Abstract
SNAI1P, a protein coded by a retrogene, is a member of the SNAI family of E2-box binding transcriptional repressors. To evaluate whether the mode of action of SNAI1P is similar to those of the other predominant members of the SNAI family, we studied its action on human claudin 7 (CLDN7) gene promoter which has seven E2-boxes. We over-expressed FLAG-tagged SNAI1P in MCF7 and MDA-MB-468 cells. SNAI1P inhibited the expression of CLDN7 in these recombinant cells. SNAI1P also inhibited cloned CLDN7 gene promoter activity in human breast cancer cells. ChIP assays revealed that SNAI1P is recruited on the CLDN7 gene promoter along with the co-repressor CtBP1 and the effector HDAC1. Treatment of the cells with trichostatin A, an inhibitor of HDAC1, abrogated the repressor activity of SNAI1P. These data suggest that SNAI1P inhibits CLDN7 gene promoter epigenetically in breast cancer cells through chromatin remodeling.
Keywords: SNAI1P, SNAIL, Claudin 7, E2-box, Transcriptional repression, CtBP1, HDAC1
Introduction
SNAIL is a member of the SNAI family of E2-box binding, C2H2-type zinc finger containing transcriptional repressor proteins in higher eukaryotes [1–6]. In addition to SNAIL, a related (~82% overall similarity) sequence was found in the human genome and classified as a retro-transcribed gene (SNAI1P or SNAI1L1) [7, 8]. Retrogenes result from the reverse transcription of an mRNA and subsequent insertion into the genome mediated by different transposable sequences such as retrovirus, LINE, or Alu elements [8]. In general, retrotransposed genes are not expressed due to the absence of promoter elements in the region of insertion. But SNAI1P gene is integrated into the intron of a functional gene and is expressed in the human breast cells constitutively at low levels [8]. SNAIL promotes tumor growth and metastasis but is highly regulated in human cells [9–18]. On the other hand, SNAI1P is not under the rigorous regulatory regimen [7, 8]. Thus, genetic or epigenetic activation of SNAI1P in human breast cells will be detrimental to the biology of the cells provided this protein has biological function similar to SNAIL. SNAI1P is similar to SNAIL but not identical in the amino acid sequences [8]. The significant differences in the primary structure of SNAI1P, particularly at the putative repressor domain, may lead to alteration in the function of SNAI1P as compared to SNAIL. In order to understand the mode of action of SNAI1P in human breast cells, we over expressed FLAG-tagged SNAI1P in the SLUG/SNAIL-deficient MCF7 and MDA-MB-468 cells.
Our ChIP-on-chip (ChIP-DSL) analysis of 20,000 human gene promoters using anti-FLAG monoclonal antibody revealed that more than 65 genes bind to SNAI1P very tightly at their promoters (M. K. Mittal and G. Chaudhuri, unpublished data). The gene for the tight junction protein claudin 7 (CLDN7) [19, 20] is one of the candidate SNAI1P-regulated genes. Here, we report that SNAI1P indeed binds in vivo to the CLDN7 gene promoter in human breast cancer cell nucleus and inhibits its expression by chromatin remodeling. But, despite this ability, SNAI1P was not able to alter the invasiveness of non-invasive breast cancer cells.
Materials and methods
Cell culture and reagents
Human breast cancer cells MCF7 and MDA-MB-468 were obtained from ATCC (Manassas, VA) and were cultured in ATCC-recommended media [21–23]. FLAG M2 antibody was purchased from Sigma Chemical Co. (St. Louis, MO). CLDN7 antibody was obtained from Invitrogen (Carlsbad, CA). CtBP1 and HDAC1 antibodies were purchased from Upstate Millipore (Burlington, MA).
Generation of stable clones
Human SNAI1P gene ORF (NG_000884) was PCR amplified from the genomic DNA isolated from MDA-MB-231 cells with Hind III and Xba I site-containing primers (5′-AAGCTTATGCCG CGCTCTTTCCTCGTCAGGAAG-3′ and 5′-TCTAGAGCGGGGACATCCTGAGCAGCCGGAC-3′). The reverse primer did not have the endogenous stop codon. The PCR product was cloned at the Hind III/Xba I sites of p3XFLAG-CMV-14 vector (Sigma). Stable clones constitutively expressing SNAI1P were selected with G418 (400 µg/ml).
Immunofluorescence analysis [23]
Cells were cultured in 8-well chamber slides for 24 h in complete media, washed with PBS, fixed and permeabilized with ice-cold methanol for 10 min. After blocking with 5% goat serum in PBS, the cells were incubated with the primary antibody followed by secondary antibody conjugated with the red fluorescent dye (Alexa Fluor R555-conjugated donkey anti mouse IgG, Invitrogen). The cells were subsequently stained with DAPI (Sigma). Finally, each slide was examined by fluorescence microscopy in a Nikon TE2000-E inverted wide-field microscope. Each representative image was examined and digitally recorded at the same cellular level and magnification [23].
