Abstract
Background
Asthma is a heterogeneous disease that is caused by the interaction of genetic susceptibility with environmental influences. Genome-wide association studies (GWAS) represent a powerful approach to investigate the association of DNA variants with disease susceptibility. To date, few GWAS for asthma have been reported.
Objectives
GWAS was performed on a population of severe or difficult-to-treat asthmatics to identify genes that are involved in the pathogenesis of asthma.
Methods
292,443 SNPs were tested for association with asthma in 473 TENOR cases and 1,892 Illumina general population controls. Asthma-related quantitative traits (total serum IgE, FEV1, FVC, and FEV1/FVC) were also tested in identified candidate regions in 473 TENOR cases and 363 phenotyped controls without a history of asthma to further analyze GWAS results. Imputation was performed in identified candidate regions for analysis with denser SNP coverage.
Results
Multiple SNPs in the RAD50-IL13 region on chromosome 5q31.1 were associated with asthma: rs2244012 in intron 2 of RAD50 (P = 3.04E-07). The HLA-DR/DQ region on chromosome 6p21.3 was also associated with asthma: rs1063355 in the 3’ UTR of HLA-DQB1 (P = 9.55E-06). Imputation identified several significant SNPs in the TH2 locus control region (LCR) 3’ of RAD50. Imputation also identified a more significant SNP, rs3998159 (P = 1.45E-06), between HLA-DQB1 and HLA-DQA2.
Conclusion
This GWAS confirmed the important role of TH2 cytokine and antigen presentation genes in asthma at a genome-wide level and the importance of additional investigation of these two regions to delineate their structural complexity and biologic function in the development of asthma.
Keywords: Asthma, GWAS, RAD50, IL13, HLA-DQB1, TENOR
INTRODUCTION
Asthma is a complex disease that is caused by the interaction of genetic susceptibility with environmental influences. Genome-wide linkage studies, candidate-gene association studies, and genome-wide association studies (GWAS) represent three major approaches to investigate the association between genetic variants and disease development.
Genome-wide linkage studies have consistently identified regions linked to asthma or asthma-related traits on chromosome 2q, 5q, 6p, 12q, and 13q [1]. The most highly replicated regions with obvious candidate genes are chromosome 5q31-33 (including interleukin (IL)5, IL13, IL4, CD14, and adrenergic beta-2-receptor (ADRB2)) and 6p21 (including lymphotoxin alpha (LTA or TNFB), tumor necrosis factor (TNF), major histocompatibility complex, class II, DQ beta 1 (HLA-DQB1), and DR beta 1 (HLA-DRB1)) [2]. In addition, a recent meta-analysis of genome-wide linkage studies of asthma, bronchial hyperresponsiveness (BHR), positive allergen skin prick test (SPT), and total immunoglobulin E (IgE) identified overlapping regions for multiple phenotypes on chromosomes 5q and 6p as well as 3p and 7p [3]. Unfortunately, genome-wide linkage studies can only identify genes with relative strong effects in broad regions that include many genes. Positional cloning studies have identified six genes for asthma: a disintegrin and metalloprotease domain 33 (ADAM33) on chromosome 20p13 [4], dipeptidyl-peptidase 10 (DPP10) on 2q14.1 [5], PHD finger protein 11 (PHF11) on 13q14.11 [6], neuropeptide S receptor 1 (NPSR1 or GPRA) on 7p14.3 [7], major histocompatibility complex, class I, G (HLA-G) on 6p21.3 [8], and cytoplasmic FMR1 interacting protein 2 (CYFIP2) on 5q33.3 [9].
Candidate-gene association studies have identified over 100 genes for asthma and asthma-related traits [2, 10, 11]. Although candidate-gene association studies have identified many genes, only a few have been replicated extensively. Thus, only 14 genes including genes on 5q and 6p (ADRB2, interleukin 4 receptor (IL4R), HLA-DRB1, IL13, CD14, TNF, membrane-spanning 4-domains, subfamily A, member 2 (MS4A2 or FCER1B), IL4, ADAM33, signal transducer and activator of transcription 6, interleukin-4 induced (STAT6), IL10, HLA-DQB1, glutathione S-transferase pi 1 (GSTP1), and LTA) that have been replicated in more than 20 independent studies [10]. Even for highly replicated genes, replication might be due to ‘winner’s bias’ and/or loose replication standard (gene as a unit and related phenotypes).
A GWAS is a hypothesis-free approach able to identify novel genes with mild/moderate effects and, thus has become the best approach for studying association between genes and common disease phenotypes. To date, only four GWAS have been performed for asthma and asthma-related traits [12]. The first GWAS of childhood asthma identified ORM1-like 3 (ORMDL3) on chromosome 17q12 [13]. The second GWAS of serum YKL-40 levels identified chitinase 3-like 1 (CHI3L1) on 1q32 [14]. The third GWAS was for a related trait, total serum IgE levels, and the most significant SNPs are in the Fc fragment of IgE, high affinity I, receptor for alpha polypeptide gene (FCER1A) on chromosome 1q23 and the second highest region observed was RAD50 on 5q31 [15]. The fourth GWAS of childhood asthma indicated phosphodiesterase 4D, cAMP-specific (phosphodiesterase E3 dunce homolog, Drosophila) (PDE4D) on chromosome 5q12 [16].
In this study, we performed GWAS of asthma in The Epidemiology and Natural History of Asthma: Outcomes and Treatment Regimens (TENOR) population of severe or difficult to treat asthmatics to search for novel genes and to confirm previously identified genes involved in asthma. The purpose of the TENOR study was to investigate the natural history of asthma in a large cohort of well characterized asthmatics with severe or difficult to treat asthma; no treatment intervention was involved and patients continued to be treated by their asthma specialist [17–19].
METHODS
Study subjects
The TENOR study was a multi-center observational and longitudinal cohort study of 4,756 asthmatics described as “severe or difficult-to-treat” by their physicians, sponsored by Genentech and Novartis [17]. Subjects were included if they had physician-characterized difficult-to-treat asthma, and met additional criteria based on frequency of urgent care visits and/or the use of multiple controller medications. The clinical sites from the original TENOR study were contacted and invited to participate in this study. Sites that agreed were mailed Oragene DNA saliva collection kits (DNA Genotek, Inc), labeled with the TENOR participant ID. Sites then mailed the kits to participating individuals, who sent their collected samples to the Center for Human Genomics at Wake Forest University School of Medicine. This process was required to maintain anonymity between investigators at Wake Forest University and the study participants. Unfortunately the TENOR study had ended (end of 2004) before this project started so it was difficult to re-contact participants. 607 samples had sufficient DNA for successful SNP genotyping. Table 1 shows the demographic data for the TENOR cases and the two control populations. The TENOR asthmatics genotyped were similar in characteristics to the larger TENOR cohort.
