Abstract
Interstrand DNA crosslinks (ICLs) are formed by natural products of metabolism and by chemotherapeutic reagents. Work in E. coli identified a two cycle repair scheme involving incisions on one strand on either side of the ICL (unhooking) producing a gapped intermediate with the incised oligonucleotide attached to the intact strand. The gap is filled by recombinational repair or lesion bypass synthesis. The remaining monoadduct is then removed by Nucleotide Excision Repair (NER). Despite considerable effort, our understanding of each step in mammalian cells is still quite limited. In part this reflects the variety of crosslinking compounds, each with distinct structural features, used by different investigators. Also, multiple repair pathways are involved, variably operative during the cell cycle. G1 phase repair requires functions from NER, although the mechanism of recognition has not been determined. Repair can be initiated by encounters with the transcriptional apparatus, or a replication fork. In the case of the latter, the reconstruction of a replication fork, stalled or broken by collision with an ICL, adds to the complexity of the repair process. The enzymology of unhooking, the identity of the lesion bypass polymerases required to fill the first repair gap, and the functions involved in the second repair cycle are all subjects of active inquiry. Here we will review current understanding of each step in ICL repair in mammalian cells.
Keywords: Chemotherapy, cisplatin, psoralen, unhooking, lesion bypass, replication arrest
INTRODUCTION
ICLs, DNA adducts that link both strands of the duplex, are among the most dangerous DNA lesions. They are obligate blockers of replication and transcription, and, unlike monoadducts, cannot be carried through a proliferative cycle without repair. If not removed they can provoke chromosomal breakage, rearrangements, or cell death (it has been estimated that 20–40 unrepaired ICLs can kill a mammalian cell (Dronkert and Kanaar, 2001; McHugh et al., 2001; Lawley and Phillips, 1996). They have been categorized as “cytotoxic” lesions, and it has been argued that their accumulation over time contributes to genomic instability and aging in tissues and organs (Mitchell et al., 2003). Hypersensitivity to crosslinking agents is a common feature of cells derived from individuals with genome instability disorders such as Fanconi Anemia (FA). Because of their heightened toxicity in proliferating cells, relative to monoadduct forming compounds, crosslinking agents have received extensive application as chemotherapeutic drugs (Vogel et al., 1998; Lawley and Phillips, 1996). Thus, ICLs can be seen as both dangerous and useful, each view contributing to the increasing interest in delineating the pathways involved in their repair. Considering the molecular challenge of repairing lesions that engage both strands of the duplex it is understandable that multiple repair pathways are involved, and that repair is more complex than for monoadducts.
Excellent comprehensive reviews on crosslink repair have been published in recent years including those by Dronkert and Kanaar (Dronkert and Kanaar, 2001), Miller and colleagues (Noll et al., 2006; Noll et al., 2004), and Lehoczky, McHugh, and Chovanec (Lehoczky et al., 2007). These authors have described the crosslinking agents employed in most studies, discussed the structure of DNA crosslinked by these compounds, and presented the state of knowledge of the repair pathways in the major experimental organisms: E. coli, yeast, and mammalian cells. More focused reviews have also been published on: the chemistry of DNA crosslinking compounds used as antitumor drugs (Rajski and Williams, 1998); the clinical significance of crosslink repair pathways (McHugh et al., 2001)); repair in E. coli (Lage et al., 2003); Fanconi Anemia and crosslink repair (Niedernhofer et al., 2005; Andreassen and Ren, 2009; Thompson and Hinz, 2009; Moldovan and D'Andrea, 2009; Patel and Joenje, 2007; de Winter and Joenje, 2009); crosslink damage and aging (Grillari et al., 2007); and crosslink repair and chromosome radial formation (McCabe et al., 2009). In this review we will focus on the repair of ICLs in mammalian cells and discuss some of the major unresolved issues. We apologize to colleagues whose work could not be referenced due to space limitations.
There are several DNA repair pathways that respond to the multiplicity of lesions introduced in DNA by radiation and reactive chemicals. These include Nucleotide Excision Repair (NER) and Transcription Coupled Repair (TCR), Base Excision Repair (BER), MisMatch Repair (MMR), the two Double Strand Break (DSB) Repair pathways-Nonhomologous End Joining (NHEJ), and Homology Directed Repair (HDR), Single Strand Break (SSB) Repair, and Translesion Synthesis (TLS), which may be viewed as an adduct tolerance pathway. We have a relatively detailed knowledge of these pathways, the result of intense investigation over several decades, utilizing technologies based in genetics, biochemistry, biophysics, and immunology. In contrast, despite considerable effort, and steady inquiry during the same several decades, our understanding of crosslink repair is less advanced. This situation is due to several factors.
The structure of crosslinked DNA differs as a function of the chemistry of the crosslinking compound. Thus some ICLs are extremely distorting, others much less so. Furthermore, the extent of distortion can be dependent on sequence context. Since helical distortion is an important determinant of DNA repair, this structural variability can be a confounding variable in comparing results from experiments with different agents. Crosslinking agents do not produce only ICLs, indeed these are usually a minority product. This can muddle the interpretation of experiments which examine the response of cells to agents that produce multiple lesions, most of which are extraneous to the purpose of the experiment. One lesson that has emerged from crosslink repair studies in yeast is that functions from multiple repair pathways are engaged (Saffran et al., 2004; Grossmann et al., 2001; Lehoczky et al., 2007). Indeed, all of the pathways listed above are thought to contribute to the repair of ICLs. Consequently, it can be difficult to distinguish the contribution to ICL repair by components of those pathways, from their response to the lesions (also present) that defined the pathway. In an effort to circumvent some of these problems a number of investigators have developed cell free systems (extracts, purified proteins) in which duplexes with defined ICLs have been introduced. This approach has a long and productive history in the DNA repair field, and has been very powerful in detailing the individual steps in the pathways listed above. However key, perhaps unknown, components may not be present in the mixtures, thus overweighting the role of some factors at the expense of others. Finally, it is clear that crosslink repair can occur at any time of the cell cycle, but some pathways are operative only in certain phases of the cycle. These distinctions may be lost in experiments performed with cells in asynchronous culture, or extracts derived from those cultures.
INTERSTRAND DNA CROSSLINKING AGENTS
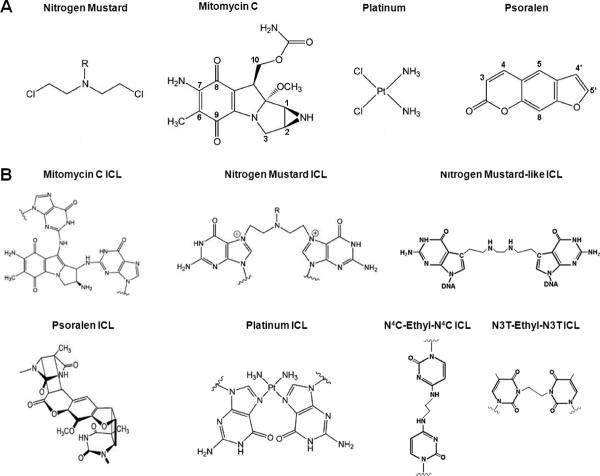
In this section we will discuss the most commonly employed experimental crosslinking compounds. As noted above these have been very capably reviewed by others, and we will not attempt to reproduce those efforts. Instead the focus will be on two features of these agents that bear on the interpretation of repair experiments: the multiplicity of adducts following the treatment of cells with these compounds; and the structural consequences for DNA carrying the crosslink. Most of the discussion will be on compounds that can be used to treat cells. However there has been recent work with synthetic substrates in which site specific ICLs have been introduced into duplex oligonucleotides. These are particularly useful for in vitro experiments and will also be described (Fig. 1a, b).
Figure 1.
DNA ICL forming agents. (A) Common crosslinking agents used in ICL experiments. (B) DNA-ICL structures including synthetic substrates.
Nitrogen mustards
Nitrogen mustards are small bifunctional alkylating compounds that contain N,N-bis-(2-chloroethyl) amine as the defining component. They react with DNA usually at the N7 of guanine (Rajski and Williams, 1998), although reactions with adenine have been described (Balcome et al., 2004). The resultant monoadduct can react with water to terminate the reaction, or there may be a second reaction with guanine (or adenine (Balcome et al., 2004)) on the opposite strand to form an interstrand DNA crosslink. Reaction with cysteines in chromosomal proteins, producing protein DNA crosslinks is also possible (Thomas et al., 1978; Loeber et al., 2009). The DNA monoadduct is the dominant product (90–95%), while of the ICLs (~5%) the N7 to N7 guanine-guanine diadduct is the major component. The reactive guanines are the distal bases in a 5'GNC step (Millard et al., 1990; Ojwang et al., 1989). The minimal distance between the guanines is 8.9 Å, while the length of the diethyleneamine 5 atom tether is only 7.5 Å (Rink and Hopkins, 1995). The helix is necessarily distorted and has a helical axis bend of 12.4–16.8° (Fan and Gold, 1999; Rink and Hopkins, 1995). Structural analysis is compromised by the instability of N7 alkylated guanines which depurinate, eliminating the crosslink. This problem has been addressed in recent clever work from the Schärer group (Angelov et al., 2009). They synthesized duplex oligonucleotides in which the crosslinked guanines were each replaced with 7-deazapurines with an aldehyde at C7. They used a post synthetic reductive double amination reaction linking the two guanines via an alkyl diamine from the aldehydes. The choice of an appropriate diamine linker allowed formation of a crosslink with a longer bridge than with a conventional nitrogen mustard, thus relieving the distortion associated with those adducts. The 7-deazapurines are more resistant to depurination than guanines, and therefore are stable substrates for subsequent experimentation.
Mitomycin C
Mitomycin C has received extensive use in the clinic as an anticancer chemotherapeutic and in the laboratory as a crosslinking agent. The compound requires reduction of its quinone ring (by oxidoreductases (Sartorelli et al., 1994)) to form a quinone methide. This reacts with the N2 exocyclic amine of guanine, yielding a monoadduct in the minor groove. Monoadducts form preferentially at 5'CpG sites, while crosslink formation has an absolute requirement for this sequence. This is the result of a specific hydrogen bond formed between the C10 oxygen atom of mitomycin and the 2-NH2 of the guanine in the strand opposite the guanine engaged in the initial monoadduct reaction (Gargiulo et al., 1995; Weidner et al., 1990). In effect, the mitomycin molecule is positioned for the two reactions required for crosslink formation, before the first, to form the monoadduct, has taken place. This remarkable structural feature not withstanding, ICLs are only one of six characterized adducts. In addition to the interstrand crosslink (about 10% of products), there are four monoadducts, as well as the intrastrand crosslink between two adjacent guanines (Palom et al., 2000). Furthermore, DNA reactive oxygen radicals are produced during the reduction (Pritsos and Sartorelli, 1986).
The mitomycin crosslink between guanines in the (C-G)•(G-C) sequence imposes minimal distortion on the duplex, with no bending (Norman et al., 1990; Rink et al., 1996). The G-mit-G linkage does not disrupt or distort the C:G and G:C base pairs involved in the crosslink. Molecular modeling suggests that the crosslinked guanines remain in parallel planes, thus stacking of adjacent bases is not perturbed. In contrast, the GpG intrastrand crosslink induces a 14.6° bend, imposing a kink in the DNA and a distortion of the adducted guanines (Rink et al., 1996).
Cisplatin
Cisplatin (cis-Diamminedichloroplatinum, cis-DDP) (Rosenberg et al., 1965; Rosenberg, 1977), has been widely used in chemotherapy with striking success against testicular cancer. The compound undergoes displacement of the chlorides by water to form the activated form, cis-[Pt(NH3)2(H2O)2]2+, which reacts with a range of cellular targets including RNA, protein, phospholipids, as well as DNA. The major DNA adducts are the 1,2 intrastrand crosslinks at GpG (65%) and ApG (25%) sites. ICLs (up to 8%) form at GpC sites linking the N7 of both guanines (Jamieson and Lippard, 1999; Malinge et al., 1999). In contrast to the nitrogen mustard adducts, the N7-platinum dG adducts are no more likely to depurinate than the unmodified dG (Baik et al., 2002). There is substantial distortion of both the intrastrand and interstrand crosslink (Poklar et al., 1996; Malinge et al., 1994; Malinge et al., 1999). In particular, DNA with a GpC ICL is unwound by 80° with a 45° bend towards the minor groove (Malinge et al., 1994). Important structural features are the flipping of the complementary cytosines into an extrahelical conformation, and the local change from B-DNA to the left handed Z-DNA (Huang et al., 1995). The ICLs are not persistently stable but can convert spontaneously to intrastrand crosslinks with a half life of 120 hrs (Perez et al., 1997).
Psoralens
Psoralens are tricyclic planar molecules with a furan ring fused to a coumarin. They are natural products and have been isolated from plant and fungal sources (see Cimino et al., 1985 for an extensive review). Reaction with DNA requires photoactivation by exposure to 320–400 nm light, and this is the basis of the well known psoralen/UVA (PUVA) therapy for psoriasis, vitiligo and other skin disorders (Stern, 2007; Millington and Levell, 2007). The requirement for photoactivation permits temporal control of the reaction, distinguishing psoralens from the compounds described above.
Psoralens intercalate into DNA with a preference for 5'TA steps (Sinden and Hagerman, 1984) (Saenz-Méndez et al., 2007) and runs of (TA) are hotspots for modification (Boyer et al., 1988). Formation of an ICL proceeds via a two step, two photon reaction. Absorption of the first photon supports cycloaddition between the ethylenic bonds in either the furan or pyrone ring with the C5–C6 double bond of thymine to form furan or pyrone monoadducts. While the pyrone monoadduct is unable to react further, the furan monoadduct may absorb another photon and react with the thymine on the other strand forming a crosslink. The frequency of pyrone vs. furan monoadducts is influenced by the peripheral substituents on the psoralen nucleus. For example, 4,5',8 trimethyl psoralen (TMP), with a 4-CH3, shows a pronounced bias towards furan addition (98%) (Kanne et al., 1984), and thus can be converted in the second step to an ICL. In contrast, 8-methoxy psoralen (8-MOP) forms 20% pyrone side, non convertible, monoadducts. The distinction between the two compounds reflects the presence of the methyl on the C-4 position of psoralen, which imposes a steric constraint on formation of the TMP pyrone monoadduct. Photoactivation with conventional laboratory UVA lamps yields mixtures of monoadducts and ICLs regardless of the psoralen derivative (Lai et al., 2008). However, with C-4 methyl psoralens it is possible to drive the adduct distribution to almost exclusively ICLs using a high powered laser for photoactivation (Johnston et al., 1977).
The structure of duplex oligonucleotides with various psoralen ICLs has been examined by gel migration, solution NMR, and X ray crystallography (Sinden and Hagerman, 1984; Haran and Crothers, 1988). NMR analysis of a duplex with an 4'-aminomethyl-4,5',8-trimethyl psoralen (AMT) crosslink revealed only minor bending (<10°) of the duplex, with distortion of the central 4 base pairs (Hwang et al., 1996). Another study of an oligonucleotide duplex crosslinked by HMT concluded that the helix was unwound by 28°, was not bent, and had a disruption of the DNA structure in the immediate vicinity of the crosslink that returned to normal within 3 base pairs. The authors suggested that the conformational flexibility of the sugar phosphate backbone in the region of the crosslink was a signal for recognition by the DNA repair apparatus (Spielmann et al., 1995b; Spielmann et al., 1995a).
X ray crystallography of HMT crosslinked duplexes demonstrated substantial distortion at the thymine linked to the pyrone ring, although there was no bending, and the structural perturbation was localized to the vicinity of the crosslink (Eichman et al., 2001). In all these analyses the duplex oligonucleotides contained only two T:A base pairs, sufficient for crosslinking. The relatively greater stability of the G:C pairs in the remainder of the duplex could influence the conclusions regarding distortion induced by the crosslink. This point was considered in a structural analysis (by electrophoretic mobility) of psoralen crosslinked duplexes containing A:T and G:C pairs, as well as mismatches. The authors concluded that the degree of local distortion imposed by a psoralen crosslink would reflect the local sequence, with inherently less stable sequences subject to greater deformation (Kumaresan et al., 1992).
In typical biological experiments crosslinking agents react throughout the cell. Thus, ICLs are introduced randomly wherever there is DNA, in both nucleus and mitochondria. However, with psoralens two strategies have been developed that offer greater control of crosslink location. Both technologies exploit the requirement for photoactivation, and the synthetic opportunities for conjugation of psoralens to other moieties, without loss of reactivity.
Targeted psoralen crosslink formation mediated by Triple Helix Forming Oligonucleotides
One approach is based on triple helix forming oligonucleotides (TFOs). The DNA triple helix consists of a third strand of nucleic acid that lies in the major groove of an intact duplex (Felsenfeld et al., 1957). The most stable structures are formed on polypurine:polypyrimidine elements, which are over represented in the human genome (Manor et al., 1988; Schroth and Ho, 1995; Behe, 1995). TFOs can be attached to psoralen via linker arms such that, following triplex formation, a crosslink can be introduced at a specific site by photoactivation (Giovannangeli et al., 1992). This demonstration was followed by a series of publications from the Glazer lab that described mutagenesis and gene conversion induced by triplex targeted psoralen in shuttle vector plasmids (Havre et al., 1993; Wang et al., 1995; Raha et al., 1996; Faruqi et al., 1996) (for reviews see Seidman and Glazer, 2003; Chin et al., 2007; Chin and Glazer, 2009). This plasmid based approach has been exploited in more recent work on the interaction of triplex –psoralen crosslinks with repair factors (Christensen et al., 2008; Lange et al., 2009). Bioactive TFOs that can introduce psoralen ICLs at specific chromosomal target sites in living cells, have also been developed (Majumdar et al., 1998; Puri et al., 2001; Puri et al., 2002; Puri et al., 2004; Seidman et al., 2005; Majumdar et al., 2003; Shahid et al., 2006). These have been used to target mutagenesis and gene conversion in the genome (Majumdar et al., 2003; Majumdar et al., 2008).
Laser localized ICLs
Laser/confocal microscopy has had a major impact on DNA repair studies. Defined subnuclear regions in living cells can be treated with laser light of specific wavelength and intensity to introduce localized DNA damage. The lesions that have received attention are base oxidation products, which can be detected by specific antibodies (Lan et al., 2004), and, of course, DSBs, for which there are surrogate protein markers such as phosphorylated histone H2AX (Rogakou et al., 1999). Psoralens would seem to lend themselves to experiments employing localization by laser photoactivation. However, there have been no reagents available for detection of psoralen ICLs in situ. This limitation was overcome by synthesizing psoralen linked to an antigenic tag, digoxigenin. This sterol has received extensive use as an immunotag, and reliable commercial antibodies are available. The equivalent derivative of angelicin, a photoreactive analogue of psoralen, which can form only monoadducts, was also synthesized. Living cells, treated with the tagged compound, were exposed to laser photactivation in a defined region of the nucleus (Thazhathveetil et al., 2007). The location of the ICLs or monoadducts was visualized by immunofluorescence following fixing and staining. Adduct removal could be monitored over time in repair proficient and deficient cells (Fig. 2). In order to distinguish phenomena due to psoralen ICLs, rather than monoadducts or ancillary damage from the laser, parallel experiments with digpsoralen and dig-angelicin were performed.
Figure 2.

Laser localized lesions and repair in G1 mammalian cells. Localized lesions (green stripe) formed by dig-tagged psoralen in WT and XPC deficient cells. Cells are stained with a cell cycle marker (NPAT, red spots) to show cells in G1 phase. NPAT shows 2 spots in G1 cells and four spots in S/G2 cells, and the spots may be present in different focal planes. A color version of this figure is available in the online version of this manuscript.
Synthetic substrates
In the preceding discussion we have considered crosslinking agents that can act directly, or after activation, on DNA in vivo and in vitro. DNA duplexes crosslinked by a variety of linkages have been constructed by chemical synthesis by a several investigators and have been the subject of biochemical and structural analysis. Incorporated into plasmids, they have been used as probes of repair and mutagenesis in living cells and extracts. Some of these ICLs will be mentioned briefly here.
N2-N2 guanine ICLs formed by trimethylene (as a malondialedyde and acrolein mimic), in either the 5'CpG or 5'GpC orientation, were synthesized by Harris and coworkers (Dooley et al., 2001). The 5'CpG crosslink increased the thermal stability of the duplex, while NMR analyses and molecular modeling indicated that the DNA was minimally distorted (Dooley et al., 2003). In contrast, the 5'GpC crosslink reduced the Tm value, and was distorting, introducing a bend and twist in the helix (Dooley et al., 2003). Duplexes and plasmids with a closely related 5'CpG crosslink were also prepared by Hecht, Moriya and colleagues (Lao and Hecht, 2005; Liu et al., 2006).
