Abstract
Acute infection leads to CD8+ T cell activation, division, and differentiation. Following clearance of infection, cells revert to two distinct subsets of memory, central (TCM) and effector (TEM) memory. Adoptive transfer of naïve T cell receptor transgenic (TCR-tg) T cells has been used to study the differentiation of these memory subsets, which are often discriminated by expression of CD62L. Naïve CD8+ T cells are CD62Lhigh, and CD62L expression is lost during the ‘effector’ phase. Adoptive transfer studies show that higher transfer frequencies result in diminished T cell expansion and a higher proportion CD62Lhigh. This suggests a relationship between CD62L expression and cell division, where division leads to conversion from CD62Lhigh to CD62Llow phenotype. To address this hypothesis we adoptively transferred graded numbers of OT-1 TCR-tg T cells from naïve donors and tracked the kinetics and phenotype of the immune response following infection. We developed a simple mathematical model of division-linked CD62L differentiation which we compared to the experimental data. Our results show that division-linked differentiation predicts the differences in proportion of cells CD62Lhigh observed between responses of different adoptive transfer number, and within individual mice. We calculate that approximately 20% of CD62Lhigh cells convert to CD62Llow during each division.
Keywords: CD8 T cells, Cell Differentiation, Cellular proliferation, Mathematical model
Introduction
The CD8+ T cell response is important for control of infection with a number of pathogens including viruses and intracellular bacteria. Prior to infection, the naïve CD8+ T cell repertoire contains a diverse array of specificities and only approximately 100 to 1000 naïve precursors are available to respond to any given pathogen epitope (1, 2). These naïve cells lack functional capacity to mediate anti-pathogen effector functions. After infection and presentation of pathogen antigens by dendritic cells, naïve CD8+ T cells are activated and divide extensively, expanding up to >10,000 fold to form a large ‘effector’ response. Coincident with this expansion, CD8+ T cells differentiate and acquire effector functions and homing characteristics that allow them to control infection. The expansion phase is followed by a contraction (death) phase, where 90–95% of antigen specific CD8 T cells are eliminated. The remaining antigen specific CD8 T cells form the initial memory pool, which can be stably maintained for life.
The mechanisms and dynamics of CD8+ T cell differentiation are not well understood. On a population level, CD8+ T cells acquire effector function following infection. However, on the individual cell level a great diversity of phenotypes is observed (3–5).
Thus, cells do not have a simple ‘on / off’ switch for activation and differentiation, where all stimulated cells follow a similar ‘program’ of behavior. Rather, individual cells acquire different phenotypes of cytokine secretion over time (3, 6). This apparently ‘individualistic’ behavior on the single cell level has been explained by factors such as asymmetric division of cell contents and receptors (7), or stochastic effects mediated by low levels of signaling (8). The ‘average’ effect of these behaviors is observed at the population level.
Another proposed explanation for acquisition of mixed individual phenotypes over time is stochastic ‘division-linked differentiation’ seen for both B cells and T cells stimulated in vitro (9–17). Lymphocytes show unique patterns of acquisition and loss of cell surface markers and effector function that correlate closely with division number rather than time of stimulation. In some systems cellular heterogeneity emerges according to simple rules of probabilistic assortment (10, 11, 18, 19).
Stochastic events are also a feature of cell division and death times. Despite the element of randomness that occurs on a single cell level, modeling of population behavior allows prediction of, for example, the proportion of cells that will commence dividing or die, or the proportion of cells that will acquire a given phenotype on each division following stimulation (18, 20, 21).
The dynamics of CD8+ T cell activation, expansion and phenotype have been studied using adoptive transfer systems in mice, where large numbers of antigen-specific T cell receptor transgenic (TCR-tg) T cells are intravenously injected into naïve mice and tracked over the course of an infection (22). In particular the division, differentiation, effector function, and survival capacity of T central (CD127high CCR7 + CD62Lhigh) (denoted TCM) and effector (CD127high CCR7-CD62Llow) (denoted TEM) memory subsets have been intensively studied. TCM cells have increased division potential, diminished effector function and increased chance of survival when compared to the TEM population (23–25). However, the precise mechanisms by which these lineages form, are not well understood. Adoptive transfer of TCR-tg T cells after sorting on CD62L expression has led to proposed differentiation pathways where the TEM population generates the TCM population, and alternatively where the TCM population generates the TEM population (25–27). On the other hand, T cell receptor (TCR) repertoire analysis and studies involving adoptive transfer of single cells show that various phenotypes can arise from a single progenitor or clonotype (28, 29).
Recently it has become clear that the number of TCR-tg T cells used in adoptive transfer studies can affect the phenotype and kinetics of responding cells (30–33). In particular, higher adoptive transfer populations exhibit reduced expansion following infection. This is in contrast to lower adoptive transfer populations whose robust expansion reflects that of the endogenous response (30, 31, 33). That is, although the total number of antigen specific cells at the peak of the response is higher in larger adoptive transfer populations, the ratio of expansion (peak number of cells divided by the adoptive transfer amount) is lower. The phenotype of populations derived from larger adoptive transfer numbers also have an increased proportion of cells retaining the CD62Lhigh and CD127high phenotype (30, 32). This suggests a mechanism whereby the extent of cell expansion and level of differentiation to the CD62Llow and CD127low phenotype maybe mechanistically linked. We have previously demonstrated how a division-linked mechanism controlling CD62L phenotype differentiation reproduces TCR repertoire structures in response to influenza in mice (34). Here, we analyze the interplay between division, differentiation and death of transgenic CD8+ T cells after adoptive transfer of different numbers of cells. We measure the rate of division-linked CD62Lhi to CD62Llo conversion, and show how this parameter alone can accurately predict the proportion of CD62Lhigh and CD62Llow cells observed between adoptive transfer groups and within individual mice in vivo. In contrast to CD62L, our analysis concludes that one-way division-linked CD127 differentiation is not compatible with the experimental results.
