Abstract
Investigation of integron carriage in a global collection of multi-drug resistant Salmonella enterica identified 3 unique class 1 integron gene cassette arrays not previously reported in this species. The present study used PCR and DNA sequence analysis to characterize the structure of these gene cassette arrays. A ~4.0 kb integron containing the gene cassette array arr2/cmlA5/blaOXA10/ aadA1 was found in isolates belonging to serovars Isangi and Typhimurium from South Africa. A ~6.0 kb integron containing the gene cassettes aac(6′)IIc/ereA2/IS1247/aac/ arr/ereA2 was found in isolates belonging to serovar Heidelberg from the Philippines. In this gene cassette array, the insertion sequence, IS1247, and two putative resistance genes, disrupt the erythromycin resistance gene cassette. Finally, a ~6.0 kb integron containing the gene cassette qacH/dfrA32/ereA1/aadA2/cmlA/aadA1 was found in serovar Stanley isolates from Taiwan. This integron, which has not been previously reported in any bacterial species, contains a new dihydrofolate reductase gene cassette sequence designated dfrA32, with only 90% sequence similarity to previously reported dfrA cassettes. The S. enterica integrons described in the present study represent novel collections of resistance genes which confer multi-drug resistance and have the potential to be widely disseminated among S. enterica as well as other bacterial species.
Introduction
Salmonella enterica is a leading cause of bacterial gastro-enteritis in humans worldwide [33]. While most S. enterica infections are self-limiting, serious cases often require treatment with antibiotics. For several decades, an increase in antibiotic resistance has been noted in S. enterica, as in other gram-negative bacteria [6]. Class 1 integrons contribute significantly to antibiotic resistance in gram-negative organisms [28]. Integrons are genetic structures capable of capturing and integrating gene cassettes that typically encode antibiotic resistance determinants; they therefore have the ability to confer novel combinations of drug resistance to the bacteria in which they reside. Integrons are frequently associated with plasmids and are therefore easily transferable among and between different bacterial species [5]. Due to their mobility and ability to quickly acquire diverse resistance determinants, integrons are uniquely adapted to transfer and disseminate antibiotic resistance.
Commonly reported integrons in S. enterica are small, usually containing 1 to 3 antibiotic resistance cassettes [16, 18, 25]. However, integrons containing larger numbers of cassettes have been reported in other bacteria [9, 19]. In a previous study, we found that the majority of integrons in a global collection of S. enterica contained 1 to 3 gene cassettes [13]. The present study describes the gene cassette arrays of 3 large, novel integrons identified from the same global collection of multi-drug resistant S. enterica.
Materials and Methods
Bacterial Isolates
Salmonella enterica isolates were obtained from laboratories in South Africa, Taiwan, and the Philippines between 2001 and 2002 as part of a large global investigation of multi-drug resistant S. enterica [13]. Other than country of origin, no epidemiological information is available for these isolates. Representative isolates from this collection that harbored integron gene cassette arrays ≥4 kb were selected for further characterization (Table 1).
Table 1.