Luciferase reporter assay
We PCR amplified 641 bp human CLDN7 gene promoter (−345 to +296) with the forward and the reverse primers 5′-CACCTGTTGGGAAGAAAGGA-3′, 5′-GCCAGGTGAATGCAAATCTT-3′, respectively, from total DNA isolated from BT549 cells. The amplified DNA was cloned into the pCR-II-TOPO vector (Invitrogen) and subsequently subcloned into the Eco RI site of pRL-Null vector (Promega). Cells were seeded on 24-well tissue culture plates and after 24 h, they were transfected with pGL3-Control and pRL-Cldn7 promoter constructs using Lipofectamine 2000 transfection reagent (Invitrogen). Luciferase activity was assayed 48 h post-transfection using a dual luciferase assay reagents (Promega), as described before [21].
Chromatin immunoprecipitation assay
Chromatin immunoprecipitation assays were performed as described previously [21, 23]. Immunoprecipitations were performed using FLAG (for SNAI1P), CtBP1 or HDAC1 antibodies. CLDN7 gene promoter DNA was amplified from the ChIP DNA using the primers described above. Trichostatin A (TSA) treatment of the cells was done as described previously [21, 23].
Immunoblot analysis
Cells from stable clones were grown in complete medium. Protein extracts were made and Western blotting was performed as described [21, 23]. Cell lysates containing equal amounts of protein were resolved by 4–12% SDS–PAGE, transferred to nitrocellulose membranes, probed with the appropriate antibodies, and detected by means of enhanced chemiluminescence [21, 22].
Wound healing assay
Cells were grown to confluence on gridded plastic dishes, and monolayers were then wounded by scratching with a 10–ll pipette tip, as described [24]. Wounds were photographed under the microscope (×20 objective) and their coordinates recorded. The same wounds were photographed again 16 h later [24].
Matrigel transwell invasion assay
Invasiveness of MDA-MB-468 and MCF-7 cells was measured with a Matrigel Invasion Chamber (Becton Dickinson Labware, Bedford, MA) following standard protocols [25]. The Matrigel-coated insert membranes were stained with crystal violet for 30 min. The membranes were washed three times with PBS, and the cells on the membranes were viewed by microscopy.
Results and discussion
Over expression of SNAI1P in the non-invasive MCF7 and MDA-MB-468 cells inhibited the expression of CLDN7 protein in these recombinant cells
Although SNAI1P is derived from SNAIL cDNA, SNAI1P protein is 6 amino acids shorter than SNAIL (Fig. 1a) [8]. SNAIL has 264 amino acids whereas SNAI1P has 254 amino acids [8]. SNAIL has two functional domains: (1) the N-terminal repressor domain containing the highly conserved SNAG subdomain (amino acid residues 1–20; Fig. 1a) [8]; and (2) the four zinc-finger containing C-terminal DNA binding domain [8]. The DNA binding domain is essential for the interaction of this repressor protein with the E2-box sequences (CAGGTG/CACCTG) at the promoters of its target genes [1–6]. These zinc finger domains also contain the nuclear localization signal [26]. SNAI1P protein is very similar to SNAIL at the SNAG domain and the DNA binding domain [8]. But due to few insertions and deletions in the SNAI1P ORF, the amino acid sequences of SNAIP in between the SNAG domain and the DNAbinding zinc fingers is about 25% different than that of SNAIL (Fig. 1a). We think this difference may be critical for determining target gene specificities of SNAIL and SNAI1P. For example, unlike SNAIL, which efficiently represses human claudin 1 gene expression in breast cells [17], over expression of SNAI1P in breast cells failed to do so (M. K. Mittal and G. Chaudhuri, unpublished data).
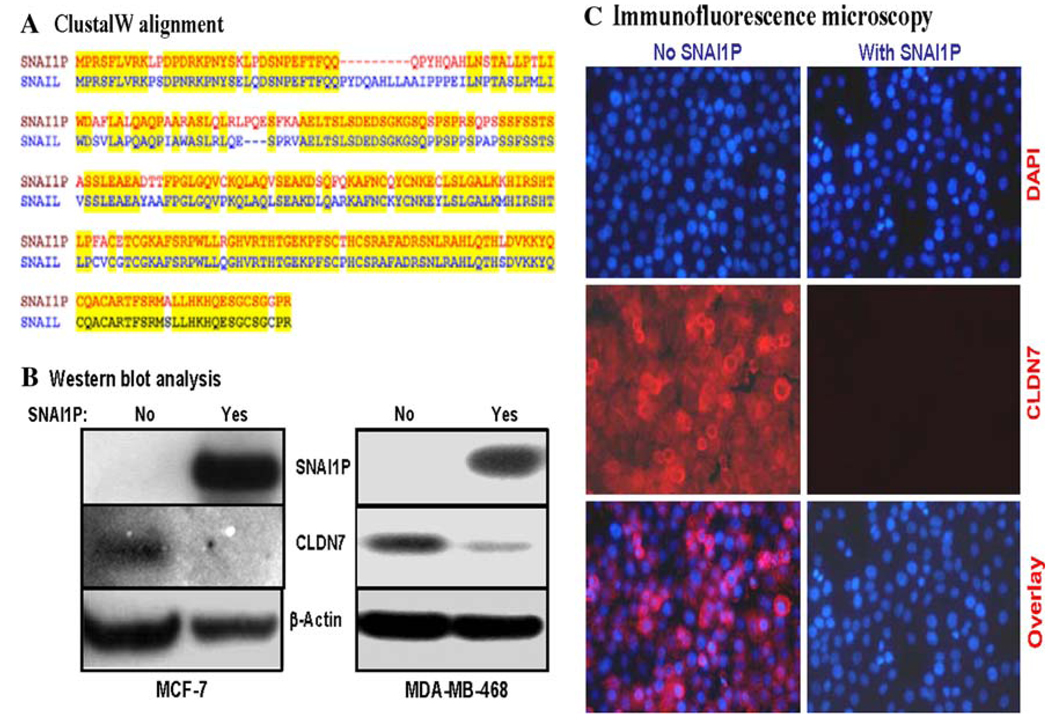
Fig. 1.