Table I.
Demographics (Mean_SD) of subjects in TENOR, Illumina, and phenotyped control
| TENOR case | Illumina control | Phenotyped control | |
|---|---|---|---|
| N | 473 | 1892 | 363 |
| Age (yrs) | 46.9 ± 18.4 | 31.4 ± 21.9 | 32.1 ± 10.3 |
| Gender (% Female) | 63.0 | 62.5 | 61.2 |
| Log total IgE (Geometric mean) | 1.9 ± 0.7 (88.5) | NA | 1.3 ± 0.7 (19.6) |
| FEV1 (%) | 78.5 ± 21.6 | NA | 97.9 ± 10.7 |
| FVC (%) | 89.5 ± 19.9 | NA | 100.8 ± 11.2 |
| FEV1/FVC | 0.72 ± 0.12 | NA | 0.82 ± 0.08 |
Illumina controls were used for GWAS.
The Wake Forest phenotyped controls were mainly recruited through the NHLBI Collaborative Study on the Genetics of Asthma and the NHLBI Severe Asthma Research Program and were genotyped as a subset of the NHLBI STAMPEED study.
General population controls were obtained using the Illumina iControlDB client to download genotypes for 3,294 Caucasian individuals with genotype data available from any of the three available HumanHap550k products (v1, v3, and −2v3). As shown in Table 1, only age and gender data are available. Additional control samples for asthma-related quantitative traits were obtained from a separate GWAS for asthma. These 363 phenotyped controls had no personal or family history of asthma and had normal pulmonary function including lack of bronchial hyperresponsiveness or bronchodilator reversibility. Testing also included measures of atopy including total serum IgE levels (Table 1). HapMap samples (N = 262) to be used for genetic ancestry check were also downloaded from the iControlDB database (Illumina, Inc.) after selecting the HumanHap300_v1 genotyping product.
DNA was isolated using the protocol described by DNA Genotek, and SNP genotyping was performed using the Illumina HumanCNV370 BeadChip. The samples were clustered by first applying Illumina’s cluster definition, removing samples with call rates less than 0.90, and then re-clustering using the samples themselves.
Statistical analysis
PLINK (version 1.06, URL: http://pngu.mgh.harvard.edu/purcell/plink/) [20] was the main software used to perform statistical analysis unless otherwise mentioned.
Quality control (QC) was applied to cases and controls separately since they were genotyped using slightly different Illumina products. Genetic ancestry of the TENOR cases was determined using the HapMap 300k dataset as a reference. Fixed 3 groups clustering and pairwise population concordance (PPC) of 1.0E-05 based on identity-by-state (IBS) were used to cross-validate ethnic group identity. Subjects were removed if they 1) were not of European white descent, 2) had low genotyping call rates (< 95%), 3) were discrepant or ambiguous for genetic sex (heterozygous haploid genotype percentage ≥ 0.01 or X chromosome homozygosity F ≥ 0.9), 4) failed the cryptic relatedness check (PI_HAT > 0.125), 5) were detected as an outlier (> 6 standard deviation for the first or se cond principal component). After subjects meeting these criteria were deleted, SNPs were deleted if the call rates were low (95%) or were inconsistent with Hardy-Weinberg Equilibrium (HWE) (P < 10E-04). QC was then applied on the subjects and SNPs of merged case-control dataset as done separately. SNPs were also deleted if the minor allele frequency (MAF) was less than 0.05 in cases and controls or the HWE P value was less than 0.01 in controls only.
Asthma susceptibility was analyzed by comparing the non-Hispanic white TENOR cases to the general population Illumina controls. To reduce population stratification, four controls were matched with every one case based on pairwise IBS. Principal components were generated using principal components analysis (PCA) in EIGENSTRAT (version 3.0, URL: http://genepath.med.harvard.edu/~reich/Software.htm) [21]. Sex, age, and significant principal components were used as covariates in the logistic additive model. Genomic control (GC) was applied on P values to reduce population stratification further [22]. A linear model was analyzed in GWAS-identified candidate regions in 473 TENOR cases and 363 phenotyped controls for asthma-related quantitative traits (total serum IgE, % predicted FEV1, FVC, and FEV1/FVC).
Haploview (URL: http://www.broad.mit.edu/mpg/haploview/) was used to generate linkage disequilibrium plots [23]. 95% confidence intervals on D’ was used to define blocks [24]. SNAP (version 2.0, URL: http://www.broad.mit.edu/mpg/snap/) was used to generate the association plots [25]. Imputation was performed based on HapMap II CEU genotype data [26] using MACH (version 1.0, URL: http://www.sph.umich.edu/csg/abecasis/MaCH/index.html) [27]. Association of candidate SNPs with nearby gene expression data in lymphocytes was performed based on GENEVAR dataset (URL: http://www.sanger.ac.uk/humgen/genevar/) [28] by using WGAViewer [29].
RESULTS
A total of 607 TENOR cases were genotyped with the HumanCNV370 BeadChip. After removal of non-white samples (see Figure E1 in the Online Repository) and removal based on the QC criteria described above, data from 474 asthmatics were carried forward to analysis. Of the 3,294 Illumina Caucasian controls downloaded from iControldb, 3,141 Illumina controls passed QC. After merging 474 TENOR cases with 3,141 Illumina controls and evaluating the combined QC metrics, 473 cases and 3,106 controls were retained. To reduce population stratification, four controls were matched with every one case based on pairwise IBS, thus 473 cases and 1,892 Illumina controls were used for GWAS (see Table I for demographics and Figure E2 in the Online Repository). After QC analysis of the 318,075 common SNPs, 292,443 SNPs were retained for the GWAS.