Origin, novel integron sequences, and resistance patterns of tested S. enterica isolates and associated plasmids
| Isolate | Serovar | Geographic origin | Integron | Resistance pattern |
|---|---|---|---|---|
| Strains | ||||
| SAL01259 | Isangi | South Africa | arr2/cmlA5/blaOXA10/aadA1 | AmpChlStrTetSamCazNalGenAtmSxtRif |
| SAL01261 | ||||
| SAL01274 | ||||
| SAL02433 | Typhimurium | South Africa | arr2/cmlA5/blaOXA10/aadA1 | AmpChlStrTetSamCazGenAtmSxtRif |
| SAL02519 | Heidelberg | Philippines | aac(6′)IIc/ereA2/IS1247/aac/arr2/ereA2 | AmpChlStrTetSamNalGenAtmSxtRif |
| SAL02520 | ||||
| SAL02521 | ||||
| SAL02530 | Stanley | Taiwan | qacH/dfrA32/ereA1/aadA2/cmlA/aadA1 | AmpChlStrTetSamSxtEry |
| SAL02533 | ||||
| SAL02536 | ||||
| Plasmids | ||||
| pSAL01259 | arr2/cmlA5/blaOXA10/aadA1 | AmpChlStrTetSamCazGenAtmSxtRif | ||
| pSAL01261 | arr2/cmlA5/blaOXA10/aadA1 | AmpChlStrTetSamCazGenAtmSxtRif | ||
| pSAL01274 | arr2/cmlA5/blaOXA10/aadA1 | AmpChlStrTetSamCazGenAtmSxtRif | ||
| pSAL02433 | arr2/cmlA5/blaOXA10/aadA1 | AmpChlStrTetSamCazGenAtmSxtRif | ||
Amp ampicillin, Chl chloramphenicol, Str streptomycin, Tet tetracycline, Sam ampicillin-sulbactam; Caz ceftazidime, Nal nalidixic acid, Gen gentamicin, Atm aztreonam, Sxt trimethoprim-sulfamethoxazole, Rif rifampin, Ery erythromycin
Gene Cassette Identification
DNA preparation and PCR amplification of integron gene cassette arrays were accomplished as previously described [13]. Primers specific for integron 5′ and 3′ conserved segments (CS) [15] were used for initial sequencing followed by additional sequencing with previously described primers for each of the integrons: arr2/cmlA5/blaOXA10/aadA1; arr2_R, cmlA5_F, cmlA_F, cmlA_R, oxa10_R, aadA1_R: aac(6′) IIc/ereA2/IS1247/aac/arr2/ereA2; aac(6′)IIc-F, ere_est_R, IS_F, TNP_F, arr_accA_R, arr_R2, ere_F2: qacH/dfrA32/ ereA1/aadA2/cmlA/aadA1; qacH-F, dfr17_F, ere2-R, aadA2_R, cmlA_R_internal, cml_R2, aadA1_R_S [13]. Sequencing was performed using the BigDye terminator 3.1 kit (Applied Biosystems) according to the manufacturer’s instructions. Capillary sequence analysis of sequencing products was performed on a 3730 DNA sequence analyzer (Applied Biosystems). Sequence analysis utilized the Lasergene 7.0.0 software package (DNAStar, Madison, WI). BLAST analysis was performed on resulting sequences to determine gene cassette homologies (http://www.ncbi.nlm.nih.gov/BLAST) [1].
To investigate the divergence of the dihydrofolate reductase encoded by the newly identified dfrA32 gene cassette, predicted amino acid sequences of 21 dfrA genes (Table 2) were obtained from GenBank (www.ncbi.nlm.nih.gov) [3] and were aligned using ClustalW2 (http://www.ebi.ac.uk/Tools/clustalw2) [17]. This alignment was used to create a rooted neighbor-joining tree by the method of Saitou and Nei [27], also using ClustalW2. The neighbor-joining tree was displayed using PhyloDraw (http://pearl.cs.pusan.ac.kr/phylodraw) [8]. ClustalW2 was also used to create an alignment of the predicted amino acid sequences of dfrA32 and its closest homolog, dfrA17 (GenBank Accession no. FJ895301), and to perform an alignment of the dfrA32 and dfrA7 attC sites.
Table 2.