Results
Cell division predicts CD62L expression at peak response
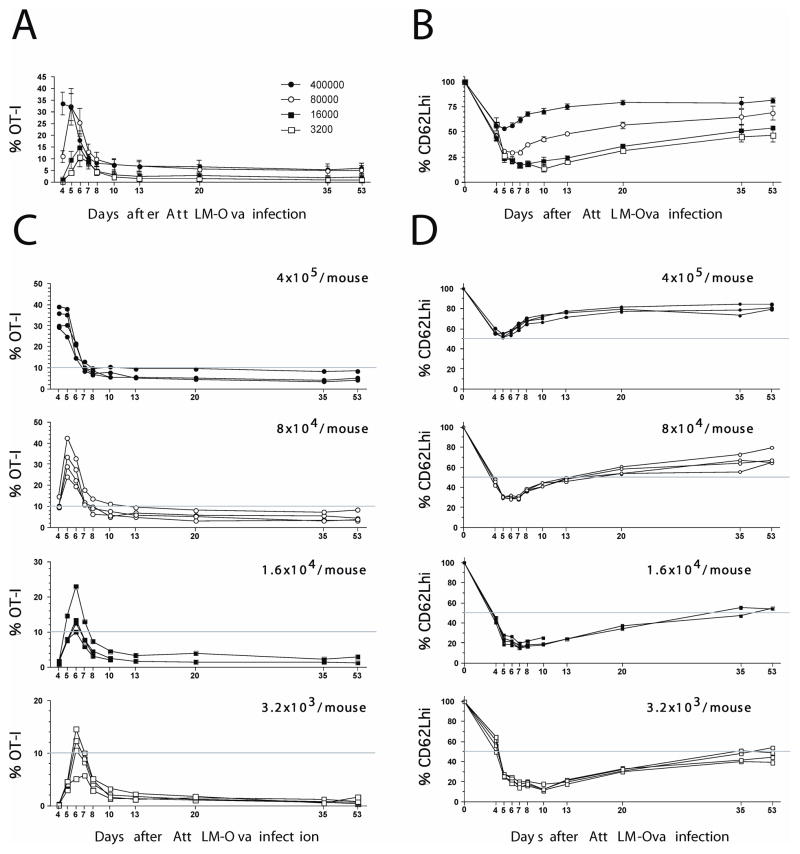
Previous studies have shown that larger adoptive transfer populations of TCR-tg T cells do not give proportionally higher levels of TCR-tg T cells at the peak of the response (30, 31). This indicates that larger transfer populations undergo reduced expansion, likely because of cells completing fewer divisions (we show later why increased death is inconsistent with the data). To study the interaction of division history on CD62L and CD127 expression, and on survival of effector cell populations, we transferred graded numbers of Thy1.1 OT-1 cells from naïve donors into C57BL/6 (B6 Thy 1.2) recipients which were subsequently infected with LM-OVA and bled at various times. This allows tracking of the levels and phenotype of the Ova-specific CD8+ T cell response longitudinally in the blood in individual mice (Sup. Fig. 1, Fig. 1). As previously reported we observed expansion of OT-1 T cells, and loss of CD62L expression during this expansion (Fig. 1). In addition, we observed that higher adoptive transfer amounts did not experience as vigorous expansion post infection as lower adoptive transfer amounts (30), and the proportion of cells remaining CD62Lhigh at the peak of the response was also higher with higher adoptive transfer amounts (Fig. 2A). Thus, there is a relationship between cell expansion and phenotype between different adoptive transfer amounts.
Figure 1. Kinetics and phenotype of OT-I CD8 T cells after infection.
Purified naïve OT-I Thy1.1 cells at the indicated numbers were transferred into B6 Thy1.2 mice, and 1 day later, the recipient mice were immunized with Att LM-Ova. Kinetics and phenotype (CD62L) of OT-I Thy1.1 cells were followed in the blood at indicated days p.i. A, C) Kinetics of OT-I CD8 T cell response after infection. B, D) Expression of CD62L on gated OT-I Thy1.1+ / CD8+ cells on indicated days after infection.
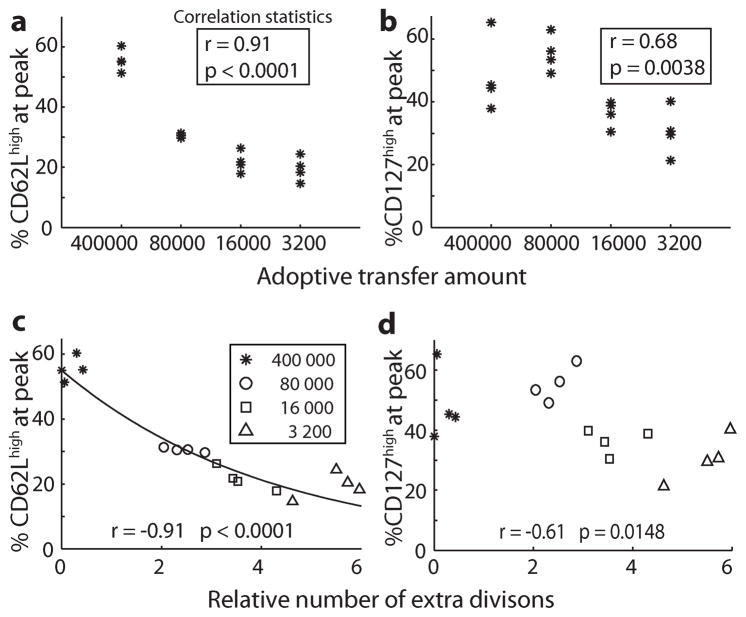
Figure 2. Adoptive transfer number and cell division affects phenotype.