GWAS of asthma was performed on 292,443 SNPs of 473 TENOR cases and 1,892 Illumina controls with sex, age, and significant principal components as covariates in the logistic additive model (see Figure 1). Genomic control (GC) was applied to P values to reduce population stratification (genomic inflation factor = 1.073 and 1.000 before and after adjustment, see Figure E3 in the Online Repository). In total, 248 SNPs had GC-adjusted P values ≤ 1.0E-03 (see Table E1 in the Online Repository). Focusing on SNPs with a GC-adjusted P values ≤ 1.0E-04 and at least two neighboring SNPs (+/− 100 kb) with GC-adjusted P values ≤ 1.0E-03, six regions were identified: RAD50-IL13 on chromosome 5q31.1, HLA-DR/DQ on 6p21.32, low density lipoprotein-related protein 1B (LRP1B) on 2q22.1-22.2, sorting nexin 10 (SNX10) on 7p15.2, carbonic anhydrase X (CA10) on 17q21.33, and potassium inwardly-rectifying channel, subfamily J, member 2 (KCNJ2) 17q24.3 (see Figure 1 and Table E1 in the Online Repository).
Figure 1.
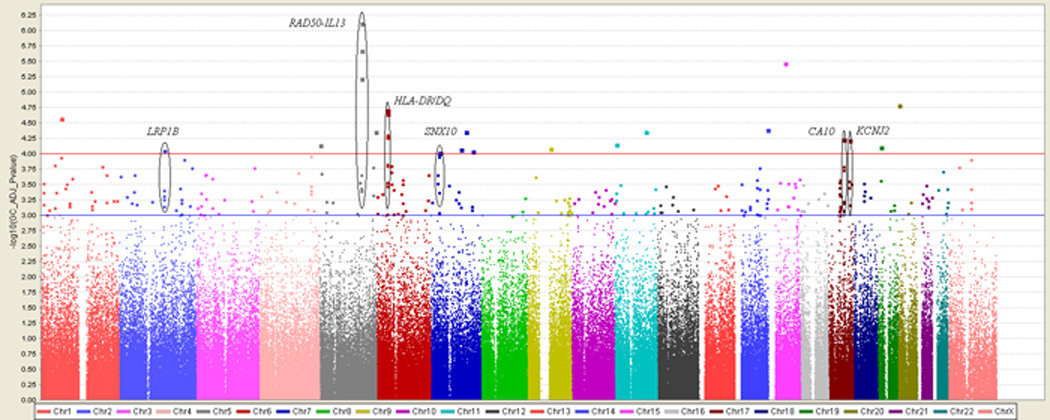
Genome-wide association of 292,443 SNPs in 473 TENOR cases and 1,892 Illumina controls. Color scale of the x-axis represents chromosomes. Negative logarithm transformed GC-adjusted P values are shown on the y-axis.
The RAD50-IL13 region had the strongest evidence for association (see Table II and Figure 2A) with multiple SNPs in this region strongly associated with asthma susceptibility (rs2244012, rs6871536, and rs2897443 in RAD50 ranked highly as 1, 2, and 4, respectively in this study). rs2244012 in intron 2 of RAD50 had an odds ratio of 1.64 (95% CI: 1.36 – 1.97; P = 3.04E-07; GC-adjusted P = 7.69E-07). Three SNPs in or near IL13 (rs2243204 (3’ downstream), rs20541 (Arg130Gln), and rs1295686 (intron 3)) were also associated with asthma (P < 0.001), but in weak LD (0.2 < r2 < 0.3) with SNPs in RAD50 (see Figure 2A and 2B). rs2243300, which is ~5kb upstream of IL4, was weakly associated with asthma (P = 0.0032). Six SNPs downstream of IL5 were not associated with asthma, although they were in weak LD with SNPs in RAD50 (see Table II, Figure 2A and 2B). Four LD blocks were identified based on 95% confidence interval of D’ (see Figure 2B) [24]. Blocks 1 and 2 were each composed of three SNPs downstream of IL5. Block 3 was composed of four SNPs in the intron of RAD50. Block 4 was composed of two SNPs in IL13.
Table II.
Association results of 14 SNPs in TH2 cytokine locus on chromosome 5
| No. | SNP | Position | Gene | Alleles (M:m) |
MAF | OR (95% CI) | P value | Log total IgE |
FEV1/FVC | FEV1 (%) |
FVC (%) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | rs4143832 | 131890876 | IL5 | C:A | 0.178 | 1.23 (1.00 – 1.51) | 5.10E-02 | 4.31E-02 | 7.01E-01 | 5.31E-01 | 8.01E-01 |
| 2 | rs11739623 | 131892051 | IL5 | C:T | 0.255 | 0.91 (0.75 – 1.10) | 3.28E-01 | 4.28E-01 | 8.14E-01 | 3.38E-01 | 3.91E-01 |
| 3 | rs2079103 | 131892405 | IL5 | G:T | 0.237 | 1.06 (0.88 – 1.28) | 5.22E-01 | 2.30E-01 | 8.84E-01 | 4.36E-01 | 6.05E-01 |
| 4 | rs2706399 | 131895601 | IL5 | A:G | 0.497 | 1.00 (0.85 – 1.17) | 9.92E-01 | 9.85E-01 | 8.28E-01 | 5.23E-01 | 4.15E-01 |
| 5 | rs743562 | 131900282 | IL5 | C:T | 0.423 | 1.06 (0.89 – 1.25) | 5.22E-01 | 5.94E-01 | 5.18E-01 | 7.35E-01 | 4.98E-01 |
| 6 | rs739719 | 131900764 | IL5 | G:T | 0.069 | 0.79 (0.57 – 1.11) | 1.72E-01 | 1.20E-01 | 1.96E-01 | 9.95E-01 | 8.02E-01 |
| 7 | rs2244012 | 131929124 | RAD50 | T:C | 0.212 | 1.64 (1.36 – 1.97) | 3.04E-07 | 5.90E-03 | 3.18E-02 | 8.64E-02 | 1.59E-01 |
| 8 | rs2897443 | 131957493 | RAD50 | C:A | 0.199 | 1.58 (1.31 – 1.92) | 2.74E-06 | 1.86E-02 | 9.75E-02 | 1.99E-01 | 2.48E-01 |
| 9 | rs6871536 | 131997773 | RAD50 | T:C | 0.208 | 1.60 (1.33 – 1.94) | 9.03E-07 | 2.61E-03 | 3.36E-02 | 1.38E-01 | 2.58E-01 |
| 10 | rs2237060 | 131998784 | RAD50 | A:C | 0.425 | 0.88 (0.74 – 1.04) | 1.22E-01 | 2.56E-01 | 1.19E-01 | 1.16E-01 | 2.79E-01 |
| 11 | rs1295686 | 132023742 | IL13 | G:A | 0.198 | 1.45 (1.19 – 1.76) | 2.21E-04 | 6.16E-02 | 2.10E-03 | 1.84E-03 | 3.77E-02 |
| 12 | rs20541 | 132023863 | IL13 | C:T | 0.191 | 1.44 (1.19 – 1.76) | 2.50E-04 | 6.06E-02 | 1.83E-03 | 2.18E-03 | 3.69E-02 |
| 13 | rs2243204 | 132027393 | IL13 | C:T | 0.086 | 1.69 (1.29 – 2.21) | 1.31E-04 | 1.67E-03 | 1.11E-02 | 1.39E-02 | 8.10E-02 |
| 14 | rs2243300 | 132031985 | IL4 | G:T | 0.08 | 1.51 (1.15 – 1.99) | 3.17E-03 | 3.14E-02 | 1.57E-03 | 7.89E-03 | 8.58E-02 |
Alleles (M:m): Major allele : minor allele.