GenBank accession numbers of dfrA genes used for alignment with dfrA32, obtained from S. enterica serovar Stanley integron gene cassette array
| Cassette | Accession number | Cassette | Accession number | Cassette | Accession number |
|---|---|---|---|---|---|
| dfrA1 | AY963803 | dfrA14 | NC_010886 | dfrA22 | FM957884 |
| dfrA2 | NZ_ACKQ01000046 | dfrA15 | FJ183470 | dfrA23 | AJ968952 |
| dfrA5 | FJ591050 | dfrA16 | AF174129 | dfrA24 | AJ972619 |
| dfrA7 | FJ854362 | dfrA17 | AF169041 | dfrA25 | FN252408 |
| dfrA10 | AY123253 | dfrA19 | NC_012556 | dfrA26 | AM403715 |
| dfrA12 | FJ950723 | dfrA20 | AJ605332 | dfrA27 | FJ976724 |
| dfrA13 | NZ_ACKX01000031 | dfrA21 | AJ870926 | dfrA28 | FN263373 |
Plasmid Analysis
Plasmids were isolated from S. enterica isolates using the Qiagen Midi Kit (Qiagen, Maryland) according to the manufacturer’s directions. Electroporation of plasmids into electrocompetent Escherichia coli Top 10 cells (Invitrogen, Carlsbad, CA) was performed using a MicroPulser (Bio-Rad, Hercules, CA). E. coli transformants were selected on Luria–Bertani agar supplemented with the appropriate antibiotic (150 μg/ml erythromycin, 50 μg/ml chloramphenicol, or 50 μg/ml rifampin). Antibiotic resistance of S. enterica isolates and E. coli transformants was determined using the disc diffusion method (Becton, Dickinson and Co, Sparks, MD) on Mueller–Hinton agar according to the guidelines of the Clinical Laboratory Standards Institute [20]. Antibiotics tested were: ampicillin (10 μg), tetracycline (30 μg), chloramphenicol (30 μg), streptomycin (10 μg), ampicillin/sulbactam (10 μg/10 μg), ceftazidime (30 μg), nalidixic acid (30 μg), gentamicin (10 μg), aztreonam (30 μg), trimethoprim-sulfamethoxazole (1.25 μg/ 23.75 μg), rifampin (5 μg), and erythromycin (15 μg). Escherichia coli ATCC25922 was included as a control in all antibiotic susceptibility testing. The plasmid preparations from E. coli transformants were PCR amplified with 5′ and 3′ CS primers to identify gene cassette arrays.
For isolates which did not generate transformants, plasmid carriage was further investigated by performing restriction endonuclease digestion on the purified plasmid preparations with 20 units of HindIII in a 10 μl volume containing 1× NEB2 buffer (New England Biolabs, Ips-wich, MA) and 0.1 μg BSA. The digestion mixture was incubated at 37°C for 2 h and subjected to electrophoresis on a 0.7% gel and stained with ethidium bromide to visualize plasmid bands.
Nucleotide Sequence Accession Numbers
The nucleotide sequences of integrons arr2/cmlA5/ blaOXA10/aadA1, aac(6′)IIc/ereA2/IS1247/aac3/arr/ereA2, and qacH/dfrA32/ereA1/aadA2/cmlA/aadA1 have been deposited in the GenBank/EMBL/DDBJ under accession nos. GU067640, GU067641, and GU067642, respectively.
Results
Antibiotic Resistance
All S. enterica isolates harboring integron cassette arrays ≥4.0 kb exhibited resistance to at least 7 antimicrobials, including ampicillin, ampicillin/sulbactam, tetracycline, chloramphenicol, streptomycin, and sulfamethoxazole. Isolates of identical serovar displayed similar resistance patterns (Table 1).
Integron Characterization
The Isangi isolates and the Typhimurium isolate from South Africa contained a gene cassette array comprised of the cassettes arr2/cmlA5/blaOXA10/aadA1. The arr2 gene cassette, which was originally described in Pseudomonas aeruginosa, encodes an ADP-ribosyl transferase and confers resistance to rifampin [29]. The cmlA5 cassette encodes a chloramphenicol efflux protein and contains its own promoter [19]. The blaOXA10 gene cassette encodes resistance to the β-lactam class of antibiotics. In this cassette array, the blaOXA10 cassette was contiguous with the aadA1 cassette due to the lack of a majority of the attC site, which usually terminates an integron gene cassette [19]. Truncations of the attC site have been reported [23]. The aadA1 gene cassette confers resistance to aminoglycoside antibiotics. All of the isolates carrying the arr2/cmlA5/ blaOXA10/aadA1 gene cassette array were resistant to rifampin, chloramphenicol, ampicillin, and streptomycin, as expected if all cassettes in this integron were expressed, and were additionally resistant to tetracycline, ceftazidime, gentamicin, aztreonam, and trimethoprim-sulfamethoxazole. Serovar Isangi isolates were also resistant to nalidixic acid.
