Abstract
The purpose of this review is to summarize the evidence that can be used to reconstruct the etiology of human cancers from mutations found in tumors. Mutational spectra of the tumor suppressor gene p53 (TP53) are tumor-specific. In several cases, these mutational spectra can be linked to exogenous carcinogens, most notably for sunlight-associated skin cancers, tobacco-associated lung cancers, and aristolochic acid-related urothelial tumors. In the TP53 gene, methylated CpG dinucleotides are sequences selectively targeted by endogenous and exogenous mutagenic processes. Recent high-throughput sequencing efforts analyzing a large number of genes in cancer genomes have so far, for the most part, produced mutational spectra similar to those in TP53 but have unveiled a previously unrecognized common G to C transversion mutation signature at GpA dinucleotides in breast cancers and several other cancers. Unraveling the origin of these G to C mutations will be of importance for understanding cancer etiology.
Introduction
Human cancer genomes contain numerous genetic and epigenetic aberrations (Chin and Gray 2008). Instability at the chromosome level manifested by chromosome deletions and rearrangements has been recognized as a signature of cancer and is referred to as aneuploidy (Rajagopalan and Lengauer 2004). Instability at the level of chromatin involves many processes that have the potential to alter gene expression patterns in cancer cells. These chromatin changes are also described as epigenetic events that, for example, lead to rearrangements of histone modifications and DNA methylation patterns often affecting specific genes or entire DNA sequence classes such as repetitive elements. Epigenetic aberrations are common in most human cancers and can affect hundreds if not thousands of genes (Rauch et al. 2008). Collectively, chromosomal, mutational, and epigenetic aberrations in cancer genomes are likely to contribute to all known hallmarks of cancer including, but not limited to, self-sufficiency in growth signals, evasion of apoptosis, unlimited replicative potential, and tissue invasion or metastasis (Hanahan and Weinberg 2000). It is generally agreed that malignant progression requires the mutational inactivation or activation, or epigenetic inactivation or activation, of only a limited number of genes but the exact number of such critical genes is not known and may be cancer type-specific.
This review deals with the instability that is observed at the nucleotide sequence level in human cancer genomes. There is an ongoing debate as to whether all or most human cancers exhibit a mutator phenotype (Bodmer et al. 2008; Loeb et al. 2008). The mutator phenotype is hypothesized to be the driving force for the accumulation of large numbers of mutations in tumors enabling the selection of tumor promoting events. There are clear examples for the existence of a mutator phenotype in certain cancers. Germline or somatic mutations of mismatch repair genes, e.g., MLH1, MSH1, MSH6, etc. are linked to a massive increase in microsatellite and other sequence alterations, and these mutations are found in hereditary forms of colorectal cancer (Edelmann and Edelmann 2004; Lynch and de la Chapelle 2003). Others have, however, argued that genetic instability is not usually required for tumor development (Bodmer et al. 2008). Initial large scale sequencing studies of human mismatch repair-stable cancer genomes failed to uncover clear evidence for a large number of sequence alterations detectable in a tumor clone (Wang et al. 2002) although subsequent studies have shown the presence of a substantial number of mutations in individual tumor samples.
The intense efforts put forward by large-scale re-sequencing approaches of cancer genomes have resulted in the identification of only a small number of genes that are commonly mutated in human tumors (Ding et al. 2008; Greenman et al. 2007; Jones et al. 2008; Sjoblom et al. 2006; Wood et al. 2007). One important discovery was that the BRAF gene, which encodes a serine/threonine kinase in the RAS signaling pathway, harbors somatic mutations in ~60% of malignant melanomas (Davies et al. 2002). Large-scale re-sequencing efforts also uncovered mutations in the PIK3CA34 gene (encoding the catalytic subunit of phosphatidylinositol-3-OH kinase) and AKT1 gene (encoding a serine/threonine kinase) in several types of cancer (Carpten et al. 2007; Samuels et al. 2004). In addition, mutations in the growth factor receptor genes ERBB2 and EGFR have been found frequently in non-small-cell lung cancers, in particular adenocarcinomas (Sharma et al. 2007). However, the relatively small number of genes that are commonly mutated in human cancers came as a surprise outcome of the large-scale sequencing efforts reported to date. These findings suggest that there are only a few genes that can effectively promote cancer formation in humans when mutated and, accordingly, are repeatedly and frequently mutated in multiple samples of the same tumor type and across different tumor types. Among these genes are the RAS genes and the TP53 gene. In this review, we will discuss some of the potential endogenous and exogenous origins of RAS and TP53 mutations. We will also discuss properties of mutations found in protein kinase genes and other genes analyzed by large-scale sequencing of human tumors.
Investigating human cancer etiology
Epidemiologic studies have the potential to identify suspect carcinogens, which may etiologically be involved in human cancers (Vineis and Perera 2007). This objective is often achieved by demonstration of a link between exposure to carcinogens and the incidence of cancer in defined human populations (Mossman et al. 2004). These observational studies examine correlations between cancer development and carcinogen exposure in humans with known exposure to cancer-causing agents (Besaratinia and Pfeifer 2006). In addition to the unavoidable exposures to food-, water- and air-borne carcinogens, humans are also exposed to specific carcinogenic agents, e.g., in occupational settings, or due to medicinal or life-style choices, e.g., tobacco smoking, alcohol drinking, etc (Carbone et al. 2004; Luch 2005). Not only can the specificity of carcinogen exposure determine the type of human cancers, but it may also influence the genetic and/or epigenetic alterations that are unique for certain types of human cancer (Besaratinia and Pfeifer 2006). For example, sunlight ultraviolet (UV) irradiation is a key determinant of non-melanoma skin cancers, and the presumed culprit of C to T transition mutations at hotspot dipyrimidines in the TP53 gene, which are characteristics of non-melanoma skin tumors (Pfeifer et al. 2005). Tobacco smoke carcinogens are linked to G to T mutations in lung cancer arising in smokers (Hainaut and Pfeifer 2001). Workplace exposure to vinyl chloride can be associated with liver tumors showing preferential mutation at A:T base pairs in TP53 (Hollstein et al. 1994), and Chinese herb nephropathy is characterized by A to T mutations in urothelial tumors (Grollman et al. 2007).
The correlative nature of human cancer development and carcinogen exposure can be used for causality inference if the observed correlation can be recapitulated experimentally (Hussain and Harris 1998; Hussain et al. 2000; Olivier et al. 2004; Thilly 1990). Obviously, experimental exposure of humans to carcinogenic agents is unethical. Thus, only in vitro or in vivo model systems can be used for inferring causality once epidemiologic studies have established a relationship between human cancer development and exposure to certain carcinogen(s). The available model systems utilize various strategies to reproduce the epidemiologic findings on carcinogen-specific genetic and/or epigenetic changes in vitro and/or in vivo (Besaratinia and Pfeifer 2006).
RAS gene mutations
Nucleotide sequence changes in human cancers were first reported in the 1980s and they affected specific cellular homologues of viral oncogenes, the RAS genes HRAS, KRAS, and NRAS (Marshall et al. 1984; Parada et al. 1982; Sukumar et al. 1983; Taparowsky et al. 1982). RAS gene mutation has been described in several human cancers including colon, lung, pancreatic, thyroid cancers, melanomas and several other types of tumor (Barbacid 1987; Bos 1989). Mutations at codon 12 of KRAS are the most common mutations found among the three RAS genes. It was soon recognized that the type of RAS mutation was tumor-specific, i.e. the KRAS gene was frequently undergoing transition mutations in colorectal cancers (e.g., GGT → GAT at codon 12) but were predominantly transversion mutations (e.g., GGT → GTT) in lung cancers. Induction of tumors in animal models was shown to be accompanied by mutation of the HRAS gene (Sukumar et al. 1983). Also, notably, when RAS gene-containing plasmids were modified in vitro with chemical carcinogens, an oncogene was produced that could transform NIH3T3 cells (Marshall et al. 1984). The nature of the oncogenic mutation depended on the mutational specificity of the carcinogen (Quintanilla et al. 1986; Zarbl et al. 1985). These experiments established an important paradigm showing that tumor initiation results from mutations arising from the binding of ultimate carcinogens to DNA. Due to the selection of point mutations producing an activated oncogene by mutation of only a few amino acids in the RAS genes (i.e. codons 12, 13, 61, and 146) only a limited number of sequence changes can be observed. Despite these limitations, however, the types of mutations in the KRAS gene are different among various types of cancer. Smoking-associated lung cancers, but to a much lesser extent lung cancers in nonsmokers, often contain activating G to T transversion mutations at codon 12 of KRAS (Husgafvel-Pursiainen et al. 1993). Convincing evidence from both epidemiologic and experimental studies has identified tobacco smoke carcinogens as initiators of lung cancer (Besaratinia and Pfeifer 2008; Hecht 1999; Pfeifer et al. 2002). It has been unclear why only one codon in one particular RAS gene is undergoing mutations in lung tumors. In an elegant study, Tang and coworkers showed that codons 12 and 14 of the KRAS gene were hotspots for DNA adduct formation by the activated tobacco smoke carcinogen benzo[a]pyrene diol epoxide (B[a]PDE), with little or no adduct formation at codons 13 and 61, respectively. The carcinogen–DNA adducts formed at codon 14 were repaired more efficiently than those formed at codon 12 (Feng et al. 2002). However, a potential carcinogen-induced origin of other human RAS gene mutations has not been clearly demonstrated, neither for pancreatic cancers nor colorectal cancers, in which these mutations are relatively frequent.
TP53 gene mutations
Inactivating mutations in the TP53 tumor suppressor gene are the most common genetic events in human cancers affecting a specific gene (Hainaut and Wiman 2005; Hofseth et al. 2004; Olivier et al. 2004; Petitjean et al. 2007; Vogelstein et al. 2000), with the vast majority arising from a single point mutation in the segment encoding the DNA-binding domain of TP53. The inactivating mutations render the mutant TP53 protein unable to carry out its normal functions, i.e., transcriptional transactivation of downstream target genes that regulate the cell cycle and apoptosis (Petitjean et al. 2007; Vogelstein et al. 2000). Several TP53 mutation databases are maintained to catalogue TP53 mutations reported in the literature (Hamroun et al. 2006; Petitjean et al. 2007).
The large number and tissue-specific diversity of mutations in the TP53 mutation databases provide indirect but compelling evidence that certain mutagens may be involved in human carcinogenesis (Hussain and Harris 1998). The TP53 gene is useful as a mutagen test system for several reasons. For many tumor suppressor genes, nonsense or frameshift mutations that lead to protein truncation and mRNA decay are most frequently reported. Examples are the BRCA1 and APC genes. These types of mutations are not particularly useful for assessing potential mutagens as initiators of carcinogenesis since the creation of a stop codon either by a point mutation or an insertion/deletion-induced frameshift event severely limits the types of mutational events that can be scored. The situation is different for TP53 in which almost 90% of all mutations are of the missense type. Thus, many different types of mutations are observed in this gene including all types of possible base substitutions. Further, the occurrence of TP53 mutations is not limited to a few particular sequences or codons along this gene (like in the RAS genes). Walker et al noted no less than 73 mutation hotspots along the TP53 gene (Walker et al. 1999). Most mutations cluster in the TP53 DNA binding domain, which encompasses exons five through eight and spans approximately 180 codons or 540 nucleotides. Although particular amino acids in direct contact with DNA may be preferentially mutated or selected (Walker et al. 1999), there are hundreds of TP53 mutants that can lead to a phenotypic change of TP53 function (Kato et al. 2003). Most TP53 missense mutations lead to the synthesis of a stable protein, which lacks its specific DNA binding and transactivation function and accumulates in the nucleus of cells. The acquisition of TP53 mutations can have two consequences: i) a dominant negative effect by hetero-oligomerization of the more stable mutant TP53 with wild-type TP53 molecules expressed from the normal remaining allele, and ii) a gain of function of mutant TP53 protein. Since there are so many different types of mutant TP53 proteins functioning in diverse pathways, it is extremely difficult to distinguish between these possibilities. There have been very few reports describing inactivation of TP53 expression by promoter hypermethylation, and missense mutations in TP53 are much more common than nonsense or frameshift mutations, which supports the idea of a function for TP53 mutants either in a dominant-negative fashion or in a gain of function pathway.
