Abstract
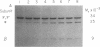
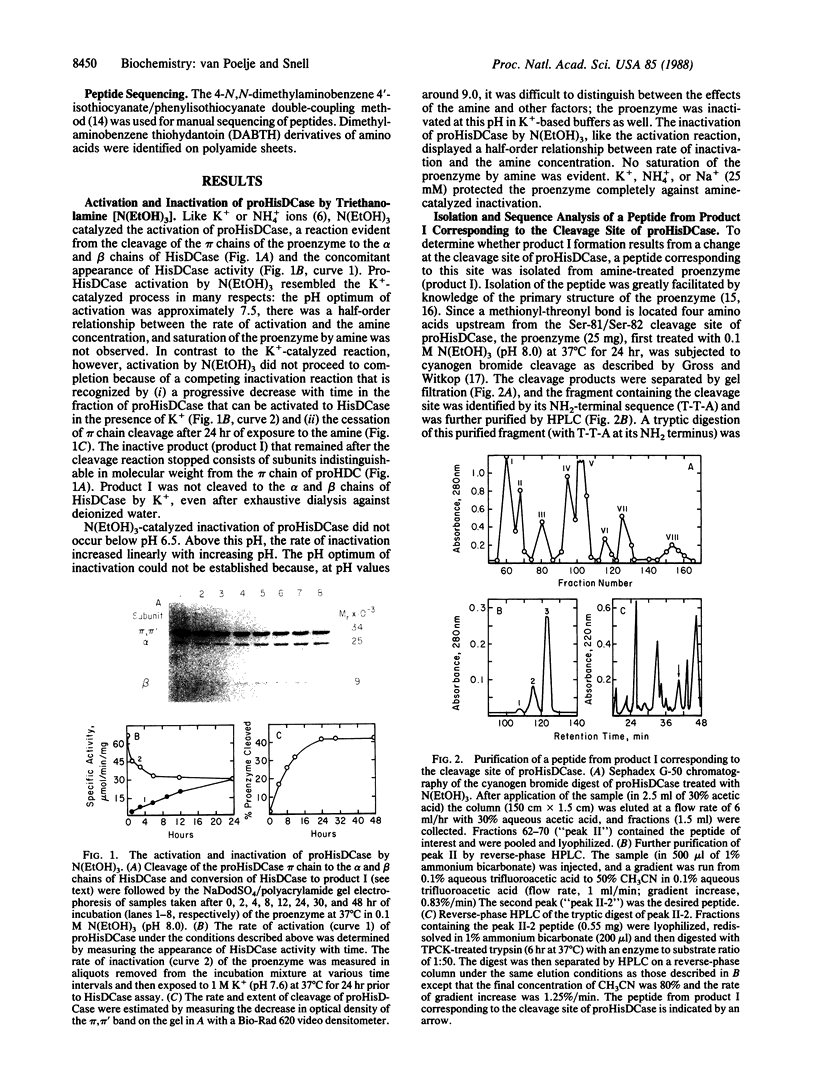
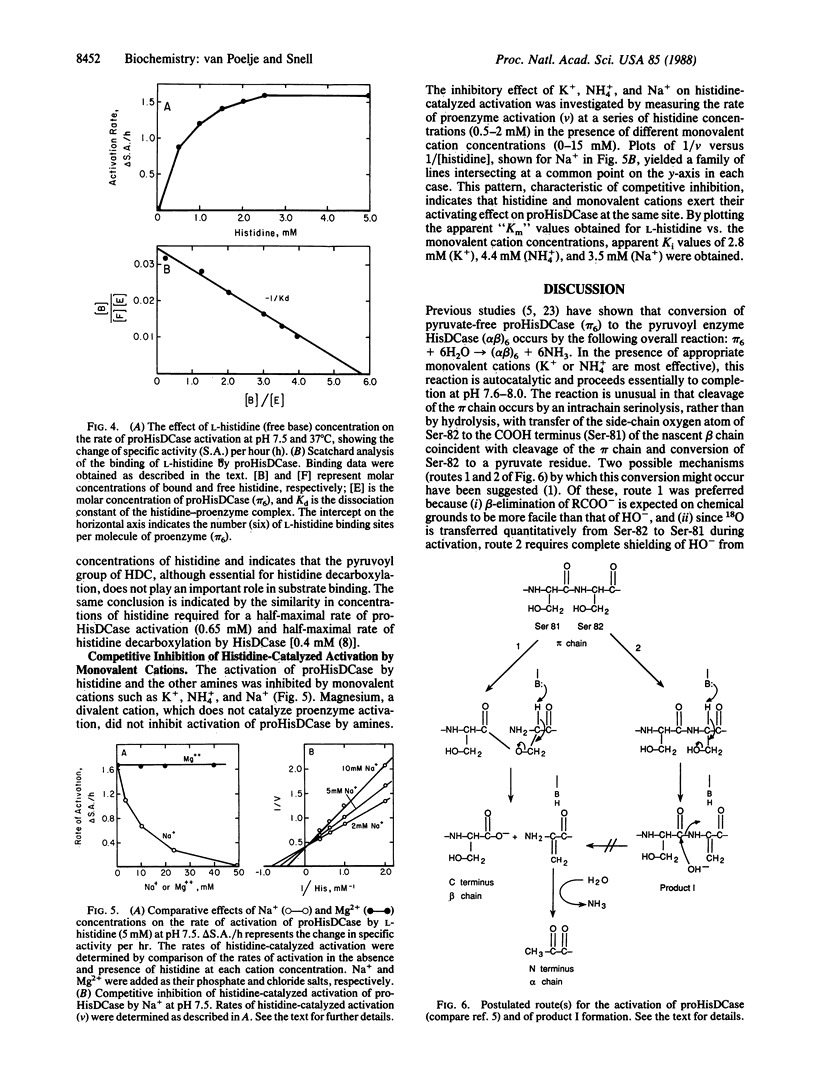
Activation of prohistidine decarboxylase (pi 6) from Lactobacillus 30a proceeds by an intramolecular, pH- and monovalent cation-dependent reaction in which its constituent pi chains are cleaved nonhydrolytically between Ser-81 and Ser-82 with loss of NH3 and conversion of Ser-82 to the pyruvoyl residue of active histidine decarboxylase (alpha beta)6. Amines with pKa values more than 7.0 substitute for K+ or NH4+ in the activation of prohistidine decarboxylase, but they also catalyze its inactivation in a competing reaction, pi 6----pi'6. Sequence analysis of the appropriate tryptic peptide from amine-inactivated prohistidine decarboxylase established that inactivation results from conversion of Ser-82 of the pi chain to an aminoacrylate residue. The inactivated proenzyme (pi'6) does not form histidine decarboxylase; this fact eliminates one of two postulated mechanisms of activation and, thus, favors activation by beta-elimination of the acyl group of an intermediate ester formed between Ser-81 and Ser-82. L-Histidine is bound by the proenzyme (Kd = 1.7 x 10(-4) M) and is an effective activator; one binding site is present per pi subunit. K+, NH4+, and Na+ competitively inhibit (Ki values = 2.8-4.4 x 10(-3) M) activation by histidine. The data suggest the presence of two classes of monovalent cation binding sites on prohistidine decarboxylase: one (near Ser-82) is readily saturable and one is unsaturable even by 2.4 M K+.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Chang G. W., Snell E. E. Histidine decarboxylase of Lactobacillus 30a. II. Purification, substrate specificity, and stereospecificity. Biochemistry. 1968 Jun;7(6):2005–2012. doi: 10.1021/bi00846a001. [DOI] [PubMed] [Google Scholar]
