Abstract
Activation of NF-κB by the pro-inflammatory cytokines tumor necrosis factor (TNF) and interleukin-1 (IL-1) requires the IκB kinase (IKK) complex, which contains two kinases named IKKα and IKKβ and a critical regulatory subunit named NEMO. Although we have previously demonstrated that NEMO associates with both IKKs, genetic studies reveal that only its interaction with IKKβ is required for TNF-induced NF-κB activation. To determine whether NEMO and IKKα can form a functional IKK complex capable of activating the classical NF-κB pathway in the absence of IKKβ, we utilized a panel of mouse embryonic fibroblasts (MEFs) lacking each of the IKK complex subunits. This confirmed that TNF-induced IκBα degradation absolutely requires NEMO and IKKβ. In contrast, we consistently observed intact IκBα degradation and NF-κB activation in response to IL-1 in two separate cell lines lacking IKKβ. Furthermore, exogenously expressed, catalytically inactive IKKβ blocked TNF-but not IL-1-induced IκBα degradation in wild-type MEFs, and reconstitution of IKKα/β double knockout cells with IKKα rescued IL-1- but not TNF-induced NF-κB activation. Finally, we have shown that incubation of IKKβ-deficient MEFs with a cell-permeable peptide that blocks the interaction of NEMO with the IKKs inhibits IL-1-induced NF-κB activation. Our results therefore demonstrate that NEMO and IKKα can form a functional IKK complex that activates the classical NF-κB pathway in response to IL-1 but not TNF. These findings further suggest NEMO differentially regulates the fidelity of the IKK subunits activated by distinct upstream signaling pathways.
NF-κB2 proteins are a family of inducible transcription factors that regulate the expression of a broad range of genes essential for innate and adaptive immune regulation, inflammation, and cell survival (1, 2). The NF-κB family consists of five members named p50 and p52 (the NH2-terminal fragments of the longer NF-κB1/p105 and NF-κB2/p100 proteins respectively), p65 (RelA), c-Rel, and RelB. These proteins homo- or heterodimerize to form either transcriptionally active (e.g. p50: p65) or repressive (e.g. p50:p50) NF-κB dimers (1, 2). NF-κB is maintained inactive in the cytosol of resting cells by members of the IκB family of inhibitory proteins that include IκBα, IκBβ, IκBε, and the COOH termini of p105 and p100. In response to a wide range of stimuli, including pro-inflammatory cytokines (e.g. TNF and IL-1), bacterial products (e.g. lipopolysaccharide (LPS), CpG DNA), and the engagement of antigen receptors on T- and B-lymphocytes, IκB proteins are rapidly phosphorylated, ubiquitinated, and degraded by the 26 S proteasome, thereby enabling NF-κB dimers to localize to the nucleus and regulate target gene transcription (2).
Signal-induced IκB phosphorylation is mediated by the high molecular weight IκB kinase (IKK) complex that contains two catalytic subunits named IKKα (IKK1) and IKKβ (IKK2) and a non-catalytic regulatory subunit named NF-κB essential modulator (NEMO) or IKKγ (3). IKKα and IKKβ share significant structural identity, and each contains an NH2-terminal catalytic domain, a central leucine zipper motif through which they heterodimerize, and a COOH-terminal helix-loop-helix domain (2, 3). IKKβ also contains a novel ubiquitin-like domain, although the function of this region is not yet known (4). We have previously demonstrated that identical hexapeptide sequences (i.e. Leu-Asp-Trp-Ser-Trp-Leu) within the extreme COOH termini of both IKKα and IKKβ facilitate their association with NEMO, and we named this region the NEMO binding domain (NBD) (5, 6). Dissociation of NEMO from the IKK complex using a cell-permeable peptide spanning the NBD effectively blocks pro-inflammatory NF-κB activation, thereby supporting the critical role of NEMO in regulating signal-induced activity of the intact IKK complex (5, 7).
Despite their significant similarities, IKKα and IKKβ play distinct roles within the overall NF-κB signaling paradigm (1, 2). In this regard, rapid and transient TNF-induced IκBα degradation occurs through a pathway that depends upon IKKβ and NEMO (1). This was definitively established in mice lacking either of these subunits that die during development from massive TNF-induced hepatocyte apoptosis due to the inability to mount an NF-κB-dependent anti-apoptotic response (8–12). This “classical” NF-κB signaling pathway is now defined as NEMO- and IKKβ-dependent IκB phosphorylation and degradation liberating canonical NF-κB complexes typified by the ubiquitous p50:p65 heterodimer. All stimuli that induce IκBα degradation, including IL-1, LPS, and antigen-receptor engagement, are considered to be activators of the NEMO- and IKKβ-dependent classical NF-κB pathway (1, 2).
Analysis of mice harboring inactive IKKα revealed an unanticipated role for this kinase in NIK (NF-κB-inducing kinase)-dependent processing of NF-κB2/p100 to generate p52 (13, 14). This processing occurs only in response to ligation of a subset of receptors, including the lymphotoxin-β receptor (LTβR), CD40, and BAFF-R (13–17). The NF-κB2/p100 targeted by IKKα is one-half of a heterodimer with RelB, thereby resulting after processing in the generation of p52:RelB NF-κB complexes (14, 17). These complexes regulate several genes encoding lymphoid chemokines (i.e. CCL19, CCL21, CXCL12, and CXCL13) and BAFF (13), and reflecting this, the major functions of this pathway are in lymphoid organogenesis and B-cell maturation. This pathway is termed the “non-canonical” or “alternative” NF-κB pathway, and studies using IKKβ- and NEMO-deficient cells have demonstrated that it functions in the absence of each of these IKK complex subunits (1).
The function of IKKα in the non-canonical pathway does not therefore require its ability to interact with NEMO via its COOH-terminal NBD (13, 15–17). Furthermore, classical NF-κB activation in response to TNF occurs in the absence of IKKα, suggesting that the interaction of IKKα with NEMO plays no role in regulating this pathway (13, 14, 18–20). We therefore sought to determine whether the ability of IKKα to interact with NEMO via its NBD plays any functional role in classical NF-κB signaling. To address this question, we examined IκBα degradation induced by TNF and IL-1 in murine embryonic fibroblasts (MEFs) lacking each of the IKK complex subunits. Remarkably we found that, although TNF-induced IκBα degradation was absolutely dependent upon NEMO and IKKβ, IL-1-induced degradation and classical NF-κB activation remained intact in cells lacking IKKβ. Furthermore, IL-1-induced NF-κB was blocked by the cell-permeable NBD peptide in IKKβ−/− MEFs, demonstrating that an IKK complex consisting of only IKKα and NEMO is sufficient for IL-1- but not TNF-induced classical NF-κB activation. Our findings therefore identify differences in the absolute requirements for the separate IKK subunits activated in a NEMO-dependent manner by distinct upstream stimuli.
Experimental Procedures
Reagents and Cell Culture
Recombinant human IL-1α was obtained from Peprotech (Rocky Hill, NJ), recombinant human TNF was from R & D Systems (Minneapolis, MN), and LPS from Salmonella typhimuri (LPS) was from Sigma. MG-132 was obtained from BIOMOL International (Plymouth Meeting, PA). Polyclonal anti-IKKα (catalog number sc-7218), anti-NEMO (sc-8330), anti-IκBα (sc-371), anti-p65/RelA (sc-372X), and anti-p50 (sc-114X) were from Santa Cruz Biotechnology (Santa Cruz, CA). Polyclonal anti-IKKβ (catalog number 2684), anti-phospho-IκBα (9241S), and anti-NF-κB2 (p100/p52) (4882) were from Cell Signaling Technology (Beverly, MA), and monoclonal anti-α-tubulin (T5168) was from Sigma. Normal rabbit IgG, used as a nonspecific antibody in immunoprecipitations, was from Santa Cruz Biotechnology (catalog number sc-2027). Protein G-Sepharose beads were from Amersham Biosciences, and horseradish peroxidase-conjugated secondary antibodies against either rabbit or mouse IgG were from Jackson ImmunoResearch Laboratories, (West Grove, PA). Murine anti-lymphotoxin-β receptor (LTβR; AC.H6) (catalog number 552939) was from BD Biosciences.