MAF: minor allele frequency.
OR (95% CI): odds ratio and 95% confidence interval.
Log total IgE, FEV1/FVC, FEV1, and FVC: Pvalues of asthma-related quantitative traits.
Figure 2.
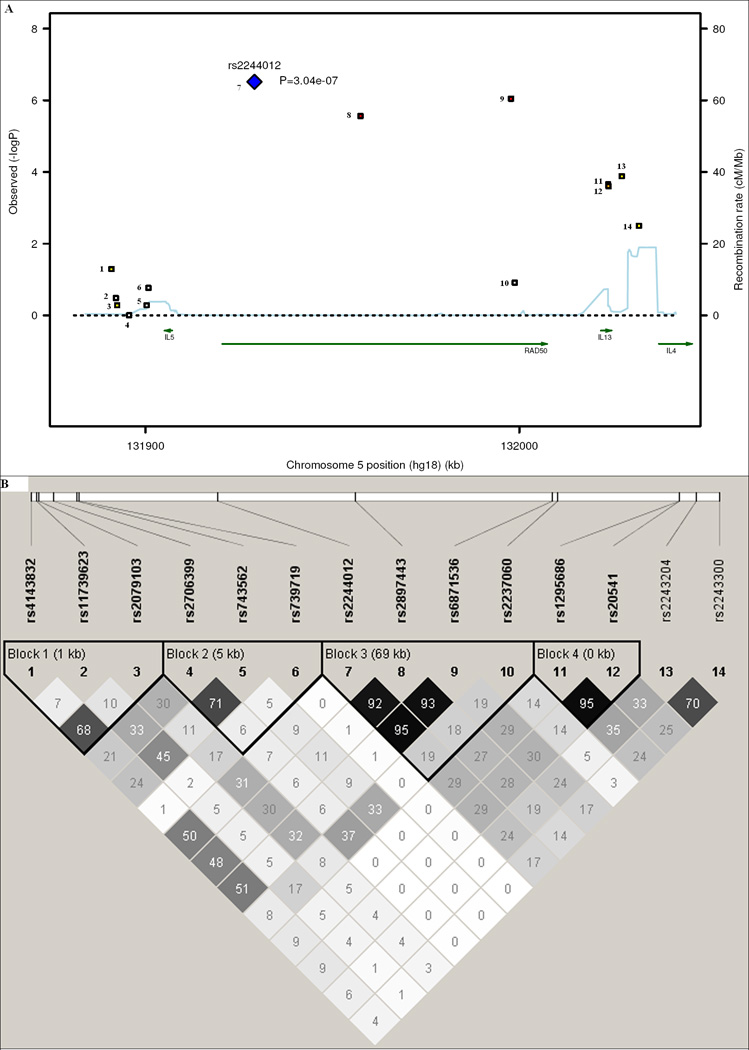
Linkage disequilibrium (LD) and association plot of 14 SNPs in the TH2 cytokine locus. (A) Association plot: negative logarithm transformed P values (left scale) and recombination rate (right scale). (B) LD plot: r2 color scheme was used and labeled. 95% confidence intervals on D’ was used to set up blocks.
Linear model analysis was performed with the 14 SNPs of the TH2 cytokine locus in 473 TENOR cases and 363 phenotyped controls for asthma-related quantitative traits (total serum IgE, FEV1, FVC, and FEV1/FVC) (see Table II). Multiple SNPs in each gene: RAD50, IL13, and IL4 but not IL5 showed significant association (P ≤ 0.05) with asthma-related quantitative traits.
The HLA-DR/DQ region (see Table III and Figure 3A) also showed consistent association with asthma. rs1063355 in the 3’ UTR of HLA-DQB1 had an odds ratio of 0.68 (95% CI: 0.58 – 0.81; P = 9.55E-06; GC-adjusted P = 1.93E-05). Ten of the 46 SNPs in the HLA-DR/DQ region had P ≤ 0.001 (see Table III and Figure 3A). Multiple SNPs in or near butyrophilin-like 2 (BTNL2), HLA-DRA, HLA-DRB1, HLA-DQB1, and HLA-DQA2 were strongly associated with asthma (P < 10E-04). LD is complicated in this region when considering all 46 SNPs (data not shown). One LD block composed of three SNPs upstream of HLA-DQB1 was formed based on 95% confidence interval of D’ of these 10 SNPs (see Figure 3B).
Table III.
Association results of 10 of 46 SNPs (with P ≤ 0.001) in the HLA-DR/DQ region
| No. | SNP | Position | Gene | Alleles (M:m) |
MAF | OR (95% CI) | P value |
|---|---|---|---|---|---|---|---|
| 1 | rs9268542 | 32492699 | BTNL2 | A:G | 0.377 | 1.40 (1.18 – 1.65) | 8.55E-05 |
| 13 | rs2239804 | 32519501 | HLA-DRA | A:G | 0.461 | 1.43 (1.21 – 1.68) | 2.80E-05 |
| 19 | rs2395185 | 32541145 | HLA-DRA | G:T | 0.331 | 1.40 (1.19 – 1.66) | 8.73E-05 |
| 20 | rs2516049 | 32678378 | HLA-DRB1 | A:G | 0.319 | 1.44 (1.22 – 1.71) | 2.62E-05 |
| 21 | rs660895 | 32685358 | HLA-DRB1 | A:G | 0.202 | 1.45 (1.19 – 1.75) | 2.02E-04 |
| 25 | rs1063355 | 32735692 | HLA-DQB1 | C:A | 0.397 | 0.68 (0.58 – 0.81) | 9.55E-06 |
| 26 | rs9275141 | 32759095 | HLA-DQB1 | T:G | 0.497 | 1.37 (1.16 – 1.61) | 1.77E-04 |
| 30 | rs5000634 | 32771542 | HLA-DQB1 | T:C | 0.386 | 1.39 (1.18 – 1.65) | 9.16E-05 |
| 35 | rs9275312 | 32773706 | HLA-DQB1 | A:G | 0.132 | 1.63 (1.31 – 2.02) | 1.13E-05 |
| 46 | rs3916765 | 32793528 | HLA-DQA2 | G:A | 0.11 | 1.50 (1.18 – 1.90) | 9.78E-04 |
Alleles (M:m): Major allele : minor allele.