Plasmids were prepared from the serovar Isangi isolate Sal01259 and the serovar Typhimurium isolate Sal02433 and these plasmids were transferred by electroporation to E. coli. The E. coli transformants exhibited resistance patterns similar to those of the S. enterica isolates (Table 1). PCR amplification of plasmids and of plasmid transformed E. coli cell lysates with 5′ and 3′ CS primers generated a product identical in size to that obtained from the original isolates. Plasmids isolated from Isangi Sal01261 and Sal01274 gave similar transfer of resistance and PCR amplification of gene cassette arrays using the 5′ and 3′ CS primers (Table 1).
These data suggest that the rifampin, sulfamethoxazole, chloramphenicol, and ampicillin resistances were transferred to E. coli on a plasmid bearing the integron gene cassette array arr2/cmlA5/blaOXA10/aadA1. Other plasmid located resistance determinants not part of the integron may be responsible for the transfer of additional resistances. Nalidixic acid resistance was not transferred from serovar Isangi isolates to E. coli; this resistance is most likely due to mutation in the chromosomally encoded topoisomerase [26].
The Heidelberg isolates contained the ~6 kb integron gene cassette array aac(6′)IIc/ereA2/IS1247/aac3/arr/ ereA2. The aac(6′)IIc gene cassette encodes a 6′-N-aminoglycoside acetyltransferase [10]. The ereA2 gene cassette, which normally confers resistance to erythromycin [4, 22], is interrupted at nucleotide position 188 by IS1247 [30] and the putative gene cassettes aac3 and arr. The aac3 and arr genes are not technically integron gene cassettes, as they both lack an attC site. The aac3 gene shows 79.8% sequence similarity to the N-acetyltransferase, aac3-Vb from Serratia marcescens [24, 31]. The arr gene displays 74% sequence identity to a rifampin ADP-ribosyl transferase from a Rhodopseudomonas palustris gene [14, 31]. The 3′ end of the ereA2 cassette, beginning at nucleotide 189, resides downstream of the arr gene.
Serovar Heidelberg isolates harboring the aac(6′)IIc/ereA2/IS1247/aac3/arr/ereA2 integron gene cassette array were resistant to rifampin, nalidixic acid, gentamicin, aztreonam, and trimethoprim-sulfamethoxazole as well as ampicillin, ampicillin/sulbactam, tetracycline, chloramphenicol, and streptomycin.
Repeated attempts to purify plasmids from Heidelberg isolates Sal02519, Sal02520, and Sal02521 failed to yield E. coli transformants on appropriate selection. Plasmids were not evident in these isolates when assessed by restriction endonuclease digestion with HindIII.
The 3 representative serovar Stanley isolates contained a gene cassette array that has not been previously described. This novel ~6 kb array contains the gene cassettes qacH/dfrA32/ereA1/aadA2/cmlA/aadA1. The Stanley isolates containing this gene cassette array were resistant to erythromycin, streptomycin, chloramphenicol, and trimethoprim/sulfamethoxazole, consistent with the resistance cassettes carried on the integron. The qacH cassette encodes an efflux pump that confers resistance to quaternary ammonium compounds and ethidium bromide [19]. Resistance for these compounds was not tested in this study.
The second gene cassette in this array exhibited only 90% sequence identity to its nearest match, dfrA17, a dihydrofolate reductase integron gene cassette, found in Shigella flexneri (Genbank accession number FJ895301, unpublished). A number of dihydrofolate reductase gene cassette variants that confer trimethoprim resistance have been described [32]. The dfrA gene cassette identified in the Stanley gene cassette array, encodes a predicted 157 amino acid protein variant which differs from dfrA17 FJ895301 at 11 amino acid residues (Fig. 1). This cassette has been named dfrA32 following the suggested naming guidelines [21]. Since the majority of amino acid changes are conservative, the trimethoprim resistance observed in all of the Stanley isolates is predicted to be due to expression of the dfrA32 cassette. A neighbor-joining tree of the predicted amino acid sequences of 21 dihydrofolate reductase genes present in GenBank, along with dfrA32, indicates that the DHFR protein product of dfrA32 is most similar to those of dfrA17 and dfrA7 (Fig. 2). The attC site of dfrA32 is most similar to that of dfrA7 (Fig. 3).