Wild-type (WT), IKKα−/−, and IKKβ−/− murine embryonic fibroblasts were generously provided by Dr. Inder Verma (The Salk Institute for Biological Studies, La Jolla, CA; MEFs 1), who also provided the IKKα/β double-deficient cells, and Dr. Michael Karin (University of California San Diego School of Medicine, La Jolla, CA; MEFs 2) who also provided the NEMO-deficient MEFs. Plat-E cells were cultured and used as previously described (21). All cells were maintained in Dulbecco's modified Eagle's medium (Invitrogen) supplemented with 10% fetal calf serum, 2 mm l-glutamine, penicillin (50 units/ml), and streptomycin (50μg/ml). For all experiments, unless otherwise indicated, cells were cultured in either 6-well tissue culture trays or 100-mm dishes and were stimulated with IL-1 (10 ng/ml) or TNF (10 ng/ml) when they reached 80% confluence.
Immunoblotting and Immunoprecipitation
Cells were washed once with phosphate-buffered saline and then incubated for 10 min at 4 °C in 100 μl of TNT lysis buffer (50 mm Tris-Cl, pH 7.5, 150 mm NaCl, and 1% Triton X-100) and a complete miniprotease inhibitor mixture (Roche Applied Science). Samples were then scraped and harvested into 1.5-ml microcentrifuge tubes, vortexed for 30 s, and then centrifuged (425 × g for 10 min). Protein levels in the supernatants were determined using a Coomassie protein assay kit (Bio-Rad), and 20 μg of protein from each sample was separated by SDS-PAGE (10%) and then transferred to a polyvinylidene difluoride membrane (Millipore, Milford, MA) and immunoblotted with primary and horseradish peroxidase-conjugated secondary antibodies. Detection of the bound antibody by enhanced chemiluminescence was performed according to the manufacturer's instructions (Pierce). For immunoprecipitations, cell extracts were incubated with 2 μg of primary antibody for 1 h at 4 °C followed by incubation (1 h at 4 °C) with 30 μl of protein G-Sepharose beads (50% slurry). A portion of each sample pre-immunoprecipitation (5%) was retained for analysis. The beads were washed three times with lysis buffer, and then the samples were analyzed by SDS-PAGE (10%) followed by immunoblotting as described above.
Densitometry
Densitometry was performed using a Gel-Doc EQ gel documentation system and the QuantityOne software package (Bio-Rad). Pixel intensity was measured in identical rectangular volumes around each band on immunoblots, and a background value in an equal rectangular volume separate from the bands was subtracted to obtain the mean pixel intensity/mm2. Statistical analysis was performed using a two-tailed paired Student's t test.
Transfections and Luciferase Reporter Assays
WT, IKKα−/−, IKKβ−/−, and NEMO-deficient MEFs grown in 12-well plates (2.5 × 105/well) were transiently transfected using FuGENE 6 (Roche Applied Science) following the manufacturer's protocol. Cells were transfected with a total of 1.0 μg of DNA/well consisting of the NF-κB-dependent firefly luciferase reporter construct pBIIx-firefly luciferase (0.2 μg/well) and a β-actin promoter Renilla luciferase reporter (0.02 μg/well) together with either control vector alone or the test DNA. Cells were stimulated with IL-1 for 5 h and then lysed in passive lysis buffer (Promega) 24–36 h after transfection. Samples were assayed using a Luminoscan 96-well automated luminometer (Thermo Labsystems, Franklin, MA), and FF:RL ratios were calculated using Ascent software (Thermo LabSystems).
Electrophoretic Mobility Shift Assays (EMSAs)
MEFs were stimulated with IL-1 (10 ng/ml) for the appropriate times and then scraped into phosphate-buffered saline at 4 °C and pelleted (425 g, 10 min). Pellets were resuspended and swollen for 30 min on ice in 100 μl of NarA buffer (10 mm HEPES, pH 7.9, 10 mm KCl, 0.1 mm EDTA, 2 mm NaF, 2 mmβ-glycerolphosphate, and complete miniprotease inhibitors), incubated a further 5 min on ice in 0.1% Nonidet P-40, and then vortexed and centrifuged (3800 × g) for 1 min. Supernatants (cytoplasmic fraction) were centrifuged (20,000 × g) for 1 h at 4 °C, and the resulting supernatants were snap frozen and retained for analysis. Pelleted nuclei were washed once with 100 μl of NarA buffer before being vortexed in 30 μl of NarC buffer (20 mm HEPES, pH 7.9, 0.4 m NaCl, pH 8, 1 mm EDTA, pH 8, 2 mm NaF, 2 mmβ-glycerolphosphate, and complete miniprotease inhibitors) for 1 h at 4 °C. Nuclear lysates were then centrifuged for 20 min (20,000 × g) at room temperature and then either used immediately or snap frozen and stored at −80 °C.
Single-stranded complimentary oligonucleotides encompassing a consensus NF-κB site (upper strand, 5′-AGTTGAGGGGACTTTCCCAGGC-3′) or the Oct-1 probe (Santa Cruz Biotechnology) were annealed and then labeled with [γ-32P]ATP using T4 polynucleotide kinase (New England Biolabs, Beverly, MA). The labeled probe was purified using mini-Quick Spin columns (Roche Applied Science) according to the manufacturer's instructions. For EMSA, 2–5 μg of nuclear extracts supplemented with 1 μg of poly(dI·dC) (Roche Applied Science) were incubated with an equal volume of 2× binding buffer (40 mm Tris-HCl, pH 7.9, 100 mm NaCl, 10 mm MgCl2, 2 mm EDTA, 20% glycerol, 0.2% Nonidet P-40, 2 mm dithiothreitol, 100 μg/ml bovine serum albumin) on ice for 10 min. After incubation, 1 μl of labeled probe was added, and then samples were incubated at room temperature for 20 min. The resulting DNA·NF-κB complexes were separated on 5% polyacrylamide non-denaturing gels by electrophoresis, and then gels were dried and visualized by autoradiography. Supershift analysis was performed following the same protocol, except that samples were incubated with antibodies (anti-p65 or anti-p50) for 2 h at 4 °C prior to the addition of labeled probe.
NBD Peptides
Small scale Fmoc (N-(9-fluorenyl)methoxycarbonyl) synthesis of the peptides was carried out on a Rainin Symphony instrument (Rainin Instruments, LLC, Oakland, CA) at the Howard Hughes Medical Institute Biopolymer-Keck Foundation Biotechnology Resource Laboratory at Yale University (New Haven, CT). Peptides were characterized by matrix-assisted laser desorption ionization mass spectrometry and analytical reverse-phase high-performance liquid chromatography analysis. Immediately prior to use, the peptides were dissolved in dimethyl sulfoxide to a stock of 50 mm. The sequences of the wild-type and mutant NBD peptides have been described previously (5, 6). The NBD peptide contains the region of IKKβ from T735 to E745 synthesized in tandem with a membrane permeabilization sequence from the Drosophila antennapedia homeodomain protein. The mutant peptide is identical, except that Trp-739 and -741 are replaced by alanines to render it biologically inactive (5, 6).
Preparation of Stable Cell Lines
All cloning procedures were performed by PCR using cloned Pfu DNA polymerase (Stratagene, La Jolla, CA). A cDNA encoding IKKβ (K44M) was subcloned into the HindIII and NotI sites of LZRS-pBMN-lacZ retroviral vector (kindly provided by Garry Nolan, Stanford University, Stanford, CA). Resulting LZRS−IKKβ(K44M) was transiently transfected using FuGENE 6 into Plat-E cells and selected for gene expression 24 h after transfection using puromycin (1 μg/ml). Puromycin-resistant cells were used to derive conditioned medium to provide a retroviral stock for MEF transduction. For cell transduction, MEFs were washed and incubated for 8 h with a retrovirus-conditioned medium containing Polybrene (8 μg/ml, Sigma). After incubation, the retrovirus was removed and replaced with normal growth medium. The transduction process was repeated a further three times until the cells became >90% GFP-positive as determined by fluorescence-activated cell sorter (FACS) analysis using Cell Quest software (FACSort, BD Biosciences, San Jose, CA).
