Abstract
Background
We previously identified ebpR, encoding a potential member of the AtxA/Mga transcriptional regulator family, and showed that it is important for transcriptional activation of the Enterococcus faecalis endocarditis and biofilm associated pilus operon, ebpABC. Although ebpR is not absolutely essential for ebpABC expression (100-fold reduction), its deletion led to phenotypes similar to those of an ebpABC mutant such as absence of pili at the cell surface and, consequently, reduced biofilm formation. A non-piliated ebpABC mutant has been shown to be attenuated in a rat model of endocarditis and in a murine urinary tract infection model, indicating an important participation of the ebpR-ebpABC locus in virulence. However, there is no report relating to the environmental conditions that affect expression of the ebpR-ebpABC locus.
Results
In this study, we examined the effect of CO2/HCO3-, pH, and the Fsr system on the ebpR-ebpABC locus expression. The presence of 5% CO2/0.1 M HCO3- increased ebpR-ebpABC expression, while the Fsr system was confirmed to be a weak repressor of this locus. The mechanism by which the Fsr system repressed the ebpR-ebpABC locus expression appears independent of the effects of CO2- bicarbonate. Furthermore, by using an ebpA::lacZ fusion as a reporter, we showed that addition of 0.1 M sodium bicarbonate to TSBG (buffered at pH 7.5), but not the presence of 5% CO2, induced ebpA expression in TSBG broth. In addition, using microarray analysis, we found 73 genes affected by the presence of sodium bicarbonate (abs(fold) > 2, P < 0.05), the majority of which belong to the PTS system and ABC transporter families. Finally, pilus production correlated with ebpA mRNA levels under the conditions tested.
Conclusions
This study reports that the ebp locus expression is enhanced by the presence of bicarbonate with a consequential increase in the number of cells producing pili. Although the molecular basis of the bicarbonate effect remains unclear, the pathway is independent of the Fsr system. In conclusion, E. faecalis joins the growing family of pathogens that regulates virulence gene expression in response to bicarbonate and/or CO2.
Background
Enterococci are part of the normal flora in human intestines and are also a leading cause of nosocomial infections [1,2]. These organisms are somehow able to migrate from the gastrointestinal tract into the bloodstream and cause systemic infections such as bacteremia and even endocarditis [2-4]. Although many strains of enterococci seem to be harmless commensals, particular subgroups of Enterococcus faecalis and Enterococcus faecium predominate among isolates from nosocomial enterococcal infections. In E. faecalis, numerous factors important for virulence have been characterized. For example, the Fsr system, a homologue of the staphylococcal Agr system, has been shown to be important for virulence due, at least in part, to its control of gelatinase and a serine protease expression via a quorum-sensing mechanism [5-7]. Microarray studies also indicated that the Fsr system regulates other genes important for virulence [8], one of which is the locus encoding Ebp pili [8], whose subunits are encoded by the ebp operon [9]. A non-piliated ebp mutant, producing much less biofilm than the parent strain, was shown to be attenuated in a rat model of endocarditis [9] and in a murine urinary tract infection model [10]. We previously described EbpR as an important activator of the ebpABC operon encoding the pili in E. faecalis OG1RF [11]. Although ebpR is not essential for ebpABC expression, we detected 100-fold less ebpABC mRNA in a ΔebpR mutant compared to the OG1RF parent strain. In addition, even in the presence of an intact ebpR gene, only 5-20% of the cells, grown aerobically in BHI or in TSBG, were found to produce pili (detected by electron microscopy or immunofluorescence) [9,11]. These results imply that other regulatory and/or environmental factors may affect pilus production.
Bicarbonate is a major element of the mammalian body for reaching and maintaining homeostasis. In equilibrium with CO2, H2CO2 and CO32-, depending on pH, temperature, and CO2 pressure, bicarbonate does not diffuse freely across the membrane and needs specific transporters [12]. In the stomach, HCO3- is secreted by the surface mucus cells, where it gets trapped in the mucus and forms part of the mucus-HCO3- barrier, thereby maintaining a pH gradient of pH 2 in the lumen to pH 7 at the mucosal epithelium interface. Interestingly, some microbial pathogens have been shown to respond in vivo to CO2 (from 5 to 20%) and/or HCO3- (10-100 mM) by enhancing production of factors important for virulence (Staphyloccocus aureus [13], Vibrio cholerae [14], group A streptococcus [15], Bacillus anthracis [16,17], Cryptococcus neoformans [18] and Citrobacter rodentium [19]). Regulatory proteins have been described which mediate the CO2/HCO3- response at the transcriptional level in B. anthracis (AtxA-like proteins [20]), in Group A streptococci (Mga [21]) and, recently, in C. rodentium with RegA [19]. For E. faecalis, except for a report showing an increase in cytolysin expression when grown in 80% H2-20% CO2 [22], we could find no other report of a CO2/HCO3- effect on known virulence-associated genes. A candidate for such study is the ebpABC operon and its regulator, ebpR, a gene encoding a transcriptional regulator affiliated with the AtxA/Mga family; as mentioned above, this family is known to have its regulon activated in response to elevated CO2 [15,23].
In the present study, we report the identification of environmental conditions affecting the expression of the ebpR-ebpABC locus and, consequently, pilus production. In addition, we found that Fsr repressed the ebpR-ebpABC locus in all conditions tested, independent of the CO2/bicarbonate effect. Finally, among the dozens of genes that are differentially expressed after being exposed to bicarbonate, the majority belong to the PTS system and ABC transporter families.
Results
ebpR and ebpA expression profiles when grown aerobically in TSBG
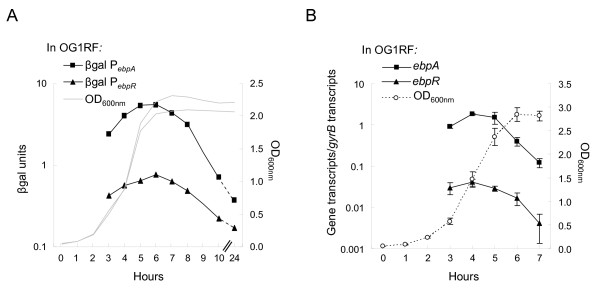
We previously identified an E. faecalis transcriptional regulator, EbpR, which positively affects the expression of the endocarditis and biofilm-associated pilus operon, ebpABC [11]. To further explore ebpR and ebpABC expression profiles, we created lacZ fusions with the ebpR and ebpA promoters (PebpR::lacZ and PebpA::lacZ). We first tested the time course of expression of ebpR and ebpA in OG1RF grown aerobically in TSBG (our standard biofilm medium) from mid-log growth phase to late stationary. In these conditions, each fusion showed the same general dome-shape pattern that reached a peak between 5 and 6 hr (Fig. 1A); specifically, the β-gal units for OG1RF carrying the ebpA promoter were 2.4, 5.4, and 0.4 at mid-log (3 hr after starting the culture), entry into stationary (5 hr) and late stationary growth phase (24 hr), respectively, while the ebpR fusion generated consistently lower β-gal units than the ebpA fusion.
Figure 1.
ebpR and ebpA expression profiles in OG1RF. A. Expression levels of ebpA and ebpR using gene promoter::lacZ fusions. OG1RF containing either PebpR::lacZ (black triangle) or PebpA::lacZ (black square) were grown in TSBG. For β-gal assays, samples were collected every hour from 3 to 8 hr, then at 10 and 24 hr after starting the culture (x axis). The left axis represents the β-gal units (OD420 nm/protein concentration in mg/ml). The right axis indicates the OD600 nm readings. All sets of cultures presented were analyzed concurrently. This figure is a representative of at least three independent experiments. B. qRT-PCR with RNA purified from OG1RF cultures grown aerobically in TSBG. The left axis represents the level of transcript normalized to gyrB transcript level. The right axis indicates the OD600 nm readings. The dashed line shows the mean (with standard deviation) of 5 independent cultures of OG1RF grown in TSBG. The transcript levels of ebpR (black triangle) and ebpA (black square) shown represent two different data sets, each tested in duplicate that were normalized using gyrB transcript levels.