Fig. 1.
Comparison of predicted amino acid sequence of dfrA32 gene cassette obtained from S. enterica serovar Stanley gene cassette array qacH/dfrA32/ereA1/aadA2/cmlA/aadA1 to predicted amino acid sequence of closest match, dfrA17 (GenBank accession number AF169041). Asterisks indicate amino acid substitutions
Fig. 2.
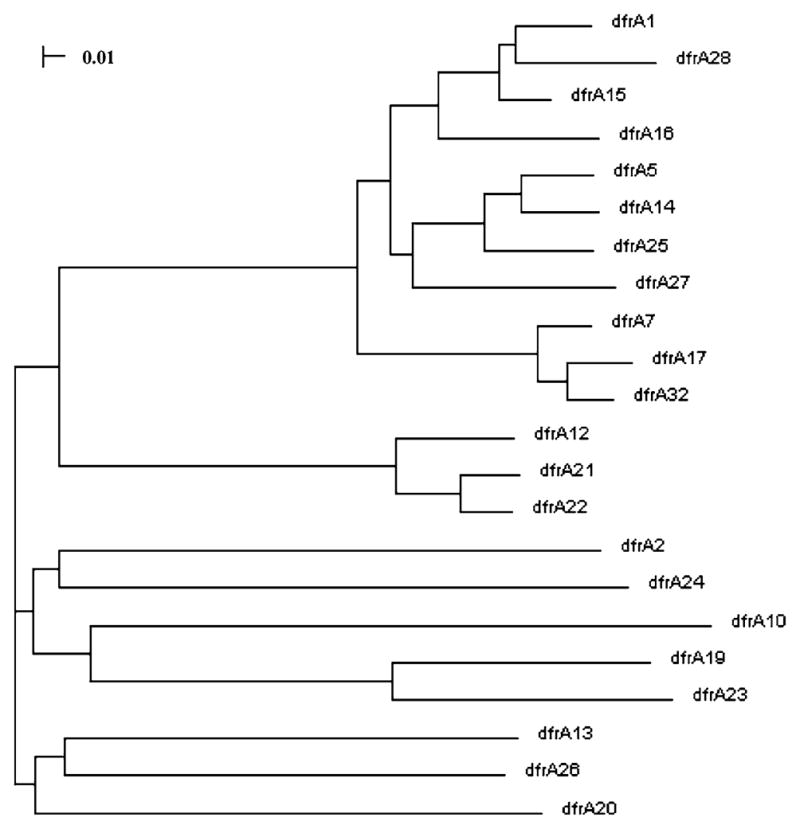
Neighbor-joining tree of aligned predicted amino acid sequences of 22 dfrA genes, showing relationship of predicted S. enterica serovar Stanley gene cassette dfrA32 product to previously described DfrA proteins
Fig. 3.
Alignment of attC sites of dfrA7 (GenBank accession number FJ854362) and dfrA32 gene cassette (obtained from S. enterica serovar Stanley isolates from Taiwan). Asterisks indicate nucleotide substitutions
The ereA1 gene cassette encodes a type 1 erythromycin esterase, which has been reported in E. coli [4]. The gene cassette series aadA2/cmlA/aadA1 has been reported in E. coli (GenBank Accession no EF113389) and S. enterica serovar Typhimurium [2]. Both aadA2 and aadA1 confer resistance to aminoglycosides while cmlA confers chloramphenicol resistance.
While a plasmid band was detected in the serovar Stanley isolates by restriction endonuclease digestion, plasmid preparations from these isolates did not amplify a 6 kb gene cassette array product nor were repeated attempts at plasmid transformations of E. coli successful, suggesting that the integron may not be located on a plasmid.
Discussion
The present study describes several S. enterica integron gene cassette arrays not previously identified in that bacterium. These cassette arrays are interesting not only for their size and content but also for their possible origins and their implications for spread of multiple antibiotic resistance.
