Abstract
The genes encoding the Ras family of small GTPases are mutated to yield constitutively active GTP-bound oncogenic proteins in one-third of all human cancers. Oncogenic Ras binds to and activates a number of proteins that promote tumorigenic phenotypes, including the family of Ral guanine nucleotide exchange factors, or RalGEFs. Activated RalGEFs convert the Ral family of small GTPases, comprised of RalA and RalB, from an inactive GDP-bound state to an active GTP-bound state. As both RalA and RalB have been implicated in a variety of tumorigenic phenotypes, we sought to determine which proteins downstream of Rals promote transformation and tumorigenesis. Here we report that shRNA-mediated knockdown of the Ral effector proteins Sec5 and Exo84, but less so in the case of RalBP1, reduced oncogenic RalGEF-mediated transformation and oncogenic Ras-driven tumorigenic growth of human cells. These results suggest that Rals promote oncogenic Ras-mediated tumorigenesis through, at least in part, Sec5 and Exo84.
Keywords: RalA, RalGEF, Ras, transformation, tumorigenesis
Introduction
Mutated and constitutively activated forms of the small GTPase Ras are found in nearly one-third of all human cancers (1). Oncogenic Ras binds to a number of effector proteins to promote transformation and tumorigenesis, including the family of Ral guanine nucleotide exchange factors (RalGEFs) (2–5). Recruiting RalGEFs to their substrates, the Ras-like GTPases RalA and RalB, converts Rals from an inactive GDP-bound state to an active GTP-bound state (6–8). Ral activation is necessary for oncogenic Ras-mediated transformation of a broad variety of human cell types (4, 9–11). Specifically, RalA is required for anchorage-independent growth and tumorigenesis, whereas RalB is important for cell survival, especially of cells in suspension, as well as for cell migration, and metastasis (9, 10, 12–14). Activated Rals bind to a limited number of effector proteins, the best-documented being RalBP1 (RLIP76), Sec5, and Exo84 (7, 15-18). RalBP1 contains a Rac/Cdc42 GAP domain (19) and is involved in the regulation of a subset of endocytic pathways (20–22). Sec5 and Exo84 constitute two members of the octameric exocyst complex, which participates in the targeting and tethering of secretory vesicles to specific plasma membrane domains, including the basolateral surface of epithelial cells, the mitotic abscission plane, and the leading edge of migrating epithelial cells (15, 23). Interactions with RalBP1, Sec5, and Exo84 link Ral proteins to receptor-mediated endocytosis, the maintenance of epithelial cell polarity, cytokinesis, and cell motility (7, 14, 15, 17, 20, 21, 23–25). The contribution of these effector proteins to Ral-dependent support of transformed and tumorigenic growth of human cells is unclear. Given the frequency of Ras mutations in human cancers, and the critical role of Rals in many different oncogenic Ras-mediated tumor phenotypes, we examined whether the Ral effectors RalBP1, Sec5 or Exo84 were required for transformation or tumorigenesis of human cells.
Results
shRNA knockdown of Sec5 or Exo84 reduce RalGEF-mediated Transformation of Human Cells
Oncogenic Ras promotes transformed cell growth through, in part, activation of RalGEFs (4, 11). Upon activation by RalGEFs, Ral proteins bind to a limited number of effectors, including RalBP1, Sec5, and Exo84, which have been implicated in receptor-mediated endocytosis, the maintenance of epithelial cell polarity, cytokinesis, and cell motility (7, 14, 15, 17, 20, 21, 23–25). To identify how oncogenic Ras promotes transformation through Rals, we first tested whether knockdown of RalBP1, Sec5, or Exo84 inhibited transformation. Transformed growth was assessed using HEK-HT cells, human embryonic kidney cells stably expressing the early region of SV40 and the hTERT catalytic subunit of telomerase. These cells are genetically defined and grow in soft agar, a transformed phenotype characteristic of many cancer cell lines, upon activation of endogenous Ral proteins by expression of Rlf-CAAX (9), a constitutively active version of the RalGEF family member Rlf (26, 27).