Since β-galactosidase assays reflect translation as well as transcription, we also directly explored the steady-state mRNA levels of transcripts of ebpR and ebpA with qRT-PCR in the same conditions used above (TSBG, aerobically) compared to the housekeeping gene gyrB. At the peak of ebpR expression, which occurred between mid- and late log phase growth, the ratio between ebpR and gyrB transcript levels was 0.04 (Fig. 1B). After entry into stationary phase, ebpR expression decreased to an ebpR/gyrB ratio of 0.004 representing a 10-fold decrease when compared to late log growth phase levels. Likewise, ebpA expression also peaked at the late log growth phase with an ebpA/gyrB ratio of 1.5 and decreased to a ratio ebpA/gyrB of 0.12 (also a 10-fold reduction when compared to ebpA expression level at late log growth phase). The ebpA steady-state mRNA levels were an average of 37-fold higher than ebpR steady-state mRNA levels. Overall, the patterns between qRT-PCR and the β-gal assays were similar except for a one-hour delay for peak expression in the β-gal assays, probably due to a delay between transcription and translation.
The CO2-NaHCO3 induction effect on ebpR and ebpA expression
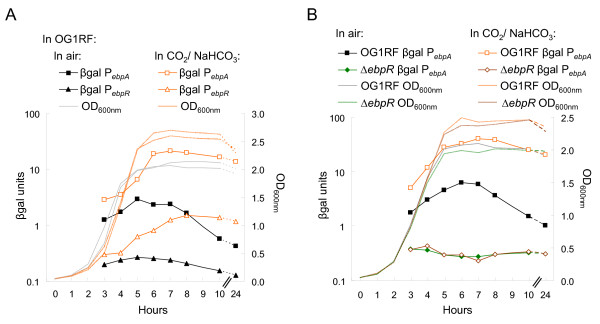
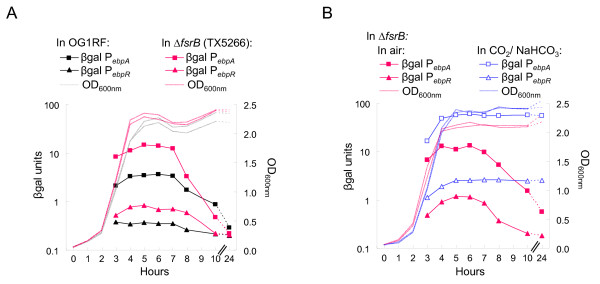
As we previously noted [11], EbpR shares some homology with transcriptional regulators of the AtxA/Mga family. In this family, it has been shown that AtxA and Mga activate their regulon from mid-log to entry into stationary phase and that their regulon is affected by the presence of 5% CO2/0.1 M NaHCO3 [15,23]. We therefore tested the effect of CO2/NaHCO3 on ebpR and ebpA expression during growth using the PebpR:: and PebpA::lacZ fusions in OG1RF as shown in Fig. 2A. For the aerobic cultures, both ebpR and ebpA β-gal profiles followed the dome-shaped pattern over time, as described above. However, the presence of CO2/NaHCO3 led to a 2-3 fold increase in the β-gal units early during growth and, after the cultures entered stationary phase, ebpR and ebpA expression levels continued to increase for two hours and then showed only a slight decrease from 8 hr to 24 hr. At 24 hr, the β-gal units for OG1RF carrying the ebpA promoter were 13.9 in the presence of CO2/NaHCO3 compared to 0.4 aerobically, a 33-fold difference. Similarly, the β-gal units for OG1RF carrying the ebpR promoter were 1.2 in presence of CO2/NaHCO3 compared to 0.13 aerobically, a 9-fold difference.
Figure 2.
CO2/NaHCO3 induction effect on ebpA expression level. Samples were collected every hour from 3 to 8 hr, then at 10 and 24 hr after starting the culture in TSBG. The left axis represents the β-gal units (OD420 nm/protein concentration in mg/ml). The right axis indicates the OD600 nm readings. All sets of cultures presented were analyzed concurrently. Each figure is a representative of at least three experiments. A. Growth curves of OG1RF are shown in gray (in air) and in orange (in the presence of 5% CO2/0.1 M NaHCO3). OG1RF containing PebpR::lacZ (triangle) or PebpA::lacZ (square) was grown in air (closed black symbol) or in the presence of 5% CO2/0.1 M NaHCO3 (open orange symbol). B. The ΔebpR mutant containing PebpR::lacZ is represented by closed green diamond when grown in air and with open brown diamond when grown in the presence of 5% CO2/0.1 M NaHCO3.
To determine whether the CO2/NaHCO3 effect on ebpA expression was dependent on the presence of ebpR, we tested ebpA expression in an ebpR deletion mutant (TX5514). Using the ebpR deletion mutant (TX5514) containing PebpA::lacZ, β-gal production was assessed in air and in the presence of 5% CO2/0.1 M NaHCO3 and β-gal production remained at the background level in both conditions (Fig. 2B). These results combined with our previously published results [11] indicate that, in air as well as in the presence of 5% CO2/0.1 M NaHCO3, ebpR is important for ebpA expression and that the 5% CO2/0.1 M NaHCO3 effect on ebpA expression level also requires the presence of ebpR.
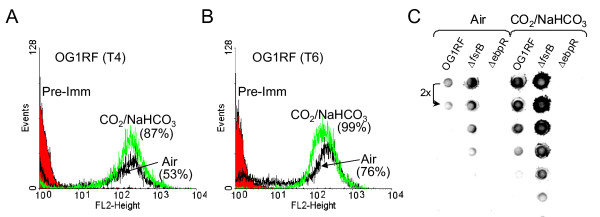
We previously reported that only a fraction of the OG1RF cells were positive for pilus expression by immunofluorescence ([11]). To examine whether the presence of CO2/NaHCO3 affected the amount of pili per cell or the percentage of cells positive for pilus production, we used flow cytometry. As early as entry into stationary growth phase, a difference in the percentage of pilus positive cell was visible (Fig. 3A) with 53% positive when grown in air compared to 87% positive when grown in the presence of CO2/NaHCO3. The difference in the percentage of positive cells remained in later stages of growth. Specifically, Fig. 3B shows that, at 6 hr, 76% of the cells were positive when grown in air compared to 99% when the cells were grown in the presence of CO2/NaHCO3. The mean fluorescence intensity, between growth conditions and growth phases, remained constant with an average of 268. We also used anti-EbpC antibodies to probe mutanolysin extracts spotted on a dot blot for pilus production. An approximately four-fold increased signal density was observed in cells grown in the presence of CO2/NaHCO3 compared to the cells grown in air (Fig. 3C). Additionally, no signal was detectable under either growth condition in the mutant lacking ebpR, confirming the importance of ebpR for ebpABC expression and pilus production aerobically as well as in the presence of 5% CO2/0.1 M NaHCO3.
Figure 3.
Detection of EbpC produced by OG1RF, ΔfsrB, and ΔebpR. A. Flow cytometry analysis of OG1RF grown in air (black) or in the presence of 5% CO2/0.1 M NaHCO3 (green) labeled with an anti-EbpC rabbit polyclonal immune serum and detected with phycoerythrin. The cells were collected at "T4", which corresponds to the entry into stationary growth phase (4 hrs after starting the culture). The percentages between brackets indicate the percentage of positive cells (WinMDI 2.9, marker set for 500-1024). In red is represented OG1RF grown in air incubated with a pre-immune serum and detected with Phycoerythrin as negative control. B. Flow cytometry analysis was done in the same conditions as above with samples collected at "T6" which corresponds to early stationary growth phase. C. An equal amount (by BCA protein assay) of mutanolysin extract preparation was 2-fold serial diluted and spotted onto a nitrocellulose membrane. Pilus presence was detected with an anti-EbpC rabbit polyclonal immune serum.
