Abstract
Mast cells produce a large amount of several chemokines after cross-linking of FcεRI and participate in the pathogenesis of allergic diseases. The objective of this study was to comprehensively investigate FcεRI-mediated chemokine induction in human mast cells and the effect of a corticosteroid (dexamethasone) and a calcineurin inhibitor (FK506). Human peripheral blood-derived mast cells were stimulated with anti-IgE Ab in the presence of dexamethasone or FK506. Gene expression profiles were evaluated using GeneChip and confirmed by real-time PCR, and chemokine concentrations were measured by cytometric bead arrays and ELISA. Expression of eight chemokines was significantly induced in mast cells by anti-IgE stimulation. Induction of CCL2, CCL7, CXCL3, and CXCL8 by anti-IgE was significantly inhibited by dexamethasone but was enhanced by FK506. In contrast, induction of CCL1, CCL3, CCL4, and CCL18 was significantly inhibited by FK506 but, with the exception of CCL1, was enhanced by dexamethasone. Combination of dexamethasone and FK506 suppressed production of all chemokines by anti-IgE stimulation. Studies using protease inhibitors indicate that mast cell proteases may degrade several of the chemokines. These results suggest that corticosteroids and calcineurin inhibitors inhibit expression of distinct subsets of chemokines, and a combination of these drugs almost completely suppresses the induction of all chemokine genes in human mast cells in response to FcεRI-dependent stimulation. This implies that a combination of a corticosteroid and a calcineurin inhibitor may be more effective than each single agent for the treatment of allergic diseases in which mast cell-derived chemokines play a major role.
Mast cells are well known to play a central role in the formation of allergic inflammation and contribute to the pathogenesis of allergic diseases, including bronchial asthma and atopic dermatitis (1, 2). After activation by cross-linking of the cell surface high-affinity IgE receptor, FcεRI, mast cells exert a wide variety of biological effects by releasing several mediators, including histamine, prostaglandins (PGs),3 leukotrienes (LTs), proteases, cytokines, and chemokines (1, 2). Among these mediators, the chemokines mainly participate in the selective recruitment of inflammatory cells into tissue sites (3). Chemokines are a large superfamily of low molecular mass, secreted, and heparin-binding molecules that can be classified into several groups based on their molecular structures (4). More than 45 human chemokines have been discovered (4), and a comprehensive transcriptome analysis has shown that human mast cells produce and release I-309 (CCL1), MCP-1 (CCL2), MIP-1α (CCL3), MIP-1β (CCL4), MCP-3 (CCL7), TARC (CCL17), LARC (CCL20), MDC (CCL22), and IL-8 (CXCL8) upon stimulation of FcεRI (5, 6). Thus, mast cells not only trigger immediate allergic reactions but also orchestrate cellular allergic inflammatory responses through release of these chemokines (1, 7).
Allergic diseases are one of the most common chronic inflammatory diseases worldwide (8, 9). Significantly elevated total IgE levels are found in the serum of most patients with asthma and atopic dermatitis, and the Ag-specific IgE level is also usually increased (10, 11). Allergen exposure triggers and exacerbates allergic inflammation and clinical symptoms in most patients with allergic diseases.
Increased numbers of mast cells and infiltrating inflammatory cells, including eosinophils, lymphocytes, and macrophages, have been reported in the asthmatic lungs and atopic dermatitis lesions (11–13). At the same time, several chemokines, including CCL2, CCL3, CCL4, RANTES (CCL5), eotaxin (CCL11), MCP-4 (CCL13), CCL17, and CCL22, which presumably attract these inflammatory cells, have also been reported to be elevated in the serum or in asthmatic lungs and atopic dermatitis lesions (7, 14–20). Some of these chemokines are thought to be involved in the selective recruitment of CCR3+ cells (eosinophils and Th2 cells) (17) or CCR4+ cells (Th2 cells) (18) and to participate in the chronic stages of allergic inflammation. Additionally, recent studies have indicated that CCL1 and PARC (CCL18) are also increased in asthmatic lungs and atopic dermatitis lesions, and may initiate and amplify allergic inflammation (19–23).
Administration of anti-IgE mAb in patients with asthma not only reduced the mean maximal fall in FEV1 during the early response after Ag challenge, but also significantly reduced the mean maximal fall in FEV1 during the late response (24). This fact clearly indicates that mediators, presumably including chemokines released from mast cells upon stimulation with FcεRI, play critical roles in the recruitment of inflammatory cells into allergic tissue sites. It is noteworthy that most of the chemokines whose level is locally or systemically elevated in allergic patients are also known to be released by mast cells upon cross-linking of cell surface IgE receptors (5, 6). Thus, it is highly possible that mast cells participate in the pathogenesis of allergic inflammation through release of these chemokines.
Topical corticosteroids have been the mainstay of antiinflammatory therapy and have been effective in the control of both acute and chronic inflammatory reactions in allergic diseases (25, 26). Corticosteroids act on several resident and infiltrating cells and reduce inflammation, primarily through suppression of inflammatory gene expression via diverse molecular mechanisms (27). However, some patients with bronchial asthma or atopic dermatitis do not respond to corticosteroid therapy because they do not adhere to the treatment regimen (28, 29), because of acquired steroid resistance (30–32), or because the induction of inflammation-causing genes itself is insensitive to the treatment by corticosteroids (33, 34).
The topical calcineurin inhibitors tacrolimus (FK506) and pimecrolimus have recently been approved for the treatment of atopic dermatitis (26, 35). Clinically, FK506 exhibits potency against atopic dermatitis that is almost equivalent to that of “mild to potent” corticosteroid ointment (35). However, the mechanisms of action of the calcineurin inhibitors are distinct from those of corticosteroids (29, 36). For instance, FK506 efficiently suppresses lymphocyte proliferation after stimulation with bacterial superantigens, whereas corticosteroids do not (32). Additionally, cyclosporin, another calcineurin inhibitor, has been shown to improve lung function in patients with severe asthma in multiple clinical trials (37).
Although corticosteroids do not inhibit the release of histamine in human mast cells (38, 39), they are known to reduce the expression of some, but not all, cytokines in human mast cells (33). In contrast, calcineurin inhibitors have been found to suppress the release of histamine, tryptase, β-hexosaminidase, and some chemokines from mast cells (40–42). However, the effect of either of these drugs on the chemokine production profile in mast cells upon stimulation of FcεRI has not yet been investigated (42).
In the present study, we comprehensively investigated FcεRI-mediated chemokine induction in human peripheral blood-derived mast cells and the effect of a corticosteroid (dexamethasone, DEX) and a calcineurin inhibitor (FK506) on the response. Additionally, we found that DEX and FK506 clearly blocked the intracellular translocation of NF-κB and NF-AT, respectively, in mast cells activated via FcεRI. We think that these results will provide valuable information concerning the pathogenesis of steroid-resistant asthma or atopic dermatitis and may also provide a rationale for the potential use of these two topical therapeutic agents to treat these allergic diseases.
Materials and Methods
Reagents
Recombinant human stem cell factor (SCF), IL-3, and IL-6 were purchased from PeproTech. FK506 and human myeloma IgE were purchased from Calbiochem. DEX, DMSO, protease inhibitor cocktail (PIC; containing 4-(2-aminoethyl)benzenesulfonyl fluoride, pepstatin A, E-64, bestatin, leupeptin, and aprotinin) and BSA were purchased from Sigma-Aldrich.
Mast cell culture and stimulation
All human subjects in this study provided written informed consent that was approved by the Ethical Review Board at the National Center for Child Health and Development, Tokyo, Japan. Human peripheral blood-derived mast cells were obtained as described previously (43, 44). Briefly, lineage-negative mononuclear cells were separated from human PBMC by using an autoMACS system (DEPLETES 0.5 program; Miltenyi Biotec) and a mixture of magnetic microbead-conjugated Abs against CD4, CD8, CD11b, CD14, CD16, and CD19 (Miltenyi Biotec) according to the manufacturer’s instructions. The cells were suspended in serum-free Iscove’s methylcellulose medium (MethoCult SFBIT H4236; StemCell Technologies) containing 200 ng/ml SCF, IL-6, 5 ng/ml IL-3, 100 U/ml penicillin, and 100 μg/ml streptomycin, and then incubated at 37°C in 5% CO2. After 2 wk of culture, fresh methylcellulose medium containing 200 ng/ml SCF, 50 ng/ml IL-6, 5 ng/ml IL-3, 100 U/ml penicillin, and 100 μg/ml streptomycin was layered over the methylcellulose cultures. At 4 wk, a 1-ml aliquot of IMDM (Invitrogen) supplemented with 200 ng/ml SCF, 50 ng/ml IL-6, insulin-transferrin-selenium (Invitrogen), 55 μM 2-ME (Invitrogen), 100 U/ml penicillin, and 100 μg/ml streptomycin was layered over the methylcellulose cultures. At 6 wk, all cells were retrieved after dissolving the methylcellulose medium with PBS. The cells were then suspended and cultured in IMDM supplemented with 100 ng/ml SCF, 50 ng/ml IL-6, 0.1% BSA, insulin-transferrin-selenium, 55 μM 2-ME, 100 U/ml penicillin, and 100 μg/ml streptomycin, and the culture medium was changed a week later. After an additional week of culture, the culture medium was switched to IMDM supplemented with 100 ng/ml SCF, 50 ng/ml IL-6, 5% FBS (Invitrogen), 55 μM 2-ME, 100 U/ml penicillin, and 100 μg/ml streptomycin. The culture medium was changed weekly thereafter, and the cells were incubated for an additional 5–7 wk. The final purity of the mast cells always exceeded 98%. The mast cells were then sensitized with 1 μg/ml human myeloma IgE (Calbiochem) at 37°C for 48 h and, after washing, the mast cells were preincubated with DEX, FK506, or DMSO for 1 h and then stimulated with 1.5 μg/ml anti-IgE Ab (Dako) for 6 h. The cultured mast cells derived from atopic donors and those from normal IgE donors have been reported to equally express FcεRI and release histamine upon stimulation with FcεRI (45). Supernatants were harvested and assayed as described below.
Oligonucleotide microarray
A comprehensive microarray analysis was performed as described previously (46). Exactly the same experiments were performed with mast cells from four individual donors, and mRNAs were mixed and hybridized with a single set of microarrays. Gene expression was measured with GeneChip Human Genome U133 Plus 2.0 probe arrays (Affymetrix). Data analysis was performed with GeneSpring software version 7.2 (Agilent Technologies). To normalize the variations in staining intensity among chips, the “average difference” values for all genes on a given chip were divided by the median value for expression of all genes on the chip. To eliminate genes containing only a background signal, genes were selected only if the raw values of the average difference were >200, and expression of the gene was judged to be “present” by the GeneChip Operating Software version 1.4 (Affymetrix). A hierarchical-clustering analysis was performed using a minimum distance value of 0.001, a separation ratio of 0.5, and the standard definition of the correlation distance.