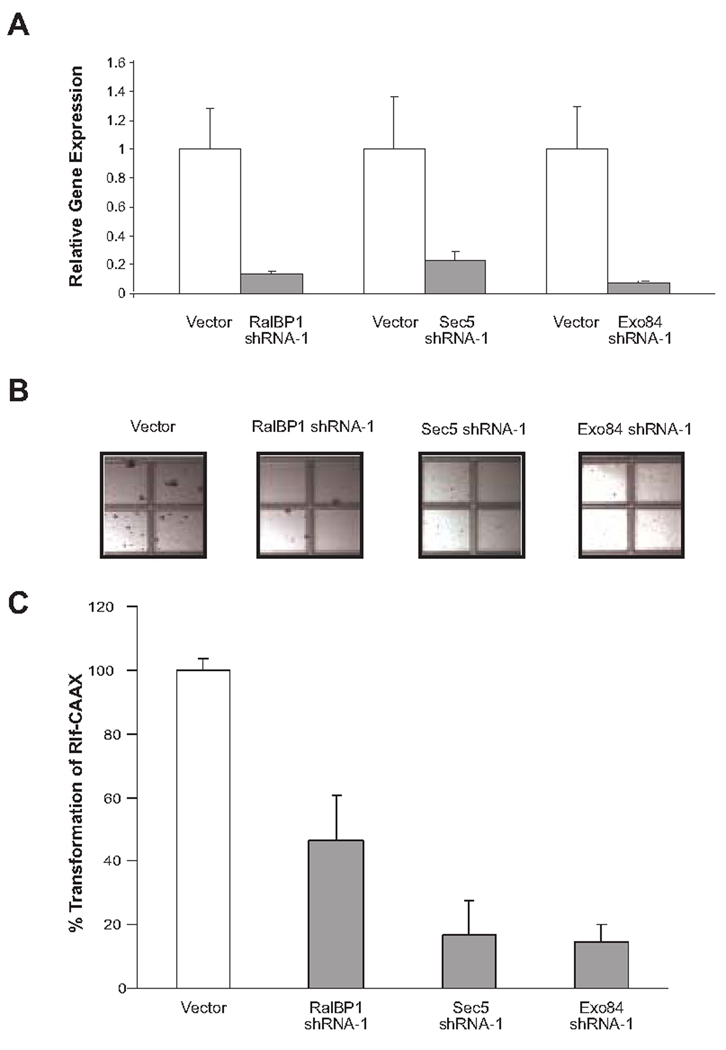
Rlf-CAAX-expressing HEK-HT cells were stably infected with retroviruses encoding shRNA (shRNA-1) against RalBP1 (22), Sec5 (28), Exo84 (18) or no shRNA to serve as a vector control. Appropriate knockdown of the targeted mRNA was confirmed by quantitative real-time RT-PCR (Fig. 1A). Specifically, RalBP1, Sec5, and Exo84 mRNA levels were reduced by 86%, 77%, and 90%, respectively. The resultant cell lines were then seeded in soft agar, and four weeks later the number of colonies were counted as a measure of transformation. shRNA-mediated knockdown of RalBP1 resulted in a 50% decrease in colonies growing in soft agar, while knockdown of either exocyst complex component Sec5 or Exo84 nearly abolished transformation, resulting in an 83% or 86% decrease in the number of colonies compared to control cells, respectively (Fig. 1, B and C).
FIGURE 1.
Knockdown of Sec5 or Exo84 decreases RalGEF-mediated transformation of human cells. A. Relative mRNA level ± standard deviation of the indicated transcripts in Rlf-CAAX-transformed HEK-HT cells stably infected with the indicated shRNAs, as determined by real-time RT-PCR. B. Photographs demonstrating representative anchorage-independent growth of Rlf-CAAX-transformed HEK-HT cells stably expressing the indicated shRNAs and C. Graphical representation expressed as the average percentage of colonies (>30 cells) ± standard deviation compared to vector control cells.
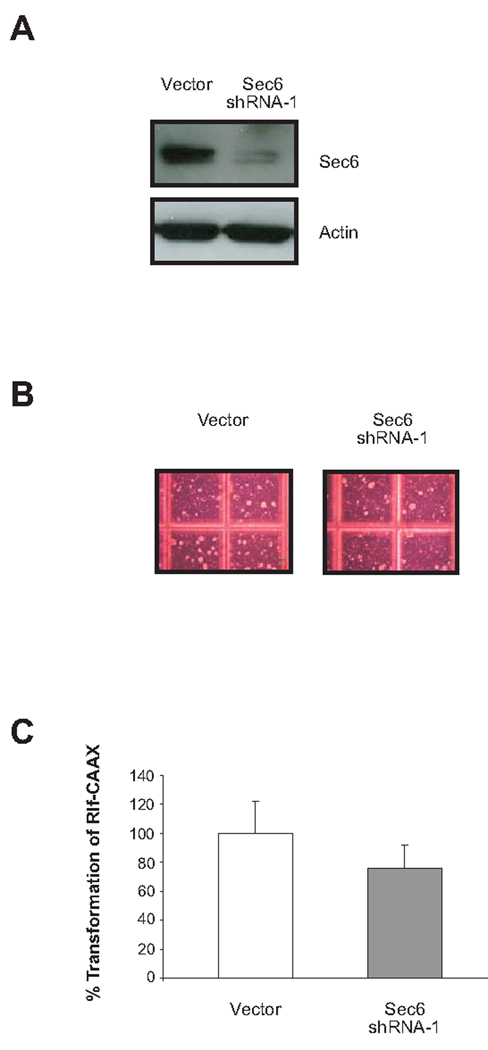
To further examine the role of the exocyst complex in RalGEF-mediated transformation, Rlf-CAAX-expressing HEK-HT cells were stably infected with retroviruses encoding shRNA against the exocyst component Sec6, which does not interact with Rals, or no shRNA as a vector control. Appropriate knockdown of Sec6 was confirmed by immunoblotting (Fig. 2A). The resultant cell lines were then seeded in soft agar, and four weeks later the number of colonies were counted as a measure of transformation. shRNA-mediated knockdown of Sec6 resulted in a modest 24% decrease in colonies growing in soft agar compared to control cells (Fig. 2, B and C). The decrease in transformation after knockdown of Sec6 was approximately 60% less than that observed following knockdown of either Sec5 or Exo84, suggesting that Ral-interacting members of the exocyst may be more important for RalGEF-mediated transformation.
FIGURE 2.