MAF: minor allele frequency.
OR (95% CI): odds ratio and 95% confidence interval.
Figure 3.
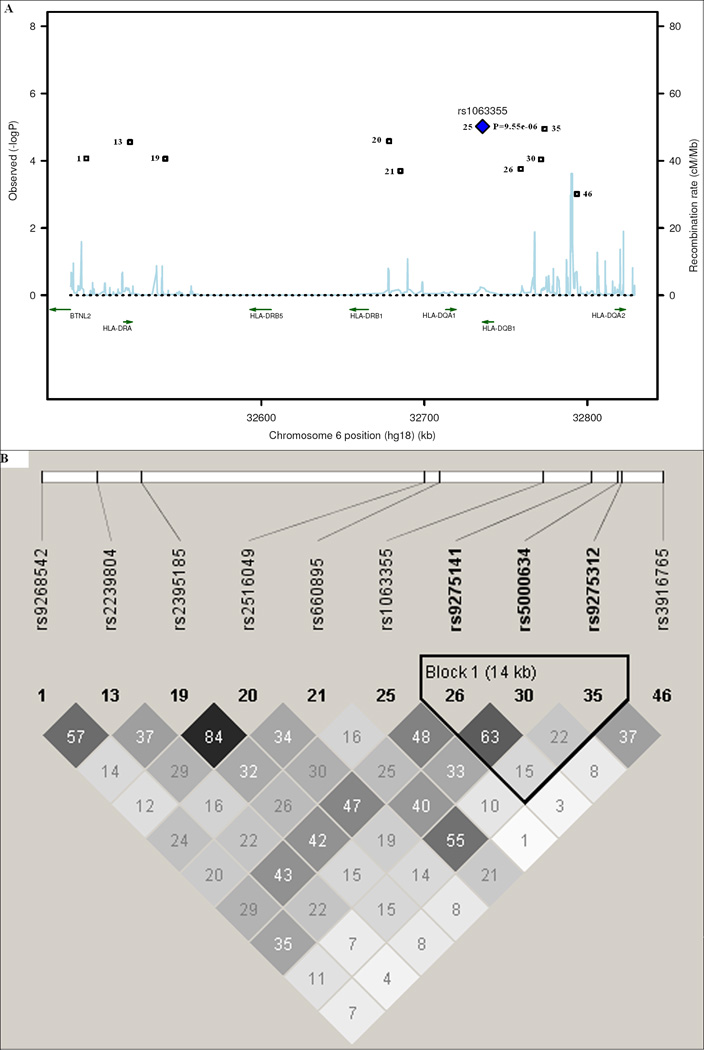
Linkage disequilibrium (LD) and association plot of 10 SNPs in the HLA-DR/DQ region. (A) Association plot: negative logarithm transformed P values (left scale) and recombination rate (right scale). (B) LD plot: r2 color scheme was used and labeled. 95% confidence intervals on D’ was used to setup blocks. Only 10 out of 46 SNPs (with P ≤ 0.001) are shown.
Linear model analysis was performed with the 10 SNPs of the HLA-DR/DQ region for asthma-related quantitative traits (see Table III). A single SNP, rs1063355, on HLA-DQB1 showed significant association with asthma-related quantitative traits (P = 0.01, 0.001, 0.007, and 0.05 for total serum IgE, FEV1, FVC, and FEV1/FVC, respectively).
DISCUSSION
The highest associated SNP identified in this study was rs2244012 in intron 2 of RAD50 (P = 3.04E-07). In addition, evidence was observed for association with multiple SNPs in the RAD50-IL13 region for asthma susceptibility and asthma related quantitative traits. The protein encoded by RAD50 is involved in DNA double-strand break repair and its expression level is constitutively low in most tissues, thus it has no known function directly related to asthma, although MER11-RAD50-NBS1 complex has been shown to be involved in somatic hypermutation and gene conversion of immunoglobulin regions [30]. On the contrary, other genes (IL4, IL5, and IL13) in the TH2 cytokines locus are better candidates based on their biologic functions. Three SNPs in IL13 in this study were associated with asthma. IL13 is critical to the pathogenesis of allergen-induced asthma and thus one of the most highly studied and replicated genes in both genome-wide linkage and candidate-gene association studies. rs20541 (Arg130Gln or IL13+4257GA), in the coding region of IL13, was also analyzed in this study and has been shown to be associated with asthma [31] and total serum IgE levels [32]. rs1800925 (IL13-1111CT), in the promoter region of IL13, has been shown to be associated with asthma [33] and total serum IgE levels [34]. In a GWAS with total serum IgE levels, four SNPs in RAD50 (rs2706347, rs3798135, rs2040704, and rs7737470), have been identified (P < 10E-04) [15]. These four SNPs in RAD50 were in strong LD with rs1800925 (0.7 < r2 < 0.8) and in weak LD with rs20541 (0.2 < r2 < 0.3) in IL13 [15]. These results are consistent with the results of this study; since many of the TENOR asthmatics were recruited from allergist’s offices and the population has increased IgE levels [18]. Since the actual functional SNPs can-not be determined purely by their P values, it is difficult to dissect the association data of RAD50 from IL13 in this study or other genetic studies due to the degree of LD present in this chromosomal region.