We have hypothesized that the primary mutagenesis process (lesion formation, DNA repair, lesion bypass) and selection for tumorigenic mutations, are the driving forces that shape the TP53 mutation spectra in different human tumors (Besaratinia and Pfeifer 2006). Scrutiny of one of the large public domain databases of TP53 gene mutations in human cancers (nearly 25,000 entries; http://www-p53.iarc.fr/index.html) has been used to find associations between various types of human cancer mutations and carcinogen exposures, e.g., from environmental, occupational, and dietary sources (Hussain and Harris 1998; Olivier et al. 2004). The putative associations identified by this approach can be validated using a wide range of experimental models, ranging from lower organisms, e.g., bacterial Ames test (Gee et al. 1994), TP53 functional assays in yeast (Fronza et al. 2000), to reporter gene-based transgenic rodents, e.g., the BigBlue® system (Lambert et al. 2005), or analysis of endogenous non-cancer-related genes, e.g., the house-keeping gene hypoxanthine phosphoribosyltransferase (HPRT) in mammalian or human tissues/cells (Albertini 2001). Although these systems lack, in one way or another, important factors that contribute to TP53 mutations and human cancers, e.g., DNA-sequence context, DNA repair capacities and fidelity of translesion DNA synthesis, which are species/cell-type dependant, they have provided invaluable information on many aspects of mutagenesis-derived carcinogenesis (Besaratinia and Pfeifer 2006).
For certain cancers, the distribution of DNA lesions along the TP53 gene caused by environmental carcinogens can be correlated well with the mutational spectra, i.e. hotspots and types of mutations (Pfeifer et al. 2002). As described below, this concept has been validated by experiments with simulated sunlight and the cigarette smoke component benzo[a]pyrene (B[a]P) representing the polycyclic aromatic hydrocarbon (PAH) class of carcinogens. The damage and repair data obtained for the respective mutagens can predict many parameters of TP53 mutagenesis in human nonmelanoma skin cancers and lung cancers from tobacco smokers, respectively. Future studies with suspected mutagens will be helpful to implicate causative agents involved in other cancers, where the exact carcinogen has not yet been identified.
In the TP53 gene, an exceptionally high percentage of mutations can be found at the dinucleotide sequence CpG (Soussi and Beroud 2003). About 3 to 4% of all cytosines in mammalian DNA are methylated by a postreplicative enzymatic process catalyzed by DNA methyltransferases. The modified base, 5-methylcytosine (5mC) is found exclusively at CpG dinucleotides in mammalian genomes. In studies with prokaryotes, 5mC was first identified as a mutational hotspot as early as 1978 (Coulondre et al. 1978). In mammalian genomes, CpG sequences are hypermutable and, as a consequence, a large fraction of all CpG sites has been lost during evolution (Pfeifer 2006). In fact, CpGs are present at only about one seventh of their expected frequency, which would be roughly one every 13 or 14 base pairs. In the human genome of 3,080,419,480 nucleotides, there are 28,163,863 CpGs, which corresponds to only 9 CpGs per kilobase (1.8% of the sequence). Attesting to the general mutability of CpG sequences, it has been estimated that up to 25% of all disease-causing human mutations in autosomal genes occur at CpG sites (Krawczak et al. 1998). Interestingly, CpG frequency is not much depleted in the human TP53 gene. There are 23 CpG dinucleotides between codons 120 and 300 of the TP53 coding sequence representing the DNA binding domain, and 22 of these are between codons 150 and 300 in the most frequently mutated region. The hypermutability of CpG sequences has generally been attributed to hydrolytic deamination of 5-methylcytosine leading to the emergence of thymine base-paired with guanine at CpG sites (Jones and Baylin 2002; Pfeifer 2006). CpG mutation leading to the formation of CpA or TpG dinucleotides is considered to be a process with no apparent involvement of exogenous mutagens.
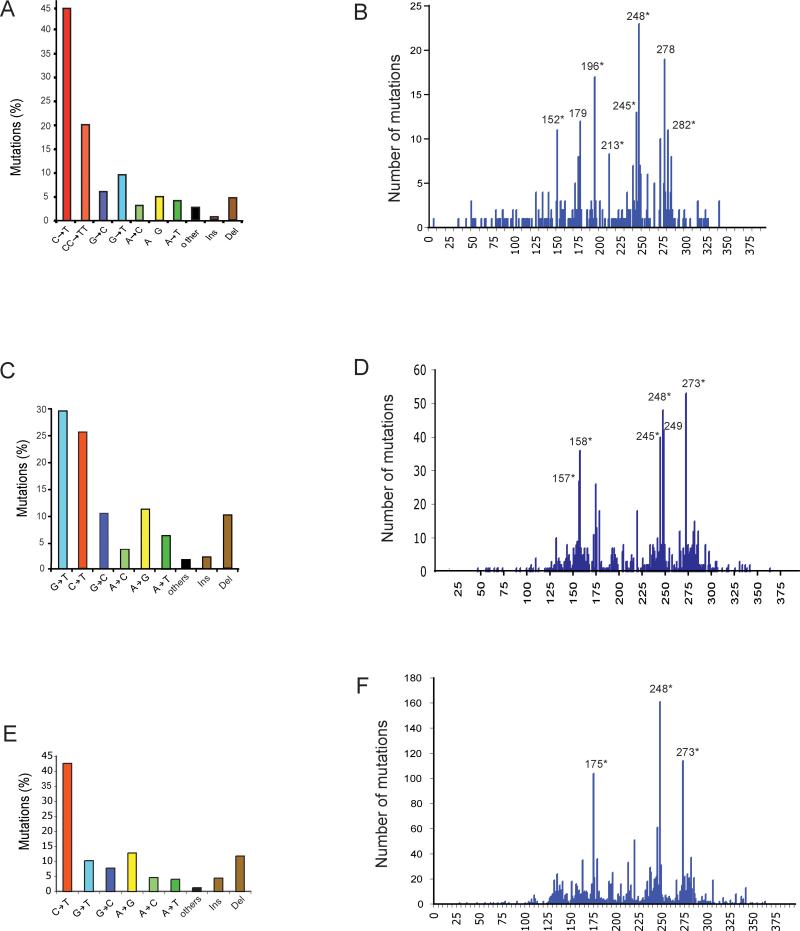
All CpG sequences analyzed in the TP53 gene are completely (at least 95% at each site) methylated in all human tissue samples examined (Rideout III et al. 1990; Tornaletti and Pfeifer 1995). The methylated CpGs (mCpGs) contain more than one third of all cancer mutations in TP53 and the vast majority of these alterations are transition mutations. Therefore, methylated CpG dinucleotides are the single most important mutational targets in TP53. Five of the six major TP53 mutational hotspots (in all cancers combined), i.e., codons 175, 245, 248, 273, and 282, all contain methylated CpG dinucleotides. C to T transitions at CpGs are particularly common in brain and colorectal cancers (http://www-p53.iarc.fr/index.html).
Endogenous deamination of 5-methylcytosine is viewed as the main source of the high frequency of CpG transitions at TP53 mutational hotspots in human internal cancers (Jones and Baylin 2002; Pfeifer 2006). We would like to point out, however that deamination of 5-methylcytosine in double-stranded DNA may not be the only mechanism that can cause transition mutations at methylated CpG sites. Both cytosine and 5-methylcytosine are subject to deamination, resulting in conversion to uracil and thymine, respectively. Hydrolytic deamination occurs at cytosines in double stranded DNA at a relatively slow rate with a half-life of about 30,000 years at 37°C and pH 7.4 (Frederico et al. 1990; Lindahl 1993; Shen et al. 1994). Methylation at the 5 position of the base ring facilitates spontaneous hydrolytic deamination, and as a result, 5-methylcytosines are deaminated two to four times more rapidly than cytosines (Ehrlich et al. 1990; Lindahl 1993; Shen et al. 1994). For double-stranded DNA the difference is only 2.2-fold, and 5-methylcytosines deaminate at a rate of 5.8 × 10-13 per second (Shen et al. 1994). From these data, it can be calculated that only two or three 5-methylcytosines deaminate per day in each cell (Pfeifer 2000; Schmutte and Jones 1998). These numbers are almost insignificant compared to steady state levels that have been measured for many endogenous and exogenous DNA adducts, which can be orders of magnitude higher. In any case, the 2-fold enhancement of the deamination rate is certainly not enough to account for the elevated mutation rate at methylated cytosines in CpG dinucleotides, which is estimated to be up to 42-fold higher than that of unmethylated cytosines at non-CpG sites (Cooper and Youssoufian 1988). The mutational effect may be augmented by the difference in repair of the resulting two mismatches. Uracil is recognized and excised efficiently by ubiquitous uracil-DNA glycosylase enzymes. Two thymine DNA glycosylase repair proteins (TDG and MBD4), which act upon deaminated mCpGs, have been identified in mammals (Hendrich et al. 1999; Neddermann et al. 1996). In fact, when MBD4 was deleted in the mouse, there was a 2-3-fold increase in CpG transition mutations in mutational reporter genes (Millar et al. 2002; Wong et al. 2002). However, the effectiveness of uracil versus thymine repair at deaminated C and 5mC bases in vivo is not known.
It is possible that certain chemicals may enhance the deamination reaction at methylated CpGs. Nitric oxide was shown to increase the rate of C to T transitions via stimulation of base deamination (Wink et al. 1991). However, nitric oxide did not cause significant 5mC-specific deamination of 5-methylcytosine containing reporter genes in other in vitro mutagenesis assays (Felley-Bosco et al. 1995; Schmutte et al. 1995). Oxidative damage to 5-methylcytosine can result in the formation of thymine glycol as one end product (Zuo et al. 1995). Thymine glycol is thought to be primarily a replication-blocking lesion, which will however pair mostly with adenine, when bypassed by a lesion-tolerant DNA polymerase (Kusumoto et al. 2002), which makes the oxidative deamination pathway a viable possibility. The symmetrical structure of a methylated CpG dinucleotide creates, of course, two possible sources of a C to T mutational event. The transition mutations may be caused by a lesion forming preferentially at guanine bases within methylated CpG sequences. This will produce G to A transition mutations indistinguishable from C to T mutations on the opposite DNA strand. Alternatively, certain mutagens may preferentially form DNA adducts at methylated cytosines and cause transition mutations by mispairing of the modified 5mC during DNA replication, or may increase the hydrolytic deamination of 5-methylcytosine. In summary, although there is only limited hard evidence to support the generally accepted idea that spontaneous hydrolytic deamination of 5-methylcytosine plays a dominant role in mammalian CpG mutagenesis, no alternative mechanism for this mutational specificity has been experimentally demonstrated so far.