Real-time PCR
Primer sets for the following nine genes were synthesized at Qiagen: CCL1 (sense, 5′-CCTGCGCCTTGGACACAGT-3′; antisense, 5′-CAGAGCCC ACAATGGAAAGAAA-3′), CCL2 (sense, 5′-TCAGCCAGATGCAATC AATGC-3′; antisense, 5′-GGACACTTGCTGCTGGTGATTC-3′), CCL3 (sense, 5′-CAGCTACACCTCCCGGCA-3′; antisense, 5′-TCGCTTGGTT AGGAAGATGACAC-3′), CCL4 (sense, 5′-CGTGTATGACCTGGAACT GAACTG-3′; antisense, 5′-TCCCTGAAGACTTCCTGTCTCTGA-3′), MCP-3 (CCL7; sense, 5′-GCCATGACTTGAGAAACAAATAATTTG-3′; antisense, 5′-AATCTCAGAACCACTCTGAGAAAGGA-3′), CCL18 (sense, 5′-ATGGCCCTCTGCTCCTGTG-3′; antisense, 5′-GGTATAGA CGAGGCAGCAGAGCT-3′), GRO3 (CXCL3; sense, 5′-GCAGGGAATT CACCTCAAGA-3′; antisense, 5′-GGTGCTCCCCTTGTTCAGTA-3′), IL-8 (CXCL8; sense, 5′TCTGCAGCTCTGTGTGAAGGTG-3′; anti-sense, 5′-AATTTCTGTGTTGGCGCAGTG-3′). and GAPDH (sense, 5′-GAAGGTGAAGGTCGGAGTC-3′; antisense, 5′-GAAGATGGTGATGG GATTTC-3′). Total RNA was extracted with RNeasy (Qiagen) and digested with DNase I (Qiagen) according to the manufacturer’s instructions. Single-strand cDNA was synthesized with SuperScript II reverse transcriptase (Invitrogen) and oligo(dT) primers. Quantitative real-time PCR was performed by using a double-stranded DNA-binding dye, SYBR Green I, and an Applied Biosystems 7700 Sequence Detection System, as previously reported (47). To determine the exact copy number of the target genes, quantified aliquots of purified PCR fragments of the target genes were serially diluted and used as standards in each experiment. Aliquots of cDNA equivalent to 5 ng of total RNA were used for real-time PCR. The mRNA expression levels were normalized to the median expression level of a housekeeping gene (GAPDH).
Cytometric bead array (CBA)
The concentrations of CCL2, CCL3, CCL4, CXCL8, and GM-CSF in cell-free supernatants were measured using a CBA human Flex Set for CCL2, CCL3, CCL4, CXCL8, and GM-CSF (BD Biosciences). In brief, 40 μl of the mixed capture beads and 50 μl of culture supernatants were incubated for 1 h at room temperature, and after adding 40 μl of the mixed PE detection reagent to the mixture, it was incubated for 2 h at room temperature. The beads were then washed with the wash buffer and analyzed with a FACSArray bioanalyzer (BD Biosciences). The CBA data were analyzed with FCAP Array software version 1.0 (BD Biosciences).
ELISA and enzyme immunoassay
The concentrations of CCL1 and CCL18 in cell-free supernatants were measured with specific ELISA kits (R&D Systems). The minimal detection limit for both kits was 7.8 pg/ml. The concentrations of PGD2 in the cell-free supernatants were measured with a specific enzyme immunoassay kit (Cayman Chemical) that has a minimal detection limit of 7.8 pg/ml.
Immunofluorescence staining
Immunofluorescence staining was used to visualize the translocation of NF-κB and NF-AT. The mast cells were sensitized with 1 μg/ml human myeloma IgE for 48 h and, after washing, the mast cells were preincubated with either 1 μM DEX, 100 nM FK506, 0.01% DMSO, or a combination of DEX (1 μM) and FK506 (100 nM) for 1 h and then stimulated with 1.5 μg/ml anti-IgE Ab for 30 min. After making cyto-centrifugation preparations by Cytospin (Shandon), cells were fixed with 3.7% formaldehyde (Fisher Biotech) in PBS for 20 min and were permeabilized by 0.3% Tween 20 (Sigma-Aldrich) in PBS for 10 min. Cells were then blocked by blocking buffer (3% normal goat serum (Santa Cruz Biotechnology), 1% normal human AB serum (MP Biomedicals), 10% Fc blocking reagent (Miltenyi Biotec), 0.3% Tween 20 in PBS) for 2 h at room temperature. After blocking, cells were incubated with 2.5 μg/ml mouse anti-NF-κB p65 mAb (IgG1, clone 20; BD Biosciences) and 2 μg/ml rabbit anti-NF-ATc3 polyclonal Ab (Santa Cruz Biotechnology; sc8321) in blocking buffer or 2.5 μg/ml mouse control IgG1 (clone P3; eBioscience) and 2 μg/ml rabbit control IgG (Santa Cruz Biotechnology; sc2027) in blocking buffer at 4°C overnight. After washing with PBS, cells were incubated with 4 μg/ml Alexa Fluor 488-conjugated goat anti-mouse IgG (Invitrogen) and 4 μg/ml Alexa Fluor 647-conjugated goat anti-rabbit IgG (Invitrogen) for 1 h at room temperature in the dark. After final washing with PBS, coverslips were mounted onto slides using SlowFade Gold antifade reagent with 4′,6-diamidino-2-phenylindole (Invitrogen) and the slides were stored in the dark at 4°C. Images from immunofluorescence slides were obtained with an Olympus IX71 inverted research microscope using ×40 objective lens and images were collected by using SlideBook software (Olympus). Five pictures were randomly taken from each slide. The average percentages of mast cells with the nuclear translocation of NF-κB or NF-AT were calculated by two independent researchers.
Statistical analysis
All data are reported as the mean ± SEM unless otherwise noted. Differences between groups were analyzed using the paired Student’s t test and considered to be significant for a p value <0.05.
Results
Identification of FcεRI-mediated chemokine induction
Increasing evidence indicates that several mediators, including chemokines, are involved in the pathogenesis of allergic diseases, including bronchial asthma and atopic dermatitis (7, 14, 15, 48, 49). To test mast cells for a possible role in the selective recruitment of inflammatory cells into sites of allergic inflammation, we measured the expression of mRNA for chemokines in unstimulated mast cells and IgE/anti-IgE-activated mast cells with a microarray system and by real-time PCR. All microarray data have been submitted to Gene Expression Omnibus as GSE15174 (“The effect of a dexamethasone and a FK506 on the induction of chemokines in human mast cells”; www.ncbi.nlm.nih.gov/geo/). The accession numbers for “Control”, “Anti-IgE + DMSO”, “Anti-IgE + DEX”, “Anti-IgE + FK506”, and “Anti-IgE + DEX + FK506” are GSM378805, GSM378807, GSM378808, GSM378809, and GSM378810, respectively.
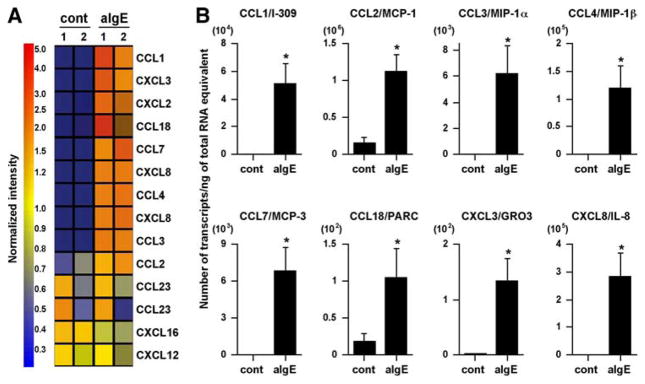
The results showed that 12 of 42 chemokines contained on the GeneChip U133 Plus 2.0 array were expressed in unstimulated or activated mast cells (Fig. 1A and Table I). Importantly, nine genes encoding CCL1, CCL2, CCL3, CCL4, CCL7, CCL18, CXCL2, CXCL3, and CXCL8 were up-regulated by FcεRI-mediated activation (Fig. 1A and Table I). We used a real-time PCR method to confirm the GeneChip data, and the results showed that eight of the nine genes were significantly up-regulated by anti-IgE stimulation (Fig. 1B). The magnitude of the increase in mRNA for CCL1, CCL2, CCL3, CCL4, CCL7, CCL18, CXCL3, and CXCL8 by FcεRI-mediated activation was 156-, 7-, 199-, 223-, 1521-, 6-, 60-, and 324-fold, respectively (Fig. 1B). We also used ELISA and CBA to measure chemokine production by anti-IgE-stimulated mast cells, and significant levels of CCL1 (17.6 ± 5.6 pg/106 cells, n = 5), CCL2 (3.2 ± 1.3 ng/106 cells, n = 5), CCL3 (1.1 ± 0.3 ng/106 cells, n = 5), CCL4 (9.4 ±2.5 ng/106 cells, n = 5), and CXCL8 (22.2 ±6.1 ng/106 cells, n = 5) were detected in the supernatant after stimulation with anti-IgE (Fig. 2A); no CCL18 was detected.
FIGURE 1.
FcεRI-mediated chemokine expression in human mast cells. Human mast cells were sensitized with 1 μg/ml human myeloma IgE for 48 h. After washing the cells, they were stimulated with 1.5 μg/ml anti-IgE Ab for 6 h. A, Gene expression was analyzed with the GeneChip Human Genome U133 Plus 2.0 probe arrays. Data were analyzed by applying a hierarchical tree algorithm to the normalized intensities. As indicated in the accompanying color bar, strongly expressed genes are represented by shades of red, and weakly expressed genes are represented by shades of blue. Exactly the same experiments were performed with mast cells from four individual donors, and mRNA were mixed and hybridized with a single set of microarrays. B, The mRNA levels of the chemokines were determined by real-time PCR. The results are shown as the means ± SEM of four independent experiments with independent donors.*, p < 0.05.
Table I.
