Abstract
There is a large amount of interindividual variability in both therapeutic and adverse responses to asthma therapies. Genetic variability may account for 50–60% of this variability. Pharmacogenomics holds out the promise of allowing clinicians to prospectively choose therapies that have the greatest likelihood to be effective for individual patients and to avoid those which may have a high likelihood of producing adverse effects. In this article we review the principals of pharmacogenomic investigation. We explore the data developed from the early pharmacogenomic studies with the most common asthma therapies. Further, we explore the potential use of pharmacogenomics and caveats in interpreting such information.
Keywords: Pharmacogenomics, Asthma, Pharmacotherapy, Genetics, ADRB2, Leukotrienes, Corticosteroid, Variability
I. Introduction
A 19-year-old woman with a history of seasonal allergies comes to your office complaining of increasing shortness of breath both with exercise and at rest. She has been on immunotherapy for her grass allergies for the past year. She has been wheezing and her internist gave her an albuterol inhaler. She has needed to use the inhaler once or twice a day at rest. There have been no environmental changes. The exam is normal and her FEV1 is 75% of predicted and increases to 90% of predicted post bronchodilator. Her mother and brother have asthma and her mother and sister have osteoporosis. National guidelines suggest that the first line of therapy might be inhaled corticosteroids. She is concerned about using corticosteroids. “Can you promise that these drugs will work for me?” she asks. “And what about my risk for developing osteoporosis?”
Although national guidelines appropriately suggest inhaled corticosteroids as the appropriate first-line therapy for this patient, 15 to 20% of patients will not respond to these highly efficacious medicines (1). Further, although inhaled steroids, on average, are likely to minimally affect bone density in this premenopausal woman, there is a wide variability in response(2). Leukotriene modifiers, while efficacious, would predictably have an even larger group of patients who might not respond (3). In this age of cost consciousness you would like to avoid having to have the patient come back to switch therapies. It would be most helpful if you could have reliable indicators that would: 1) predict the classes of medications to which this patient would respond; 2) predict the likelihood that she might develop toxicity. Isn't there some type of test that will supply that information? Wouldn't it be nice to draw a blood test and predict her likelihood of responding to inhaled corticosteroids, developing osteoporosis from corticosteroids, responding to a leukotriene modifier, or developing adverse effects from her use of beta agonists for that matter? Evolving data from the field of pharmacogenomics may be of help.
Multiple factors can play role in affecting what appear to be inconsistencies in the therapeutic response. Environmental factors contribute significantly to this variability. However, the evolving understanding of the variability of the genetic code incorporated into our DNA, and the sequencing of the human genome, have allowed us to understand that genetic variability accounts for up to 50% of our inter-individual differences in response to therapy (4). Pharmacogenomics is a field of study that examines the relationship between genetic variability (or polymorphisms) and the response (both therapeutic and/or adverse) to therapeutic interventions. Such information would obviously allow us to better manage the asthma of the 19-year-old above. What type of information is available? What information will be available? Will we be able to use this information to manage asthma patients?
In an attempt to answer these questions we will review pharmacogenomics as it relates to asthma. Below we will: 1) briefly review the nomenclature for the variability of the genetic code that is examined in the area of pharmacogenomics, pharmacogenomic mechanisms and methods of investigation; 2) discuss pharmacogenomic information as it relates to the three principal components of asthma therapy -- beta agonist therapy, leukotriene modifier therapy, and inhaled corticosteroids; 3) explore the implications of pharmacogenomics and caveats and limitations in relationship to interpreting pharmacogenomic data.
II. Pharmacogenomics - Terms and Approaches
As mentioned above, pharmacogenomics looks at the association between variability in the genome and outcomes in response to therapeutic interventions. Table 1 lists and defines terms used in characterizing the genomic variability and approaches to detecting the variability.
Table 1.
Genetic Terminology
| Gene - An ordered sequence of nucleotide bases that, via mRNA, encodes one polypeptide chain. The gene includes leader and trailer untranslated regions (UTR) preceding and following the coding region respectively, as well as intervening sequences (introns) between individual coding segments (exons). The ends of an individual strand of DNA are designated as 3' prime and 5' prime based on the position on the deoxyribose sugar. |
| Polymorphism - One of two or more alternate forms (alleles) of a chromosomal locus that differ in nucleotide sequence or have variable numbers of repeated nucleotide units |
| SNP - Single Nucleotide Polymorphism - DNA sequence variations that occur when a single nucleotide in the genome sequence is altered. These are designated by abbreviations for the possible amino acids before and after the position on the protein, for example Gly16Arg. |
| Synonymous SNP - a change in the SNP that does not alter amino acid coded for in the sequence. |
| Non-synonymous SNP - a change in the SNP that does alter the amino acid coded for in the sequence |
| Haplotype - A set of closely linked genetic polymorphisms present on one chromosome. |
| Coding Region Polymorphisms - Alterations in the genetic code of the area of a gene that encodes the polypeptide chain |
| Non-coding Region Polymorphisms - Alterations in areas in the genetic code that are not translated into the final protein but may affect the type or degree of transcription of the gene |
| Copy Number Variant - Deletions and duplications of DNA segments that are present in variable copy number compared with a reference genome |
| Insertion - Addition of one or more nucleotide base pairs into a DNA sequence. |
| Deletion - Genetic aberration in which a part of a chromosome or a sequence of DNA is missing. |
| Candidate Gene Studies - Studies of a specific gene in which variation might influence the risk of a specific disease or response to medication, usually because the gene is part of a biological pathway that is plausibly related to the disease. |
| Family-based Studies - Study of genetic variations between an affected individual and his parents or siblings in an attempt to identify the aberrant gene. This allows to account for the confounding environmental effects. |
| Pathway Studies - Simultaneous study of polymorphisms in multiple genes involved in a biological pathway |
| Genome-Wide Association Studies (GWAS) - An approach to gene mapping that involves scanning markers across the entire genome to find associations between a particular phenotype and allelic variation in a population. This methodology relies on the fact that the markers will be in linkage disequilibrium (see next) with polymorphisms truly associated with the phenotype. |
| Linkage Disequilibrium - The occurrence of combinations of genetic variants at different loci at a frequency that varies from what would be accounted for by chance. Also known as “allelic association” For example if variants A and B occur at one locus and X and Y occur at another. If each time X is detected, A is detected, these variants X and A are in linkage disequilibrium. |
It is worthwhile understanding how one goes about trying to identify polymorphisms that may relate to altered drug responses. Classically, one would adopt a candidate gene approach. In this approach, a gene thought to be related to the biological response to an agent would be examined for polymorphisms and those polymorphisms would be examined for associations to phenotypic outcomes. For example, since beta-agonists were known to exert their effects through the beta-2-adrenergic receptor (ADRB2) this gene was sequenced and investigations conducted to examine whether particular phenotypic outcomes were related to a single polymorphism, or to a group of polymorphisms, as described below.
