a critical role in facilitating the translocation but it has not been cloned yet. Several difficulties have been encountered in PATRR research, such as difficulties with PCR amplification, palindrome sequencing, and cloning. We recently made significant technical progress and overcame many of these obstacles to perform detailed investigation of the PATRR17 [17]. Further characterization of the dynamics of the PATRRs should aid in the elucidation of the complex biology of palindrome-mediated translocations in the near future.
Acknowledgments
The author wishes to thank Dr. T.H. Shaikh for providing materials, Dr. M. Taniguchi for helpful discussion, and Miss H. Kowa, K. Nagaoka, T. Mori, and E. Hosoba for technical assistance. These studies were supported by a grant-in-aid for Scientific Research, Genome, and 21st Century COE program from the Ministry of Education, Culture, Sports, Science, and Technology of Japan to H.K. (16390102). Support was also provided by grant from the National Institutes of Health to B.S.E. (CA39926).
Footnotes
Publisher's Disclaimer: This article was originally published in a journal published by Elsevier, and the attached copy is provided by Elsevier for the author’s benefit and for the benefit of the author’s institution, for non-commercial research and educational use including without limitation use in instruction at your institution, sending it to specific colleagues that you know, and providing a copy to your institution’s administrator.
References
- 1.Zackai EH, Emanuel BS. Site-specific reciprocal translocation, t(11;22)(q23;q11), in several unrelated families with 3:1 meiotic disjunction. Am J Med Genet. 1980;7:507–521. doi: 10.1002/ajmg.1320070412. [DOI] [PubMed] [Google Scholar]
- 2.Shaikh TH, Budarf ML, Celle L, Zackai EH, Emanuel BS. Clustered 11q23 and 22q11 breakpoints and 3:1 meiotic malsegregation in multiple unrelated t(11;22) families. Am J Hum Genet. 1999;65:1595–1607. doi: 10.1086/302666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Funke B, Edelmann L, McCain N, Pandita RK, Ferreira J, Merscher S, Zohouri M, Cannizzaro L, Shanske A, Morrow BE. Der(22) syndrome and velo-cardio-facial syndrome/Di George syndrome share a 1.5-Mb region of overlap on chromosome 22q11. Am J Hum Genet. 1999;64:747–758. doi: 10.1086/302284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Edelmann L, Spited E, McCain N, Goldberg R, Pandita RK, Duong S, Fox J, Blumenthal D, Lalani SR, Shaffer LG, Morrow BE. A common breakpoint on 11q23 in carriers of the constitutional t(11;22) translocation. Am J Hum Genet. 1999;65:1608–1616. doi: 10.1086/302689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tapia-Paez I, O’Brien KP, Kost-Alimova M, Sahlen S, Kedra D, Bruder CE, Andersson B, Roe BA, Hu P, Imreh S, Blennow E, Dumanski JP. Fine mapping of the constitutional translocation t(11;22) (q23;q11) Hum Genet. 2000;106:506–516. doi: 10.1007/s004390000287. [DOI] [PubMed] [Google Scholar]
- 6.Kurahashi H, Shaikh TH, Hu P, Roe BA, Emanuel BS, Budarf ML. Regions of genomic instability on 22q11 and 11q23 as the etiology for the recurrent constitutional t(11;22) Hum Mol Genet. 2000;9:1665–1670. doi: 10.1093/hmg/9.11.1665. [DOI] [PubMed] [Google Scholar]
- 7.Kurahashi H, Emanuel BS. Long AT-rich palindromes and the constitutional t(11;22) breakpoint. Hum Mol Genet. 2001;10:2605–2617. doi: 10.1093/hmg/10.23.2605. [DOI] [PubMed] [Google Scholar]