To generate double knock-out (DKO) MEFs stably transduced with IKKα, full-length IKKα cDNA was cloned into pCR-Blunt II-TOPO vector (Invitrogen) and then subcloned into the EcoRI restriction site of retroviral GFP-MIGR1 vector (kindly provided by Dr. Warren Pear, University of Pennsylvania, Philadelphia, PA). The resulting MIG−IKKα was transiently transfected using FuGENE 6 into Plat-E cells to produce ecotropic virus that was derived from conditioned medium containing Polybrene (8 μg/ml).
For cell sorting, transduced cells were trypsinized and washed in FACS buffer (sterile phosphate-buffered saline, 0.5 mm EDTA, and 0.5% bovine serum albumin). Evaluation of GFP was performed on a three-laser (argon (488 nm), krypton (407 nm), and dye laser (tuned to 600 nm)), 10-parameter FACSVantage™ cell sorter from BD Biosciences Immunocytometry Systems. Compensation and data analyses were performed using FlowJo software (Tree Star, Ashland, OR).
Results
IL-1 (but Not TNF or LPS) Induces IκBα Degradation in the Absence of IKKβ
To identify the components of the IKK complex required for classical NF-κB activation in response to distinct stimuli, we compared TNF, IL-1, and LPS signaling in wild-type (WT), IKKα−/−, IKKβ−/−, and NEMO-deficient MEFs. The phenotype of each MEF line was verified by immunoblotting (Fig. 1A) and confirmed for each experiment performed. As described previously (13, 14, 18–20), incubation with TNF led to rapid degradation of IκBα that returned to basal levels within 60 min in both WT and IKKα−/− MEFs (Fig. 1B, lanes 1–4 and 9–11). In contrast, IκBα levels remained constant in NEMO-deficient and IKKβ−/− MEFs (Fig. 1B, lanes 5–8 and 13–16), confirming that TNF-induced classical NF-κB activation requires both IKKβ and NEMO and occurs in the absence of IKKα (8–12). Similarly, IL-1-induced IκBα degradation was intact in WT and IKKα−/− MEFs but not in NEMO-deficient cells (Fig. 1C, lanes 1–12). To our surprise, however, IκBα was degraded and reappeared in IL-1-stimulated IKKβ−/− MEFs (Fig. 1C, lanes 13–16) with similar kinetics to WT cells. Furthermore, IL-1 stimulation rapidly induced IκBα phosphorylation at the critical serine residues (Ser-32 and -36) required to trigger its degradation by the proteasome in both WT (Fig. 1D, compare lanes 1 and 2) and IKKβ−/− cells (compare lanes 5 and 6).
FIGURE 1. IL-1 (but not TNF or LPS) induces IκBα degradation in IKKβ−/− MEFs.
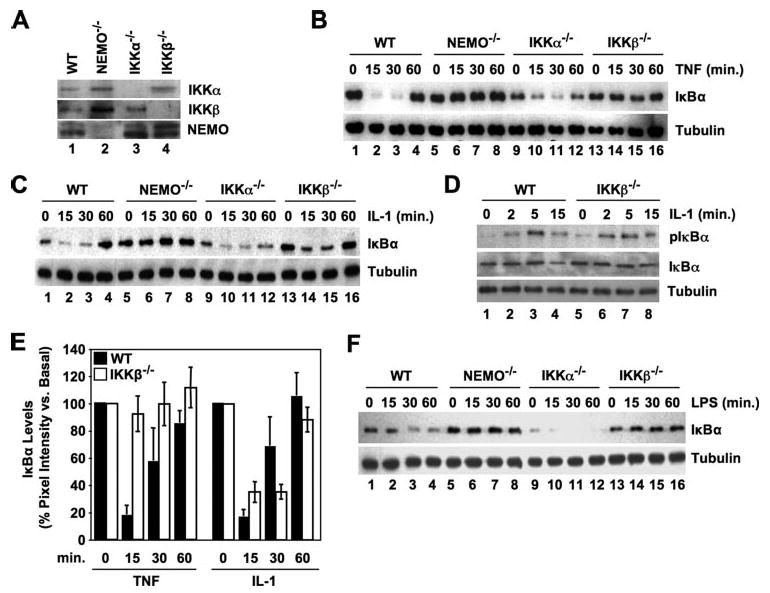
A, lysates from wild-type (WT), NEMO-deficient, IKKα−/−, and IKKβ−/− MEFs were analyzed by immunoblotting using the antibodies indicated. B, C, and F, the MEFs indicated (top) were either untreated or incubated with TNF (10 ng/ml) (B), IL-1 (10 ng/ml) (C), or LPS (100 ng/ml) (F) for the times shown, and then lysates were prepared and immunoblotted using anti-IκBα (upper panels). The same blots were also probed using anti-tubulin as a loading control (lower panels). D, WT and IKKβ−/− MEFs were incubated with MG132 (10 μm) for 15 min and then either untreated or stimulated with IL-1 (10 ng/ml) for the times shown. Lysates were immunoblotted and then probed with either anti-phospho-IκBα (pIκBα), anti-IκBα, or anti-tubulin as indicated. E, densitometry was performed as described under “Experimental Procedures.” Immunoblots from eleven separate TNF and IL-1 time course experiments comparing IκBα degradation in WT (blackbars) and IKKβ−/− MEFs (white bars) were analyzed. To normalize the data, pixel intensities for each band were determined as a percentage of the basal (untreated) value for each cell type in each experiment. Mean values for each time point were then calculated for all experiments and the data presented as the means ± S.E.A Student's t test was performed to compare data for each time point between the WT and IKKβ−/− cells, and only those data sets that demonstrated significant differences are labeled (*, p < 0.05;**, p < 0.001).
Densitometric analysis of data from eleven separate time course experiments confirmed that IL-1 (but not TNF) consistently induced IκBα degradation in MEFs lacking IKKβ (Fig. 1E, white bars). Significant differences between WT and IKKβ−/− cells were only observed at the 15- and 30-min time points in the TNF-treated samples (60 min of TNF, p = 0.17) (indicated on Fig. 1D). No significant differences were observed between WT and IKKβ−/− MEFs treated with IL-1 for any time point tested (15 min, p = 0.17; 30 min, p = 0.42; 60 min, p = 0.91). Collectively therefore, the data in panels C-E of Fig. 1 demonstrate that IL-1 induces the phosphorylation and degradation of IκBα in cells lacking IKKβ with similar kinetics to that in WT cells.
Activation of the classical NF-κB pathway by LPS occurs via ligation of the Toll-like receptor TLR4 (22). As some of the signaling intermediates downstream of TLR4 are shared with the IL-1 signaling pathway (i.e. MyD88, IRAK, and TRAF6), we questioned whether LPS could also activate classical NF-κB signaling in cells lacking IKKβ. As shown in Fig. 1F, LPS induced IκBα degradation in WT and IKKα−/− cells (lanes 1–4 and 9–11) but not in either NEMO-deficient or IKKβ−/− MEFs (lanes 5–8 and 13–16). These findings therefore confirm that LPS- and TNF-induced IκBα degradation require NEMO and IKKβ and occur in the absence of IKKα. In contrast, our data clearly demonstrate that, although IL-1-induced IκBα degradation does depend upon NEMO, it has no absolute requirement for either kinase and occurs in both IKKα- and IKKβ-deficient MEFs.
IL-1 Activates Classical NF-κB in IKKβ−/− MEFs
Consistent with its inability to stimulate IκBα degradation, previous studies have demonstrated that TNF cannot induce NF-κB DNA binding or transcriptional activity in cells lacking IKKβ−/− (8, 9). We therefore questioned whether the IL-1-induced IκBα degradation we observed in IKKβ−/− MEFs precedes NF-κB activation, nuclear translocation, and transcriptional activity. Nuclear lysates from IL-1-stimulated WT, NEMO-deficient, IKKα−/−, and IKKβ−/− MEFs were subjected to EMSAs using an NF-κB-specific probe, and as shown in Fig. 2A, NF-κB DNA binding was maximal in WT cells after 15 min and returned to basal levels after 60 min (lanes 1–4). As expected, DNA binding was absent in NEMO-deficient cells (Fig. 2A, lanes 5–9) but occurred in IKKα−/− MEFs (lanes 9–12), although this was less robust than the levels seen in WT cells. Consistent with our findings in Fig. 1, C and D, NF-κB DNA binding occurred in IKKβ−/− cells to the same level and with identical kinetics to that observed in WT MEFs (Fig. 2A, lanes 13–16).