FcεRI-mediated chemokine expression in human mast cellsa
| Control |
Anti-IgE |
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lot 1 |
Lot 2 |
Lot 1 |
Lot 2 |
||||||||||||
| Probe ID | Symbol | Alternate | GenBankb | Norm | Raw | Fl | Norm | Raw | Fl | Norm | Raw | Fl | Norm | Raw | Fl |
| 207533_at | CCL1 | I-309 | NM_002981 | 0.01 | 14 | A | 0.01 | 2 | A | 3.60 | 8382 | P | 2.00 | 2006 | P |
| 216598_s_at | CCL2 | MCP-1 | S69738 | 0.44 | 4852 | P | 0.63 | 2781 | P | 1.37 | 11637 | P | 1.72 | 6311 | P |
| 205114_s_at | CCL3 | MIP-1α | NM_002983 | 0.01 | 60 | P | 0.02 | 43 | P | 2.11 | 9456 | P | 1.98 | 3821 | P |
| 204103_at | CCL4 | MIP-1β | NM_002984 | 0.02 | 168 | A | 0.01 | 20 | A | 1.98 | 12603 | P | 2.25 | 6169 | P |
| 1555759_a_at | CCL5 | RANTES | AF043341 | 0.89 | 15 | A | 1.11 | 8 | A | 4.68 | 62 | A | 0.59 | 3 | A |
| 1405_i_at | CCL5 | RANTES | M21121 | 0.99 | 5 | A | 0.91 | 2 | A | 10.47 | 38 | A | 0.39 | 1 | A |
| 235100_at | CCL5 | RANTES | BG435715 | 2.65 | 34 | A | 1.17 | 6 | A | 0.49 | 5 | A | 0.83 | 4 | A |
| 204655_at | CCL5 | RANTES | NM_002985 | 0.38 | 15 | A | 1.54 | 25 | A | 1.44 | 46 | A | 0.56 | 8 | A |
| 1561006_at | CCL5 | RANTES | AF147386 | 0.85 | 9 | A | 0.79 | 4 | A | 1.15 | 10 | A | 2.59 | 10 | A |
| 208075_s_at | CCL7 | MCP-3 | NM_006273 | 0.01 | 5 | A | 0.11 | 22 | A | 1.89 | 709 | P | 2.93 | 474 | P |
| 214038_at | CCL8 | MCP-2 | AI984980 | 0.13 | 8 | A | 1.08 | 25 | A | 1.34 | 59 | A | 0.92 | 18 | A |
| 210133_at | CCL11 | Eotaxin | D49372 | 1.19 | 49 | A | 0.81 | 14 | A | 2.52 | 80 | A | 0.30 | 4 | A |
| 206407_s_at | CCL13 | MCP-4 | NM_005408 | 0.20 | 8 | A | 1.44 | 24 | A | 0.56 | 18 | A | 2.24 | 30 | A |
| 216714_at | CCL13 | MCP-4 | Z77651 | 0.85 | 7 | A | 1.15 | 4 | A | 0.80 | 5 | A | 1.38 | 4 | A |
| 205392_s_at | CCL14 | HCC-1 | NM_004166 | 1.13 | 100 | A | 0.13 | 5 | A | 1.67 | 114 | A | 0.87 | 26 | A |
| 210390_s_at | CCL15 | HCC-2 | AF031587 | 0.91 | 28 | A | 0.83 | 10 | A | 1.09 | 26 | A | 1.51 | 16 | A |
| 207354_at | CCL16 | LEC | NM_004590 | 2.56 | 49 | A | 0.45 | 4 | A | 1.00 | 15 | A | 1.00 | 6 | A |
| 207900_at | CCL17 | TARC | NM_002987 | 1.13 | 76 | A | 1.10 | 30 | A | 0.63 | 33 | A | 0.90 | 20 | A |
| 209924_at | CCL18 | PARC | AB000221 | 0.66 | 110 | A | 0.16 | 11 | A | 2.50 | 325 | P | 1.34 | 75 | A |
| 32128_at | CCL18 | PARC | 4864840_RC | 0.13 | 10 | A | 0.40 | 13 | A | 5.06 | 301 | P | 1.60 | 41 | P |
| 210072_at | CCL19 | MIP-3β | U88321 | 0.72 | 38 | A | 1.13 | 24 | A | 0.87 | 36 | A | 1.21 | 21 | A |
| 205476_at | CCL20 | LARC | NM_004591 | 0.67 | 6 | A | 1.04 | 4 | A | 0.96 | 7 | A | 1.15 | 3 | A |
| 204606_at | CCL21 | SLC, ECL | NM_002989 | 0.76 | 9 | A | 1.20 | 6 | A | 1.39 | 13 | A | 0.80 | 3 | A |
| 207861_at | CCL22 | MDC | NM_002990 | 0.90 | 11 | A | 0.98 | 5 | A | 1.37 | 13 | A | 1.02 | 4 | A |
| 210548_at | CCL23 | MPIF-1 | U58913 | 1.80 | 1503 | P | 0.50 | 168 | P | 1.50 | 973 | P | 0.35 | 97 | P |
| 210549_s_at | CCL23 | MPIF-1 | U58913 | 1.47 | 1348 | P | 0.58 | 215 | P | 1.31 | 932 | P | 0.69 | 211 | P |
| 221463_at | CCL24 | Eotaxin-2 | NM_002991 | 1.46 | 32 | A | 0.85 | 8 | A | 0.94 | 16 | A | 1.06 | 8 | A |
| 206988_at | CCL25 | TECK | NM_005624 | 1.01 | 14 | A | 1.60 | 9 | A | 0.99 | 10 | A | 0.94 | 4 | A |
| 223710_at | CCL26 | Eotaxin-3 | AF096296 | 0.92 | 8 | A | 1.08 | 4 | A | 2.66 | 19 | A | 0.52 | 2 | A |
| 207955_at | CCL27 | CTACK | NM_006664 | 0.91 | 81 | A | 1.09 | 39 | A | 0.84 | 57 | A | 1.30 | 38 | A |
| 230327_at | CCL27 | CTACK | AI203673 | 0.94 | 100 | A | 1.06 | 46 | A | 0.12 | 10 | A | 1.11 | 40 | A |
| 224240_s_at | CCL28 | MEC | AF266504 | 0.89 | 94 | P | 0.72 | 31 | P | 1.11 | 91 | P | 1.13 | 40 | A |
| 224027_at | CCL28 | MEC | AF110384 | 0.96 | 65 | P | 0.88 | 24 | A | 1.13 | 59 | A | 1.04 | 23 | A |
| 204470_at | CXCL1 | GRO1 | NM_001511 | 0.78 | 30 | A | 1.04 | 16 | A | 0.96 | 29 | A | 1.13 | 15 | A |
| 209774_x_at | CXCL2 | GRO2 | M57731 | 0.20 | 30 | A | 0.07 | 4 | A | 2.54 | 290 | P | 1.80 | 88 | P |
| 230101_at | CXCL2 | GRO2 | AV648479 | 1.08 | 65 | A | 1.10 | 27 | A | 0.50 | 23 | A | 0.92 | 19 | A |
| 1569203_at | CXCL2 | GRO2 | BC005276 | 0.47 | 2 | A | 0.70 | 1 | A | 0.53 | 2 | A | 0.84 | 1 | A |
| 207850_at | CXCL3 | GRO3 | NM_002090 | 0.12 | 54 | A | 0.16 | 31 | A | 2.89 | 1051 | P | 1.84 | 288 | P |
| 206390_x_at | CXCL4 | PF4 | NM_002619 | 3.00 | 124 | A | 0.49 | 8 | A | 0.46 | 15 | A | 1.51 | 21 | A |
| 207815_at | CXCL4 | PF4 | NM_002620 | 0.76 | 6 | A | 1.24 | 4 | A | 0.08 | 1 | A | 1.96 | 6 | P |
| 215101_s_at | CXCL5 | ENA-78 | BG166705 | 0.24 | 4 | A | 2.15 | 13 | A | 0.28 | 3 | A | 1.72 | 9 | A |
| 214974_x_at | CXCL5 | ENA-78 | AK026546 | 0.81 | 22 | A | 1.19 | 13 | A | 0.81 | 17 | P | 1.69 | 16 | P |
| 207852_at | CXCL5 | ENA-78 | NM_002994 | 0.17 | 1 | A | 1.07 | 2 | A | 1.17 | 4 | A | 0.45 | 1 | A |
| 206336_at | CXCL6 | GCP-2 | NM_002993 | 0.89 | 25 | A | 0.49 | 6 | A | 1.11 | 24 | A | 1.29 | 12 | A |
| 214146_s_at | CXCL7 | PPBP | R64130 | 0.63 | 5 | A | 1.29 | 4 | A | 1.47 | 9 | A | 0.71 | 2 | A |
| 202859_x_at | CXCL8 | IL-8 | NM_000584 | 0.02 | 136 | P | 0.02 | 74 | M | 1.98 | 11837 | P | 2.44 | 6312 | P |
| 211506_s_at | CXCL8 | IL-8 | AF043337 | 0.01 | 35 | A | 0.01 | 23 | A | 1.99 | 12539 | P | 2.09 | 5665 | P |
| 203915_at | CXCL9 | MIG | NM_002416 | 1.11 | 77 | A | 1.11 | 31 | A | 0.89 | 48 | A | 0.45 | 10 | A |
| 204533_at | CXCL10 | IP-10 | NM_001565 | 0.96 | 79 | A | 1.04 | 34 | A | 0.26 | 16 | A | 1.25 | 35 | A |
| 210163_at | CXCL11 | I-TAC | AF030514 | 0.86 | 6 | A | 0.65 | 2 | A | 2.08 | 11 | A | 1.14 | 3 | A |
| 211122_s_at | CXCL11 | I-TAC | AF002985 | 0.39 | 3 | A | 0.26 | 1 | A | 1.61 | 9 | A | 2.27 | 6 | A |
| 203666_at | CXCL12 | SDF-1 | NM_000609 | 1.12 | 164 | P | 0.96 | 57 | P | 1.04 | 117 | P | 0.75 | 37 | P |
| 209687_at | CXCL12 | SDF-1 | U19495 | 0.85 | 45 | A | 0.82 | 18 | A | 1.15 | 48 | A | 1.76 | 31 | A |
| 205242_at | CXCL13 | BLC | NM_006419 | 0.42 | 3 | A | 1.05 | 3 | A | 1.07 | 6 | A | 0.95 | 2 | A |
| 218002_s_at | CXCL14 | BMAC | NM_004887 | 0.38 | 3 | A | 1.11 | 3 | A | 0.89 | 5 | A | 1.97 | 5 | A |
| 237038_at | CXCL14 | BMAC | AI927990 | 1.12 | 14 | A | 0.43 | 2 | A | 0.90 | 8 | A | 1.10 | 4 | A |
| 222484_s_at | CXCL14 | BMAC | AF144103 | 1.01 | 5 | A | 1.45 | 3 | A | 0.99 | 4 | A | 0.97 | 2 | A |
| 223454_at | CXCL16 | SR-PSOX | AF275260 | 1.29 | 3837 | P | 1.19 | 1420 | P | 0.81 | 1858 | P | 0.72 | 712 | P |
| 203687_at | CX3CL1 | Fractalkine | NM_002996 | 1.20 | 17 | A | 1.22 | 7 | A | 0.80 | 9 | A | 0.73 | 4 | A |
| 823_at | CX3CL1 | Fractalkine | U84487 | 0.74 | 52 | A | 1.09 | 31 | A | 1.02 | 56 | P | 0.98 | 23 | A |
| 206366_x_at | XCL1 | Lymphotactin-α | U23772 | 1.02 | 14 | A | 0.35 | 2 | A | 1.87 | 20 | A | 0.98 | 4 | A |
| 206365_at | XCL1 | Lymphotactin-α | NM_002995 | 0.94 | 10 | A | 1.06 | 4 | A | 3.50 | 28 | A | 0.85 | 3 | A |
| 214567_s_at | XCL2 | Lymphotactin-β | NM_003175 | 0.42 | 3 | A | 1.04 | 3 | A | 1.32 | 6 | A | 0.96 | 2 | A |
Norm is normalized data. Raw is raw data (average difference value) of microarray. Fl is flag, which is judged to be “P (Present), M (Marginal) or A (Absent)” by the GeneChip operating software version 1.4.
GenBank accession nos. (www.ncbi.nlm.nih.gov).
FIGURE 2.
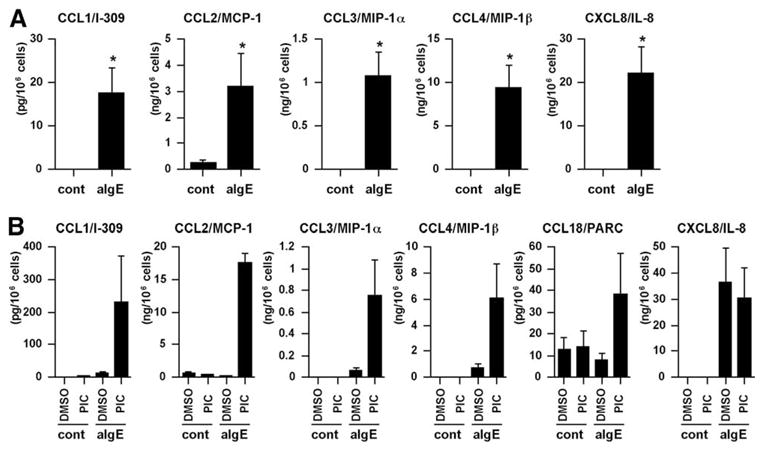
FcεRI-mediated chemokine production in human mast cells. Concentrations of chemokine proteins were determined in the culture supernatant of mast cells by CBA and ELISA. A, IgE-sensitized human mast cells were stimulated with medium control (cont) or 1.5 μg/ml anti-IgE Ab for 6 h. B, IgE-sensitized human mast cells were stimulated with buffer of 1.5 μg/ml anti-IgE Ab in the presence of 0.1% DMSO or 0.1% PIC for 48 h. The results are shown as the means ± SEM of five (A) or three (B) independent experiments with independent donors.*, p < 0.05.
The results of recent studies have suggested that cytokines and chemokines are degraded by purified mast cell proteases, such as tryptase, chymase, and cathepsin (50, 51). To determine whether the CCL18 produced by mast cells is degraded by proteases, mast cells were exposed to the PIC or DMSO (vehicle control) for 1 h, and then stimulated with anti-IgE Ab for 48 h. High concentrations of CCL18 were detected only in the PIC-treated cells after stimulation with anti-IgE Ab (Fig. 2B). Interestingly, higher levels of other CC chemokines (i.e., CCL1, CCL2, CCL3, and CCL4) were also detected in the supernatant of the PIC-treated mast cells after stimulation with anti-IgE Ab compared with the supernatant of DMSO-treated mast cells (Fig. 2B). In contrast, high concentrations of CXCL8 protein were detected in both DMSO- and PIC-treated mast cells. These results suggest that the CC chemokines CCL1, CCL2, CCL3, CCL4, and CCL18 may be sensitive to mast cell proteases.