The Fsr system effect on the ebp locus
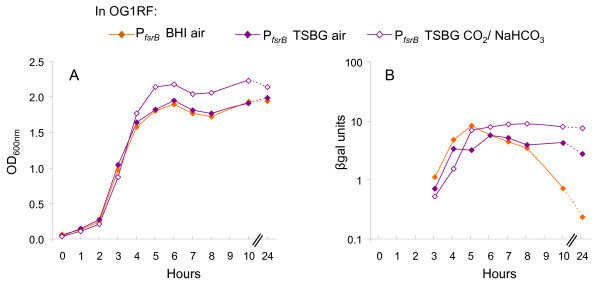
We previously presented data in our microarray study suggesting that Fsr repressed the ebpR-ebpABC locus. However, the Fsr effect was only seen at one time point (during late log growth phase) using BHI grown cells [8]; in this medium, fsrB expression increased from mid-log to entry into stationary phase and then decreased rapidly [6]. Since our current study used mainly TSBG (our biofilm medium) as growth medium, we investigated the fsrB expression profile in TSBG. fsrB expression also increased until entry into stationary growth phase, reaching 66% of the expression detected in BHI broth, but then remained relatively constant throughout stationary phase (Fig. 4). These results indicate that fsr expression is variable in different conditions.
Figure 4.
fsrB expression profile in OG1RF. For β-gal assays, samples were collected every hour from 3 to 8 hr, then at 10 and 24 hr after starting the culture (x axis). All sets of cultures presented were analyzed concurrently. The figure is a representative of at least two experiments. The growth curves are represented in brown for cells grown in BHI-air and purple for cells grown in TSBG (thin line when grown in air, dense line when grown in the presence of 5% CO2/0.1 M NaHCO3). OG1RF containing PfsrB::lacZ was grown in BHI air (brown closed diamond), in TSBG- air (purple closed diamond) or in TSBG-5% CO2/0.1 M NaHCO3 (purple open diamond). A. OD600 nm readings. B. β-gal assays (β-gal units = OD420 nm/protein concentration in mg/ml).
We next tested ebpR and ebpA expression using the PebpR:: and PebpA::lacZ fusions in OG1RF and TX5266 (ΔfsrB mutant), grown in parallel in TSBG aerobically. Both ebpR and ebpA gene expression profiles followed the same pattern in TX5266 as they did in OG1RF with an increase in expression until the culture reached stationary phase followed by a slow decrease (Fig. 5A). However, ebpR expression was 2-fold lower in OG1RF with 0.3 β-gal units compared to 0.8 β-gal units in TX5266 at entry into stationary phase. Similarly, ebpA expression was 4-fold lower in OG1RF with 3.7 β-gal units compared to 14.1 β-gal units in TX5266 early in stationary phase. These results confirm the role of the Fsr system as a repressor of the ebpR-ebpABC locus in TSBG, adding to the results obtained by microarray at one specific growth phase using cells grown in BHI.
Figure 5.
ebpR and ebpA expression profiles in TX5266 (ΔfsrB mutant). For β-gal assays, samples were collected every hour from 3 to 8 hr, then at 10 and 24 hr after starting the culture (x axis). The left axis represents the β-gal units (OD420nm/protein concentration in mg/ml). The right axis indicates the OD600 nm readings. All sets of cultures presented were analyzed concurrently. Each figure is a representative of at least three experiments. A. OG1RF containing either PebpR::lacZ (black triangle) or PebpA::lacZ (black square) and ΔfsrB containing either PebpR::lacZ (pink triangle) or PebpA::lacZ (pink square) were grown in TSBG aerobically. B. The ΔfsrB mutant (TX5266) containing either PebpR::lacZ (triangle) or PebpA::lacZ (square) was grown in TSBG aerobically (pink closed symbol) or in the presence of 5% CO2/0.1 M NaHCO3 (open blue symbol).
To determine whether the CO2/NaHCO3 effect on ebpA and ebpR expression is mediated through Fsr, we looked at ebpR and ebpA expression in TX5266 in air and in the presence of 5% CO2/0.1 M NaHCO3. As shown in Fig. 5B, the ebpA and ebpR expression profiles in TX5266 grown aerobically and in the presence of 5% CO2/0.1 M NaHCO3 presented the same general profile as in OG1RF (Fig. 2A). That is, ebpA expression increased from 6.8 β-gal units at mid-log growth phase to 13.8 β-gal units at late log growth phase and decreased gradually to 0.6 β-gal units by 24 hr (late stationary). In the presence of 5% CO2/0.1 M NaHCO3, ebpA expression increased from 16.8 β-gal units at mid-log growth phase to 56.5 β-gal units (5-fold more than with cultures grown in air) at 6 hr and remained stable with 55.3 β-gal units at 24 hr. ebpR expression profile in TX5266 also remained higher in the presence of 5% CO2/0.1 M NaHCO3 vs. in aerobic conditions with 0.2 and 2.6 β-gal units, respectively, at 24 hr. Finally, we also examined the effect of CO2/NaHCO3 on fsrB expression by transferring the PfsrB::lacZ fusion into OG1RF and followed expression in air and in the presence of CO2/NaHCO3. In those conditions, fsrB expression was not significantly affected by the presence of CO2/NaHCO3 (Fig. 4). Our observation of a further increase in ebpR and ebpA expression in TX5266 in the presence of CO2/NaHCO3 as was observed in OG1RF (Fig. 2A and 5B), together with the lack of an effect of CO2/NaHCO3 on fsr expression, indicate that HCO3- is not stimulating ebpR and ebpA expression via an effect on the Fsr system.
Finally, at the protein level, pilus production from the ΔfsrB mutant was compared with that of OG1RF. Cells were grown in TSBG aerobically or in presence of 5% CO2/0.1 M NaHCO3, and collected at 7 hr (stationary phase). As shown in Fig. 3C, a 3-5 fold increase in pilus production was observed in the ΔfsrB mutant compared to OG1RF with cells grown aerobically or in presence of 5% CO2/0.1 M NaHCO3. Similarly, 3-5 fold increase in pilus production was also seen with cells grown in the presence of 5% CO2/0.1 M NaHCO3 versus cells grown aerobically for both OG1RF and the ΔfsrB mutant. In conclusion, the differences observed in ebp mRNA expression levels between OG1RF and the ΔfsrB mutant and between the conditions used in this study (growth in air versus in the presence of 5% CO2/0.1 M NaHCO3) translated into comparable variations in pilus production at the surface of the cells.
ebpR threshold level
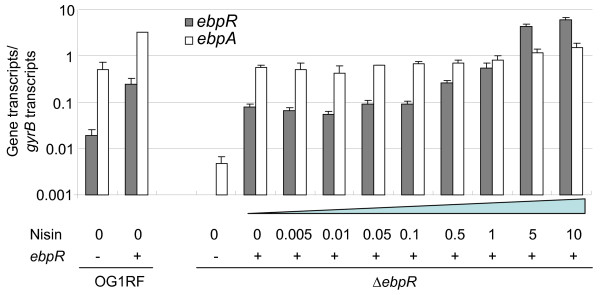
In the results obtained above, the ebpR and ebpA steady-state mRNA levels followed a similar pattern with ebpA expression being 7- to 37-fold higher than ebpR expression, depending on the technique. To investigate whether ebpA expression was directly related to the ebpR expression level, we introduced our previously cloned ebpR under a nisin inducible promoter (pTEX5515) into wild type OG1RF and into its ΔebpR mutant, TX5514 [11]. Our previous experiments showed that, even without nisin induction, pilus production was detected at the surface of the cells of the ebpR-complemented ΔebpR mutant, but not when the ebpR mutant carried the empty plasmid [11]. In this study, we investigated the steady-state mRNA level of ebpR and ebpA in different constructs with or without increasing amounts of nisin, compared to their respective levels in OG1RF carrying the empty vector, using qRT-PCR. The ebpR expression level in the ebpR-complemented ΔebpR mutant was 0.08 (normalized to the gyrB expression level) without induction, increased 4-fold with 0.5 ng/ml nisin to 0.26 and reached 9.33 with 10 ng/ml nisin (Fig. 6), representing a 65-fold increase from 0 to 10 ng/ml nisin. In the same background, ebpA steady-state mRNA levels were only slightly affected with a basal expression level without nisin of 0.6 up to 1.5 with 10 ng/ml nisin (Fig. 6), a less than a 3-fold increase. However, as expected from our previous results, ebpA expression was 100-fold lower in the ΔebpR mutant carrying the empty vector than in OG1RF carrying the empty vector or in the ebpR-complemented ΔebpR mutant. We conclude from these experiments that, above the ebpR expression level provided by ebpR copy on pTEX5515 without induction, there is not a strong direct relationship between ebpR expression and ebpA expression.