It is widely accepted [though not universally; see (Thilly 2003)] that mutagenesis induced by endogenous and exogenous agents is an important component of tumorigenesis. This concept has been well proven in animal experiments in which exposure of animals to specific mutagens led to the formation of tumors harboring carcinogen-specific (“fingerprint-type”) mutations in RAS genes (Barbacid 1987) or in the TP53 gene (Ruggeri et al. 1993). While RAS mutations are very frequently observed in carcinogen-induced tumors in rodents, mutations in TP53 are much more infrequent. Therefore, for reproducing the TP53 mutational spectrum that occurs in human cancers, more indirect approaches have been developed.
One approach involves identification of sequence-specific DNA lesions generated by carcinogens in the TP53 gene, and correlation of these “fingerprints” with TP53 mutations collected from human cancer databases (Besaratinia and Pfeifer 2006; Pfeifer et al. 2002). This approach is based on mapping of DNA damage at the nucleotide resolution level by the ligation-mediated PCR (LMPCR) technique (Denissenko et al. 1996; Pfeifer et al. 1991). Using this technique, we have compared the distribution of DNA damage in the TP53 gene of human cells exposed to sunlight ultraviolet (UV) radiation, benzo[a]pyrene diolepoxide (BPDE), or aflatoxin B1 (AFB1) with the distribution of TP53 mutations in human cancers of the skin (non-melanoma), lung, and liver (Denissenko et al. 1998b; Denissenko et al. 1996; Tommasi et al. 1997). These experiments revealed a previously unrecognized role of methylated CpG sites as preferential targets for physical and chemical genotoxic agents.
Base changes characteristic for skin cancer, i.e. transitions at CC or TC dipyrimidine sequences, show a strong association with methylated CpGs (Tommasi et al. 1997; You et al. 1999). The relative contribution of TP53 mutations affecting dipyrimidines within mCpG sequences is ~35% of the total mutations, despite the fact that 5’CCG and 5’TCG occur only 20 times in the 1,080 bp double-stranded target sequence between codons 120 and 300 (Fig. 1). Importantly, all these CpG sequences are methylated in human keratinocytes (Tornaletti and Pfeifer 1995). For skin cancer, it was found that mutational hotspots that contain 5-methylcytosine at dipyrimidines are much more susceptible to pyrimidine dimer formation after exposure of cells to natural sunlight rather than to 254 nm UVC. Methylation of cytosine enhances pyrimidine dimer formation by sunlight by up to 15-fold and methylated cytosines are preferentially mutated by sunlight (Tommasi et al. 1997; You et al. 1999).
Figure 1. Mutation spectra (A, C, E) and codon distribution (B, D, F) of the TP53 tumor suppressor gene in human non-melanoma skin tumors (basal cell and squamous cell carcinomas), lung tumors of smokers, and breast cancers.
A, B. Skin cancer; C, D. Lung cancer of smokers; E, F. Breast cancer. Data were obtained from the TP53 mutation database of the International Agency for Research on Cancer (http://www-p53.iarc.fr/index.html; R13 version). Ins: Insertions; Del: Deletions. Codons containing methylated CpG sequences are indicated by asterisks (*).
Tobacco smoking is a strong risk factor for the development of lung cancer (Hecht 1999). The characteristic signature of TP53 lung tumor mutations in smokers is the G to T transversion (Fig. 1). Ninety percent of the guanines undergoing these transversion events in lung cancer are located on the nontranscribed DNA strand (Hussain and Harris 1998; Pfeifer et al. 2002). Of note, five of the six most prominent mutation hotspots in the TP53 gene are represented by G to T mutations at codons containing methylated CpG sequences, including codons 157, 158, 245, 248, and 273 (Fig. 1B) (Pfeifer et al. 2002). G to T transversions are typical for bulky adduct-producing mutagens including the class of polycyclic aromatic hydrocarbons (PAHs). Benzo[a]pyrene is a widely studied member of the PAH class. Upon metabolic activation to benzo[a]pyrene diolepoxide (B[a]PDE), it induces G to T mutations (Luch 2005). The distribution of B[a]PDE adducts along the TP53 gene was mapped, at nucleotide resolution level, in carcinogen-treated normal human bronchial epithelial cells (Denissenko et al. 1996). Selective adduct formation sites were major mutational hotspots in human lung cancers, i.e. there was an excellent correlation between the benzo[a]pyrene adduct spectrum and the mutation spectrum in lung cancer (Pfeifer et al. 2002). The mechanistic basis for the selective occurrence of these PAH-damage hotspots is related to patterns of cytosine methylation in the TP53 gene (Denissenko et al. 1997). The distribution of B[a]PDE-DNA adducts differed drastically in CpG-methylated DNA compared to non-methylated DNA. Guanines flanked by 5-methylcytosines were the preferentially adducted positions. Therefore, CpG dinucleotides, which are methylated in the human TP53 gene in all human tissues examined, in addition to being an endogenous promutagenic factor, represent a preferential target for exogenous chemical carcinogens as well. The extent by which enhanced binding of an individual carcinogen at methylated CpGs affects mutagenesis at the same location has been studied in mouse cells carrying the lacI and cII transgenes. These cells were treated with B[a]PDE and the mutations were scored. A dominant fraction of the mutations (58-77% of all G to T mutations) occurred at methylated CpG sequences (Yoon et al. 2001). In summary, the PAH-DNA adduct patterns in the TP53 gene in bronchial epithelial cells coincide with G to T mutational hotspots in tobacco-smoking associated lung cancers (Denissenko et al. 1996; Smith et al. 2000), and this mutational pathway is faithfully reproduced with a tobacco smoke carcinogen (B[a]PDE) in mutational reporter genes rich in methylated CpG sites (Yoon et al. 2001).
The Hupki mouse model
More recently, a novel model system has been developed to investigate experimentally induced mutations in the human TP53 gene. The human p53 knock-in (Hupki) mouse model has addressed the issue of DNA sequence context by replacing exons 4-9 of the endogenous mouse TP53 allele with the homologous normal human TP53 gene sequence (Luo et al. 2001b). The Hupki mouse model has the capacity to detect both spontaneously arisen and carcinogen-induced mutations in the human TP53 gene in vitro (Liu et al. 2004; Liu et al. 2005; Luo et al. 2001a; Reinbold et al. 2008; Vom Brocke et al. 2008) or in vivo (Luo et al. 2001b; Tong et al. 2006).
The Hupki mouse model system was constructed using gene-targeting technology to create a mouse strain that harbors human wild-type TP53 DNA sequences from exons 4 to 9 in place of the homologous murine DNA sequences in both copies of the mouse Tp53 gene (Luo et al. 2001b). The substituted segment encodes the polyproline domain and DNA-binding domain of wild-type human TP53, and the chimeric TP53 gene remains under normal transcriptional regulation at the mouse locus. The Hupki mice develop normally, exhibit no apparent defects, remain fertile, and show no susceptibility to spontaneous lymphomas, sarcomas, or other neoplasms, which are common in TP53-deficient mice (Luo et al. 2001b). The Hupki mice retain a variety of normal TP53 functions and characteristics, including nuclear accumulation of TP53 protein after exposure to DNA-damaging agents, transcriptional activation of known TP53 downstream targets, and induction of apoptosis in thymocytes after gamma-irradiation, an outcome modulated by a functional TP53 gene (Luo et al. 2001a; Luo et al. 2001b).
In addition to its application for in vivo animal studies, the Hupki model system is also amenable to in vitro cell culture experiments. Murine fibroblasts, in contrast to human cells, spontaneously undergo immortalization during in vitro culturing, and require only one key genetic defect, such as loss of TP53 function, thus allowing the selection of TP53 mutant cells in vitro. Primary embryonic fibroblasts from the Hupki mice readily undergo immortalization during in vitro passaging, which allows for dysfunctional TP53 point mutations that are characteristic of human tumors, to be selected for (Feldmeyer et al. 2006; Liu et al. 2004; Liu et al. 2005; Luo et al. 2001a; Reinbold et al. 2008; Vom Brocke et al. 2008).
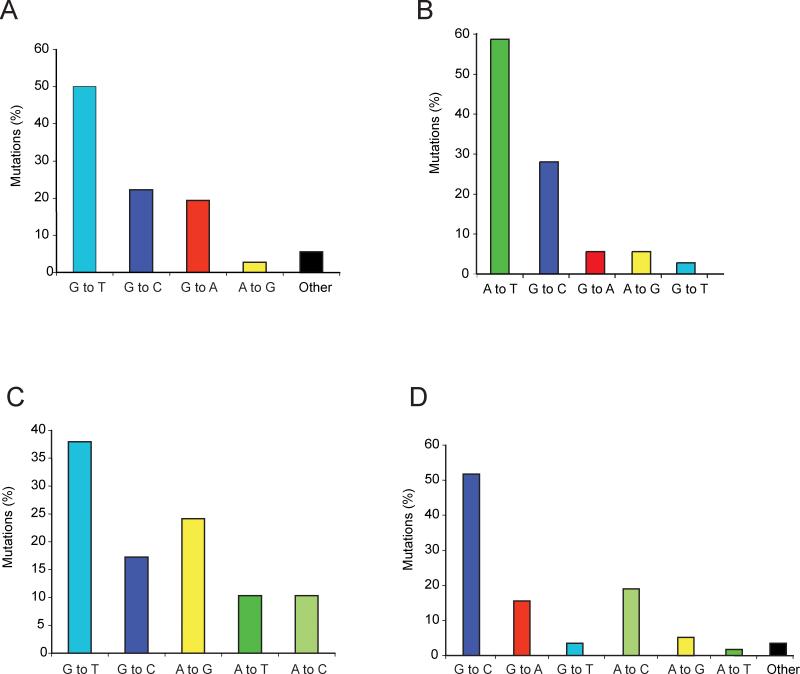
Hupki mouse embryonic fibroblasts treated with benzo[a]pyrene (B[a]P), a tobacco-derived carcinogen, harbored TP53 mutations comprised of predominantly single base substitutions in the DNA-binding domain of this gene [29 out of 36 (~81%) of all mutations] (Feldmeyer et al. 2006; Liu et al. 2005; Reinbold et al. 2008). G to T transversion mutations constituted half of all B[a]P-induced mutations, of which all but one (17 out of 18) occurred at sites where the mutated guanines were positioned on the non-transcribed strand of the TP53 gene. Distribution of the twenty-nine B[a]P-induced mutations in the DNA-binding domain of the TP53 gene revealed codons 157, 158 and 273 as the most frequently mutated sites. The overall pattern and distribution of B[a]P-induced mutations in the Hupki mouse model system (Feldmeyer et al. 2006; Liu et al. 2005; Reinbold et al. 2008) resembled the characteristic features of TP53 mutations in lung tumors of smokers (see, Fig. 2A and Fig. 1C) (Besaratinia and Pfeifer 2008; Hainaut and Pfeifer 2001; Toyooka et al. 2003) and the distribution of B[a]P-DNA adducts in human bronchial epithelial cells (Pfeifer et al. 2002).