One can broaden this approach by taking a “pathway approach” to such investigations. In this approach, genes thought to be involved in the entire pathway of a response to an agent are examined. The pathway genes are interrogated for polymorphisms and then examined for associations to the drug response. Such an approach is described for the leukotriene modifiers below in which the major synthetic genes, transport genes, and receptors were examined.
Lastly, an even broader approach can be undertaken utilizing genome wide association studies (GWAS). Using this methodology, hundreds of thousands of relatively evenly spaced markers across the entire genome are scanned for associations to phenotypic outcomes. These markers are not necessarily in a gene or translated sections of the genome. If markers are associated with particular outcomes, genes nearby these markers are examined to see if polymorphisms in these genes may actually be the polymorphisms of interest. This non-heuristic approach to finding associations may lead to discovering associations with genes that may not have been thought to be associated with the outcomes of interest. Such an approach led to the discovery of the association of the ADAM proteins (membrane-anchored metalloproteases) with asthma (5). A precise mechanism for this association has still not been definitively identified.
Why aren't all studies conducted now performed using GWAS? While the GWAS approach is quite powerful, due to the large number of variants being examined it requires very large sample sizes with well characterized outcomes. Further, it requires that the polymorphism under examination be quite common and, due to correction for multiple comparisons, it can miss true and potentially important associations or identify spurious ones.
Whether we utilize a candidate gene approach, pathway analysis, or GWAS, pharmacogenomic associations are accumulating rapidly to the point that there are now databases that rank order genes most likely to be important for a drug responses (6). In the following sections, we discuss progress in regards to asthma pharmacogenomics utilizing all of the above techniques.
III. Beta Agonists
Beta-agonists are the most commonly used medications in the treatment of asthma. Short-acting and long-acting beta-agonists act by binding to the beta2-adrenergic receptor (ADRB2). Receptor binding results in activation of adenylyl cyclase via stimulatory G proteins that activate protein kinase A. The latter phosphorylates several target proteins resulting in a decrease in intracellular calcium causing smooth muscle relaxation in the airways.
The ADRB2 gene has been sequenced and has been found to be intronless with over 80 reported SNPs identified in the coding region of the gene, in the 5' and 3' untranslated regions, and in the adjacent leader and trailer genomic regions (8, 9). More than 40 of these SNPs have been validated in the dbSNP database. Two of these polymorphisms, at amino acid positions 16 (Gly16 Arg) and 27 (Gln27Glu) have been found to alter receptor function, ligand binding and signal transduction in vitro (10–12). Additionally, a variant in the ADRB2 promoter region has been reported to affect receptor transcription and density (13). Further, a variable-length poly-C polymorphism in the 3' UTR appears to alter messenger RNA expression, degradation, and in vitro downregulation (14) but has not been shown to affect salmeterol-induced responses (15).
The Gly16Arg polymorphism has been the focus of most clinical studies in relation to use of both short-acting and long-acting beta-agonists. At this locus, individuals may harbor polymorphisms that code for arginine or glycine. Approximately 1/6 of Caucasians and 1/5 of Black Americans are Arg/Arg.
A. Short-acting beta-agonists
A.1. Acute bronchodilator response
The association between genetic polymorphisms and an acute bronchodilator response has been inconsistent. An initial study in children with asthma found that 60% of those homozygous for arginine at the B16 amino acid position of ADRB2 (B16 Arg/Arg) had ≥ 15.3% increase in predicted FEV1 after albuterol, compared with only 13% in individuals homozygous for glycine at that position (16) and similar signals were detected in another small study(17). However, subsequent studies have not detected such associations in adults or children (8, 18–22). Of interest, while family-based studies in Latinos found an association between Arg16Gly and bronchodilator response in Puerto Ricans (23), no such association was observed in Mexicans, highlighting the importance of assessing genetic findings in different populations. Recent studies have suggested that B16 polymorphisms may affect responses to β-agonists administered during acute exacerbations of asthma in that Gly/Gly patients appeared to have a more rapid response to repeated albuterol administration (ICU length of stay 43 hours in Gly/Gly vs. 74 hours in Arg/Arg) (24) and another suggesting that amino acid 27 position polymorphisms may affect the rapidity of response (25).
A.2. Regular Use
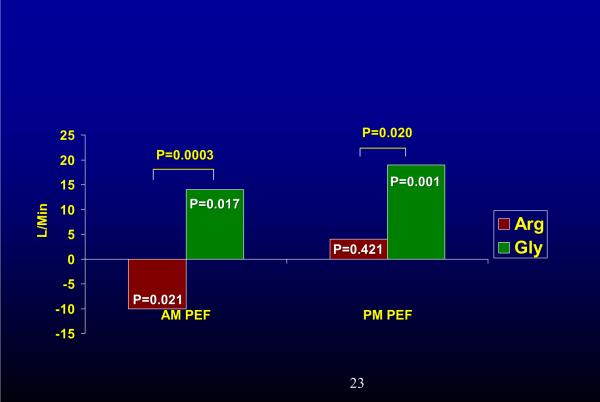
Several studies have suggested that regular use of short-acting β-agonists appears to be associated with poorer outcomes in those who were B16 Arg/Arg. A retrospective analysis of >250 patients with mild asthma showed that B16 Arg homozygosity was associated with a decline in peak expiratory flow with regular use of the beta-agonist albuterol (26). In another retrospective study this polymorphism was associated with an increased frequency of asthma exacerbations in those using albuterol regularly (1.9 vs 0.81 exacerbations per year) (27). A prospective study confirmed a decline in morning peak expiratory flow rate, lack of benefit in evening peak flow rate, worsening symptoms and increased β-agonist use in Arg/Arg patients treated with regular short-acting β-agonists as compared to Gly/Gly patients (Fig 1). In another report, investigators revealed an association between B16 locus and airway hyperresponsiveness in non-smokers (28)
Fig 1.