Miller and coworkers synthesized a group of ICLs differing in extent of duplex distortion (reviewed in (Noll et al., 2004). Three constructs reflect the different disposition of the N4C-ethyl-N4C linkage: a direct C-C mismatch, or staggered in the 5'CpG or 5'GpC orientation. Of these, the 5'GpC crosslink is the most distorting because the ethyl linker is not long enough to span the distance between the N4 amino groups of the cytosine, while in the 5'CpG orientation there is little effect on the helix (Noll et al., 2005). The C-C “mispaired” crosslink causes bending toward the major groove by an angle of ~27°. There is perturbation of stacking of adjacent bases although this is localized and fully accommodated within three bases on either side of the lesion (Webba da et al., 2002). This group also synthesized oligonucleotide duplexes containing an N3T-ethyl-N3T crosslink as a T-T mispair, somewhat analogous to the crosslinked C-C mispair. Structural analysis indicated that the ethyl crosslink can be accommodated in the duplex with little distortion and no hydrogen-bond disruptions (da Silva et al., 2004). Thus the two “mispair” ICLs differed in extent of local distortion. Another important difference is the influence of the modifications on hydrogen bonding with complementary bases. The attachment of the ethyl linker to the N3 on the thymine interferes with hydrogen bonding, while linkage of the ethyl group to the exocyclic amines of the cytosines does not preclude their participation in hydrogen bonding.
CROSSLINK REPAIR: THE BASIC MODEL
The problem of crosslink repair was addressed many years ago in experiments in E. coli. This work resulted in the well known “Cole” model which accounted for the sensitivity to crosslinking agents of strains deficient in NER and HR, and the virtual intolerance of strains with deficiencies in both pathways (Cole, 1973; Cole et al., 1976). This model was further modified by work from other laboratories (Sladek et al., 1989a; Sladek et al., 1989b; Van et al., 1986; Cheng et al., 1991) (see Lage et al., 2003; Vidal et al., 2006 for an appraisal of the role of individual components of the E. coli NER apparatus).
In this scheme the NER proteins recognize a crosslink and then incise one strand on either side of the ICL. This produces an “unhooked” substrate with the excised fragment still attached to the non-incised strand by the crosslinking agent. A gap is introduced by the exonuclease activity of DNA pol I in the region adjacent to the 3' side of the crosslink. This is the substrate for a RecA mediated strand exchange with an undamaged homologous chromosome. The still crosslinked oligonucleotide incision product is forced out of the restored duplex, forming a large monoadduct, which can be repaired by conventional NER. In the absence of a functional recombination pathway, the gap may be filled by lesion bypass, by pol II in the case of a nitrogen mustard N7-N7 guanine crosslink (Berardini et al., 1997; Berardini et al., 1999), and pol IV with an acrolein N2-N2 guanine crosslink (Kumari et al., 2008). These are the elements of a major two cycle repair pathway found in all organisms- recognition followed by unhooking, gap formation, then gap repair by recombination or lesion bypass synthesis (Fig. 3). This process converts the crosslink to a monoadduct, after which the second cycle of repair can occur, by, it is presumed, conventional NER. Although this scheme would seem to provide a general solution to crosslink repair, there are quite significant differences and variations in yeast and vertebrate cells.
Figure 3.
The `Cole' model. A schematic of ICL repair in E. coli. After the 1st cycle of incision, the gap is either filled by strand exchange or repair synthesis bypassing the adducted base. A color version of this figure is available in the online version of this manuscript.
CROSSLINK REPAIR IN MAMMALIAN CELLS
Although the process in mammalian cells is more complex, as in bacteria, the essential step is the unhooking event. This can be regarded as the central node of a multi entry flow chart. In this discussion we will follow the crosslink through the stages in this schematic (Fig. 4). We will first consider repair in the absence of replication, and then repair triggered by the encounter at a replication fork.
Figure 4.
Entry into an ICL repair pathway depends on the mode of recognition. (A) In the context of a non-replicating DNA, the distortion of the DNA helical structure caused by the lesion attracts protein(s) involved in the global damage surveillance of DNA. This process has been shown to involve proteins of the NER pathway, with XPC leading the initial recognition. The first incision step on either side of the lesion on one strand of the duplex by XPF-ERCC1 complex (and perhaps XPG) generates a gapped structure which serves as a substrate for bypass polymerases. The now flipped out monoadduct-like structure can be recognized by DDB2 and perhaps also by glycosylases such as MPG or Neil1. Again, the NER pathway will initiate the second cycle of repair and remove the remaining adduct on the opposite strand. (B) Stalling of RNA polymerases at the site of lesion during transcription can also serve as a means of ICL recognition in non-replicating DNA. (C) and (D), a stalled replication fork, either due to a single or dual fork encounter, is attractive to proteins of the Fanconi Anemia (FA) pathway and proteins such as Mus81-EME1/2, Snm1B, and MRN. Initial recognition is thought to be mediated by the FancM-FAAP24 complex, which then becomes part of the FA core complex. The FA core complex is a prerequisite to recruit the FancD2 and FancI proteins which are modified via ubiquitination and phosphorylation. The ICL is incised by XPF-ERCC1 and Mus81-EME1 on the leading strand, generating a DSB at the fork. In the case of converging forks (D), the first incision cycle may occur on either strand. The gapped structure will be filled in by lesion bypass polymerases, including polζ, polκ, polι, polN, polη and Rev1. When a single fork is stalled by the ICL polymerase(s) will extend a parental strand to fill the gap. When two forks converge on the ICL a leading daughter strand is extended to bypass the lesion. Upon removal of the remaining single adduct on the opposite strand, in an NER dependent pathway, the broken fork will be reconstructed by recombinational repair. A color version of this figure is available in the online version of this manuscript.
In contrast to the apparent situation in E. coli there are three routes for crosslink detection in mammalian cells. Adducts can be recognized in otherwise unperturbed duplex DNA, by factors that recognize DNA damage. Crosslink discovery might also be via encounter with the transcription machinery. Finally, ICLs could block a replication fork, triggering a repair response that would remove the crosslink and restore replication. Brief consideration of these scenarios points to the relevance of cell cycle status, with the latter pathway option available only in S phase. The influence of the cell cycle on crosslink repair has been emphasized in insightful work from McHugh and colleagues (Barber et al., 2005; McHugh and Sarkar, 2006).
Crosslink recognition in the absence of replication
DNA lesions are recognized as distortions in the local structure of the duplex due to destabilization of base stacking, with an attendant increase in helical flexibility (Isaacs and Spielmann, 2004; Maillard et al., 2007a; Maillard et al., 2008; Yang, 2008). This appears to be the common denominator for recognition by the BER, NER, and MMR damage detection proteins, although there are important structural distinctions that underlie the lesion specificity of each repair system. Thus, while MMR can recognize certain base adducts such as cisplatin and aminofluorene and acetylaminofluorene, NER does not recognized mismatched bases (see Isaacs and Spielmann, 2004; Yang, 2008 for discussion). Binding of damaged DNA by recognition proteins typically accentuates the distortion inherent to the lesion. Crystal structures of glycosylases bound to their substrates reveal nucleotide flipping, discontinuous base stacking, and DNA kinking as characteristic of these complexes (Fuxreiter et al., 2002). DNA containing a single mismatched base pair is bent by about 45° on binding by the human MutSα complex (the heterodimer of Msh2:Msh6)
Entry into the global genome repair pathway of NER begins with adduct recognition by the XPC-HR23B-Centrin complex. Crystallographic analysis of a truncation product of the yeast XPC homolog, Rad4, bound to a duplex oligonucleotide containing a cyclopyrimidine dimer indicates the DNA is highly kinked with a 42° bend. This is consistent with previous reports of bending of damaged DNA by XPC (Janicijevic et al., 2003). The protein binds 11 consecutive base pairs of the adjacent undamaged sequence, making contacts with the phosphodiester and ribose group. At the damage site the protein inserts a β-hairpin through the DNA duplex, forcing both the intrastrand crosslinked bases, and the undamaged complementary bases, out of the helix. The flipped out bases of the undamaged strand are bound by the protein, while the adducted nucleotides are disordered and are not engaged by the protein (Min and Pavletich, 2007). The binding of XPC protein to undamaged single stranded DNA, rather than to the actual lesion is the basis of the well known versatility of damage recognition by the XPC complex (Sugasawa et al., 2002; Sugasawa and Hanaoka, 2007). The interaction of XPC with both the adjacent undamaged duplex and the region distorted by the lesion has been emphasized by Naegeli and colleagues, who have developed a bipartite binding model of damage recognition and binding (Maillard et al., 2007b; Maillard et al., 2008; Camenisch et al., 2009).
The other early recognition factor associated with NER, the DDB1-DDB2 complex, binds to UV lesions and a variety of other forms of DNA damage. The current view is that DDB1-DDB2 assists the recruitment of XPC to DNA damage that might otherwise be overlooked by XPC (Fitch et al., 2003; Wang et al., 2004). DDB1-DDB2 exists in vivo as a member of a CUL4-E3 ubiquitin ligase. This complex polyubiquitinates XPC, DDB2 and CUL4 (Li et al., 2006) as a key step in what has been termed a lesion “hand off” transfer from DDB1-DDB2 to XPC (Sugasawa et al., 2005). The crystal structures of DDB1-DDB2 bound to a 6–4 UV photoproduct shows that DDB2 makes contacts with the damaged DNA, inserting a three residue hairpin into the duplex from the minor groove. This widens the minor groove, unwinds the DNA in the vicinity of the adduct, flips the adducted bases out of the helix, and kinks the DNA by about 40° (Scrima et al., 2008).
While such substantial distortions of DNA would face a serious energy barrier in undamaged DNA, the damage eases the deformation of the DNA by the recognition proteins, reducing the energetic cost of complex formation (Isaacs and Spielmann, 2004; Fuxreiter et al., 2002; Min and Pavletich, 2007). Thus, recognition by XPC and subsequent repair, of lesions that introduce greater distortion and reduce the Tm value of the adducted duplex, is more efficient than that of lesions that introduce little or no distortion (Geacintov et al., 2002). The sequence context can influence the structural consequences of DNA adducts and modulate the efficiency of recognition and repair (Kropachev et al., 2009) (Cai et al., 2009).
So how are ICLs, in the absence of transcription or replication, recognized? The work on monoadduct recognition emphasizes the importance of helical distortion-local denaturation, disruption of base stacking, etc. However, while ICLs may perturb the duplex (see above), affected bases obviously cannot flip out of the helix. Furthermore, crosslinks, with some exceptions (Noronha et al., 2002; Dooley et al., 2003), typically stabilize duplexes. Assuming helical distortions are central to the process, it would seem likely that perturbation of the helix adjacent to the crosslinked bases would be the attractant for recognition proteins. This interpretation would merge current thinking about adduct recognition with the special challenge presented by crosslinks. To pursue this reasoning we might then ask: which recognition proteins are involved in the early stages of repair of ICLs, and is there the expected correspondence between efficiency of recognition and extent of ICL induced distortion? As will become apparent, there is very little direct information on this point and what is available is not always in accord with expectation.
The role of damage recognition factors in ICL repair can be considered experimentally in several different ways. A traditional approach is to measure the effect on survival of cells with deficiencies in genes involved in lesion recognition following treatment with crosslinking compounds. Typically these experiments employ unsynchronized cultures and take a few days for an endpoint. Cells vary in the time they spend in each phase of the cell cycle, and this can influence the outcome and confuse comparisons of results with different cells. For example, the popular DT40 chicken cells divide rapidly, and asynchronous populations have only 15% of the cells in G1 phase, reducing their utility for studies of genes involved in repair in the absence of replication (Nojima et al., 2005). The results of survival assays are a summation of the capacity of the multiple crosslink repair pathways to protect cells from the killing effects of ICLs, as well as the monoadducts also introduced by the compounds. The influence of a defect in a gene involved in one pathway can be masked by other pathways. This point was emphasized in a thoughtful study in yeast from the Doetsch group which showed that multiple repair pathways were involved in resistance to crosslinking agents. They found different contributions of individual pathways to the response to the different compounds, indicating that conclusions based on one compound were not necessarily applicable to another (Beljanski et al., 2004).
An alternative approach is to construct plasmids with a defined crosslink at a specific site, and introduce these into repair proficient or deficient cells. The readout of these assays may require transcription (expression of a reporter gene) or replication (plasmid yield, mutagenesis). Plasmid constructs or duplex oligonucleotides have also been incubated in cell free extracts in which incision can be directly monitored. These experiments report the activity of the functions that survive preparation, and are silent on those that do not. Each assay system has strengths and weaknesses, and contradictory results are not uncommon. In the following section we will discuss the role of damage recognition proteins in the response to crosslinking agents, as reported by these different assays.
XPC
In yeast genetic evidence indicates a requirement for Rad4 (the XPC equivalent) during ICL repair (Wu et al., 2004), including G1 phase repair (Sarkar et al., 2006). In mammalian cells, although it has been known for many years that NER functions are involved in crosslink repair (Kaye et al., 1980; Hoy et al., 1985a), there has been relatively little focus on XPC. XPC deficient cells treated with cisplatin were no more sensitive than wild type cells, while cells deficient in the transcriptional coupled repair factors CSA and CSB, and the post recognition NER factors, XPA, XPD, XPF, and XPG were sensitive (Furuta et al., 2002; McKay et al., 2001). These results implied that the XPC recognition function was not important for cell survival after cisplatin treatment, although this assay cannot identify the toxic adduct(s) (see De Silva et al., 2002 for a discussion of this issue). The response of the NER incision apparatus to a cisplatin crosslinked duplex oligonucleotide was examined in a cell free extract system. While incisions were observed with an intrastrand crosslink, none were detected with the interstrand crosslink (Zamble et al., 1996). Furthermore, the “futile cycle” incisions (below), seen with psoralen crosslinked substrates (Mu et al., 2000) were not observed. These results, although clearly limited, suggest that the XPC complex does not recognize cisplatin ICLs despite the strong distortion imposed by this adduct. In contrast, RPA binds cisplatin ICLs with greater affinity than the intrastrand crosslink (Patrick et al., 2008), although the biological significance of this observation is unclear.
The involvement of XPC in the repair of psoralen ICLs has been addressed in relatively few experiments. In recent work, a modest sensitivity to TMP/UVA was shown by cells with a deficiency in XPC. The XPC protein was recruited rapidly to sites of laser localized TMP ICLs in wild type G1 phase cells, while the psoralen ICLs were not removed in XPC deficient G1 phase cells (Muniandy et al., 2009). Host cell reactivation experiments with a reporter plasmid carrying psoralen ICLs demonstrated a major contribution of XPC to the repair process (Chen et al., 2003). Legerski, Li, and colleagues constructed plasmids containing a single psoralen crosslink placed between a promoter and a reporter gene. The plasmids were introduced into wild type and repair deficient cells and the expression of the reporter gene measured as a reflection of repair competence. They found that NER factors were required, although the influence of the deficiency of XPC was not as pronounced as that of the other NER functions (XPA, ERCC1 etc.). This result was consistent with roles for both transcription coupled as well as global genome repair of the psoralen ICL (Wang et al., 2001). In an analogous experiment the repair of a plasmid carrying a defined mitomycin crosslink was examined in repair proficient and deficient cells. Interestingly, with this construct the decline in repair due to the XPC deficiency was dramatic and equivalent to that in the other NER mutant cells. Repair was also reduced in cells with mutations in the CSA or CSB protein (involved in TCR), although not as strongly as in the XPC deficient cells (Zheng et al., 2003). The authors called attention to the involvement of a TCR pathway (see below). However their data also indicated role for XPC, which is not a component of TCR.
A comparison of these results from different experimental systems suggests that the requirement for XPC was at variance with expectation. There was no apparent involvement in the repair of ICLs formed by cisplatin, the most distorting lesion, while there was a clear requirement with the MMC crosslink, the least distorting. In the instances where XPC is involved in recognition it is not clear what structural features of an ICL are essential for binding. Although the XPC complex can bind a triplex-psoralen crosslink (Thoma et al., 2005), it is not known how the complex would respond to psoralen crosslinks without the conjugation to the triplex oligonucleotide. Nor is it known whether crosslinks formed by other compounds would be bound by the XPC complex, or how these results would correlate to the data from biological experiments.
XPE
XPE deficient cells are not sensitive to psoralen (Muniandy et al., 2009). Binding of DNA with different lesions by the XPE binding factor (as it was then known) was reported by (Payne and Chu, 1994). No binding to 8-MOP ICLs or monoadducts was observed (see also Reardon et al., 1993). Furthermore, XPE deficient cells are proficient in crosslink unhooking activity (Bredberg and Soderhall, 1985; Muniandy et al., 2009). DDB2 was recruited slowly to laser localized psoralen ICLs, in contrast to the rapid accumulation of XPC (Muniandy et al., 2009). It seems that the DDB1-DDB2 complex is not involved in recognition of psoralen ICLs.
MMR
The role of mismatch repair functions in the repair of ICLs has yet to be resolved. Cells deficient in MSH2, the common component of the MutSα (MSH2-MSH6) and MutSß (MSH2-MSH3) mismatch recognition complexes) are sensitive to MMC (Fiumicino et al., 2000) and psoralen (Wu et al., 2005). Recently the Lippard group demonstrated binding of an oligonucleotide duplex with a cisplatin interstrand crosslink by MutSβ, as well as PARP-1, DNA Ligase III, XRCC1, and Ku80, Ku70 (Zhu and Lippard, 2009). The Legerski group showed PCNA stimulated binding of a TMP crosslinked duplex oligonucleotide by the MutSß complex. Incision of the crosslinked substrate in cell free extracts was dependent on MSH2, as well as ERCC1, and XPF (see below), suggesting activity of the MutSβ complex in the absence of replication (Zhang et al., 2002). These results contrast with those from experiments in yeast in which a role for mismatch repair factors was assigned to a post unhooking step in S phase (Barber et al., 2005). In the experiments with laser localized psoralen ICLs in G1 phase cells the removal of the psoralen adducts was unaffected by deficiency in MSH2. Furthermore, recruitment of MSH3 was dependent on functional XPC, suggesting that it entered the repair pathway after recognition by XPC (Muniandy et al., 2009). The linkage between mismatch repair and replication raises the possibility that the involvement of MMR functions in ICL repair might be coupled to repair synthesis, rather than recognition.
Other recognition candidates
Binding of psoralen crosslinked DNA by a complex of human α Spectrin II and the Fanconi Anemia proteins FANCA, FANCC, and FANCG has been shown by the Lambert group (McMahon et al., 2001). The key member of this complex is α Spectrin II which bound the crosslinked DNA with specificity as compared to non crosslinked DNA, but could not distinguish monoadducted DNA and undamaged DNA. Cells treated with siRNA against α Spectrin II showed an increase in chromosome abnormalities, and were modestly more sensitive to MMC than control cultures (McMahon et al., 2009). However, the biological the significance of crosslink binding by α Spectrin II has yet to be established.
It should be apparent from this overview of crosslink recognition that there is not a consensus on the role of recognition proteins in crosslink repair. The multiplicity of assay systems coupled with the disparate crosslinking agents has resulted in considerable uncertainty. It is quite possible that the mode of recognition could vary as a function of the structural characteristics of crosslinks formed by different compounds. Systematic experiments to address this possibility have not been performed.
Transcription coupled repair
The experiments showing impaired reactivation of crosslinked reporter plasmids in cells with defects in TCR (Zheng et al., 2003) were in agreement with earlier work from the Hanawalt laboratory, which first demonstrated TCR of psoralen ICLs (Vos and Hanawalt, 1987; Islas et al., 1994). As mentioned above, the genetics of cisplatin ICL repair are consistent with an involvement of TCR (Furuta et al., 2002; McKay et al., 2001). Furthermore, an analysis of mutations induced by MMC or psoralen in the plasmid reporter systems indicated a strong bias for the non transcribed strand, reflecting preferential incision in the transcribed strand (Zheng et al., 2003; Wang et al., 2001). Earlier experiments in yeast also argued for crosslink repair by TCR (Meniel et al., 1995a; Meniel et al., 1995b). However, while the results with plasmids are in accord with TCR of ICLs, there is an inherent bias in the design of the experiments. Typically, the plasmids express a reporter gene and the lesion is placed between the (invariably strong) promoter and translation start site. Transcription, required for TCR, is also required for the endpoint of the assay. Thus, only those molecules that are transcribed can contribute to the readout, and they also become candidates for TCR. Repair by non TCR pathways, of plasmids that do not become transcription templates, is not reported in these assays.
After recognition, in the absence of replication
It is generally assumed that, following recognition, NER functions are involved in further processing, much as with monoadducts. As mentioned above, cells with deficiencies in NER functions are not markedly sensitive to crosslinking agents such as nitrogen mustard (De Silva et al., 2000), although there may be some variation in sensitivity as a function of the compound (Kaye et al., 1980). A clearer indication of a role for NER activities comes from the experiments with crosslinked reporter plasmids transfected into NER proficient and deficient cells. As discussed in the preceding paragraph the results of experiments with mitomycin or psoralen crosslinked plasmids demonstrated a role for NER and TCR functions in supporting expression of the reporter gene. In both sets of experiments sharp declines in reporter activity were seen with host cells with deficiencies in XPA, XPD, and XPB (Wang et al., 2001; Zheng et al., 2003). An implication of results with cells vs. plasmids is that the crosslinked plasmids are repaired by NER dependent pathways, while genomic ICLs have access to additional, NER independent, pathways.