Effect of over-expression of SNAI1P in MCF7 and MDA-MB-468 cells on the expressions of CLDN7 in these cells. a CLUSTAL W alignment of the amino acid sequences of human SNAIL and SNAI1P. b Western blotting analysis data showing the expressions of 3xFLAG-tagged SNAI1P and CLDN7 in the recombinant MCF7 and MDA-MB-468 cells. The recombinant SNAI1P protein was detected using FLAG antibody. β-actin was used as a loading control in this analysis. c Immunofluorescence analysis showing repression of CLDN7 levels in the recombinant MDA-MB-468 cells. Upper panel, the nuclei of the cells were stained with DAPI (blue); middle panel, CLDN7 protein was tagged with red Alexafluor dye; and, lower panel, the superimposed photograph
To study further the binding of SNAI1P to its target gene promoters, we expressed recombinant SNAI1P in SNAIL/SLUG-deficient human breast cancer cells, e.g., MCF-7 and MDA-MB-468 [21, 23]. The 3xFLAG-tagged SNAI1P protein was abundantly expressed both in the recombinant MDA-MB-468 and MCF-7 cells (Fig. 1b). To understand the mode of action of SNAI1P in human breast cancer cells, we are studying SNAI1P-binding gene promoters in these cells using these recombinant MDA-MB-468 and MCF-7 cells employing the ChIP-DSL technique [27] with the reagents and human 20,000 gene promoter chip from Aviva Systems Biology (San Diego, CA). Through these experiments we discovered that the promoter of the tight junction protein claudin 7 (CLDN7) can bind strongly with the SNAI1P protein (data not shown). Since CLDN7 is relevant in the etiology of human breast and other cancers [28–30], we characterized further the interactions of SNAI1P and the CLDN7 gene promoter in the human breast cells. When we over expressed SNAI1P in MDAMB- 468 and MCF-7 cells, the levels of the CLDN7 protein decreased significantly (Fig. 1b). Our immunofluorescence microscopy data further verified the down regulation of CLDN7 gene expression in the presence of SNAI1P in the recombinant MDA-MB-468 cells (Fig. 1c). These data strongly suggest that SNAI1P inhibits the expression of CLDN7 gene in human breast cells.
SNAI1P works through the E2-box containing minimal promoter sequence of human CLDN7 gene to repress it
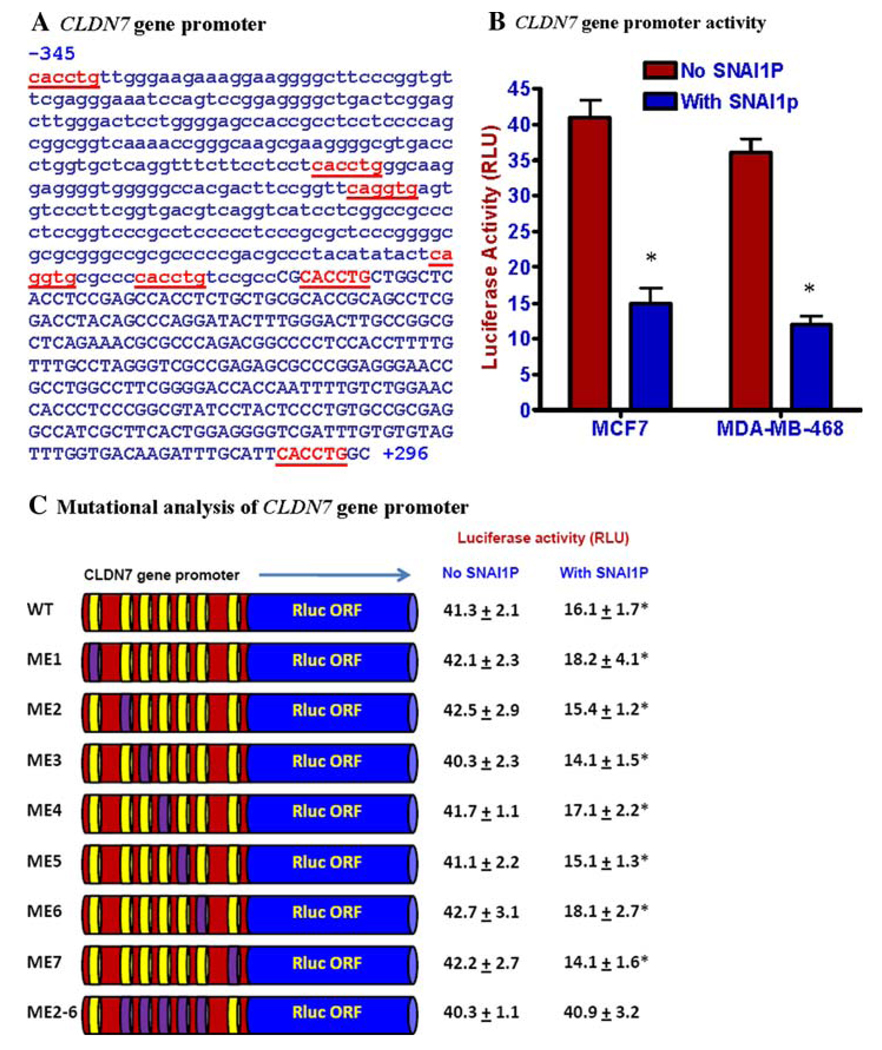
We then evaluated the effect of SNAI1P over expression on the activity of the cloned human CLDN7 gene promoter. We amplified a 641 bp (−345 to +296) promoter sequence (Fig. 2a) from human CLDN7 gene and cloned that in front of Renilla luciferase gene in pRL-Null vector. This promoter sequence has seven E2-boxes, five at the upstream and two at the downstream of the transcription start site (Fig. 2a). SNAI1P expression in recombinant MDA-MB-468 and MCF-7 cells showed down regulation of CLDN7 gene promoter activities (Fig. 2b). The E2-boxes 2–6 are highly conserved between the mouse and the human CLDN7 gene promoters. While mutation of individual E2-boxes did not affect significantly the repressive action of SNAI1P on the CLDN7 gene promoter, constructs with mutations in all of the E2-boxes 2–6 (ME2-6) abrogated the activity of SNAI1P on this promoter (Fig. 2c), suggesting that SNAI1P perhaps is capable of repressing this promoter through either of the E2-boxes 2–6.