Change in AM and PM PEF with regular use of albuterol from placebo (X axis) in B16 Arg/Arg vs. B16 Gly/Gly patients. Regular use of albuterol resulted in a decline in AM PEF in Arg/Arg patients while it produced an improvement in AM PEF in Gly/Gly individuals. Similar deteriorations in FEV1, symptoms, and reliever medication use were seen in Arg/Arg patients, while Gly/Gly patients improved (data not shown). Reference (18)
B. Long-acting Beta-agonists
The findings of poorer outcomes in B16 Arg/Arg patients taking short acting β-agonists has generated much interest in whether such effects carry-over to the long-acting β-agonists (LABAs). This interest has been heightened by the questions about rare life-threatening events associated with salmeterol use (29). While it might be attractive to associate these outcomes with the B16 polymorphisms, no information has been available regarding the genotypes of the patients who sustained such adverse events. Initial analyses have suggested that suggested that LABA use might lead to poorer outcomes in asthmatics with the B16 Arg/Arg genotype (30, 31). One of the studies, a retrospective analysis of patients who had been in studies with the NHLBI's Asthma Clinical Research Network (ACRN), suggested that a genotype-specific effect (B16 Arg/Arg showing a decline in lung function over time) might occur even when LABAs are used with inhaled corticosteroids (30). This concern was reinforced by a cross sectional survey in Scotland that examined 546 children and young adults attending asthma clinics that also found a relationship between asthma exacerbations and long-acting β-agonist use and B16 Arg/Arg even in patients using concomitant inhaled corticosteroids (32). However, retrospective analyses of other studies did not detect a genotype-specific effect (33–35). While the studies utilized in these retrospective analyses, for the most part, included patients screened for a positive bronchodilator response, the ACRN recently completed a prospective study in which patients were not screened in this manner (7). The latter included subjects who demonstrated bronchial hyper responsiveness to inhaled methacholine while on inhaled steroid therapy, even in the absence of a 12% reversibility in FEV1. This study did not detect a genotype-specific difference in peak flow between Arg/Arg and Gly/Gly patients using salmeterol with moderate doses of inhaled corticosteroids. Interestingly, Gly/Gly patients experienced an improvement in methacholine responsiveness when salmeterol was added to inhaled corticosteroids but Arg/Arg did not. A post-hoc analysis in Blacks suggested that Arg/Arg Blacks may not experience an improvement in peak flow from the addition of salmeterol to inhaled corticosteroids.
C. Additional Polymorphisms and Associations
Genome wide association studies have only now been undertaken in relation to β-agonist responses. A recent pathway analysis suggested a possible association between promoter polymorphism in Arginase 1 and bronchodilator responses (36). Arginase may be involved in airway responses by depleting stores of L-arginine, leading to decreased production of nitric oxide which relaxes bronchial smooth muscle.
D. Summary
Taken together, these data suggest that coding polymorphisms at the 16th amino-acid position of ADRB2 identify patients with adverse responses to regular use of short-acting β-agonists. There is conflicting data on whether or not these polymorphisms identify patients who have a diminished response to LABAs when used concurrently with moderate doses of inhaled corticosteroids. Whether these polymorphisms are of greater significance in particular racial or ethnic groups using LABAs remains to be confirmed. Further, pathway analysis and genome wide association studies (GWAS) may identify further predictors related to response and/or adverse effects related to the use of these common drugs used by patients with asthma.
IV. Leukotriene Modifiers
Leukotriene modifiers (LTMs) (a category meant to include both synthesis inhibitors and receptor antagonists (LTRAs) exert their effects by inhibiting the formation or action of leukotrienes which are products of the metabolism of arachidonic acid. While these agents are effective in the treatment of asthma there is significant heterogeneity in the therapeutic response to these medications (3, 37). Since the leukotriene biosynthetic pathway has been well characterized, leukotriene pharmacogenetic responses lend themselves to candidate gene and pathway analyses. The results of these studies, reviewed below, suggest that variation in these genes can exert an effect on the response to these agents.
A. LTC4 Synthase
The most consistent association between differential responses to LTMs and a genetic polymorphism arise in relation to a polymorphism in the LTC4 synthase gene promoter, (A-444C). The frequency of the variant C allele is 0.32 in Caucasians. In vitro, the C variant is associated with increased production of leukotriene C4 by blood eosinophils stimulated with calcium ionophore (38) and increased expression of leukotriene C4 synthase in blood eosinophils (39, 40). In 23 asthmatics treated with a LTRA (leukotriene receptor antagonist) there was a trend toward greater improvement in lung function in those with at least one copy of the C variant(38). In children, a greater decline in nitric oxide in response to a LTRA occurred in those possessing the variant (41). A LTRA produced a 14% improvement in FEV1 in those with the C variant compared with a 3% improvement in those without the C allele (p=0.01) (42). In a leukotriene pathway analysis, this same variant was associated with a 76% reduction in exacerbations in response to montelukast in those who possessed the variant as compared with those who did not. (43). Taken together, these studies suggest that this promoter variant in leukotriene C4 synthase is associated with differential responsiveness to a leukotriene receptor antagonist.
B. 5-lipoxygenase
Drazen and colleagues first described an addition/deletion variant in the promoter region of 5-lipoxygenase (ALOX5) that was associated with variation in the FEV1 response to a 5-lipoxygenase inhibitor. The 3% of individuals who did not possess at least one copy of the wild type number of copy variants (five copies) had a negligible response to the 5-LO inhibitor as compared to 15–20% improvement in those who did possess the wild-type copies (44). While another study suggested an influence of this region on the response to an LTM its signal was in the opposite direction. Lima and colleagues reported a 73% reduction in asthma exacerbations in carriers of the mutant allele (2, 3, 4, 6, or 7 copies of ALOX5 promoter) as opposed to the homozygous wild-type (5 copies) (43). Another study in 61 patients reported a signal consistent with that reported initially, in that montelukast selectively improved FEV1 and reduced exacerbations in those who had at least one 5 repeat variant as compared with those patients who were homozygous for 4 repeat variants (45).
Additional SNPs in ALOX5 have been associated with variation in responses to LTMs. Klotsman and colleagues identified 2 SNPs in ALOX5 that were associated with an 18–25% improvement in FEV1 compared with a 8–10% improvement in those bearing the wild alleles treated with montelukast (46). Lima (43) also found associations between a different SNP in ALOX5 (rs2115189) and differential FEV1 response to montelukast. This SNP was also associated with a differential FEV1 responses to montelukast. This SNP was also associated with a differential FEV1 response in a study of zileuton, the 5-lipoxygenase inhibitor, in over 550 patients (47). The persistent association of ALOX5 variants with differential treatment outcomes to both leukotriene synthesis inhibitors and leukotriene receptor antagonists confirms a role for this enzyme in relation to therapeutic outcomes for this class of drugs.