Post recognition: unhooking
The key event in crosslink repair is the unhooking step, and thus the identification of the critical factors is clearly important. Given the requirement for the NER pathways in bacteria and yeast the roles of the incision nucleases of the mammalian NER pathway, XPG (3' of the adduct) and ERCC1-XPF (5'), are of particular interest. Cells with deficiencies in XPF or ERCC1 are extremely sensitive to crosslinking agents, while those with defective XPG, like cells with defects in the other NER genes, generally show modest sensitivity (but see below) (Hoy et al., 1985b; Hoy et al., 1985a; Collins, 1993; Damia et al., 1996; De Silva et al., 2000; Clingen et al., 2007).
Unhooking in living cells can be monitored by the well known alkaline “comet” assay. The assay provides a measure of the electrophoretic behavior of DNA from single cells embedded in agarose (Collins, 2004). DNA in cells is organized in large supercoiled loops. Strand breaks relax the DNA, which can be unwound in an alkaline environment. The relaxed and unwound DNA will migrate towards the anode in an electrophoretic field. Although breaks can be introduced deliberately by radiation or other treatments, there are sufficient spontaneous breaks such that alkaline electrophoresis of untreated cells reveals a “head” (compact DNA in the vestige of the nucleus) and a “tail” (the more rapidly migrating nicked and relaxed material). This has the appearance of a comet. The pattern from cells that have been exposed to crosslinking agents is markedly different in that there is no tail, as the DNA is bound by the crosslinks. Unhooking of the ICLs restores the tail, and this serves as an assay of cellular competence to perform this step. While the assay does require care and attention to detail to perform reproducibly, it has several major advantages over other whole cell assays. The most important is that monoadducts do not interfere with detection of ICLs (Wu et al., 2009). Consequently it is possible to measure the response of cells to ICLs, regardless of the monoadducts formed by the crosslinking agent. Unhooking in wild type cells may take a few hours (with judicious dosing), and so it is feasible to perform experiments in which synchronized cells are treated and monitored in a particular phase of the cell cycle. Note that if an agent also forms or induces appreciable levels of single or double strand breaks this can compromise the analysis of crosslink repair (De Silva et al., 2000). Thus, if cells become apoptotic as a result of treatment, the interpretation of comet tail recovery can be confounded (Kumaresan et al., 2007).
Unhooking of ICLs formed by nitrogen mustard (Murray and Meyn, 1986) and psoralen (Rothfuss and Grompe, 2004) in G1 phase synchronized cells has been reported. In the psoralen experiments Rothfuss and Grompe made the interesting observation that the process in synchronized G1 phase cells was substantially faster than in asynchronous cells. Unhooking in psoralen/UVA treated G1 phase cells deficient in XPD was greatly reduced relative to wild type cells (Richards et al., 2005) indicating a requirement for NER activities in G1. However, most studies of the influence of repair gene deficiency on comet tail recovery have used asynchronous cultures. Deficiencies in XPG, and XPB did not alter the kinetics of unhooking in cells treated with nitrogen mustard relative to wild type cells (De Silva et al., 2000). Similarly, non synchronized human cells deficient in XPG also showed wild type kinetics of unhooking following treatment with psoralen/UVA (Rothfuss and Grompe, 2004). Cells with mutant XPF or ERCC1 genes were unable to unhook nitrogen mustard ICLs (De Silva et al., 2000). Thus unhooking of nitrogen mustard and psoralen ICLs was clearly dependent on XPF/ERCC1. The other NER functions, including XPG, were not required, insofar as asynchronous cultures were concerned. Rather different conclusions were reached in experiments with cisplatin ICLs. XPF/ERCC1 deficient cells were much more sensitive than those with defects in XPB, XPD, and XPG, but all were equally impaired in unhooking (De Silva et al., 2002). The authors suggested that the cisplatin crosslink was not the lesion critical to the survival assay, since inability to unhook was not directly matched by the impact on survival. This conclusion emphasizes the problems of elucidating the dynamics of crosslink repair with compounds that produce ICLs as minor products, while the more abundant single strand adducts can be highly toxic.
Incision of crosslinked model substrates has been examined in cell free extracts and in incubations with purified proteins. In a well known experiment, linear duplexes, or a plasmid, with a psoralen crosslink were incubated in human cell free extracts. The activities in the extract incised the substrates at two sites, both 5' of the crosslink, releasing a 22–28 base long fragment, without removing the ICL. In effect the repair reaction had occurred to one side of the crosslink. The reaction was also performed with purified NER factors, XPA, RPA, TFIIH, XPC, XPG, ERCC1-XPF. These were necessary and sufficient for the dual incisions adjacent to the crosslink (Bessho et al., 1997). In a follow-up study the same group found that the resultant gap was filled in by what was termed “futile repair synthesis” (Mu et al., 2000). Adjacent dual incision products were also recovered in recent work from the Miller laboratory in which duplexes with ICLs of differing degrees of distortion were incubated in cell free extracts (Smeaton et al., 2008). The appearance of the dual incision fragments was a function of the distortion of the ICL and dependent on NER functions. In light of the discussion above, it would seem that the results from the two labs reflect an effort by the repair apparatus to repair the distortion adjacent to the ICL, rather than removing the cause of the distortion. What remains to be determined is whether this is a peculiarity of the in vitro conditions, perhaps missing some key factor(s), or an accurate representation of events that occur, at least some of the time,in vivo.
A different endpoint was measured in another cell free extract system by Legerski and colleagues. They found that DNA synthesis on both a psoralen crosslinked plasmid and undamaged plasmid in the same extract was dependent on the ICL and ERCC1-XPF, and the recombinational repair genes XRCC2 and XRCC3. Synthesis was not affected by the absence of XPA, XPC, and XPG in the extracts. The author suggested that synthesis was the result of Break Induced Replication (BIR) (Malkova et al., 1996). This would be stimulated by incision of the crosslinked plasmid by the ERCC1-XPF complex followed by strand transfer to the undamaged plasmid and extension by DNA synthesis. The authors used this assay to identify a requirement for MutSβ in the recognition and unhooking of psoralen ICLs (Zhang et al., 2002). Neither XPC nor XPG were necessary for incision. RPA was required for both incision and DNA synthesis, while PCNA was essential only for synthesis, although stimulatory for incision (Li et al., 2000; Zhang et al., 2003). An involvement of additional factors was indicated as the result of fractionation of the extracts. A complex containing the Pso4 pre-mRNA splicing factor, and the Werner Syndrome (WS) helicase was found to stimulate ICL induced repair synthesis. This was supported by a reduced reactivation of crosslinked expression reporter plasmids in cells with siRNA knockdown of members of the Pso4 complex (Zhang et al., 2005). Extracts from WS cell line were inactive in the extract repair synthesis assay. As noted by the authors of this report, Pso4 is bound to the nuclear matrix, perhaps reflecting earlier indications that DNA repair factories are associated with the nuclear matrix (Koehler and Hanawalt, 1996).
Unhooking of psoralen crosslinked linear duplexes has been studied in extracts of chromatin associated proteins (Kumaresan et al., 1995; Kumaresan et al., 2007). The 3' and 5' incisions were 9 nucleotides apart, and were dependent on XPF. These incisions were reduced in extracts from cells deficient in FANCA (Kumaresan and Lambert, 2000), or in extracts in which antibodies against FANCA were added (Kumaresan et al., 2007). Similar results were obtained with extracts from cells with defects in other Fanconi proteins, including FANCB, C, D2, F, and G. Suppression was generally not complete, with the 3' incision more resistant to the defect in Fanconi protein. It should be noted that a role for Fanconi proteins in unhooking is controversial. There are reports indicating partial (Papadopoulo et al., 1987; Averbeck et al., 1988) (Matsumoto et al., 1989; Li et al., 1999), or no (Poll et al., 1984; Pichierri et al., 2002; Zhang et al., 2002; Rothfuss and Grompe, 2004) requirement, with some variability depending on complementation group.
Unhooking in cell extracts has also been characterized by Smeaton et al. (Smeaton et al., 2008). They prepared duplex oligonucleotides with different ICLs that varied in the degree of distortion imposed on the DNA. As mentioned above they observed the NER dependent dual 5' incisions described in earlier work by Bessho et al. (Bessho et al., 1997). However, remarkably, they demonstrated NER independent unhooking of the crosslinked substrates, also stimulated by crosslink induced distortion. This unhooking reaction was unaffected by the absence of ERCC1-XPF.
Although the work of Smeaton et al. suggests that there may be an alternative to ERCC1-XPF, most current models assume a fundamental contribution of this complex to unhooking (but see below (Bergstralh and Sekelsky, 2008)). This view is based in part on the in vivo data, but also on a very influential publication by the Wood group, who showed that a complex of the purified proteins could unhook a psoralen crosslinked oligonucleotide in the vicinity of a fork (Kuraoka et al., 2000). The complex was not active on an intact duplex with a crosslink, an interesting contrast to the incision activities in various cell extracts discussed above (Mu et al., 2000; Kumaresan et al., 2002; Smeaton et al., 2008). The dependence on a forked structure prompted speculation that unhooking of ICLs at stalled replication forks could occur in an ERCC1-XPF dependent, but otherwise NER independent, fashion. The incision product was a single strand crosslinked to 4 nucleotides. Similar results were obtained in a later study (Fisher et al., 2008). Recent work shows that ERCC1-XPF does not function alone. Instead it is found associated with the SLX4 protein, which acts as a scaffold for this complex as well as two other structure specific nucleases, MUS81-EME1 and SLX1. The SLX4 association enhances the activity of the nucleases, XPF, MUS81, SLX1, in these complexes (Munoz et al., 2009; Fekairi et al., 2009; Andersen et al., 2009). Cells treated with siRNA to knock down SLX4 expression are sensitive to crosslinking agents, although the sensitivity is not as profound as that seen in the XPF or ERCC1 deficient cells. On the other hand the knock down cells are not sensitive to UV light arguing that SLX4 is involved in the transactions of ERCC1-XPF outside of classical NER. However, ERCC1-XPF is involved in recombination (Sargent et al., 1997; Adair et al., 2000; Niedernhofer et al., 2001), double strand break repair (Ahmad et al., 2008), as well as adduct repair. Thus, whether SLX4 plays a role in unhooking, or other pathways associated with crosslink repair (reconstruction of blocked replication forks, DSB repair, etc.) remains to be determined.
Based on their biochemical studies the Sancar group made the interesting proposal that ERCC1-XPF in combination with RPA could act as a 3'-5' exonuclease on a linear duplex containing a crosslink. The product of this reaction was one strand of the original duplex attached to a single thymine via the psoralen crosslink (Mu et al., 2000). Digestion of one of the strands past the crosslink would unhook the crosslink and generate a stretch of single stranded DNA. It seems reasonable to consider that this could be done by other nucleases, and could be an alternative to incision based unhooking.
Recognition by replication fork encounter
The demonstration of unhooking by ERCC1-XPF of crosslinked forked substrates by the Wood group solidified the concept of crosslink recognition by replication fork collision. This recognition pathway and its implications have been the subject of much discussion since then. An aspect of this research has been the shift from an experimental concern for repair/removal of ICLs as chemical entities, to a focus on the resolution of the consequences of the encounter- stalled and collapsed replication forks. This reflects the strong general interest in recombinational repair, which is essential for efficient and faithful replication and protects against genomic rearrangements that may have clinical consequences (Thompson and Schild, 2002; Thompson and Hinz, 2009). It is also well established that cells with defects in recombination repair functions are sensitive to crosslinking agents, and mutations in some of these genes are linked to cancer (Liu et al., 1998; Sasaki et al., 2004; De Silva et al., 2000). Double strand breaks induced by crosslinking compounds figure prominently in most of these studies. DSB formation can be monitored by pulsed field electrophoresis, and, of course, by the appearance of γH2AX foci (Rogakou et al., 1999), which has been widely used as a surrogate marker. However, it appears that there are other inducers of γH2AX foci, such as single strand DNA at repair foci and chromatin swelling (Clingen et al., 2008; Baure et al., 2009; Marti et al., 2006). Consequently, other measurements of DSB formation are important to avoid misinterpretation of γH2AX foci.
In the following section we will discuss experiments intended to address crosslink repair in the context of a replication fork. As discussed above, while experiments may be designed with this as the explicit focus, other pathways of discovery and repair are functional throughout the cell cycle (Barber et al., 2005). Thus results of experiments in cells synchronized in S phase may reflect the activity of multiple repair pathways.
Grompe and colleagues showed that synchronized cells treated with psoralen/UVA required passage through S phase to elicit chromosome breakage or cell cycle delay. They proposed that replication fork arrest at a crosslink triggers cellular responses. They further argued that at least the initial steps of crosslink recognition and repair occurred exclusively in S phase (Akkari et al., 2000) (but see below). A similar proposal was made by McHugh, Hartley and colleagues, who called attention to the DSBs generated only during S phase in cells treated with crosslinkers. They developed a model in which double strand break formation at a crosslink stalled fork was followed by unhooking by ERCC1-XPF, and recombinational repair of the resultant gap (De Silva et al., 2000; McHugh et al., 2001). This model accounted for the double strand breaks found in S phase cells treated with crosslinkers and, reminiscent of the Cole model, suggested that recombination was a pathway for gap repair, as well as fork reconstruction. Of course with models that invoke HDR, there is a need for homologous sequences that would span the crosslink site. This could not come from the daughter duplexes at the fork, since neither could replicate through the crosslink. Instead gap repair by recombination might be expected in G2 phase with one sister chromatid supplying information to repair the other. However, when this issue was considered in synchronized yeast cells it was found that recombinational repair was not a major contributor to crosslink repair in G2 phase (Barber et al., 2005).
Double strand breaks were provoked by nitrogen mustard and cisplatin in replicating, but not stationary phase, yeast cells (McHugh et al., 2000). Similar observations were made in CHO cells treated with nitrogen mustard (De Silva et al., 2000). Two scenarios could account for this. In one report (Rothfuss and Grompe, 2004) the authors suggested that unhooking could occur in G1 phase cells. In the ensuing S phase the encounter of replication forks with unrepaired nicks and gaps generated by unhooking would produce single sided double strand breaks. This process would not require a specific nuclease for production of the DSBs, which would occur well after unhooking. The alternative view, very much in current vogue, is that there is a repair pathway in S phase, initiated by collision of a crosslink with a fork. Support for this comes from experiments with crosslinked replicating plasmids (Liu et al., 2006) and actively dividing cells (Niedernhofer et al., 2004). Cleavage by a nuclease at a fork, of a template strand upstream of the crosslink, would release one arm of the replication fork. This would generate a DSB, and a substrate that would require a second incision to complete unhooking (Kuraoka et al., 2000; De Silva et al., 2000; Bessho, 2003).
The enzymology of DSB formation has been considered in several studies. Although one report described a requirement for ERCC1-XPF for γH2AX focus formation in response to psoralen/UVA (Mogi and Oh, 2006), other investigators, using electrophoretic techniques, found that ERCC1-XPF was not necessary for nitrogen mustard or MMC induced DSB formation (De Silva et al., 2000; Niedernhofer et al., 2004). Instead it has been proposed that the Mus81-Eme1 nuclease, a structure specific endonuclease with a preference for replication fork like structures, is required. This was shown in experiments with wild type and Mus81 deficient cells treated with for 24–30 hrs with MMC (Hanada et al., 2006). The cells accumulated in S phase and the appearance of double strand breaks was dependent on Mus81-Eme1. This was in accord with proposals that this nuclease is involved in converting replication blocking structures to DSBs, which can then enter a recombinational repair pathway to restore the replication fork (Hanada et al., 2007). It is noteworthy that when cells were treated for 1 hr with MMC, breaks appeared without requirement for Mus81-Eme1 (Dendouga et al., 2005; Hanada et al., 2006). Following exposure to MMC Rad51 foci appeared in both wild type and Mus81 knock out cells. In wild type cells the foci declined over time while they persisted in the mutant cells. This argued that Mus81-Eme1 was involved in resolving a late step in the recombination pathways that repaired crosslink induced DSBs (Dendouga et al., 2005). It was suggested that the discrepancy between the results of experiments with long and short exposure to MMC was reflection of the small fraction of ICLs in comparison to monoadducts, such that the monoadducts were the actual agents of replication fork block and DSB formation in the short exposure experiments (Hanada et al., 2006). The longer exposure would allow greater crosslink accumulation which would then block replication forks, now requiring Mus81-Eme1 for cleavage. This explanation requires a qualitative difference between forks blocked by monoadducts and ICLs, and overlooks the continued production of monoadducts during the longer exposure. The recovery of conflicting results from two versions of an experiment with the same crosslinking agent, which produces several other adducts, is an excellent example of the challenge facing investigators in the field. It would be of interest to address the role of Mus81-Eme1 using psoralen/UVA as the damaging agent, as the production of adducts is immediate to the light exposure.
Mus81-Eme1 recruitment at a stalled fork requires SNM1B, one of three mammalian orthologues of the SNM1 gene in S. cerevisiae. SNM1B deficient cells are sensitive to crosslinking agents such as MMC and cisplatin (Demuth et al., 2004), but not UV (Bae et al., 2008). SNM1B interacts with the MRN complex, indirectly with FANCD2, and is required for activation of ATM and Chk2 after MMC treatment. SNM1B binds Mus81 in vitro, and is necessary for the production of DSBs in cells treated with MMC (Bae et al., 2008), and these authors have proposed that SNM1B is essential for the conversion of a crosslink stalled fork to a broken fork via cleavage by Mus81-Eme1.
Although not required for DSB formation, and thus no longer seen as the exclusive agent of unhooking during fork related repair, ERCC1-XPF is required for the resolution of the breaks as monitored by the decline in γH2AX foci (Niedernhofer et al., 2004). Results from a plasmid based assay support this conclusion. A DSB, adjacent to a crosslink, stimulated repair via a homology dependent pathway. Repair was dependent on ERCC1-XPF, Rev3, and components of the FA and MMR pathways (Zhang et al., 2007). Data from these experiments support a model in which the template strand for leading strand synthesis, at a crosslink stalled fork, is cleaved by Mus81-Eme1. The cleavage, in response to a stalled fork rather than the crosslink as such, would be the first of the two incisions required for unhooking. This generates a single sided DSB (the leading side daughter duplex) and a still crosslinked parental duplex with gap adjacent to the crosslink. This would be the substrate for incision on the 5' side of the crosslink by ERCC1-XPF (Niedernhofer et al., 2004). Although not a feature of the original models, presumably this could also be engaged by an exonuclease(s) that could digest one strand of the parental duplex through the crosslink, leaving a single base crosslinked to the other strand (Mu et al., 2000). Extended exonuclease digestion would generate the single strand patches that have been shown to appear following exposure of cells to crosslinking agents such as MMC (Lee et al., 2006).
A contrary role for ERCC1-XPF was advanced by Bergstralh and Sekelsky. They argued that this complex was not involved in unhooking associated with replicative repair. Instead they proposed that unhooking at a blocked replication fork was dependent on Mus81-Eme1 and MutSβ/MutL(Bergstralh and Sekelsky, 2008). In their proposal the requirement for ERCC1-XPF would be after the actual repair of the crosslink, during the reconstruction of the fork by recombination. This scenario would remove ERCC1-XPF from a direct involvement in formation of the repair gap, the filling of which by TLS would generate base substitution mutations. However, in experiments with a triplex targeted crosslink it was found that the ERCC1-XPF complex was required for the appearance of point mutations at the crosslink site (Richards et al., 2005). This observation would appear to support the more conventional view of ERCC1-XPF as essential for unhooking.
The Fanconi Anemia pathway
Aside from the question of the enzymology of incision, crosslink discovery by replication has received considerable elaboration in the last few years particularly as events at the blocked fork are concerned. In effect, the models for crosslink discovery and unhooking at replication forks have become embedded in larger schemes that address the cellular response to stalled forks. How blocked forks are broken and resolved has been the subject of recent excellent reviews and primary publications, and is beyond the scope of this article (Collis et al., 2008; Youds et al., 2009; Thompson and Hinz, 2009) (Osman and Whitby, 2007; Budzowska and Kanaar, 2009; Sun et al., 2008). The FA proteins play prominent roles in multiple steps in the process. FANCM and FAAP24 are thought to recognize blocked forks and, via fork reversal, enable access to a cavalcade of enzymes and proteins that repair the damage (Gari et al., 2008a; Gari et al., 2008b). The activity of the core complex ((A–C, E–G, L, FAAP100) is required for the central event of the FA pathway, the monoubiquitination of FANCD2 and FANCI. Core complex proteins are additionally important for lesion bypass synthesis (Mirchandani et al., 2008). FANCD2 and FANCI, in concert with the chromatin remodeler Tip60 (Hejna et al., 2008), are involved in the reconstruction of the broken fork via HDR which also engages FANCD1/BRCA2 and FANCN/PALB2 (see (Thompson and Hinz, 2009; Moldovan and D'Andrea, 2009)).