The integron gene cassette array arr2/cmlA5/blaOXA10/ aadA1 identified in Isangi and Typhimurium S. enterica isolates contains a series of gene cassettes identical to those in an integron found on a plasmid in P. aeruginosa in China (unpublished, GenBank accession number EU886979) and as a segment of a previously reported larger, complex class 1 integron in Acinetobacter baumanii strain AYE from France [9]. This sequence of gene cassettes has not been reported in S. enterica. As was previously reported [13], the serovar Isangi and Typhimurium isolates differ in genetic lineage as defined by MLST as well as by serovar. The successful transfer of this integron gene cassette array to E. coli by electroporation provides evidence that the array is mobile and therefore may have been horizontally transferred between the 2 different genetic lineages of S. enterica or transferred to the Isangi and Typhimurium serovars from another common source.
The aac(6′)IIc/ereA2/IS1247/aac/arr/ereA2 integron contains an insertion sequence interrupting the gene cassette ereA2, which normally confers erythromycin resistance. A potential ancestor of this integron may be the integron aac(6′)IIc/ereA2, found on a plasmid in the newly emerged S. enterica serovar Keurmassar from Senegal in 2000 [11]. The aac(6′)IIc/ereA2/IS1247/aac/arr/ereA2 integron has previously been reported in a Klebsiella oxytoca isolate from a single patient in Paris [31] and on an Enterobacter cloacae plasmid [7]. Although plasmid localization was not demonstrated for the K. oxytoca integron, the existence of this identical and unusual integron in these diverse bacteria isolated in different geographic locations suggests that this unique structure can be mobilized across bacterial species.
To our knowledge, the qacH/dfrA32/ereA1/aadA2/cmlA/ aadA1 integron found in serovar Stanley isolates has not been reported in any bacterial species. The 3′ end of this S. enterica integron resembles a plasmid-borne 3.2 kb aadA2/cmlA/aadA1 integron reported in E. coli [19]. Since integrons are believed to preferentially add cassettes at the first position [12], this E. coli integron could be a progenitor of the S. enterica serovar Stanley integron described here. The dfrA32 cassette in this integron is novel in that it demonstrates only 90% sequence similarity to any reported dihydrofolate reductase genes. While the exact origin of the dfrA32 cassette is unknown, its similarity to dfrA17 and dfrA7, which have been found in E. coli, Shigella, and Salmonella suggest that this unique cassette may have evolved from these potential early progenitors. Since E. coli, S. flexneri, and S. enterica share environmental niches, it is conceivable that dfrA32 arose through lateral gene transfer and mutation.
The integron gene cassette arrays described in this report have not been previously observed in Salmonella. These findings are unique and have important implications for development of multidrug resistance in S. enterica. The integron’s capacity for creating and disseminating complex collections of resistance elements and the importance of horizontal gene transfer in the expansion of antibiotic resistance are reinforced by these data. Integrons and the gene cassettes they contain exist as a common pool which has the capacity to be exchanged by many bacterial species. Multiple drug resistance determinants in one genetic structure exacerbate the problem of antibiotic resistance as selection for any gene cassette within an integron selects for all of the cassettes. The present study further emphasizes the vital role integrons play in the spread of antibiotic resistance throughout the microbial community.
Acknowledgments
We thank Stanley M. Reynolds and Nkuchia M. M’ikanatha (Pennsylvania Department of Health) for assistance with serotyping and Tersia Kruger and Karen H. Keddy (Center for Enteric Diseases, Johannesburg, South Africa), Wen Chien Ko (National Cheng Kung Hospital, Tainan, Taiwan), and Celia C. Carlos (Research Institute for Tropical Medicine, Manila, Philippines) for providing isolates. This study was supported by NIAID R56 AI059385. The isolate collection was assembled with the support of CDC grant R01-C1322404.
Contributor Information
Mary Krauland, Infectious Diseases Epidemiology Research Unit, University of Pittsburgh, Pittsburgh, USA.
Lee Harrison, Infectious Diseases Epidemiology Research Unit, University of Pittsburgh, Pittsburgh, USA.
David Paterson, UQ Centre for Clinical Research, University of Queensland, Brisbane, Australia.