C. Additional Genes
A polymorphism in ABCC1 (ATP-binding cassette, subfamily C, member 1), a member of the multidrug resistance-associated protein family that is responsible for transporting leukotrienes outside of the cell, has been associated with differential changes in FEV1 in the LTRA pathway analysis (43). This same SNP was also associated with differential improvement in FEV1 in response to zileuton (47). The pathway analysis has also found associations between variation in LTA4 Hydrolase and change in rates of exacerbation in response to montelukast (43). CysLTR2 genes have been associated with differential changes in peak flow rates in response to montelukast (46).
Recently, montelukast levels have been shown to be associated with variants in gene SLCO2B1 (solute carrier organic ion transporter family, member 2B1) (48) which codes for the organic anion transporter 2B1, OATP2B1 which appears to have a role in montelukast transport across the gut. Heterozygotes at SNP rs12422149 had 30% lower levels of montelukast compared to those with the wild type alleles. Heterozygosity was also associated with poorer improvement on one of the symptom indices used in the trial.
D. Summary
The LTC4 synthase polymorphism appears to be consistently associated with differential responses to LTM therapy. ALOX5 polymorphisms play a role in response to LTM therapy but the directionality of this response remains to be determined. Additional candidates will still require replication.
V. ICS
Candidate gene and pathway approaches have been utilized to investigate pharmacogenomic associations for corticosteroid responses. Most of these associations have not yet been replicated in independent populations. GWAS studies are currently underway. The genes associated with alterations in responses to inhaled corticosteroids are discussed below.
A. CRHR1
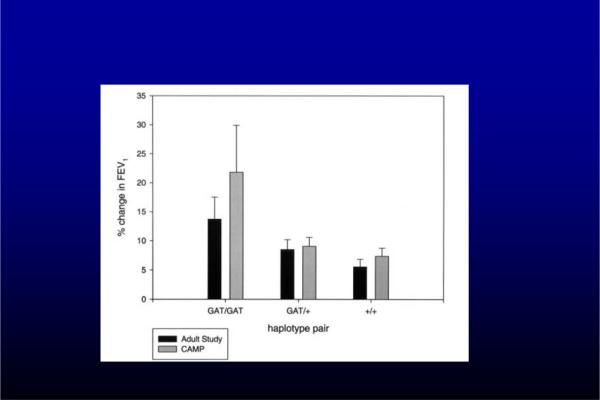
SNPs in 14 candidate genes were examined for relationship to response to 2 months of inhaled corticosteroid therapy (49). SNPs in CRHR1 (corticotropin-releasing hormone receptor 1) were associated with a more than a two times greater improvement in FEV1 in response to inhaled corticosteroids in pediatric cohort treated with inhaled corticosteroids in the CAMP (Children's Asthma Management Program) Study. A haplotype consisting of combination of three SNPs in this gene showed even greater differential responses to therapy (Fig 2). In the same study, a different SNP in CRHR1 was associated with a four fold difference in FEV1 response to inhaled corticosteroid in a replication population of adults in the NHLBI Asthma Clinical Research Network cohort. Of note, a small sample size study was not able to replicate these associations with CRHR1 (50). Thus, the clinical significance of polymorphisms in CRHR1 remains unclear.
Figure 2.
Improvement in FEV1 in association with different haplotypes of CRHR1. (49)
B. T-bet
TBX21, a gene that encodes for the transcription factor T-bet (T-box expressed in T cells), is responsible for the induction of T-helper (TH1) cells and the suppression of TH2 cells from naïve T lymphocytes. T-bet knockout mice develop asthma-like inflammation. A non-synonymous single nucleotide polymorphism was associated with a large improvement in airway hyperresponsiveness in the CAMP population in response to corticosteroid therapy (51). However, the minor allele frequency of this variant is less than 5%, suggesting that unless this large effect is reproducible and accurate, the finding may not be clinically useful in and of itself..
C. FCER2
A polymorphism in FCER2 (the low affinity IgE receptor gene) has recently been associated with variation in protection from asthma exacerbations in response to inhaled corticosteroids (52). A 3–4 fold SNP-associated increased risk of asthma exacerbations was seen in Blacks and Caucasians receiving inhaled corticosteroids. The variant was also associated with increased IgE levels.
D. Adenylyl Cyclase 9
Adenylyl cyclase type 9 (AC9), an enzyme activated by the β2 -adrenergic receptor, has also been implicated in affecting the response to inhaled corticosteroids. Using the CAMP cohort, an interaction was found bet the non-synonymous polymorphism at amino acid 772 and improved bronchodilator response over the 4 years of inhaled corticosteroid treatment (53).
E. Summary
The corticosteroid biological action and effect pathway is complex and it is unlikely that common polymorphisms in any one gene control a large enough part of the pathway to be determinative in and of themselves. It is likely that combinations of polymorphisms in multiple genes affect the therapeutic response to corticosteroids.
VI. IMPLICATIONS AND APPLICABILITY OF PHARMACOGENOMIC FINDINGS
A. Utility
What are the potential uses of pharmacogenomic information? The ability to predict which patients are more likely to respond to therapy and which patients are more likely to develop adverse effects would allow us to customize therapy. What are the practical outcomes of being able to customize therapy? They include: 1) Choosing the therapy more likely to be effective the first time resulting in more rapid and effective outcomes for the patient that result in decreased patient return visits and potentially improved compliance; 2) Choosing therapy that has less likelihood of producing side effects for the particular patient or possibly being able to alert the patient about particular side-effects to which he/she might be susceptible; 3) Choosing the most appropriate dose for a patient and thus minimizing underdosing, overdosing, and resultant increased use of medical resources and delayed and/or less effective outcomes. Additional benefits of pharmacogenomic information relate to development and introduction of new drugs. Many new pharmacotherapies do not reach consumers due to toxicities that occur in a small percentage of patients. In cases where pharmacogenomics allow us to prospectively identify those patients who are at risk for side effects development and use of such drugs can proceed for the vast majority of patients who are at minimal risk of developing these toxicities.