A linkage between the activity of ERCC1-XPF and the activation and localization of FANCD2 has been suggested by recent work from Moses, Grompe and their colleagues (McCabe et al., 2008). They reported a decrease in monoubiquitination of, and focus formation by, FANCD2 in ERCC1 deficient cells after MMC or hydroxyurea treatment. They proposed that double strand break formation (provoked by both compounds) was the critical event that triggered the FANCD2 response, and that this required the activity of ERCC1-XPF. This question has also been addressed by McHugh and associates. Although they found that MMC induced monoubiquitination of FANCD2 was not affected by ERCC1 deficiency, they did find reduced focus formation and a reduction in chromatin associated FANCD2 (Bhagwat et al., 2009). They suggested that unhooking by ERCC1-XPF was required for stable chromatin association of FANCD2, and subsequent double strand break repair. It should be noted that these results describe FANCD2 recruitment after the action of ERCC1-XPF, in contrast to models in which FANCD2 accumulation at stalled forks precedes ERCC1-XPF (Thompson and Hinz, 2009).
This focus on double strand breaks, subsequent to a stalled fork, as the inducer of the Fanconi pathway is consistent with work showing activation of FANCD2 in response to agents that introduce breaks independent of replication (Nakanishi et al., 2002; Roques et al., 2009) The possibility that a blocked, but unbroken, fork is also an inducer cannot be considered with most compounds in general use- aphidicolin, hydroxyurea, MMC etc.,- because they induce double strand breaks. However, FANCD2 is monoubiquitinated and appears in foci in cellssynchronized in S phase by double thymidine block, which does not provoke double strand breaks (Bolderson et al., 2004; Taniguchi et al., 2002). This would imply multiple entry points for FANCD2- at stalled forks, as well as the breaks that can form at stalled forks-and could reconcile differences in current models of fork reconstruction.
Role of recombinational repair
There has been a shift in the recent models away from positing recombinational pathways as involved in both repair of the incision gap and the replication fork. The current view is that gap filling is accomplished exclusively by lesion bypass polymerases. Recombinational repair to rebuild the fork proceeds only after both cycles of repair of the crosslink adducts (Niedzwiedz et al., 2004; Patel and Joenje, 2007) (Thompson and Hinz, 2009; Niedernhofer et al., 2005; Mirchandani and D'Andrea, 2006). The models account for the sensitivity of HDR deficient cells to crosslinking agents by placing these functions downstream of the unhooking/gap repair events, and do not invoke them as engaged directly in these processes (Sasaki et al., 2004; Clingen et al., 2008) (although there may be an exception in the instance of unhooking of cisplatin in which XRCC3, a Rad51 paralogue, has been implicated (De Silva et al., 2002)). Thus, for example, in experiments with a replicating crosslinked plasmid, unhooked, but incompletely repaired, intermediates accumulated in cell extracts derived from BRCA2 deficient cells (Cipak et al., 2006). Cell extracts prepared from BRCA2 complemented cells were able to complete the repair process. It should be noted that all the models describe events at a single replication fork as it encounters a crosslink.
The double fork collision model
An alternative view has been proposed by Walter and colleagues who examined replication and repair in a Xenopus egg extract of a plasmid with ICLs formed by either the non distorting 7-deaza-guanine nitrogen mustard analogue from the Schärer group (Angelov et al., 2009) or by cisplatin. In this system the NER pathway appears to be inactive. However the plasmid is replicated bi-directionally and both forks collide with the crosslink. They followed the progress of replication and length of the daughter strands by 32P-α-dATP incorporation. The daughter strands stopped 20 (cisplatin) or 24 (nitrogen mustard) bases from the crosslink. The authors suggested that the DNA around the cisplatin crosslink was more easily unwound, and thus the fork could come somewhat closer. Eventually the leading strands advanced to within 1 nucleotide of the crosslink. This was followed by unhooking by unidentified nucleases, creating a structure in which translesion synthesis occurred via extension of the nascent leading strand past the crosslinked base. The collision of the forks with the crosslink induced Chk1 phosphorylation activating the ATR signaling pathway, and ubiquitination of FANCD2. The authors of this report called attention to two features of their model that distinguished it from earlier proposals. The first is the collision of two replication forks instead of one. The second is the use of a nascent leading strand as the primer for lesion bypass synthesis, rather than the parental strand, as in the single fork models. In this scenario the unhooking incisions yield the equivalent of a double sided double strand break. This is the classic substrate for repair by NHEJ, and a failure to keep the broken ends sequestered could result in joining with deletion. Furthermore, although the FA pathway was activated, their model does not invoke a fork reversal step, one of the functions proposed for FANCM (Gari et al., 2008a).
After unhooking: processing and gap filling
The intermediate unhooked by incisions is likely to require further processing prior to gap synthesis and the start of the second repair cycle that removes the resultant monoadduct. These events must occur regardless of cell cycle status. Unhooking of the psoralen crosslinked substrate by purified ERCC1-XPF released a 4 base oligonucleotide crosslinked to the unhooked strand (Kuraoka et al., 2000). In the NER independent unhooking reactions described by the Miller group the incised oligonucleotide varied from 4 to 11 bases depending on the chemistry of the crosslink (Smeaton et al., 2008). The structural chemistry of the crosslink (extent of distortion) can reduce the stability of the hybrid of the incised oligonucleotide and the non incised strand, or it can enhance the stability, relative to the non crosslinked hybrid (Noronha et al., 2002). Consequently there may be a processing step required for elaboration of a repair “gap” on the incised strand. This could require a helicase. Unwinding of the unhooked product in G1 phase cells would presumably rely on the NER helicases. In the context of a blocked or broken replication fork and NER independent repair, one candidate for this putative activity would be the FANCJ helicase (Peng et al., 2007; Gupta et al., 2007; Wu and Brosh, Jr., 2009; Thompson and Hinz, 2009). FANCJ deficient cells are sensitive to crosslinking agents, and it has the right polarity, 5'-3', to allow it to load on the un-cleaved parental strand and unwind, at least to the crosslinked base. Experiments in C. elegans indicated that the FANCJ equivalent protein was not required for FANCD2 ubiquitination, but did not address a more specific role (Youds et al., 2008).
Digestion of the oligonucleotide to a crosslinked di- or mono-nucleotide is probably necessary to enable repair synthesis by lesion bypass polymerases and PCNA. A candidate nuclease, Pso2 (Snm1) (Li et al., 2005), was identified in experiments in yeast as important in S phase repair, although it has been assigned a role in G1 phase as well (Barber et al., 2005; McHugh and Sarkar, 2006). The human SNM1 protein is a 5'-exonuclease, specific for single stranded DNA (Hejna et al., 2007). It is a member of a family that includes, among others, SNM1B/Apollo, involved in replication fork collapse ((Bae et al., 2008), see above), and SNM1C/Artemis, involved in NHEJ and V(D)J recombination (Ma et al., 2002). Mouse cells deficient in SNM1 were sensitive to MMC (Dronkert et al., 2000), as were human cells in which SNM1 expression was suppressed by siRNA (Hemphill et al., 2008). Unlike yeast, the mouse SNM1−/− ES cells are not sensitive to other crosslinking agents, such as cisplatin, psoralen, or nitrogen mustard (Dronkert et al., 2000). This may reflect the relatively non distorting nature of MMC ICLs, perhaps suggesting that alternative functions process the other, more distorting adducts, but a definitive explanation has not been established. Curiously, SNM1A has been shown to partially rescue yeast pso2/snm1 mutants exposed to nitrogen mustard and cisplatin (Hazrati et al., 2008). Unfortunately MMC was not employed in those experiments. Digestion of the incised oligonucleotide in the 3'-5' direction would also be expected to enhance lesion bypass synthesis (Minko et al., 2008). This function might be supplied by the ERCC1-XPF complex as discussed above, other nucleases, or it has also been suggested that polδ could provide this activity (Nick McElhinny et al., 2006).
Repair synthesis to fill the gap produced by unhooking and subsequent processing is an essential step that is required to complete the first repair cycle and restore the duplex. Since synthesis past an adducted base is unavoidable, there is a requirement for recruitment of error prone lesion bypass polymerases. These polymerases, many in the Y family, have been the subject of intense investigation since their discovery, and have been described in recent reviews (Yang and Woodgate, 2007; Budzowska and Kanaar, 2009; Broyde et al., 2008; Lehmann, 2006). Understandably, most work with these polymerases has been with monoadducts, which, if unrepaired, pose a problem for replication fork progression during S phase. The Lehmann group described the switch from replicative polymerases to the translesion polymerases via mono-ubiquitination of the replication processivity factor PCNA (Kannouche and Lehmann, 2004). All Y family TLS polymerases bind the mono-ubiquitinated form of PCNA, and different polymerases are specific for different forms of DNA damage (Lehmann et al., 2007). The signal for this modification is a stalled replication fork (Kannouche et al., 2003). However, crosslink repair in any cell cycle phase requires translesion bypass synthesis, and PCNA monoubiquitination in G1 phase obviously cannot be triggered by a stalled replication fork. This issue was addressed in experiments in yeast by Sarkar at al, who showed rapid mono-ubiquitination of PCNA in G1 phase yeast cells treated with crosslinking agents, but not UV (Sarkar et al., 2006). PCNA modification required a functional pol32, a component of the replicative polymerase polδ, which is also involved in repair. PCNA mono-ubiquitination was not dependent on Rev3, a component of polζ. Thus it appears that the encounter by polδ of the monadduct product of crosslink unhooking is the signal for PCNA mono-ubiquitination and the recruitment of TLS polymerases. These would then complete synthesis of the repair gap.
While accurate bypass of UV induced thymine dimers can be accomplished by polη alone, many lesions require the action of two polymerases: one to insert a base across from the site of the lesion, and polζ to extend that insertion (Johnson et al., 2000). Recent elegant work with a plasmid model showed that bypass of benzopyrene or cisplatin-GG (intrastrand) adducts required a variable combination of two polymerases. Depending on the adduct, polκ or polη were responsible for insertion of correct or mutating bases while polζ was required for extension (Shachar et al., 2009). These studies were with classical monoadducts and the equivalent studies have not been performed on monoadducts formed by unhooking of ICLs of different compounds. However some information about the relevant polymerases is available from disparate experimental systems.
DT40 cells deficient in RAD18, which regulates TLS as a ubiquitin E3 ligase for PCNA, and Rev3, the catalytic subunit of polζ, were very sensitive to nitrogen mustard, MMC, and cisplatin (Nojima et al., 2005). Rev1, is a deoxycytidyl transferase that interacts with other Y family polymerases and with Rev7, a subunit of polζ (Guo et al., 2003; Masuda et al., 2003; Akagi et al., 2009). Cells deficient in Rev1 show increased chromosome breakage and radial formation after treatment with MMC, as compared to controls (Mirchandani et al., 2008). Interestingly, chromosome radials, which are the fusion of broken sister chromatids of disparate chromosomes, are a hallmark of FA cells treated with crosslinking agents. A direct connection between Rev1 and the FA pathway is suggested by the reduction in Rev1 focus formation after DNA damage in FANCG deficient cells (Mirchandani et al., 2008)(see below).
A role for polη was shown in experiments with both psoralen and MMC ICLs. Early work demonstrated the sensitivity of cells from an XP variant donor (deficient in polη) to psoralen/UVA (Misra and Vos, 1993). Plasmids carrying a triple helix targeted psoralen crosslink showed an elevated frequency and altered mutation pattern after passage through cells deficient in polη (Raha et al., 1996). The Legerski group showed a substantial reduction in reactivation of an expression plasmid with a defined mitomycin crosslink after transfection into polη deficient cells (Zheng et al., 2003). Psoralen/UVA treated Polη deficient cells show an enhanced production of γH2AX foci, suggesting that this polymerase contributes to repair of crosslinks and reduces the probability that they will encounter a fork, resulting in a DSB (Mogi et al., 2008). While this observation is consistent with that scenario, as noted above and by the authors of the report, there are inducers of γH2AX foci other than DSBs, and they may be activated in the absence of polη.
Bypass in model systems has been examined with substrates that mimic the product of unhooking. Minko et al. found that plasmids carrying an G-G interstrand acrolein crosslink (similar to that formed by MMC) were replicated in mammalian cells with very low levels of mutagenesis, suggesting that most bypass events were error free (Minko et al., 2008). Human polκ could synthesize past a substrate designed to mimic the unhooked acrolein, incorporating dCTP across from the adducted G. The most efficient bypass was obtained with a substrate with a dinucleotide attached via the crosslink to the template strand. The authors concluded that polκ could accurately bypass the G-N2-N2-G crosslink. In contrast, bypass by the combination of Rev1 and polζ was inefficient. Knockdown of polκ in cultured human cells followed by treatment with MMC resulted in an enhanced frequency of radial chromosomes, again reminiscent of the phenotype of FA cells.
Replication bypass of model unhooked substrates has been examined from another point of view in the work of Smeaton et al. (Smeaton et al., 2009). They prepared oligonucleotide substrates with a base crosslinked to another base. However, the chemistry of the crosslink was such that in some constructs hydrogen bonding was obstructed (N3T-ethyl-N3T), while in others it was not (N4C-ethyl-N4C). Bypass of the non obstructing lesion in mammalian cell extracts was efficient and error free. Bypass was not detected with the obstructing substrates. These data emphasize the relationship between the chemical structure of the crosslink and the efficiency of lesion bypass synthesis. Furthermore, they suggest that, depending on the structure of the crosslink remnant, ICL repair could be blocked even after unhooking.
Analysis of the dynamics of lesion bypass in the context of crosslink repair is a relatively recent development, and not all candidate polymerases have been considered experimentally. For example, Drosophila cells deficient in the polymerases encoded by the mus308 gene are sensitive to crosslinking agents (Harris et al., 1996; Oshige et al., 1999). There are two mammalian homologues, polN and polQ(θ) (Seki et al., 2003; Marini et al., 2003; Takata et al., 2006; Seki and Wood, 2008). While a recent study indicates a role for polN (Zietlow et al., 2009), the participation of polQ(θ) in gapfilling during crosslink repair has not been addressed. Given the different structures of the unhooked crosslinks formed by the compounds discussed here it is likely that there will be some variety in the combinations of bypass polymerases involved in completion of the first repair cycle. The mechanistic role of the FA proteins in bypass is very much an open question. Current models place FA functions at blocked replication forks, and their contribution to lesion bypass and polζ dependent mutagenesis as an S phase function. However, bypass must occur during G1 and G2 phase repair. Whether there are differences in the functions involved in gap filling in the context of a blocked fork or in the absence of replication remains to be determined.
Another issue that has been raised in recent work is the extent of repair synthesis. In classical NER the repair gap is about 30 nucleotides. However, the Gautier group, working with a plasmid with a single ICL in a Xenopus egg extract, observed synthesis corresponding to about 300 nucleotides (Ben-Yehoyada et al., 2009). Whether this reflects fill-in of an extended repair gap (which would be preceded by the exposure of a similarly sized single strand region), or “nick translation” that would replace intact sequence was not determined.
Crosslink mutagenesis
Bypass of an adduct can have mutagenic consequences. It is well established that polζ is required for monoadduct mutagenesis, as explained by the two step/two polymerase model of lesion bypass (Johnson et al., 2000; Lawrence et al., 2000). Similarly, it plays a role in the mutagenesis of ICLs. Thus, the frequency of base substitutions at the site of a triplex targeted crosslink was markedly reduced in cells in which the expression of polζ was suppressed (Richards et al., 2005). Furthermore, mutagenesis of crosslinked plasmids declined in Rev1 and Rev3 mutant host cells (Shen et al., 2006). There is a connection between components of the FA pathway and mutagenesis of both monoadducts and ICLs (Niedzwiedz et al., 2004; Thompson et al., 2005). It has been known for some time that FA cells are hypomutagenic in response to psoralen (Papadopoulo et al., 1990a; Papadopoulo et al., 1990b; Hinz et al., 2006). Recent work shows that mutagenesis requires the function of the FA core complex, but not FANCD2 (Mirchandani et al., 2008). What the molecular interaction between the FA proteins and mutagenic bypass is not clear. As noted above the FA core complex makes a contribution to Rev1 focus formation, but interactions between FA core complex members and Rev1 have not been detected (Mirchandani et al., 2008).
Although many details of lesion bypass during crosslink repair remain obscure, the pattern of mutagenesis can be informative. The “diagonal” nature of crosslinking agents such as psoralen makes it possible to deduce, from the mutation spectra, the identity of the incised strand and the strand carrying the crosslinked base during lesion bypass synthesis. One might imagine that if unhooking occurred randomly such that there was an equal probability that either strand could be incised, then bypass synthesis would also occur on either strand at a particular crosslink site averaged over a population of cells. Thus it would be expected that mutations at both of the bases in a crosslink would be recovered. However, in several studies the pattern of mutations induced by psoralen showed a remarkable asymmetry (Sage et al., 1993; Laquerbe et al., 1995; Yang et al., 1994; Chiou and Yang, 1995; Sage et al., 1993). The pattern was consistent with preferential incision of the transcribed strand, such that the unhooked crosslink would be on the non-transcribed strand during bypass synthesis. The identical asymmetry in base substitution was recovered in experiments with a triplex targeted crosslink at a genomic site (Richards et al., 2005). With one exception (Sage et al., 1993) these reports described experiments with HPRT as the mutation marker gene. There is an origin of replication located in the region of the HPRT promoter (Cohen et al., 2002), and so the direction of transcription and the leading strand of replication would be the same. Thus, the mutation pattern is consistent with initiation of repair by the encounter of ICLs with the transcriptional apparatus or a single replication fork. The data are not consistent with the pattern of mutations-at both positions of the crosslink- that would be expected if two forks had collided on opposite sides of the same crosslink (Raschle et al., 2008). However it should be noted that the data set is quite limited, in that mutation patterns in only two genes have been analyzed. It is reasonable to expect that there will be examples of the dual fork model operative at the level of the genome, and that the mutation pattern will be indicative of that pathway.
The second repair cycle
The completion of repair synthesis restores the duplex and forces the crosslinked base, linked to the unhooked strand, out of the helix. This is the substrate for the second cycle of repair. Recent evidence suggests that the unhooked extra helical psoralen crosslink is recognized by the XPE complex. Recruitment of DDB2 to laser localized ICLs occurred much later than XPC protein, and was dependent on XPC and repair synthesis (Muniandy et al., 2009). These data support a scenario in which entry into an NER dependent pathway in G1 phase cells converts a psoralen ICL, which is not recognized by the XPE complex (Payne and Chu, 1994), to a structure that is. Although the XPE complex is not required for any stage of psoralen crosslink repair, it serves as a marker of the completion of the first repair cycle and the start of the second. At this point it is generally assumed that this is a task for the NER pathway, although there is only limited evidence, from an in vitro system, in direct support of this view (Cipak et al., 2006).
An alternative suggestion for the second repair cycle has been proposed in work with the NEIL1 glycosylase. Cells deficient in this activity are sensitive to psoralen/UVA (Couve-Privat et al., 2007). Recently, this group prepared three stranded oligonucleotide structures designed to mimic an unhooked psoralen crosslink attached to a 9-mer oligonucleotide. These constructs were substrates for NEIL1, which was able to cleave the glycosidic bond between the psoralen-thymine adduct and the sugar, producing a thymine base crosslinked to the 9-mer oligonucleotide (Couve et al., 2009). They suggested that the glycosylase, which has a relatively small active site, would bind only the ejected thymine, and not the bulky crosslinked oligonucleotide. Glycoslylases other than NEIL1 might also be involved in crosslink repair. Samson and colleagues found that cells derived from knock-out mice in which the alkyladenine glycosylase (also known as methylpurine glycosylase, MPG (Miao et al., 2000)) gene was inactivated were sensitive to psoralen (Allan et al., 1998), and had reduced and delayed formation of γH2AX foci in comparison to wild type cells. However, the purified protein did not bind a psoralen crosslinked oligonucleotide duplex, arguing against a direct role in crosslink recognition (Maor-Shoshani et al., 2008). If these enzymes do remove the crosslinked monoadduct that appears at the start of the second repair cycle they would provide an alternative to the NER pathway, perhaps explaining the relatively mild sensitivity of NER deficient cells (other than ERCC1-XPF) to crosslinking agents. If there is an involvement of a glycosylase then it might expected that other members of the BER pathway, AP endonuclease, polβ, etc, would also be engaged. This possibility has not received attention.