Figure 2. Spontaneous and induced mutation spectra of the TP53 tumor suppressor gene in the Hupki mouse model system.
A. Mutations in B[a]P-treated Hupki mouse embryonic fibroblasts (n = 36) (data were compiled from (Feldmeyer et al. 2006; Liu et al. 2005; Reinbold et al. 2008).
B. Mutations in aristolochic acid-treated Hupki mouse embryonic fibroblasts (n = 36) (data were compiled from (Feldmeyer et al. 2006; Liu et al. 2004; Nedelko et al. 2009; vom Brocke et al. 2006).
C. Mutations in 3-nitrobenzanthrone-treated Hupki mouse embryonic fibroblasts (n = 29) (data were obtained from (Vom Brocke et al. 2008).
D. Spontaneously derived (control) mutations from Hupki mouse embryonic fibroblasts (n = 58) (data were compiled from refs. (Feldmeyer et al. 2006; Liu et al. 2004; Liu et al. 2005; Luo et al. 2001a; Reinbold et al. 2008; Vom Brocke et al. 2008; vom Brocke et al. 2006).
The Hupki mouse embryonic fibroblasts were treated with aristolochic acid (AA) (Feldmeyer et al. 2006; Liu et al. 2004; Nedelko et al. 2009), a plant extract potentially involved in Chinese herb nephropathy and possibly leading to urothelial cancer development (Nortier et al. 2000). Twenty-one out of the 36 AA-induced TP53 mutations (~56%) were A to T transversion mutations (Feldmeyer et al. 2006; Liu et al. 2004; Nedelko et al. 2009) (see, Fig. 2B), an otherwise rare type of mutation but reflecting the hallmark mutation detected in urothelial tumors from patients with documented AA exposure (Grollman et al. 2007; Lord et al. 2004). The induced A to T transversion mutations were due to the adducted adenines located almost exclusively on the non-transcribed strand of the TP53 gene consistent with the fact that 20 of the 21 mutations were A to T and only one of the 21 mutations was T to A (Feldmeyer et al. 2006; Liu et al. 2004; Nedelko et al. 2009). This finding is consistent with the preferential formation of AA-adenine adducts found in the DNA of AA-treated cells and nephropathy patients (Arlt et al. 2001; Arlt et al. 2002; Lord et al. 2001; Lord et al. 2004; Nortier et al. 2000), as well as in the DNA from target organs of AA-exposed rats (Kohara et al. 2002; Pfau et al. 1990). Collectively, the data on aristolochic acid-induced DNA damage and mutations support a role of this compound in the etiology of tumors linked to Chinese herb nephropathy.
In other experiments, the Hupki mouse embryonic fibroblasts were treated with 3-nitrobenzanthrone (3-NBA) (Vom Brocke et al. 2008), a member of the class of nitropolycyclic aromatic hydrocarbons, present in the particulate fraction of diesel exhaust (US-EPA (Gilman 2002), and a ubiquitous urban air pollutant (Arlt 2005). The established cultures of 3-NBA-treated cells harbored TP53 mutations in the DNA-binding domain of this gene, which consisted mainly of base substitutions (22 out of 29, ~76%) (Vom Brocke et al. 2008). Of these, G to T transversions were the major type of mutations (10 out of 22 (~46%) followed by A to T transversions (3 out of 22, (~14%) (Fig. 2). This ratio of G to T to A to T transversions (3:1) perfectly mirrored the ratio of dG/dA adduct formation (75:25%) determined in similarly treated cells with 3-NBA or its reactive metabolite, N-hydroxy-3-aminobenzanthrone (N-OH-3-ABA) (Vom Brocke et al. 2008). A similar correlation in ratios of 3-NBA-derived purine adducts to transversion mutations was previously found in liver tissues of the MutaMouse™, where the proportion of induced dG to dA adducts was 6 to 1 and that of corresponding G to T and A to T mutations was 5 to 1 (Arlt 2005).
Luo et al. have demonstrated that UVB-irradiated Hupki mice exhibit characteristic molecular pathology features of sunlight-associated human skin cancers, including (i) development of clones of epidermal cell patches with TP53-immunoreactive nuclei, (ii) formation of UV-induced cyclobutane pyrimidine dimers at skin cancer mutational hotspots in the TP53 gene, which co-localize with the respective lesions induced in UVB-exposed human keratinocytes, and (iii) induction of signature C to T transition mutations in the respective TP53 mutational hotspots found in human skin cancers (Luo et al. 2001a).
Tong et al. (Tong et al. 2006) have used the Hupki mice to investigate the effect of local DNA sequence on TP53 codon 249 mutation, a prevalent occurrence in human hepatocellular carcinoma associated with synergistic exposure to aflatoxin B1 (AFB1) and hepatitis B virus (HBV) infection (Montesano et al. 1997). A single intraperitoneal injection of AFB1 to the Hupki mice and counterpart wild-type animals showed that the mice expressing the humanized TP53 gene were more prone to hepatocellular carcinoma development and death, compared to mice expressing the murine TP53, without acquiring any mutations in the TP53 gene (Tong et al. 2006). These findings support the notion that the specificity of TP53 codon 249 mutation in human hepatocellular carcinoma is not solely dependent upon DNA sequence context of this gene (Denissenko et al. 1998b), and that other determining factors, e.g., concomitant HBV infection, may synergistically be involved in this specific mutational process (Hussain et al. 2007). Also, despite the overall conservation in evolution of DNA repair mechanisms, differences exist between humans and mice, such as the efficiency of the global genomic repair sub-pathway of nucleotide excision repair (Hanawalt 2002). Such discrepancies may set some limitations because promutagenic lesions in the Hupki TP53 gene are subject to the murine DNA repair machinery. Nonetheless, the Hupki TP53 model system has recapitulated many aspects of TP53 mutagenesis in human tumors (Feldmeyer et al. 2006; Jaworski et al. 2005; Liu et al. 2004; Liu et al. 2005; Luo et al. 2001a; Luo et al. 2001b; Reinbold et al. 2008; Tong et al. 2006; Vom Brocke et al. 2008; vom Brocke et al. 2006). Future studies will determine the accuracy of its portrayal of these events in other types of human cancers.
Mutational spectra obtained by large scale DNA sequencing of human tumors
Large-scale and high-throughput DNA sequencing is now being used to find almost any genome alteration in individual tumors. In addition to the TP53 mutation databases, the catalogue of somatic mutations in cancer (COSMIC) database is currently the most comprehensive resource available for information on cancer-associated DNA sequence changes (Forbes et al. 2008). It combines information from the scientific literature with resequencing data of the Sanger Institute.
Initial efforts of systematic genome-wide screening for genes mutated in cancer have led to the discovery that the BRAF gene, which encodes a serine/threonine kinase in the RAS signaling pathway, frequently contains somatic mutations in a large fraction of malignant melanomas and other tumors (Davies et al. 2002). Large-scale re-sequencing efforts also uncovered common mutations in the PI3 kinase pathway in several types of human cancer (Carpten et al. 2007; Samuels et al. 2004). Recently, cancer genome sequencing has focused on a collection of ~500 genes encoding protein kinases (Greenman et al. 2007). The frequent inactivation of a particular biochemical pathway by mutation or epigenetic inactivation of any one of the critical pathway components rather than by inactivation of only a single gene is becoming a common theme that is currently explored. Exceptions, however, are still the TP53 and KRAS genes, which despite of the sequence analysis of hundreds or even thousands of genes, still stand at the top of the list of the most frequently mutated cancer genes (Ding et al. 2008; Parsons et al. 2008; Sjoblom et al. 2006). On average, large-scale re-sequencing of large sets human genes has identified generally between 10 and 100 mutations in each individual tumor. The percentage of silent mutations is often quite high and can be almost as high as if one would expect that none of the observed non-synonymous mutations would lead to a phenotypic change selected in the tumor and almost all such mutations are innocuous passenger mutations. However, careful analysis has led to the prediction that at least a limited number of the newly identified mutations other than TP53, KRAS, etc., are biologically significant (Wood et al. 2007).
Importantly, the large-scale sequencing data have generally confirmed the cancer-specific mutation data obtained earlier for the TP53 gene (see Figures 3 and 4). For example, sequencing of 518 protein kinase genes in six melanoma samples uncovered 144 mutations and more than 90% of these mutations were C to T transitions at dipyrimidine sites (Greenman et al. 2007). These data strongly support UVB-induced pyrimidine dimer lesions as the cause of these mutations in melanoma, at least in those six samples analyzed. Another interesting observation is that tumors from glioma patients treated with the chemotherapeutic agent temozolomide contained a vast excess (>95%) of G to A transition mutations (Greenman et al. 2007). Temozolomide is an alkylating agent that produces O6-methyl-guanine adducts. These adducts can mispair with thymine during DNA replication leading to the observed mutational change from G to A.
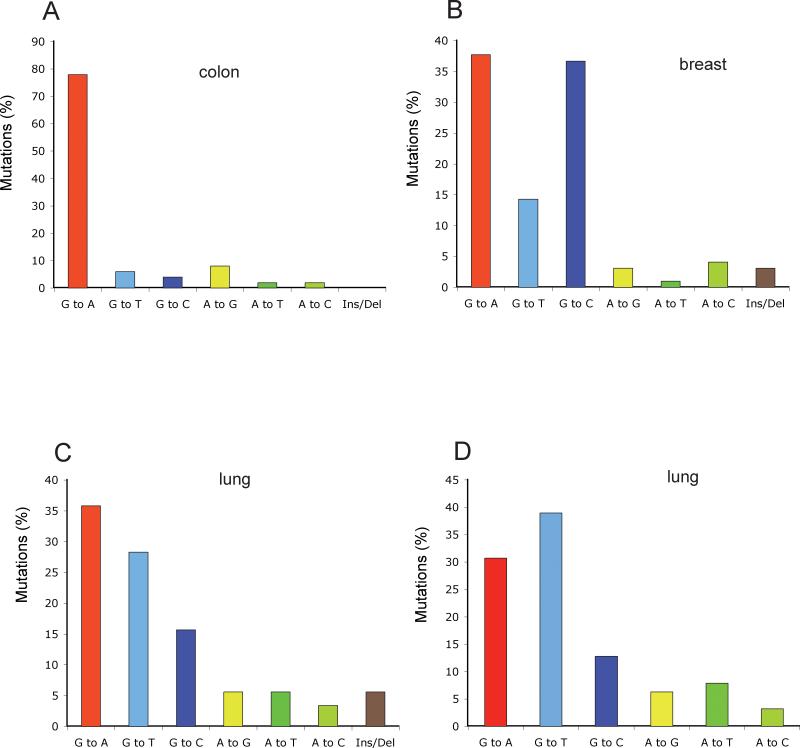
Figure 3. Mutation spectra of protein kinase genes and other genes of interest in lung, colorectal, and breast cancers.
Data for 518 protein kinase-encoding genes were obtained from Greenman et al (Greenman et al. 2007) (panels A, B, C). Data for 623 genes with a known or potential relationship to cancer were obtained from Ding et al (Ding et al. 2008) (panel D).