B. Ability to Influence Therapy
When are pharmacogenomic findings most likely to influence therapy? Several factors determine the utility of a pharmacogenomic predictions (see Table 2): These include: 1) the frequency of the variant allele (the more frequent the more likely the test will have economic utility (54); 2) the strength of the association between the polymorphism and the drug effect: 3) the impact of making the correct choice more quickly in terms of either the implications of the failure to obtain an initial therapeutic response and/or the impact and potential for an adverse effect due to the drug. The cost and availability of the test also influence the utility of the test but since the cost of genomic testing is falling rapidly, the contribution of these factors will be increasingly diminished.
Table 2.
Factors Affecting the Likelihood that a Pharmacogenomic Association will Alter Practice
| 1. Frequency of the variant(s) of interest in the population |
| -Increased frequency increases the likelihood that testing for the association will make economic sense and is worthy of consideration. |
| 2. Strength of the association between the polymorphism and the drug effect |
| -The stronger the predictive value of the association the greater the potential utility of the test. |
| 3. Impact of the making the “right” vs. “wrong” choice |
| -If the consequences of the choice are associated with high morbidity then even if the frequency of the polymorphism is low, it may still make sense to test especially if the strength of the association is high. |
C. Caveats
What prevents us from widely applying the information developed so far in guiding therapy for our patients? These are summarized in Table 3. First as seen above, many of the pharmacogenomic associations detected thus far account for only a small proportion of the variability in response. For instance, the CRHR1 variants account for less than 3% of the variability in response. Secondly, pharmacogenomic associations may be confounded by several factors. Such factors can include gene-gene interactions, i.e. coincidental variants in other parts of the gene, or in other genes, that may modify the observed effects. For example, if a series of alleles identifies patients with a diminished response to β-agonists but a patient bearing those alleles has a variant of CRHR1 that improves the response to an inhaled corticosteroid, the outcome when that patient uses a combined β-agonist-inhaled corticosteroid may not be correctly predicted by either allele alone. Another form of gene-gene interaction that needs to be considered is the effect of genetic background and ancestry. While a particular gene may predict certain responses in Caucasians, the gene may not have the same significance in Asians for example. Different patterns of response to ADRB2-related polymorphisms in Mexican Hispanics as compared to Hispanics from Puerto Rico (55) may be due to such differences in “genetic background”.
Table 3.
Impediments to Use of Pharmacogenomic Information
| 1. Detection of single polymorphism effects with only small effects on outcomes. |
| 2. Variability in associations due to the effects of differing genetic backgrounds eg race or ethnicity. |
| 3. Inadequate reproducibility in multiple populations. |
| 4. Inadequate informatics support to clinicians. |
D. Future
Can we overcome these limitations and use the pharmacogenomic information obtained from retrospective and prospective studies? We remain cautiously optimistic. First, it possible that there will be variants whose effect is large enough that either singly, or in combination with just a few other variants, these polymorphisms may predict a clinically important therapeutic outcome. Additionally, and more likely, we will most likely use information from panels of many gene variants, which, with the aid of informatics, can be used to develop patient profiles of likelihood of responding to, or developing adverse effects from, a particular drug (56). The time-limiting step for the realization of this scenario is not limited by the technology or the cost of gene ascertainment. Chip technology now permits several thousand variants to be tested at once for costs that keep dropping and are now well below one thousand dollars. The current limitation is the development of adequate reproducible information, in populations of interest, so that reliable and generalizable information can be used to construct predictive algorithms. The development of such information is dependent on large, well-characterized cohorts of subjects of interest with appropriate therapeutic outcomes. In particular, as mentioned above, attention needs to be paid to the genetic background of the subjects included in the study so that one can understand whether the outcomes will apply to many groups, or just one. Further, the phenotypes in which the outcomes were observed can also be critical. For example, pharmacogenomic associations in severe asthmatic may not apply to mild asthmatics. For this reason, in many cases, such associations may require prospective validation.
In summary, all of the different genetic approaches -- candidate gene, family-based, pathway, and GWAS-- will identify increasing numbers of associations between genetic variants and inter-individual responses to therapeutic interventions. With appropriate validation (currently and yet to be undertaken) and adequate informatics support, these associations will allow us to choose amongst asthma therapies most likely to produce intended outcomes and to minimize unintended side effects. Such validation will need to include confirmation of associations in multiple cohorts and explication of the genetic background of the populations in which the associations have been verified. In many cases the associations will require prospective confirmation. As the information becomes available we will approach our goal of using the right medications for the right patient.