Conclusions
It is customary to introduce discussions of crosslink repair by noting our poor understanding of the process. Indeed, in this review we have called attention to inconsistencies, contradictions, and uncertainties in the literature. In large part this is because of the multiplicity of crosslinking agents used by investigators, and the differing properties of those agents and their products. This is unlike the experience in the elucidation of a pathway such as NER in which so much of the inquiry was with UV photoproducts. Conclusions from that work could serve as a reference for studies on monoadducts formed by other agents. This situation obviously does not apply to the crosslink field. However, this state of affairs is not likely to be long lived. The maturation of the monoadduct repair fields and the migration of insight and technology to ICL repair will have a major impact. Advances in the Fanconi Anemia field have had a significant influence on, and expanded the interest in, ICL repair. Additionally, the value of DNA repair analyses for the development of improved chemotherapy regimens has been recognized. The prominence of crosslinking agents in the clinic adds the potential of practical application to the rationale for these studies. In the not too distant future we can expect considerable clarification of what has proven to be a very challenging problem.
ACKNOWLEDGEMENT
The writing of this article was supported entirely by the intramural research program of the NIH, National Institute on Aging (AG000746-02).
Footnotes
Declaration of interest The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
REFERENCES
- Adair GM, Rolig RL, Moore-Faver D, Zabelshansky M, Wilson JH, Nairn RS. Role of ERCC1 in removal of long non-homologous tails during targeted homologous recombination. EMBO J. 2000;19:5552–5561. doi: 10.1093/emboj/19.20.5552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmad A, Robinson AR, Duensing A, van DE, Beverloo HB, Weisberg DB, Hasty P, Hoeijmakers JH, Niedernhofer LJ. ERCC1-XPF endonuclease facilitates DNA double-strand break repair. Mol. Cell Biol. 2008;28:5082–5092. doi: 10.1128/MCB.00293-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akagi J, Masutani C, Kataoka Y, Kan T, Ohashi E, Mori T, Ohmori H, Hanaoka F. Interaction with DNA polymerase eta is required for nuclear accumulation of REV1 and suppression of spontaneous mutations in human cells. DNA Repair (Amst) 2009;8:585–599. doi: 10.1016/j.dnarep.2008.12.006. [DOI] [PubMed] [Google Scholar]
- Akkari YM, Bateman RL, Reifsteck CA, Olson SB, Grompe M. DNA replication is required to elicit cellular responses to psoralen-induced DNA interstrand cross-links. Mol. Cell Biol. 2000;20:8283–8289. doi: 10.1128/mcb.20.21.8283-8289.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allan JM, Engelward BP, Dreslin AJ, Wyatt MD, Tomasz M, Samson LD. Mammalian 3-methyladenine DNA glycosylase protects against the toxicity and clastogenicity of certain chemotherapeutic DNA cross-linking agents. Cancer Res. 1998;58:3965–3973. [PubMed] [Google Scholar]
- Andersen SL, Bergstralh DT, Kohl KP, LaRocque JR, Moore CB, Sekelsky J. Drosophila MUS312 and the vertebrate ortholog BTBD12 interact with DNA structure-specific endonucleases in DNA repair and recombination. Mol. Cell. 2009;35:128–135. doi: 10.1016/j.molcel.2009.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreassen PR, Ren K. Fanconi anemia proteins, DNA interstrand crosslink repair pathways, and cancer therapy. Curr. Cancer Drug Targets. 2009;9:101–117. doi: 10.2174/156800909787314011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angelov T, Guainazzi A, Scharer OD. Generation of DNA interstrand cross-links by post-synthetic reductive amination. Org. Lett. 2009;11:661–664. doi: 10.1021/ol802719a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Averbeck D, Papadopoulo D, Moustacchi E. Repair of 4,5',8-trimethylpsoralen plus light-induced DNA damage in normal and Fanconi's anemia cell lines. Cancer Res. 1988;48:2015–2020. [PubMed] [Google Scholar]
- Bae JB, Mukhopadhyay SS, Liu L, Zhang N, Tan J, Akhter S, Liu X, Shen X, Li L, Legerski RJ. Snm1B/Apollo mediates replication fork collapse and S Phase checkpoint activation in response to DNA interstrand cross-links. Oncogene. 2008;27:5045–5056. doi: 10.1038/onc.2008.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baik MH, Friesner RA, Lippard SJ. Theoretical study on the stability of N-glycosyl bonds: why does N7-platination not promote depurination? J. Am. Chem Soc. 2002;124:4495–4503. doi: 10.1021/ja017588+. [DOI] [PubMed] [Google Scholar]
- Balcome S, Park S, Quirk Dorr DR, Hafner L, Phillips L, Tretyakova N. Adenine-containing DNA-DNA cross-links of antitumor nitrogen mustards. Chem Res. Toxicol. 2004;17:950–962. doi: 10.1021/tx0499463. [DOI] [PubMed] [Google Scholar]
- Barber LJ, Ward TA, Hartley JA, McHugh PJ. DNA interstrand cross-link repair in the Saccharomyces cerevisiae cell cycle: overlapping roles for PSO2 (SNM1) with MutS factors and EXO1 during S phase. Mol. Cell Biol. 2005;25:2297–2309. doi: 10.1128/MCB.25.6.2297-2309.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baure J, Izadi A, Suarez V, Giedzinski E, Cleaver JE, Fike JR, Limoli CL. Histone H2AX phosphorylation in response to changes in chromatin structure induced by altered osmolarity. Mutagenesis. 2009;24:161–167. doi: 10.1093/mutage/gen064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behe MJ. An overabundance of long oligopurine tracts occurs in the genome of simple and complex eukaryotes. Nucleic Acids Res. 1995;23:689–695. doi: 10.1093/nar/23.4.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beljanski V, Marzilli LG, Doetsch PW. DNA damage-processing pathways involved in the eukaryotic cellular response to anticancer DNA cross-linking drugs. Mol. Pharmacol. 2004;65:1496–1506. doi: 10.1124/mol.65.6.1496. [DOI] [PubMed] [Google Scholar]
- Ben-Yehoyada M, Wang LC, Kozekov ID, Rizzo CJ, Gottesman ME, Gautier J. Checkpoint signaling from a single DNA interstrand crosslink. Mol. Cell. 2009;35:704–715. doi: 10.1016/j.molcel.2009.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berardini M, Foster PL, Loechler EL. DNA polymerase II (polB) is involved in a new DNA repair pathway for DNA interstrand cross-links in Escherichia coli. J. Bacteriol. 1999;181:2878–2882. doi: 10.1093/gao/9781884446054.article.t031385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berardini M, Mackay W, Loechler EL. Evidence for a recombination-independent pathway for the repair of DNA interstrand cross-links based on a site-specific study with nitrogen mustard. Biochemistry. 1997;36:3506–3513. doi: 10.1021/bi962778w. [DOI] [PubMed] [Google Scholar]
- Bergstralh DT, Sekelsky J. Interstrand crosslink repair: can XPF-ERCC1 be let off the hook? Trends Genet. 2008;24:70–76. doi: 10.1016/j.tig.2007.11.003. [DOI] [PubMed] [Google Scholar]
- Bessho T. Induction of DNA replication-mediated double strand breaks by psoralen DNA interstrand cross-links. J. Biol. Chem. 2003;278:5250–5254. doi: 10.1074/jbc.M212323200. [DOI] [PubMed] [Google Scholar]
- Bessho T, Mu D, Sancar A. Initiation of DNA interstrand cross-link repair in humans: the nucleotide excision repair system makes dual incisions 5' to the cross- linked base and removes a 22- to 28-nucleotide-long damage-free strand. Mol. Cell Biol. 1997;17:6822–6830. doi: 10.1128/mcb.17.12.6822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhagwat N, Olsen AL, Wang AT, Hanada K, Stuckert P, Kanaar R, D'Andrea A, Niedernhofer LJ, McHugh PJ. XPF-ERCC1 participates in the Fanconi anemia pathway of crosslink repair. Mol. Cell Biol. 2009 doi: 10.1128/MCB.00086-09. MCB.00086-09v1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolderson E, Scorah J, Helleday T, Smythe C, Meuth M. ATM is required for the cellular response to thymidine induced replication fork stress. Hum. Mol. Genet. 2004;13:2937–2945. doi: 10.1093/hmg/ddh316. [DOI] [PubMed] [Google Scholar]
- Boyer V, Moustacchi E, Sage E. Sequence specificity in photoreaction of various psoralen derivatives with DNA: role in biological activity. Biochemistry. 1988;27:3011–3018. doi: 10.1021/bi00408a052. [DOI] [PubMed] [Google Scholar]
- Bredberg A, Soderhall S. Normal rate of DNA breakage in xeroderma pigmentosum complementation group E cells treated with 8-methoxypsoralen plus near-ultraviolet radiation. Biochim. Biophys. Acta. 1985;824:268–271. doi: 10.1016/0167-4781(85)90058-2. [DOI] [PubMed] [Google Scholar]
- Broyde S, Wang L, Rechkoblit O, Geacintov NE, Patel DJ. Lesion processing: high-fidelity versus lesion-bypass DNA polymerases. Trends Biochem. Sci. 2008;33:209–219. doi: 10.1016/j.tibs.2008.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budzowska M, Kanaar R. Mechanisms of dealing with DNA damage-induced replication problems. Cell Biochem. Biophys. 2009;53:17–31. doi: 10.1007/s12013-008-9039-y. [DOI] [PubMed] [Google Scholar]
- Cai Y, Patel DJ, Geacintov NE, Broyde S. Differential nucleotide excision repair susceptibility of bulky DNA adducts in different sequence contexts: hierarchies of recognition signals. J. Mol. Biol. 2009;385:30–44. doi: 10.1016/j.jmb.2008.09.087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camenisch U, Trautlein D, Clement FC, Fei J, Leitenstorfer A, Ferrando-May E, Naegeli H. Two-stage dynamic DNA quality check by xeroderma pigmentosum group C protein. EMBO J. 2009;28:2387–2399. doi: 10.1038/emboj.2009.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Xu XS, Yang J, Wang G. Defining the function of XPC protein in psoralen and cisplatin-mediated DNA repair and mutagenesis. Carcinogenesis. 2003;24:1111–1121. doi: 10.1093/carcin/bgg051. [DOI] [PubMed] [Google Scholar]
- Cheng S, Sancar A, Hearst JE. RecA-dependent incision of psoralen-crosslinked DNA by (A)BC excinuclease. Nucleic Acids Res. 1991;19:657–663. doi: 10.1093/nar/19.3.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin JY, Glazer PM. Repair of DNA lesions associated with triplex-forming oligonucleotides. Mol. Carcinog. 2009;48:389–399. doi: 10.1002/mc.20501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin JY, Schleifman EB, Glazer PM. Repair and recombination induced by triple helix DNA. Front Biosci. 2007;12:4288–4297. doi: 10.2741/2388. [DOI] [PubMed] [Google Scholar]
- Chiou CC, Yang JL. Mutagenicity and specific mutation spectrum induced by 8- methoxypsoralen plus a low dose of UVA in the hprt gene in diploid human fibroblasts. Carcinogenesis. 1995;16:1357–1362. doi: 10.1093/carcin/16.6.1357. [DOI] [PubMed] [Google Scholar]
- Christensen LA, Wang H, Van Houten B, Vasquez KM. Efficient processing of TFO-directed psoralen DNA interstrand crosslinks by the UvrABC nuclease. Nucleic Acids Res. 2008;36:7136–7145. doi: 10.1093/nar/gkn880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimino GD, Gamper H, Isaacs ST, Hearst JE. Psoralens as photoactive probes of nucleic acid structure and function: organic chemistry, photochemistry, and biochemistry. Annu. Rev. Biochem. 1985;54:1151–1193. doi: 10.1146/annurev.bi.54.070185.005443. [DOI] [PubMed] [Google Scholar]
- Cipak L, Watanabe N, Bessho T. The role of BRCA2 in replication-coupled DNA interstrand cross-link repair in vitro. Nat. Struct. Mol. Biol. 2006;13:729–733. doi: 10.1038/nsmb1120. [DOI] [PubMed] [Google Scholar]
- Clingen PH, Arlett CF, Hartley JA, Parris CN. Chemosensitivity of primary human fibroblasts with defective unhooking of DNA interstrand cross-links. Exp. Cell Res. 2007;313:753–760. doi: 10.1016/j.yexcr.2006.11.007. [DOI] [PubMed] [Google Scholar]
- Clingen PH, Wu JY, Miller J, Mistry N, Chin F, Wynne P, Prise KM, Hartley JA. Histone H2AX phosphorylation as a molecular pharmacological marker for DNA interstrand crosslink cancer chemotherapy. Biochem. Pharmacol. 2008;76:19–27. doi: 10.1016/j.bcp.2008.03.025. [DOI] [PubMed] [Google Scholar]
- Cohen SM, Brylawski BP, Cordeiro-Stone M, Kaufman DG. Mapping of an origin of DNA replication near the transcriptional promoter of the human HPRT gene. J. Cell Biochem. 2002;85:346–356. doi: 10.1002/jcb.10136. [DOI] [PubMed] [Google Scholar]
- Cole RS. Repair of DNA containing interstrand crosslinks in Escherichia coli: sequential excision and recombination. Proc. Natl. Acad. Sci. U. S. A. 1973;70:1064–1068. doi: 10.1073/pnas.70.4.1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole RS, Levitan D, Sinden RR. Removal of psoralen interstrand cross-links from DNA of Escherichia coli: mechanism and genetic control. J. Mol. Biol. 1976;103:39–59. doi: 10.1016/0022-2836(76)90051-6. [DOI] [PubMed] [Google Scholar]
- Collins AR. Mutant rodent cell lines sensitive to ultraviolet light, ionizing radiation and cross-linking agents: a comprehensive survey of genetic and biochemical characteristics. Mutat. Res. 1993;293:99–118. doi: 10.1016/0921-8777(93)90062-l. [DOI] [PubMed] [Google Scholar]
- Collins AR. The comet assay for DNA damage and repair: principles, applications, and limitations. Mol. Biotechnol. 2004;26:249–261. doi: 10.1385/MB:26:3:249. [DOI] [PubMed] [Google Scholar]
- Collis SJ, Ciccia A, Deans AJ, Horejsi Z, Martin JS, Maslen SL, Skehel JM, Elledge SJ, West SC, Boulton SJ. FANCM and FAAP24 function in ATR-mediated checkpoint signaling independently of the Fanconi anemia core complex. Mol. Cell. 2008;32:313–324. doi: 10.1016/j.molcel.2008.10.014. [DOI] [PubMed] [Google Scholar]
- Couve S, Mace-Aime G, Rosselli F, Saparbaev MK. The human oxidative DNA glycosylase NEIL1 excises psoralen-induced interstrand DNA cross-links in a three-stranded DNA structure. J. Biol. Chem. 2009;284:11963–11970. doi: 10.1074/jbc.M900746200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couve-Privat S, Mace G, Rosselli F, Saparbaev MK. Psoralen-induced DNA adducts are substrates for the base excision repair pathway in human cells. Nucleic Acids Res. 2007;35:5672–5682. doi: 10.1093/nar/gkm592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Silva MW, Wilds CJ, Noronha AM, Colvin OM, Miller PS, Gamcsik MP. Accommodation of mispair aligned N3T-ethyl-N3T DNA interstrand cross link. Biochemistry. 2004;43:12549–12554. doi: 10.1021/bi0486435. [DOI] [PubMed] [Google Scholar]
- Damia G, Imperatori L, Stefanini M, D'Incalci M. Sensitivity of CHO mutant cell lines with specific defects in nucleotide excision repair to different anti-cancer agents. Int. J. Cancer. 1996;66:779–783. doi: 10.1002/(SICI)1097-0215(19960611)66:6<779::AID-IJC12>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- De Silva IU, McHugh PJ, Clingen PH, Hartley JA. Defining the roles of nucleotide excision repair and recombination in the repair of DNA interstrand cross-links in mammalian cells. Mol. Cell Biol. 2000;20:7980–7990. doi: 10.1128/mcb.20.21.7980-7990.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Silva IU, McHugh PJ, Clingen PH, Hartley JA. Defects in interstrand cross-link uncoupling do not account for the extreme sensitivity of ERCC1 and XPF cells to cisplatin. Nucleic Acids Res. 2002;30:3848–3856. doi: 10.1093/nar/gkf479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Winter JP, Joenje H. The genetic and molecular basis of Fanconi anemia. Mutat. Res. 2009;668:11–19. doi: 10.1016/j.mrfmmm.2008.11.004. [DOI] [PubMed] [Google Scholar]
- Demuth I, Digweed M, Concannon P. Human SNM1B is required for normal cellular response to both DNA interstrand crosslink-inducing agents and ionizing radiation. Oncogene. 2004;23:8611–8618. doi: 10.1038/sj.onc.1207895. [DOI] [PubMed] [Google Scholar]
- Dendouga N, Gao H, Moechars D, Janicot M, Vialard J, McGowan CH. Disruption of murine Mus81 increases genomic instability and DNA damage sensitivity but does not promote tumorigenesis. Mol. Cell Biol. 2005;25:7569–7579. doi: 10.1128/MCB.25.17.7569-7579.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dooley PA, Tsarouhtsis D, Korbel GA, Nechev LV, Shearer J, Zegar IS, Harris CM, Stone MP, Harris TM. Structural studies of an oligodeoxynucleotide containing a trimethylene interstrand cross-link in a 5'-(CpG) motif: model of a malondialdehyde cross-link. J. Am. Chem Soc. 2001;123:1730–1739. doi: 10.1021/ja003163w. [DOI] [PubMed] [Google Scholar]
- Dooley PA, Zhang M, Korbel GA, Nechev LV, Harris CM, Stone MP, Harris TM. NMR determination of the conformation of a trimethylene interstrand cross-link in an oligodeoxynucleotide duplex containing a 5'-d(GpC) motif. J. Am. Chem Soc. 2003;125:62–72. doi: 10.1021/ja0207798. [DOI] [PubMed] [Google Scholar]
- Dronkert ML, de Wit J, Boeve M, Vasconcelos ML, van Steeg H, Tan TL, Hoeijmakers JH, Kanaar R. Disruption of mouse SNM1 causes increased sensitivity to the DNA interstrand cross-linking agent mitomycin C. Mol. Cell Biol. 2000;20:4553–4561. doi: 10.1128/mcb.20.13.4553-4561.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dronkert ML, Kanaar R. Repair of DNA interstrand cross-links. Mutat. Res. 2001;486:217–247. doi: 10.1016/s0921-8777(01)00092-1. [DOI] [PubMed] [Google Scholar]
- Eichman BF, Mooers BH, Alberti M, Hearst JE, Ho PS. The crystal structures of psoralen cross-linked DNAs: drug-dependent formation of Holliday junctions. J. Mol. Biol. 2001;308:15–26. doi: 10.1006/jmbi.2001.4567. [DOI] [PubMed] [Google Scholar]
- Fan Y-H, Gold B. Sequence-Specificity for DNA Interstrand Cross-Linking by α ώ-Alkanediol Dimethylsulfonate Esters: Evidence for DNA Distortion by the Initial Monofunctional Lesion. J. Am. Chem Soc. 1999;121:11942–11946. [Google Scholar]
- Faruqi AF, Seidman MM, Segal DJ, Carroll D, Glazer PM. Recombination induced by triple-helix-targeted DNA damage in mammalian cells. Mol. Cell Biol. 1996;16:6820–6828. doi: 10.1128/mcb.16.12.6820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fekairi S, Scaglione S, Chahwan C, Taylor ER, Tissier A, Coulon S, Dong MQ, Ruse C, Yates JR, III, Russell P, Fuchs RP, McGowan CH, Gaillard PH. Human SLX4 is a Holliday junction resolvase subunit that binds multiple DNA repair/recombination endonucleases. Cell. 2009;138:78–89. doi: 10.1016/j.cell.2009.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenfeld G, Davies DR, Rich A. Formation of a three stranded polynucleotide molecule. J. Am. Chem. Soc. 1957;79:2023–2024. [Google Scholar]