- 8.Kurahashi H, Shaikh TH, Zackai EH, Celle L, Driscoll DA, Budarf ML, Emanuel BS. Tightly clustered 11q23 and 22q11 breakpoints permit PCR-based detection of the recurrent constitutional t(11;22) Am J Hum Genet. 2000;67:763–768. doi: 10.1086/303054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Edelmann L, Spited E, Koren K, Pulijaal V, Bialer MG, Shanske A, Goldberg R, Morrow BE. AT-rich palindromes mediate the constitutional t(11;22) translocation. Am J Hum Genet. 2001;68:1–13. doi: 10.1086/316952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tapia-Paez I, Kost-Alimova M, Hu P, Roe BA, Blennow E, Fedorova L, Imreh S, Dumanski JP. The position of t(11;22)(q23;q11) constitutional translocation breakpoint is conserved among its carriers. Hum Genet. 2001;109:167–177. doi: 10.1007/s004390100560. [DOI] [PubMed] [Google Scholar]
- 11.Budarf ML, Eckman B, Michaud D, McDonald T, Gavigan S, Buetow KH, Tatsumura Y, Liu Z, Hilliard C, Driscoll D, Goldmuntz E, Meese E, Zwarthoff EC, Williams S, McDermid H, Dumanski JP, Biegel J, Bell CJ, Emanuel BS. Regional localization of over 300 loci on human chromosome 22 using a somatic cell hybrid mapping panel. Genomics. 1996;35:275–288. doi: 10.1006/geno.1996.0358. [DOI] [PubMed] [Google Scholar]
- 12.Kehrer-Sawatzki H, Haussler J, Krone W, Bode H, Jenne DE, Mehnert KU, Tummers U, Assum G. The second case of a t(17;22) in a family with neurofibromatosis type 1: sequence analysis of the breakpoint regions. Hum Genet. 1997;99:237–247. doi: 10.1007/s004390050346. [DOI] [PubMed] [Google Scholar]
- 13.Rhodes CH, Call KM, Budarf ML, Barnoski BL, Bell CJ, Emanuel BS, Bigner SH, Park JP, Mohandas TK. Molecular studies of an ependymoma-associated constitutional t(1;22)(p22;q11.2) Cytogenet Cell Genet. 1997;78:247–252. doi: 10.1159/000134667. [DOI] [PubMed] [Google Scholar]
- 14.Debeer P, Mols R, Huysmans C, Devriendt K, Van de Ven WJ, Fryns JP. Involvement of a palindromic chromosome 22-specific low-copy repeat in a constitutional t(X; 22)(q27;q11) Clin Genet. 2002;62:410–414. doi: 10.1034/j.1399-0004.2002.620510.x. [DOI] [PubMed] [Google Scholar]
- 15.Spiteri E, Babcock M, Kashork CD, Wakui K, Gogineni S, Lewis DA, Williams KM, Minoshima S, Sasaki T, Shimizu N, Potocki L, Pulijaal V, Shanske A, Shaffer LG, Morrow BE. Frequent translocations occur between low copy repeats on chromosome 22q11.2 (LCR22s) and telomeric bands of partner chromosomes. Hum Mol Genet. 2003;12:1823–1837. doi: 10.1093/hmg/ddg203. [DOI] [PubMed] [Google Scholar]
- 16.Kurahashi H, Shaikh T, Takata M, Toda T, Emanuel BS. The constitutional t(17;22): another translocation mediated by palindromic AT-rich repeats. Am J Hum Genet. 2003;72:733–738. doi: 10.1086/368062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Inagaki H, Ohye T, Kogo H, Yamada K, Kowa H, Shaikh TH, Emanuel BS, Kurahashi H. A palindromic AT-rich repeat in the NF1 gene is hypervariable in humans and evolutionarily conserved among primates. Hum Mutat. 2005;26:332–342. doi: 10.1002/humu.20228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nimmakayalu MA, Gotter AL, Shaikh TH, Emanuel BS. A novel sequence-based approach to localize translocation breakpoints identifies the molecular basis of a t(4;22) Hum Mol Genet. 2003;12:2817–2825. doi: 10.1093/hmg/ddg301. [DOI] [PubMed] [Google Scholar]
- 19.Gotter AL, Shaikh TH, Budarf ML, Rhodes CH, Emanuel BS. A palindrome-mediated mechanism distinguishes translocations involving LCR-B of chromosome 22q11.2. Hum Mol Genet. 2004;13:103–115. doi: 10.1093/hmg/ddh004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kuroda-Kawaguchi T, Skaletsky H, Brown LG, Minx PJ, Cordum HS, Waterston RH, Wilson RK, Silber S, Oates R, Rozen S, Page DC. The AZFc region of the Y chromosome features massive palindromes and uniform recurrent deletions in infertile men. Nat Genet. 2001;29:279–286. doi: 10.1038/ng757. [DOI] [PubMed] [Google Scholar]