Distinct inhibition of FcεRI-mediated chemokine induction by FK506 and DEX
We initially used the GeneChip system to examine the effect of a corticosteroid (DEX) and a calcineurin inhibitor (FK506) on chemokine expression in human mast cells. Hierarchical clustering analysis of the gene expression profiles of the 11 chemokines found to be present in unstimulated or stimulated mast cells with the GeneChip system revealed three distinct gene clusters (Fig. 3A and Table II). The first gene cluster contained the genes for four CC chemokines, CCL1, CCL3, CCL4, and CCL18; expression of these genes was inhibited by FK506 and not by DEX (Fig. 3A). In contrast, the second gene cluster contained the genes for two CC chemokines, CCL2 and CCL7, and three CXC chemokines, CXCL2, CXCL3, and CXCL8; expression of these genes was inhibited by DEX and not by FK506 (Fig. 3A). The third gene cluster contained the genes for two chemokines, CCL23 and CXCL16, which were unaffected by anti-IgE or by either of the drugs tested (Fig. 3A).
FIGURE 3.
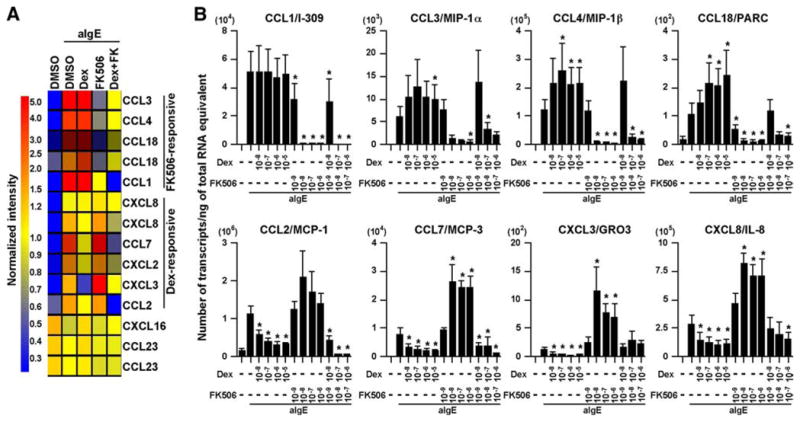
Effect of FK506 and DEX on the up-regulation of chemokines in human mast cells by anti-IgE Ab. IgE-sensitized human mast cells were preincubated with 1 μM DEX, 100 nM FK506, or 0.01% DMSO for 1 h and then stimulated with 1.5 μg/ml anti-IgE Ab for 6 h. A, The gene expression profile was analyzed with the GeneChip Human Genome U133 Plus 2.0 probe arrays. See Fig. 1 for information regarding the data analysis and the color code. B, The chemokine mRNA levels were determined by real-time PCR. The results are shown as the means ± SEM of four independent experiments with independent donors.*, p < 0.05.
Table II.
Effect of FK506 and DEX on the up-regulation of chemokines in human mast cells by FcεRI-mediated stimulationa
| Control |
Anti-IgE |
Anti-IgE + DEX |
Anti-IgE + FK506 |
Anti-IgE + Dex + FK506 |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Probe ID | Symbol | Alternate | Norm | Raw | Fl | Norm | Raw | Fl | Norm | Raw | Fl | Norm | Raw | Fl | Norm | Raw | Fl |
| 207533_at | CCL1 | I-309 | 0.03 | 14 | A | 24.78 | 8,382 | P | 20.50 | 6,423 | P | 1.00 | 321 | P | 0.13 | 41 | A |
| 216598_s_at | CCL2 | MCP-1 | 0.50 | 4,852 | P | 1.54 | 11,637 | P | 1.00 | 7,001 | P | 1.66 | 11,909 | P | 0.18 | 1,266 | P |
| 205114_s_at | CCL3 | MIP-1α | 0.03 | 60 | P | 5.93 | 9,456 | P | 6.13 | 9,054 | P | 0.57 | 856 | P | 1.00 | 1,514 | P |
| 204103_at | CCL4 | MIP-1β | 0.03 | 168 | A | 3.35 | 12,603 | P | 3.69 | 12,843 | P | 0.62 | 2,214 | P | 1.00 | 3,573 | P |
| 1555759_a_at | CCL5 | RANTES | 0.64 | 15 | A | 3.37 | 62 | A | 1.71 | 29 | A | 0.71 | 12 | A | 1.00 | 18 | A |
| 1405_i_at | CCL5 | RANTES | 0.99 | 5 | A | 10.47 | 38 | A | 4.99 | 17 | A | 0.82 | 3 | A | 0.59 | 2 | A |
| 235100_at | CCL5 | RANTES | 2.45 | 34 | A | 0.45 | 5 | A | 3.51 | 35 | A | 1.00 | 10 | A | 0.72 | 7 | A |
| 204655_at | CCL5 | RANTES | 0.27 | 15 | A | 1.05 | 46 | A | 2.12 | 85 | A | 0.40 | 17 | A | 1.00 | 41 | A |
| 1561006_at | CCL5 | RANTES | 0.75 | 9 | A | 1.02 | 10 | A | 1.75 | 16 | A | 0.96 | 9 | A | 1.00 | 9 | A |
| 208075_s_at | CCL7 | MCP-3 | 0.02 | 5 | A | 3.61 | 709 | P | 1.00 | 182 | P | 9.96 | 1,856 | P | 0.49 | 91 | P |
| 214038_at | CCL8 | MCP-2 | 0.14 | 8 | A | 1.47 | 59 | A | 0.70 | 26 | A | 1.00 | 38 | A | 1.02 | 39 | A |
| 210133_at | CCL11 | Eotaxin | 0.59 | 49 | A | 1.25 | 80 | A | 1.00 | 60 | A | 1.11 | 68 | A | 0.53 | 33 | A |
| 206407_s_at | CCL13 | MCP-4 | 0.17 | 8 | A | 0.48 | 18 | A | 1.00 | 34 | A | 1.45 | 51 | A | 1.61 | 57 | A |
| 216714_at | CCL13 | MCP-4 | 0.95 | 7 | A | 0.89 | 5 | A | 1.00 | 5 | A | 1.27 | 7 | A | 1.45 | 8 | A |
| 205392_s_at | CCL14 | HCC-1 | 1.00 | 100 | A | 1.47 | 114 | A | 1.53 | 110 | A | 0.10 | 7 | A | 0.79 | 58 | A |
| 210390_s_at | CCL15 | HCC-2 | 0.90 | 28 | A | 1.07 | 26 | A | 0.59 | 13 | A | 1.58 | 37 | P | 1.00 | 23 | A |
| 207354_at | CCL16 | LEC | 2.31 | 49 | A | 0.90 | 15 | A | 1.00 | 15 | A | 0.95 | 15 | A | 3.45 | 54 | A |
| 207900_at | CCL17 | TARC | 1.78 | 76 | A | 1.00 | 33 | A | 0.67 | 21 | A | 0.86 | 27 | A | 1.93 | 61 | A |
| 209924_at | CCL18 | PARC | 0.50 | 110 | A | 1.90 | 325 | P | 3.43 | 544 | P | 0.56 | 90 | A | 1.00 | 163 | P |
| 32128_at | CCL18 | PARC | 0.11 | 10 | A | 4.18 | 301 | P | 8.84 | 589 | P | 0.36 | 25 | A | 1.00 | 68 | P |
| 210072_at | CCL19 | MIP-3β | 0.83 | 38 | A | 1.00 | 36 | A | 0.45 | 15 | A | 1.42 | 48 | A | 1.92 | 65 | A |
| 205476_at | CCL20 | LARC | 1.01 | 6 | A | 1.46 | 7 | A | 1.00 | 4 | A | 0.98 | 4 | A | 0.56 | 2 | A |
| 204606_at | CCL21 | SLC, ECL | 0.94 | 9 | A | 1.73 | 13 | A | 0.56 | 4 | A | 2.52 | 18 | A | 1.00 | 7 | A |
| 207861_at | CCL22 | MDC | 0.66 | 11 | A | 1.00 | 13 | A | 1.31 | 16 | A | 1.19 | 15 | A | 0.90 | 11 | A |
| 210548_at | CCL23 | MPIF-1 | 1.20 | 1,503 | P | 1.00 | 973 | P | 1.09 | 984 | P | 0.94 | 863 | P | 0.93 | 863 | P |
| 210549_s_at | CCL23 | MPIF-1 | 1.12 | 1,348 | P | 1.00 | 932 | P | 1.04 | 895 | P | 0.89 | 784 | P | 0.91 | 804 | P |
| 221463_at | CCL24 | Eotaxin-2 | 1.50 | 32 | A | 0.96 | 16 | A | 1.00 | 16 | A | 1.64 | 26 | A | 0.69 | 11 | A |
| 206988_at | CCL25 | TECK | 0.66 | 14 | A | 0.65 | 10 | A | 2.47 | 36 | A | 1.05 | 16 | A | 1.00 | 15 | A |
| 223710_at | CCL26 | Eotaxin-3 | 1.00 | 8 | A | 2.91 | 19 | A | 0.78 | 5 | A | 0.41 | 3 | A | 1.13 | 7 | A |
| 207955_at | CCL27 | CTACK | 1.07 | 81 | A | 0.98 | 57 | A | 1.00 | 54 | A | 0.61 | 34 | A | 1.02 | 57 | A |
| 230327_at | CCL27 | CTACK | 1.00 | 100 | A | 0.12 | 10 | A | 0.97 | 69 | P | 1.16 | 85 | A | 1.04 | 76 | A |
| 224240_s_at | CCL28 | MEC | 0.80 | 94 | P | 1.00 | 91 | P | 1.08 | 91 | P | 0.74 | 63 | P | 1.16 | 100 | P |
| 224027_at | CCL28 | MEC | 1.00 | 65 | P | 1.17 | 59 | A | 1.62 | 75 | P | 0.89 | 42 | A | 0.97 | 46 | A |
| 204470_at | CXCL1 | GRO1 | 1.00 | 30 | A | 1.22 | 29 | A | 0.83 | 18 | A | 1.27 | 28 | A | 0.45 | 10 | A |
| 209774_x_at | CXCL2 | GRO2 | 0.14 | 30 | A | 1.77 | 290 | P | 1.00 | 152 | P | 1.70 | 264 | P | 0.75 | 117 | P |
| 230101_at | CXCL2 | GRO2 | 0.85 | 65 | A | 0.39 | 23 | A | 1.19 | 65 | P | 1.00 | 56 | P | 1.14 | 64 | A |
| 1569203_at | CXCL2 | GRO2 | 0.47 | 2 | A | 0.53 | 2 | A | 0.63 | 2 | A | 1.09 | 4 | A | 0.67 | 2 | A |
| 207850_at | CXCL3 | GRO3 | 0.06 | 54 | A | 1.38 | 1,051 | P | 0.40 | 286 | P | 5.25 | 3,799 | P | 1.00 | 725 | P |
| 206390_x_at | CXCL4 | PF4 | 1.50 | 124 | A | 0.23 | 15 | A | 1.68 | 99 | A | 1.00 | 61 | A | 0.97 | 59 | A |
| 207815_at | CXCL4 | PF4 | 1.00 | 6 | A | 0.10 | 1 | A | 2.00 | 9 | A | 0.72 | 3 | A | 1.27 | 6 | A |