FIGURE 2. IL-1 activates NEMO-dependent classical NF-κB in IKKβ−/− MEFs.
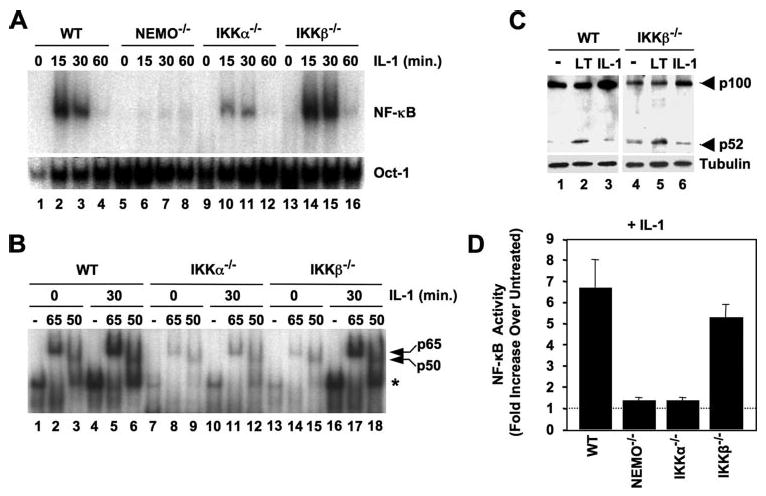
A, WT, NEMO-deficient, IKKα−/−, and IKKβ−/− MEFs were either untreated or incubated with IL-1 for the times indicated, and then nuclear extracts were prepared and used for EMSA. Assays were performed using either a consensus NF-κB binding site probe (upper panel) or an Oct1 probe as a loading control (lower panel). B, the MEFs shown (top) were either untreated or incubated with IL-1 for 30 min, and then nuclear lysates were prepared. For supershift analysis, samples were incubated prior to the EMSA reaction either in the absence of antibodies (−) or with anti-p65 or -p50 as shown. The positions of the shifted NF-κB complex (*) and supershifted p65- and p50-containing complexes are indicated (right). C, WT and IKKβ−/− MEFs were either untreated (−) or stimulated with anti-LTβR (LT) or IL-1 (10 ng/ml) for 8 h, and then lysates were immunoblotted and probed with either anti-p100/p52 (upper panel) or anti-tubulin (lower panel). D, MEFs were transiently transfected with the NF-κB-dependent reporter pBIIx-firefly luciferase together with β-actin Renilla luciferase. Twenty-four hours later, the cells were either untreated or incubated for a further 5 h with IL-1 (10 ng/ml), and then NF-κB activity was determined by dual luciferase assay. The data are expressed for each MEF line as fold values relative to the basal activity in untreated cells that was normalized between the cell lines (dotted line).
To identify the NF-κB complexes activated by IL-1 in the distinct cell types, we performed supershift analysis using antibodies against the classical NF-κB subunits p65 and p50. This analysis revealed that both basal and IL-1-induced DNA-bound NF-κB in WT, IKKα-, and IKKβ-deficient MEFS was completely shifted using anti-p65 (Fig. 2B, lanes 2, 5, 8, 11, 14, and 17). In addition, anti-p50 up-shifted approximately half of the NF-κB complexes (Fig. 2B, lanes 3, 6, 9, 12, 15, and 18), demonstrating that IL-1-induced IκBα degradation leads to nuclear translocation and DNA binding of the classical NF-κB p50:p65 heterodimer and a separate p65-containing dimer in the absence of either IKKα or IKKβ.
Our data strongly suggest that IKKα can activate the classical NF-κB pathway in response to IL-1 in the absence of IKKβ. As the well described role of IKKα is the kinase critical for non-canonical NF-κB signaling (13, 14), we questioned whether IL-1 activates the non-canonical NF-κB pathway in IKKβ−/− MEFs. As shown in Fig. 2C, incubation of both WT and IKKβ−/− MEFs with anti-LTβR for 8 h induced p100 processing to generate p52 (Fig. 2C, lanes 2 and 5) as previously described (13, 17, 23). In contrast, IL-1 did not induce any increase in p52 levels above basal in either cell type (Fig. 2C, lanes 3 and 6), confirming that IL-1 does not activate the non-canonical pathway in the absence of IKKβ.
We next performed NF-κB-dependent luciferase reporter assays to determine whether IL-1 can induce NF-κB transcriptional activity in cells lacking IKKβ. As expected, IL-1 induced NF-κB transcriptional activity in WT MEFs but not in the cells lacking NEMO (Fig. 2D), again confirming the key role for NEMO in classical NF-κB signaling. Surprisingly, IL-1 stimulation did not lead to NF-κB transcriptional activity in IKKα-deficient MEFs, despite inducing IκBα phosphorylation and degradation (Fig. 1, C–E) and NF-κB DNA binding (Fig. 2, A and B). This observation therefore supports the model proposed in several previous reports that IKKα plays a crucial downstream or subsidiary role in maintaining the full classical NF-κB transcriptional response that is independent of its function as an IκB kinase (24–29). Consistent with our IκBα phosphorylation, degradation, and NF-κB DNA binding data (Fig. 1, C–E; Fig. 2, A and B), IL-1-induced NF-κB transcriptional activity was intact in IKKβ−/− cells (Fig. 2D), leading us to surmise that IKKα can facilitate IL-1-induced classical NF-κB signaling and transcriptional activation in the absence of IKKβ. In contrast, our data suggest that IKKβ is unable to support the full NF-κB transcriptional response to this cytokine in the absence of IKKα.
To confirm that these findings were not unique to one line of IKKβ−/− MEFS, we repeated our experiments using a separate IKKβ-deficient cell line and appropriate WT controls (MEFs 2). We first confirmed that these IKKβ−/− cells lack IKKβ but contain IKKα and NEMO (Fig. 3A), and this was verified for each experiment performed. As shown in Fig. 3B, TNF was unable to induce IκBα degradation in these IKKβ−/− MEFs (Fig. 3B, lanes 6–8), whereas IL-1 stimulation induced transient degradation that was maximal at 15 min and returned to unstimulated levels after 60 min (Fig. 3B, lanes 9 and 10). To determine whether IL-1 could activate NF-κB in these cells, we performed EMSA analysis, and as shown in Fig. 3C, IL-1 induced significant mobility shift indicating NF-κB activation in both lines of IKKβ−/− cells. Luciferase reporter assays confirmed that, although the basal level of NF-κB activity was reduced in these IKKβ−/− cells compared with WT MEFs, incubation with IL-1 induced NF-κB transcriptional activity in the absence of IKKβ (Fig. 3D).
FIGURE 3. IL-1 activates NF-κB in two separate lines of IKKβ−/− MEFs.
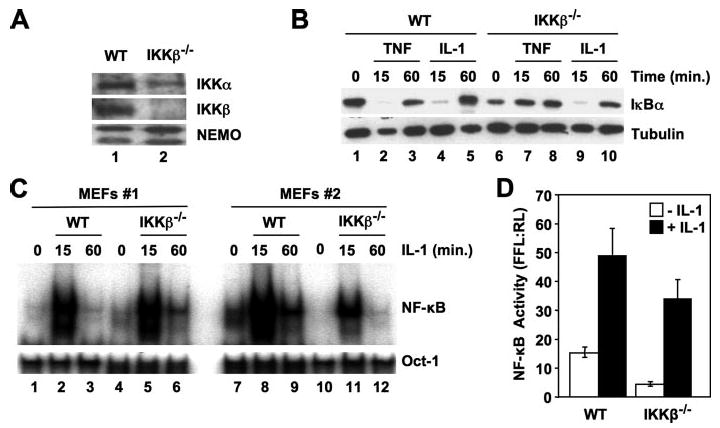
A, separately derived WT and IKKβ−/− MEFs (MEFs 2) from those used in Figs. 1 and 2 were lysed, and then samples were immunoblotted using the antibodies indicated. B, WT and IKKβ−/− MEFs (MEFs 2) were either untreated or incubated for the times indicated with TNF (10 ng/ml) or IL-1 (10 ng/ml), and then lysates were immunoblotted using either anti-IκBα or anti-tubulin. C, WT and IKKβ−/− MEFs (MEFs 1 and 2) were incubated with IL-1 (10 ng/ml) for the times indicated, and then nuclear extracts were prepared for EMSA. Assays were performed using either a consensus NF-κB probe (upper panel) or an Oct1 probe as a loading control (lower panel). D, MEFs 2 were transiently transfected with the NF-κB-dependent reporter pBIIx-firefly luciferase together with β-actin Renilla luciferase. Twenty-four hours later, the cells were either untreated or incubated for a further 5 h with IL-1 (10 ng/ml), and then NF-κB activity was determined by dual luciferase assay. The data are expressed as the mean ratio ± S.E. of the firefly:Renilla (FFL:RL) luciferase activity from three separate experiments, each performed in triplicate.