Jane Marsh, Email: jwmarsh@pitt.edu, Infectious Diseases Epidemiology Research Unit, University of Pittsburgh, Pittsburgh, USA.
References
- 1.Altschul SF, Gish W, et al. Basic local alignment search tool. J Mol Biol. 1990;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Antunes P, Machado J, et al. Dissemination of sul3-containing elements linked to class 1 integrons with an unusual 3′ conserved sequence region among Salmonella isolates. Antimicrob Agents Chemother. 2007;51(4):1545–1548. doi: 10.1128/AAC.01275-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Benson DA, Karsch-Mizrachi I, et al. GenBank. Nucleic Acids Res. 2008;36(Database issue):D25–D30. doi: 10.1093/nar/gkm929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Biskri L, Mazel D. Erythromycin esterase gene ere(A) is located in a functional gene cassette in an unusual class 2 integron. Antimicrob Agents Chemother. 2003;47(10):3326–3331. doi: 10.1128/AAC.47.10.3326-3331.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carattoli A. Plasmid-mediated antimicrobial resistance in Salmonella enterica. Curr Issues Mol Biol. 2003;5(4):113–122. [PubMed] [Google Scholar]
- 6.Centers for Disease Control and Prevention. National Antimicrobial Resistance Monitoring System for Enteric Bacteria (NARMS): Human Isolates Final Report, 2004. U.S. Department of Health and Human Services, CDC; Atlanta, Georgia: 2007. [Google Scholar]
- 7.Chen YT, Liao TL, et al. Mobilization of qnrB2 and ISCR1 in plasmids. Antimicrob Agents Chemother. 2009;53(3):1235–1237. doi: 10.1128/AAC.00970-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Choi JH, Jung HY, et al. PhyloDraw: a phylogenetic tree drawing system. Bioinformatics. 2000;16(11):1056–1058. doi: 10.1093/bioinformatics/16.11.1056. [DOI] [PubMed] [Google Scholar]
- 9.Fournier PE, Vallenet D, et al. Comparative genomics of multidrug resistance in Acinetobacter baumannii. PLoS Genet. 2006;2(1):e7. doi: 10.1371/journal.pgen.0020007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Galani I, Souli M, et al. Characterization of a new integron containing bla(VIM-1) and aac(6′)-IIc in an Enterobacter cloacae clinical isolate from Greece. J Antimicrob Chemother. 2005;55(5):634–638. doi: 10.1093/jac/dki073. [DOI] [PubMed] [Google Scholar]
- 11.Gassama-Sow A, Aidara-Kane A, et al. Integrons in Salmonella Keurmassar, Senegal. Emerg Infect Dis. 2004;10(7):1339–1341. doi: 10.3201/eid1007.030666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hall RM, Collis CM. Mobile gene cassettes and integrons: capture and spread of genes by site-specific recombination. Mol Microbiol. 1995;15(4):593–600. doi: 10.1111/j.1365-2958.1995.tb02368.x. [DOI] [PubMed] [Google Scholar]
- 13.Krauland MG, Marsh JW, et al. Integron-mediated multidrug resistance in a global collection of nontyphoidal Salmonella enterica isolates. Emerg Infect Dis. 2009;15(3):388–396. doi: 10.3201/eid1503.081131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Larimer FW, Chain P, et al. Complete genome sequence of the metabolically versatile photosynthetic bacterium Rhodo-pseudomonas palustris. Nat Biotechnol. 2004;22(1):55–61. doi: 10.1038/nbt923. [DOI] [PubMed] [Google Scholar]
- 15.Levesque C, Piche L, et al. PCR mapping of integrons reveals several novel combinations of resistance genes. Antimicrob Agents Chemother. 1995;39(1):185–191. doi: 10.1128/aac.39.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martinez N, Mendoza MC, et al. Detailed structure of integrons and transposons carried by large conjugative plasmids responsible for multidrug resistance in diverse genomic types of Salmonella enterica serovar Brandenburg. J Antimicrob Chemother. 2007;60(6):1227–1234. doi: 10.1093/jac/dkm336. [DOI] [PubMed] [Google Scholar]