400000, 8000, 1600 and 3200 naïve OT-1 T cells were transferred into naïve mice which were subsequently infected with LM-OVA. The percentage OT-1 cells and phenotype were measured at the peak of the response in each animal. There was a significant positive correlation between the proportion of cells CD62Lhigh and adoptive transfer amount (A), and the percentage of cells CD127high and adoptive transfer amount (B) (Spearman’s tied-rank correlation). The relative number of extra divisions required by different mice to achieve the peak of their response (compensating for different adoptive transfer numbers) was calculated (C, D). There was a significant negative correlation between the relative number of extra divisions, and both the percentage of cells CD62Lhigh (C) and the percentage of cells CD127high (D) (Spearman’s rank correlation). The predicted relationship between CD62L expression and division number using a division-linked mechanism where 21.2% (median value from between mice, figure 3) of the CD62Lhigh population becomes CD62Llow with each division is overlaid in panel (solid line in panel C).
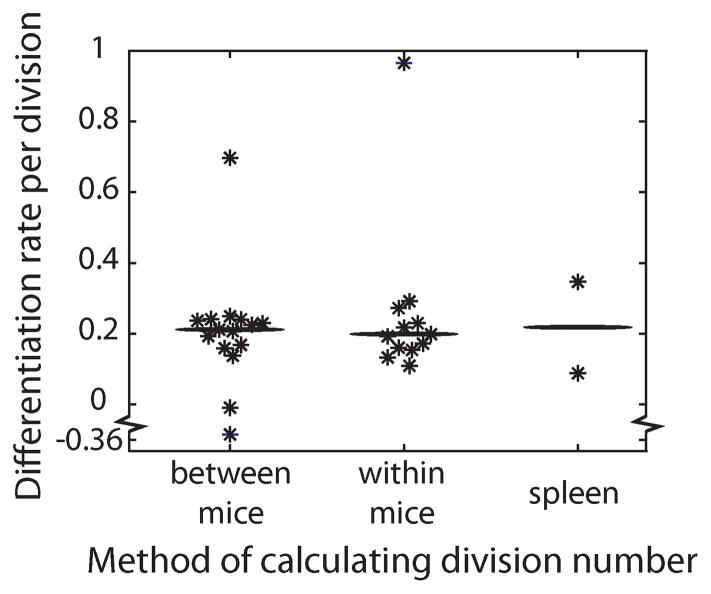
Using this data, we investigated the accuracy of a division-linked mechanism of differentiation to predict differentiation of OT-1 cells from CD62Lhigh to CD62Llow phenotype during the expansion phase of infection. Individual animal data on the adoptive transfer amount and size of the response at peak, was used to estimate the relative number of divisions cells had undergone in each mouse. We found that the relative number of divisions was well correlated with the observed proportion of cells remaining CD62Lhigh (Fig. 2C). Using the relative number of divisions and the differences in CD62L expression for each mouse and equation 1, we estimated the rate of differentiation from CD62Lhigh to CD62Llow per division (c) for each mouse. The median differentiation rate over all mice was calculated as 21.2% per division (Fig. 3). That is, approximately 20% of CD62Lhigh cells become CD62Llow upon each division. Using this median differentiation rate per division, a division-linked model of CD62L differentiation accurately predicts the proportion of cells CD62Lhigh among different mice at the peak of infection (Fig. 2C).
Figure 3. Estimating the proportion of CD62Lhigh cells that become CD62Llow on each division.
The rate of differentiation (c) from CD62Lhigh to CD62Llow per division was calculated with three methods. In the first, the relative extra number of divisions to reach the peak proportion of OT-1 cells calculated between mice taking into consideration the adoptive transfer amount (n = 15, median c value = 0.212 [indicated by horizontal bar]). The second method calculates the number of divisions from the first day of measurement to the peak of infection in individual mice (n = 12, median c value = 0.199). The third calculates the number of divisions cells undergo in the spleen (n = 2, mean c value = 0.218).
Progressive differentiation during the acute response
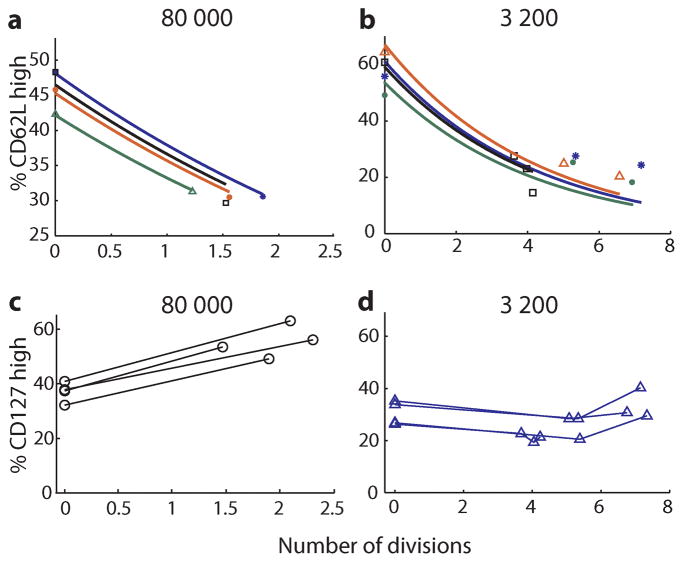
The results above demonstrate that division linked differentiation can explain the observed relationship between cell expansion and cell phenotype in different mice measured cross-sectionally at the peak of the response. However, this may have arisen due to differences in the cellular environment following transfer of different numbers of cells that fortuitously conform to the model. It does not demonstrate that differentiation occurs progressively with division in individual animals over the course of the response. Therefore, we analyzed the longitudinal data for each mouse to look at the relationship between expansion and CD62L expression over time within an individual. For each mouse we identified the first day when OT-1 cells and phenotype were measureable in blood, and then compared this with the levels and phenotype at the peak of the response. The difference in OT-1 T cell levels was used to calculate the number of divisions in this time, and the difference in CD62L expression was directly observed. Using these data the differentiation rate (c) was then computed for each animal over these days. This technique of measuring the differentiation rate gave very similar values to when relative division number was calculated across transfer groups (median of median value was 19.9% and 21.2% for longitudinal vs. cross-sectional measurements respectively)(Fig. 3), indicating that division linked differentiation also explains the dynamics of differentiation within individual mice over time. This is demonstrated in figure 4A and 4B, where we use the previous median differentiation rate (calculated from the cross sectional data), and show that this predicts CD62L phenotype levels in individual animals over time during the expansion phase of infection.