Dominant Negative IKKβ Blocks TNF- but Not IL-1-induced IκBα Degradation in MEFs
The data described above were generated using cells lacking distinct components of the IKK complex. We therefore wished to determine whether exogenous inhibition of IKKβ could block IL-1-induced classical NF-κB signaling in cells expressing an intact IKK complex. To accomplish this, we stably transduced WT MEFs with a retroviral construct encoding a catalytically inactive dominant negative version of IKKβ (K44M). Concomitant introduction of an EGFP-tagged version of the vector (LZRS−EGFP) confirmed that >96% of cells were transduced using our approach (Fig. 4A). As shown in Fig. 4B, TNF-induced IκBα degradation was completely blocked in cells expressing IKKβ(K44M) compared with cells transduced with the LZRS−EGFP vector alone (Fig. 4B, compare lanes 2 and 6). In contrast, IL-1-induced IκBα degradation remained detectable in IKKβ(K44M)-expressing cells (Fig. 4C). These data therefore demonstrate that exogenous expression of a dominant negative inhibitor of IKKβ blocks TNF- but not IL-1-induced IκBα degradation in WT MEFs.
FIGURE 4. Dominant negative IKKβ (K44M) blocks TNF- but not IL-1-induced IκBα degradation.
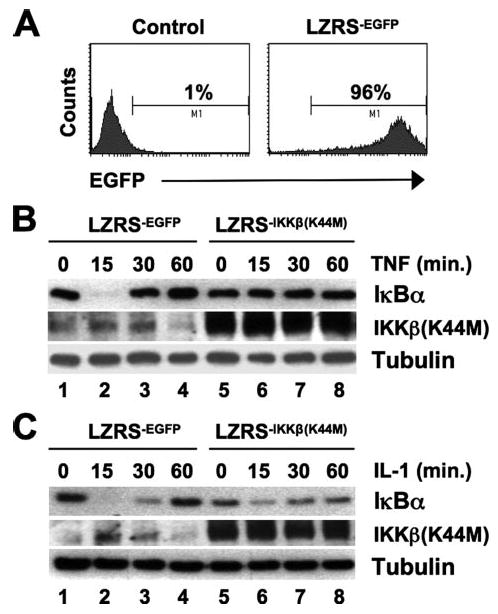
A, WT MEFs (MEFs 1) were either mock-transduced (Control, left panel) or stably transduced with LZRS−EGFP (right panel), and the percentage of EGFP-positive cells was determined by FACS analysis. B and C, stably transduced WT LZRS−EGFPor LZRS−IKKβ(K44M) MEFs were treated for the times indicated with TNF (B) or IL-1 (C), and then lysates were immunoblotted using antibodies against IκBα, IKKβ, and α-tubulin as shown.
Reconstitution of IKKα/β Double-deficient Cells with IKKα Re-establishes IL-1 but Not TNF-induced IκBα Degradation
Previous studies have clearly demonstrated that IκBα degradation does not occur in MEFs lacking both IKKα and IKKβ in response to either TNF or IL-1 (30). Our findings to date strongly suggest that IKKα is sufficient to transduce IL-1- but not TNF-induced signaling to NF-κB in the absence of IKKβ. To further explore this hypothesis, we therefore questioned whether reintroduction of IKKα into IKK DKO MEFs could reestablish IL-1-induced IκBα degradation in these cells. We first verified that the DKO cells lacked both IKKα and IKKβ but contained NEMO (Fig. 5A), and then the cells were retrovirally transduced with IKKα in the MIGR1 vector expressing GFP (Fig. 5B), and positive cells were sorted for use in experiments. As expected, incubation of DKO cells with either TNF or IL-1 did not induce IκBα degradation (Fig. 5, C and D, lanes 5–8). Similarly, IκBα degradation was absent in response to TNF in DKO cells stably transduced with IKKα (Fig. 5C, lanes 9–12). Consistent with our findings in IKKβ-deficient cells, however, re-introduction of IKKα into DKO MEFs re-established IL-1-induced IκBα degradation in these cells (Fig. 5D, lanes 9–12).
FIGURE 5. Reconstitution of IKKα/β double knock-out MEFs with IKKα restores IL-1- but not TNF-induced IκBα degradation.
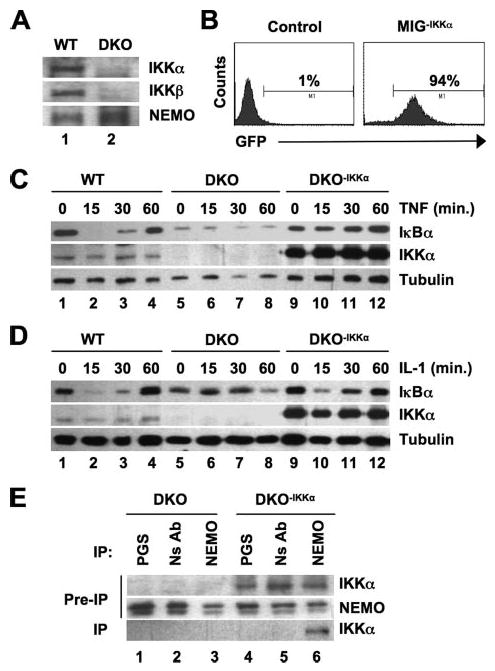
A, WT and IKKα/β DKO MEFs were lysed, and samples were immunoblotted with the antibodies indicated. B, IKKα/β DKO MEFs were either mock-transduced (Control; left) or transduced with MIGR1−IKKα (right), and then GFP-positive cells (94% of the population) were sorted and stable cultures of DKO−IKKα were generated. C and D, WT, DKO, and DKO−IKKα MEFs were either untreated or incubated TNF (C) or IL-1 (10 ng/ml each) (D) for the times indicated. Lysates were prepared and immunoblotted using antibodies against IκBα, IKKα, or α-tubulin as shown. E, lysates of DKO or DKO−IKKα MEFS were incubated with either protein G-Sepharose alone (PGS), a nonspecific antibody (NS Ab), or anti-NEMO, and immunoprecipitation (IP) was performed as described under “Experimental Procedures.” Immunoprecipitated material was immunoblotted using anti-IKKα, and samples of lysates retained prior to immunoprecipitation (Pre-IP) were probed using anti-IKKα and NEMO.
We conclude from our accumulated data that, in the absence of IKKβ, IKKα can facilitate IL-1-induced IκBα degradation and NF-κB activation. Our findings using NEMO-deficient cells (Figs. 1 and 2) also lead us to surmise that NEMO is necessary for IKKα-dependent signaling. However, a direct role for NEMO in regulating the function of IKKα in the classical NF-κB pathway has not yet been described. We previously demonstrated that NEMO associates with IKKα via the NEMO binding domain (NBD) within the COOH terminus of the kinase (5, 6), and we verified that NEMO and IKKα formed a complex in our DKO-IKKα cells (Fig. 5E, lane 6). Our results therefore suggest that NEMO and IKKα form a complex in cells lacking IKKβ that responds to IL-1 (but not TNF) to phosphorylate IκBα, leading to its degradation.