Acknowledgments
Supported by the National Heart, Lung, and Blood Institute/National Institutes of Health Grant U10 HL74227
Abbreviations
- B16 Arg/Arg (B16 Gly/Gly)
Homozygosity for arginine (or glycine) at the 16 amino acid position of ADRB2
- LABAs
Long-acting β-agonists
- ACRN
Asthma Clinical Research Network
- GWAS
Genome wide association studies
- LTMs
Leukotriene modifiers
- LTRAs
Leukotriene receptor antagonists
- ALOX5
5-Lipoxygenase enzyme
- CRHR1
Corticotropin-releasing hormone receptor 1
- ADRB2
Beta-2-adrenergic receptor
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Szefler SJ, Martin RJ, King TS, Boushey HA, Cherniack RM, Chinchilli VM, et al. Significant variability in response to inhaled corticosteroids for persistent asthma. J Allergy Clin Immunol. 2002 Mar;109(3):410–8. doi: 10.1067/mai.2002.122635. [DOI] [PubMed] [Google Scholar]
- 2.Israel E, Banerjee TR, Fitzmaurice GM, Kotlov TV, LaHive K, LeBoff MS. Effects of inhaled glucocorticoids on bone density in premenopausal women. N Engl J Med. 2001 Sep 27;345(13):941–7. doi: 10.1056/NEJMoa002304. [DOI] [PubMed] [Google Scholar]
- 3.Szefler SJ, Phillips BR, Martinez FD, Chinchilli VM, Lemanske RF, Strunk RC, et al. Characterization of within-subject responses to fluticasone and montelukast in childhood asthma. J Allergy Clin Immunol. 2005 Feb;115(2):233–42. doi: 10.1016/j.jaci.2004.11.014. [DOI] [PubMed] [Google Scholar]
- 4.Palmer LJ, Silverman ES, Weiss ST, Drazen JM. Pharmacogenetics of asthma. Am J Respir Crit Care Med. 2002 Apr 1;165(7):861–6. doi: 10.1164/ajrccm.165.7.2109096. [DOI] [PubMed] [Google Scholar]
- 5.Van Eerdewegh P, Little RD, Dupuis J, Del Mastro RG, Falls K, Simon J, et al. Association of the ADAM33 gene with asthma and bronchial hyperresponsiveness. Nature. 2002 Jul 25;418(6896):426–30. doi: 10.1038/nature00878. [DOI] [PubMed] [Google Scholar]
- 6.Hansen NT, Brunak S, Altman RB. Generating genome-scale candidate gene lists for pharmacogenomics. Clin Pharmacol Ther. 2009 Aug;86(2):183–9. doi: 10.1038/clpt.2009.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wechsler ME, Kunselman SJ, Chinchilli VM, Bleecker E, Boushey HA, Calhoun WJ, Ameredes BT, Castro M, Craig TJ, Denlinger L, Fahy JV, Jarjour N, Kazani S, Kim S, Kraft M, Lazarus SC, Lemanske RF, Jr, Markezich A, Martin RJ, Permaul P, Peters SP, Ramsdell J, Sorkness CA, Sutherland ER, Szefler SJ, Walter MJ, Wasserman SI, Israel E, National Heart, Lung and Blood Institute's Asthma Clinical Research Network Effect of beta2-adrenergic receptor polymorphism on response to longacting beta2 agonist in asthma (LARGE trial): a genotype-stratified, randomised, placebo-controlled, crossover trial. Lancet. 2009 Nov 21;374(9703):1754–64. doi: 10.1016/S0140-6736(09)61492-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hawkins GA, Tantisira K, Meyers DA, Ampleford EJ, Moore WC, Klanderman B, et al. Sequence, haplotype, and association analysis of ADRbeta2 in a multiethnic asthma case-control study. Am J Respir Crit Care Med. 2006 Nov 15;174(10):1101–9. doi: 10.1164/rccm.200509-1405OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weiss ST, Litonjua AA, Lange C, Lazarus R, Liggett SB, Bleecker ER, et al. Overview of the pharmacogenetics of asthma treatment. Pharmacogenomics J. 2006 Sep-Oct;6(5):311–26. doi: 10.1038/sj.tpj.6500387. [DOI] [PubMed] [Google Scholar]
- 10.Green SA, Cole G, Jacinto M, Innis M, Liggett SB. A polymorphism of the human beta 2-adrenergic receptor within the fourth transmembrane domain alters ligand binding and functional properties of the receptor. J BiolChem. 1993;268:23116–21. 1993. [PubMed] [Google Scholar]
- 11.Green SA, Turki J, Innis M, Liggett SB. Amino-terminal polymorphisms of the human beta 2-adrenergic receptor impart distinct agonist-promoted regulatory properties. Biochemistry. 1994;33:9414–9. doi: 10.1021/bi00198a006. 1994. [DOI] [PubMed] [Google Scholar]
- 12.Green SA, Turki J, Bejarano P, Hall IP, Liggett SB. Influence of beta(2)-adrenergic receptor genotypes on signal transduction in human airway smooth muscle cells. AmJRespir Cell MolBiol. 1995;13:25–33. doi: 10.1165/ajrcmb.13.1.7598936. 1995. [DOI] [PubMed] [Google Scholar]
- 13.McGraw DW, Forbes SL, Kramer LA, Liggett SB. Polymorphisms of the 5' leader cistron of the human beta 2-adrenergic receptor regulate receptor expression. J ClinInvest. 1998;102:1927–32. doi: 10.1172/JCI4862. 1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Panebra A, Schwarb MR, Swift SM, Weiss ST, Bleecker ER, Hawkins GA, et al. Variable-length poly-C tract polymorphisms of the beta2-adrenergic receptor 3'-UTR alter expression and agonist regulation. Am J Physiol Lung Cell Mol Physiol. 2008 Feb;294(2):L190–5. doi: 10.1152/ajplung.00277.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Anderson WBS, Bleecker ER, et al. A prospective haplotype analysis of beta2-adrenergic receptor polymorphisms and clinical response to salmeterol and salmeterol/fluticasone propionate [Abstract] Am J Respir Crit Care Med. 2008;(177):A775. [Google Scholar]
- 16.Martinez FD, Graves PE, Baldini M, Solomon S, Erickson R. Association between genetic polymorphisms of the beta 2-adrenoceptor and response to albuterol in children with and without a history of wheezing. J ClinInvest. 1997;100:3184–8. doi: 10.1172/JCI119874. 1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lima JJ, Thomason DB, Mohamed MH, Eberle LV, Self TH, Johnson JA. Impact of genetic polymorphisms of the beta 2-adrenergic receptor on albuterol bronchodilator pharmacodynamics. ClinPharmacolTher. 1999;65:519–25. doi: 10.1016/S0009-9236(99)70071-8. 1999. [DOI] [PubMed] [Google Scholar]