Figure 4. Predicting differentiation during the acute response.
The relationship between the number of cell divisions and cell phenotype is studied. (A, B) The number of divisions is plotted against the percentage of cells CD62Lhigh for the 80 000 (A) and 3 200 (B) adoptive transfer amounts. Overlaid is the prediction of a mechanism whereby 21.2% (median differentiation rate in cross-sectional study, figure 3) of the CD62Lhigh population becomes CD62Llow with each division (−). Different symbols and colors are used to distinguish individual mice. The percentage cells CD127high increased with division in the 80 000 adoptive transfer group (C), but did not consistently increase or decrease with division in the 3 200 adoptive transfer group (D). Lines connect points from individual animals.
Calculating the CD62L conversion rate in the spleen
Both methods described above provide similar estimates for the conversion rate per division from CD62Lhigh to CD62Llow status, indicating that division-linked differentiation explains phenotypic variation within a mouse and across adoptive transfer groups. However, a major limitation of these calculations is that they assume that blood lymphocytes proportions accurately reflect the total cell numbers in other areas of the body. The spleen would provide a better estimate of the total size of the OT-1 population. However, splenic sampling has the problem that it precludes longitudinal sampling within an animal. To investigate whether the blood provides a good measure of total OT-1 cells in the body, we studied cell numbers in the spleen of mice which were adoptively transferred with the highest and lowest levels (400 000 and 3200 TCR-tg T cells) and were subsequently infected. We also used a control group of mice receiving 400000 cells but remaining uninfected. Spleens were extracted the day before, the day of, and the day after the peak of the response, with OT-1 levels and the proportion CD62Lhigh recorded. For uninfected mice, spleens were extracted on day 4 post transfer.
By comparing the number of OT-1 cells in the spleen of infected and uninfected mice on day five that received 400 000 cells, we were able to estimate the total expansion of OT-1 cells (and therefore number of divisions undergone) in infected animals. However, since this occurred in different mice, we took the average number of cells in uninfected animals (n=3) and in infected animals (n=3) to derive a single value for the average number of divisions undergone. Similarly, for animals receiving 3200 OT-1 cells, we compared the average number of OT-1 cells on day five (n=3) and day six (the peak of infection)(n=3) to obtain a single value for the number of divisions undergone. Comparing the number of divisions and phenotype in different animals we estimated the differentiation rate per division in each animal. The results are shown in figure 3. The mean conversion rate per division was 21.8%, in close agreement with the estimates from blood (Fig. 3).
More complex regulation of CD127 phenotype
The expression of CD62L appears predictable based on a simple model of division-linked differentiation. Expression of the interleukin-7 receptor CD127 is high on naïve and memory T cells, but down-regulated during the ‘effector’ phase of the immune response. CD127 expression is high on both TCM and TEM memory T cells. We next sought to determine whether the pattern of expression of CD127 could be explained with a similar division linked process. As with CD62L, transfer of higher numbers of OT-1 cells resulted in a significantly higher proportion of cells remaining CD127high at the peak of infection (Fig. 2B)(p=0.0038). However, although the proportion of CD127high cells at peak was significantly correlated with division number (p = 0.0148), the relationship between division number and CD127 expression was much weaker than that observed with CD62L (r2 = 0.37 versus 0.83, respectively) (Fig. 2D). Moreover, when we attempted to correlate expansion with CD127 expression longitudinally in individual animals during the acute phase of the response, the fit was extremely poor since CD127 expression actually increased in the high transfer amounts (Fig. 4C). This indicates that a simple one-way division-linked CD127 differentiation model is not compatible with the experimental results.
Higher adoptive transfer results in better memory formation
The modeling above assumes that the difference in OT-1 expansion in different animals is determined by the number of divisions the cells have undergone, rather than an increased death rate of cells with increasing transfer amounts. To investigate the death rate of cells, we looked at the contraction phase of the response after peak. As shown in Figure 5A, larger adoptive transfer populations experienced reduced contraction from peak to memory. Although the death rate of T cells after the peak is most likely different to the death rate during expansion, the fact that the death rates after the peak is lower in the high adoptive transfer group does not support a model where reduced contraction (before the peak) is due to increased death. This supports our assumption that reduced expansion is most likely due to diminished proliferation. The differences in contraction raises questions about the basis of memory formation in different adoptive transfer groups.
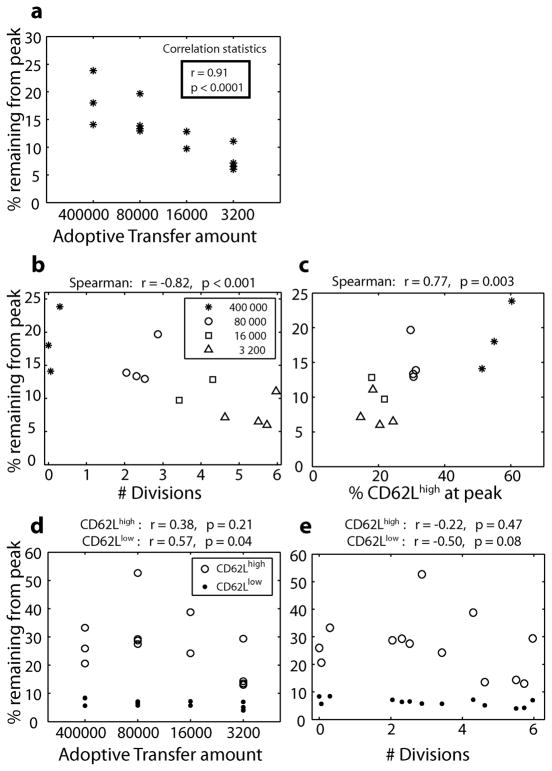
Figure 5. Division history, CD62L expression, and memory formation in vivo.