Fig. 2.
Repression of CLDN7 gene promoter activity in SNAI1P-expressing human breast cells. a Nucleotide sequence of human CLDN7 gene promoter showing (underscored) the SLUG-binding E2-box (CAGGTG/CACCTG) elements. The upstream sequences are shown in lower case letters. The 5′-end of the transcribed sequences is in uppercase letters. b Dual luciferase assay data showing the repression of the function of CLDN7 gene promoter in the recombinant MCF7 and MDA-MB-468 cells. Results are mean ± SE (n = 6). The differences in the luciferase activities between the control and the SNAI1P-expressing cells were statistically significant (P < 0.001; shown by an ‘*’). c Dual luciferase assay data showing the function of CLDN7 gene promoter mutants in the recombinant MCF7 and MDA-MB-468 cells. The wild-type (WT) E2-boxes are shown as yellow discs. The first and the last nucleotide of the E2-box sequence were mutated to ‘A’ by overlap extension PCR [21, 23]. The mutant E-boxes are shown as purple discs. Results are mean ± SE (n = 6). The differences in the luciferase activities between the control and the SNAI1P-expressing cells with the WT or the single E2-box mutants (ME1–ME7) of the CLDN7 gene promoter constructs were statistically significant (P < 0.001; shown by an ‘*’)
In vivo binding of SNAI1P, CtBP1 and HDAC1 proteins to the CLDN7 gene promoter in human breast cells
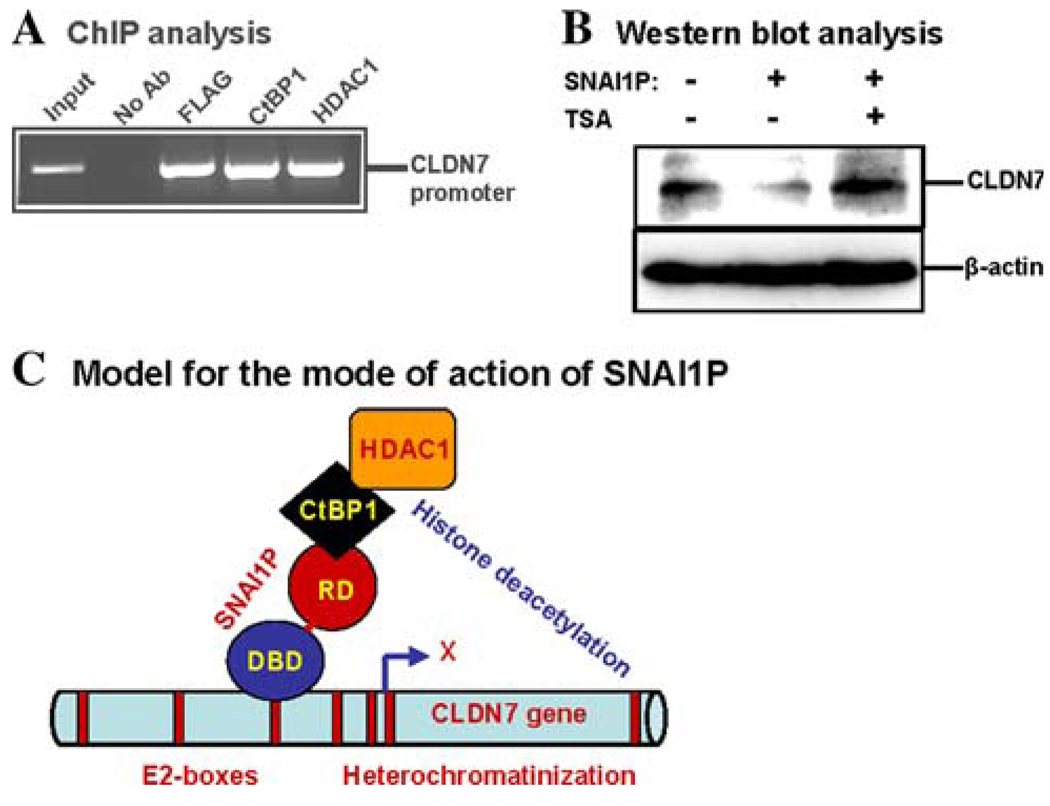
Due to the high homology of SNAI1P with SNAIL, it is assumed that SNAI1P also binds to its target gene promoters in vivo and thus mediates its repressor function [8]. To evaluate whether SNAI1P indeed binds to the CLDN7 gene promoter in the human breast cell nuclei, we performed chromatin immunoprecipitation assay using anti-FLAG monoclonal antibody with the 3xFLAG-tagged-SNAI1P over-expressing MDA-MB-468 cells. Our data further verified that SNAI1P tightly binds to the human CLDN7 gene promoter (Fig. 3a).