- 21.Barbouti A, Stankiewicz P, Nusbaum C, Cuomo C, Cook A, Hoglund M, Johansson B, Hagemeijer A, Park SS, Mitelman F, Lupski JR, Fioretos T. The breakpoint region of the most common isochromosome, i(17q), in human neoplasia is characterized by a complex genomic architecture with large, palindromic, low-copy repeats. Am J Hum Genet. 2004;74:1–10. doi: 10.1086/380648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tanaka H, Bergstrom DA, Yao MC, Tapscott SJ. Widespread and nonrandom distribution of DNA palindromes in cancer cells provides a structural platform for subsequent gene amplification. Nat Genet. 2005;37:320–327. doi: 10.1038/ng1515. [DOI] [PubMed] [Google Scholar]
- 23.Kurahashi H, Inagaki H, Yamada K, Ohye T, Taniguchi M, Emanuel BS, Toda T. Cruciform DNA structure underlies the etiology for palindrome-mediated human chromosomal translocations. J Biol Chem. 2004;279:35377–35383. doi: 10.1074/jbc.M400354200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zheng GX, Kochel T, Hoepfner RW, Timmons SE, Sinden RR. Torsionally tuned cruciform and Z-DNA probes for measuring unrestrained supercoiling at specific sites in DNA of living cells. J Mol Biol. 1991;221:107–122. doi: 10.1016/0022-2836(91)80208-c. [DOI] [PubMed] [Google Scholar]
- 25.Raghavan SC, Swanson PC, Wu X, Hsieh CL, Lieber MR. A non-B-DNA structure at the Bcl-2 major breakpoint region is cleaved by the RAG complex. Nature. 2004;428:88–93. doi: 10.1038/nature02355. [DOI] [PubMed] [Google Scholar]
- 26.Raghavan SC, Chastain P, Lee JS, Hegde BG, Houston S, Langen R, Hsieh CL, Haworth IS, Lieber MR. Evidence for a triplex DNA conformation at the bcl-2 major breakpoint region of the t(14;18) translocation. J Biol Chem. 2005;280:22749–22760. doi: 10.1074/jbc.M502952200. [DOI] [PubMed] [Google Scholar]
- 27.Trinh TQ, Sinden RR. Preferential DNA secondary structure mutagenesis in the lagging strand of replication in E. coli. Nature. 1991;352:544–547. doi: 10.1038/352544a0. [DOI] [PubMed] [Google Scholar]
- 28.Leach DR. Long DNA palindromes, cruciform structures, genetic instability and secondary structure repair. Bioessays. 1994;16:893–900. doi: 10.1002/bies.950161207. [DOI] [PubMed] [Google Scholar]
- 29.Bzymek M, Lovett ST. Evidence for two mechanisms of palindrome-stimulated deletion in Escherichia coli: single-strand annealing and replication slipped mispairing. Genetics. 2001;158:527–540. doi: 10.1093/genetics/158.2.527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leach DR, Okely EA, Pinder DJ. Repair by recombination of DNA containing a palindromic sequence. Mol Microbiol. 1997;26:597–606. doi: 10.1046/j.1365-2958.1997.6071957.x. [DOI] [PubMed] [Google Scholar]
- 31.Doherty JP, Lindeman R, Trent RJ, Graham MW, Woodcock DM. Escherichia coli host strains SURE and SRB fail to preserve a palindrome cloned in lambda phage: improved alternate host strains. Gene. 1993;124:29–35. doi: 10.1016/0378-1119(93)90758-u. [DOI] [PubMed] [Google Scholar]
- 32.Nag DK, White MA, Petes TD. Palindromic sequences in heteroduplex DNA inhibit mismatch repair in yeast. Nature. 1989;340:318–320. doi: 10.1038/340318a0. [DOI] [PubMed] [Google Scholar]
- 33.Nag DK, Kurst A. A 140-bp-long palindromic sequence induces double-strand breaks during meiosis in the yeast Saccharomyces cerevisiae. Genetics. 1997;146:835–847. doi: 10.1093/genetics/146.3.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nasar F, Jankowski C, Nag DK. Long palindromic sequences induce double-strand breaks during meiosis in yeast. Mol Cell Biol. 2000;20:3449–3458. doi: 10.1128/mcb.20.10.3449-3458.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gordenin DA, Lobachev KS, Degtyareva NP, Malkova AL, Perkins E, Resnick MA. Inverted DNA repeats: a source of eukaryotic genomic instability. Mol Cell Biol. 1993;13:5315–5322. doi: 10.1128/mcb.13.9.5315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tran H, Degtyareva N, Gordenin D, Resnick MA. Altered replication and inverted repeats induce mismatch repair-independent recombination between highly diverged DNAs in yeast. Mol Cell Biol. 1997;17:1027–1036. doi: 10.1128/mcb.17.2.1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lobachev KS, Shor BM, Tran HT, Taylor W, Keen JD, Resnick MA, Gordenin DA. Factors affecting inverted repeat stimulation of recombination and deletion in Saccharomyces cerevisiae. Genetics. 