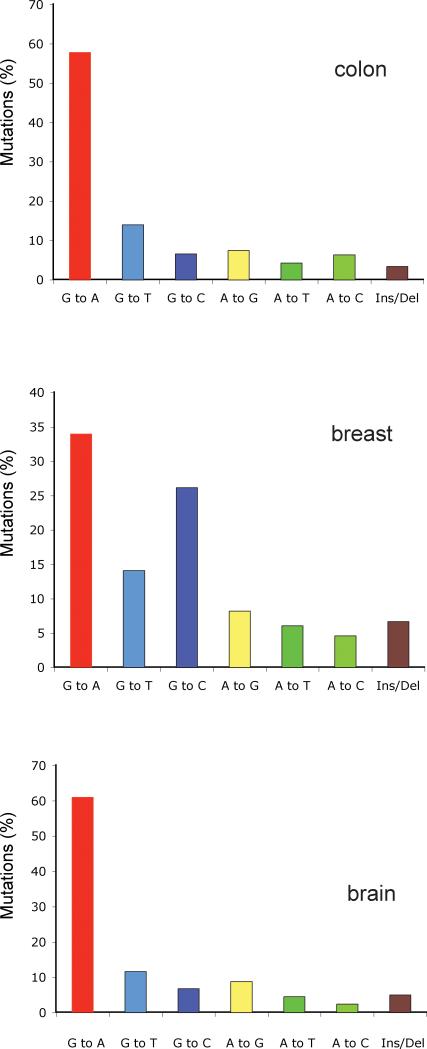
Figure 4. Mutation spectra of brain, colorectal, and breast cancers derived from large-scale sequencing of cancer genomes.
Data from 20,661 protein-coding genes were obtained from Jones et al (Jones et al. 2008).
Colorectal cancers are characterized by a high percentage of G:C to A:T mutations (Figure 3A, Figure 4A). Also, intriguingly, the high preponderance of G:C to A:T transitions at CpG dinucleotides, previously recognized in the TP53 gene of colorectal cancers, is also found in the protein kinase genes (Greenman et al. 2007). These transition mutations represent roughly 50% of all mutations. Other investigators determined the sequences of over 23,000 transcripts in colorectal, pancreatic, breast, and brain tumors. The percentage of C to T transitions at CpG sites was 47.8% for colon cancer, 43.1% for brain cancer, and 37.9% for pancreatic cancer (Jones et al. 2008). Overall, these data are very similar to what has been observed in the TP53 gene (Soussi and Beroud 2003).
Lung cancer is a particularly illustrative example. As was shown earlier, the TP53 mutation spectrum in lung cancer is different from those of other cancers, perhaps with the exception of aflatoxin-associated liver cancers. Lung tumors are characterized by a large percentage of G to T transversions thought to be derived from mutagenic agents in tobacco smoke (Hainaut and Pfeifer 2001). The same situation was recapitulated when 623 genes with a known or potential relationship to cancer were sequenced in 188 lung adenocarcinomas (Ding et al. 2008). Of 1013 non-synonymous mutations, 41% were G to T transversions, a percentage even higher than that for TP53 (Fig. 1). In smokers, 43% of the mutations were G to T transversions but this number dropped to 13% in never-smokers (Ding et al. 2008). Again, this difference between smokers and nonsmokers is similar to that in the TP53 gene (Hainaut and Pfeifer 2001) and suggests a mutagenic role of tobacco-smoke carcinogens in lung carcinogenesis.
The large scale sequencing of breast cancer genomes has provided an unexpected and unusual result. As shown by independent groups sequencing either 518 protein kinase genes (Greenman et al. 2007), or the exons of more than 20,000 genes (Jones et al. 2008), breast cancers are characterized by a low fraction of C to T transitions at CpG sites, and by a high frequency of G to C transversions (Figures 3, 4). Interesting, a large fraction of the G to C transversions occur at the dinucleotide sequence 5’GpA, which is equivalent to a C to G transversion at 5’-TpC on the opposite DNA strand, so that this unique type of mutation accounts for >20-30% of all mutations in breast tumors. This data suggests that breast cancers are caused by an etiological agent that induces this particular type of mutation. There are few known mutagens that specifically induce G/C to C/G transversions, let alone selectively at a particular dinucleotide sequence. Polycyclic aromatic hydrocarbons containing a cyclopentane ring, such as cyclopenta[c,d]pyrene and benz[j]aceanthrylene have been shown to induce predominantly G to C transversions in the KRAS gene of mouse lung tumors but the sequence context is different (Jackson et al. 2006). Surprisingly, however, analysis of G to C transversions in the TP53 gene in breast cancer shows that this type of mutation is quite rare (7.8%) (Fig. 1). When 181 breast cancer TP53 G to C transversions were analyzed for sequence context, we found that 55 of them (=30%) were in the sequence context 5’GpA, which is just slightly more than would be expected from a random distribution of flanking bases. One possible interpretation for the different results in the TP53 gene versus the other genes is of course that the frequency of TpC/GpA dinucleotides that produce a mutant TP53 protein after G to C transversion may be low in the TP53 gene. Although there are several common hotspot codons in TP53 (248, 249, 273) that can frequently be mutated by a G to C transversion, these codons do not contain GpA sequences. However, a G to C mutation at 5’GpA sites can mutate several other commonly mutated TP53 codons, including codons 278 and 280, producing a mutant TP53 protein. After large scale sequence analysis of additional types of tumors, it was reported that the G/C to C/G transversions are targeted to 5’GpA dinucleotides not only in breast cancers but also commonly in lung cancers and other cancers but G to C mutations do not have a dinucleotide sequence preference among germ line variants (Greenman et al. 2007). Little strand bias for the G to C transversion was observed arguing against the possibility that this mutation is caused by a bulky DNA adduct subject to transcription-coupled DNA repair. The unique sequence preference of G to C transversions, in particular in breast and lung cancers, is puzzling, as is the fact that this pattern is not readily found in the TP53 gene. One possibility is that there are unknown exogenous or endogenous mutagens, in particular in breast, and perhaps other tissues, that effectively induce this unique type of mutation. Candidate mutagens, once identified, can be tested in the various in vitro and in vivo mutation reporter systems described earlier in this review with the goal of identifying a causative agent for these tumors.
Conclusions and future perspectives
Initial studies on human tumor-specific mutation spectra have laid the groundwork for finding etiological connections between specific mutagens/carcinogens and tumor development. These connections were made possible by the design and application of various in vitro and in vivo DNA damage detection and mutation reporter systems that can be used to score the DNA damaging effects, mutagenic competence and mutational specificity of suspected human carcinogens. Recent large-scale sequencing efforts of cancer genomes have confirmed that several previously well-studied genes are indeed the most commonly mutated ones (e.g. TP53, KRAS) but have expanded the catalogue of tumor-associated DNA sequence changes. These new sequencing data have confirmed established relationships between UV exposure and melanoma and tobacco smoke carcinogens and lung cancer, and the new studies have generally found similar mutation spectra in the TP53 gene and the many other genes that have now been sequenced. A notable exception is the identification of common G to C transversion mutations at a unique dinucleotide sequence in breast cancer and other cancers, which may provide an important signature for identification of a suspected human mutagen. It can be expected that the continued efforts aimed at sequencing the genomes of different types of cancer in many individuals will lead to new information on tumor-specific mutation spectra important for deciphering the etiology of human cancer.
Acknowledgements
Work of the authors is supported by NIH grant CA084469 to G.P.P.
REFERENCES
- Albertini RJ. HPRT mutations in humans: biomarkers for mechanistic studies. Mutat Res. 2001;489:1–16. doi: 10.1016/s1383-5742(01)00064-3. [DOI] [PubMed] [Google Scholar]
- Arlt VM. 3-Nitrobenzanthrone, a potential human cancer hazard in diesel exhaust and urban air pollution: a review of the evidence. Mutagenesis. 2005;20:399–410. doi: 10.1093/mutage/gei057. [DOI] [PubMed] [Google Scholar]
- Arlt VM, Schmeiser HH, Pfeifer GP. Sequence-specific detection of aristolochic acid-DNA adducts in the human p53 gene by terminal transferase-dependent PCR. Carcinogenesis. 2001;22:133–140. doi: 10.1093/carcin/22.1.133. [DOI] [PubMed] [Google Scholar]
- Arlt VM, Stiborova M, Schmeiser HH. Aristolochic acid as a probable human cancer hazard in herbal remedies: a review. Mutagenesis. 2002;17:265–277. doi: 10.1093/mutage/17.4.265. [DOI] [PubMed] [Google Scholar]
- Barbacid M. ras genes. Annu Rev Biochem. 1987;56:779–827. doi: 10.1146/annurev.bi.56.070187.004023. [DOI] [PubMed] [Google Scholar]
- Besaratinia A, Pfeifer GP. Investigating human cancer etiology by DNA lesion footprinting and mutagenicity analysis. Carcinogenesis. 2006;27:1526–1537. doi: 10.1093/carcin/bgi311. [DOI] [PubMed] [Google Scholar]
- Besaratinia A, Pfeifer GP. Second-hand smoke and human lung cancer. Lancet Oncol. 2008;9:657–666. doi: 10.1016/S1470-2045(08)70172-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodmer W, Bielas JH, Beckman RA. Genetic instability is not a requirement for tumor development. Cancer Res. 2008;68:3558–3560. doi: 10.1158/0008-5472.CAN-07-6544. discussion 3560-3551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bos JL. ras oncogenes in human cancer: a review. Cancer Res. 1989;49:4682–4689. [PubMed] [Google Scholar]
- Carbone M, Klein G, Gruber J, Wong M. Modern criteria to establish human cancer etiology. Cancer Res. 2004;64:5518–5524. doi: 10.1158/0008-5472.CAN-04-0255. [DOI] [PubMed] [Google Scholar]
- Carpten JD, Faber AL, Horn C, Donoho GP, Briggs SL, Robbins CM, Hostetter G, Boguslawski S, Moses TY, Savage S, Uhlik M, Lin A, Du J, Qian YW, Zeckner DJ, Tucker-Kellogg G, Touchman J, Patel K, Mousses S, Bittner M, Schevitz R, Lai MH, Blanchard KL, Thomas JE. A transforming mutation in the pleckstrin homology domain of AKT1 in cancer. Nature. 2007;448:439–444. doi: 10.1038/nature05933. [DOI] [PubMed] [Google Scholar]