Figure 6.
Effect of nisin induction on ebpR and ebpA expression. Cells were grown to an OD600 nm of ~0.8 (3 hr, late log exponential growth phase) and at this point cells were left untreated (0) or treated with increasing concentration of nisin (from 0.005 to 10 ng/ml). Then, cells were collected and RNA extracted. After reverse transcription, ebpA and ebpR cDNA was quantified by real time PCR. The strains were OG1RF or ΔebpR (TX5514) carrying either the empty plasmid (-) or ebpR in trans under the nisin promoter (+). ebpR (gray bars) and ebpA (white bars) transcript levels were normalized with gyrB transcript levels. The data correspond to the mean of two independent experiments.
Bicarbonate effect on ebpA expression
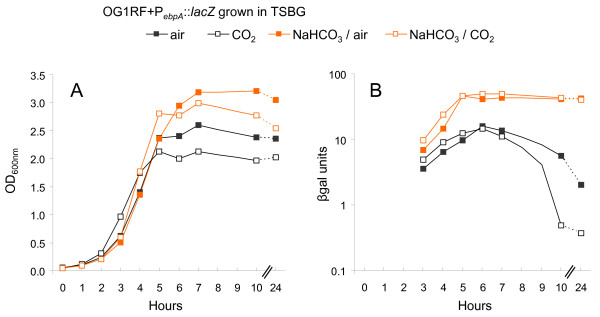
Studies using H. pylori have shown independent effects of pH, CO2, and bicarbonate on gene expression (these three environmental elements being interconnected in vivo) where pH appears to be responsible for H. pylori orientation [24]. In contrast, bicarbonate and not CO2 appears to be the inducer of expression of the B. anthracis toxins [25]. Using the PebpA::lacZ fusion in OG1RF, we first investigated the independent effect of CO2 and NaHCO3 on ebpA in buffered TSBG with or without the presence of 0.1 M NaHCO3 and/or 5% CO2. pH was controlled during the experiment and remained at pH 7.5 ± 0.25. As shown in Fig. 7, ebpA expression in TSBG-air did not differ appreciably from that in TSBG- 5% CO2, reaching a peak of expression early in stationary phase (15.8 and 14.5 β-gal units, respectively); expression then decreased to 2 and 0.4 β-gal units, respectively, at 24 hr. In the presence of NaHCO3, ebpA expression peak was ~4-fold higher with 46.5 β-gal units for the NaHCO3-air culture at entry into stationary phase (5 hr) compared to 9.8 β-gal when the cells were grown without NaHCO3, and 46.0 β-gal units for the 5% CO2 plus NaHCO3 culture compared to 12.5 β-gal when grown in presence of CO2 only. The bicarbonate effect persisted late into stationary phase with 42.5 and 40.7 β-gal units when grown in air-NaHCO3 and CO2-NaHCO3 respectively. A similar profile with increased ebpR expression in the presence of bicarbonate but not in presence of CO2 was also observed (data not shown). Furthermore, the differential effect of CO2 and NaHCO3 was also detected in BHI or when potassium bicarbonate was used as a source for HCO3- (data not shown). Taken together, these results demonstrate that the increase in ebpR and ebpA expression is caused by the addition of HCO3- and not CO2.
Figure 7.
ebpA expression affected by NaHCO3, and not CO2. For β-gal assays, samples were collected every hour from 3 to 8 hr, then at 10 and 24 hr after starting the culture (x axis). Growth curves of OG1RF containing PebpA::lacZ are shown in air with a thin gray line, in NaHCO3/air with thin orange line, in CO2 with a dense gray line, and in NaHCO3/CO2 with a dense orange line. The β-gal assays for OG1RF containing PebpA::lacZ are represented with closed black square, closed orange square, open black square, and open orange square when the cells were grown in air, 5% CO2, NaHCO3-air, and NaHCO3-5% CO2, respectively. All sets of cultures presented were analyzed concurrently. This figure is a representative of at least two experiments. A. OD600 nm readings. B. β-gal assays (β-gal units = OD420 nm/protein concentration in mg/ml).
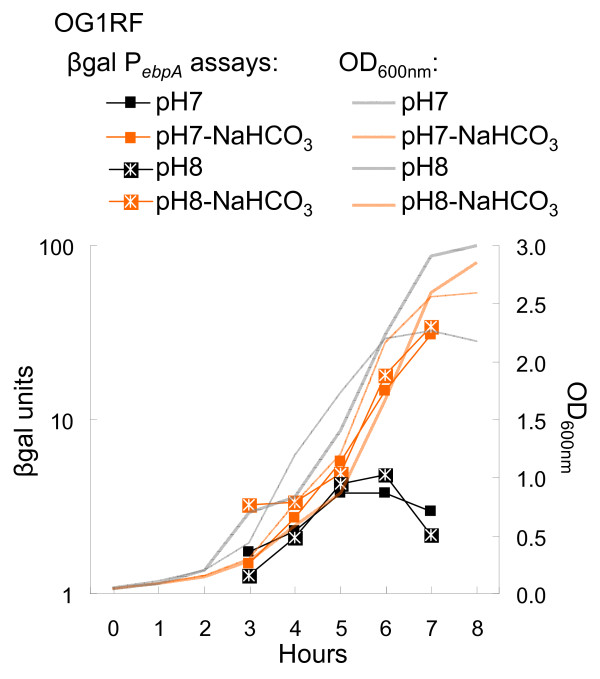
Since NaHCO3 is in equilibrium with H2CO3, HCO3-, and CO32- depending of the pH, temperature and partial pressure of CO2, we next tested a possible pH effect on ebpA expression when cells were grown in buffered TSBG. In a preliminary experiment, OG1RF (PebpA::lacZ) was grown in buffered TSBG with pH ranging from 5 to 9. Severe growth inhibition was observed at pH 5 and 9 with mild growth inhibition at pH 6, compared to unaffected growth at pH 7 and 8 (data not shown). Consequently, further experiments were conducted with buffered media with pH 7 and 8 only. Without the addition of sodium bicarbonate, ebpA expression levels of cells grown at pH 8 ± 0.25 were comparable with the levels in cells grown at pH 7 ± 0.25 (Fig. 8). However, adding NaHCO3 led to a 4- to 5-fold increase in β-gal production at either pH (pH was controlled during the experiment and remained constant with a ± 0.25 variation). For example, β-gal units were 9.4 at 6 hr for cells grown at pH 7-air, while at the same time point and pH, β-gal units were 40.1 when grown in the presence of NaHCO3. In conclusion, between pH (range 7-8), CO2 and bicarbonate, bicarbonate appears to be the main environmental inducer of the ebpABC operon.