| 215101_s_at | CXCL5 | ENA-78 | 0.78 | 4 | A | 0.89 | 3 | A | 0.72 | 2 | A | 0.41 | 1 | A | 1.00 | 3 | A |
| 214974_x_at | CXCL5 | ENA-78 | 0.85 | 22 | A | 0.85 | 17 | P | 1.25 | 24 | P | 1.00 | 19 | P | 1.53 | 30 | P |
| 207852_at | CXCL5 | ENA-78 | 0.15 | 1 | A | 1.00 | 4 | A | 0.33 | 1 | A | 1.23 | 5 | A | 5.44 | 22 | A |
| 206336_at | CXCL6 | GCP-2 | 0.80 | 25 | A | 1.00 | 24 | A | 1.18 | 27 | P | 0.46 | 11 | A | 1.33 | 31 | A |
| 214146_s_at | CXCL7 | PPBP | 0.43 | 5 | A | 1.00 | 9 | A | 2.12 | 18 | A | 0.65 | 6 | A | 2.79 | 24 | A |
| 202859_x_at | CXCL8 | IL-8 | 0.01 | 136 | P | 0.97 | 11,837 | P | 1.02 | 11,482 | P | 1.06 | 12,238 | P | 1.00 | 11,600 | P |
| 211506_s_at | CXCL8 | IL-8 | 0.01 | 35 | A | 1.39 | 12,539 | P | 1.00 | 8,353 | P | 1.63 | 13,942 | P | 0.74 | 6,367 | P |
| 203915_at | CXCL9 | MIG | 0.92 | 77 | A | 0.73 | 48 | A | 1.28 | 77 | A | 1.00 | 62 | A | 1.09 | 67 | A |
| 204533_at | CXCL10 | IP-10 | 2.14 | 79 | A | 0.57 | 16 | A | 0.63 | 17 | A | 1.00 | 27 | A | 3.47 | 95 | A |
| 210163_at | CXCL11 | I-TAC | 0.65 | 6 | A | 1.56 | 11 | A | 0.72 | 5 | A | 1.00 | 7 | A | 2.12 | 14 | A |
| 211122_s_at | CXCL11 | I-TAC | 0.63 | 3 | A | 2.62 | 9 | A | 0.15 | 1 | A | 7.08 | 24 | A | 0.62 | 2 | A |
| 203666_at | CXCL12 | SDF-1 | 1.00 | 164 | P | 0.92 | 117 | P | 0.80 | 94 | P | 1.19 | 143 | P | 1.19 | 143 | P |
| 209687_at | CXCL12 | SDF-1 | 0.87 | 45 | A | 1.19 | 48 | A | 0.87 | 32 | A | 1.32 | 50 | A | 1.00 | 38 | A |
| 205242_at | CXCL13 | BLC | 0.50 | 3 | A | 1.27 | 6 | A | 1.23 | 5 | A | 0.71 | 3 | A | 1.00 | 4 | A |
| 218002_s_at | CXCL14 | BMAC | 0.43 | 3 | A | 1.00 | 5 | A | 0.77 | 4 | A | 5.18 | 24 | A | 1.27 | 6 | A |
| 237038_at | CXCL14 | BMAC | 1.00 | 14 | A | 0.80 | 8 | A | 1.14 | 11 | A | 0.99 | 10 | A | 2.08 | 21 | A |
| 222484_s_at | CXCL14 | BMAC | 1.02 | 5 | A | 1.00 | 4 | A | 0.68 | 2 | A | 0.89 | 3 | A | 1.02 | 4 | A |
| 223454_at | CXCL16 | SR-PSOX | 1.34 | 3,837 | P | 0.84 | 1,858 | P | 1.08 | 2,221 | P | 0.88 | 1,855 | P | 1.00 | 2,102 | P |
| 203687_at | CX3CL1 | Fractalkine | 1.26 | 17 | A | 0.83 | 9 | A | 0.49 | 5 | A | 1.00 | 10 | A | 1.12 | 11 | A |
| 823_at | CX3CL1 | Fractalkine | 0.81 | 52 | A | 1.11 | 56 | P | 1.01 | 47 | A | 0.68 | 32 | A | 1.00 | 48 | A |
| 206366_x_at | XCL1 | Lymphotactin-α | 0.54 | 14 | A | 1.00 | 20 | A | 0.69 | 12 | A | 2.58 | 48 | A | 1.11 | 21 | A |
| 206365_at | XCL1 | Lymphotactin-α | 0.27 | 10 | A | 1.00 | 28 | A | 1.22 | 32 | A | 0.47 | 12 | A | 1.78 | 47 | A |
| 214567_s_at | XCL2 | Lymphotactin-β | 0.54 | 3 | A | 1.70 | 6 | A | 1.11 | 4 | A | 0.65 | 2 | A | 0.97 | 3 | A |
Norm is normalized data. Raw is raw data (average difference value) of microarray. Fl is flag, which is judged to be “P (Present), M (Marginal) or A (Absent)” by the GeneChip operating software version 1.4.
We further confirmed the effects of DEX and FK506 on the expression of chemokines in mast cells by real-time PCR. Induction of CCL1, CCL3, CCL4 and CCL18 by anti-IgE Ab was significantly and dose-dependently inhibited by FK506 and not by DEX (Fig. 3B, top), whereas induction of CCL2, CCL7, CXCL,3 and CXCL8 by anti-IgE Ab was significantly and dose-dependently inhibited by DEX and not by FK506 (Fig. 3B, bottom). Surprisingly, the induction of CCL3, CCL4, and CCL18 by anti-IgE Ab was significantly up-regulated by DEX (Fig. 3B). Additionally, the induction of CCL7, CXCL3, and CXCL8 by anti-IgE Ab was significantly up-regulated by FK506 (Fig. 3B). The up-regulation of these chemokines by each of these two drugs was completely abrogated when both DEX and FK506 were used in combination (Fig. 3B). These results were further confirmed by measuring the concentration of the chemokine proteins in the culture supernatant. Production of CCL1, CCL3, and CCL4 in response to anti-IgE Ab was significantly inhibited by FK506, whereas the production of CCL3 and CCL4 in response to anti-IgE Ab was significantly enhanced by DEX (Fig. 4A). In contrast, production of CCL2 and CXCL8 in response to anti-IgE Ab was significantly inhibited by DEX but enhanced by FK506 (Fig. 4B). In contrast to the expression profiles of the chemokines, induction of the proinflammatory cytokines M-CSF, GM-CSF, IL-3, and IL-5 and the eicosanoid metabolites PGD2 and LTC4 by FcεRI-dependent stimulation was significantly inhibited by DEX alone or by FK506 alone (Fig. 4C and data not shown). Additionally, histamine release was only inhibited by FK506 but not affected by DEX (Fig. 5), as previously reported (52).
FIGURE 4.
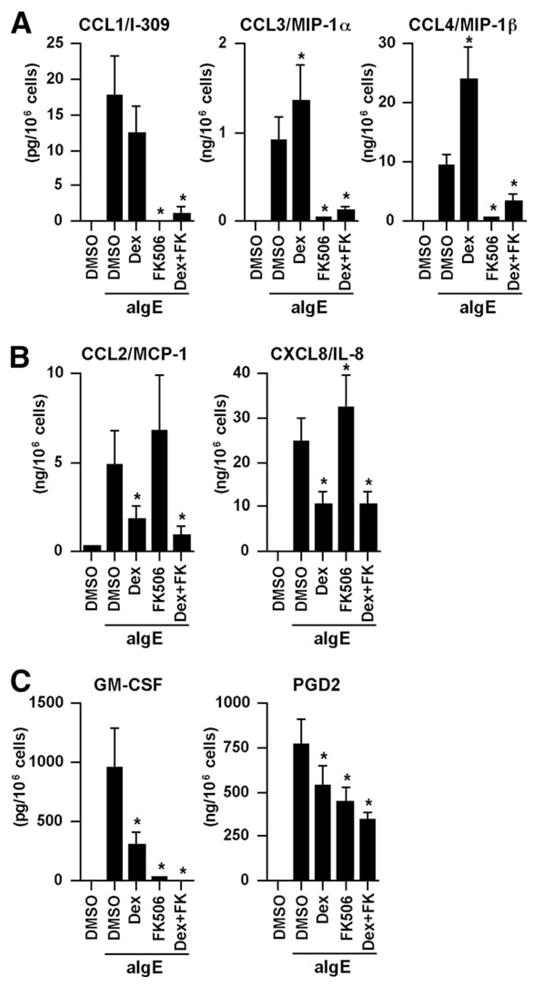
Effect of FK506 and DEX on the production of chemokines and other mediators in human mast cells in response to anti-IgE Ab. IgE-sensitized human mast cells were preincubated with 1 μM DEX, 100 nM FK506, or 0.01% DMSO for 1 h and then stimulated with 1.5 μg/ml anti-IgE Ab for 6 h. Concentrations of the chemokines, GM-CSF, and PGD2 in the culture supernatant were measured by CBA, ELISA, and enzyme immunoassay. The results are shown as the means ± SEM of five independent experiments with independent donors.*, p < 0.05.
FIGURE 5.
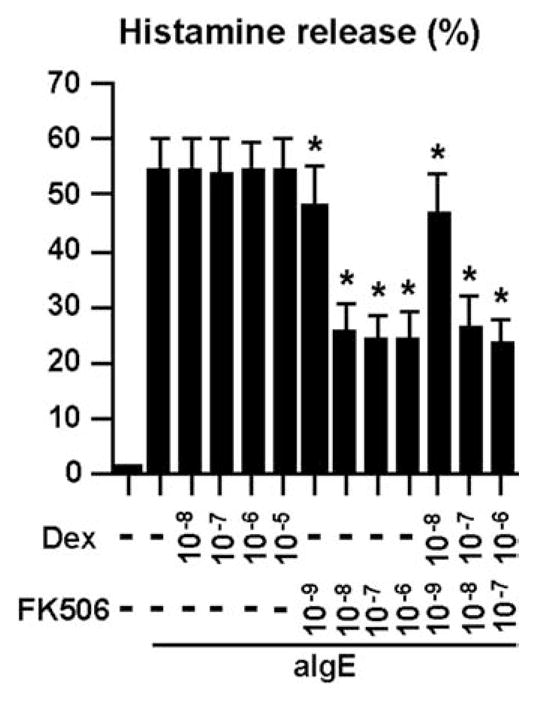
Effect of FK506 and DEX on the degranulation of human mast cells by anti-IgE Ab. IgE-sensitized human mast cells were preincubated with 1 μM DEX, 100 nM FK506, or 0.01% DMSO for 1 h and then stimulated with 1.5 μg/ml anti-IgE Ab for 6 h. Concentrations of histamine in the culture supernatant were measured by ELISA. The results are shown as the means ± SEM of five independent experiments with independent donors.*, p < 0.05.
Effect of DEX and FK506 on the intracellular translocation of NF-κB and NF-AT in mast cells
To clarify the molecular mechanisms by which DEX and FK506 inhibit release of distinct subsets of chemokines from mast cells, we analyzed the intracellular translocation of two transcription factors, NF-κB and NF-AT, after activation via FcεRI in the presence or absence of these immunosuppressants. Using a confocal fluorescence microscope, we found that both NF-κB and NF-AT were located in the cytoplasm of mast cells before stimulation or treatment with vehicle control (Fig. 6A, upper panel, top row); however, 30 min after stimulation with anti-IgE, both NF-κB and NF-AT translocated into the nuclei of the mast cells (Fig. 6A, lower panel, top row). Treatment with DEX or FK506 significantly reduced the number of mast cells with nuclear translocation of NF-κB or NF-AT, respectively (Fig. 6B). Additionally, a combination of DEX and FK506 reduced the number of mast cells with nuclear translocation of both NF-κB and NF-AT (Fig. 6).
FIGURE 6.