- 18.Israel E, Chinchilli VM, Ford JG, Boushey HA, Cherniack R, Craig TJ, et al. Use of regularly scheduled albuterol treatment in asthma: genotype-stratified, randomised, placebo-controlled cross-over trial. Lancet. 2004 Oct 23–29;364(9444):1505–12. doi: 10.1016/S0140-6736(04)17273-5. [DOI] [PubMed] [Google Scholar]
- 19.Silverman EK, Kwiatkowski DJ, Sylvia JS, Lazarus R, Drazen JM, Lange C, et al. Family-based association analysis of beta2-adrenergic receptor polymorphisms in the childhood asthma management program. J Allergy Clin Immunol. 2003 Nov;112(5):870–6. doi: 10.1016/s0091-6749(03)02023-2. [DOI] [PubMed] [Google Scholar]
- 20.Taylor DR, Epton MJ, Kennedy MA, Smith AD, Iles S, Miller AL, et al. Bronchodilator response in relation to beta2-adrenoceptor haplotype in patients with asthma. Am J Respir Crit Care Med. 2005 Sep 15;172(6):700–3. doi: 10.1164/rccm.200501-092OC. [DOI] [PubMed] [Google Scholar]
- 21.Hall IP, Blakey JD, Al Balushi KA, Wheatley A, Sayers I, Pembrey ME, et al. Beta2-adrenoceptor polymorphisms and asthma from childhood to middle age in the British 1958 birth cohort: a genetic association study. Lancet. 2006 Aug 26;368(9537):771–9. doi: 10.1016/S0140-6736(06)69287-8. [DOI] [PubMed] [Google Scholar]
- 22.Tsai HJ, Shaikh N, Kho JY, Battle N, Naqvi M, Navarro D, et al. Beta 2-adrenergic receptor polymorphisms: pharmacogenetic response to bronchodilator among African American asthmatics. Hum Genet. 2006 Jun;119(5):547–57. doi: 10.1007/s00439-006-0169-2. [DOI] [PubMed] [Google Scholar]
- 23.Choudhry S, Ung N, Avila PC, Ziv E, Nazario S, Casal J, et al. Pharmacogenetic Differences in Response to Albuterol between Puerto Ricans and Mexicans with Asthma. Am J Respir Crit Care Med. 2005 March 15;171(6):563–70. doi: 10.1164/rccm.200409-1286OC. 2005. [DOI] [PubMed] [Google Scholar]
- 24.Carroll CL, Stoltz P, Schramm CM, Zucker AR. Beta2-adrenergic receptor polymorphisms affect response to treatment in children with severe asthma exacerbations. Chest. 2009 May;135(5):1186–92. doi: 10.1378/chest.08-2041. [DOI] [PubMed] [Google Scholar]
- 25.Martin AC, Zhang G, Rueter K, Khoo SK, Bizzintino J, Hayden CM, et al. Beta2-adrenoceptor polymorphisms predict response to beta2-agonists in children with acute asthma. J Asthma. 2008 Jun;45(5):383–8. doi: 10.1080/02770900801971792. [DOI] [PubMed] [Google Scholar]
- 26.Israel E, Drazen JM, Liggett SB, Boushey HA, Cherniack RM, Chinchilli VM, et al. The effect of polymorphisms of the beta2-adrenergic receptor on the response to regular use of albuterol in asthma. Am J Respir Crit Care Med. 2000;162:75–80. doi: 10.1164/ajrccm.162.1.9907092. 2000. [DOI] [PubMed] [Google Scholar]
- 27.Taylor DR, Drazen JM, Herbison GP, Yandava CN, Hancox RJ, Town GI. Asthma exacerbations during long term beta-agonist use: influence of beta2 adrenoceptor polymorphism. Thorax. 2000;55:762–7. doi: 10.1136/thorax.55.9.762. 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Litonjua AA, Silverman EK, Tantisira KG, Sparrow D, Sylvia JS, Weiss ST. Beta 2-adrenergic receptor polymorphisms and haplotypes are associated with airways hyperresponsiveness among nonsmoking men. Chest. 2004 Jul;126(1):66–74. doi: 10.1378/chest.126.1.66. [DOI] [PubMed] [Google Scholar]
- 29.Nelson HS, Weiss ST, Bleecker ER, Yancey SW, Dorinsky PM. The Salmeterol Multicenter Asthma Research Trial: a comparison of usual pharmacotherapy for asthma or usual pharmacotherapy plus salmeterol. Chest. 2006 Jan;129(1):15–26. doi: 10.1378/chest.129.1.15. [DOI] [PubMed] [Google Scholar]
- 30.Wechsler ME, Lehman E, Lazarus SC, Lemanske RF, Jr., Boushey HA, Deykin A, et al. beta-Adrenergic receptor polymorphisms and response to salmeterol. Am J Respir Crit Care Med. 2006 Mar 1;173(5):519–26. doi: 10.1164/rccm.200509-1519OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee DK, Currie GP, Hall IP, Lima JJ, Lipworth BJ. The arginine-16 beta2-adrenoceptor polymorphism predisposes to bronchoprotective subsensitivity in patients treated with formoterol and salmeterol. Br J Clin Pharmacol. 2004 Jan;57(1):68–75. doi: 10.1046/j.1365-2125.2003.01955.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Palmer CN, Lipworth BJ, Lee S, Ismail T, Macgregor DF, Mukhopadhyay S. Arginine-16 {beta}2 adrenoceptor genotype predisposes to exacerbations in young asthmatics taking regular salmeterol. Thorax. 2006 Nov;61(11):940–4. doi: 10.1136/thx.2006.059386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bleecker ER, Postma DS, Lawrance RM, Meyers DA, Ambrose HJ, Goldman M. Effect of ADRB2 polymorphisms on response to longacting beta2-agonist therapy: a pharmacogenetic analysis of two randomised studies. Lancet. 2007 Dec 22;370(9605):2118–25. doi: 10.1016/S0140-6736(07)61906-0. [DOI] [PubMed] [Google Scholar]
- 34.Bleecker ER, Yancey SW, Baitinger LA, Edwards LD, Klotsman M, Anderson WH, et al. Salmeterol response is not affected by beta2-adrenergic receptor genotype in subjects with persistent asthma. J Allergy Clin Immunol. 2006 Oct;118(4):809–16. doi: 10.1016/j.jaci.2006.06.036. [DOI] [PubMed] [Google Scholar]
- 35.Bleecker ER, Nelson H, Corren J, Kraft M, Yancey S, Ortega H, et al. Arg16Gly Polymorphism of the [beta]2-adrenergic Receptor Gene Does Not Modulate Clinical Response to Salmeterol in Subjects with Asthma. Journal of Allergy and Clinical Immunology. 2008;121(2 Supplement 1):S143–S. [Google Scholar]
- 36.Litonjua AA, Lasky-Su J, Schneiter K, Tantisira KG, Lazarus R, Klanderman B, et al. ARG1 is a novel bronchodilator response gene: screening and replication in four asthma cohorts. Am J Respir Crit Care Med. 2008 Oct 1;178(7):688–94. doi: 10.1164/rccm.200709-1363OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Malmstrom K, Rodriguez-Gomez G, Guerra J, Villaran C, Pineiro A, Wei LX, et al. Oral montelukast, inhaled beclomethasone, and placebo for chronic asthma. AnnInternMed. 1999;130:487–95. doi: 10.7326/0003-4819-130-6-199903160-00005. 1999. [DOI] [PubMed] [Google Scholar]