Figure 8.
pH and NaHCO3 effect on ebpA expression. OG1RF containing PebpA::lacZ was used in these experiments. Growth curves are represented in thin gray line for pH 7 aerobically, thin orange line for pH 7-Air/NaHCO3, dense gray line for pH 8 aerobically, and dense orange line for pH 8-Air/NaHCO3. All sets of cultures presented were analyzed concurrently. This figure is a representative of at least two experiments. A. OD600 nm readings. B. β-gal assays (β-gal units = OD420 nm/protein concentration in mg/ml).
Effect of bicarbonate exposure on the OG1RF transcriptome
In an effort to begin to delineate the "bicarbonate regulon", we used microarray analysis with cells grown to late exponential growth phase (3 hr) and then submitted to a 15 min exposure with 0.1 M NaHCO3. Our goal was to define the first set of genes affected by the presence of bicarbonate. Out of the 73 genes that were differentially expressed (abs(fold)>2, P < 0.05, data deposited at ArrayExpress, additional file 1), only two genes were repressed by the presence of bicarbonate more than 5-fold (EF0082 and EF0083 with 9.9- and 7-fold, respectively) while four genes were activated more than 5-fold (EF0411-3 with ~10-fold, and EF2642 with 6.5-fold). EF0082 is part of the ers regulon (ers encodes a PrfA-like protein involved in the E. faecalis stress response [26,27]), but its function remains unknown, as is also true for EF0083. The EF0411-3 genes appear to be organized as an operon and encode proteins with the characteristics of a mannitol PTS system. EF2642 also appeared to be expressed in an operon with EF2641, which was also activated (4.1-fold, P < 0.05). EF2641 and EF2642 encode a putative glycine betaine/L-proline ABC transporter ATP-binding protein and permease protein, respectively. Those results were confirmed by qRT-PCR with a decrease of 32-fold for EF0082 in the presence of bicarbonate while EF0411 and EF2641 expression levels increased in the presence of bicarbonate by 24-fold and 8.5-fold, respectively (results not shown). The ebpR-ebpABC locus did not appear to be affected in these conditions (late log growth phase following a 15 min. incubation time with 0.1 M NaHCO3), suggesting that the bicarbonate effect on the ebpR-ebpABC locus may be indirect, requiring a cascade of events.
Discussion
We previously noted that EbpR shares homology with the AtxA/Mga family [11]. Regulators in this family have been shown to be active toward their target(s) in the presence of CO2 or CO2/HCO3-. While atxA is constitutively expressed, acpA and acpB (also members of the AtxA/Mga family) as well as mga are activated by the presence of CO2. In the work described here, we present evidence that bicarbonate is a strong inducer of the ebpR-ebpABC locus and consequently of pilus presence. Among the other environmental conditions tested, pH appears to have a weak effect in the limited conditions tested, while CO2 had no effect. Although ebpR and ebpA expression levels share a similar pattern, we were not able to show that an increase in ebpR expression, beyond a certain level, resulted in a proportional further increase of ebpA expression. Finally, the Fsr system affects expression of the ebpR-ebpABC locus independently of either the growth phase or the presence of bicarbonate.
It is interesting that ebpABC, also shown to be important for E. faecalis virulence, responded to bicarbonate. Bicarbonate influences expression of adcA (encoding an adhesin [28]) and kfc (encoding a factor important for gut colonization) in C. rodentium, which are controlled by the bicarbonate regulator RegA [19], as well as the three toxin genes in B. anthracis [25]. Bicarbonate-mediated transcriptional activation may be a system to sense a change in the environment. For example, the proximal portion of the duodenum is exposed to intermittent pulses of gastric H(+) discharged by the stomach. To protect the epithelial surface, at least two HCO3-/Cl- anion exchangers have been described as being responsible for the release of HCO3- into the duodenum lumen [29]. We postulate that E. faecalis may be sensing this signal and consequently produces adhesin structures like the ebpABC-encoded pili to favor colonization of the intestinal track, similar to adcA in C. rodentium, the expression of which is controlled by bicarbonate and whose gene product has been shown to be involved in adherence to mammalian cells [28].
From the various results obtained in this study where expression of ebpA followed the same expression profile as the ebpR expression, we postulated that the ebpA expression level was proportionally linked to the ebpR expression. To investigate our hypothesis, we used an ebpR construct under the control of a nisin regulated promoter. However, as shown in Fig. 6, the ebpR expression level was already 2-fold higher in the complemented ΔebpR strain (in the absence of nisin) when compared to its native level in wild type OG1RF (0.06 vs. 0.03) and was not detected (with a detection level of 10-5 the level of gyrB) in the ebpR deletion mutant with the empty plasmid. We did not observe a strong effect on ebpA expression after nisin induction, leading to the conclusion that ebpR expression was already above the threshold required to significantly increase ebpA expression. We tried another construct pCJK96 (rhamnose induction [30]), but faced the same issues (data not shown). Thus, although we did not determine the threshold necessary for the ebpA expression, the presence of ebpR was confirmed to be critical for ebpA expression.
One difference between ebpR and ebpA expression profiles in the presence of bicarbonate (vs. absence of bicarbonate) occurred after entry into stationary phase. ebpR and ebpA expression without bicarbonate begins to decrease, while it remained constant in the presence of bicarbonate. This difference may be explained either by an induction pathway that remains active (in the presence of HCO3-) in stationary phase or by inhibition early in stationary phase of a repression pathway (e.g., quorum sensing or phase dependent regulator). The first mechanism would also explain the slight difference observed in the presence of HCO3- during log growth phase. A potential candidate is a RegA homologue, an AraC/XylS-like regulator from C. rodentium [19]. Among the E. faecalis AraC/XylS-like regulators, none shares additional significant similarity with RegA. A second possibility would be a quorum sensing mechanism. A likely candidate would be the Fsr system [6]. However, the Fsr system, although a weak repressor of ebpR, does not appear to mediate the bicarbonate effect, since a similar ebpA expression pattern compared to OG1RF was observed in an fsrB mutant in the presence or absence of bicarbonate. Finally, we looked at the stress response pathway including ers and its regulon [26,27]. Interestingly, several members of the ers regulon were affected by a 15 min bicarbonate exposure, including EF0082-3 and EF0104-6. However, although both operons are activated by ers, EF0082-3 were strongly repressed (-8 fold), while EF0104-6 were activated (3 fold) by bicarbonate exposure. In addition, ers was not affected. In conclusion, the regulation pathways in E. faecalis resemble a network with several targets genes being under the control of independent regulation pathways illustrated by ebpR-ebpABC being independently a member of the bicarbonate and the fsr regulon, and EF0082 a member of the bicarbonate and ers regulon.
We also showed using microarray profiling that expression of many other genes (mostly PTS systems and ABC transporters) was altered in response to HCO3-. Among those genes are EF2641 and EF2642, which encode a putative glycine betaine/L-proline ABC transporter and permease protein, respectively. Interestingly, this ABC transporter shares some homology with the bicarbonate transporter described in B. anthracis (Tau family of ABC transporters) [25]. However, we did not find a TauA motif, that has been proposed as the bicarbonate binding motif, associated with the EF2641-2 locus or in available E. faecalis genomes including OG1RF. Interestingly, expression of ebpR-ebpABC was not affected by the 15 minutes bicarbonate exposure. Those results could be explained by the need of a cascade of events for a bicarbonate effect on ebpR-ebpABC expression or that the cells need an unknown factor, not present at the growth phase tested. Indeed, as seen in Fig. 2, Fig. 7, and Fig. 8, the greatest difference in ebpR-ebpABC expression was observed from mid stationary to late stationary growth phases (conditions that we found unsuited for microarray due to low and unstable mRNA expression). In conclusion, although we did not detect an effect of 15 minutes bicarbonate exposure on ebpR-ebpABC by microarray, the bicarbonate regulon was shown to share some components with the ers regulon and a later bicarbonate effect on ebp expression was shown by β-gal assays, qRT-PCR and western blot.