The Interaction of NEMO with IKKα Is Required for IL-1-induced NF-κB Activation in IKKβ−/− MEFs
To definitively determine whether the interaction of NEMO with IKKα is required for IL-1-induced NF-κB activation in the absence of IKKβ, we first verified that NEMO and IKKα associate in IKKβ−/− MEFs. As shown in Fig. 6A, IKKα was present in immune complexes precipitated from both WT and IKKβ−/− MEFs using anti-NEMO, confirming that these proteins do form a complex in IKKβ-deficient cells. We have previously reported that the interaction of IKKβ and IKKα with NEMO can be blocked using a cell-permeable peptide encompassing the NEMO binding domain (NBD) present in each of the kinases (5, 6). We therefore questioned whether the NBD peptide would inhibit IL-1-induced NF-κB activation in IKKβ−/− MEFs, and as shown in Fig. 6B, the WT (NBDWT, lanes 3–5) but not the inactive mutant peptide (NBDMUT, lanes 6 and 7) dose-dependently blocked IL-1-induced NF-κB activity measured by EMSA. These results therefore demonstrate that disruption of the NEMO-IKKα interaction blocks IL-1-induced NF-κB activation in cells lacking IKKβ.
FIGURE 6. IL-1-induced NF-κB activation in IKKβ−/− MEFs requires the association of IKKα with NEMO.
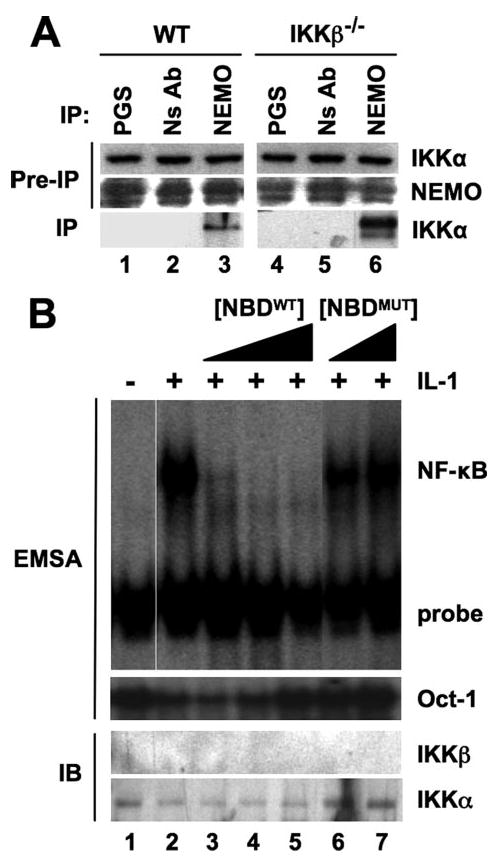
A, lysates of WT or IKKβ−/− MEFS were incubated with either protein G-Sepharose alone (PGS), a nonspecific antibody (NS Ab), or anti-NEMO, and then immunoprecipitation (IP) and immunoblotting were performed as described in the legend to Fig. 5E. B, IKKβ−/− MEFs were either untreated or incubated for 15 min with a range of concentrations of the WT (NBDWT; 50, 100, and 200 μm) or inactive mutant (NBDMUT; 100 and 200 μm) NBD peptides. Cells were incubated a further 15 min with (+) or without (−) IL-1 (10 ng/ml; +), and then cytoplasmic and nuclear extracts were prepared for immunoblotting (IB) and EMSA, respectively. EMSAs were performed as described under “Experimental Procedures” using NF-κB and Oct-1 consensus binding site probes, and cytoplasmic extracts were immunoblotted using anti-IKKα and -IKKβ as indicated.
Discussion
Genetic studies have established that TNF-induced classical NF-κB activation depends upon NEMO and IKKβ, whereas IKKα is considered to play no role in the upstream activation events of this pathway (1, 2). Consistent with this, we found that TNF-, IL-1- and LPS-induced IκBα degradation is intact in IKKα−/− fibroblasts but absent in NEMO-deficient cells. However, we found that, although TNF- and LPS-induced IκBα degradation absolutely required IKKβ, IL-1-induced classical NF-κB activation remained intact in two separate IKKβ-deficient fibroblast cell lines. We provide comprehensive evidence that IL-1-induced IκBα phosphorylation, degradation, p50:p65 nuclear translocation and NF-κB-dependent luciferase reporter expression occurs in IKKβ−/− fibroblasts, and we have demonstrated that reconstitution of IKKα/b double knock-out MEFs with IKKα is sufficient to re-establish IL-1- but not TNF-induced IκBα degradation. We have also shown that this activation requires the association of NEMO with IKKα, as the cell-permeable NBD peptide blocks IL-1-induced NF-κB in IKKβ−/− MEFs. Our results therefore demonstrate that IL-1-induced IκBα degradation can occur via either NEMO·IKKβ or NEMO·IKKα complexes, whereas TNF signaling is non-promiscuous and depends solely on NEMO and IKKβ for transduction (see model in Fig. 7). This leads us to conclude that, although classical NF-κB activation by all stimuli critically depends upon NEMO, separate upstream signaling pathways differ in their categorical requirements for the effector subunits of the IKK complex.
FIGURE 7. Distinct modes of IKK complex activity transduce TNF and IL-1 signaling to NF-κB.
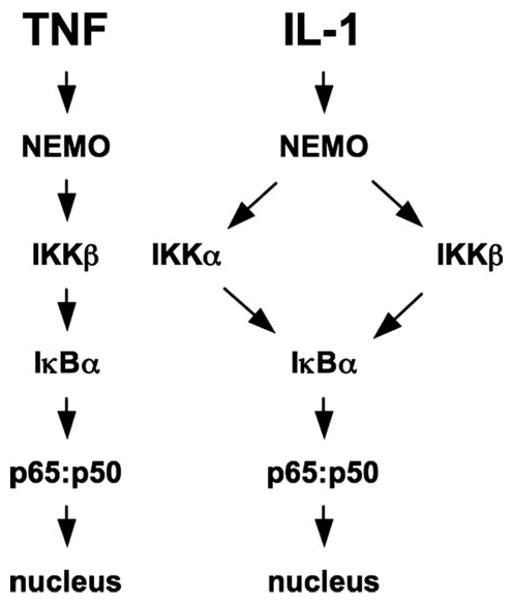
Our findings verify that TNF-induced IκBα degradation and classical NF-κB activation is critically dependent upon NEMO and IKKβ (left). Similarly, IL-1 signaling absolutely requires intact NEMO (right). However, IL-1-induced IκBα degradation, NF-κB nuclear translocation, and NF-κB transcriptional activity occurs in the absence of IKKβ, demonstrating that NEMO and IKKα can form a signaling complex capable of activating the classical NF-κB pathway in response to certain pro-inflammatory stimuli.
Although our findings appear to conflict with the current model of classical NF-κB activation, they are consistent with a number of previous studies. In the original report of mice lacking IKKβ, TNF-induced IκBα degradation and NF-κB activity were significantly reduced in IKKβ−/− fibroblasts, whereas IL-1 signaling remained comparatively unaffected (8). Using different IKKβ-deficient MEFs, Schmidt-Supprian et al. (31) later reported profoundly defective TNF signaling, although they demonstrated a less dramatic effect on IL-1-induced NF-κB activation. Tang et al. (32) have also observed that JNK inactivation, which requires NF-κB-dependent gene expression in response to TNF, was intact in IKKβ−/− MEFs stimulated with IL-1. Furthermore Schmidt-Supprian et al. (31) have demonstrated that neither TNF nor LPS could induce IL-6 expression in IKKβ−/− MEFS, whereas IL-1 did so in the absence of IKKβ. Tanaka et al. (33) also demonstrated that IL-1 induced low level IL-6 expression in IKKβ−/− MEFs; however, contrary to our study, they did not observe any difference between IL-1- and TNF-induced NF-κB in their cells. Similarly, Li et al. (9) have reported no differences between the lack of TNF and IL-1 signaling in IKKβ-deficient MEFs. We performed our studies using two separate IKKβ−/− MEF cell lines and found that IL-1 (but not TNF) induced IκB degradation and NF-κB activation in each. The reasons for the discrepancies among the separate reports are therefore unclear; however, our data coupled with that of previous studies (8, 31, 32) lead us to question the overall conclusion that IKKβ is absolutely necessary for all classical NF-κB-inducing stimuli.