1998;148:1507–1524. doi: 10.1093/genetics/148.4.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Farah JA, Hartsuiker E, Mizuno K, Ohta K, Smith GR. A 160-bp palindrome is a Rad50. Rad32-dependent mitotic recombination hotspot in Schizosaccharomyces pombe. Genetics. 2002;161:461–468. doi: 10.1093/genetics/161.1.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Farah JA, Cromie G, Steiner WW, Smith GR. A novel recombination pathway initiated by the MRN complex eliminates palindromes during meiosis in Schizosaccharomyces pombe. Genetics. 2005;169:1261–1274. doi: 10.1534/genetics.104.037515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Collide A, Drew J, Penberth J, Bois P, Luckett J, Scaerou F, Jeffreys A, Reik W. Instability of long inverted repeats within mouse transgenes. EMBO J. 1996;15:1163–1171. [PMC free article] [PubMed] [Google Scholar]
- 41.Akgun E, Zahn J, Baumes S, Brown G, Liang F, Romanienko PJ, Lewis S, Jasin M. Palindrome resolution and recombination in the mammalian germ line. Mol Cell Biol. 1997;17:5559–5570. doi: 10.1128/mcb.17.9.5559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cunningham LA, Cote AG, Cam-Ozdemir C, Lewis SM. Rapid, stabilizing palindrome rearrangements in somatic cells by the center-break mechanism. Mol Cell Biol. 2003;23:8740–8750. doi: 10.1128/MCB.23.23.8740-8750.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lobachev KS, Stenger JE, Kozyreva OG, Jurka J, Gordenin DA, Resnick MA. Inverted Alu repeats unstable in yeast are excluded from the human genome. EMBO J. 2000;19:3822–3830. doi: 10.1093/emboj/19.14.3822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stenger JE, Lobachev KS, Gordenin D, Darden TA, Jurka J, Resnick MA. Biased distribution of inverted and direct Alus in the human genome: implications for insertion, exclusion, and genome stability. Genome Res. 2001;11:12–27. doi: 10.1101/gr.158801. [DOI] [PubMed] [Google Scholar]
- 45.Kurahashi H, Emanuel BS. Unexpectedly high rate of de novo constitutional t(11;22) translocations in sperm from normal males. Nat Genet. 2001;29:139–140. doi: 10.1038/ng1001-139. [DOI] [PubMed] [Google Scholar]
- 46.Lemoine FJ, Degtyareva NP, Lobachev K, Petes TD. Chromosomal translocations in yeast induced by low levels of DNA polymerase: a model for chromosome fragile sites. Cell. 2005;120:587–598. doi: 10.1016/j.cell.2004.12.039. [DOI] [PubMed] [Google Scholar]
- 47.Kovtun IV, McMurray CT. Trinucleotide expansion in haploid germ cells by gap repair. Nat Genet. 2001;27:407–411. doi: 10.1038/86906. [DOI] [PubMed] [Google Scholar]
- 48.Lobachev KS, Gordenin DA, Resnick MA. The Mrell complex is required for repair of hairpin-capped double-strand breaks and prevention of chromosome rearrangements. Cell. 2002;108:183–193. doi: 10.1016/s0092-8674(02)00614-1. [DOI] [PubMed] [Google Scholar]
- 49.Elborough KM, West SC. Resolution of synthetic Holliday junctions in DNA by an endonuclease activity from calf thymus. EMBO J. 1990;9:2931–2936. doi: 10.1002/j.1460-2075.1990.tb07484.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gellert M. V(D)J recombination: RAG proteins, repair factors, and regulation. Annu Rev Biochem. 2002;71:101–132. doi: 10.1146/annurev.biochem.71.090501.150203. [DOI] [PubMed] [Google Scholar]
- 51.Richardson C, Jasin M. Frequent chromosomal translocations induced by DNA double-strand breaks. Nature. 2000;405:697–700. doi: 10.1038/35015097. [DOI] [PubMed] [Google Scholar]
- 52.Emanuel BS, Conforto D, Cohen MM. The recurrent t(11;22): FISH analysis of 11q23 and 22q11 in oogenesis. Am J Hum Genet. 2001;69:216. [Google Scholar]