- Chang J. Y. The destruction of serine and threonine thiohydantoins during the sequence determination of peptides by 4-N,N-dimethylaminoazobenzene 4'-isothiocyanate. Biochim Biophys Acta. 1979 May 23;578(1):175–187. doi: 10.1016/0005-2795(79)90125-9. [DOI] [PubMed] [Google Scholar]
- Fairbanks G., Steck T. L., Wallach D. F. Electrophoretic analysis of the major polypeptides of the human erythrocyte membrane. Biochemistry. 1971 Jun 22;10(13):2606–2617. doi: 10.1021/bi00789a030. [DOI] [PubMed] [Google Scholar]
- GROSS E., WITKOP B. Nonenzymatic cleavage of peptide bonds: the methionine residues in bovine pancreatic ribonuclease. J Biol Chem. 1962 Jun;237:1856–1860. [PubMed] [Google Scholar]
- Huynh Q. K., Recsei P. A., Vaaler G. L., Snell E. E. Histidine decarboxylase of Lactobacillus 30a. Sequences of the overlapping peptides, the complete alpha chain, and prohistidine decarboxylase. J Biol Chem. 1984 Mar 10;259(5):2833–2839. [PubMed] [Google Scholar]
- Ichiyama A., Nakamura S., Nishizuka Y., Hayaishi O. Enzymic studies on the biosynthesis of serotonin in mammalian brain. J Biol Chem. 1970 Apr 10;245(7):1699–1709. [PubMed] [Google Scholar]
- KOEPSELL H. J., SHARPE E. S. Micro-determination of pyruvic and alpha-keto-glutaric acids. Arch Biochem Biophys. 1952 Jul;38:443–449. doi: 10.1016/0003-9861(52)90050-7. [DOI] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lever J. E. Quantitative assay of the binding of small molecules to protein: comparison of dialysis and membrane filter assays. Anal Biochem. 1972 Nov;50(1):73–83. doi: 10.1016/0003-2697(72)90487-3. [DOI] [PubMed] [Google Scholar]