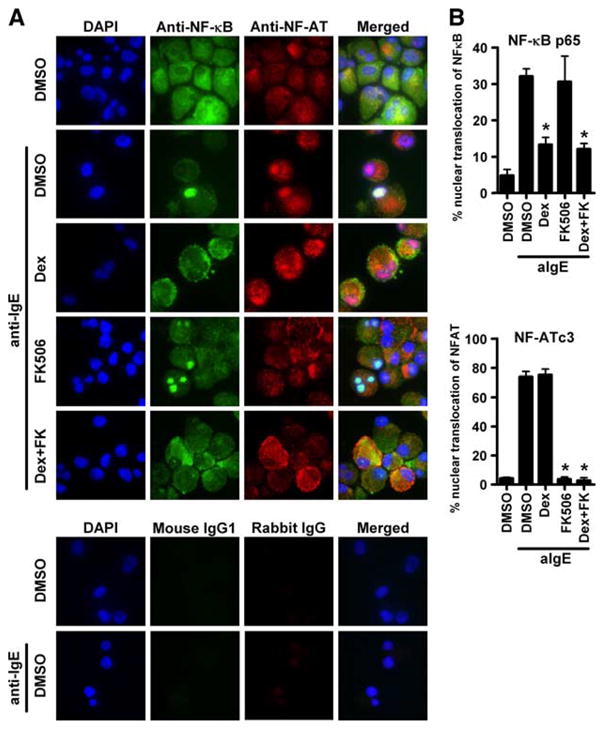
Effect of FK506 and DEX on the translocation of NF-κB and NF-AT by anti-IgE treatment in human mast cells. IgE-sensitized human mast cells were preincubated with either 1 μM DEX, 100 nM FK506, or 0.01% DMSO for 1 h and then stimulated with 1.5 μg/ml anti-IgE Ab for 30 min. A, Immunofluorescence images were showing the distribution of NF-κB and NF-AT. Mast cells were treated with mouse anti-NF-κB p65 and rabbit anti-NF-ATc3 for localization of endogenous NF-κB (green fluorescence) and NF-AT (red fluorescence). Nuclei were counterstained with 4′,6-diamidino-2-phenylindole (DAPI) (blue fluorescence). The images are representative of four independent preparations. B, Summary of the percentage of mast cells in which NF-κB p65 (upper panel) and NF-ATc3 (lower panel) has localized within the nuclei. The results are shown as the means ± SEM of four independent experiments.*, p < 0.05.
Discussion
Chemokines play an important role in the selective recruitment of inflammatory cells and regulate immune responses. Despite the importance of mast cell-derived chemokines in allergic diseases, no studies have comprehensively investigated the effect of corticosteroids and calcineurin inhibitors on the production of FcεRI-mediated chemokines in human mast cells (42). In the present study, we used human peripheral blood progenitor cell-derived cultured mast cells (44) that have been known to express a higher amount of FcεRI and to release higher amounts of histamine and cytokines than do cord blood-derived mast cells upon stimulation of FcεRI (43), and we determined their chemokine expression profiles after cross-linking of cell surface IgE receptors in the presence or absence of DEX or FK506.
In the first series of experiments, the chemokine expression profiles of human mast cells before and after cross-linking of cell-surface IgE receptors were determined with a microarray system. Among the 42 genes for human chemokines measurable by the GeneChip system, 12 genes (14 probes) were found to be expressed in human mast cells (Fig. 1A), and mRNA expression of nine chemokines was found to be up-regulated after stimulation. Some of these results are consistent with those of previous studies (5, 6). Significant induction of mRNA expression of eight genes was confirmed by real-time PCR (Fig. 1B), and the mRNA data were confirmed by measuring chemokine proteins in the supernatant of cultured mast cells (Fig. 2A). The protein levels of all chemokines measured, except CCL18, correlated well with the mRNA levels (Figs. 1B and 2A). Production of CCL18 is discussed below.
In the next series of experiments we assessed the effect of DEX alone and FK506 alone on chemokine mRNA expression by mast cells. A hierarchical clustering analysis of the expression profiles of the genes encoding 11 chemokines revealed three distinct gene clusters based on differences in susceptibility to DEX and FK506 (Fig. 3A and Table II). Expression of the chemokines in the first cluster was inhibited by FK506 and not by DEX, whereas the expression of chemokines in the second cluster was inhibited by DEX and not by FK506. Expression of the chemokines in the third cluster was unaffected by any of the stimuli or drugs tested (Fig. 3A). We then confirmed the GeneChip data by real-time PCR and discovered significant up-regulation of several chemokine genes by these drugs (Fig. 3B). We further confirmed the mRNA data by measuring chemokine proteins in the supernatant of mast cells by ELISA or CBA (Fig. 4). Thus, DEX and FK506 inhibited the expression of some specific chemokines in mast cells after stimulation with anti-IgE Ab.
Unexpectedly, induction of CCL3, CCL4, and CCL18 by anti-IgE Ab was enhanced by DEX, and induction of CCL7, CXCL3, and CXCL8 was enhanced by FK506 (Figs. 3 and 4). The failure of these drugs to inhibit, and tendency to enhance, the release of certain chemokines from mast cells may underlie the pathogenesis of drug-resistant forms of allergic diseases observed clinically (28–32). The clinical phenotypes caused by the unresponsiveness or the overexpression of these chemokines is worthy of future investigation (53).
Several different signal transduction pathways in mast cells are known to be activated upon stimulation of cell-surface FcεRI (54, 55). Cross-linking of FcεRI triggers phosphorylation of several kinases and other signaling molecules, which in turn leads to release of prestored proteins, synthesis of arachidonic acid metabolites, and induction of genes encoding cytokines and chemokines. However, it is still unknown which chemokines are regulated by which individual signal transduction pathway(s) or transcription factor(s) in mast cells.
On the other hand, the mechanisms of the antiinflammatory effects of corticosteroids and calcineurin inhibitors have been well documented. Upon binding by glucocorticoids, the cytoplasmic glucocorticoid receptor (GR) translocates into the nucleus after dissociation of accessory proteins. GR interacts with, and/or inhibits activation of, transcription factors such as NF-κB and AP-1 and thereby represses expression of genes regulated by these transcription factors. GR can also diminish expression of inflammatory genes by accelerating the decay of gene-specific mRNA (56). Additionally, the activated GR forms homodimers, binds to glucocorticoid response elements (GRE), and then activates transcription of several genes that can regulate inflammation, including phosphatases that inhibit signal transduction and IκB (27, 36, 57–59). In sharp contrast, calcineurin inhibitors act by binding to the 12-kDa macrophilin and inhibit the phosphatase activity of calcineurin, thereby blocking translocation of the transcription factor NF-AT into the nucleus. Thus, calcineurin inhibitors mainly repress NF-AT-regulated genes (60, 61).
Our results suggest that the suppression and induction of the chemokines by FK506 or DEX in mast cells is at least in part transcriptional because mRNA levels of these chemokines were significantly altered by these drugs (Fig. 3B). We thus investigated the NF-AT, NF-κB binding sites in the proximal promoter region (up to 2000 bp upstream of the transcription starting point) and GRE in the first intron of the chemokine genes using a directed software (TRANSFAC professional version 8.1; BIOBASE Biological Databases) (62). As a result, multiple NF-AT and NF-κB binding sites were found in the promoter regions of most chemokine genes with very few exceptions (Table III). Additionally, multiple GRE were also found in the first intron of the chemokine genes with very few exceptions (Table III). The presence or absence of these transcription factor binding sites, however, could not explain clearly the increasing or decreasing effects of FK506 and DEX on chemokine expression found in our study.
Table III.
Number of putative NF-AT, NF-κ, and GRE binding sites within proximal 2000 bp of the promoter region and the first intron, and the effect of FK506 and DEX
| No. of Putative Binding Sites |
Effect on FcεRI Signalb |
|||||
|---|---|---|---|---|---|---|
| Gene Name | Ref. Seq.a | NF-ATc | NF-κBc | GREd | FK506 | DEX |
| CCL2 | NM_002982 | 9 (2) | 4 (2) | 4 | ↑ | ↓ |
| CCL7 | NM_006273 | 8 (3) | 3 (1) | 2 | ↑ | ↓ |
| CXCL3 | NM_002090 | 7 (4) | 2 (0) | 0 | ↑ | ↓ |
| CXCL8 | NM_000584 | 5 (1) | 2 (1) | 3 | ↑ | ↓ |
| CCL1 | NM_002981 | 3 (1) | 5 (1) | 7 | ↓ | No effect |
| CCL3 | NM_002983 | 8 (2) | 0 (0) | 2 | ↓ | ↑ |
| CCL4 | NM_002984 | 7 (0) | 6 (3) | 4 | ↓ | ↑ |
| CCL18 | NM_002988 | 6 (3) | 7 (1) | 23 | ↓ | ↑ |
GenBank accession number of the reference sequences (www.ncbi.nlm.nih.gov/Genbank/index.html).
Effect of the drugs on the chemokine mRNA expression after stimulation via FcεRI.
Number of putative NF-AT and NF-κB binding sites within proximal 2000 bp (and within 500 bp) of the promoter region is shown.
Number of putative GRE binding sites in the first intron of the genes is shown.
Therefore, to clarify the molecular mechanisms by which DEX and FK506 inhibit release of distinct subsets of chemokines from mast cells, we analyzed the translocation of two transcription factors, NF-κB and NF-AT, after activation via FcεRI in the presence or absence of these immunosuppressants. Using a confocal fluorescence microscope and specific Abs against NF-κB and NF-AT, we found that treatment with DEX or FK506 significantly inhibited the nuclear translocation of NF-κB or NF-AT, respectively (Fig. 6). Additionally, a combination of DEX and FK506 inhibited nuclear translocation of both NF-κB and NF-AT. Thus, we concluded that the inhibitory effect of DEX and FK506 is caused at least in part by the inhibition of intracellular signal transduction pathways involving NF-κB and NF-AT, respectively.
Importantly, the combination of a corticosteroid and calcineurin inhibitor almost completely abolished the induction of chemokine gene expression in mast cells by FcεRI cross-linking, even though expression of some of them were up-regulated by one of these drugs alone (Figs. 3, A and B, and 4, A and B). Additionally, the combination of DEX and FK506 additively suppressed expression of other inflammatory mediators, including PGD2 and GM-CSF, which are critical to the pathogenesis of inflammatory diseases (Fig. 4C). These findings strongly suggest the superiority of a combination therapy of a corticosteroid and a calcineurin inhibitor over monotherapy (35) or sequential therapy with these drugs (63).
Mast cell granular proteins have recently been shown to exhibit strong protease activity that is capable of cleaving several cytokines (51) and chemokines, including CCL5 (RANTES) and CCL11 (eotaxin), but not CXCL8 (50). This finding suggests that mast cell proteases may also cleave other chemokines. Our data showed high levels of mRNA for CCL18 (Fig. 1, A and B), whereas the concentration of CCL18 protein in the culture supernatant was almost below the detection limit (Fig. 2B). In the presence of the protease inhibitor cocktail, mast cells were demonstrated to produce and release CCL18, suggesting that CCL18 may ordinarily be degraded by endogenous proteases.
Our data clearly showed that several other CC chemokines, CCL1, CCL2, CCL3 and CCL4, in addition to CCL18, were also likely to be cleaved by mast cell protease (Fig. 2B). In the presence of a protease inhibitor cocktail, we observed a 9- to 85-fold increase in the concentration of these CC chemokines in the mast cell supernatant. This finding indicates that mast cell proteases may regulate inflammatory cell recruitment by limiting local levels of some chemokines. Upon stimulation with Th2 cytokines, bronchial epithelial cells have been reported to produce a large amount of serine protease inhibitors (64) that are capable of inhibiting the protease activity of a major mite allergen, Der p 1 (65). If such protease inhibitors from epithelial cells are also capable of inhibiting mast cell proteases, the concentrations of these CC chemokines in tissue would dramatically increase, and these chemokines may play a critical role in the pathogenesis of allergic diseases. Pang et al. found that purified human tryptase and chymase failed to degrade CCL2, suggesting that other protease(s) released by mast cells may be involved in the cleavage of CCL2 (50). Further study is needed to identify the proteases involved in the degradation of mast cell-derived CC chemokines. In sharp contrast to CC chemokines, the protein levels of CXCL8 were elevated by stimulation via FcεRI and were unchanged by the presence of PIC (Fig. 2B). This observation confirmed a previous study (50), but it remains unknown whether other CXC chemokines are resistant to the mast cell proteases or not.