Knockdown of Sec6 does not greatly decrease RalGEF-mediated transformation of human cells. A. Detection of Sec6 by immunoblot analysis in Rlf-CAAX-transformed HEK-HT cells stably infected with vector control or Sec6 shRNA. Actin serves as a loading control. B. Photographs demonstrating representative anchorage-independent growth of Rlf-CAAX-transformed HEK-HT cells stably expressing the indicated shRNAs and C. graphical representation expressed as the average percentage of colonies (>30 cells) ± standard deviation compared to vector control cells.
shRNA knockdown of Sec5 or Exo84 reduce Oncogenic Ras-mediated Tumorigenesis of Human Cells
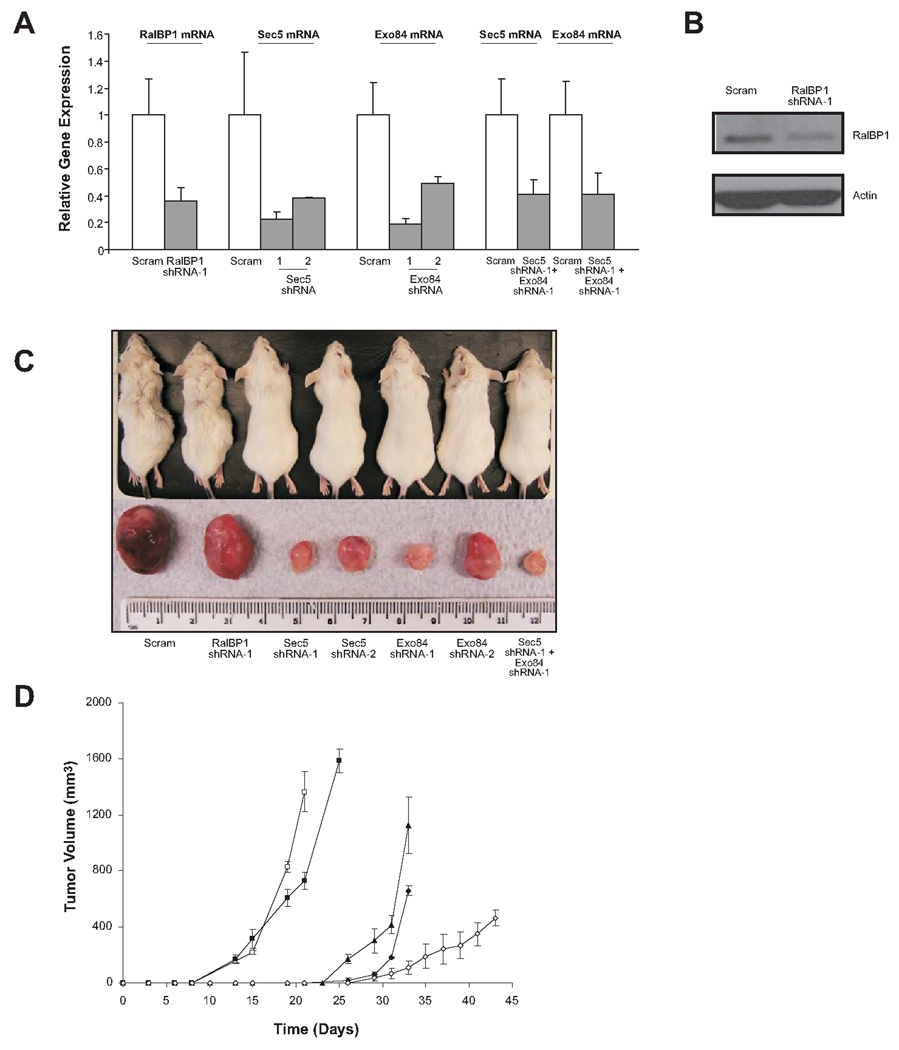
To address whether the decrease of in vitro soft agar growth of Rlf-CAAX-transformed HEK-HT cells reflected a decrease in tumorigenic growth in vivo, expression of the same three Ral effector proteins was knocked down by shRNA in the tumorigenic version of HEK-HT cells in which Rlf-CAAX was replaced with oncogenic RasG12V (29). Specifically, RasG12V-expressing HEK-HT cells were stably infected with the aforementioned retroviruses encoding shRNA (shRNA-1) targeting RalBP1, Sec5, Exo84, or a scramble sequence as a control. Reduced expression of RalBP1, Sec5, and Exo84 was assessed by real-time RT-PCR (Fig. 3A). RalBP1, Sec5, and Exo84 mRNA levels were decreased by 64%, 78%, and 80%, respectively. The resultant stable cell lines were then injected subcutaneously into the flank of four immunocompromised mice each, and tumor volume was recorded at regular intervals. Scramble control cells generated tumors with a latency of 13 days, and reached maximum tumor volume by 3 weeks, as previously reported (9). Knockdown of RalBP1 had little effect on the tumorigenic growth of HEK-HT RasG12V cells, as tumors formed at the same time and grew with nearly identical kinetics as those derived from scramble control cells (Fig. 3, C and D). To confirm that RalBP1 was knocked down at the protein level as well as at the mRNA level, we performed immunoblotting on RalBP1 knockdown RasG12V-transformed HEK-HT cells, and confirmed RalBP1 protein knockdown (Fig. 3B). This suggests that RalBP1 is either not involved in mediating Ras-induced tumorigenesis of HEK-HT cells, or that a reduced amount of RalBP1 protein is still sufficient to support Ras-induced tumorigenesis. However, consistent with the soft agar results (Fig. 1C), knockdown of either Sec5 or Exo84 prolonged the latency period of tumorigenesis and impeded subsequent tumor growth. Specifically, tumors arising from cells in which Sec5 or Exo84 were knocked down had a latency of 26 days, twice as long as scramble control cells, and reached maximum volume by around 5 weeks, 2 weeks longer than it took scramble control cells to reach the same size (Fig. 3, C and D). Knockdown of Sec5 or Exo84 thus reduced the tumorigenic potential of oncogenic Ras-transformed human cells.