Fig. 3.
Recruitment of SNAI1P on the CLDN7 gene promoter along with the co-repressor CtBP1 and the effector HDAC1 in vivo. a ChIP analysis data showing the in vivo binding of SNAI1P, CtBP1, and HDAC1 at the CLDN7 gene promoter in the SNAI1P-over expressing MDA-MB-468 cells. Anti-FLAG antibody was used to pull down the SNAI1P complex. b Western blot data showing that the inhibition of HDAC1 activity by trichostatin A (100 nM, 24 h) abrogated the CLDN7 gene repressor activity of SNAI1P. c Model showing the possible mode of action of SNAI1P to repress CLDN7 gene expression in human breast cells. For simplicity, SNAI1P binding to only one of the E2-boxes is shown
The co-repressors involved in the histone modification modality of the SNAI family of transcriptional repressors are not clearly known. Recently, the AJUBA family of LIM proteins is shown to function as co-repressors for SNAIL [10, 31]. These proteins serve as platforms for the assembly of chromatin-modifying factor such as arginine methyl-transferase 5 (PRMT5) [10]. We previously found that CtBP1 is the co-repressor for repression of human BRCA2 gene by another member of the SNAI family, the SLUG protein [21]. We also detected HDAC1 as the effector for the heterochromatinization of human BRCA2 gene promoter [21]. We tested whether, like SLUG, SNAI1P can also induce the binding of the co-repressor CtBP1 and the effector HDAC1 at the CLDN7 gene promoter in vivo. We found by ChIP analysis that human CLDN7 gene promoter indeed binds to CtBP1 and HDAC1 when SNAI1P was expressed (Fig. 3a). These bindings were dependent on the presence of over expressed SNAI1P protein, as non-recombinant MDA-MB-468 cells, which express very low levels of SNAI1P, did not show any recruitment of CtBP1 at the CLDN7 gene promoter (data not shown). These data suggest that 3xFLAG-tagged SNAI1P binding to the CLDN7 gene promoter is a prerequisite for CtBP1 and HDAC1 bindings to this promoter. The involvement of HDAC1 in the SNAI1P-mediated repression of human CLDN7 gene promoter was further verified by the use of the HDAC1 inhibitor trichostatin A (TSA). In the absence of TSA and SNAI1P over expression, CLDN7 protein is expressed significantly in the MDA-MB-468 cells (Fig. 3b). On the other hand, in the presence of TSA, SNAI1P-mediated repression of the CLDN7 protein level was abrogated (Fig. 3b). These data further suggest that HDAC1 is involved in the SNAI1P-mediated repression of the human CLDN7 gene promoter. We did not test yet whether AJUBA or PRMT1 is SNAI1P-dependently recruited at the CLDN7 gene promoter.
Co-recruitment of CtBP1 to the SNAI1P-binding promoter is interesting because SNAI1P does not have the classical CtBP1 binding PLSDSSK domain. We propose that CtBP1 is recruited by SNAI1P at the CLDN7 gene promoter indirectly through another yet to be identified protein. Based on our observations, we propose a model for SNAI1P-mediated down regulation of human CLDN7 gene expression by chromatin remodeling (Fig. 3c). According to this model, SNAI1P is recruited to any or all of the E2-box sequences at the CLDN7 gene promoter through its DNA binding domain. Assisted perhaps by other transcription regulator(s), CtBP1 is recruited by SNAI1P. CtBP1 then recruits HDAC1, and perhaps other effectors (e.g., HMT1), to catalyze histone modification and silencing of the CLDN7 gene promoter (Fig. 3c).