The dynamics of the T cell response to infection were tracked from day 4 out to day 53 post-infection. Memory formation was studied by measuring the proportion of OT-1 cells at the peak that remained at day 53 post-infection. Survival of total OT-1 cells is positively correlated with adoptive transfer frequency (A) (Spearman’s tied rank correlation test), indicating that higher transfer amounts lead to better memory formation. This may have occurred because these cells underwent fewer divisions (were ‘younger’) or because a higher proportion of these cells were CD62Lhigh. Both division number (B) and CD62L expression (C) are significantly correlated with survival (Spearman’s rank correlation test). The average survival of the CD62Lhigh population (mean survival = 27.01%) was significantly higher than the CD62Llow population (mean survival = 6.25%, p < 0.0001 paired t-test), regardless of the adoptive transfer amount (D, E). When the CD62Lhigh and CD62Llow populations were analyzed separately, the survival of the CD62Lhigh population was not significantly correlated with adoptive transfer amount. However, there was a significant positive correlation between the survival of the CD62Llow population and adoptive transfer amount (p = 0.04, r = 0.57 Spearman tied-rank correlation) (D). There was also a trend towards reduced survival of CD62Llow cells that had divided more times (E), although this was not significant (p=0.08).
Memory formation is affected by division history and CD62L phenotype
The variation in contraction between adoptive transfer frequencies could have arisen from either (i) differences in the number of divisions the cells had completed (with lower adoptive transfer amounts undergoing more divisions and perhaps being ‘senescent’ and dying faster) or (ii) the phenotype of OT-1 cells or some other property altered by adoptive transfer (for example lower adoptive transfer amounts having a higher proportion of cells CD62Llow). To investigate these possibilities we first looked at the correlation between the number of divisions and memory formation. The proportion of cells remaining from the peak is significantly negatively correlated with the relative number of divisions in different animals (Figure 5B, p = 0.001), which would appear to support the ‘senescence’ argument. However, a higher proportion of CD62Lhigh cells is also associated with a higher proportion of cells surviving into memory (figure 5C, p = 0.003), in support of the ‘phenotypic’ argument. To attempt to tease apart the effects of cell phenotype versus number of divisions undergone, we looked at the contraction of CD62Lhigh and CD62Llow cells, and how this was affected by the number of divisions. The survival of OT-1 cells from peak to memory was very different in the CD62Lhigh and CD62Llow compartments, with significantly less contraction in the CD62Lhigh compartment (mean 27.01% of peak remaining at day 53) compared to the CD62Llow (mean 6.25%, p < 0.0001 paired t-test).
We then analyzed the effect of cell division on the CD62Lhigh and CD62Llow compartments independently. The contraction of CD62Lhigh cells was not significantly correlated with adoptive transfer amount or division number (Figures 5D and 5E). However, the contraction of CD62Llow cells was significantly correlated with the adoptive transfer amount (p ≈ 0.04, r ≈ −0.57 Spearman tied-rank correlation), and had a trend towards correlation with division number (r ≈ −0.50, p ≈ 0.08). Thus, although it is clear that the adoptive transfer amount and the number of cell divisions the transferred cells have undergone affects the ability of cells to enter the memory compartment, whether this occurs directly as a result of the ‘age’ of the cells, or is mediated by some phenotypic characteristic that is dependent on division history (such as CD62L expression) is unclear.
Unrecruited cells do not impact on calculations
Our analysis assumes that all cells are recruited into the immune response and undergo division-linked differentiation. However, it is possible that only a small fraction of cells are recruited and the remaining cells undergo no division and remain ‘naïve’. As these cells are CD62Lhigh and are likely to be long lived, they have the potential to effect both the division-linked differentiation and memory formation analysis. We find that even at the highest adoptive transfer amount, less than 0.1% of the total OT-1 population is comprised of unrecruited cells (calculated from the difference in OT-1 cell numbers in the spleen in uninfected and infected mice, Supplementary Figure 2E). Since the percentage of CD62Lhigh cells varies from approximately 10–50%, these unrecruited cells cannot make any significant contribution to this level.
Discussion
The study of CD8+ T cell memory differentiation has produced a number of proposed lineages (27, 34). These include linear relationships, where cells progress though effector (CD127low/CD62Llow), effector memory (CD127high/CD62Llow) and central memory (CD127high/CD62Lhigh) phenotypes, and divergent relationships, where a single progenitor gives rise to multiple phenotypes early on which then maintain this commitment (35). A significant limitation of these heuristic lineage hypotheses is that they only describe general population dynamics and do not make quantitative predictions about cellular behavior. The deterministic ‘switch’ between phenotypes, or between ‘effector’ and ‘memory’ stages of an immune response, often proposed in these models is increasingly being replaced by a more complex understanding of random events at the molecular level leading to diverse behaviors on a single cell level, and ultimately predictable behaviors on a cell population level (8, 36). As the understanding of these mechanisms grow, more rigorous analysis must be applied so that competing hypothesis can be distinguished and tested. The use of mathematical models allows us to make and test quantitative predictions about cell behavior. The present analysis gives insight into how the same CD8+ T cell clonotype can give rise to both ‘central memory’ and ‘effector memory’ populations (28, 29), and how these populations evolve during the course of an infection. It also demonstrates the link between population size and phenotype, and how this can be predicted in vivo.
Adoptive transfer studies allow the tracking of antigen specific TCR-tg T cell levels and phenotype over the course of an immune response, and have been applied to help understand memory T cell lineages. Much of this work has employed a high adoptive transfer frequency, resulting in 1 000 – 10 000 fold greater precursors than naïve antigen specific endogenous estimates (30–33). Although these studies do show that adoptive transfer can dramatically change the immune response, this does not diminish the usefulness of the adoptive transfer procedure. Rather it emphasizes the need for careful experimental design and data analysis. We adoptively transfer TCR-tg T cells in 4 quantities ranging from 3 200 to 400000 donor cells. This allows us to study the kinetics and phenotype of an immune response under various conditions. We observe that CD62L expression is lost during the acute expansion phase of the response and that higher adoptive transfer population’s experience a diminished expansion (and in turn, an increased proportion of cells CD62Lhigh). A simple mathematical model, where differentiation from CD62Lhigh to CD62Llow occurs on division and the reverse conversion does not occur accurately predicts the proportion of CD62Lhigh and CD62Llow cells between mice and within individual mice during the expansion phase of the response. In addition, this model of division linked differentiation is also consistent with the observed distribution of CD8+ T cell clonotype sizes and phenotypes in the endogenous repertoire following influenza virus infection in vivo (34). Interestingly, such a model is inconsistent with the kinetics of the CD127 phenotype, which has been shown to be more dependent on the activation stimulus provided (37).