Finally, we have previously shown in the rat endocarditis model that an fsrB mutant is less attenuated than a gelE mutant [31]. Since, in the absence of the Fsr system, weak transcription of gelE was detected, it was postulated that the increase in virulence of the fsrB mutant compared to the gelE mutant might be a consequence of the residual production of gelatinase. However, since pilus production is also important in the rat endocarditis model [9], we can now postulate that, in the absence of the Fsr system as well as in presence of bicarbonate (by far the most important buffer for maintaining acid-base balance in the blood), pilus production increases, potentially causing the increased virulence of the fsrB mutant compared to the gelE mutant.
Conclusion
Considering that bicarbonate is an activator of the ebpR-ebpABC locus and that this locus is ubiquitous among E. faecalis isolates (animal, commensal, and clinical isolates) [9], these results seem to suggest an intrinsic aptitude of this species for pilus production which could play an important role in colonization of both commensal and pathogenic niches. Future studies should assess expression of the ebpR-ebpABC locus and the role of pili in a gut colonization model.
Methods
Strains, media, growth conditions
The strains used in this study are listed in Table 1. All strains were routinely grown in brain heart infusion broth (BHI broth; Difco Laboratories, Detroit, Mich.) at 150-200 rpm aerobically or on BHI agar at 37°C, unless otherwise indicated. Tryptic soy broth (Difco Laboratories, Detroit, Mich.) with 0.25% glucose (TSBG) was used to test strains for biofilm production, one of the assays where both ebpR and ebpA mutants are attenuated compared to OG1RF [9,11].
Table 1.
Strains and plasmids used in this study
| Strain or Plasmid | Relevant characteristics | Source or reference |
|---|---|---|
| E. coli strains | ||
| TG1 | E. coli general cloning host | [35] |
| E. faecalis strains | ||
| OG1RF | E. faecalis. FusR, RifR | [36] |
| TX5266 | OG1RF fsrB deletion mutant, deletion from bp 79 to 684 of fsrB. FusR, RifR | [6] |
| TX5514 | OG1RF ebpR deletion mutant, deletion from -5 bp to +1337 bp of ebpR. FusR, RifR | [11] |
| TX5584 | TX5514(pMSP3535). ErmR, FusR, RifR | [11] |
| TX5582 | TX5514(pTEX5515); ebpR mutant containing ebpR gene cloned into pMSP3535. ErmR, FusR , RifR | [11] |
| TX5583 | OG1RF(pMSP3535). ErmR, FusR, RifR | This study |
| TX5581 | OG1RF(pTEX5515); ebpR mutant containing ebpR gene cloned into pMSP3535. ErmR, FusR , RifR | This study |
| Plasmids | ||
| pTCV-lacZ | Shuttle vector containing promoterless lacZ. ErmR | [32] |
| pMSP3535 | Nisin inducible expression shuttle vector with pAMβ1 and ColE1 replicons. ErmR | [37] |
| pTEX5269 | fsrB promoter cloned upstream of lacZ in pTCV-lacZ (PfsrB::lacZ), from bp -110 to -8 (103 bp) relative to fsrB start codon; ErmR | [6] |
| pTEX5585 | ebpA promoter cloned upstream of lacZ in pTCV-lacZ (PebpA::lacZ), from -221 bp to +80 bp (301 bp) relative to ebpA start codon. ErmR | This study |
| pTEX5586 | ebpR promoter cloned upstream of lacZ in pTCV-lacZ (PebpR::lacZ), from -248 to + 53 bp (301 bp) relative to ebpR start codon. ErmR | [11] |
| pTEX5515 | pMSP3535 with ebpR from -20 bp to +1561 bp from the ATG. This ebpR fragment contains the full ORF and the RBS of ebpR. ErmR | [11] |
For all assays, strains were first streaked on BHI agar with the appropriate antibiotics, as needed. Five to ten colonies were inoculated into BHI broth and grown overnight (with antibiotics when appropriate), then cells were diluted so that the starting optical density at 600 nm was 0.05. For cultures grown in the presence of bicarbonate, a solution of 9% sodium bicarbonate was freshly prepared, filtered, and added for a final concentration of 0.8% (0.1 M final). The cultures were buffered with 100 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES) for a final pH of 7.5 ± 0.25 or as indicated. For comparison between cultures grown with and without bicarbonate, an equal volume of water was added to the culture without added bicarbonate. The cultures were then placed on a rotating platform set at 150 rpm at 37°C aerobically or in a 5% CO2 atmosphere. The pH was monitored during growth and remained at 7.5 ± 0.25. For each set of results, the cultures and following assays were analyzed concurrently. The presence of none of the four lacZ constructs (PTCV, PebpA, PebpR, and PfsrB) affected the growth of their host (OG1RF, ΔebpR, or Δfsr) in the conditions tested. To obtain accurate readings, cultures from 3 hr to 24 hr were diluted 5-fold before determining the OD.
Construction of the ef1091 promotor fusion
The same protocol was used to create the PebpA::lacZ fusion as previously described for the PebpR::lacZ fusion [11]. The primers cgggatccaagactacgccgaaaacc (introduced restriction sites are highlighted in bold) and ggaattcacacgaatgatttcttcca were used to amplify from 221 bp upstream to 80 bp downstream of the ebpA start codon (301 bp total). The fragment was amplified by PCR, cloned into pGEM-T-Easy vector (Promega, Madison, WI), sequenced, and then subcloned into pTCV-lacZ [32] using EcoRI and BamHI sites. After transfer into OG1RF, TX5266 (ΔfsrB), and TX5514 (ΔebpR), the plasmids were then purified and confirmed again by sequencing using previously published primers Vlac1 and Vlac2, which are located upstream and downstream of the promoter area [32].
β-galactosidase assay
Assays were performed according to the protocol of Hancock et al. [33] with some modifications. Following growth in the designated culture conditions and at each time point mentioned, a sample was collected (~2 × 109 CFU), centrifuged, and the pellet frozen until used. Cell pellets were resuspended in 1 ml of 1/10 Z buffer (Z buffer: 60 mM Na2HPO4, 40 mM NaH2PO4, 10 mM KCl, 1 mM MgSO4, [pH 7.0]). The cell suspension was transferred to a 2.0-ml tube containing a 0.5 ml volume of 0.1 mm diameter zirconia beads (BioSpec Products, Bartlesville, Okla.). The cells were disrupted using a vortex adapter for 5 min, then centrifuged at 13.6 K rpm for 1 min. Serial dilution of the aqueous layer was used in a β-galactosidase assay as described by Miller [34] with a final volume of 200 μl (96-wells microtiter plate). Twenty-five μl were assayed for total protein using the BCA protein assay kit (Pierce, Rockford, IL). Due to day to day variability, only data obtained within the same experiment (with cultures grown and samples assayed in parallel) were used for comparisons. To normalize the samples assayed in parallel, we used the total protein content as described in [33]. Experiments were repeated on at least two independent occasions and β-gal units for each experiment corresponded to OD420 nm/protein concentration in mg/ml. The figures show data from one representative experiment.