FIGURE 3.
Sec5 and Exo84 are critical for oncogenic Ras-mediated tumorigenesis of human cells. A. Relative mRNA level ± standard deviation of the indicated transcripts in RasG12V-transformed HEK-HT cells stably infected with the indicated shRNAs, as determined by real-time RT-PCR. B. Detection of RalBP1 by immunoblot analysis in RasG12V-transformed HEK-HT cells stably infected with scramble control or RalBP1 shRNA. Actin serves as a loading control. C. Photograph of a representative mouse flank and excised tumor resulting from injection of RasG12V-transformed HEK-HT cells expressing the indicated shRNAs, taken when scramble control cells reached maximum tumor volume. D. Average tumor volume ± standard error versus time, of at least three mice from a representative experiment, injected with RasG12V-transformed HEK-HT cells stably expressing a scramble control shRNA (□), RalBP1 shRNA-1 (■), Sec5 shRNA-1 (▲), Exo84 shRNA-1 (♦), or Sec5 shRNA-1 and Exo84 shRNA-1 (◊).
To independently validate that knockdown of Sec5 or Exo84 reduced tumor growth we generated retroviruses encoding a second shRNA (shRNA-2) against a different region of the Sec5 (28) or Exo84 mRNA (Fig. 3A). RasG12V-transformed HEK-HT cells were infected with these retroviruses and stable cell lines were assayed for Sec5 and Exo84 expression. These second shRNAs reduced Sec5 expression by 62% and Exo84 expression by 51%, as assessed by real-time RT-PCR (Fig. 3A). Thus, these shRNAs knocked down the expression of the desired target mRNA, although not as efficiently as the first shRNAs. The resultant stable cell lines were injected into immunocompromised mice as before, and tumor volume was recorded at regular intervals. Consistent with the previous findings, cells expressing these independently generated Sec5 and Exo84 shRNAs exhibited a clear reduction in tumorigenic growth (Fig. 3C). Moreover, there was a correlation between tumor size and the degree of Sec5 or Exo84 expression (Fig. 3, A and C, compare knockdown level and subsequent tumor size of Sec5 and Exo84 shRNA-1 to shRNA-2). Sec5 and Exo84 thus act as critical mediators of oncogenic Ras-induced tumorigenesis.
Since suppression of Sec5 and Exo84 expression had the greatest effects on transformation and tumorigenesis, and both proteins are members of the exocyst complex, we tested whether simultaneously reducing the expression of both of these exocyst components would further reduce the level of tumor growth. Specifically, RasG12V-transformed HEK-HT cells were sequentially infected with the aforementioned retroviruses encoding Sec5 shRNA-1 and Exo84 shRNA-1. Appropriate knockdown of the targeted mRNA was confirmed by quantitative real-time RT-PCR (Fig. 3A). Sec5 and Exo84 expression levels were reduced by 59% and 58%, respectively. Tumors arising in mice injected with this cell line grew the slowest, with a latency of 29 days, and displayed impaired growth kinetics (Fig. 3, C and D). Thus the concerted loss of Sec5 and Exo84 function further reduced tumor growth.
Knockdown of Sec5 and/or Exo84 is Associated with a Minor Proliferation Defect in vivo
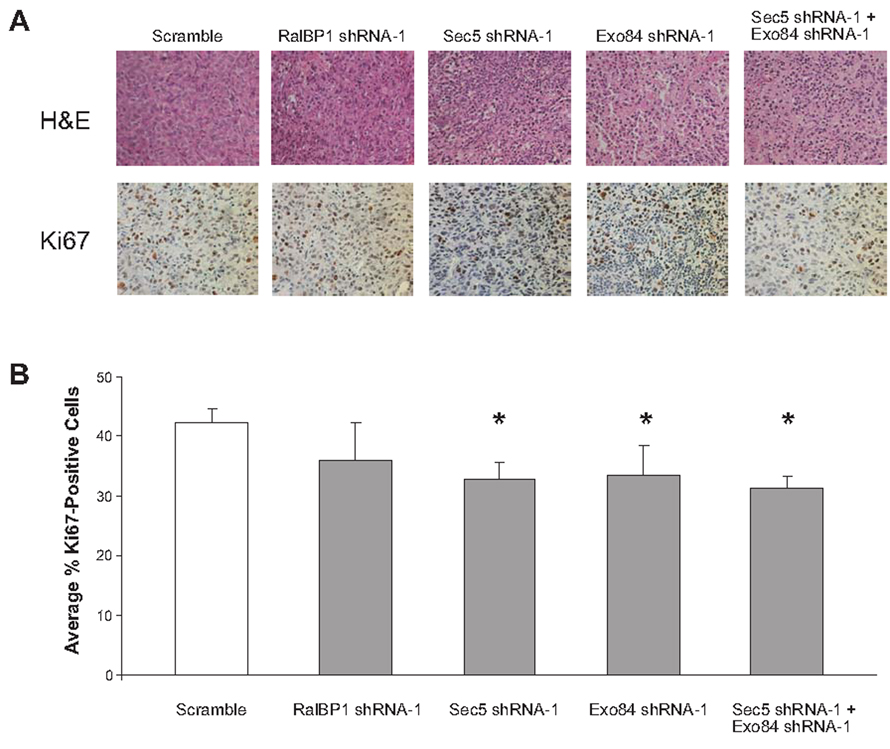
To ascertain the nature of the defect in tumor growth upon knockdown of either Sec5 or Exo84, tumors arising from RasG12V-transformed HEK-HT cells stably expressing RalBP1, Sec5, Exo84, or both Sec5 and Exo84 shRNA were removed and immunohistologically compared to tumors arising from scramble control cells. Specifically, tumors arising from the aforementioned cells were excised, formalin fixed, paraffin embedded, and sectioned. H&E staining of the tumors did not reveal any gross histological differences compared to scramble control tumors (Fig. 4A, top). Since Ral proteins have been previously linked to proliferation (7), changes in cell proliferation were assessed by Ki67 immunohistochemical staining. Tumors derived from RalBP1 knockdown cells had no statistically significant difference in the number of Ki67-positive cells (P>0.05) compared to scramble control tumors. (Fig. 4A, bottom and 4B) However, tumors eventually arising from Sec5, Exo84, or Sec5 and Exo84 double knockdown cells exhibited a small, but statistically significant 10% decrease in the number of Ki67-positive cells (P<0.05) compared to scramble control tumors (Fig. 4A bottom and 4B). The relatively minor effect may be a consequence of tumors overcoming the loss of Sec5 and Exo84 by the time they can be excised and examined. Nevertheless, the detected decrease in Ki67 staining does suggest that knockdown of Sec5 and/or Exo84 reduces proliferation of tumorigenic cells in vivo.