- Fisher LA, Bessho M, Bessho T. Processing of a psoralen DNA interstrand cross-link by XPF-ERCC1 complex in vitro. J. Biol. Chem. 2008;283:1275–1281. doi: 10.1074/jbc.M708072200. [DOI] [PubMed] [Google Scholar]
- Fitch ME, Nakajima S, Yasui A, Ford JM. In vivo recruitment of XPC to UV-induced cyclobutane pyrimidine dimers by the DDB2 gene product. J. Biol. Chem. 2003;278:46906–46910. doi: 10.1074/jbc.M307254200. [DOI] [PubMed] [Google Scholar]
- Fiumicino S, Martinelli S, Colussi C, Aquilina G, Leonetti C, Crescenzi M, Bignami M. Sensitivity to DNA cross-linking chemotherapeutic agents in mismatch repair-defective cells in vitro and in xenografts. Int. J. Cancer. 2000;85:590–596. doi: 10.1002/(sici)1097-0215(20000215)85:4<590::aid-ijc23>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- Furuta T, Ueda T, Aune G, Sarasin A, Kraemer KH, Pommier Y. Transcription-coupled nucleotide excision repair as a determinant of cisplatin sensitivity of human cells. Cancer Res. 2002;62:4899–4902. [PubMed] [Google Scholar]
- Fuxreiter M, Luo N, Jedlovszky P, Simon I, Osman R. Role of base flipping in specific recognition of damaged DNA by repair enzymes. J. Mol. Biol. 2002;323:823–834. doi: 10.1016/s0022-2836(02)00999-3. [DOI] [PubMed] [Google Scholar]
- Gargiulo D, Kumar GS, Musser SS, Tomasz M. Structural and function modification of DNA by mitomycin C. Mechanism of the DNA sequence specificity of mitomycins. Nucleic Acids Symp. Ser. 1995;34:169–170. [PubMed] [Google Scholar]
- Gari K, Decaillet C, Delannoy M, Wu L, Constantinou A. Remodeling of DNA replication structures by the branch point translocase FANCM. Proc. Natl. Acad. Sci. U. S. A. 2008a;105:16107–16112. doi: 10.1073/pnas.0804777105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gari K, Decaillet C, Stasiak AZ, Stasiak A, Constantinou A. The Fanconi anemia protein FANCM can promote branch migration of Holliday junctions and replication forks. Mol. Cell. 2008b;29:141–148. doi: 10.1016/j.molcel.2007.11.032. [DOI] [PubMed] [Google Scholar]
- Geacintov NE, Broyde S, Buterin T, Naegeli H, Wu M, Yan S, Patel DJ. Thermodynamic and structural factors in the removal of bulky DNA adducts by the nucleotide excision repair machinery. Biopolymers. 2002;65:202–210. doi: 10.1002/bip.10239. [DOI] [PubMed] [Google Scholar]
- Giovannangeli C, Thuong NT, Helene C. Oligodeoxynucleotide-directed photo-induced cross-linking of HIV proviral DNA via triple-helix formation. Nucleic Acids Res. 1992;20:4275–4281. doi: 10.1093/nar/20.16.4275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grillari J, Katinger H, Voglauer R. Contributions of DNA interstrand cross-links to aging of cells and organisms. Nucleic Acids Res. 2007;35:7566–7576. doi: 10.1093/nar/gkm1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossmann KF, Ward AM, Matkovic ME, Folias AE, Moses RE. S. cerevisiae has three pathways for DNA interstrand crosslink repair. Mutat. Res. 2001;487:73–83. doi: 10.1016/s0921-8777(01)00106-9. [DOI] [PubMed] [Google Scholar]
- Guo C, Fischhaber PL, Luk-Paszyc MJ, Masuda Y, Zhou J, Kamiya K, Kisker C, Friedberg EC. Mouse Rev1 protein interacts with multiple DNA polymerases involved in translesion DNA synthesis. EMBO J. 2003;22:6621–6630. doi: 10.1093/emboj/cdg626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta R, Sharma S, Sommers JA, Kenny MK, Cantor SB, Brosh RM., Jr. FANCJ (BACH1) helicase forms DNA damage inducible foci with replication protein A and interacts physically and functionally with the single-stranded DNA-binding protein. Blood. 2007;110:2390–2398. doi: 10.1182/blood-2006-11-057273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanada K, Budzowska M, Davies SL, van DE, Onizawa H, Beverloo HB, Maas A, Essers J, Hickson ID, Kanaar R. The structure-specific endonuclease Mus81 contributes to replication restart by generating double-strand DNA breaks. Nat. Struct. Mol. Biol. 2007;14:1096–1104. doi: 10.1038/nsmb1313. [DOI] [PubMed] [Google Scholar]
- Hanada K, Budzowska M, Modesti M, Maas A, Wyman C, Essers J, Kanaar R. The structure-specific endonuclease Mus81-Eme1 promotes conversion of interstrand DNA crosslinks into double-strands breaks. EMBO J. 2006;25:4921–4932. doi: 10.1038/sj.emboj.7601344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haran TE, Crothers DM. Phased psoralen cross-links do not bend the DNA double helix. Biochemistry. 1988;27:6967–6971. doi: 10.1021/bi00418a044. [DOI] [PubMed] [Google Scholar]
- Harris PV, Mazina OM, Leonhardt EA, Case RB, Boyd JB, Burtis KC. Molecular cloning of Drosophila mus308, a gene involved in DNA cross- link repair with homology to prokaryotic DNA polymerase I genes. Mol. Cell Biol. 1996;16:5764–5771. doi: 10.1128/mcb.16.10.5764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havre PA, Gunther EJ, Gasparro FP, Glazer PM. Targeted mutagenesis of DNA using triple helix-forming oligonucleotides linked to psoralen. Proc. Natl. Acad. Sci. U. S. A. 1993;90:7879–7883. doi: 10.1073/pnas.90.16.7879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazrati A, Ramis-Castelltort M, Sarkar S, Barber LJ, Schofield CJ, Hartley JA, McHugh PJ. Human SNM1A suppresses the DNA repair defects of yeast pso2 mutants. DNA Repair (Amst) 2008;7:230–238. doi: 10.1016/j.dnarep.2007.09.013. [DOI] [PubMed] [Google Scholar]
- Hejna J, Holtorf M, Hines J, Mathewson L, Hemphill A, Al-Dhalimy M, Olson SB, Moses RE. Tip60 is required for DNA interstrand cross-link repair in the Fanconi anemia pathway. J. Biol. Chem. 2008;283:9844–9851. doi: 10.1074/jbc.M709076200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hejna J, Philip S, Ott J, Faulkner C, Moses R. The hSNM1 protein is a DNA 5'-exonuclease. Nucleic Acids Res. 2007;35:6115–6123. doi: 10.1093/nar/gkm530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemphill AW, Bruun D, Thrun L, Akkari Y, Torimaru Y, Hejna K, Jakobs PM, Hejna J, Jones S, Olson SB, Moses RE. Mammalian SNM1 is required for genome stability. Mol. Genet. Metab. 2008;94:38–45. doi: 10.1016/j.ymgme.2007.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinz JM, Nham PB, Salazar EP, Thompson LH. The Fanconi anemia pathway limits the severity of mutagenesis. DNA Repair (Amst) 2006;5:875–884. doi: 10.1016/j.dnarep.2006.05.039. [DOI] [PubMed] [Google Scholar]
- Hoy CA, Thompson LH, Mooney CL, Salazar EP. Defective DNA cross-link removal in Chinese hamster cell mutants hypersensitive to bifunctional alkylating agents. Cancer Res. 1985a;45:1737–1743. [PubMed] [Google Scholar]
- Hoy CA, Thompson LH, Salazar EP, Stewart SA. Different genetic alterations underlie dual hypersensitivity of CHO mutant UV-1 to DNA methylating and cross-linking agents. Somat. Cell Mol. Genet. 1985b;11:523–532. doi: 10.1007/BF01534718. [DOI] [PubMed] [Google Scholar]
- Huang H, Zhu L, Reid BR, Drobny GP, Hopkins PB. Solution structure of a cisplatin-induced DNA interstrand cross-link. Science. 1995;270:1842–1845. doi: 10.1126/science.270.5243.1842. [DOI] [PubMed] [Google Scholar]
- Hwang GS, Kim JK, Choi BS. The solution structure of a psoralen cross-linked DNA duplex by NMR and relaxation matrix refinement. Biochem. Biophys. Res. Commun. 1996;219:191–197. doi: 10.1006/bbrc.1996.0204. [DOI] [PubMed] [Google Scholar]
- Isaacs RJ, Spielmann HP. A model for initial DNA lesion recognition by NER and MMR based on local conformational flexibility. DNA Repair (Amst) 2004;3:455–464. doi: 10.1016/j.dnarep.2004.01.004. [DOI] [PubMed] [Google Scholar]
- Islas AL, Baker FJ, Hanawalt PC. Transcription-coupled repair of psoralen cross-links but not monoadducts in Chinese hamster ovary cells. Biochemistry. 1994;33:10794–10799. doi: 10.1021/bi00201a029. [DOI] [PubMed] [Google Scholar]
- Jamieson ER, Lippard SJ. Structure, Recognition, and Processing of Cisplatin-DNA Adducts. Chem Rev. 1999;99:2467–2498. doi: 10.1021/cr980421n. [DOI] [PubMed] [Google Scholar]
- Janicijevic A, Sugasawa K, Shimizu Y, Hanaoka F, Wijgers N, Djurica M, Hoeijmakers JH, Wyman C. DNA bending by the human damage recognition complex XPC-HR23B. DNA Repair (Amst) 2003;2:325–336. doi: 10.1016/s1568-7864(02)00222-7. [DOI] [PubMed] [Google Scholar]
- Johnson RE, Washington MT, Haracska L, Prakash S, Prakash L. Eukaryotic polymerases iota and zeta act sequentially to bypass DNA lesions. Nature. 2000;406:1015–1019. doi: 10.1038/35023030. [DOI] [PubMed] [Google Scholar]
- Johnston BH, Johnson MA, Moore CB, Hearst JE. Psoralen-DNA photoreaction: controlled production of mono- and diadducts with nanosecond ultraviolet laser pulses. Science. 1977;197:906–908. doi: 10.1126/science.887929. [DOI] [PubMed] [Google Scholar]
- Kanne D, Rapoport H, Hearst JE. 8-Methoxypsoralen-nucleic acid photoreaction. Effect of methyl substitution on pyrone vs. furan photoaddition. J. Med. Chem. 1984;27:531–534. doi: 10.1021/jm00370a017. [DOI] [PubMed] [Google Scholar]
- Kannouche P, Fernandez de Henestrosa AR, Coull B, Vidal AE, Gray C, Zicha D, Woodgate R, Lehmann AR. Localization of DNA polymerases eta and iota to the replication machinery is tightly co-ordinated in human cells. EMBO J. 2003;22:1223–1233. doi: 10.1093/emboj/cdf618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kannouche PL, Lehmann AR. Ubiquitination of PCNA and the Polymerase Switch in Human Cells. Cell Cycle. 2004;3:1011–1013. [PubMed] [Google Scholar]
- Kaye J, Smith CA, Hanawalt PC. DNA repair in human cells containing photoadducts of 8-methoxypsoralen or angelicin. Cancer Res. 1980;40:696–702. [PubMed] [Google Scholar]
- Koehler DR, Hanawalt PC. Recruitment of damaged DNA to the nuclear matrix in hamster cells following ultraviolet irradiation. Nucleic Acids Res. 1996;24:2877–2884. doi: 10.1093/nar/24.15.2877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kropachev K, Kolbanovskii M, Cai Y, Rodriguez F, Kolbanovskii A, Liu Y, Zhang L, Amin S, Patel D, Broyde S, Geacintov NE. The sequence dependence of human nucleotide excision repair efficiencies of benzo[a]pyrene-derived DNA lesions: insights into the structural factors that favor dual incisions. J. Mol. Biol. 2009;386:1193–1203. doi: 10.1016/j.jmb.2008.12.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumaresan KR, Hang B, Lambert MW. Human endonucleolytic incision of DNA 3' and 5' to a site-directed psoralen monoadduct and interstrand cross-link. J. Biol. Chem. 1995;270:30709–30716. doi: 10.1074/jbc.270.51.30709. [DOI] [PubMed] [Google Scholar]
- Kumaresan KR, Hwang M, Thelen MP, Lambert MW. Contribution of XPF functional domains to the 5' and 3' incisions produced at the site of a psoralen interstrand cross-link. Biochemistry. 2002;41:890–896. doi: 10.1021/bi011614z. [DOI] [PubMed] [Google Scholar]
- Kumaresan KR, Lambert MW. Fanconi anemia, complementation group A, cells are defective in ability to produce incisions at sites of psoralen interstrand cross-links. Carcinogenesis. 2000;21:741–751. doi: 10.1093/carcin/21.4.741. [DOI] [PubMed] [Google Scholar]
- Kumaresan KR, Ramaswamy M, Yeung AT. Structure of the DNA interstrand cross-link of 4,5',8-trimethylpsoralen. Biochemistry. 1992;31:6774–6783. doi: 10.1021/bi00144a018. [DOI] [PubMed] [Google Scholar]
- Kumaresan KR, Sridharan DM, McMahon LW, Lambert MW. Deficiency in incisions produced by XPF at the site of a DNA interstrand cross-link in Fanconi anemia cells. Biochemistry. 2007;46:14359–14368. doi: 10.1021/bi7015958. [DOI] [PubMed] [Google Scholar]
- Kumari A, Minko IG, Harbut MB, Finkel SE, Goodman MF, Lloyd RS. Replication bypass of interstrand cross-link intermediates by Escherichia coli DNA polymerase IV. J. Biol. Chem. 2008;283:27433–27437. doi: 10.1074/jbc.M801237200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuraoka I, Kobertz WR, Ariza RR, Biggerstaff M, Essigmann JM, Wood RD. Repair of an interstrand DNA crosslink initiated by ERCC1-XPF repair/recombination nuclease. J. Biol. Chem. 2000;275:26632–26636. doi: 10.1074/jbc.C000337200. [DOI] [PubMed] [Google Scholar]
- Lage C, de PM, De Alencar TA, da FG, da S, V, Cabral-Neto J, Leitao AC. New insights on how nucleotide excision repair could remove DNA adducts induced by chemotherapeutic agents and psoralens plus UV-A (PUVA) in Escherichia coli cells. Mutat. Res. 2003;544:143–157. doi: 10.1016/j.mrrev.2003.06.003. [DOI] [PubMed] [Google Scholar]
- Lai C, Cao H, Hearst JE, Corash L, Luo H, Wang Y. Quantitative analysis of DNA interstrand cross-links and monoadducts formed in human cells induced by psoralens and UVA irradiation. Anal. Chem. 2008;80:8790–8798. doi: 10.1021/ac801520m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan L, Nakajima S, Oohata Y, Takao M, Okano S, Masutani M, Wilson SH, Yasui A. In situ analysis of repair processes for oxidative DNA damage in mammalian cells. Proc. Natl. Acad. Sci. U. S. A. 2004;101:13738–13743. doi: 10.1073/pnas.0406048101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange SS, Reddy MC, Vasquez KM. Human HMGB1 directly facilitates interactions between nucleotide excision repair proteins on triplex-directed psoralen interstrand crosslinks. DNA Repair (Amst) 2009;8:865–872. doi: 10.1016/j.dnarep.2009.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lao Y, Hecht SS. Synthesis and properties of an acetaldehyde-derived oligonucleotide interstrand cross-link. Chem Res. Toxicol. 2005;18:711–721. doi: 10.1021/tx0497292. [DOI] [PubMed] [Google Scholar]
- Laquerbe A, Guillouf C, Moustacchi E, Papadopoulo D. The mutagenic processing of psoralen photolesions leaves a highly specific signature at an endogenous human locus. J. Mol. Biol. 1995;254:38–49. doi: 10.1006/jmbi.1995.0597. [DOI] [PubMed] [Google Scholar]
- Lawley PD, Phillips DH. DNA adducts from chemotherapeutic agents. Mutat. Res. 1996;355:13–40. doi: 10.1016/0027-5107(96)00020-6. [DOI] [PubMed] [Google Scholar]
- Lawrence CW, Gibbs PE, Murante RS, Wang XD, Li Z, McManus TP, McGregor WG, Nelson JR, Hinkle DC, Maher VM. Roles of DNA polymerase zeta and Rev1 protein in eukaryotic mutagenesis and translesion replication. Cold Spring Harb. Symp. Quant. Biol. 2000;65:61–69. doi: 10.1101/sqb.2000.65.61. [DOI] [PubMed] [Google Scholar]
- Lee YJ, Park SJ, Ciccone SL, Kim CR, Lee SH. An in vivo analysis of MMC-induced DNA damage and its repair. Carcinogenesis. 2006;27:446–453. doi: 10.1093/carcin/bgi254. [DOI] [PubMed] [Google Scholar]
- Lehmann AR. Translesion synthesis in mammalian cells. Exp. Cell Res. 2006;312:2673–2676. doi: 10.1016/j.yexcr.2006.06.010. [DOI] [PubMed] [Google Scholar]
- Lehmann AR, Niimi A, Ogi T, Brown S, Sabbioneda S, Wing JF, Kannouche PL, Green CM. Translesion synthesis: Y-family polymerases and the polymerase switch. DNA Repair (Amst) 2007;6:891–899. doi: 10.1016/j.dnarep.2007.02.003. [DOI] [PubMed] [Google Scholar]
- Lehoczky P, McHugh PJ, Chovanec M. DNA interstrand cross-link repair in Saccharomyces cerevisiae. FEMS Microbiol. Rev. 2007;31:109–133. doi: 10.1111/j.1574-6976.2006.00046.x. [DOI] [PubMed] [Google Scholar]
- Li J, Wang QE, Zhu Q, El-Mahdy MA, Wani G, Praetorius-Ibba M, Wani AA. DNA damage binding protein component DDB1 participates in nucleotide excision repair through DDB2 DNA-binding and cullin 4A ubiquitin ligase activity. Cancer Res. 2006;66:8590–8597. doi: 10.1158/0008-5472.CAN-06-1115. [DOI] [PubMed] [Google Scholar]
- Li L, Peterson CA, Lu X, Wei P, Legerski RJ. Interstrand cross-links induce DNA synthesis in damaged and undamaged plasmids in mammalian cell extracts. Mol. Cell Biol. 1999;19:5619–5630. doi: 10.1128/mcb.19.8.5619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Peterson CA, Zhang X, Legerski RJ. Requirement for PCNA and RPA in interstrand crosslink-induced DNA synthesis. Nucleic Acids Res. 2000;28:1424–1427. doi: 10.1093/nar/28.6.1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Hejna J, Moses RE. The yeast Snm1 protein is a DNA 5'-exonuclease. DNA Repair (Amst) 2005;4:163–170. doi: 10.1016/j.dnarep.2004.08.012. [DOI] [PubMed] [Google Scholar]
- Liu N, Lamerdin JE, Tebbs RS, Schild D, Tucker JD, Shen MR, Brookman KW, Siciliano MJ, Walter CA, Fan W, Narayana LS, Zhou ZQ, Adamson AW, Sorensen KJ, Chen DJ, Jones NJ, Thompson LH. XRCC2 and XRCC3, new human Rad51-family members, promote chromosome stability and protect against DNA cross-links and other damages. Mol. Cell. 1998;1:783–793. doi: 10.1016/s1097-2765(00)80078-7. [DOI] [PubMed] [Google Scholar]
- Liu X, Lao Y, Yang IY, Hecht SS, Moriya M. Replication-coupled repair of crotonaldehyde/acetaldehyde-induced guanine-guanine interstrand cross-links and their mutagenicity. Biochemistry. 2006;45:12898–12905. doi: 10.1021/bi060792v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loeber RL, Michaelson-Richie ED, Codreanu SG, Liebler DC, Campbell CR, Tretyakova NY. Proteomic analysis of DNA-protein cross-linking by antitumor nitrogen mustards. Chem Res. Toxicol. 2009;22:1151–1162. doi: 10.1021/tx900078y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Pannicke U, Schwarz K, Lieber MR. Hairpin opening and overhang processing by an Artemis/DNA-dependent protein kinase complex in nonhomologous end joining and V(D)J recombination. Cell. 2002;108:781–794. doi: 10.1016/s0092-8674(02)00671-2. [DOI] [PubMed] [Google Scholar]
- Maillard O, Camenisch U, Blagoev KB, Naegeli H. Versatile protection from mutagenic DNA lesions conferred by bipartite recognition in nucleotide excision repair. Mutat. Res. 2008;658:271–286. doi: 10.1016/j.mrrev.2008.01.007. [DOI] [PubMed] [Google Scholar]
- Maillard O, Camenisch U, Clement FC, Blagoev KB, Naegeli H. DNA repair triggered by sensors of helical dynamics. Trends Biochem. Sci. 2007a;32:494–499. doi: 10.1016/j.tibs.2007.08.008. [DOI] [PubMed] [Google Scholar]
- Maillard O, Solyom S, Naegeli H. An aromatic sensor with aversion to damaged strands confers versatility to DNA repair. PLoS. Biol. 2007b;5:e79. doi: 10.1371/journal.pbio.0050079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majumdar A, Khorlin A, Dyatkina N, Lin FL, Powell J, Liu J, Fei Z, Khripine Y, Watanabe KA, George J, Glazer PM, Seidman MM. Targeted gene knockout mediated by triple helix forming oligonucleotides. Nat. Genet. 1998;20:212–214. doi: 10.1038/2530. [DOI] [PubMed] [Google Scholar]