In conclusion, mast cells produce several chemokines upon stimulation of the cell surface IgE receptor and putative mast cell proteases were found to diminish the levels of some chemokines. The chemokines produced by mast cells can be classified into three groups based on differences in transcriptional regulation (NF-κB and NF-AT) and susceptibility to DEX and FK506.
Acknowledgments
We thank Noriko Hashimoto and Yuri Nakamura (National Research Institute for Child Health and Development) for their skillful technical assistance. We also thank Dr. Joan Cook-Mills (Northwestern University Feinberg School of Medicine) for helpful advice in immunofluorescence staining.
Footnotes
Abbreviations used in this paper: PG, prostaglandin; CBA, cytometric bead array; DEX, dexamethasone; FK506, tacrolimus; GR, glucocorticoid receptor; GRE, glucocorticoid response element; LT, leukotriene; PIC, protease inhibitor cocktail; SCF, stem cell factor.
Disclosures
The authors have no financial conflicts of interest.
This work was supported in part by grants from the National Institute of Biomedical Innovation (ID05-24 and ID05-41), the Japan Health Science Foundation (KH51046), and the National Institutes of Health (R01 HL068546).
References
- 1.Galli SJ, Kalesnikoff J, Grimbaldeston MA, Piliponsky AM, Williams CM, Tsai M. Mast cells as “tunable” effector and immunoregulatory cells: recent advances. Annu Rev Immunol. 2005;23:749–786. doi: 10.1146/annurev.immunol.21.120601.141025. [DOI] [PubMed] [Google Scholar]
- 2.Kawakami T, Galli SJ. Regulation of mast-cell and basophil function and survival by IgE. Nat Rev Immunol. 2002;2:773–786. doi: 10.1038/nri914. [DOI] [PubMed] [Google Scholar]
- 3.Ono SJ, Nakamura T, Miyazaki D, Ohbayashi M, Dawson M, Toda M. Chemokines: roles in leukocyte development, trafficking, and effector function. J Allergy Clin Immunol. 2003;111:1185–1199. doi: 10.1067/mai.2003.1594. [DOI] [PubMed] [Google Scholar]
- 4.Zlotnik A, Yoshie O. Chemokines: a new classification system and their role in immunity. Immunity. 2000;12:121–127. doi: 10.1016/s1074-7613(00)80165-x. [DOI] [PubMed] [Google Scholar]
- 5.Wakahara S, Fujii Y, Nakao T, Tsuritani K, Hara T, Saito H, Ra C. Gene expression profiles for FcεRI, cytokines and chemokines upon FcεRI activation in human cultured mast cells derived from peripheral blood. Cytokine. 2001;16:143–152. doi: 10.1006/cyto.2001.0958. [DOI] [PubMed] [Google Scholar]
- 6.Nakajima T, Inagaki N, Tanaka H, Tanaka A, Yoshikawa M, Tamari M, Hasegawa K, Matsumoto K, Tachimoto H, Ebisawa M, et al. Marked increase in CC chemokine gene expression in both human and mouse mast cell transcriptomes following Fcε receptor I cross-linking: an interspecies comparison. Blood. 2002;100:3861–3868. doi: 10.1182/blood-2002-02-0602. [DOI] [PubMed] [Google Scholar]
- 7.Homey B, Steinhoff M, Ruzicka T, Leung DY. Cytokines and chemokines orchestrate atopic skin inflammation. J Allergy Clin Immunol. 2006;118:178–189. doi: 10.1016/j.jaci.2006.03.047. [DOI] [PubMed] [Google Scholar]
- 8.Worldwide variation in prevalence of symptoms of asthma, allergic rhinoconjunctivitis, and atopic eczema: ISAAC. The International Study of Asthma and Allergies in Childhood (ISAAC) Steering Committee. Lancet. 1998;351:1225–1232. [PubMed] [Google Scholar]
- 9.Pearce N, Ait-Khaled N, Beasley R, Mallol J, Keil U, Mitchell E, Robertson C. Worldwide trends in the prevalence of asthma symptoms: phase III of the International Study of Asthma and Allergies in Childhood (ISAAC) Thorax. 2007;62:758–766. doi: 10.1136/thx.2006.070169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weinmayr G, Weiland SK, Bjorksten B, Brunekreef B, Buchele G, Cookson WO, Garcia-Marcos L, Gotua M, Gratziou C, van Hage M, et al. Atopic sensitization and the international variation of asthma symptom prevalence in children. Am J Respir Crit Care Med. 2007;176:565–574. doi: 10.1164/rccm.200607-994OC. [DOI] [PubMed] [Google Scholar]
- 11.Leung DY, Harbeck R, Bina P, Reiser RF, Yang E, Norris DA, Hanifin JM, Sampson HA. Presence of IgE antibodies to staphylococcal exotoxins on the skin of patients with atopic dermatitis: evidence for a new group of allergens. J Clin Invest. 1993;92:1374–1380. doi: 10.1172/JCI116711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brightling CE, Bradding P, Symon FA, Holgate ST, Wardlaw AJ, Pavord ID. Mast-cell infiltration of airway smooth muscle in asthma. N Engl J Med. 2002;346:1699–1705. doi: 10.1056/NEJMoa012705. [DOI] [PubMed] [Google Scholar]
- 13.Carroll NG, Mutavdzic S, James AL. Distribution and degranulation of airway mast cells in normal and asthmatic subjects. Eur Respir J. 2002;19:879–885. doi: 10.1183/09031936.02.00275802. [DOI] [PubMed] [Google Scholar]
- 14.Smit JJ, Lukacs NW. A closer look at chemokines and their role in asthmatic responses. Eur J Pharmacol. 2006;533:277–288. doi: 10.1016/j.ejphar.2005.12.064. [DOI] [PubMed] [Google Scholar]
- 15.Medina-Tato DA, Watson ML, Ward SG. Leukocyte navigation mechanisms as targets in airway diseases. Drug Discov Today. 2006;11:866–879. doi: 10.1016/j.drudis.2006.08.008. [DOI] [PubMed] [Google Scholar]
- 16.Kaburagi Y, Shimada Y, Nagaoka T, Hasegawa M, Takehara K, Sato S. Enhanced production of CC-chemokines (RANTES, MCP-1, MIP-1α, MIP-1β, and eotaxin) in patients with atopic dermatitis. Arch Dermatol Res. 2001;293:350–355. doi: 10.1007/s004030100230. [DOI] [PubMed] [Google Scholar]
- 17.Taha RA, Minshall EM, Leung DY, Boguniewicz M, Luster A, Muro S, Toda M, Hamid QA. Evidence for increased expression of eotaxin and monocyte chemotactic protein-4 in atopic dermatitis. J Allergy Clin Immunol. 2000;105:1002–1007. doi: 10.1067/mai.2000.106483. [DOI] [PubMed] [Google Scholar]
- 18.Fujisawa T, Fujisawa R, Kato Y, Nakayama T, Morita A, Katsumata H, Nishimori H, Iguchi K, Kamiya H, Gray PW, et al. Presence of high contents of thymus and activation-regulated chemokine in platelets and elevated plasma levels of thymus and activation-regulated chemokine and macrophage-derived chemokine in patients with atopic dermatitis. J Allergy Clin Immunol. 2002;110:139–146. doi: 10.1067/mai.2002.126079. [DOI] [PubMed] [Google Scholar]
- 19.Nomura I, Gao B, Boguniewicz M, Darst MA, Travers JB, Leung DY. Distinct patterns of gene expression in the skin lesions of atopic dermatitis and psoriasis: a gene microarray analysis. J Allergy Clin Immunol. 2003;112:1195–1202. doi: 10.1016/j.jaci.2003.08.049. [DOI] [PubMed] [Google Scholar]
- 20.Gombert M, Dieu-Nosjean MC, Winterberg F, Bunemann E, Kubitza RC, Da Cunha L, Haahtela A, Lehtimaki S, Muller A, Rieker J, et al. CCL1-CCR8 interactions: an axis mediating the recruitment of T cells and Langerhans-type dendritic cells to sites of atopic skin inflammation. J Immunol. 2005;174:5082–5091. doi: 10.4049/jimmunol.174.8.5082. [DOI] [PubMed] [Google Scholar]
- 21.Pivarcsi A, Gombert M, Dieu-Nosjean MC, Lauerma A, Kubitza R, Meller S, Rieker J, Muller A, Da Cunha L, Haahtela A, et al. CC chemokine ligand 18, an atopic dermatitis-associated and dendritic cell-derived chemokine, is regulated by staphylococcal products and allergen exposure. J Immunol. 2004;173:5810–5817. doi: 10.4049/jimmunol.173.9.5810. [DOI] [PubMed] [Google Scholar]
- 22.Zou J, Young S, Zhu F, Gheyas F, Skeans S, Wan Y, Wang L, Ding W, Billah M, McClanahan T, et al. Microarray profile of differentially expressed genes in a monkey model of allergic asthma. Genome Biol. 2002;3:research0020. doi: 10.1186/gb-2002-3-5-research0020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gunther C, Bello-Fernandez C, Kopp T, Kund J, Carballido-Perrig N, Hinteregger S, Fassl S, Schwarzler C, Lametschwandtner G, Stingl G, et al. CCL18 is expressed in atopic dermatitis and mediates skin homing of human memory T cells. J Immunol. 2005;174:1723–1728. doi: 10.4049/jimmunol.174.3.1723. [DOI] [PubMed] [Google Scholar]
- 24.Fahy JV, Fleming HE, Wong HH, Liu JT, Su JQ, Reimann J, Fick RB, Jr, Boushey HA. The effect of an anti-IgE monoclonal antibody on the early- and late-phase responses to allergen inhalation in asthmatic subjects. Am J Respir Crit Care Med. 1997;155:1828–1834. doi: 10.1164/ajrccm.155.6.9196082. [DOI] [PubMed] [Google Scholar]
- 25.Busse WW. National Asthma Education and Prevention Program Expert Panel Report 3: Guidelines for the Diagnosis and Management of Asthma, Summary Report 2007. J Allergy Clin Immunol. 2007;120:S94–S138. doi: 10.1016/j.jaci.2007.09.043. [DOI] [PubMed] [Google Scholar]
- 26.Ellis C, Luger T, Abeck D, Allen R, Graham-Brown RA, De Prost Y, Eichenfield LF, Ferrandiz C, Giannetti A, Hanifin J, et al. International Consensus Conference on Atopic Dermatitis II (ICCAD II): clinical update and current treatment strategies. Br J Dermatol. 2003;148(Suppl 63):3–10. doi: 10.1046/j.1365-2133.148.s63.1.x. [DOI] [PubMed] [Google Scholar]
- 27.Rhen T, Cidlowski JA. Antiinflammatory action of glucocorticoids: new mechanisms for old drugs. N Engl J Med. 2005;353:1711–1723. doi: 10.1056/NEJMra050541. [DOI] [PubMed] [Google Scholar]
- 28.Horne R. Compliance, adherence, and concordance: implications for asthma treatment. Chest. 2006;130:65S–72S. doi: 10.1378/chest.130.1_suppl.65S. [DOI] [PubMed] [Google Scholar]
- 29.Boguniewicz M, Eichenfield LF, Hultsch T. Current management of atopic dermatitis and interruption of the atopic march. J Allergy Clin Immunol. 2003;112:S140–S150. doi: 10.1016/j.jaci.2003.09.031. [DOI] [PubMed] [Google Scholar]