RNA purification for qRT-PCR
To follow gene expression in OG1RF during growth in TSBG at 37°C, 150 rpm, samples were collected every hour from three to 7 hr after starting the culture. For the nisin induction assay, cells were grown to an OD600 nm of ~0.8 (3 hr, late log exponential growth phase), and at this point cells were left untreated or treated with increasing concentration of nisin (from 0.005 ng/ml to 10 ng/ml). In each case, an equivalent of OD600 nm ~ 1 of cells was centrifuged, and the pellet was conserved at -80°C. RNA and cDNA were prepared using the methods described before [8]. Quantitative PCR on cDNA was performed using SYBR green PCR master mix kit (Applied Biosystems, Foster City, CA) and a 7500 Real-Time PCR system (Applied Biosystems). ebpA was selected for those experiments because it is the first gene of the ebpABC operon. The following primers were used: gyrB, accaacaccgtgcaagcc and caagccaaaacaggtcgcc; ebpA, aaaaatgattcggctccagaa and tgccagattcgctctcaaag; ebpR, acggatatggcaaaaacg and agaagagcgactaatattgatgg; EF0082, aaactccttgaactgattgg and ccagataaagaatgcccata; EF0411, agctgaactaacggaacaag and tcttttaagagcgaaaccac; and EF2641, attcgtggtgttcctaaaga and catcccaccagataattgac. For each primer set, a reference curve was established using a known amount of gDNA purified from OG1RF. The amount (in ng/ml) obtained for the gene of interest transcripts were normalized with the amount of gyrB transcripts.
Microarray analysis
The BHI cultures of OG1RF were started as described above. Cultures were grown to an OD600 nm of ~0.8 (3 hr, late log exponential growth phase), and at this point 25 ml of culture were centrifuged and resuspended in either BHI-buffered or BHI-buffered with 0.1 M bicarbonate, incubated for 15 min at 37°C @ 150 rpm, then centrifuged and the pellet conserved at -80°C until use. The microarray consists of 70-mer oligonucleotides that were printed on a GAPS II slide (Corning Incorporated, Corning, NY) at the University of Texas Medical School Microarray Core Laboratory. The RNA preparation, probe labeling, hybridization, data acquisition and statistical analysis were performed following the same methods as described previously [8]. The results of the bicarbonate induction are deposited at ArrayExpress http://www.ebi.ac.uk/microarray-as/ae/ under accession number E-MEXP-2518.
Flow cytometry analysis
An equivalent of ~ 1 OD600 nm of culture was collected for flow cytometry analysis, centrifuged and the pellet frozen until used. The pellet was then washed twice with 1 ml of PBS (80 mM Na2HPO4, 20 mM NaH2PO4, 100 mM NaCl, pH 7.5), resuspended in 0.5 ml of paraformaldehyde buffer (4.4% w/v paraformaldehyde, 30 mM Na2HPO4, 30 mM NaH2PO4), and incubated at RT for 15 min. The cells were pelleted and resuspended in 0.5 ml of PBS-2% BSA, and subsequently placed at -80°C for at least an hour. Before labeling, the cells were washed twice in PBS. A pellet corresponding to 108 CFU was resuspended in 100 μl of PBS with the anti-EbpC polyclonal rabbit serum at a 1:1000 dilution, and incubated at 4°C for 2 h. After centrifugation and two washes with PBS, the cells were resuspended in 100 μl of PBS with R-Phycoerythrin-conjugated affinipure F(ab')2 goat anti-Rabbit IgG (H+L) (Jackson ImmunoResearch Laboratories, Inc) at a dilution of 1:100, and incubated at 4°C for 2 h. The cells were then washed twice, resuspended in 1 ml PBS, and conserved at 4°C until they were analyzed with a BD FACSCalibur™ system (BD Biosciences, San Jose, CA).
Protein extraction and dot blot
Surface protein extracts from E. faecalis OG1RF and derivatives were prepared using mutanolysin (Sigma Chemical Co., St. Louis, MO). Cells grown at 37°C in specified conditions were collected at 7 hr after starting the culture. The cells were washed and resuspended in 1/100 volume of 0.02 M Tris-HCl (pH 7.0)-0.01 M MgSO4 buffer. Mutanolysin was added to a final concentration of 5 U for an equivalent of 1 OD600 nm of cells and incubated at 37°C for 1 hr. The supernatants were collected after centrifugation at 13.6 K rpm for 5 min. An equal amount of mutanolysin extract preparation (quantified using the BCA protein assay kit) was 2-fold serial diluted and was spotted onto NitroPure (GE Water and Process Tech., Watertown, MA) using the Bio-Dot® Microfiltration Apparatus (Biorad, Hercules, CA). The membranes were incubated with anti-EbpC rabbit polyclonal antiserum [9] at a dilution of 1:2000, followed by protein A-horseradish peroxidase conjugate (1:5000). Pilus production was then revealed using chemiluminescence (Amersham, Piscataway, NJ).
Authors' contributions
AB and BEM designed the study. AB performed the experiments except the beta-galactose assays done also by LCT. AB wrote the draft of the manuscript. BEM assisted in critical review of the manuscript. All authors read and approved the final manuscript.
Supplementary Material
Microarray results following 15 minutes bicarbonate induction. Define the first set of genes affected shortly after addition of bicarbonate to the medium.
Contributor Information
Agathe Bourgogne, Email: agathe.bourgogne@uth.tmc.edu.
L Charlene Thomson, Email: lizette.thomson@uth.tmc.edu.
Barbara E Murray, Email: barbara.e.murray@uth.tmc.edu.
Acknowledgements
We are grateful to K.V. Singh, T. M. Koehler, D. A. Garsin, J.R. Galloway-Pena and S. R. Nallapareddy for helpful discussions. This study was supported by grant NIH R37 AI47923 from the Division of Microbiology and Infectious Diseases, NIAID, to B.E.M.
References
- Murray BE. The life and times of the Enterococcus. Clin Microbiol Rev. 1990;3(1):46–65. doi: 10.1128/cmr.3.1.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogier JC, Serror P. Safety assessment of dairy microorganisms: The Enterococcus genus. Int J Food Microbiol. 2008;3:291–301. doi: 10.1016/j.ijfoodmicro.2007.08.017. [DOI] [PubMed] [Google Scholar]
- Murray BE. In: Infectious diseases. 2. Gorbach SL, Bartlett JG, Blacklow NR, editor. W. B. Saunders Company, Philadelphia, Pa; 1998. Enterococci; pp. 1723–1730. [Google Scholar]
- Edmond MB, Wallace SE, McClish DK, Pfaller MA, Jones RN, Wenzel RP. Nosocomial bloodstream infections in United States hospitals: a three-year analysis. Clin Infect Dis. 1999;29(2):239–244. doi: 10.1086/520192. [DOI] [PubMed] [Google Scholar]
- Qin X, Singh KV, Weinstock GM, Murray BE. Effects of Enterococcus faecalis fsr genes on production of gelatinase and a serine protease and virulence. Infect Immun. 2000;68(5):2579–2586. doi: 10.1128/IAI.68.5.2579-2586.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin X, Singh KV, Weinstock GM, Murray BE. Characterization of fsr, a regulator controlling expression of gelatinase and serine protease in Enterococcus faecalis OG1RF. J Bacteriol. 2001;183(11):3372–3382. doi: 10.1128/JB.183.11.3372-3382.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama J, Chen S, Oyama N, Nishiguchi K, Azab EA, Tanaka E, Kariyama R, Sonomoto K. Revised model for Enterococcus faecalis fsr quorum-sensing system: the small open reading frame fsrD encodes the gelatinase biosynthesis-activating pheromone propeptide corresponding to staphylococcal agrD. J Bacteriol. 2006;188(23):8321–8326. doi: 10.1128/JB.00865-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourgogne A, Hilsenbeck SG, Dunny GM, Murray BE. Comparison of OG1RF and an isogenic fsrB deletion mutant by transcriptional analysis: the Fsr system of Enterococcus faecalis is more than the activator of gelatinase and serine protease. J Bacteriol. 2006;188(8):2875–2884. doi: 10.1128/JB.188.8.2875-2884.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nallapareddy SR, Singh KV, Sillanpaa J, Garsin DA, Hook M, Erlandsen SL, Murray BE. Endocarditis and biofilm-associated pili of Enterococcus faecalis. J Clin Invest. 2006;116(10):2799–2807. doi: 10.1172/JCI29021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh KV, Nallapareddy SR, Murray BE. Importance of the ebp (Endocarditis- and Biofilm-Associated Pilus) locus in the pathogenesis of Enterococcus faecalis ascending urinary tract infection. J Infect Dis. 2007;195(11):1671–1677. doi: 10.1086/517524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourgogne A, Singh KV, Fox KA, Pflughoeft KJ, Murray BE, Garsin DA. EbpR is important for biofilm formation by activating expression of the endocarditis and biofilm-associated pilus operon (ebpABC) of Enterococcus faecalis OG1RF. J Bacteriol. 2007;189(17):6490–6493. doi: 10.1128/JB.00594-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchiyama H, Hayashi H, Suzuki Y. Functional characterization of Cl-/HCO3- exchange in villous cells of the mouse ileum. Biomed Res. 2006;27(6):265–274. doi: 10.2220/biomedres.27.265. [DOI] [PubMed] [Google Scholar]