An inherent limitation of our study is that it implicitly assumes that all cells divide the same amount over time. That is, if the total population of cells double, it assumes all cells divided once. However, in vitro studies of cell division have suggested that cells are not recruited synchronously into division, and also have a distribution in their time to divide(20). An important question is whether such asynchronous recruitment and division would produce a different outcome in terms of the observed division-linked differentiation. To further investigate this, we applied a well established model of cell division (the ‘Cyton model’) that has been developed to incorporate factors such as a distribution in the time to first division and subsequent division and differentiation rates (36). By incorporating a known rate of division linked differentiation into this explicit model, we then tested how well a simple estimate of differentiation based upon total cell number and cell differentiation state would perform. We find that estimates of differentiation rate based upon total cell populations are remarkably accurate, and thus that our estimates of division-linked differentiation based on total population number are unlikely to be affected by asynchronous recruitment and division within the T cell population.
Quantifying CD8+ T cell division, differentiation, and memory formation is important for vaccine design and viral control in chronic infection. Here we measure how the extent of cell division determines CD62L expression, and observe that division is associated with the extent of memory formation in vivo. It seems highly likely that other phenotypic characteristics such as cytokine expression or cytolytic function may also be determined by the extent of cell division. The possibility that the regulation of immune effector functions such as cytokine production and immunoglobulin isotypes have evolved to change with division to optimize the effectiveness of the immune response has been proposed (10, 11, 14, 15, 38) and if correct would explain the existence of stochastic division-linked processes inherent to lymphocyte behavior. Recent work has focused on how the ‘quality’ (functional characteristics) of a response may be more important determinants of outcome than the simple ‘quantity’ of the response (39). Thus, understanding and predicting how different approaches to vaccination affect both number and phenotype of cells may be critical to optimizing vaccine efficacy.
Materials and Methods
Adoptive transfer and experimental infection of mice
Recipient mice were 6–10 week old C57BL/6 mice from the National Cancer Institute. They were given graded numbers of Thy1.1 OT-1 cells (400 000, 80 000, 16 000, 3 200 cells) purified from the spleens of naïve donors. Mice were infected with8×106 CFU of Att LM-Ova bacteria (prepared as previously described (37)) i.v. one day after cell transfer. Mice were bled by small tail snips to obtain ~20μl blood on various days after infection, and the transferred population identified with Thy1.1 (OX-7, Pharmingen), and stained with antibodies for CD62L (MEL-14, eBioscience) or CD127 (A7R34, eBioscience). Experimental protocols were approved by the University of Iowa Animal Care and Use committee.
Modeling division-linked differentiation
The simplest model of division-linked differentiation assumes that a fixed proportion of CD62Lhigh cells differentiate to the CD62Llow phenotype upon each division and that the reverse conversion does not take place. If P0 is the proportion of cells that are CD62Lhigh at the start of division, and P(n) is the proportion of cells CD62Lhigh after n divisions, then we can calculate the rate of differentiation (c) to CD62Llow phenotype per division as
| (1) |
P0 and P(n) are easily read from the experimental data. However, calculating the absolute number of divisions that cells have undergone following transfer is difficult, as it requires assumptions about the ‘take’ of transferred cells and the total number of cells in the whole animal. Instead we calculated the relative number of divisions in different animals or in individual animals across time based on the sequential sampling in blood.
We first calculated the relative number of divisions at the peak of the response in different mice. To do this we determined which mouse had undergone the least divisions (our ‘baseline mouse’, B) – that is, the mouse that received the highest adoptive transfer dose and had the smallest peak level of OT-1 cells. We then computed the relative extra number of divisions required by the remaining mice to achieve the peak of their response. For example, a mouse with the same peak level of cells that received half the adoptive transfer amount of mouse B required one extra division to reach peak. Similarly, if a mouse received the same adoptive transfer number as mouse B but had twice amount at peak, then it required one extra division to do so.
We also estimated the number of divisions by comparing the change in number of cells in an individual mouse over time. That is, if the proportion of OT-1 cells quadrupled from day 4 to 5, we assumed two extra divisions, etc. Note that as the experimental data measured the OT-1 population is a percentage of blood lymphocytes, the calculation of the number of divisions the cells have undergone (n) requires additional assumptions about these populations (discussed in Extended Methods), which however had little impact on the estimated number of divisions.
Supplementary Material
Acknowledgments
MPD and TS wish to thank Philip Hodgkin for his long-term input and inspiration in studying division-linked differentiation and helpful comments on the manuscript. We also thank Cameron Wellard for his extensive help in understanding the mechanics of the Cyton model. We thank Vanessa Venturi for critical reading of the manuscript. This work was supported by the James S. McDonnell Foundation 21st Century Research Award/Studying Complex Systems, the Australian Research Council (ARC), the National Health and Medical Research Council (NHMRC), the National Institutes of Health (MPD and JTH), and start-up funds from the University of Iowa – Department of Pathology (VPB). MPD is a Sylvia and Charles Viertel Senior Medical Research Fellow.