FIGURE 4.
Tumors derived from cells expressing Sec5 shRNA and/or Exo84 shRNA exhibit reduced Ki67 staining. A. Representative H&E (top) and Ki67 (bottom) staining of tumors arising in mice from injection of RasG12V-transformed HEK-HT cells expressing the indicated shRNAs (Magnification, X400). B. Average percentage of Ki67-positive cells ± standard deviation from at least three independent fields consisting of a total of at least one thousand cells from two different tumors. * p < 0.05, as assessed by Student’s t-test.
Knockdown of Sec5 and Exo84 is Associated with a Proliferation Defect in vitro
To further examine the effects of Sec5 and Exo84 knockdown on cell proliferation observed by immunohistochemical staining, we performed in vitro cell proliferation assays in which the previously described RasG12V-transformed HEK-HT cells stably expressing scramble, RalBP1, Sec5, Exo84, or both Sec5 and Exo84 shRNA were seeded at the same density under normal culture conditions. Cells were stained with trypan blue and counted in triplicate at timepoints 0, 48 and 96 hours. Knockdown of either Sec5, Exo84 or both significantly reduced cell number at 48 and/or 96 hours after seeding. Specifically at the 96 h timepoint, shRNA-mediated knockdown of either Sec5, Exo84, or both resulted in a 40–50% reduction in cell number, whereas knockdown of RalBP1 results in slightly less pronounces decrease (30%) compared to scramble control treated cells (Fig. 5A). As the number of nonviable (Trypan blue-positive) cells was very low in the cell lines (not shown), this difference in cell number was attributed to a decrease in proliferation.
FIGURE 5.
Knockdown of Sec5 or Exo84 decreases the proliferation of Ras-transformed human cells. A. Cell number ± standard deviation versus time of cultured RasG12V-transformed HEK-HT cells stably expressing a scramble control shRNA (□), RalBP1 shRNA-1 (■), Sec5 shRNA-1 (▲), Exo84 shRNA-1 (♦), or Sec5 shRNA-1 and Exo84 shRNA-1 (◊). B. Relative mRNA level ± standard deviation of the indicated transcripts in HEK-HT cells stably infected with the indicated shRNAs, as determined by real-time RT-PCR. C. Cell number ± standard deviation versus time of cultured HEK-HT cells stably expressing a scramble control shRNA (□), RalBP1 shRNA-1 (■), Sec5 shRNA-1 (▲), or Exo84 shRNA-1 (♦).
We also tested whether knockdown of the individual Ral effectors decreased proliferation of untransformed HEK-HT cells. Specifically, HEK-HT cells were stably infected with the aforementioned retroviruses encoding shRNA (shRNA-1) targeting RalBP1, Sec5, Exo84, or a scramble sequence as a control. Reduced expression of RalBP1, Sec5, and Exo84 was assessed by real-time RT-PCR. RalBP1, Sec5, and Exo84 mRNA levels were decreased by approximately 90%, 82%, and 90%, respectively (Fig. 5B). The proliferation rates of the resultant stable cell lines were then determined by cell proliferation assays, as previously described. Unlike the situation in RasG12V-transformed HEK-HT cells, knockdown of RalBP1, Sec5, or Exo84 did not decrease proliferation at 48 or 96 hours after seeding, compared to scramble control treated cells (Fig. 5C). Thus, it is not that knockdown of Sec5, Exo84, or to a somewhat lesser extent RalBP1, is generally anti-proliferative, but rather that tumorigenic RasG12V-transformed HEK-HT cells are more sensitive to loss of these proteins than their untransformed counterparts, at least with respect to proliferation.
Discussion
Inappropriate activation of the small GTPase Ras plays a critical role in oncogenesis. Mutated and constitutively activated forms of Ras are found in nearly one-third of all human cancers (1). Consequently, there has been considerable effort to define the effector pathway downstream of Ras signaling responsible for mediating its oncogenic effects. To this end, a critical role for the RalGEF/Ral effector pathway in Ras-mediated transformed and tumorigenic growth of a variety of human cell types has been demonstrated (4, 9–11). RalGTPases signal through a small set of effector proteins, the most documented being RalBP1 and the exocyst complex components Sec5 and Exo84. In the current study we have examined which Ral effectors mediate Ras-driven oncogenesis.