- Wong AC, Bergdoll MS. Effect of environmental conditions on production of toxic shock syndrome toxin 1 by Staphylococcus aureus. Infect Immun. 1990;58(4):1026–1029. doi: 10.1128/iai.58.4.1026-1029.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwanaga M, Yamamoto K. New medium for the production of cholera toxin by Vibrio cholerae O1 biotype El Tor. J Clin Microbiol. 1985;22(3):405–408. doi: 10.1128/jcm.22.3.405-408.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caparon MG, Geist RT, Perez-Casal J, Scott JR. Environmental regulation of virulence in group A streptococci: transcription of the gene encoding M protein is stimulated by carbon dioxide. J Bacteriol. 1992;174(17):5693–5701. doi: 10.1128/jb.174.17.5693-5701.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koehler TM. Bacillus anthracis genetics and virulence gene regulation. Curr Top Microbiol Immunol. 2002;271:143–164. doi: 10.1007/978-3-662-05767-4_7. [DOI] [PubMed] [Google Scholar]
- Drysdale M, Bourgogne A, Koehler TM. Transcriptional analysis of the Bacillus anthracis capsule regulators. J Bacteriol. 2005;187(15):5108–5114. doi: 10.1128/JB.187.15.5108-5114.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogensen EG, Janbon G, Chaloupka J, Steegborn C, Fu MS, Moyrand F, Klengel T, Pearson DS, Geeves MA, Buck J. Cryptococcus neoformans senses CO2 through the carbonic anhydrase Can2 and the adenylyl cyclase Cac1. Eukaryot Cell. 2006;5(1):103–111. doi: 10.1128/EC.5.1.103-111.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Hart E, Tauschek M, Price GD, Hartland EL, Strugnell RA, Robins-Browne RM. Bicarbonate-mediated transcriptional activation of divergent operons by the virulence regulatory protein, RegA, from Citrobacter rodentium. Mol Microbiol. 2008;68(2):314–327. doi: 10.1111/j.1365-2958.2008.06171.x. [DOI] [PubMed] [Google Scholar]
- Hoffmaster AR, Koehler TM. The anthrax toxin activator gene atxA is associated with CO2-enhanced non-toxin gene expression in Bacillus anthracis. Infect Immun. 1997;65(8):3091–3099. doi: 10.1128/iai.65.8.3091-3099.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hondorp ER, McIver KS. The Mga virulence regulon: infection where the grass is greener. Mol Microbiol. 2007;66(5):1056–1065. doi: 10.1111/j.1365-2958.2007.06006.x. [DOI] [PubMed] [Google Scholar]
- Day AM, Cove JH, Phillips-Jones MK. Cytolysin gene expression in Enterococcus faecalis is regulated in response to aerobiosis conditions. Mol Genet Genomics. 2003;269(1):31–39. doi: 10.1007/s00438-003-0819-1. [DOI] [PubMed] [Google Scholar]
- Dai Z, Koehler TM. Regulation of anthrax toxin activator gene (atxA) expression in Bacillus anthracis: temperature, not CO2/bicarbonate, affects AtxA synthesis. Infect Immun. 1997;65(7):2576–2582. doi: 10.1128/iai.65.7.2576-2582.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber S, Konradt M, Groll C, Scheid P, Hanauer G, Werling HO, Josenhans C, Suerbaum S. The spatial orientation of Helicobacter pylori in the gastric mucus. Proc Natl Acad Sci USA. 2004;101(14):5024–5029. doi: 10.1073/pnas.0308386101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson AC, Soyer M, Hoch JA, Perego M. The bicarbonate transporter is essential for Bacillus anthracis lethality. PLoS Pathog. 2008;4(11):e1000210. doi: 10.1371/journal.ppat.1000210. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Giard JC, Riboulet E, Verneuil N, Sanguinetti M, Auffray Y, Hartke A. Characterization of Ers, a PrfA-like regulator of Enterococcus faecalis. FEMS Immunol Med Microbiol. 2006;46(3):410–418. doi: 10.1111/j.1574-695X.2005.00049.x. [DOI] [PubMed] [Google Scholar]
- Riboulet-Bisson E, Sanguinetti M, Budin-Verneuil A, Auffray Y, Hartke A, Giard JC. Characterization of the Ers (PrfA-like) regulon of Enterococcus faecalis. Infect Immun. 2008;76(7):3064–3074. doi: 10.1128/IAI.00161-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart E, Yang J, Tauschek M, Kelly M, Wakefield MJ, Frankel G, Hartland EL, Robins-Browne RM. RegA, an AraC-like protein, is a global transcriptional regulator that controls virulence gene expression in Citrobacter rodentium. Infect Immun. 2008;76(11):5247–5256. doi: 10.1128/IAI.00770-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konturek SJ, Konturek PC, Pawlik T, Sliwowski Z, Ochmanski W, Hahn EG. Duodenal mucosal protection by bicarbonate secretion and its mechanisms. J Physiol Pharmacol. 2004;55(Suppl 2):5–17. [PubMed] [Google Scholar]
- Kristich CJ, Wells CL, Dunny GM. A eukaryotic-type Ser/Thr kinase in Enterococcus faecalis mediates antimicrobial resistance and intestinal persistence. Proc Natl Acad Sci USA. 2007;104(9):3508–3513. doi: 10.1073/pnas.0608742104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh KV, Nallapareddy SR, Nannini EC, Murray BE. Fsr-independent production of protease(s) may explain the lack of attenuation of an Enterococcus faecalis fsr mutant versus a gelE-sprE mutant in induction of endocarditis. Infect Immun. 2005;73(8):4888–4894. doi: 10.1128/IAI.73.8.4888-4894.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poyart C, Trieu-Cuot P. A broad-host-range mobilizable shuttle vector for the construction of transcriptional fusions to beta-galactosidase in gram-positive bacteria. FEMS Microbiol Lett. 1997;156(2):193–198. doi: 10.1111/j.1574-6968.1997.tb12726.x. [DOI] [PubMed] [Google Scholar]
- Hancock LE, Shepard BD, Gilmore MS. Molecular analysis of the Enterococcus faecalis serotype 2 polysaccharide determinant. J Bacteriol. 2003;185(15):4393–4401. doi: 10.1128/JB.185.15.4393-4401.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JH. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, US; 1972. [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: a Laboratory Manual. Cold Spring Harbor Laboratory Press, US; 1989. [Google Scholar]
- Murray BE, Singh KV, Ross RP, Heath JD, Dunny GM, Weinstock GM. Generation of restriction map of Enterococcus faecalis OG1 and investigation of growth requirements and regions encoding biosynthetic function. J Bacteriol. 1993;175(16):5216–5223. doi: 10.1128/jb.175.16.5216-5223.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryan EM, Bae T, Kleerebezem M, Dunny GM. Improved vectors for nisin-controlled expression in gram-positive bacteria. Plasmid. 2000;44(2):183–190. doi: 10.1006/plas.2000.1484. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Microarray results following 15 minutes bicarbonate induction. Define the first set of genes affected shortly after addition of bicarbonate to the medium.