- Chin L, Gray JW. Translating insights from the cancer genome into clinical practice. Nature. 2008;452:553–563. doi: 10.1038/nature06914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper DN, Youssoufian H. The CpG dinucleotide and human genetic disease. Hum Genet. 1988;78:151–155. doi: 10.1007/BF00278187. [DOI] [PubMed] [Google Scholar]
- Coulondre C, Miller JH, Farabaugh PJ, Gilbert W. Molecular basis of base substitution hotspots in Escherichia coli. Nature. 1978;274:775–780. doi: 10.1038/274775a0. [DOI] [PubMed] [Google Scholar]
- Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S, Teague J, Woffendin H, Garnett MJ, Bottomley W, Davis N, Dicks E, Ewing R, Floyd Y, Gray K, Hall S, Hawes R, Hughes J, Kosmidou V, Menzies A, Mould C, Parker A, Stevens C, Watt S, Hooper S, Wilson R, Jayatilake H, Gusterson BA, Cooper C, Shipley J, Hargrave D, Pritchard-Jones K, Maitland N, Chenevix-Trench G, Riggins GJ, Bigner DD, Palmieri G, Cossu A, Flanagan A, Nicholson A, Ho JW, Leung SY, Yuen ST, Weber BL, Seigler HF, Darrow TL, Paterson H, Marais R, Marshall CJ, Wooster R, Stratton MR, Futreal PA. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- Denissenko MF, Chen JX, Tang M-s, Pfeifer GP. Cytosine methylation determines hot spots of DNA damage in the human P53 gene. Proc. Natl. Acad. Sci. U.S.A. 1997;94:3893–3898. doi: 10.1073/pnas.94.8.3893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denissenko MF, Koudriakova TB, Smith L, O'Connor TR, Riggs AD, Pfeifer GP. The P53 codon 249 mutational hotspot in hepatocellular carcinoma is not related to selective formation or persistence of aflatoxin B1 adducts. Oncogene. 1998b;17:3007–3013. doi: 10.1038/sj.onc.1202214. [DOI] [PubMed] [Google Scholar]
- Denissenko MF, Pao A, Tang M-s, Pfeifer GP. Preferential formation of benzo[a]pyrene adducts at lung cancer mutational hotspots in P53. Science. 1996;274:430–432. doi: 10.1126/science.274.5286.430. [DOI] [PubMed] [Google Scholar]
- Ding L, Getz G, Wheeler DA, Mardis ER, McLellan MD, Cibulskis K, Sougnez C, Greulich H, Muzny DM, Morgan MB, Fulton L, Fulton RS, Zhang Q, Wendl MC, Lawrence MS, Larson DE, Chen K, Dooling DJ, Sabo A, Hawes AC, Shen H, Jhangiani SN, Lewis LR, Hall O, Zhu Y, Mathew T, Ren Y, Yao J, Scherer SE, Clerc K, Metcalf GA, Ng B, Milosavljevic A, Gonzalez-Garay ML, Osborne JR, Meyer R, Shi X, Tang Y, Koboldt DC, Lin L, Abbott R, Miner TL, Pohl C, Fewell G, Haipek C, Schmidt H, Dunford-Shore BH, Kraja A, Crosby SD, Sawyer CS, Vickery T, Sander S, Robinson J, Winckler W, Baldwin J, Chirieac LR, Dutt A, Fennell T, Hanna M, Johnson BE, Onofrio RC, Thomas RK, Tonon G, Weir BA, Zhao X, Ziaugra L, Zody MC, Giordano T, Orringer MB, Roth JA, Spitz MR, Wistuba, Ozenberger B, Good PJ, Chang AC, Beer DG, Watson MA, Ladanyi M, Broderick S, Yoshizawa A, Travis WD, Pao W, Province MA, Weinstock GM, Varmus HE, Gabriel SB, Lander ES, Gibbs RA, Meyerson M, Wilson RK. Somatic mutations affect key pathways in lung adenocarcinoma. Nature. 2008;455:1069–1075. doi: 10.1038/nature07423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelmann L, Edelmann W. Loss of DNA mismatch repair function and cancer predisposition in the mouse: animal models for human hereditary nonpolyposis colorectal cancer. Am J Med Genet C Semin Med Genet. 2004;129C:91–99. doi: 10.1002/ajmg.c.30021. [DOI] [PubMed] [Google Scholar]
- Ehrlich M, Zhang X-Y, Inamdar NM. Spontaneous deamination of cytosine and 5-methylcytosine residues in DNA and replacement of 5-methylcytosine residues with cytosine residues. Mutation Res. 1990;238:277–286. doi: 10.1016/0165-1110(90)90019-8. [DOI] [PubMed] [Google Scholar]
- Feldmeyer N, Schmeiser HH, Muehlbauer KR, Belharazem D, Knyazev Y, Nedelko T, Hollstein M. Further studies with a cell immortalization assay to investigate the mutation signature of aristolochic acid in human p53 sequences. Mutat Res. 2006;608:163–168. doi: 10.1016/j.mrgentox.2006.02.017. [DOI] [PubMed] [Google Scholar]
- Felley-Bosco E, Mirkovitch J, Ambs S, Mace K, Pfeifer A, Keefer LK, Harris CC. Nitric oxide and ethylnitrosourea: relative mutagenicity in the p53 tumor suppressor and hypoxanthine-phosphoribosyltransferase genes. Carcinogenesis. 1995;16:2069–2074. doi: 10.1093/carcin/16.9.2069. [DOI] [PubMed] [Google Scholar]
- Feng Z, Hu W, Chen JX, Pao A, Li H, Rom W, Hung MC, Tang MS. Preferential DNA damage and poor repair determine ras gene mutational hotspot in human cancer. J Natl Cancer Inst. 2002;94:1527–1536. doi: 10.1093/jnci/94.20.1527. [DOI] [PubMed] [Google Scholar]
- Forbes SA, Bhamra G, Bamford S, Dawson E, Kok C, Clements J, Menzies A, Teague JW, Futreal PA, Stratton MR. The Catalogue of Somatic Mutations in Cancer (COSMIC). Curr Protoc Hum Genet Chapter. 2008;10 doi: 10.1002/0471142905.hg1011s57. Unit 10 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frederico LA, Kunkel TA, Shaw BR. A sensitive genetic assay for the detection of cytosine deamination: determination of rate constants and the activation energy. Biochemistry. 1990;29:2532–2537. doi: 10.1021/bi00462a015. [DOI] [PubMed] [Google Scholar]
- Fronza G, Inga A, Monti P, Scott G, Campomenosi P, Menichini P, Ottaggio L, Viaggi S, Burns PA, Gold B, Abbondandolo A. The yeast p53 functional assay: a new tool for molecular epidemiology. Hopes and facts. Mutat Res. 2000;462:293–301. doi: 10.1016/s1383-5742(00)00011-9. [DOI] [PubMed] [Google Scholar]
- Gee P, Maron DM, Ames BN. Detection and classification of mutagens: a set of base-specific Salmonella tester strains. Proc Natl Acad Sci U S A. 1994;91:11606–11610. doi: 10.1073/pnas.91.24.11606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenman C, Stephens P, Smith R, Dalgliesh GL, Hunter C, Bignell G, Davies H, Teague J, Butler A, Stevens C, Edkins S, O'Meara S, Vastrik I, Schmidt EE, Avis T, Barthorpe S, Bhamra G, Buck G, Choudhury B, Clements J, Cole J, Dicks E, Forbes S, Gray K, Halliday K, Harrison R, Hills K, Hinton J, Jenkinson A, Jones D, Menzies A, Mironenko T, Perry J, Raine K, Richardson D, Shepherd R, Small A, Tofts C, Varian J, Webb T, West S, Widaa S, Yates A, Cahill DP, Louis DN, Goldstraw P, Nicholson AG, Brasseur F, Looijenga L, Weber BL, Chiew YE, DeFazio A, Greaves MF, Green AR, Campbell P, Birney E, Easton DF, Chenevix-Trench G, Tan MH, Khoo SK, Teh BT, Yuen ST, Leung SY, Wooster R, Futreal PA, Stratton MR. Patterns of somatic mutation in human cancer genomes. Nature. 2007;446:153–158. doi: 10.1038/nature05610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grollman AP, Shibutani S, Moriya M, Miller F, Wu L, Moll U, Suzuki N, Fernandes A, Rosenquist T, Medverec Z, Jakovina K, Brdar B, Slade N, Turesky RJ, Goodenough AK, Rieger R, Vukelic M, Jelakovic B. Aristolochic acid and the etiology of endemic (Balkan) nephropathy. Proc Natl Acad Sci U S A. 2007;104:12129–12134. doi: 10.1073/pnas.0701248104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hainaut P, Pfeifer GP. Patterns of p53 G-->T transversions in lung cancers reflect the primary mutagenic signature of DNA-damage by tobacco smoke. Carcinogenesis. 2001;22:367–374. doi: 10.1093/carcin/22.3.367. [DOI] [PubMed] [Google Scholar]
- Hainaut P, Wiman KG. 25 years of p53 research. Springer; Dordrecht, Netherlands: 2005. [Google Scholar]
- Hamroun D, Kato S, Ishioka C, Claustres M, Beroud C, Soussi T. The UMD TP53 database and website: update and revisions. Hum Mutat. 2006;27:14–20. doi: 10.1002/humu.20269. [DOI] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- Hanawalt PC. Subpathways of nucleotide excision repair and their regulation. Oncogene. 2002;21:8949–8956. doi: 10.1038/sj.onc.1206096. [DOI] [PubMed] [Google Scholar]
- Hecht SS. Tobacco smoke carcinogens and lung cancer. J. Natl. Cancer Inst. 1999;91:1194–1210. doi: 10.1093/jnci/91.14.1194. [DOI] [PubMed] [Google Scholar]
- Hendrich B, Hardeland U, Ng H-H, Jiricny J, Bird A. The thymine glycosylase MBD4 can bind to the product of deamination at methylated CpG sites. Nature. 1999;401:301–304. doi: 10.1038/45843. [DOI] [PubMed] [Google Scholar]
- Hofseth LJ, Hussain SP, Harris CC. p53: 25 years after its discovery. Trends Pharmacol Sci. 2004;25:177–181. doi: 10.1016/j.tips.2004.02.009. [DOI] [PubMed] [Google Scholar]
- Hollstein M, Marion MJ, Lehman T, Welsh J, Harris CC, Martel-Planche G, Kusters I, Montesano R. p53 mutations at A:T base pairs in angiosarcomas of vinyl chloride-exposed factory workers. Carcinogenesis. 1994;15:1–3. doi: 10.1093/carcin/15.1.1. [DOI] [PubMed] [Google Scholar]
- Husgafvel-Pursiainen K, Hackman P, Ridanpaa M, Anttila S, Karjalainen A, Partanen T, Taikina-Aho O, Heikkila L, Vainio H. K-ras mutations in human adenocarcinoma of the lung: association with smoking and occupational exposure to asbestos. Int J Cancer. 1993;53:250–256. doi: 10.1002/ijc.2910530213. [DOI] [PubMed] [Google Scholar]
- Hussain SP, Harris CC. Molecular epidemiology of human cancer: contribution of mutation spectra studies of tumor suppressor genes. Cancer Res. 1998;58:4023–4037. [PubMed] [Google Scholar]
- Hussain SP, Hollstein MH, Harris CC. p53 tumor suppressor gene: at the crossroads of molecular carcinogenesis, molecular epidemiology, and human risk assessment. Ann N Y Acad Sci. 2000;919:79–85. doi: 10.1111/j.1749-6632.2000.tb06870.x. [DOI] [PubMed] [Google Scholar]