In a transgenic mouse study, a TH2 locus control region (LCR) was identified as the 25 kb fragment at the 3’ end of Rad50 [35]. An LCR is defined experimentally as regulating the expression of linked genes in a copy number dependent and tissue-specific manner. The TH2 LCR is involved in the chromatin configuration to re-organize promoters of IL4, IL5, IL13 in proximity and co-regulation of TH2 cytokine expression [36]. Seven Rad50 DNase I hypersensitive sites (RHS1-7) were identified, where RHS4-7 formed the core of the LCR [37]. LCR-C (RHS7) and LCR-B (RHS6) were possible TH2 cytokine expression enhancers; LCR-A (RHS6) and LCR-O (RHS5) were likely insulators [38]. RHS7 is essential for TH2 cytokine expression by showing TH2 specific demethylation after allergen stimulation and intrachromosomal interactions between LCR and the promoters of TH2 cytokines [39]. Furthermore, RHS6, Rad50 promoter (RHS2), and IL5 promoter interacted with interferon gamma (Ifng) on a different chromosome, which suggests an interchromosomal regulation of the expression of TH1/TH2 cytokines [40]. Although all the above experiments were done in mouse, the RAD50 sequence is highly conserved in the LCR between human and mouse. With imputation, multiple significant SNPs were found in the LCR (see Table E2 in the Online Repository): rs3798135 (P = 1.49E-06, in RHS5/LCR-O), rs12653750 (P = 1.49E-06, in RHS6/LCR-A), rs2040704 (P = 1.33E-06, in RHS6/LCR-B), and rs2240032 (P = 6.68E-06, in RHS7/LCR-C). The association of rs2244012 with the expression levels of IL13 in lymphocytes from white adults based on GENEVAR dataset was not significant (P = 0.176), but may be due to small sample size.
Since both a previous GWAS for total serum IgE levels and our GWAS of asthma identified RAD50, it appears to be a new candidate gene for asthma. Although it is still possible the signal from RAD50 is purely due to its LD with the promoter of IL13, RAD50 deserves to be carefully studied when considering TH2 cytokine locus.
HLA-DR/DQ also showed consistent association with asthma, for example, rs1063355 in the 3’ UTR of HLA-DQB1 (P = 9.55E-06), rs2239804 in intron of HLA-DRA (P = 2.80E-05), and rs2516049 5’ upstream of HLA-DRB1 (P = 2.62E-05). HLA-DR/DQ is part of the HLA class II region, which is one of the most gene/variant dense regions in the human genome and is associated with many diseases [41]. HLA-DQB1 and HLA-DRB1 have been shown to be associated with asthma in multiple independent studies [42–44]. Genetic variants in the HLA-DR/DQ region have also been shown to be highly associated with HLA-DR/DQ gene expression, indicating that the association of HLA-DR/DQ with disease might be due to gene expression levels in addition to antigen recognition [45, 46]. The association of rs2516049 with asthma in our study and with the expression levels of HLA-DRB1 (P = 1.25E-04) in lymphocytes from white adults based on GENEVAR dataset indicated that the variant might function through expression level changes (see Figure E4 in the Online Repository) [28, 29]. Imputation identified a SNP with a more significant P value, rs3998159 (P = 1.45E-06), between HLA-DQB1 and HLA-DQA2. It is difficult to determine the functional genes/SNPs in the HLA-DR/DQ region in our study due to the complicated LD pattern in this region. The long-range LD and haplotype analysis based on the MHC Haplotype Project may solve the issue [47].
Using a GWAS approach, this study is the first to confirm the association of RAD50-IL13 and HLA-DR/DQ regions with asthma susceptibility, regions which have been identified by multiple candidate-gene association studies and one genome-wide association study on total serum IgE levels. Our results weakly replicated the findings of the other GWAS: ORMDL3 and gasdermin B (GSDML) (rs7216389) with asthma (P = 0.057); FCER1A (rs2251746) with total serum IgE (P = 0.040); CHI3L1 (rs880633) with FEV1 (P = 0.003), FVC (P = 0.031), and FEV1/FVC (P = 0.040). rs1588265 (P = 0.507) and rs1544791 (P = 0.678) in PDE4D with asthma were not replicated. GWAS of total serum IgE by Weidinger [15] identified several SNPs in RAD50 (P < 10E-04). In our study, the most significant SNP in RAD50 for total serum IgE is rs6871536 (P = 2.61E-03). The geometric mean of total serum IgE in Weidinger’s study is 42.41 (95% CI: 39.56 – 45.47). In our study, the geometric mean of total serum IgE is higher, 48.94 (95% CI: 43.04 – 55.65). The difference in the total serum IgE distribution and relatively small sample size in our study may lead to the difference of significant levels between these two studies.
The potential for false negative results could not be avoided in this study due to the relatively small sample size (473 cases) which may also be the reason that although significance levels of 10-7 were observed, no SNP reached Bonferroni adjusted multiple test criterion (p=0.05/292,443 = 1.71E-07. However, evidence for multiple SNPs was observed in our results in this comprehensively phenotyped relatively homogeneous cohort of difficult-to-treat asthmatics from the larger TENOR study. Our control datasets (general population and phenotyped controls) both have some limitations. They were both significantly younger (see Table I) than TENOR cases, making our results a little conservative because some controls might become asthma cases in the future. Genotyping confirmation and fine-mapping of candidate regions were impossible since the Illumina controls were from a public database, but our approach compensated for this by using imputation. Population stratification was relatively strong between TENOR cases and Illumina 550k controls.
This GWAS confirmed the important role of TH2 cytokine and antigen presentation genes in asthma at a genome-wide level. Furthermore, these findings will stimulate more comprehensive research (e.g., re-sequencing, long-range LD, epistasis, epigenetics, copy number variant, and function) on these two regions due to their functional importance and structural complexity.
Key messages
GWAS of asthma identifies RAD50-IL13 and HLA-DR/DQ. These findings will stimulate more comprehensive research on these genes because of their structural complexity and functional importance in the pathogenesis of asthma.
Supplementary Material
ACKNOWLEDGMENTS
We would like to thank Dr. Elizabeth J. Ampleford for analytical assistance. We would also like to acknowledge the TENOR/SARP/CSGA/STAMPEED Study Group and the TENOR/SARP/CSGA/STAMPEED participants who contributed DNA samples.
Declaration of all sources of funding: The clinical TENOR study was supported by Genentech, Inc. and Novartis Pharmaceuticals Corporation, and this genetic study was funded by NIH HL76285 and HL87665.
Abbreviations used
- GC
Genomic control
- GWAS
Genome-wide association study
- HLA
Human leukocyte antigen
- LCR
Locus control region
- LD
Linkage disequilibrium
- QC
quality control
- SNP
Single nucleotide polymorphism
- TENOR
The Epidemiology and Natural History of Asthma: Outcomes and Treatment Regimens
- UTR
Untranslated region
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Capsule summary
This study confirmed the association of the candidate genes: RAD50-IL13 and HLA-DR/DQ with asthma susceptibility at the genome-wide level and provides confirmation of the important role of TH2 cytokine and antigen presentation genes in asthma.