- Majumdar A, Muniandy PA, Liu J, Liu JL, Liu ST, Cuenoud B, Seidman MM. Targeted Gene Knock In and Sequence Modulation Mediated by a Psoralen-linked Triplex-forming Oligonucleotide. J. Biol. Chem. 2008;283:11244–11252. doi: 10.1074/jbc.M800607200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majumdar A, Puri N, Cuenoud B, Natt F, Martin P, Khorlin A, Dyatkina N, George AJ, Miller PS, Seidman MM. Cell Cycle Modulation of Gene Targeting by a Triple Helix-forming Oligonucleotide. J. Biol. Chem. 2003;278:11072–11077. doi: 10.1074/jbc.M211837200. [DOI] [PubMed] [Google Scholar]
- Malinge JM, Giraud-Panis MJ, Leng M. Interstrand cross-links of cisplatin induce striking distortions in DNA. J. Inorg. Biochem. 1999;77:23–29. doi: 10.1016/s0162-0134(99)00148-8. [DOI] [PubMed] [Google Scholar]
- Malinge JM, Perez C, Leng M. Base sequence-independent distorsions induced by interstrand cross-links in cis-diamminedichloroplatinum (II)-modified DNA. Nucleic Acids Res. 1994;22:3834–3839. doi: 10.1093/nar/22.19.3834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malkova A, Ivanov EL, Haber JE. Double-strand break repair in the absence of RAD51 in yeast: a possible role for break-induced DNA replication. Proc. Natl. Acad. Sci. U. S. A. 1996;93:7131–7136. doi: 10.1073/pnas.93.14.7131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manor H, Rao BS, Martin RG. Abundance and degree of dispersion of genomic d(GA)n.d(TC)n sequences. J. Mol. Evol. 1988;27:96–101. doi: 10.1007/BF02138367. [DOI] [PubMed] [Google Scholar]
- Maor-Shoshani A, Meira LB, Yang X, Samson LD. 3-Methyladenine DNA glycosylase is important for cellular resistance to psoralen interstrand cross-links. DNA Repair (Amst) 2008;7:1399–1406. doi: 10.1016/j.dnarep.2008.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marini F, Kim N, Schuffert A, Wood RD. POLN, a nuclear PolA family DNA polymerase homologous to the DNA cross-link sensitivity protein Mus308. J. Biol. Chem. 2003;278:32014–32019. doi: 10.1074/jbc.M305646200. [DOI] [PubMed] [Google Scholar]
- Marti TM, Hefner E, Feeney L, Natale V, Cleaver JE. H2AX phosphorylation within the G1 phase after UV irradiation depends on nucleotide excision repair and not DNA double-strand breaks. Proc. Natl. Acad. Sci. U. S. A. 2006;103:9891–9896. doi: 10.1073/pnas.0603779103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda Y, Ohmae M, Masuda K, Kamiya K. Structure and enzymatic properties of a stable complex of the human REV1 and REV7 proteins. J. Biol. Chem. 2003;278:12356–12360. doi: 10.1074/jbc.M211765200. [DOI] [PubMed] [Google Scholar]
- Matsumoto A, Vos JM, Hanawalt PC. Repair analysis of mitomycin C-induced DNA crosslinking in ribosomal RNA genes in lymphoblastoid cells from Fanconi's anemia patients. Mutat. Res. 1989;217:185–192. doi: 10.1016/0921-8777(89)90070-0. [DOI] [PubMed] [Google Scholar]
- McCabe KM, Hemphill A, Akkari Y, Jakobs PM, Pauw D, Olson SB, Moses RE, Grompe M. ERCC1 is required for FANCD2 focus formation. Mol. Genet. Metab. 2008;95:66–73. doi: 10.1016/j.ymgme.2008.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCabe KM, Olson SB, Moses RE. DNA interstrand crosslink repair in mammalian cells. J. Cell Physiol. 2009;220:569–573. doi: 10.1002/jcp.21811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McHugh PJ, Sarkar S. DNA Interstrand Cross-Link Repair in the Cell Cycle: A Critical Role for Polymerase zeta in G(1) Phase. Cell Cycle. 2006;5:1044–1047. doi: 10.4161/cc.5.10.2763. [DOI] [PubMed] [Google Scholar]
- McHugh PJ, Sones WR, Hartley JA. Repair of intermediate structures produced at DNA interstrand cross- links in Saccharomyces cerevisiae. Mol. Cell Biol. 2000;20:3425–3433. doi: 10.1128/mcb.20.10.3425-3433.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McHugh PJ, Spanswick VJ, Hartley JA. Repair of DNA interstrand crosslinks: molecular mechanisms and clinical relevance. Lancet Oncol. 2001;2:483–490. doi: 10.1016/S1470-2045(01)00454-5. [DOI] [PubMed] [Google Scholar]
- McKay BC, Becerril C, Ljungman M. P53 plays a protective role against UV- and cisplatin-induced apoptosis in transcription-coupled repair proficient fibroblasts. Oncogene. 2001;20:6805–6808. doi: 10.1038/sj.onc.1204901. [DOI] [PubMed] [Google Scholar]
- McMahon LW, Sangerman J, Goodman SR, Kumaresan K, Lambert MW. Human alpha spectrin II and the FANCA, FANCC, and FANCG proteins bind to DNA containing psoralen interstrand cross-links. Biochemistry. 2001;40:7025–7034. doi: 10.1021/bi002917g. [DOI] [PubMed] [Google Scholar]
- McMahon LW, Zhang P, Sridharan DM, Lefferts JA, Lambert MW. Knockdown of alphaII spectrin in normal human cells by siRNA leads to chromosomal instability and decreased DNA interstrand cross-link repair. Biochem. Biophys. Res. Commun. 2009;381:288–293. doi: 10.1016/j.bbrc.2009.02.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meniel V, Magana-Schwencke N, Averbeck D. Preferential repair in Saccharomyces cerevisiae rad mutants after induction of interstrand cross-links by 8-methoxypsoralen plus UVA. Mutagenesis. 1995a;10:543–548. doi: 10.1093/mutage/10.6.543. [DOI] [PubMed] [Google Scholar]
- Meniel V, Magana-Schwencke N, Averbeck D. Preferential repair in yeast after induction of interstrand cross-links by 8-methoxypsoralen plus UVA. Mutat. Res. 1995b;329:121–130. doi: 10.1016/0027-5107(95)00023-c. [DOI] [PubMed] [Google Scholar]
- Miao F, Bouziane M, Dammann R, Masutani C, Hanaoka F, Pfeifer G, O'Connor TR. 3-Methyladenine-DNA glycosylase (MPG protein) interacts with human RAD23 proteins. J. Biol. Chem. 2000;275:28433–28438. doi: 10.1074/jbc.M001064200. [DOI] [PubMed] [Google Scholar]
- Millard JT, Raucher S, Hopkins PB. Mechlorethamine cross-links deoxyguanosine residues at 5'GNC residues in duplex DNA fragments. J. Am. Chem Soc. 1990;112:2549–2560. [Google Scholar]
- Millington GW, Levell NJ. Vitiligo: the historical curse of depigmentation. Int. J. Dermatol. 2007;46:990–995. doi: 10.1111/j.1365-4632.2007.03195.x. [DOI] [PubMed] [Google Scholar]
- Min JH, Pavletich NP. Recognition of DNA damage by the Rad4 nucleotide excision repair protein. Nature. 2007;449:570–575. doi: 10.1038/nature06155. [DOI] [PubMed] [Google Scholar]
- Minko IG, Harbut MB, Kozekov ID, Kozekova A, Jakobs PM, Olson SB, Moses RE, Harris TM, Rizzo CJ, Lloyd RS. Role for DNA polymerase kappa in the processing of N2-N2-guanine interstrand cross-links. J. Biol. Chem. 2008;283:17075–17082. doi: 10.1074/jbc.M801238200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirchandani KD, D'Andrea AD. The Fanconi anemia/BRCA pathway: a coordinator of cross-link repair. Exp. Cell Res. 2006;312:2647–2653. doi: 10.1016/j.yexcr.2006.06.014. [DOI] [PubMed] [Google Scholar]
- Mirchandani KD, McCaffrey RM, D'Andrea AD. The Fanconi anemia core complex is required for efficient point mutagenesis and Rev1 foci assembly. DNA Repair (Amst) 2008;7:902–911. doi: 10.1016/j.dnarep.2008.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misra RR, Vos JM. Defective replication of psoralen adducts detected at the gene-specific level in xeroderma pigmentosum variant cells. Mol. Cell Biol. 1993;13:1002–1012. doi: 10.1128/mcb.13.2.1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell JR, Hoeijmakers JH, Niedernhofer LJ. Divide and conquer: nucleotide excision repair battles cancer and ageing. Curr. Opin. Cell Biol. 2003;15:232–240. doi: 10.1016/s0955-0674(03)00018-8. [DOI] [PubMed] [Google Scholar]
- Mogi S, Butcher CE, Oh DH. DNA polymerase eta reduces the gamma-H2AX response to psoralen interstrand crosslinks in human cells. Exp. Cell Res. 2008;314:887–895. doi: 10.1016/j.yexcr.2007.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogi S, Oh DH. gamma-H2AX formation in response to interstrand crosslinks requires XPF in human cells. DNA Repair (Amst) 2006;5:731–740. doi: 10.1016/j.dnarep.2006.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moldovan GL, D'Andrea AD. How the Fanconi Anemia Pathway Guards the Genome. Annu. Rev. Genet. 2009;43:223–249. doi: 10.1146/annurev-genet-102108-134222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu D, Bessho T, Nechev LV, Chen DJ, Harris TM, Hearst JE, Sancar A. DNA interstrand cross-links induce futile repair synthesis in mammalian cell extracts. Mol. Cell Biol. 2000;20:2446–2454. doi: 10.1128/mcb.20.7.2446-2454.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muniandy PA, Thapa D, Thazhathveetil AK, Liu ST, Seidman MM. Repair of laser localized DNA interstrand crosslinks in G1 phase mammalian cells. J. Biol. Chem. 2009 doi: 10.1074/jbc.M109.029025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz IM, Hain K, Declais AC, Gardiner M, Toh GW, Sanchez-Pulido L, Heuckmann JM, Toth R, Macartney T, Eppink B, Kanaar R, Ponting CP, Lilley DM, Rouse J. Coordination of structure-specific nucleases by human SLX4/BTBD12 is required for DNA repair. Mol. Cell. 2009;35:116–127. doi: 10.1016/j.molcel.2009.06.020. [DOI] [PubMed] [Google Scholar]
- Murray D, Meyn RE. Cell cycle-dependent cytotoxicity of alkylating agents: determination of nitrogen mustard-induced DNA cross-links and their repair in Chinese hamster ovary cells synchronized by centrifugal elutriation. Cancer Res. 1986;46:2324–2329. [PubMed] [Google Scholar]
- Nakanishi K, Taniguchi T, Ranganathan V, New HV, Moreau LA, Stotsky M, Mathew CG, Kastan MB, Weaver DT, D'Andrea AD. Interaction of FANCD2 and NBS1 in the DNA damage response. Nat. Cell Biol. 2002;4:913–920. doi: 10.1038/ncb879. [DOI] [PubMed] [Google Scholar]
- Nick McElhinny SA, Pavlov YI, Kunkel TA. Evidence for extrinsic exonucleolytic proofreading. Cell Cycle. 2006;5:958–962. doi: 10.4161/cc.5.9.2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niedernhofer LJ, Essers J, Weeda G, Beverloo B, de Wit J, Muijtjens M, Odijk H, Hoeijmakers JH, Kanaar R. The structure-specific endonuclease Ercc1-Xpf is required for targeted gene replacement in embryonic stem cells. EMBO J. 2001;20:6540–6549. doi: 10.1093/emboj/20.22.6540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niedernhofer LJ, Lalai AS, Hoeijmakers JH. Fanconi anemia (cross)linked to DNA repair. Cell. 2005;123:1191–1198. doi: 10.1016/j.cell.2005.12.009. [DOI] [PubMed] [Google Scholar]
- Niedernhofer LJ, Odijk H, Budzowska M, van Drunen E, Maas A, Theil AF, de Wit J, Jaspers NG, Beverloo HB, Hoeijmakers JH, Kanaar R. The structure-specific endonuclease Ercc1-Xpf is required to resolve DNA interstrand cross-link-induced double-strand breaks. Mol. Cell Biol. 2004;24:5776–5787. doi: 10.1128/MCB.24.13.5776-5787.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niedzwiedz W, Mosedale G, Johnson M, Ong CY, Pace P, Patel KJ. The Fanconi Anaemia Gene FANCC Promotes Homologous Recombination and Error-Prone DNA Repair. Mol. Cell. 2004;15:607–620. doi: 10.1016/j.molcel.2004.08.009. [DOI] [PubMed] [Google Scholar]
- Nojima K, Hochegger H, Saberi A, Fukushima T, Kikuchi K, Yoshimura M, Orelli BJ, Bishop DK, Hirano S, Ohzeki M, Ishiai M, Yamamoto K, Takata M, Arakawa H, Buerstedde JM, Yamazoe M, Kawamoto T, Araki K, Takahashi JA, Hashimoto N, Takeda S, Sonoda E. Multiple repair pathways mediate tolerance to chemotherapeutic cross-linking agents in vertebrate cells. Cancer Res. 2005;65:11704–11711. doi: 10.1158/0008-5472.CAN-05-1214. [DOI] [PubMed] [Google Scholar]
- Noll DM, Mason TM, Miller PS. Formation and repair of interstrand cross-links in DNA. Chem Rev. 2006;106:277–301. doi: 10.1021/cr040478b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noll DM, Noronha AM, Wilds CJ, Miller PS. Preparation of interstrand cross-linked DNA oligonucleotide duplexes. Front Biosci. 2004;9:421–437. doi: 10.2741/1246. [DOI] [PubMed] [Google Scholar]
- Noll DM, Webba da SM, Noronha AM, Wilds CJ, Colvin OM, Gamcsik MP, Miller PS. Structure, flexibility, and repair of two different orientations of the same alkyl interstrand DNA cross-link. Biochemistry. 2005;44:6764–6775. doi: 10.1021/bi050014n. [DOI] [PubMed] [Google Scholar]
- Norman D, Live D, Sastry M, Lipman R, Hingerty BE, Tomasz M, Broyde S, Patel DJ. NMR and computational characterization of mitomycin cross-linked to adjacent deoxyguanosines in the minor groove of the d(T-A-C-G-T-A).d(T-A-C-G-T-A) duplex. Biochemistry. 1990;29:2861–2875. doi: 10.1021/bi00463a032. [DOI] [PubMed] [Google Scholar]
- Noronha AM, Noll DM, Wilds CJ, Miller PS. N(4)C-ethyl-N(4)C cross-linked DNA: synthesis and characterization of duplexes with interstrand cross-links of different orientations. Biochemistry. 2002;41:760–771. doi: 10.1021/bi011610u. [DOI] [PubMed] [Google Scholar]
- Ojwang JO, Grueneberg DA, Loechler EL. Synthesis of a duplex oligonucleotide containing a nitrogen mustard interstrand DNA-DNA cross-link. Cancer Res. 1989;49:6529–6537. [PubMed] [Google Scholar]
- Oshige M, Aoyagi N, Harris PV, Burtis KC, Sakaguchi K. A new DNA polymerase species from Drosophila melanogaster: a probable mus308 gene product. Mutat. Res. 1999;433:183–192. doi: 10.1016/s0921-8777(99)00005-1. [DOI] [PubMed] [Google Scholar]
- Osman F, Whitby MC. Exploring the roles of Mus81-Eme1/Mms4 at perturbed replication forks. DNA Repair (Amst) 2007;6:1004–1017. doi: 10.1016/j.dnarep.2007.02.019. [DOI] [PubMed] [Google Scholar]
- Palom Y, Belcourt MF, Musser SM, Sartorelli AC, Rockwell S, Tomasz M. Structure of adduct X, the last unknown of the six major DNA adducts of mitomycin C formed in EMT6 mouse mammary tumor cells. Chem Res. Toxicol. 2000;13:479–488. doi: 10.1021/tx000024j. [DOI] [PubMed] [Google Scholar]
- Papadopoulo D, Averbeck D, Moustacchi E. The fate of 8-methoxypsoralen-photoinduced DNA interstrand crosslinks in Fanconi's anemia cells of defined genetic complementation groups. Mutat. Res. 1987;184:271–280. doi: 10.1016/0167-8817(87)90026-5. [DOI] [PubMed] [Google Scholar]
- Papadopoulo D, Guillouf C, Porfirio B, Moustacchi E. Decreased mutagenicity in Fanconi's anemia lymphoblasts following treatment with photoactivated psoralens. Prog. Clin. Biol. Res. 1990a;340A:241–248. [PubMed] [Google Scholar]
- Papadopoulo D, Porfirio B, Moustacchi E. Mutagenic response of Fanconi's anemia cells from a defined complementation group after treatment with photoactivated bifunctional psoralens. Cancer Res. 1990b;50:3289–3294. [PubMed] [Google Scholar]
- Patel KJ, Joenje H. Fanconi anemia and DNA replication repair. DNA Repair (Amst) 2007;6:885–890. doi: 10.1016/j.dnarep.2007.02.002. [DOI] [PubMed] [Google Scholar]
- Patrick SM, Tillison K, Horn JM. Recognition of cisplatin-DNA interstrand cross-links by replication protein A. Biochemistry. 2008;47:10188–10196. doi: 10.1021/bi800460d. [DOI] [PubMed] [Google Scholar]
- Payne A, Chu G. Xeroderma pigmentosum group E binding factor recognizes a broad spectrum of DNA damage. Mutat. Res. 1994;310:89–102. doi: 10.1016/0027-5107(94)90012-4. [DOI] [PubMed] [Google Scholar]
- Peng M, Litman R, Xie J, Sharma S, Brosh RM, Jr., Cantor SB. The FANCJ/MutLalpha interaction is required for correction of the cross-link response in FA-J cells. EMBO J. 2007;26:3238–3249. doi: 10.1038/sj.emboj.7601754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez C, Leng M, Malinge JM. Rearrangement of interstrand cross-links into intrastrand cross-links in cis-diamminedichloroplatinum(II)-modified DNA. Nucleic Acids Res. 1997;25:896–903. doi: 10.1093/nar/25.4.896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichierri P, Averbeck D, Rosselli F. DNA cross-link-dependent RAD50/MRE11/NBS1 subnuclear assembly requires the Fanconi anemia C protein. Hum. Mol. Genet. 2002;11:2531–2546. doi: 10.1093/hmg/11.21.2531. [DOI] [PubMed] [Google Scholar]
- Poklar N, Pilch DS, Lippard SJ, Redding EA, Dunham SU, Breslauer KJ. Influence of cisplatin intrastrand crosslinking on the conformation, thermal stability, and energetics of a 20-mer DNA duplex. Proc. Natl. Acad. Sci. U. S. A. 1996;93:7606–7611. doi: 10.1073/pnas.93.15.7606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poll EH, Arwert F, Kortbeek HT, Eriksson AW. Fanconi anaemia cells are not uniformly deficient in unhooking of DNA interstrand crosslinks, induced by mitomycin C or 8-methoxypsoralen plus UVA. Hum. Genet. 1984;68:228–234. doi: 10.1007/BF00418393. [DOI] [PubMed] [Google Scholar]
- Pritsos CA, Sartorelli AC. Generation of reactive oxygen radicals through bioactivation of mitomycin antibiotics. Cancer Res. 1986;46:3528–3532. [PubMed] [Google Scholar]
- Puri N, Majumdar A, Cuenoud B, Miller PS, Seidman MM. Importance of Clustered 2'-O-(2-Aminoethyl) Residues for the Gene Targeting Activity of Triple Helix-Forming Oligonucleotides. Biochemistry. 2004;43:1343–1351. doi: 10.1021/bi035808l. [DOI] [PubMed] [Google Scholar]
- Puri N, Majumdar A, Cuenoud B, Natt F, Martin P, Boyd A, Miller PS, Seidman MM. Targeted gene knockout by 2'-O-aminoethyl modified triplex forming oligonucleotides. J. Biol. Chem. 2001;276:28991–28998. doi: 10.1074/jbc.M103409200. [DOI] [PubMed] [Google Scholar]
- Puri N, Majumdar A, Cuenoud B, Natt F, Martin P, Boyd A, Miller PS, Seidman MM. Minimum Number of 2'-O-(2-Aminoethyl) Residues Required for Gene Knockout Activity by Triple Helix Forming Oligonucleotides. Biochemistry. 2002;41:7716–7724. doi: 10.1021/bi025734y. [DOI] [PubMed] [Google Scholar]
- Raha M, Wang G, Seidman MM, Glazer PM. Mutagenesis by third-strand-directed psoralen adducts in repair- deficient human cells: high frequency and altered spectrum in a xeroderma pigmentosum variant. Proc. Natl. Acad. Sci. U. S. A. 1996;93:2941–2946. doi: 10.1073/pnas.93.7.2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajski SR, Williams RM. DNA Cross-Linking Agents as Antitumor Drugs. Chem Rev. 1998;98:2723–2796. doi: 10.1021/cr9800199. [DOI] [PubMed] [Google Scholar]
- Raschle M, Knipsheer P, Enoiu M, Angelov T, Sun J, Griffith JD, Ellenberger TE, Scharer OD, Walter JC. Mechanism of replication-coupled DNA interstrand crosslink repair. Cell. 2008;134:969–980. doi: 10.1016/j.cell.2008.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reardon JT, Nichols AF, Keeney S, Smith CA, Taylor JS, Linn S, Sancar A. Comparative analysis of binding of human damaged DNA-binding protein (XPE) and Escherichia coli damage recognition protein (UvrA) to the major ultraviolet photoproducts: T[c,s]T, T[t,s]T, T[6-4]T, and T[Dewar]T. J. Biol. Chem. 1993;268:21301–21308. [PubMed] [Google Scholar]