We now demonstrate that the Ral effectors Sec5 and Exo84 are critical mediators of oncogenic Ras signaling. Stable knockdown of either Sec5 or Exo84 in RalGEF-transformed cells led to a greater decrease in anchorage-independent colony formation than RalBP1 or Sec6 knockdown. The decrease in anchorage-independent growth was similar to that previously observed upon knockdown of RalA in the same cells (9), arguing that these effectors mediate much of the transforming signal downstream of Rals. In support of our finding, it was previously shown that a mutation that reduced the binding of RalA to Sec5 and Exo84 resulted in a greater reduction of transformation by activated RalA than a mutation that reduced binding of RalA to RalBP1 (9). With regard to Ras-mediated tumorigenesis, we show that stable knockdown of Sec5, Exo84, or both in highly tumorigenic RasG12V-transformed cells prolonged the latency period of tumorigenesis and impeded subsequent tumor growth. Conversely, stable knockdown of RalBP1 did not significantly affect tumorigenesis of the same cells. Immunohistochemical analysis revealed that the outgrowing tumors formed from Sec5 and/or Exo84 knockdown cells displayed slightly less Ki67-positive cells than those derived from scramble control or RalBP1 knockdown cells. Similarly, knockdown of Sec5, Exo84 or both reduced the proliferation of RasG12V-transformed cells slightly more than knockdown of RalBP1, and clearly more than scramble control treated cells, an effect that was not observed in the same cells in the absence of RasG12V. Taken together, these data are suggestive of a defect in cell proliferation inflicted by the knockdown of Sec5, Exo84 or both in Ras-trasnformed cells.
While Rals have been implicated in the regulation of a number of diverse cellular processes, our current findings suggest that interactions with the exocyst complex components Sec5 and Exo84 may contribute to their oncogenic activity. Ral GTPases have been proposed to regulate a Sec5-Exo84 interaction interface for dynamic control of exocyst complex assembly and function in mammalian cells (17, 18, 30). Interactions with these proteins link Rals to basolateral membrane protein targeting and maintenance of epithelial cell polarity, the maintenance of cytokinesis, and cell migration (14, 17, 24, 25). While deregulation of these processes would likely have significant repercussions, the exact mechanism by which the effects of Ral proteins on exocyst function may be linked to Ras-mediated transformation of human cells remains to be elucidated.
Materials and Methods
Cell Lines
Human embryonic kidney cells stably expressing the early region of SV40, the catalytic subunit of telomerase hTERT (HEK-HT), and either H-RasG12V or Rlf-CAAX were previously described (4, 9, 29, 31), and have not subsequently been verified to express T-Ag, t-Ag, hTERT or, when appropriate, H-RasG12V or Rlf-CAAX. Cell lines were stably infected with retroviruses generated from the described vectors encoding the indicated shRNAs. Specifically, Rlf-CAAX-transformed HEK-HT cells were stably infected with a retrovirus derived from pSUPER-retro-puro encoding RalBP1 shRNA-1, Sec5 shRNA-1, Exo84 shRNA-1, Sec6 shRNA-1, or a vector control. RasG12V-transformed HEK-HT cells were stably infected with a retrovirus derived from pSUPER-retro-puro encoding RalBP1 shRNA-1, Sec5 shRNA-1, Sec5 shRNA-2, Exo84 shRNA-1, Exo84 shRNA-2, or a scramble control. RasG12V-transfected HEK-HT cells expressing Sec5 shRNA-1 were further infected with pSUPER-retro-GFP/neo Exo84 shRNA-1 to generate cells expressing Sec5 shRNA-1 and Exo84 shRNA-1. Untransformed HEK-HT cells were stably infected with a retrovirus derived from pSUPER-retro-puro encoding RalBP1 shRNA-1, Sec5 shRNA-1, Exo84 shRNA-1, or a scramble control. Stable polyclonal populations were enriched by either culture in media supplemented with puromycin, as previously described (31), or sorting for GFP-positive cells by FACS analysis. To measure proliferation rate, the indicated HEK-HT and RasG12V-transfected HEK-HT cell lines were seeded at 20,000 and 15,000 cells, respectively, per well of 6-well plates, in duplicate, and viable (Trypan blue-negative) cells were counted 48 and 96 hours later.
Plasmids
pSUPER-retro-puro was engineered to encode RalBP1 shRNA-1, Sec5 shRNA-1 and 2, Exo84 shRNA-1, and Sec6 shRNA-1. pSUPER-retro-GFP/neo was engineered to encode Exo84 shRNA-1. pSM2c encoding Exo84 shRNA-2 was obtained from the Duke University RNA Interference Facility. shRNAs targeted human sequences and were as follows: RalBP1 shRNA-1 (5’-GTA GAG AGG ACC ATG ATG T) (22), Sec5 shRNA-1 (5’-GG TCG GAA AGA CAA GGC AGA T) (28), Sec5 shRNA-2 (5’-CGG CAG AAT GGA TGT CTG C) (28), Exo84 shRNA-1 (5’-GGT GCC ACT TTA CTC TAT A) (18), Exo84 shRNA-2 (5’- ACA ATA TAA TTT GAA TGG CTA A), Sec6 shRNA-1 (5’-GGG AAG AGA AAA TTG ACA G) (32).