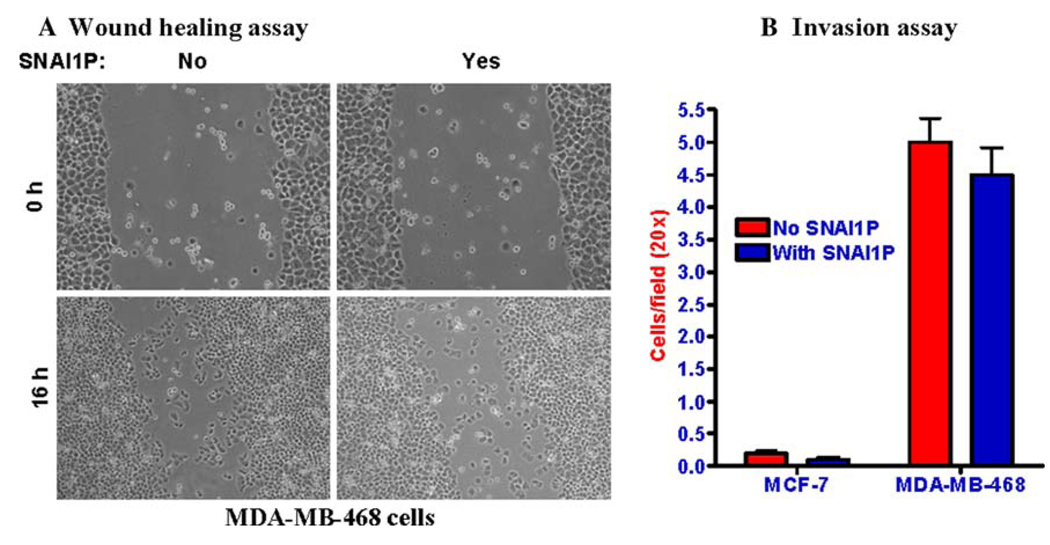
Over expression of SNAI1P in MDA-MB-468 and MCF-7 cells did not significantly alter their invasiveness
Induced expression of SNAIL in different types of cells has been shown to be directly involved in the metastatic transformation of those cells [1–6, 9–18]. It is thought that SNAIL mediates such transformation through concerted inhibitions of the expressions of a yet to be identified cluster of genes. SNAI1P over expression in the dog kidney epithelial cell line MDCK altered their morphology and increased their invasiveness in in vitro assays [8]. Whether SNAI1P will act on human breast epithelial cells is not known. We tested whether over expression of SNAI1P in the non-invasive MCF-7 or marginally invasive MDAMB-468 cells will also induce their invasiveness. We performed wound healing [24] as well as Matrigel invasion [25] assays with the native and recombinant SNAI1P-over expressing cells. Our data suggest that unlike SNAIL, SNAI1P over expression does not induce invasiveness in these cells (Fig. 4a, b). It thus appears that SNAI1P, in spite of its ability to repress some E2-box containing gene promoters, perhaps fails to repress certain key genes which are critical for the prevention of metastatic transformation of human breast cells. Decreased level of CLDN7 is associated with invasive human breast and other cancers [28–30]. Since CLDN7 is a tight junction protein, its down regulation should make the cells loosely bound to each other giving them mesenchymal phenotype and thus favoring increased migratory properties. Our data suggests that down regulations of a cluster of genes including the CLDN7 gene in human breast cancer cells by SNAI1P are not enough in determining their invasiveness, at least in vitro.
Fig. 4.
Over expression of SNAI1P in MDA-MB-468 and MCF-7 cells did not significantly alter their invasiveness. a Wound healing assay showing that with or without SNAI1P expression, MDA-MB-468 cells failed to repopulate the ‘wound’ created on the culture dish. b Matrigel invasion assay shows that over expression of SNAI1P in the MCF-7 or MDA-MB-468 cells did not alter the invasiveness of these cells. MDA-MB-231 cells were assayed in parallel as a control. It showed 8–10 fold more invasiveness that the MDA-MB-468 cells. Results are mean ± SE (n = 6). The differences in the invasiveness between the control and the SNAI1P-over expressing cells were not statistically significant
We conclude that SNAI1P inhibits CLDN7 gene promoter epigenetically through chromatin remodeling. CtBP1 and HDAC1 are involved as the co-repressor and the effector in the mediation of this SNAI1P-mediated chromatin remodeling at the CLDN7 gene promoter. Even though SNAI1P was shown to modulate invasiveness of dog kidney epithelial cell MDCK in vitro [8], inhibition of the expression of CLDN7 and perhaps several other genes in MCF-7 and MDA-MB-468 cells by SNAI1P were apparently not enough to alter the invasiveness of these cells in in vitro assays. Decreased level of CLDN7 is associated with invasive human breast and other cancers [28–30]. Why the down regulations of CLDN7 gene along with other genes by SNAI1P in human breast cancer cells do not alter their invasive properties need further in vitro and in vivo analysis.
Acknowledgments
This work was supported by the DOD-CDMRP IDEA Grant # W81XWH-06-1-0466 to GC. Immunofluorescence analysis was performed in the MMC Morphology Core facility (supported by NIH grants U54NS041071-06, G12RR03032-19, U54CA91408, and U54RR019192-04).