Abbreviations
- TCR-tg
T cell receptor transgenic
- TCM
T central memory
- TEM
T effector memory
- TCR
T cell receptor
Footnotes
Conflict of interest: The authors declare no financial or commercial conflict of interest
References
- 1.Moon JJ, Chu HH, Pepper M, McSorley SJ, Jameson SC, Kedl RM, Jenkins MK. Naïve CD4(+) T cell frequency varies for different epitopes and predicts repertoire diversity and response magnitude. Immunity. 2007;27:203–213. doi: 10.1016/j.immuni.2007.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Obar JJ, Khanna KM, Lefrancois L. Endogenous naïve CD8+ T cell precursor frequency regulates primary and memory responses to infection. Immunity. 2008;28:859–869. doi: 10.1016/j.immuni.2008.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jenkins MR, Kedzierska K, Doherty PC, Turner SJ. Heterogeneity of effector phenotype for acute phase and memory influenza A virus-specific CTL. J Immunol. 2007;179:64–70. doi: 10.4049/jimmunol.179.1.64. [DOI] [PubMed] [Google Scholar]
- 4.Johnson BJ, Costelloe EO, Fitzpatrick DR, Haanen JB, Schumacher TN, Brown LE, Kelso A. Single-cell perforin and granzyme expression reveals the anatomical localization of effector CD8+ T cells in influenza virus-infected mice. Proc Natl Acad Sci U S A. 2003;100:2657–2662. doi: 10.1073/pnas.0538056100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kelso A, Groves P. A single peripheral CD8+ T cell can give rise to progeny expressing type 1 and/or type 2 cytokine genes and can retain its multipotentiality through many cell divisions. Proc Natl Acad Sci U S A. 1997;94:8070–8075. doi: 10.1073/pnas.94.15.8070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fitzpatrick DR, Shirley KM, McDonald LE, Bielefeldt-Ohmann H, Kay GF, Kelso A. Distinct methylation of the interferon gamma (IFN-gamma) and interleukin 3 (IL-3) genes in newly activated primary CD8+ T lymphocytes: regional IFN-gamma promoter demethylation and mRNA expression are heritable in CD44(high)CD8+ T cells. J Exp Med. 1998;188:103–117. doi: 10.1084/jem.188.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chang JT, V, Palanivel R, Kinjyo I, Schambach F, Intlekofer AM, Banerjee A, Longworth SA, Vinup KE, Mrass P, Oliaro J, Killeen N, Orange JS, Russell SM, Weninger W, Reiner SL. Asymmetric T lymphocyte division in the initiation of adaptive immune responses. Science (New York, N Y) 2007;315:1687–1691. doi: 10.1126/science.1139393. [DOI] [PubMed] [Google Scholar]
- 8.Feinerman O, Veiga J, Dorfman JR, Germain RN, Altan-Bonnet G. Variability and robustness in T cell activation from regulated heterogeneity in protein levels. Science (New York, N Y) 2008;321:1081–1084. doi: 10.1126/science.1158013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bird JJ, Brown DR, Mullen AC, Moskowitz NH, Mahowald MA, Sider JR, Gajewski TF, Wang CR, Reiner SL. Helper T cell differentiation is controlled by the cell cycle. Immunity. 1998;9:229–237. doi: 10.1016/s1074-7613(00)80605-6. [DOI] [PubMed] [Google Scholar]
- 10.Deenick EK, Hasbold J, Hodgkin PD. Switching to IgG3, IgG2b, and IgA is division linked and independent, revealing a stochastic framework for describing differentiation. J Immunol. 1999;163:4707–4714. [PubMed] [Google Scholar]
- 11.Gett AV, Hodgkin PD. Cell division regulates the T cell cytokine repertoire, revealing a mechanism underlying immune class regulation. Proc Natl Acad Sci U S A. 1998;95:9488–9493. doi: 10.1073/pnas.95.16.9488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gudmundsdottir H, Wells AD, Turka LA. Dynamics and requirements of T cell clonal expansion in vivo at the single-cell level: effector function is linked to proliferative capacity. J Immunol. 1999;162:5212–5223. [PubMed] [Google Scholar]
- 13.Hasbold J, Hong JS, Kehry MR, Hodgkin PD. Integrating signals from IFN-gamma and IL-4 by B cells: positive and negative effects on CD40 ligand-induced proliferation, survival, and division-linked isotype switching to IgG1, IgE, and IgG2a. J Immunol. 1999;163:4175–4181. [PubMed] [Google Scholar]
- 14.Hasbold J, Lyons AB, Kehry MR, Hodgkin PD. Cell division number regulates IgG1 and IgE switching of B cells following stimulation by CD40 ligand and IL-4. Eur J Immunol. 1998;28:1040–1051. doi: 10.1002/(SICI)1521-4141(199803)28:03<1040::AID-IMMU1040>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 15.Hodgkin PD, Lee JH, Lyons AB. B cell differentiation and isotype switching is related to division cycle number. J Exp Med. 1996;184:277–281. doi: 10.1084/jem.184.1.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Murali-Krishna K, Ahmed R. Cutting edge: naïve T cells masquerading as memory cells. J Immunol. 2000;165:1733–1737. doi: 10.4049/jimmunol.165.4.1733. [DOI] [PubMed] [Google Scholar]
- 17.Tangye SG, Ferguson A, Avery DT, Ma CS, Hodgkin PD. Isotype switching by human B cells is division-associated and regulated by cytokines. J Immunol. 2002;169:4298–4306. doi: 10.4049/jimmunol.169.8.4298. [DOI] [PubMed] [Google Scholar]
- 18.Hasbold J, Corcoran LM, Tarlinton DM, Tangye SG, Hodgkin PD. Evidence from the generation of immunoglobulin G-secreting cells that stochastic mechanisms regulate lymphocyte differentiation. Nat Immunol. 2004;5:55–63. doi: 10.1038/ni1016. [DOI] [PubMed] [Google Scholar]