- Hussain SP, Schwank J, Staib F, Wang XW, Harris CC. TP53 mutations and hepatocellular carcinoma: insights into the etiology and pathogenesis of liver cancer. Oncogene. 2007;26:2166–2176. doi: 10.1038/sj.onc.1210279. [DOI] [PubMed] [Google Scholar]
- Jackson MA, Lea I, Rashid A, Peddada SD, Dunnick JK. Genetic alterations in cancer knowledge system: analysis of gene mutations in mouse and human liver and lung tumors. Toxicol Sci. 2006;90:400–418. doi: 10.1093/toxsci/kfj101. [DOI] [PubMed] [Google Scholar]
- Jaworski M, Hailfinger S, Buchmann A, Hergenhahn M, Hollstein M, Ittrich C, Schwarz M. Human p53 knock-in (hupki) mice do not differ in liver tumor response from their counterparts with murine p53. Carcinogenesis. 2005;26:1829–1834. doi: 10.1093/carcin/bgi142. [DOI] [PubMed] [Google Scholar]
- Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat Rev Genet. 2002;3:415–428. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- Jones S, Zhang X, Parsons DW, Lin JC, Leary RJ, Angenendt P, Mankoo P, Carter H, Kamiyama H, Jimeno A, Hong SM, Fu B, Lin MT, Calhoun ES, Kamiyama M, Walter K, Nikolskaya T, Nikolsky Y, Hartigan J, Smith DR, Hidalgo M, Leach SD, Klein AP, Jaffee EM, Goggins M, Maitra A, Iacobuzio-Donahue C, Eshleman JR, Kern SE, Hruban RH, Karchin R, Papadopoulos N, Parmigiani G, Vogelstein B, Velculescu VE, Kinzler KW. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science. 2008;321:1801–1806. doi: 10.1126/science.1164368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato S, Han SY, Liu W, Otsuka K, Shibata H, Kanamaru R, Ishioka C. Understanding the function-structure and function-mutation relationships of p53 tumor suppressor protein by high-resolution missense mutation analysis. Proc Natl Acad Sci U S A. 2003;100:8424–8429. doi: 10.1073/pnas.1431692100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohara A, Suzuki T, Honma M, Ohwada T, Hayashi M. Mutagenicity of aristolochic acid in the lambda/lacZ transgenic mouse (MutaMouse). Mutat Res. 2002;515:63–72. doi: 10.1016/s1383-5718(01)00350-3. [DOI] [PubMed] [Google Scholar]
- Krawczak M, Ball EV, Cooper DN. Neighboring-nucleotide effects on the rates of germ-line single-base-pair substitution in human genes. Am J Hum Genet. 1998;63:474–488. doi: 10.1086/301965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusumoto R, Masutani C, Iwai S, Hanaoka F. Translesion synthesis by human DNA polymerase eta across thymine glycol lesions. Biochemistry. 2002;41:6090–6099. doi: 10.1021/bi025549k. [DOI] [PubMed] [Google Scholar]
- Lambert IB, Singer TM, Boucher SE, Douglas GR. Detailed review of transgenic rodent mutation assays. Mutat Res. 2005;590:1–280. doi: 10.1016/j.mrrev.2005.04.002. [DOI] [PubMed] [Google Scholar]
- Lindahl T. Instability and decay of the primary structure of DNA. Nature. 1993;362:709–715. doi: 10.1038/362709a0. [DOI] [PubMed] [Google Scholar]
- Liu Z, Hergenhahn M, Schmeiser HH, Wogan GN, Hong A, Hollstein M. Human tumor p53 mutations are selected for in mouse embryonic fibroblasts harboring a humanized p53 gene. Proc Natl Acad Sci U S A. 2004;101:2963–2968. doi: 10.1073/pnas.0308607101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Muehlbauer KR, Schmeiser HH, Hergenhahn M, Belharazem D, Hollstein MC. p53 mutations in benzo(a)pyrene-exposed human p53 knock-in murine fibroblasts correlate with p53 mutations in human lung tumors. Cancer Res. 2005;65:2583–2587. doi: 10.1158/0008-5472.CAN-04-3675. [DOI] [PubMed] [Google Scholar]
- Loeb LA, Bielas JH, Beckman RA. Cancers exhibit a mutator phenotype: clinical implications. Cancer Res. 2008;68:3551–3557. doi: 10.1158/0008-5472.CAN-07-5835. discussion 3557. [DOI] [PubMed] [Google Scholar]
- Lord GM, Cook T, Arlt VM, Schmeiser HH, Williams G, Pusey CD. Urothelial malignant disease and Chinese herbal nephropathy. Lancet. 2001;358:1515–1516. doi: 10.1016/s0140-6736(01)06576-x. [DOI] [PubMed] [Google Scholar]
- Lord GM, Hollstein M, Arlt VM, Roufosse C, Pusey CD, Cook T, Schmeiser HH. DNA adducts and p53 mutations in a patient with aristolochic acid-associated nephropathy. Am J Kidney Dis. 2004;43:e11–17. doi: 10.1053/j.ajkd.2003.11.024. [DOI] [PubMed] [Google Scholar]
- Luch A. Nature and nurture - lessons from chemical carcinogenesis. Nat Rev Cancer. 2005;5:113–125. doi: 10.1038/nrc1546. [DOI] [PubMed] [Google Scholar]
- Luo JL, Tong WM, Yoon JH, Hergenhahn M, Koomagi R, Yang Q, Galendo D, Pfeifer GP, Wang ZQ, Hollstein M. UV-induced DNA damage and mutations in Hupki (human p53 knock-in) mice recapitulate p53 hotspot alterations in sun-exposed human skin. Cancer Res. 2001a;61:8158–8163. [PubMed] [Google Scholar]
- Luo JL, Yang Q, Tong WM, Hergenhahn M, Wang ZQ, Hollstein M. Knock-in mice with a chimeric human/murine p53 gene develop normally and show wild-type p53 responses to DNA damaging agents: a new biomedical research tool. Oncogene. 2001b;20:320–328. doi: 10.1038/sj.onc.1204080. [DOI] [PubMed] [Google Scholar]
- Lynch HT, de la Chapelle A. Hereditary colorectal cancer. N Engl J Med. 2003;348:919–932. doi: 10.1056/NEJMra012242. [DOI] [PubMed] [Google Scholar]
- Marshall CJ, Vousden KH, Phillips DH. Activation of c-Ha-ras-1 proto-oncogene by in vitro modification with a chemical carcinogen, benzo(a)pyrene diol-epoxide. Nature. 1984;310:586–589. doi: 10.1038/310586a0. [DOI] [PubMed] [Google Scholar]
- Millar CB, Guy J, Sansom OJ, Selfridge J, MacDougall E, Hendrich B, Keightley PD, Bishop SM, Clarke AR, Bird A. Enhanced CpG mutability and tumorigenesis in MBD4-deficient mice. Science. 2002;297:403–405. doi: 10.1126/science.1073354. [DOI] [PubMed] [Google Scholar]
- Montesano R, Hainaut P, Wild CP. Hepatocellular carcinoma: from gene to public health. J Natl Cancer Inst. 1997;89:1844–1851. doi: 10.1093/jnci/89.24.1844. [DOI] [PubMed] [Google Scholar]
- Mossman BT, Klein G, Zur Hausen H. Modern criteria to determine the etiology of human carcinogens. Semin Cancer Biol. 2004;14:449–452. doi: 10.1016/j.semcancer.2004.06.008. [DOI] [PubMed] [Google Scholar]
- Neddermann P, Gallinari P, Lettieri T, Schmid D, Truong O, Hsuan JJ, Wiebauer K, Jiricny J. Cloning and expression of human G/T mismatch-specific thymine-DNA glycosylase. J. Biol. Chem. 1996;271:12767–12774. doi: 10.1074/jbc.271.22.12767. [DOI] [PubMed] [Google Scholar]
- Nedelko T, Arlt VM, Phillips DH, Hollstein M. TP53 mutation signature supports involvement of aristolochic acid in the aetiology of endemic nephropathy-associated tumours. Int J Cancer. 2009;124:987–990. doi: 10.1002/ijc.24006. [DOI] [PubMed] [Google Scholar]
- Nortier JL, Martinez MC, Schmeiser HH, Arlt VM, Bieler CA, Petein M, Depierreux MF, De Pauw L, Abramowicz D, Vereerstraeten P, Vanherweghem JL. Urothelial carcinoma associated with the use of a Chinese herb (Aristolochia fangchi). N Engl J Med. 2000;342:1686–1692. doi: 10.1056/NEJM200006083422301. [DOI] [PubMed] [Google Scholar]
- Olivier M, Hussain SP, Caron de Fromentel C, Hainaut P, Harris CC. TP53 mutation spectra and load: a tool for generating hypotheses on the etiology of cancer. IARC Sci Publ. 2004:247–270. [PubMed] [Google Scholar]
- Parada LF, Tabin CJ, Shih C, Weinberg RA. Human EJ bladder carcinoma oncogene is homologue of Harvey sarcoma virus ras gene. Nature. 1982;297:474–478. doi: 10.1038/297474a0. [DOI] [PubMed] [Google Scholar]
- Parsons DW, Jones S, Zhang X, Lin JC, Leary RJ, Angenendt P, Mankoo P, Carter H, Siu IM, Gallia GL, Olivi A, McLendon R, Rasheed BA, Keir S, Nikolskaya T, Nikolsky Y, Busam DA, Tekleab H, Diaz LA, Jr., Hartigan J, Smith DR, Strausberg RL, Marie SK, Shinjo SM, Yan H, Riggins GJ, Bigner DD, Karchin R, Papadopoulos N, Parmigiani G, Vogelstein B, Velculescu VE, Kinzler KW. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petitjean A, Achatz MI, Borresen-Dale AL, Hainaut P, Olivier M. TP53 mutations in human cancers: functional selection and impact on cancer prognosis and outcomes. Oncogene. 2007;26:2157–2165. doi: 10.1038/sj.onc.1210302. [DOI] [PubMed] [Google Scholar]
- Pfau W, Schmeiser HH, Wiessler M. Aristolochic acid binds covalently to the exocyclic amino group of purine nucleotides in DNA. Carcinogenesis. 1990;11:313–319. doi: 10.1093/carcin/11.2.313. [DOI] [PubMed] [Google Scholar]
- Pfeifer GP. p53 mutational spectra and the role of methylated CpG sequences. Mutat Res. 2000;450:155–166. doi: 10.1016/s0027-5107(00)00022-1. [DOI] [PubMed] [Google Scholar]
- Pfeifer GP. Mutagenesis at methylated CpG sequences. Curr Top Microbiol Immunol. 2006;301:259–281. doi: 10.1007/3-540-31390-7_10. [DOI] [PubMed] [Google Scholar]
- Pfeifer GP, Denissenko MF, Olivier M, Tretyakova N, Hecht SS, Hainaut P. Tobacco smoke carcinogens, DNA damage and p53 mutations in smoking-associated cancers. Oncogene. 2002;21:7435–7451. doi: 10.1038/sj.onc.1205803. [DOI] [PubMed] [Google Scholar]
- Pfeifer GP, Drouin R, Riggs AD, Holmquist GP. In vivo mapping of a DNA adduct at nucleotide resolution: detection of pyrimidine (6-4) pyrimidone photoproducts by ligation-mediated polymerase chain reaction. Proc. Natl. Acad. Sci. U.S.A. 1991;88:1374–1378. doi: 10.1073/pnas.88.4.1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeifer GP, You YH, Besaratinia A. Mutations induced by ultraviolet light. Mutat Res. 2005;571:19–31. doi: 10.1016/j.mrfmmm.2004.06.057. [DOI] [PubMed] [Google Scholar]