- Recsei P. A., Huynh Q. K., Snell E. E. Conversion of prohistidine decarboxylase to histidine decarboxylase: peptide chain cleavage by nonhydrolytic serinolysis. Proc Natl Acad Sci U S A. 1983 Feb;80(4):973–977. doi: 10.1073/pnas.80.4.973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Recsei P. A., Snell E. E. Histidine decarboxylase of Lactobacillus 30a. VI. Mechanism of action and kinetic properties. Biochemistry. 1970 Mar 31;9(7):1492–1497. doi: 10.1021/bi00809a003. [DOI] [PubMed] [Google Scholar]
- Recsei P. A., Snell E. E. Prohistidine decarboxylase from Lactobacillus 30a. A new type of zymogen. Biochemistry. 1973 Jan 30;12(3):365–371. doi: 10.1021/bi00727a001. [DOI] [PubMed] [Google Scholar]
- Recsei P. A., Snell E. E. Pyruvoyl enzymes. Annu Rev Biochem. 1984;53:357–387. doi: 10.1146/annurev.bi.53.070184.002041. [DOI] [PubMed] [Google Scholar]
- Recsei P. A., Snell E. E. Pyruvoyl-dependent histidine decarboxylases. Mechanism of cleavage of the proenzyme from Lactobacillus buchneri. J Biol Chem. 1985 Mar 10;260(5):2804–2806. [PubMed] [Google Scholar]
- Riley W. D., Snell E. E. Histidine decarboxylase of Lactobacillus 30a. V. Origin of enzyme-bound pyruvate and separation of nonidentical subunits. Biochemistry. 1970 Mar 31;9(7):1485–1491. doi: 10.1021/bi00809a002. [DOI] [PubMed] [Google Scholar]
- Rosenbusch J. P., Weber K. Subunit structure of aspartate transcarbamylase from Escherichia coli. J Biol Chem. 1971 Mar 25;246(6):1644–1657. [PubMed] [Google Scholar]
- Suelter C. H. Enzymes activated by monovalent cations. Science. 1970 May 15;168(3933):789–795. doi: 10.1126/science.168.3933.789. [DOI] [PubMed] [Google Scholar]
- Tabor C. W., Tabor H. The speEspeD operon of Escherichia coli. Formation and processing of a proenzyme form of S-adenosylmethionine decarboxylase. J Biol Chem. 1987 Nov 25;262(33):16037–16040. [PubMed] [Google Scholar]
- Vanderslice P., Copeland W. C., Robertus J. D. Cloning and nucleotide sequence of wild type and a mutant histidine decarboxylase from Lactobacillus 30a. J Biol Chem. 1986 Nov 15;261(32):15186–15191. [PubMed] [Google Scholar]