REFERENCES
- 1.Hoffjan S, Ober C. Present status on the genetic studies of asthma. Curr Opin Immunol. 2002;14:709–717. doi: 10.1016/s0952-7915(02)00393-x. [DOI] [PubMed] [Google Scholar]
- 2.Ober C, Hoffjan S. Asthma genetics 2006: the long and winding road to gene discovery. Genes Immun. 2006;7:95–100. doi: 10.1038/sj.gene.6364284. [DOI] [PubMed] [Google Scholar]
- 3.Denham S, Koppelman GH, Blakey J, Wjst M, Ferreira MA, Hall IP, et al. Meta-analysis of genome-wide linkage studies of asthma and related traits. Respir Res. 2008;9:38. doi: 10.1186/1465-9921-9-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Van Eerdewegh P, Little RD, Dupuis J, Del Mastro RG, Falls K, Simon J, et al. Association of the ADAM33 gene with asthma and bronchial hyperresponsiveness. Nature. 2002;418:426–430. doi: 10.1038/nature00878. [DOI] [PubMed] [Google Scholar]
- 5.Allen M, Heinzmann A, Noguchi E, Abecasis G, Broxholme J, Ponting CP, et al. Positional cloning of a novel gene influencing asthma from chromosome 2q14. Nat Genet. 2003;35:258–263. doi: 10.1038/ng1256. [DOI] [PubMed] [Google Scholar]
- 6.Zhang Y, Leaves NI, Anderson GG, Ponting CP, Broxholme J, Holt R, et al. Positional cloning of a quantitative trait locus on chromosome 13q14 that influences immunoglobulin E levels and asthma. Nat Genet. 2003;34:181–186. doi: 10.1038/ng1166. [DOI] [PubMed] [Google Scholar]
- 7.Laitinen T, Polvi A, Rydman P, Vendelin J, Pulkkinen V, Salmikangas P, et al. Characterization of a common susceptibility locus for asthma-related traits. Science. 2004;304:300–304. doi: 10.1126/science.1090010. [DOI] [PubMed] [Google Scholar]
- 8.Nicolae D, Cox NJ, Lester LA, Schneider D, Tan Z, Billstrand C, et al. Fine mapping and positional candidate studies identify HLA-G as an asthma susceptibility gene on chromosome 6p21. Am J Hum Genet. 2005;76:349–357. doi: 10.1086/427763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Noguchi E, Yokouchi Y, Zhang J, Shibuya K, Shibuya A, Bannai M, et al. Positional identification of an asthma susceptibility gene on human chromosome 5q33. Am J Respir Crit Care Med. 2005;172:183–188. doi: 10.1164/rccm.200409-1223OC. [DOI] [PubMed] [Google Scholar]
- 10.Vercelli D. Discovering susceptibility genes for asthma and allergy. Nat Rev Immunol. 2008;8:169–182. doi: 10.1038/nri2257. [DOI] [PubMed] [Google Scholar]
- 11.Zhang J, Pare PD, Sandford AJ. Recent advances in asthma genetics. Respir Res. 2008;9:4. doi: 10.1186/1465-9921-9-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Willis-Owen SA, Cookson WO, Moffatt MF. Genome-wide association studies in the genetics of asthma. Curr Allergy Asthma Rep. 2009;9:3–9. doi: 10.1007/s11882-009-0001-x. [DOI] [PubMed] [Google Scholar]
- 13.Moffatt MF, Kabesch M, Liang L, Dixon AL, Strachan D, Heath S, et al. Genetic variants regulating ORMDL3 expression contribute to the risk of childhood asthma. Nature. 2007;448:470–473. doi: 10.1038/nature06014. [DOI] [PubMed] [Google Scholar]
- 14.Ober C, Tan Z, Sun Y, Possick JD, Pan L, Nicolae R, et al. Effect of variation in CHI3L1 on serum YKL-40 level, risk of asthma, and lung function. N Engl J Med. 2008;358:1682–1691. doi: 10.1056/NEJMoa0708801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weidinger S, Gieger C, Rodriguez E, Baurecht H, Mempel M, Klopp N, et al. Genome-wide scan on total serum IgE levels identifies FCER1A as novel susceptibility locus. PLoS Genet. 2008;4:e1000166. doi: 10.1371/journal.pgen.1000166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Himes BE, Hunninghake GM, Baurley JW, Rafaels NM, Sleiman P, Strachan DP, et al. Genome-wide association analysis identifies PDE4D as an asthma-susceptibility gene. Am J Hum Genet. 2009;84:581–593. doi: 10.1016/j.ajhg.2009.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dolan CM, Fraher KE, Bleecker ER, Borish L, Chipps B, Hayden ML, et al. Design and baseline characteristics of the epidemiology and natural history of asthma: Outcomes and Treatment Regimens (TENOR) study: a l arge cohort of patients with severe or difficult-to-treat asthma. Ann Allergy Asthma Immunol. 2004;92:32–39. doi: 10.1016/S1081-1206(10)61707-3. [DOI] [PubMed] [Google Scholar]
- 18.Borish L, Chipps B, Deniz Y, Gujrathi S, Zheng B, Dolan CM. Total serum IgE levels in a large cohort of patients with severe or difficult-to-treat asthma. Ann Allergy Asthma Immunol. 2005;95:247–253. doi: 10.1016/S1081-1206(10)61221-5. [DOI] [PubMed] [Google Scholar]
- 19.Miller MK, Johnson C, Miller DP, Deniz Y, Bleecker ER, Wenzel SE. Severity assessment in asthma: An evolving concept. J Allergy Clin Immunol. 2005;116:990–995. doi: 10.1016/j.jaci.2005.08.018. [DOI] [PubMed] [Google Scholar]
- 20.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 22.Clayton DG, Walker NM, Smyth DJ, Pask R, Cooper JD, Maier LM, et al. Population structure, differential bias and genomic control in a large-scale, case-control association study. Nat Genet. 2005;37:1243–1246. doi: 10.1038/ng1653. [DOI] [PubMed] [Google Scholar]
- 23.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 24.Gabriel SB, Schaffner SF, Nguyen H, Moore JM, Roy J, Blumenstiel B, et al. The structure of haplotype blocks in the human genome. Science. 2002;296:2225–2229. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- 25.Johnson AD, Handsaker RE, Pulit SL, Nizzari MM, O'Donnell CJ, de Bakker PI. SNAP: a web-based tool for identification and annotation of proxy SNPs using HapMap. Bioinformatics. 2008;24:2938–2939. doi: 10.1093/bioinformatics/btn564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Frazer KA, Ballinger DG, Cox DR, Hinds DA, Stuve LL, Gibbs RA, et al. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–861. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li Y, Abecasis GR. Mach 1.0: Rapid Haplotype Reconstruction and Missing Genotype Inference. Am J Hum Genet. 2006;S79:2290. [Google Scholar]