- Quintanilla M, Brown K, Ramsden M, Balmain A. Carcinogen-specific mutation and amplification of Ha-ras during mouse skin carcinogenesis. Nature. 1986;322:78–80. doi: 10.1038/322078a0. [DOI] [PubMed] [Google Scholar]
- Rajagopalan H, Lengauer C. Aneuploidy and cancer. Nature. 2004;432:338–341. doi: 10.1038/nature03099. [DOI] [PubMed] [Google Scholar]
- Rauch TA, Zhong X, Wu X, Wang M, Kernstine KH, Wang Z, Riggs AD, Pfeifer GP. High-resolution mapping of DNA hypermethylation and hypomethylation in lung cancer. Proc Natl Acad Sci U S A. 2008;105:252–257. doi: 10.1073/pnas.0710735105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinbold M, Luo JL, Nedelko T, Jerchow B, Murphy ME, Whibley C, Wei Q, Hollstein M. Common tumour p53 mutations in immortalized cells from Hupki mice heterozygous at codon 72. Oncogene. 2008;27:2788–2794. doi: 10.1038/sj.onc.1210932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rideout WM, III, Coetzee GA, Olumi AF, Jones PA. 5-Methylcytosine as an endogenous mutagen in the human LDL receptor and p53 genes. Science. 1990;249:1288–1290. doi: 10.1126/science.1697983. [DOI] [PubMed] [Google Scholar]
- Ruggeri B, DiRado M, Zhang SY, Bauer B, Goodrow T, Klein-Szanto AJP. Benzo[a]pyrene-induced murine skin tumors exhibit frequent and characteristic G to T mutations in the p53 gene. Proc. Natl. Acad. Sci. USA. 1993;90:1013–1017. doi: 10.1073/pnas.90.3.1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuels Y, Wang Z, Bardelli A, Silliman N, Ptak J, Szabo S, Yan H, Gazdar A, Powell SM, Riggins GJ, Willson JK, Markowitz S, Kinzler KW, Vogelstein B, Velculescu VE. High frequency of mutations of the PIK3CA gene in human cancers. Science. 2004;304:554. doi: 10.1126/science.1096502. [DOI] [PubMed] [Google Scholar]
- Schmutte C, Jones PA. Involvement of DNA methylation in human carcinogenesis. Biol. Chem. 1998;379:377–388. doi: 10.1515/bchm.1998.379.4-5.377. [DOI] [PubMed] [Google Scholar]
- Schmutte C, Yang AS, Beart RW, Jones PA. Base excision repair of U:G mismatches at a mutational hotspot in the p53 gene is more efficient than base excision repair of T:G mismatches in extracts of human colon tumors. Cancer Res. 1995;55:3742–3746. [PubMed] [Google Scholar]
- Sharma SV, Bell DW, Settleman J, Haber DA. Epidermal growth factor receptor mutations in lung cancer. Nat Rev Cancer. 2007;7:169–181. doi: 10.1038/nrc2088. [DOI] [PubMed] [Google Scholar]
- Shen JC, Rideout WM, III, Jones PA. The rate of hydrolytic deamination of 5-methylcytosine in double-stranded DNA. Nucleic Acids Res. 1994;22:972–976. doi: 10.1093/nar/22.6.972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjoblom T, Jones S, Wood LD, Parsons DW, Lin J, Barber TD, Mandelker D, Leary RJ, Ptak J, Silliman N, Szabo S, Buckhaults P, Farrell C, Meeh P, Markowitz SD, Willis J, Dawson D, Willson JK, Gazdar AF, Hartigan J, Wu L, Liu C, Parmigiani G, Park BH, Bachman KE, Papadopoulos N, Vogelstein B, Kinzler KW, Velculescu VE. The consensus coding sequences of human breast and colorectal cancers. Science. 2006;314:268–274. doi: 10.1126/science.1133427. [DOI] [PubMed] [Google Scholar]
- Smith LE, Denissenko MF, Bennett WP, Li H, Amin S, Tang M-s, Pfeifer GP. Targeting of lung cancer mutational hotspots by polycyclic aromatic hydrocarbons. J. Natl. Cancer Inst. 2000;92:803–811. doi: 10.1093/jnci/92.10.803. [DOI] [PubMed] [Google Scholar]
- Soussi T, Beroud C. Significance of TP53 mutations in human cancer: a critical analysis of mutations at CpG dinucleotides. Hum Mutat. 2003;21:192–200. doi: 10.1002/humu.10189. [DOI] [PubMed] [Google Scholar]
- Sukumar S, Notario V, Martin-Zanca D, Barbacid M. Induction of mammary carcinomas in rats by nitroso-methylurea involves malignant activation of H-ras-1 locus by single point mutations. Nature. 1983;306:658–661. doi: 10.1038/306658a0. [DOI] [PubMed] [Google Scholar]
- Taparowsky E, Suard Y, Fasano O, Shimizu K, Goldfarb M, Wigler M. Activation of the T24 bladder carcinoma transforming gene is linked to a single amino acid change. Nature. 1982;300:762–765. doi: 10.1038/300762a0. [DOI] [PubMed] [Google Scholar]
- Thilly WG. Mutational spectrometry in animal toxicity testing. Annu Rev Pharmacol Toxicol. 1990;30:369–385. doi: 10.1146/annurev.pa.30.040190.002101. [DOI] [PubMed] [Google Scholar]
- Thilly WG. Have environmental mutagens caused oncomutations in people? Nat Genet. 2003;34:255–259. doi: 10.1038/ng1205. [DOI] [PubMed] [Google Scholar]
- Tommasi S, Denissenko MF, Pfeifer GP. Sunlight induces pyrimidine dimers preferentially at 5-methylcytosine bases. Cancer Res. 1997;57:4727–4730. [PubMed] [Google Scholar]
- Tong WM, Lee MK, Galendo D, Wang ZQ, Sabapathy K. Aflatoxin-B exposure does not lead to p53 mutations but results in enhanced liver cancer of Hupki (human p53 knock-in) mice. Int J Cancer. 2006;119:745–749. doi: 10.1002/ijc.21890. [DOI] [PubMed] [Google Scholar]
- Tornaletti S, Pfeifer GP. Complete and tissue-independent methylation of CpG sites in the p53 gene: implications for mutations in human cancers. Oncogene. 1995;10:1493–1499. [PubMed] [Google Scholar]
- Toyooka S, Tsuda T, Gazdar AF. The TP53 gene, tobacco exposure, and lung cancer. Hum Mutat. 2003;21:229–239. doi: 10.1002/humu.10177. [DOI] [PubMed] [Google Scholar]
- US-EPA. Gilman P. In: Health Assessment Document for Diesel Engine Exhaust. U.S. Environmental Protection Agency NCfEA, editor. Washington, DC: 2002. [Google Scholar]
- Vineis P, Perera F. Molecular epidemiology and biomarkers in etiologic cancer research: the new in light of the old. Cancer Epidemiol Biomarkers Prev. 2007;16:1954–1965. doi: 10.1158/1055-9965.EPI-07-0457. [DOI] [PubMed] [Google Scholar]
- Vogelstein B, Lane D, Levine AJ. Surfing the p53 network. Nature. 2000;408:307–310. doi: 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- Vom Brocke J, Krais A, Whibley C, Hollstein MC, Schmeiser HH. The carcinogenic air pollutant 3-nitrobenzanthrone induces GC to TA transversion mutations in human p53 sequences. Mutagenesis. 2008 doi: 10.1093/mutage/gen049. [DOI] [PubMed] [Google Scholar]
- vom Brocke J, Schmeiser HH, Reinbold M, Hollstein M. MEF immortalization to investigate the ins and outs of mutagenesis. Carcinogenesis. 2006;27:2141–2147. doi: 10.1093/carcin/bgl101. [DOI] [PubMed] [Google Scholar]
- Walker DR, Bond JP, Tarone RE, Harris CC, Makalowski W, Boguski MS, Greenblatt MS. Evolutionary conservation and somatic mutation hotspot maps of p53: correlation with p53 protein structural and functional features. Oncogene. 1999;18:211–218. doi: 10.1038/sj.onc.1202298. [DOI] [PubMed] [Google Scholar]
- Wang TL, Rago C, Silliman N, Ptak J, Markowitz S, Willson JK, Parmigiani G, Kinzler KW, Vogelstein B, Velculescu VE. Prevalence of somatic alterations in the colorectal cancer cell genome. Proc Natl Acad Sci U S A. 2002;99:3076–3080. doi: 10.1073/pnas.261714699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wink DA, Kasprzak KS, Maragos CM, Elespuru RK, Misra M, Dunams TM, Cebula TA, Koch WH, Andrews AW, Allen JS, Keefe LK. DNA deaminating ability and genotoxicity of nitric oxide and its progenitors. Science. 1991;254:1001–1003. doi: 10.1126/science.1948068. [DOI] [PubMed] [Google Scholar]
- Wong E, Yang K, Kuraguchi M, Werling U, Avdievich E, Fan K, Fazzari M, Jin B, Brown AM, Lipkin M, Edelmann W. Mbd4 inactivation increases C to T transition mutations and promotes gastrointestinal tumor formation. Proc Natl Acad Sci U S A. 2002;99:14937–14942. doi: 10.1073/pnas.232579299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood LD, Parsons DW, Jones S, Lin J, Sjoblom T, Leary RJ, Shen D, Boca SM, Barber T, Ptak J, Silliman N, Szabo S, Dezso Z, Ustyanksky V, Nikolskaya T, Nikolsky Y, Karchin R, Wilson PA, Kaminker JS, Zhang Z, Croshaw R, Willis J, Dawson D, Shipitsin M, Willson JK, Sukumar S, Polyak K, Park BH, Pethiyagoda CL, Pant PV, Ballinger DG, Sparks AB, Hartigan J, Smith DR, Suh E, Papadopoulos N, Buckhaults P, Markowitz SD, Parmigiani G, Kinzler KW, Velculescu VE, Vogelstein B. The genomic landscapes of human breast and colorectal cancers. Science. 2007;318:1108–1113. doi: 10.1126/science.1145720. [DOI] [PubMed] [Google Scholar]
- Yoon JH, Smith LE, Feng Z, Tang M, Lee CS, Pfeifer GP. Methylated CpG dinucleotides are the preferential targets for G-to-T transversion mutations induced by benzo[a]pyrene diol epoxide in mammalian cells: similarities with the p53 mutation spectrum in smoking-associated lung cancers. Cancer Res. 2001;61:7110–7117. [PubMed] [Google Scholar]
- You YH, Li C, Pfeifer GP. Involvement of 5-methylcytosine in sunlight-induced mutagenesis. J Mol Biol. 1999;293:493–503. doi: 10.1006/jmbi.1999.3174. [DOI] [PubMed] [Google Scholar]
- Zarbl H, Sukumar S, Arthur AV, Martin-Zanca D, Barbacid M. Direct mutagenesis of Ha-ras-1 oncogenes by N-nitroso-N-methylurea during initiation of mammary carcinogenesis in rats. Nature. 1985;315:382–385. doi: 10.1038/315382a0. [DOI] [PubMed] [Google Scholar]
- Zuo S, Boorstein RJ, Teebor GW. Oxidative damage to 5-methylcytosine in DNA. Nucleic Acids Res. 1995;25:3239–3243. doi: 10.1093/nar/23.16.3239. [DOI] [PMC free article] [PubMed] [Google Scholar]