Real-time RT-PCR
Total RNA was isolated from cells (Qiagen RNeasy mini kit, Qiagen Inc., Valencia, CA), and reverse-transcribed (iScript cDNA synthesis kit, Bio-Rad, Hercules, CA). Real-time PCR analysis of described mRNA levels was performed on triplicate samples using iTaq SYBR Green Supermix with ROX (Bio-Rad). The primers used were: RalBP1 5’-ACT GTG CAG ATC AGC AAT CG (forward) and 5’-CCT GAT CTC CTC CTT GAT GC (reverse), Exo84 5’-CTG CTT GAG AAG GTG GAA GG (forward) and 5’-GGT AGC CAC CAA CAA GCA AT (reverse), and Sec5 5’-GAT CCT TCA GCT CAT GCA CA (forward) and 5’- GAC TGA GAT GGC CCA ACA CT (reverse). Pre-validated primers and probes against 18S RNA were included as controls (Applied Biosystems, Foster City, CA). Measurements were performed on an ABI Prism 7000 sequence detection system.
Protein detection
Clarified total cellular lysates were immunoblotted with α-RalBP1 (Santa Cruz, Santa Cruz, CA), α-Sec6 (Stressgen, Ann Arbor, MI), or α-actin (Sigma, St. Louis, MO) antibodies using standard procedures to detect endogenous RalBP1, Sec6, and actin, respectively.
Soft agar
50,000 cells per 3 cm plate were suspended in soft agar as previously described (9, 31) and colonies greater than 30 cells were scored after 4 weeks. Assays were performed in triplicate and at least twice independently.
Tumor growth
7 × 106 cells mixed with Matrigel (BD, Franklin Lakes, NJ) were injected subcutaneously into the flank of 4 SCID/beige mice per cell line, after which tumor volumes were determined at regular intervals as described previously (9, 31). All procedures with mice were done according to a protocol approved by the Duke University Institutional Animal Care and Use Committee,
Immunohistochemistry
Tumors arising in mice from injection of RasG12V-transformed HEK-HT cells expressing the indicated shRNAs were excised, fixed in formalin, paraffin embedded, sectioned, and H&E stained, or used for immunohistochemical detection of Ki67. Ki67-positive cells were counted around the proliferative tumor periphery. Statistical significance was determined by Student’s t-test. A p value of < 0.05 was considered significant.
Acknowledgements
We thank Sarah Ronnebaum and Stacey Adam for technical assistance, and Sarah Ronnebaum and Corinne Linardic for critical review of the manuscript.
Grant support: This work was funded by NIH grant CA094184. K. Lim was a Department of Defense Breast Cancer Research Pre-doctoral Scholar.
References
- 1.Bos JL. ras oncogenes in human cancer: a review. Cancer Res. 1989;49:4682–4689. [PubMed] [Google Scholar]
- 2.Feig LA, Urano T, Cantor S. Evidence for a Ras/Ral signaling cascade. Trends Biochem Sci. 1996;21:438–441. doi: 10.1016/s0968-0004(96)10058-x. [DOI] [PubMed] [Google Scholar]
- 3.Shields JM, Pruitt K, McFall A, Shaub A, Der CJ. Understanding Ras: 'it ain't over 'til it's over'. Trends Cell Biol. 2000;10:147–154. doi: 10.1016/s0962-8924(00)01740-2. [DOI] [PubMed] [Google Scholar]
- 4.Hamad NM, Elconin JH, Karnoub AE, et al. Distinct requirements for Ras oncogenesis in human versus mouse cells. Genes Dev. 2002;16:2045–2057. doi: 10.1101/gad.993902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gonzalez-Garcia A, Pritchard CA, Paterson HF, Mavria G, Stamp G, Marshall CJ. RalGDS is required for tumor formation in a model of skin carcinogenesis. Cancer Cell. 2005;7:219–226. doi: 10.1016/j.ccr.2005.01.029. [DOI] [PubMed] [Google Scholar]
- 6.Chardin P, Tavitian A. The ral gene: a new ras related gene isolated by the use of a synthetic probe. Embo J. 1986;5:2203–2208. doi: 10.1002/j.1460-2075.1986.tb04485.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Feig LA. Ral-GTPases: approaching their 15 minutes of fame. Trends Cell Biol. 2003;13:419–425. doi: 10.1016/s0962-8924(03)00152-1. [DOI] [PubMed] [Google Scholar]
- 8.Takai Y, Sasaki T, Matozaki T. Small GTP-binding proteins. Physiol Rev. 2001;81:153–208. doi: 10.1152/physrev.2001.81.1.153. [DOI] [PubMed] [Google Scholar]
- 9.Lim KH, Baines AT, Fiordalisi JJ, et al. Activation of RalA is critical for Ras-induced tumorigenesis of human cells. Cancer Cell. 2005;7:533–545. doi: 10.1016/j.ccr.2005.04.030. [DOI] [PubMed] [Google Scholar]
- 10.Lim KH, O'Hayer K, Adam SJ, et al. Divergent roles for RalA and RalB in malignant growth of human pancreatic carcinoma cells. Curr Biol. 2006;16:2385–2394. doi: 10.1016/j.cub.2006.10.023. [DOI] [PubMed] [Google Scholar]