- 19.Ma CS, Hodgkin PD, Tangye SG. Automatic generation of lymphocyte heterogeneity: Division-dependent changes in the expression of CD27, CCR7 and CD45 by activated human naïve CD4+ T cells are independently regulated. Immunol Cell Biol. 2004;82:67–74. doi: 10.1046/j.0818-9641.2003.01206.x. [DOI] [PubMed] [Google Scholar]
- 20.Gett AV, Hodgkin PD. A cellular calculus for signal integration by T cells. Nat Immunol. 2000;1:239–244. doi: 10.1038/79782. [DOI] [PubMed] [Google Scholar]
- 21.Hawkins ED, Hommel M, Turner ML, Battye FL, Markham JF, Hodgkin PD. Measuring lymphocyte proliferation, survival and differentiation using CFSE time-series data. NatProtoc. 2007;2:2057–2067. doi: 10.1038/nprot.2007.297. [DOI] [PubMed] [Google Scholar]
- 22.Kearney ER, Pape KA, Loh DY, Jenkins MK. Visualization of peptide-specific T cell immunity and peripheral tolerance induction in vivo. Immunity. 1994;1:327–339. doi: 10.1016/1074-7613(94)90084-1. [DOI] [PubMed] [Google Scholar]
- 23.Sallusto F, Geginat J, Lanzavecchia A. Central memory and effector memory T cell subsets: function, generation, and maintenance. Annu Rev Immunol. 2004;22:745–763. doi: 10.1146/annurev.immunol.22.012703.104702. [DOI] [PubMed] [Google Scholar]
- 24.Sallusto F, Lenig D, Forster R, Lipp M, Lanzavecchia A. Two subsets of memory T lymphocytes with distinct homing potentials and effector functions. Nature. 1999;401:708–712. doi: 10.1038/44385. [DOI] [PubMed] [Google Scholar]
- 25.Wherry EJ, Teichgraber V, Becker TC, Masopust D, Kaech SM, Antia R, von Andrian UH, Ahmed R. Lineage relationship and protective immunity of memory CD8 T cell subsets. Nat Immunol. 2003;4:225–234. doi: 10.1038/ni889. [DOI] [PubMed] [Google Scholar]
- 26.Fearon DT, Manders P, Wagner SD. Arrested differentiation, the self-renewing memory lymphocyte, and vaccination. Science (New York, N Y) 2001;293:248–250. doi: 10.1126/science.1062589. [DOI] [PubMed] [Google Scholar]
- 27.Lefrancois L, Marzo AL. The descent of memory T-cell subsets. Nat Rev Immunol. 2006;6:618–623. doi: 10.1038/nri1866. [DOI] [PubMed] [Google Scholar]
- 28.Kedzierska K, Venturi V, Field K, Davenport MP, Turner SJ, Doherty PC. Early establishment of diverse T cell receptor profiles for influenza-specific CD8+CD62Lhi memory T cells. Proc Natl Acad Sci U S A. 2006;103:9184–9189. doi: 10.1073/pnas.0603289103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stemberger C, Huster KM, Koffler M, Anderl F, Schiemann M, Wagner H, Busch DH. A Single Naïve CD8(+) T Cell Precursor Can Develop into Diverse Effector and Memory Subsets. Immunity. 2007;27:985–997. doi: 10.1016/j.immuni.2007.10.012. [DOI] [PubMed] [Google Scholar]
- 30.Badovinac VP, Haring JS, Harty JT. Initial T cell receptor transgenic cell precursor frequency dictates critical aspects of the CD8(+) T cell response to infection. Immunity. 2007;26:827–841. doi: 10.1016/j.immuni.2007.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kemp RA, Powell TJ, Dwyer DW, Dutton RW. Cutting edge: regulation of CD8+ T cell effector population size. J Immunol. 2004;173:2923–2927. doi: 10.4049/jimmunol.173.5.2923. [DOI] [PubMed] [Google Scholar]
- 32.Marzo AL, Klonowski KD, Le Bon A, Borrow P, Tough DF, Lefrancois L. Initial T cell frequency dictates memory CD8+ T cell lineage commitment. Nat Immunol. 2005;6:793–799. doi: 10.1038/ni1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stock AT, Mueller SN, Kleinert LM, Heath WR, Carbone FR, Jones CM. Optimization of TCR transgenic T cells for in vivo tracking of immune responses. Immunol Cell Biol. 2007;85:394–396. doi: 10.1038/sj.icb.7100076. [DOI] [PubMed] [Google Scholar]
- 34.Schlub TE, Venturi V, Kedzierska K, Wellard C, Doherty PC, Turner SJ, Ribeiro RM, Hodgkin PD, Davenport MP. Division-linked differentiation can account for CD8(+) T-cell phenotype in vivo. EurJ Immunol. 2009;39:67–77. doi: 10.1002/eji.200838554. [DOI] [PubMed] [Google Scholar]
- 35.Baron V, Bouneaud C, Cumano A, Lim A, Arstila TP, Kourilsky P, Ferradini L, Pannetier C. The repertoires of circulating human CD8(+) central and effector memory T cell subsets are largely distinct. Immunity. 2003;18:193–204. doi: 10.1016/s1074-7613(03)00020-7. [DOI] [PubMed] [Google Scholar]
- 36.Hawkins ED, Turner ML, Dowling MR, van Gend C, Hodgkin PD. A model of immune regulation as a consequence of randomized lymphocyte division and death times. Proc Natl Acad Sci U S A. 2007;104:5032–5037. doi: 10.1073/pnas.0700026104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Badovinac VP, Messingham KA, Jabbari A, Haring JS, Harty JT. Accelerated CD8+ T-cell memory and prime-boost response after dendritic-cell vaccination. Nat Med. 2005;11:748–756. doi: 10.1038/nm1257. [DOI] [PubMed] [Google Scholar]
- 38.Tangye SG, Hodgkin PD. Divide and conquer: the importance of cell division in regulating B-cell responses. Immunology. 2004;112:509–520. doi: 10.1111/j.1365-2567.2004.01950.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Darrah PA, Patel DT, De Luca PM, Lindsay RW, Davey DF, Flynn BJ, Hoff ST, Andersen P, Reed SG, Morris SL, Roederer M, Seder RA. Multifunctional TH1 cells define a correlate of vaccine-mediated protection against Leishmania major. Nat Med. 2007;13:843–850. doi: 10.1038/nm1592. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.