Abbreviations
- SNAI1L1
SNAIL-like 1
- CLDN7
Claudin 7
- TSA
Trichostatin A
- CtBP1
C-terminal binding protein 1
- HDAC1
Histone deacetylase 1
- PRMT1
Protein arginine methyl transferase 1
- HMT1
Histone methyl transferase 1
References
- 1.Nieto MA. The Snail superfamily of zinc-finger transcription factors. Nat Rev Mol Cell Biol. 2002;3:155–166. doi: 10.1038/nrm757. doi: 10.1038/nrm757. [DOI] [PubMed] [Google Scholar]
- 2.Barrallo-Gimeno A, Nieto MA. The Snail genes as inducers of cell movement and survival: implications in development and cancer. Development. 2005;132:3151–3161. doi: 10.1242/dev.01907. doi: 10.1242/dev.01907. [DOI] [PubMed] [Google Scholar]
- 3.Peinado H, Olmeda D, Cano A. Snail, Zeb and bHLH factors in tumour progression: an alliance against the epithelial phenotype? Nat Rev Cancer. 2007;7:415–428. doi: 10.1038/nrc2131. doi: 10.1038/nrc2131. [DOI] [PubMed] [Google Scholar]
- 4.Moreno-Bueno G, Portillo F, Cano A. Transcriptional regulation of cell polarity in EMT and cancer. Oncogene. 2008;27:6958–6969. doi: 10.1038/onc.2008.346. doi: 10.1038/onc.2008.346. [DOI] [PubMed] [Google Scholar]
- 5.Dhasarathy A, Kajita M, Wade PA. The transcription factor snail mediates epithelial to mesenchymal transitions by repression of estrogen receptor-alpha. Mol Endocrinol. 2007;21:2907–2918. doi: 10.1210/me.2007-0293. doi: 10.1210/me.2007-0293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Côme C, Magnino F, Bibeau F, De Santa Barbara P, Becker KF, Theillet C, Savagner P. Snail and slug play distinct roles during breast carcinoma progression. Clin Cancer Res. 2006;12:5395–5402. doi: 10.1158/1078-0432.CCR-06-0478. doi: 10.1158/1078-0432.CCR-06-0478. [DOI] [PubMed] [Google Scholar]
- 7.Paznekas WA, Okajima K, Schertzer M, Wood S, Jabs EW. Genomic organization, expression, and chromosome location of the human SNAIL gene (SNAI1) and a related processed pseudogene (SNAI1P) Genomics. 1999;62:42–49. doi: 10.1006/geno.1999.6010. doi: 10.1006/geno.1999.6010. [DOI] [PubMed] [Google Scholar]
- 8.Locascio A, Vega S, de Frutos CA, Manzanares M, Nieto MA. Biological potential of a functional human SNAIL retrogene. J Biol Chem. 2002;277:38803–38809. doi: 10.1074/jbc.M205358200. doi: 10.1074/jbc.M205358200. [DOI] [PubMed] [Google Scholar]
- 9.Ko H, Kim HS, Kim NH, Lee SH, Kim KH, Hong SH, Yook JI. Nuclear localization signals of the E-cadherin transcriptional repressor Snail. Cells Tissues Organs. 2007;185:66–72. doi: 10.1159/000101305. doi: 10.1159/000101305. [DOI] [PubMed] [Google Scholar]
- 10.Hou Z, Peng H, Ayyanathan K, Yan KP, Langer EM, Longmore GD, Rauscher FJ., 3rd The LIM protein AJUBA recruits protein arginine methyltransferase 5 to mediate SNAIL-dependent transcriptional repression. Mol Cell Biol. 2008;28:3198–3207. doi: 10.1128/MCB.01435-07. doi: 10.1128/MCB.01435-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yook JI, Li XY, Ota I, Hu C, Kim HS, Kim NH, Cha SY, Ryu JK, Choi YJ, Kim J, Fearon ER, Weiss SJ. A Wnt-Axin2-GSK3beta cascade regulates Snail1 activity in breast cancer cells. Nat Cell Biol. 2006;8:1398–1406. doi: 10.1038/ncb1508. doi: 10.1038/ncb1508. [DOI] [PubMed] [Google Scholar]
- 12.Twigg SR, Wilkie AO. Characterisation of the human snail (SNAI1) gene and exclusion as a major disease gene in craniosynostosis. Hum Genet. 1999;105:320–326. doi: 10.1007/s004399900143. doi: 10.1007/s004390051108. [DOI] [PubMed] [Google Scholar]
- 13.Okubo T, Truong TK, Yu B, Itoh T, Zhao J, Grube B, Zhou D, Chen S. Down-regulation of promoter 1.3 activity of the human aromatase gene in breast tissue by zinc-finger protein, snail (SnaH) Cancer Res. 2001;61:1338–1346. [PubMed] [Google Scholar]
- 14.Moody SE, Perez D, Pan TC, Sarkisian CJ, Portocarrero CP, Sterner CJ, Notorfrancesco KL, Cardiff RD, Chodosh LA. The transcriptional repressor Snail promotes mammary tumor recurrence. Cancer Cell. 2005;8:197–209. doi: 10.1016/j.ccr.2005.07.009. doi: 10.1016/j.ccr.2005.07.009. [DOI] [PubMed] [Google Scholar]
- 15.Davidson NE, Sukumar S. Of Snail, mice, and women. Cancer Cell. 2005;8:173–174. doi: 10.1016/j.ccr.2005.08.006. doi: 10.1016/j.ccr.2005.08.006. [DOI] [PubMed] [Google Scholar]