- 11.Rangarajan A, Hong SJ, Gifford A, Weinberg RA. Species- and cell type-specific requirements for cellular transformation. Cancer Cell. 2004;6:171–183. doi: 10.1016/j.ccr.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 12.Chien Y, White MA. RAL GTPases are linchpin modulators of human tumour-cell proliferation and survival. EMBO Rep. 2003;4:800–806. doi: 10.1038/sj.embor.embor899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oxford G, Owens CR, Titus BJ, et al. RalA and RalB: antagonistic relatives in cancer cell migration. Cancer Res. 2005;65:7111–7120. doi: 10.1158/0008-5472.CAN-04-1957. [DOI] [PubMed] [Google Scholar]
- 14.Rosse C, Hatzoglou A, Parrini MC, White MA, Chavrier P, Camonis J. RalB mobilizes the exocyst to drive cell migration. Mol Cell Biol. 2006;26:727–734. doi: 10.1128/MCB.26.2.727-734.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bodemann BO, White MA. Ral GTPases and cancer: linchpin support of the tumorigenic platform. Nat Rev Cancer. 2008;8:133–140. doi: 10.1038/nrc2296. [DOI] [PubMed] [Google Scholar]
- 16.Cantor SB, Urano T, Feig LA. Identification and characterization of Ral-binding protein 1, a potential downstream target of Ral GTPases. Mol Cell Biol. 1995;15:4578–4584. doi: 10.1128/mcb.15.8.4578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moskalenko S, Henry DO, Rosse C, Mirey G, Camonis JH, White MA. The exocyst is a Ral effector complex. Nat Cell Biol. 2002;4:66–72. doi: 10.1038/ncb728. [DOI] [PubMed] [Google Scholar]
- 18.Moskalenko S, Tong C, Rosse C, et al. Ral GTPases regulate exocyst assembly through dual subunit interactions. J Biol Chem. 2003;278:51743–51748. doi: 10.1074/jbc.M308702200. [DOI] [PubMed] [Google Scholar]
- 19.Jullien-Flores V, Dorseuil O, Romero F, et al. Bridging Ral GTPase to Rho pathways. RLIP76, a Ral effector with CDC42/Rac GTPase-activating protein activity. J Biol Chem. 1995;270:22473–22477. doi: 10.1074/jbc.270.38.22473. [DOI] [PubMed] [Google Scholar]
- 20.Jullien-Flores V, Mahe Y, Mirey G. RLIP76, an effector of the GTPase Ral, interacts with the AP2 complex: involvement of the Ral pathway in receptor endocytosis. J Cell Sci. 2000;113(Pt 16):2837–2844. doi: 10.1242/jcs.113.16.2837. [DOI] [PubMed] [Google Scholar]
- 21.Nakashima S, Morinaka K, Koyama S, et al. Small G protein Ral and its downstream molecules regulate endocytosis of EGF and insulin receptors. Embo J. 1999;18:3629–3642. doi: 10.1093/emboj/18.13.3629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rosse C, L'Hoste S, Offner N, Picard A, Camonis J. RLIP, an effector of the Ral GTPases, is a platform for Cdk1 to phosphorylate epsin during the switch off of endocytosis in mitosis. J Biol Chem. 2003;278:30597–30604. doi: 10.1074/jbc.M302191200. [DOI] [PubMed] [Google Scholar]
- 23.Shipitsin M, Feig LA. RalA but not RalB enhances polarized delivery of membrane proteins to the basolateral surface of epithelial cells. Mol Cell Biol. 2004;24:5746–5756. doi: 10.1128/MCB.24.13.5746-5756.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cascone I, Selimoglu R, Ozdemir C, et al. Distinct roles of RalA and RalB in the progression of cytokinesis are supported by distinct RalGEFs. Embo J. 2008;27:2375–2387. doi: 10.1038/emboj.2008.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen XW, Inoue M, Hsu SC, Saltiel AR. RalA-exocyst-dependent recycling endosome trafficking is required for the completion of cytokinesis. J Biol Chem. 2006;281:38609–38616. doi: 10.1074/jbc.M512847200. [DOI] [PubMed] [Google Scholar]
- 26.Wolthuis RM, Bauer B, van 't Veer LJ, et al. RalGDS-like factor (Rlf) is a novel Ras and Rap 1A-associating protein. Oncogene. 1996;13:353–362. [PubMed] [Google Scholar]
- 27.Wolthuis RM, de Ruiter ND, Cool RH, Bos JL. Stimulation of gene induction and cell growth by the Ras effector Rlf. Embo J. 1997;16:6748–6761. doi: 10.1093/emboj/16.22.6748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Prigent M, Dubois T, Raposo G, et al. ARF6 controls post-endocytic recycling through its downstream exocyst complex effector. J Cell Biol. 2003;163:1111–1121. doi: 10.1083/jcb.200305029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hahn WC, Counter CM, Lundberg AS, Beijersbergen RL, Brooks MW, Weinberg RA. Creation of human tumour cells with defined genetic elements. Nature. 1999;400:464–468. doi: 10.1038/22780. [DOI] [PubMed] [Google Scholar]
- 30.Camonis JH, White MA. Ral GTPases: corrupting the exocyst in cancer cells. Trends Cell Biol. 2005;15:327–332. doi: 10.1016/j.tcb.2005.04.002. [DOI] [PubMed] [Google Scholar]
- 31.O'Hayer KM, Counter CM. A genetically defined normal human somatic cell system to study ras oncogenesis in vivo and in vitro. Methods Enzymol. 2006;407:637–647. doi: 10.1016/S0076-6879(05)07050-3. [DOI] [PubMed] [Google Scholar]
- 32.Chien Y, Kim S, Bumeister R, et al. RalB GTPase-mediated activation of the IkappaB family kinase TBK1 couples innate immune signaling to tumor cell survival. Cell. 2006;127:157–170. doi: 10.1016/j.cell.2006.08.034. [DOI] [PubMed] [Google Scholar]