- 38.Sampson AP, Siddiqui S, Buchanan D, Howarth PH, Holgate ST, Holloway JW, et al. Variant LTC(4) synthase allele modifies cysteinyl leukotriene synthesis in eosinophils and predicts clinical response to zafirlukast. Thorax. 2000 Oct;55(Suppl 2):S28–31. doi: 10.1136/thorax.55.suppl_2.S28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sanak M, Simon H-U, Szczeklik A. Leukotriene C4 synthase promoter polymorphism and risk of aspirin-induced asthma. Lancet. 1997;350:1599–600. doi: 10.1016/s0140-6736(05)64015-9. 1997. [DOI] [PubMed] [Google Scholar]
- 40.Sanak M, Pierzchalska M, Bazan-Socha S, Szcezklik A. Enhanced expression of the leukotriene C4 synthase due to overactive transcription of an allelic variant associated with aspirin-intolerant asthma. AmJRespir Cell MolBiol. 2000;23:290–6. doi: 10.1165/ajrcmb.23.3.4051. 2000. [DOI] [PubMed] [Google Scholar]
- 41.Whelan GJ, Blake K, Kissoon N, Duckworth LJ, Wang J, Sylvester JE, et al. Effect of montelukast on time-course of exhaled nitric oxide in asthma: influence of LTC4 synthase A(−444)C polymorphism. Pediatr Pulmonol. 2003 Nov;36(5):413–20. doi: 10.1002/ppul.10385. [DOI] [PubMed] [Google Scholar]
- 42.Asano K, Shiomi T, Hasegawa N, Nakamura H, Kudo H, Matsuzaki T, et al. Leukotriene C4 synthase gene A(−444)C polymorphism and clinical response to a CYS-LT(1) antagonist, pranlukast, in Japanese patients with moderate asthma. Pharmacogenetics. 2002 Oct;12(7):565–70. doi: 10.1097/00008571-200210000-00009. [DOI] [PubMed] [Google Scholar]
- 43.Lima JJ, Zhang S, Grant A, Shao L, Tantisira KG, Allayee H, et al. Influence of leukotriene pathway polymorphisms on response to montelukast in asthma. Am J Respir Crit Care Med. 2006 Feb 15;173(4):379–85. doi: 10.1164/rccm.200509-1412OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Drazen JM, Yandava CN, Dube L, Szczerback N, Hippensteel R, Pillari A, et al. Pharmacogenetic association between ALOX5 promoter genotype and the response to anti-asthma treatment. NatGenet. 1999;22:168–70. doi: 10.1038/9680. 1999. [DOI] [PubMed] [Google Scholar]
- 45.Telleria JJ, Blanco-Quiros A, Varillas D, Armentia A, Fernandez-Carvajal I, Jesus Alonso M, et al. ALOX5 promoter genotype and response to montelukast in moderate persistent asthma. Respir Med. 2008 Jun;102(6):857–61. doi: 10.1016/j.rmed.2008.01.011. [DOI] [PubMed] [Google Scholar]
- 46.Klotsman M, York TP, Pillai SG, Vargas-Irwin C, Sharma SS, van den Oord EJ, et al. Pharmacogenetics of the 5-lipoxygenase biosynthetic pathway and variable clinical response to montelukast. Pharmacogenet Genomics. 2007 Mar;17(3):189–96. doi: 10.1097/FPC.0b013e3280120043. [DOI] [PubMed] [Google Scholar]
- 47.Tantisira KG, Lima J, Sylvia J, Klanderman B, Weiss ST. 5-lipoxygenase pharmacogenetics in asthma: overlap with Cys-leukotriene receptor antagonist loci. Pharmacogenet Genomics. 2009 Mar;19(3):244–7. doi: 10.1097/FPC.0b013e328326e0b1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mougey EB, Feng H, Castro M, Irvin CG, Lima JJ. Absorption of montelukast is transporter mediated: a common variant of OATP2B1 is associated with reduced plasma concentrations and poor response. Pharmacogenet Genomics. 2009 Feb;19(2):129–38. doi: 10.1097/FPC.0b013e32831bd98c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tantisira KG, Lake S, Silverman ES, Palmer LJ, Lazarus R, Silverman EK, et al. Corticosteroid pharmacogenetics: association of sequence variants in CRHR1 with improved lung function in asthmatics treated with inhaled corticosteroids. Hum Mol Genet. 2004 Jul 1;13(13):1353–9. doi: 10.1093/hmg/ddh149. [DOI] [PubMed] [Google Scholar]
- 50.Dijkstra A, Koppelman GH, Vonk JM, Bruinenberg M, Schouten JP, Postma DS. Pharmacogenomics and outcome of asthma: no clinical application for long-term steroid effects by CRHR1 polymorphisms. J Allergy Clin Immunol. 2008 Jun;121(6):1510–3. doi: 10.1016/j.jaci.2008.04.015. [DOI] [PubMed] [Google Scholar]
- 51.Tantisira KG, Hwang ES, Raby BA, Silverman ES, Lake SL, Richter BG, et al. TBX21: a functional variant predicts improvement in asthma with the use of inhaled corticosteroids. Proc Natl Acad Sci U S A. 2004 Dec 28;101(52):18099–104. doi: 10.1073/pnas.0408532102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tantisira KG, Silverman ES, Mariani TJ, Xu J, Richter BG, Klanderman BJ, et al. FCER2: a pharmacogenetic basis for severe exacerbations in children with asthma. J Allergy Clin Immunol. 2007 Dec;120(6):1285–91. doi: 10.1016/j.jaci.2007.09.005. [DOI] [PubMed] [Google Scholar]
- 53.Tantisira KG, Small KM, Litonjua AA, Weiss ST, Liggett SB. Molecular properties and pharmacogenetics of a polymorphism of adenylyl cyclase type 9 in asthma: interaction between beta-agonist and corticosteroid pathways. Hum Mol Genet. 2005 Jun 15;14(12):1671–7. doi: 10.1093/hmg/ddi175. [DOI] [PubMed] [Google Scholar]
- 54.Stallings SC, Huse D, Finkelstein SN, Crown WH, Witt WP, Maguire J, et al. A framework to evaluate the economic impact of pharmacogenomics. Pharmacogenomics. 2006 Sep;7(6):853–62. doi: 10.2217/14622416.7.6.853. [DOI] [PubMed] [Google Scholar]
- 55.Burchard EG, Ziv E, Coyle N, Gomez SL, Tang H, Karter AJ, et al. The Importance of Race and Ethnic Background in Biomedical Research and Clinical Practice. N Engl J Med. 2003 March 20;348(12):1170–5. doi: 10.1056/NEJMsb025007. 2003. [DOI] [PubMed] [Google Scholar]
- 56.Himes BE, Wu AC, Duan QL, Klanderman B, Litonjua AA, Tantisira K, et al. Predicting response to short-acting bronchodilator medication using Bayesian networks. Pharmacogenomics. 2009 Sep;10(9):1393–412. doi: 10.2217/pgs.09.93. [DOI] [PMC free article] [PubMed] [Google Scholar]