- 30.Ito K, Chung KF, Adcock IM. Update on glucocorticoid action and resistance. J Allergy Clin Immunol. 2006;117:522–543. doi: 10.1016/j.jaci.2006.01.032. [DOI] [PubMed] [Google Scholar]
- 31.Clayton MH, Leung DY, Surs W, Szefler SJ. Altered glucocorticoid receptor binding in atopic dermatitis. J Allergy Clin Immunol. 1995;96:421–423. doi: 10.1016/s0091-6749(95)70062-5. [DOI] [PubMed] [Google Scholar]
- 32.Hauk PJ, Hamid QA, Chrousos GP, Leung DY. Induction of corticosteroid insensitivity in human PBMCs by microbial superantigens. J Allergy Clin Immunol. 2000;105:782–787. doi: 10.1067/mai.2000.105807. [DOI] [PubMed] [Google Scholar]
- 33.Okumura S, Sagara H, Fukuda T, Saito H, Okayama Y. FcεRI-mediated amphiregulin production by human mast cells increases mucin gene expression in epithelial cells. J Allergy Clin Immunol. 2005;115:272–279. doi: 10.1016/j.jaci.2004.10.004. [DOI] [PubMed] [Google Scholar]
- 34.Matsuda A, Fukuda S, Matsumoto K, Saito H. Th1/Th2 Cytokines reciprocally regulate in vitro pulmonary angiogenesis via CXC chemokine synthesis. Am J Respir Cell Mol Biol. 2007;38:168–175. doi: 10.1165/rcmb.2007-0162OC. [DOI] [PubMed] [Google Scholar]
- 35.Ashcroft DM, Dimmock P, Garside R, Stein K, Williams HC. Efficacy and tolerability of topical pimecrolimus and tacrolimus in the treatment of atopic dermatitis: meta-analysis of randomised controlled trials. Br Med J. 2005;330:516. doi: 10.1136/bmj.38376.439653.D3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pratt WB, Toft DO. Steroid receptor interactions with heat shock protein and immunophilin chaperones. Endocr Rev. 1997;18:306–360. doi: 10.1210/edrv.18.3.0303. [DOI] [PubMed] [Google Scholar]
- 37.Niven AS, Argyros G. Alternate treatments in asthma. Chest. 2003;123:1254–1265. doi: 10.1378/chest.123.4.1254. [DOI] [PubMed] [Google Scholar]
- 38.Schleimer RP, Schulman ES, MacGlashan DW, Jr, Peters SP, Hayes EC, Adams GK, 3rd, Lichtenstein LM, Adkinson NF., Jr Effects of dexamethasone on mediator release from human lung fragments and purified human lung mast cells. J Clin Invest. 1983;71:1830–1835. doi: 10.1172/JCI110938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Smith SJ, Piliponsky AM, Rosenhead F, Elchalal U, Nagler A, Levi-Schaffer F. Dexamethasone inhibits maturation, cytokine production and FcεRI expression of human cord blood-derived mast cells. Clin Exp Allergy. 2002;32:906–913. doi: 10.1046/j.1365-2745.2002.01418.x. [DOI] [PubMed] [Google Scholar]
- 40.de Paulis A, Stellato C, Cirillo R, Ciccarelli A, Oriente A, Marone G. Anti-inflammatory effect of FK-506 on human skin mast cells. J Invest Dermatol. 1992;99:723–728. doi: 10.1111/1523-1747.ep12614216. [DOI] [PubMed] [Google Scholar]
- 41.Zuberbier T, Chong SU, Grunow K, Guhl S, Welker P, Grassberger M, Henz BM. The ascomycin macrolactam pimecrolimus (Elidel, SDZ ASM 981) is a potent inhibitor of mediator release from human dermal mast cells and peripheral blood basophils. J Allergy Clin Immunol. 2001;108:275–280. doi: 10.1067/mai.2001.116865. [DOI] [PubMed] [Google Scholar]
- 42.Holm M, Kvistgaard H, Dahl C, Andersen HB, Hansen TK, Schiotz PO, Junker S. Modulation of chemokine gene expression in CD133 cord blood-derived human mast cells by cyclosporin A and dexamethasone. Scand J Immunol. 2006;64:571–579. doi: 10.1111/j.1365-3083.2006.01835.x. [DOI] [PubMed] [Google Scholar]
- 43.Iida M, Matsumoto K, Tomita H, Nakajima T, Akasawa A, Ohtani NY, Yoshida NL, Matsui K, Nakada A, Sugita Y, et al. Selective down-regulation of high-affinity IgE receptor (FcεRI) α-chain messenger RNA among transcriptome in cord blood-derived versus adult peripheral blood-derived cultured human mast cells. Blood. 2001;97:1016–1022. doi: 10.1182/blood.v97.4.1016. [DOI] [PubMed] [Google Scholar]
- 44.Saito H, Kato A, Matsumoto K, Okayama Y. Culture of human mast cells from peripheral blood progenitors. Nat Protocol. 2006;1:2178–2183. doi: 10.1038/nprot.2006.344. [DOI] [PubMed] [Google Scholar]
- 45.Nomura I, Katsunuma T, Matsumoto K, Iida M, Tomita H, Tomikawa M, Kawahara H, Akasawa A, Pawankar R, Saito H. Human mast cell progenitors in peripheral blood from atopic subjects with high IgE levels. Clin Exp Allergy. 2001;31:1424–1431. doi: 10.1046/j.1365-2222.2001.01181.x. [DOI] [PubMed] [Google Scholar]
- 46.Kato A, Homma T, Batchelor J, Hashimoto N, Imai S, Wakiguchi H, Saito H, Matsumoto K. Interferon-α/β receptor-mediated selective induction of a gene cluster by CpG oligodeoxynucleotide 2006. BMC Immunol. 2003;4:8. doi: 10.1186/1471-2172-4-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kato A, Ogasawara T, Homma T, Saito H, Matsumoto K. Lipopolysaccharide-binding protein critically regulates lipopolysaccharide-induced IFN-β signaling pathway in human monocytes. J Immunol. 2004;172:6185–6194. doi: 10.4049/jimmunol.172.10.6185. [DOI] [PubMed] [Google Scholar]
- 48.Williams HC. Clinical practice: atopic dermatitis. N Engl J Med. 2005;352:2314–2324. doi: 10.1056/NEJMcp042803. [DOI] [PubMed] [Google Scholar]
- 49.Leung DY, Boguniewicz M, Howell MD, Nomura I, Hamid QA. New insights into atopic dermatitis. J Clin Invest. 2004;113:651–657. doi: 10.1172/JCI21060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pang L, Nie M, Corbett L, Sutcliffe A, Knox AJ. Mast cell β-tryptase selectively cleaves eotaxin and RANTES and abrogates their eosinophil chemotactic activities. J Immunol. 2006;176:3788–3795. doi: 10.4049/jimmunol.176.6.3788. [DOI] [PubMed] [Google Scholar]
- 51.Zhao W, Oskeritzian CA, Pozez AL, Schwartz LB. Cytokine production by skin-derived mast cells: endogenous proteases are responsible for degradation of cytokines. J Immunol. 2005;175:2635–2642. doi: 10.4049/jimmunol.175.4.2635. [DOI] [PubMed] [Google Scholar]
- 52.Sengoku T, Kishi S, Sakuma S, Ohkubo Y, Goto T. FK506 inhibition of histamine release and cytokine production by mast cells and basophils. Int J Immunopharmacol. 2000;22:189–201. doi: 10.1016/s0192-0561(99)00076-4. [DOI] [PubMed] [Google Scholar]
- 53.Caproni M, Torchia D, Antiga E, Terranova M, Volpi W, del Bianco E, D’Agata A, Fabbri P. The comparative effects of tacrolimus and hydrocortisone in adult atopic dermatitis: an immunohistochemical study. Br J Dermatol. 2007;156:312–319. doi: 10.1111/j.1365-2133.2006.07609.x. [DOI] [PubMed] [Google Scholar]
- 54.Siraganian RP. Mast cell signal transduction from the high-affinity IgE receptor. Curr Opin Immunol. 2003;15:639–646. doi: 10.1016/j.coi.2003.09.010. [DOI] [PubMed] [Google Scholar]
- 55.Rivera J, Cordero JR, Furumoto Y, Luciano-Montalvo C, Gonzalez-Espinosa C, Kovarova M, Odom S, Parravicini V. Macromolecular protein signaling complexes and mast cell responses: a view of the organization of IgE-dependent mast cell signaling. Mol Immunol. 2002;38:1253–1258. doi: 10.1016/s0161-5890(02)00072-x. [DOI] [PubMed] [Google Scholar]
- 56.Stellato C, Matsukura S, Fal A, White J, Beck LA, Proud D, Schleimer RP. Differential regulation of epithelial-derived C-C chemokine expression by IL-4 and the glucocorticoid budesonide. J Immunol. 1999;163:5624–5632. [PubMed] [Google Scholar]
- 57.Poon M, Liu B, Taubman MB. Identification of a novel dexamethasone-sensitive RNA-destabilizing region on rat monocyte chemoattractant protein 1 mRNA. Mol Cell Biol. 1999;19:6471–6478. doi: 10.1128/mcb.19.10.6471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tobler A, Meier R, Seitz M, Dewald B, Baggiolini M, Fey MF. Glucocorticoids downregulate gene expression of GM-CSF, NAP-1/IL-8, and IL-6, but not of M-CSF in human fibroblasts. Blood. 1992;79:45–51. [PubMed] [Google Scholar]
- 59.Scheinman RI, Cogswell PC, Lofquist AK, Baldwin AS., Jr Role of transcriptional activation of IκB alpha in mediation of immunosuppression by glucocorticoids. Science. 1995;270:283–286. doi: 10.1126/science.270.5234.283. [DOI] [PubMed] [Google Scholar]
- 60.Rao A, Luo C, Hogan PG. Transcription factors of the NFAT family: regulation and function. Annu Rev Immunol. 1997;15:707–747. doi: 10.1146/annurev.immunol.15.1.707. [DOI] [PubMed] [Google Scholar]
- 61.Martinez-Martinez S, Redondo JM. Inhibitors of the calcineurin/NFAT pathway. Curr Med Chem. 2004;11:997–1007. doi: 10.2174/0929867043455576. [DOI] [PubMed] [Google Scholar]
- 62.Kel A, Voss N, Jauregui R, Kel-Margoulis O, Wingender E. Beyond microarrays: finding key transcription factors controlling signal transduction pathways. BMC Bioinformatics. 2006;7(Suppl 2):S13. doi: 10.1186/1471-2105-7-S2-S13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nakahara T, Koga T, Fukagawa S, Uchi H, Furue M. Intermittent topical corticosteroid/tacrolimus sequential therapy improves lichenification and chronic papules more efficiently than intermittent topical corticosteroid/emollient sequential therapy in patients with atopic dermatitis. J Dermatol. 2004;31:524–528. doi: 10.1111/j.1346-8138.2004.tb00548.x. [DOI] [PubMed] [Google Scholar]
- 64.Yuyama N, Davies DE, Akaiwa M, Matsui K, Hamasaki Y, Suminami Y, Yoshida NL, Maeda M, Pandit A, Lordan JL, et al. Analysis of novel disease-related genes in bronchial asthma. Cytokine. 2002;19:287–296. doi: 10.1006/cyto.2002.1972. [DOI] [PubMed] [Google Scholar]
- 65.Sakata Y, Arima K, Takai T, Sakurai W, Masumoto K, Yuyama N, Suminami Y, Kishi F, Yamashita T, Kato T, et al. The squamous cell carcinoma antigen 2 inhibits the cysteine proteinase activity of a major mite allergen, Der p 1. J Biol Chem. 2004;279:5081–5087. doi: 10.1074/jbc.M311585200. [DOI] [PubMed] [Google Scholar]