- 17.Larkin MA, Blackshields G, et al. ClustalW and ClustalX version 2. Bioinformatics. 2007;23(21):2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 18.Murphy BP, O’Mahony R, et al. Investigation of a global collection of nontyphoidal Salmonella of various serotypes cultured between 1953 and 2004 for the presence of class 1 integrons. FEMS Microbiol Lett. 2007;266(2):170–176. doi: 10.1111/j.1574-6968.2006.00537.x. [DOI] [PubMed] [Google Scholar]
- 19.Naas T, Mikami Y, et al. Characterization of In53, a class 1 plasmid- and composite transposon-located integron of Escherichia coli which carries an unusual array of gene cassettes. J Bacteriol. 2001;183(1):235–249. doi: 10.1128/JB.183.1.235-249.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.National Committee for Clinical Laboratory Standards. Performance standards for antimicrobial disk susceptibility tests. 8. NCCLS; Wayne, PA: 2003. Approved standard M2–A8. [Google Scholar]
- 21.Partridge SR, Tsafnat G, et al. Gene cassettes and cassette arrays in mobile resistance integrons. FEMS Microbiol Rev. 2009;33(4):757–784. doi: 10.1111/j.1574-6976.2009.00175.x. [DOI] [PubMed] [Google Scholar]
- 22.Peters ED, Leverstein-van Hall MA, et al. Novel gene cassettes and integrons. Antimicrob Agents Chemother. 2001;45(10):2961–2964. doi: 10.1128/AAC.45.10.2961-2964.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ramirez MS, Parenteau TR, et al. Functional characterization of Tn1331 gene cassettes. J Antimicrob Chemother. 2008;62(4):669–673. doi: 10.1093/jac/dkn279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rather PN, Mierzwa R, et al. Cloning and DNA sequence analysis of an aac(3)-Vb gene from Serratia marcescens. Antimicrob Agents Chemother. 1992;36(10):2222–2227. doi: 10.1128/aac.36.10.2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rodriguez I, Martin MC, et al. Class 1 and class 2 integrons in non-prevalent serovars of Salmonella enterica: structure and association with transposons and plasmids. J Antimicrob Chemother. 2006;58(6):1124–1132. doi: 10.1093/jac/dkl400. [DOI] [PubMed] [Google Scholar]
- 26.Ruiz J. Mechanisms of resistance to quinolones: target alterations, decreased accumulation and DNA gyrase protection. J Antimicrob Chemother. 2003;51(5):1109–1117. doi: 10.1093/jac/dkg222. [DOI] [PubMed] [Google Scholar]
- 27.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4(4):406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 28.Toleman MA, Bennett PM, et al. Common regions e.g. orf513 and antibiotic resistance: IS91-like elements evolving complex class 1 integrons. J Antimicrob Chemother. 2006;58(1):1–6. doi: 10.1093/jac/dkl204. [DOI] [PubMed] [Google Scholar]
- 29.Tribuddharat C, Fennewald M. Integron-mediated rifampin resistance in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1999;43(4):960–962. doi: 10.1128/aac.43.4.960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van der Ploeg J, Willemsen M, et al. Adaptation of Xanthobacter autotrophicus GJ10 to bromoacetate due to activation and mobilization of the haloacetate dehalogenase gene by insertion element IS1247. J Bacteriol. 1995;177(5):1348–1356. doi: 10.1128/jb.177.5.1348-1356.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Verdet C, Benzerara Y, et al. Emergence of DHA-1-producing Klebsiella spp. in the Parisian region: genetic organization of the ampC and ampR genes originating from Morganella morganii. Antimicrob Agents Chemother. 2006;50(2):607–617. doi: 10.1128/AAC.50.2.607-617.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wei Q, Jiang X, et al. dfrA27, a new integron-associated trimethoprim resistance gene from Escherichia coli. J Antimicrob Chemother. 2009;63(2):405–406. doi: 10.1093/jac/dkn474. [DOI] [PubMed] [Google Scholar]
- 33.World Health Organization. Drug-resistant Salmonella Fact Sheet N 139. 2005 http://www.who.int/mediacentre/factsheets/fs139/en/