It has been proposed that NF-κB activity previously observed in IKKβ-deficient MEFs occurs because IKKα compensates for IKKβ in the IKK complex (31). Our data argue against this interpretation, as we consistently observed complete inhibition of TNF signaling in the absence of IKKβ, whereas IL-1-induced IKK activity remained intact. A more accurate conclusion from these combined studies is therefore that IL-1 and TNF signaling to NF-κB have different absolute requirements for the IKK subunits necessary for phosphorylating IκBα. A consistent finding between our study and many previous reports is that IκBα degradation induced by both cytokines requires NEMO (10–12, 34). We therefore speculate that the molecular components of the separate upstream signaling cascades differentially interface with NEMO, resulting in distinct downstream IKK activation profiles. This hypothesis is supported by the fact that specific mutations in the COOH-terminal zinc finger domain of NEMO prevent TNF-induced NF-κB activation but have no effect on IL-1-stimulated IKK activation (34, 35). Furthermore, the COP9 signalosome component CSN3 has been identified as a NEMO-interacting protein that specifically blocks TNF- but not IL-1-induced NF-κB activation (36). It is therefore possible that the terminal signaling components of the TNF and IL-1 pathways associate with distinct regions of NEMO and that this association directs the activation of either IKKβ alone in response to TNF or, in the case of IL-1, either IKKβ or IKKα. Further studies exploring the relationship between the distinct domains of NEMO and the downstream kinases activated by separate signaling cascades are therefore required and may identify novel mechanisms to selectively disrupt pro-inflammatory signaling via the IKK complex.
Our results suggest that classical NF-κB-inducing stimuli belong to two separate categories, those that that depend solely upon IKKβ (e.g. TNF) and stimuli, such as IL-1, that can utilize either kinase in the IKK complex. As LPS signaling via TLR4 shares several adapter molecules with the IL-1 receptor-induced pathway (i.e. MyD88, IRAK, and TRAF6), we therefore expected LPS to mirror IL-1 and activate NF-κB in IKKβ-deficient MEFs. However, LPS did not activate NF-κB in IKKβ−/− cells and behaved identically to TNF in all of our experiments. Despite sharing several components, closer examination of IL-1- and LPS-induced signaling pathways reveals differences in the other adapter proteins required for each. Unlike IL-1 signaling, which requires only MyD88, TLR4 engagement recruits additional adapters, including TIRAP/MAL, TRAM, and TRIF (22). It is therefore possible that other TLRs, such as TLRs 5, 7, 8, 9, and 11, which signal solely through MyD88, may more closely resemble IL-1 and activate NF-κB via either IKKα or IKKβ. Although further work will be required to address this hypothesis, it is interesting to note that the same mutations in NEMO that block TNF-but not IL-1-induced NF-κB activation also inhibit LPS signaling (34, 35). This therefore strengthens the hypothesis that the differential IKK requirements of distinct stimuli involves separate domains of NEMO and, together with our findings, places LPS in the same category of stimuli as TNF that depends completely on IKKβ for downstream NF-κB activation. Intriguingly, Schmidt-Supprian et al. (31) report that T-cell receptor-induced NF-κB activation and proliferation is intact in T-cells lacking IKKβ, suggesting that similar to IL-1 in our study, TCR signaling can utilize either IKKα or IKKβ.
A major goal of this study was to explore the physiological function of the interaction of IKKα with NEMO. In pursuing this, we have demonstrated that NEMO and IKKα can form a signaling unit capable of transducing a subset of classical NF-κB signals. However, IKKα has been ascribed a number of separate roles in classical NF-κB signaling that are distinct from IκBα phosphorylation. In this regard, it has been proposed that IKKα regulates NF-κB-dependent transcription by selectively phosphorylating histones associated with NF-κB target genes (24, 29) or by directly phosphorylating NF-κB proteins on residues required for transcriptional competency (28). Our data support a transcriptional regulatory role for IKKα, as we did not observe NF-κB-dependent luciferase activity in IKKα−/− MEFs (Fig. 2D), although IκBα degradation and NF-κB DNA binding occurred in these cells (Fig. 1, B, C, and F; Fig. 2, A and B). As histone and possibly NF-κB phosphorylation are nuclear events and NEMO has been shown to localize to the nucleus (37–39), it will be intriguing to determine whether the association with NEMO plays a role in any of these transcriptional regulatory functions of IKKα.
Intact IKKα is absolutely critical for non-canonical NF-κB activation in response to a range of stimuli, including ligation of the LTβR (13, 14). Because our data indicated that IL-1 stimulation leads to IKKα activation in the absence of IKKβ, it was therefore conceivable that IL-1 could induce the non-canonical pathway in IKKβ−/− MEFs. However, as shown in Fig. 2C, we found that, similar to TNF (13, 14, 17, 23), IL-1 did not activate the non-canonical pathway in either WT or IKKβ-deficient cells. This finding therefore broadens our understanding of the range of activities of IKKα in NF-κB signaling and separates the roles of IKKα activated in response to both classical and non-canonical stimuli.
IKKα has also been reported to negatively regulate the classical NF-κB pathway, and two separate studies have demonstrated that IκBα degradation is prolonged in IKKα-deficient cells following stimulation (26, 27). We also observed protracted IκBα degradation in IKKα−/− cells treated with TNF, IL-1, or LPS (Fig. 1, A, B, and F). The mechanisms proposed for this include hyperactivity of IKKβ, because of the absence of direct negative regulation by IKKα (27) or by lack of IKKα-dependent NF-κB protein turnover and removal from pro-inflammatory gene promoters (26). Although it remains to be determined whether either or both of these mechanisms fully account for the negatively regulatory role of IKKα, it is exciting to speculate on the potential requirement of the IKKα-NEMO interaction for this function. Notably, we had demonstrated previously that, although the NBD peptide blocks pro-inflammatory NF-κB activation by disrupting the association of IKKα and IKKβ with NEMO, it also modestly increases basal NF-κB activity in unstimulated cells (5). Consistent with this, versions of zebrafish IKKα that lack the NBD do not negatively regulate NF-κB activity when stably expressed in IKKα−/− MEFs (25). These findings therefore suggest that the suppressive function of IKKα in the classical NF-κB pathway requires its ability to interact with NEMO; however, further direct studies of the IKKα NBD in mammalian systems are clearly required.
In conclusion, we have demonstrated that, although IL-1 and TNF both require NEMO for classical NF-κB activation in MEFs, only TNF absolutely requires IKKβ. Our finding that IL-1-induced IκBα phosphorylation and NF-κB activation are intact in IKKβ−/− MEFs leads us to propose a model in which NEMO “interprets” and differentially relays distinct upstream signals to the downstream IKK subunits (Fig. 7). These results also suggest that drugs targeting IKKβ alone may not completely block pro-inflammatory NF-κB activity in diseases in which IL-1 plays a major pathological role. However, a deeper understanding of the precise mechanisms by which the IKK complex responds to distinct upstream signals and activates the separate IKK subunits may identify more useful pathway-specific targets for effective therapeutic drugs.
Acknowledgments
We are grateful to Janet Crawford for peptide synthesis and to Kelly McCorkell and Catherine Wharry for helpful discussion.
Footnotes
This work was supported by grants from the National Institutes of Health (1RO1HL080612-01A1 and NO1AI-22070).
The abbreviations used are: NF-κB, nuclear factor-κB; IKK, IκB kinase; IκB, inhibitor of NF-κB; LTβR, lymphotoxin-β receptor; MEF, murine embryonic fibroblast; NEMO, NF-κB essential modulator; NBD, NEMO binding domain; IL, interleukin; LPS, lipopolysaccharide; WT, wild type; EMSA, electrophoretic mobility shift assay; FACS, fluorescence-activated cell sorter; GFP, green fluorescent protein; EGFP, enhanced GFP; TLR, Toll-like receptor; TNF, tumor necrosis factor; DKO, double knock-out.