- 16.De Craene B, Berx G. Snail in the frame of malignant tumor recurrence. Breast Cancer Res. 2006;8:105. doi: 10.1186/bcr1521. doi: 10.1186/bcr1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martínez-Estrada OM, Cullerés A, Soriano FX, Peinado H, Bolós V, Martínez FO, Reina M, Cano A, Fabre M, Vilaró S. The transcription factors Slug and Snail act as repressors of Claudin-1 expression in epithelial cells. Biochem J. 2006;394:449–457. doi: 10.1042/BJ20050591. doi: 10.1042/BJ20050591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Martin TA, Goyal A, Watkins G, Jiang WG. Expression of the transcription factors snail, slug, and twist and their clinical significance in human breast cancer. Ann Surg Oncol. 2005;12:488–496. doi: 10.1245/ASO.2005.04.010. doi: 10.1245/ASO.2005.04.010. [DOI] [PubMed] [Google Scholar]
- 19.Chiba H, Osanai M, Murata M, Kojima T, Sawada N. Transmembrane proteins of tight junctions. Biochim Biophys Acta. 2008;1778:588–600. doi: 10.1016/j.bbamem.2007.08.017. doi: 10.1016/j.bbamem.2007.08.017. [DOI] [PubMed] [Google Scholar]
- 20.Krause G, Winkler L, Mueller SL, Haseloff RF, Piontek J, Blasig IE. Structure and function of claudins. Biochim Biophys Acta. 2008;1778:631–645. doi: 10.1016/j.bbamem.2007.10.018. [DOI] [PubMed] [Google Scholar]
- 21.Tripathi MK, Misra S, Khedkar SV, Hamilton N, Irvin-Wilson C, Sharan C, Sealy L, Chaudhuri G. Regulation of BRCA2 gene expression by the SLUG repressor protein in human breast cells. J Biol Chem. 2005;280:17163–17171. doi: 10.1074/jbc.M501375200. doi: 10.1074/jbc.M501375200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bailey CK, Misra S, Mittal MK, Chaudhuri G. Human SLUG does not directly bind to CtBP1. Biochem Biophys Res Commun. 2007;353:661–664. doi: 10.1016/j.bbrc.2006.12.097. doi: 10.1016/j.bbrc.2006.12.097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mittal M, Myers JN, Misra S, Bailey CK, Chaudhuri G. In vivo binding to and functional repression of the VDR gene promoter by SLUG in human breast cells. Biochem Biophys Res Commun. 2008;372:30–34. doi: 10.1016/j.bbrc.2008.04.187. doi: 10.1016/j.bbrc.2008.04.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bennett RD, Mauer AS, Strehler EE. Calmodulin-like protein increases filopodia-dependent cell motility via up-regulation of myosin-10. J Biol Chem. 2007;282:3205–3212. doi: 10.1074/jbc.M607174200. doi: 10.1074/jbc.M607 174200. [DOI] [PubMed] [Google Scholar]
- 25.Xia Y, Wen HY, Kellems RE. Angiotensin II inhibits human trophoblast invasion through AT1 receptor activation. J Biol Chem. 2002;277:24601–24608. doi: 10.1074/jbc.M201369200. doi: 10.1074/jbc.M201369200. [DOI] [PubMed] [Google Scholar]
- 26.Yamasaki H, Sekimoto T, Ohkubo T, Douchi T, Nagata Y, Ozawa M, Yoneda Y. Zinc finger domain of Snail functions as a nuclear localization signal for importin beta-mediated nuclear import pathway. Genes Cells. 2005;10:455–464. doi: 10.1111/j.1365-2443.2005.00850.x. doi: 10.1111/j.1365-2443.2005.00850.x. [DOI] [PubMed] [Google Scholar]
- 27.Kwon YS, Garcia-Bassets I, Hutt KR, Cheng CS, Jin M, Liu D, Benner C, Wang D, Ye Z, Bibikova M, Fan JB, Duan L, Glass CK, Rosenfeld MG, Fu XD. Sensitive ChIP-DSL technology reveals an extensive estrogen receptor alpha-binding program on human gene promoters. Proc Natl Acad Sci USA. 2007;104:4852–4857. doi: 10.1073/pnas.0700715104. doi: 10.1073/pnas.0700715104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kominsky SL, Argani P, Korz D, Evron E, Raman V, Garrett E, Rein A, Sauter G, Kallioniemi OP, Sukumar S. Loss of the tight junction protein claudin-7 correlates with histological grade in both ductal carcinoma in situ and invasive ductal carcinoma of the breast. Oncogene. 2003;22:2021–2033. doi: 10.1038/sj.onc.1206199. doi: 10.1038/sj.onc.1206199. [DOI] [PubMed] [Google Scholar]
- 29.Oshima T, Kunisaki C, Yoshihara K, Yamada R, Yamamoto N, Sato T, Makino H, Yamagishi S, Nagano Y, Fujii S, Shiozawa M, Akaike M, Wada N, Rino Y, Masuda M, Tanaka K, Imada T. Reduced expression of the claudin-7 gene correlates with venous invasion and liver metastasis in colorectal cancer. Oncol Rep. 2008;19:953–959. [PubMed] [Google Scholar]
- 30.Lioni M, Brafford P, Andl C, Rustgi A, El-Deiry W, Herlyn M, Smalley KS. Dysregulation of claudin-7 leads to loss of E-cadherin expression and the increased invasion of esophageal squamous cell carcinoma cells. Am J Pathol. 2007;170:709–721. doi: 10.2353/ajpath.2007.060343. doi: 10.2353/ajpath.2007.060343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ayyanathan K, Peng H, Hou Z, Fredericks WJ, Goyal RK, Langer EM, Longmore GD, Rauscher FJ., 3rd The Ajuba LIM domain protein is a corepressor for SNAG domain mediated repression and participates in nucleocytoplasmic shuttling. Cancer Res. 2007;67:9097–9106. doi: 10.1158/0008-5472.CAN-07-2987. doi: 10.1158/0008-5472.CAN-07-2987. [DOI] [PubMed] [Google Scholar]