- Richards S, Liu ST, Majumdar A, Liu JL, Nairn RS, Bernier M, Maher V, Seidman MM. Triplex targeted genomic crosslinks enter separable deletion and base substitution pathways. Nucleic Acids Res. 2005;33:5382–5393. doi: 10.1093/nar/gki851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rink SM, Hopkins PB. A mechlorethamine-induced DNA interstrand cross-link bends duplex DNA. Biochemistry. 1995;34:1439–1445. doi: 10.1021/bi00004a039. [DOI] [PubMed] [Google Scholar]
- Rink SM, Lipman R, Alley SC, Hopkins PB, Tomasz M. Bending of DNA by the mitomycin C-induced, GpG intrastrand cross-link. Chem Res. Toxicol. 1996;9:382–389. doi: 10.1021/tx950156q. [DOI] [PubMed] [Google Scholar]
- Rogakou EP, Boon C, Redon C, Bonner WM. Megabase chromatin domains involved in DNA double-strand breaks in vivo. J. Cell Biol. 1999;146:905–916. doi: 10.1083/jcb.146.5.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roques C, Coulombe Y, Delannoy M, Vignard J, Grossi S, Brodeur I, Rodrigue A, Gautier J, Stasiak AZ, Stasiak A, Constantinou A, Masson JY. MRE11-RAD50-NBS1 is a critical regulator of FANCD2 stability and function during DNA double-strand break repair. EMBO J. 2009;28:2400–2413. doi: 10.1038/emboj.2009.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg B. Noble metal complexes in cancer chemotherapy. Adv. Exp. Med. Biol. 1977;91:129–150. doi: 10.1007/978-1-4684-0796-9_10. [DOI] [PubMed] [Google Scholar]
- Rosenberg B, VanCamp L, KRIGAS T. Inhibition of cell division in E. coli by electrolysis products from a platinum electrode. Nature. 1965;205:698–699. doi: 10.1038/205698a0. [DOI] [PubMed] [Google Scholar]
- Rothfuss A, Grompe M. Repair Kinetics of Genomic Interstrand DNA Cross-Links: Evidence for DNA Double-Strand Break-Dependent Activation of the Fanconi Anemia/BRCA Pathway. Mol. Cell Biol. 2004;24:123–134. doi: 10.1128/MCB.24.1.123-134.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saenz-Méndez P, Guedes RC, dos Santos D, Eriksson LA. Theoretical Study of Sequence Selectivity and Preferred Binding Mode of Psoralen with DNA. Research Letters in Physical Chemistry. 2007;2007 Article ID 60623, 5 pages, 2007.doi:10.1155/2007/60623. [Google Scholar]
- Saffran WA, Ahmed S, Bellevue S, Pereira G, Patrick T, Sanchez W, Thomas S, Alberti M, Hearst JE. DNA repair defects channel interstrand DNA cross-links into alternate recombinational and error-prone repair pathways. J. Biol. Chem. 2004;279:36462–36469. doi: 10.1074/jbc.M402323200. [DOI] [PubMed] [Google Scholar]
- Sage E, Drobetsky EA, Moustacchi E. 8-Methoxypsoralen induced mutations are highly targeted at crosslinkable sites of photoaddition on the non-transcribed strand of a mammalian chromosomal gene. EMBO J. 1993;12:397–402. doi: 10.1002/j.1460-2075.1993.tb05671.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sargent RG, Rolig RL, Kilburn AE, Adair GM, Wilson JH, Nairn RS. Recombination-dependent deletion formation in mammalian cells deficient in the nucleotide excision repair gene ERCC1. Proc. Natl. Acad. Sci. U. S. A. 1997;94:13122–13127. doi: 10.1073/pnas.94.24.13122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar S, Davies AA, Ulrich HD, McHugh PJ. DNA interstrand crosslink repair during G1 involves nucleotide excision repair and DNA polymerase zeta. EMBO J. 2006;25:1285–1294. doi: 10.1038/sj.emboj.7600993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sartorelli AC, Hodnick WF, Belcourt MF, Tomasz M, Haffty B, Fischer JJ, Rockwell S. Mitomycin C: a prototype bioreductive agent. Oncol. Res. 1994;6:501–508. [PubMed] [Google Scholar]
- Sasaki MS, Takata M, Sonoda E, Tachibana A, Takeda S. Recombination repair pathway in the maintenance of chromosomal integrity against DNA interstrand crosslinks. Cytogenet. Genome Res. 2004;104:28–34. doi: 10.1159/000077463. [DOI] [PubMed] [Google Scholar]
- Schroth GP, Ho PS. Occurrence of potential cruciform and H-DNA forming sequences in genomic DNA. Nucleic Acids Res. 1995;23:1977–1983. doi: 10.1093/nar/23.11.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scrima A, Konickova R, Czyzewski BK, Kawasaki Y, Jeffrey PD, Groisman R, Nakatani Y, Iwai S, Pavletich NP, Thoma NH. Structural basis of UV DNA-damage recognition by the DDB1-DDB2 complex. Cell. 2008;135:1213–1223. doi: 10.1016/j.cell.2008.10.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidman MM, Glazer PM. The potential for gene repair via triple helix formation. J. Clin. Invest. 2003;112:487–494. doi: 10.1172/JCI19552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidman MM, Puri N, Majumdar A, Cuenoud B, Miller PS, Alam R. The development of bioactive triple helix-forming oligonucleotides. Ann. N. Y. Acad. Sci. 2005;1058:119–127. doi: 10.1196/annals.1359.020. [DOI] [PubMed] [Google Scholar]
- Seki M, Marini F, Wood RD. POLQ (Pol theta), a DNA polymerase and DNA-dependent ATPase in human cells. Nucleic Acids Res. 2003;31:6117–6126. doi: 10.1093/nar/gkg814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki M, Wood RD. DNA polymerase theta (POLQ) can extend from mismatches and from bases opposite a (6-4) photoproduct. DNA Repair (Amst) 2008;7:119–127. doi: 10.1016/j.dnarep.2007.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shachar S, Ziv O, Avkin S, Adar S, Wittschieben J, Reissner T, Chaney S, Friedberg EC, Wang Z, Carell T, Geacintov N, Livneh Z. Two-polymerase mechanisms dictate error-free and error-prone translesion DNA synthesis in mammals. EMBO J. 2009;28:383–393. doi: 10.1038/emboj.2008.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahid KA, Majumdar A, Alam R, Liu ST, Kuan JY, Sui X, Cuenoud B, Glazer PM, Miller PS, Seidman MM. Targeted cross-linking of the human beta-globin gene in living cells mediated by a triple helix forming oligonucleotide. Biochemistry. 2006;45:1970–1978. doi: 10.1021/bi0520986. [DOI] [PubMed] [Google Scholar]
- Shen X, Jun S, O'Neal LE, Sonoda E, Bemark M, Sale JE, Li L. REV3 and REV1 play major roles in recombination-independent repair of DNA interstrand cross-links mediated by monoubiquitinated proliferating cell nuclear antigen (PCNA) J. Biol. Chem. 2006;281:13869–13872. doi: 10.1074/jbc.C600071200. [DOI] [PubMed] [Google Scholar]
- Sinden RR, Hagerman PJ. Interstrand psoralen cross-links do not introduce appreciable bends in DNA. Biochemistry. 1984;23:6299–6303. doi: 10.1021/bi00321a002. [DOI] [PubMed] [Google Scholar]
- Sladek FM, Melian A, Howard-Flanders P. Incision by UvrABC excinuclease is a step in the path to mutagenesis by psoralen crosslinks in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 1989a;86:3982–3986. doi: 10.1073/pnas.86.11.3982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sladek FM, Munn MM, Rupp WD, Howard-Flanders P. In vitro repair of psoralen-DNA cross-links by RecA, UvrABC, and the 5'- exonuclease of DNA polymerase I. J. Biol. Chem. 1989b;264:6755–6765. [PubMed] [Google Scholar]
- Smeaton MB, Hlavin EM, McGregor MT, Noronha AM, Wilds CJ, Miller PS. Distortion-Dependent Unhooking of Interstrand Cross-Links in Mammalian Cell Extracts. Biochemistry. 2008;47:9920–9930. doi: 10.1021/bi800925e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smeaton MB, Hlavin EM, Noronha AM, Murphy SP, Wilds CJ, Miller PS. Effect of cross-link structure on DNA interstrand cross-link repair synthesis. Chem Res. Toxicol. 2009;22:1285–1297. doi: 10.1021/tx9000896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spielmann HP, Dwyer TJ, Hearst JE, Wemmer DE. Solution structures of psoralen monoadducted and cross-linked DNA oligomers by NMR spectroscopy and restrained molecular dynamics. Biochemistry. 1995a;34:12937–12953. [PubMed] [Google Scholar]
- Spielmann HP, Dwyer TJ, Sastry SS, Hearst JE, Wemmer DE. DNA structural reorganization upon conversion of a psoralen furan-side monoadduct to an interstrand cross-link: implications for DNA repair. Proc. Natl. Acad. Sci. U. S. A. 1995b;92:2345–2349. doi: 10.1073/pnas.92.6.2345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern RS. Psoralen and ultraviolet a light therapy for psoriasis. N. Engl. J. Med. 2007;357:682–690. doi: 10.1056/NEJMct072317. [DOI] [PubMed] [Google Scholar]
- Sugasawa K, Hanaoka F. Sensing of DNA damage by XPC/Rad4: one protein for many lesions. Nat. Struct. Mol. Biol. 2007;14:887–888. doi: 10.1038/nsmb1007-887. [DOI] [PubMed] [Google Scholar]
- Sugasawa K, Okuda Y, Saijo M, Nishi R, Matsuda N, Chu G, Mori T, Iwai S, Tanaka K, Tanaka K, Hanaoka F. UV-induced ubiquitylation of XPC protein mediated by UV-DDB-ubiquitin ligase complex. Cell. 2005;121:387–400. doi: 10.1016/j.cell.2005.02.035. [DOI] [PubMed] [Google Scholar]
- Sugasawa K, Shimizu Y, Iwai S, Hanaoka F. A molecular mechanism for DNA damage recognition by the xeroderma pigmentosum group C protein complex. DNA Repair (Amst) 2002;1:95–107. doi: 10.1016/s1568-7864(01)00008-8. [DOI] [PubMed] [Google Scholar]
- Sun W, Nandi S, Osman F, Ahn JS, Jakovleska J, Lorenz A, Whitby MC. The FANCM ortholog Fml1 promotes recombination at stalled replication forks and limits crossing over during DNA double-strand break repair. Mol. Cell. 2008;32:118–128. doi: 10.1016/j.molcel.2008.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takata K, Shimizu T, Iwai S, Wood RD. Human DNA polymerase N (POLN) is a low fidelity enzyme capable of error-free bypass of 5S-thymine glycol. J. Biol. Chem. 2006;281:23445–23455. doi: 10.1074/jbc.M604317200. [DOI] [PubMed] [Google Scholar]
- Taniguchi T, Garcia-Higuera I, Andreassen PR, Gregory RC, Grompe M, D'Andrea AD. S-phase-specific interaction of the Fanconi anemia protein, FANCD2, with BRCA1 and RAD51. Blood. 2002;100:2414–2420. doi: 10.1182/blood-2002-01-0278. [DOI] [PubMed] [Google Scholar]
- Thazhathveetil AK, Liu ST, Indig FE, Seidman MM. Psoralen conjugates for visualization of genomic interstrand cross-links localized by laser photoactivation. Bioconjug. Chem. 2007;18:431–437. doi: 10.1021/bc060309t. [DOI] [PubMed] [Google Scholar]
- Thoma BS, Wakasugi M, Christensen J, Reddy MC, Vasquez KM. Human XPC-hHR23B interacts with XPA-RPA in the recognition of triplex-directed psoralen DNA interstrand crosslinks. Nucleic Acids Res. 2005;33:2993–3001. doi: 10.1093/nar/gki610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas CB, Kohn KW, Bonner WM. Characterization of DNA-protein cross-links formed by treatment of L1210 cells and nuclei with bis(2-chloroethyl)methylamine (nitrogen mustard) Biochemistry. 1978;17:3954–3958. doi: 10.1021/bi00612a012. [DOI] [PubMed] [Google Scholar]
- Thompson LH, Hinz JM. Cellular and molecular consequences of defective Fanconi anemia proteins in replication-coupled DNA repair: Mechanistic insights. Mutat. Res. 2009;668:54–72. doi: 10.1016/j.mrfmmm.2009.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson LH, Hinz JM, Yamada NA, Jones NJ. How Fanconi anemia proteins promote the four Rs: replication, recombination, repair, and recovery. Environ. Mol. Mutagen. 2005;45:128–142. doi: 10.1002/em.20109. [DOI] [PubMed] [Google Scholar]
- Thompson LH, Schild D. Recombinational DNA repair and human disease. Mutat. Res. 2002;509:49–78. doi: 10.1016/s0027-5107(02)00224-5. [DOI] [PubMed] [Google Scholar]
- Van HB, Gamper H, Holbrook SR, Hearst JE, Sancar A. Action mechanism of ABC excision nuclease on a DNA substrate containing a psoralen crosslink at a defined position. Proc. Natl. Acad. Sci. U. S. A. 1986;83:8077–8081. doi: 10.1073/pnas.83.21.8077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal LS, Santos LB, Lage C, Leitao AC. Enhanced sensitivity of Escherichia coli uvrB mutants to mitomycin C points to a UV-C distinct repair for DNA adducts. Chem Res. Toxicol. 2006;19:1351–1356. doi: 10.1021/tx060035y. [DOI] [PubMed] [Google Scholar]
- Vogel EW, Barbin A, Nivard MJ, Stack HF, Waters MD, Lohman PH. Heritable and cancer risks of exposures to anticancer drugs: inter-species comparisons of covalent deoxyribonucleic acid-binding agents. Mutat. Res. 1998;400:509–540. doi: 10.1016/s0027-5107(98)00060-8. [DOI] [PubMed] [Google Scholar]
- Vos JM, Hanawalt PC. Processing of psoralen adducts in an active human gene: repair and replication of DNA containing monoadducts and interstrand cross-links. Cell. 1987;50:789–799. doi: 10.1016/0092-8674(87)90337-0. [DOI] [PubMed] [Google Scholar]
- Wang G, Levy DD, Seidman MM, Glazer PM. Targeted mutagenesis in mammalian cells mediated by intracellular triple helix formation. Mol. Cell Biol. 1995;15:1759–1768. doi: 10.1128/mcb.15.3.1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang QE, Zhu Q, Wani G, Chen J, Wani AA. UV radiation-induced XPC translocation within chromatin is mediated by damaged-DNA binding protein, DDB2. Carcinogenesis. 2004;25:1033–1043. doi: 10.1093/carcin/bgh085. [DOI] [PubMed] [Google Scholar]
- Wang X, Peterson CA, Zheng H, Nairn RS, Legerski RJ, Li L. Involvement of nucleotide excision repair in a recombination- independent and error-prone pathway of DNA interstrand cross-link repair. Mol. Cell Biol. 2001;21:713–720. doi: 10.1128/MCB.21.3.713-720.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webba da SM, Noronha AM, Noll DM, Miller PS, Colvin OM, Gamcsik MP. Solution structure of a DNA duplex containing mispair-aligned N4C-ethyl-N4C interstrand cross-linked cytosines. Biochemistry. 2002;41:15181–15188. doi: 10.1021/bi026368l. [DOI] [PubMed] [Google Scholar]
- Weidner MF, Sigurdsson ST, Hopkins PB. Sequence preferences of DNA interstrand cross-linking agents: dG-to-dG cross-linking at 5'-CG by structurally simplified analogues of mitomycin C. Biochemistry. 1990;29:9225–9233. doi: 10.1021/bi00491a017. [DOI] [PubMed] [Google Scholar]
- Wu HI, Brown JA, Dorie MJ, Lazzeroni L, Brown JM. Genome-wide identification of genes conferring resistance to the anticancer agents cisplatin, oxaliplatin, and mitomycin C. Cancer Res. 2004;64:3940–3948. doi: 10.1158/0008-5472.CAN-03-3113. [DOI] [PubMed] [Google Scholar]
- Wu JH, Wilson JB, Wolfreys AM, Scott A, Jones NJ. Optimization of the comet assay for the sensitive detection of PUVA-induced DNA interstrand cross-links. Mutagenesis. 2009;24:173–181. doi: 10.1093/mutage/gen068. [DOI] [PubMed] [Google Scholar]
- Wu Q, Christensen LA, Legerski RJ, Vasquez KM. Mismatch repair participates in error-free processing of DNA interstrand crosslinks in human cells. EMBO Rep. 2005;6:551–557. doi: 10.1038/sj.embor.7400418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Brosh RM., Jr. FANCJ helicase operates in the Fanconi Anemia DNA repair pathway and the response to replicational stress. Curr. Mol. Med. 2009;9:470–482. doi: 10.2174/156652409788167159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang SC, Lin JG, Chiou CC, Chen LY, Yang JL. Mutation specificity of 8-methoxypsoralen plus two doses of UVA irradiation in the hprt gene in diploid human fibroblasts. Carcinogenesis. 1994;15:201–207. doi: 10.1093/carcin/15.2.201. [DOI] [PubMed] [Google Scholar]
- Yang W. Structure and mechanism for DNA lesion recognition. Cell Res. 2008;18:184–197. doi: 10.1038/cr.2007.116. [DOI] [PubMed] [Google Scholar]
- Yang W, Woodgate R. What a difference a decade makes: insights into translesion DNA synthesis. Proc. Natl. Acad. Sci. U. S. A. 2007;104:15591–15598. doi: 10.1073/pnas.0704219104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youds JL, Barber LJ, Boulton SJ. C. elegans: A model of Fanconi anemia and ICL repair. Mutat. Res. 2009;668:103–116. doi: 10.1016/j.mrfmmm.2008.11.007. [DOI] [PubMed] [Google Scholar]
- Youds JL, Barber LJ, Ward JD, Collis SJ, O'Neil NJ, Boulton SJ, Rose AM. DOG-1 is the Caenorhabditis elegans BRIP1/FANCJ homologue and functions in interstrand cross-link repair. Mol. Cell Biol. 2008;28:1470–1479. doi: 10.1128/MCB.01641-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamble DB, Mu D, Reardon JT, Sancar A, Lippard SJ. Repair of cisplatin--DNA adducts by the mammalian excision nuclease. Biochemistry. 1996;35:10004–10013. doi: 10.1021/bi960453+. [DOI] [PubMed] [Google Scholar]
- Zhang N, Kaur R, Lu X, Shen X, Li L, Legerski RJ. The Pso4 mRNA splicing and DNA repair complex interacts with WRN for processing of DNA interstrand cross-links. J. Biol. Chem. 2005;280:40559–40567. doi: 10.1074/jbc.M508453200. [DOI] [PubMed] [Google Scholar]
- Zhang N, Liu X, Li L, Legerski R. Double-strand breaks induce homologous recombinational repair of interstrand cross-links via cooperation of MSH2, ERCC1-XPF, REV3, and the Fanconi anemia pathway. DNA Repair (Amst) 2007;6:1670–1678. doi: 10.1016/j.dnarep.2007.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang N, Lu X, Legerski RJ. Partial reconstitution of human interstrand cross-link repair in vitro: characterization of the roles of RPA and PCNA. Biochem. Biophys. Res. Commun. 2003;309:71–78. doi: 10.1016/s0006-291x(03)01535-3. [DOI] [PubMed] [Google Scholar]
- Zhang N, Lu X, Zhang X, Peterson CA, Legerski RJ. hMutSbeta is required for the recognition and uncoupling of psoralen interstrand cross-links in vitro. Mol. Cell Biol. 2002;22:2388–2397. doi: 10.1128/MCB.22.7.2388-2397.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng H, Wang X, Warren AJ, Legerski RJ, Nairn RS, Hamilton JW, Li L. Nucleotide excision repair- and polymerase eta-mediated error-prone removal of mitomycin C interstrand cross-links. Mol. Cell Biol. 2003;23:754–761. doi: 10.1128/MCB.23.2.754-761.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu G, Lippard SJ. Photoaffinity labeling reveals nuclear proteins that uniquely recognize cisplatin-DNA interstrand cross-links. Biochemistry. 2009;48:4916–4925. doi: 10.1021/bi900389b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zietlow L, Smith LA, Bessho M, Bessho T. Evidence for the involvement of human DNA polymerase N in the repair of DNA interstrand cross-links. Biochemistry. 2009 doi: 10.1021/bi9015346. DOI: 10.1021/bi9015346. [DOI] [PMC free article] [PubMed] [Google Scholar]