References
- 1.Bonizzi G, Karin M. Trends Immunol. 2004;25:280–288. doi: 10.1016/j.it.2004.03.008. [DOI] [PubMed] [Google Scholar]
- 2.Hayden MS, Ghosh S. Genes Dev. 2004;18:2195–2224. doi: 10.1101/gad.1228704. [DOI] [PubMed] [Google Scholar]
- 3.Karin M, Ben-Neriah Y. Annu Rev Immunol. 2000;18:621–663. doi: 10.1146/annurev.immunol.18.1.621. [DOI] [PubMed] [Google Scholar]
- 4.May MJ, Larsen SE, Shim JH, Madge LA, Ghosh S. J Biol Chem. 2004;279:45528–45539. doi: 10.1074/jbc.M408579200. [DOI] [PubMed] [Google Scholar]
- 5.May MJ, D'Acquisto F, Madge LA, Glockner J, Pober JS, Ghosh S. Science. 2000;289:1550–1554. doi: 10.1126/science.289.5484.1550. [DOI] [PubMed] [Google Scholar]
- 6.May MJ, Marienfeld RB, Ghosh S. J Biol Chem. 2002;277:45992–46000. doi: 10.1074/jbc.M206494200. [DOI] [PubMed] [Google Scholar]
- 7.Jimi E, Aoki K, Saito H, D'Acquisto F, May MJ, Nakamura I, Sudo T, Kojima T, Okamoto F, Fukushima H, Okabe K, Ohya K, Ghosh S. Nat Med. 2004;10:617–624. doi: 10.1038/nm1054. [DOI] [PubMed] [Google Scholar]
- 8.Li Q, Van Antwerp D, Mercurio F, Lee KF, Verma IM. Science. 1999;284:321–325. doi: 10.1126/science.284.5412.321. [DOI] [PubMed] [Google Scholar]
- 9.Li ZW, Chu W, Hu Y, Delhase M, Deerinck T, Ellisman M, Johnson R, Karin M. J Exp Med. 1999;189:1839–1845. doi: 10.1084/jem.189.11.1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Makris C, Godfrey VL, Krahn-Senftleben G, Takahashi T, Roberts JL, Schwarz T, Feng L, Johnson RS, Karin M. Mol Cell. 2000;5:969–979. doi: 10.1016/s1097-2765(00)80262-2. [DOI] [PubMed] [Google Scholar]
- 11.Rudolph D, Yeh WC, Wakeham A, Rudolph B, Nallainathan D, Potter J, Elia AJ, Mak TW. Genes Dev. 2000;14:854–862. [PMC free article] [PubMed] [Google Scholar]
- 12.Schmidt-Supprian M, Bloch W, Courtois G, Addicks K, Israel A, Rajewsky K, Pasparakis M. Mol Cell. 2000;5:981–992. doi: 10.1016/s1097-2765(00)80263-4. [DOI] [PubMed] [Google Scholar]
- 13.Dejardin E, Droin NM, Delhase M, Haas E, Cao Y, Makris C, Li ZW, Karin M, Ware CF, Green DR. Immunity. 2002;17:525–535. doi: 10.1016/s1074-7613(02)00423-5. [DOI] [PubMed] [Google Scholar]
- 14.Senftleben U, Cao Y, Xiao G, Greten FR, Krahn G, Bonizzi G, Chen Y, Hu Y, Fong A, Sun SC, Karin M. Science. 2001;293:1495–1499. doi: 10.1126/science.1062677. [DOI] [PubMed] [Google Scholar]
- 15.Claudio E, Brown K, Park S, Wang H, Siebenlist U. Nat Immunol. 2002;3:958–965. doi: 10.1038/ni842. [DOI] [PubMed] [Google Scholar]
- 16.Coope HJ, Atkinson PG, Huhse B, Belich M, Janzen J, Holman MJ, Klaus GG, Johnston LH, Ley SC. EMBO J. 2002;21:5375–5385. doi: 10.1093/emboj/cdf542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Muller JR, Siebenlist U. J Biol Chem. 2003;278:12006–12012. doi: 10.1074/jbc.M210768200. [DOI] [PubMed] [Google Scholar]
- 18.Hu Y, Baud V, Delhase M, Zhang P, Deerinck T, Ellisman M, Johnson R, Karin M. Science. 1999;284:316–320. doi: 10.1126/science.284.5412.316. [DOI] [PubMed] [Google Scholar]
- 19.Hu Y, Baud V, Oga T, Kim KI, Yoshida K, Karin M. Nature. 2001;410:710–714. doi: 10.1038/35070605. [DOI] [PubMed] [Google Scholar]
- 20.Li Q, Lu Q, Hwang JY, Buscher D, Lee KF, Izpisua-Belmonte JC, Verma IM. Genes Dev. 1999;13:1322–1328. doi: 10.1101/gad.13.10.1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Orange JS, Harris KE, Andzelm MM, Valter MM, Geha RS, Strominger JL. Proc Natl Acad Sci U S A. 2003;100:14151–14156. doi: 10.1073/pnas.1835830100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kawai T, Akira S. Cell Death Differ. 2006;13:816–825. doi: 10.1038/sj.cdd.4401850. [DOI] [PubMed] [Google Scholar]
- 23.Derudder E, Dejardin E, Pritchard LL, Green DR, Korner M, Baud V. J Biol Chem. 2003;278:23278–23284. doi: 10.1074/jbc.M300106200. [DOI] [PubMed] [Google Scholar]
- 24.Anest V, Hanson JL, Cogswell PC, Steinbrecher KA, Strahl BD, Baldwin AS. Nature. 2003;423:659–663. doi: 10.1038/nature01648. [DOI] [PubMed] [Google Scholar]
- 25.Correa RG, Matsui T, Tergaonkar V, Rodriguez-Esteban C, Izpisua-Belmonte JC, Verma IM. Curr Biol. 2005;15:1291–1295. doi: 10.1016/j.cub.2005.06.023. [DOI] [PubMed] [Google Scholar]
- 26.Lawrence T, Bebien M, Liu GY, Nizet V, Karin M. Nature. 2005;434:1138–1143. doi: 10.1038/nature03491. [DOI] [PubMed] [Google Scholar]
- 27.Li Q, Lu Q, Bottero V, Estepa G, Morrison L, Mercurio F, Verma IM. Proc Natl Acad Sci U S A. 2005;102:12425–12430. doi: 10.1073/pnas.0505997102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sizemore N, Lerner N, Dombrowski N, Sakurai H, Stark GR. J Biol Chem. 2002;277:3863–3869. doi: 10.1074/jbc.M110572200. [DOI] [PubMed] [Google Scholar]
- 29.Yamamoto Y, Verma UN, Prajapati S, Kwak YT, Gaynor RB. Nature. 2003;423:655–659. doi: 10.1038/nature01576. [DOI] [PubMed] [Google Scholar]
- 30.Li Q, Estepa G, Memet S, Israel A, Verma IM. Genes Dev. 2000;14:1729–1733. [PMC free article] [PubMed] [Google Scholar]
- 31.Schmidt-Supprian M, Courtois G, Tian J, Coyle AJ, Israel A, Rajewsky K, Pasparakis M. Immunity. 2003;19:377–389. doi: 10.1016/s1074-7613(03)00237-1. [DOI] [PubMed] [Google Scholar]
- 32.Tang G, Minemoto Y, Dibling B, Purcell NH, Li Z, Karin M, Lin A. Nature. 2001;414:313–317. doi: 10.1038/35104568. [DOI] [PubMed] [Google Scholar]
- 33.Tanaka M, Fuentes ME, Yamaguchi K, Durnin MH, Dalrymple SA, Hardy KL, Goeddel DV. Immunity. 1999;10:421–429. doi: 10.1016/s1074-7613(00)80042-4. [DOI] [PubMed] [Google Scholar]
- 34.Makris C, Roberts JL, Karin M. Mol Cell Biol. 2002;22:6573–6581. doi: 10.1128/MCB.22.18.6573-6581.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huang TT, Feinberg SL, Suryanarayanan S, Miyamoto S. Mol Cell Biol. 2002;22:5813–5825. doi: 10.1128/MCB.22.16.5813-5825.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hong X, Xu L, Li X, Zhai Z, Shu H. FEBS Lett. 2001;499:133–136. doi: 10.1016/s0014-5793(01)02535-2. [DOI] [PubMed] [Google Scholar]
- 37.Birbach A, Gold P, Binder BR, Hofer E, de Martin R, Schmid JA. J Biol Chem. 2002;277:10842–10851. doi: 10.1074/jbc.M112475200. [DOI] [PubMed] [Google Scholar]
- 38.Huang TT, Wuerzberger-Davis SM, Wu ZH, Miyamoto S. Cell. 2003;115:565–576. doi: 10.1016/s0092-8674(03)00895-x. [DOI] [PubMed] [Google Scholar]
- 39.Verma UN, Yamamoto Y, Prajapati S, Gaynor RB. J Biol Chem. 2004;279:3509–3515. doi: 10.1074/jbc.M309300200. [DOI] [PubMed] [Google Scholar]