- 28.Stranger BE, Forrest MS, Dunning M, Ingle CE, Beazley C, Thorne N, et al. Relative impact of nucleotide and copy number variation on gene expression phenotypes. Science. 2007;315:848–853. doi: 10.1126/science.1136678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ge D, Zhang K, Need AC, Martin O, Fellay J, Urban TJ, et al. WGAViewer: software for genomic annotation of whole genome association studies. Genome Res. 2008;18:640–643. doi: 10.1101/gr.071571.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yabuki M, Fujii MM, Maizels N. The MRE11-RAD50-NBS1 complex accelerates somatic hypermutation and gene conversion of immunoglobulin variable regions. Nat Immunol. 2005;6:730–736. doi: 10.1038/ni1215. [DOI] [PubMed] [Google Scholar]
- 31.Heinzmann A, Mao XQ, Akaiwa M, Kreomer RT, Gao PS, Ohshima K, et al. Genetic variants of IL-13 signalling and human asthma and atopy. Hum Mol Genet. 2000;9:549–559. doi: 10.1093/hmg/9.4.549. [DOI] [PubMed] [Google Scholar]
- 32.Graves PE, Kabesch M, Halonen M, Holberg CJ, Baldini M, Fritzsch C, et al. A cluster of seven tightly linked polymorphisms in the IL-13 gene is associated with total serum IgE levels in three populations of white children. J Allergy Clin Immunol. 2000;105:506–513. doi: 10.1067/mai.2000.104940. [DOI] [PubMed] [Google Scholar]
- 33.van der Pouw Kraan TC, van Veen A, Boeije LC, van Tuyl SA, de Groot ER, Stapel SO, et al. An IL-13 promoter polymorphism associated with increased risk of allergic asthma. Genes Immun. 1999;1:61–65. doi: 10.1038/sj.gene.6363630. [DOI] [PubMed] [Google Scholar]
- 34.Liu X, Beaty TH, Deindl P, Huang SK, Lau S, Sommerfeld C, et al. Associations between total serum IgE levels and the 6 potentially functional variants within the genes IL4, IL13, and IL4RA in German children: the German Multicenter Atopy Study. J Allergy Clin Immunol. 2003;112:382–388. doi: 10.1067/mai.2003.1635. [DOI] [PubMed] [Google Scholar]
- 35.Lee GR, Fields PE, Griffin TJ, Flavell RA. Regulation of the Th2 cytokine locus by a locus control region. Immunity. 2003;19:145–153. doi: 10.1016/s1074-7613(03)00179-1. [DOI] [PubMed] [Google Scholar]
- 36.Spilianakis CG, Flavell RA. Long-range intrachromosomal interactions in the T helper type 2 cytokine locus. Nat Immunol. 2004;5:1017–1027. doi: 10.1038/ni1115. [DOI] [PubMed] [Google Scholar]
- 37.Fields PE, Lee GR, Kim ST, Bartsevich VV, Flavell RA. Th2-specific chromatin remodeling and enhancer activity in the Th2 cytokine locus control region. Immunity. 2004;21:865–876. doi: 10.1016/j.immuni.2004.10.015. [DOI] [PubMed] [Google Scholar]
- 38.Lee DU, Rao A. Molecular analysis of a locus control region in the T helper 2 cytokine gene cluster: a target for STAT6 but not GATA3. Proc Natl Acad Sci U S A. 2004;101:16010–16015. doi: 10.1073/pnas.0407031101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee GR, Spilianakis CG, Flavell RA. Hypersensitive site 7 of the TH2 locus control region is essential for expressing TH2 cytokine genes and for long-range intrachromosomal interactions. Nat Immunol. 2005;6:42–48. doi: 10.1038/ni1148. [DOI] [PubMed] [Google Scholar]
- 40.Spilianakis CG, Lalioti MD, Town T, Lee GR, Flavell RA. Interchromosomal associations between alternatively expressed loci. Nature. 2005;435:637–645. doi: 10.1038/nature03574. [DOI] [PubMed] [Google Scholar]
- 41.Shiina T, Hosomichi K, Inoko H, Kulski JK. The HLA genomic loci map: expression, interaction, diversity and disease. J Hum Genet. 2009;54:15–39. doi: 10.1038/jhg.2008.5. [DOI] [PubMed] [Google Scholar]
- 42.Torio A, Sanchez-Guerrero I, Muro M, Villar LM, Minguela A, Marin L, et al. HLA class II genotypic frequencies in atopic asthma: association of DRB1*01-DQB1*0501 genotype with Artemisia vulgaris allergic asthma. Hum Immunol. 2003;64:811–815. doi: 10.1016/s0198-8859(03)00109-5. [DOI] [PubMed] [Google Scholar]
- 43.Lin YC, Lu CC, Su HJ, Shen CY, Lei HY, Guo YL. The association between tumor necrosis factor, HLA-DR alleles, and IgE-mediated asthma in Taiwanese adolescents. Allergy. 2002;57:831–834. doi: 10.1034/j.1398-9995.2002.23719.x. [DOI] [PubMed] [Google Scholar]
- 44.Cho SH, Kim YK, Oh HB, Jung JW, Son JW, Lee MH, et al. Association of HLA-DRB1(*)07 and DRB1(*)04 to citrus red mite (Panonychus citri) and house dust mite sensitive asthma. Clin Exp Allergy. 2000;30:1568–1575. doi: 10.1046/j.1365-2222.2000.00915.x. [DOI] [PubMed] [Google Scholar]
- 45.Dixon AL, Liang L, Moffatt MF, Chen W, Heath S, Wong KC, et al. A genome-wide association study of global gene expression. Nat Genet. 2007;39:1202–1207. doi: 10.1038/ng2109. [DOI] [PubMed] [Google Scholar]
- 46.Beaty JS, West KA, Nepom GT. Functional effects of a natural polymorphism in the transcriptional regulatory sequence of HLA-DQB1. Mol Cell Biol. 1995;15:4771–4782. doi: 10.1128/mcb.15.9.4771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Horton R, Gibson R, Coggill P, Miretti M, Allcock RJ, Almeida J, et al. Variation analysis and gene annotation of eight MHC haplotypes: the MHC Haplotype Project. Immunogenetics. 2008;60:1–18. doi: 10.1007/s00251-007-0262-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


