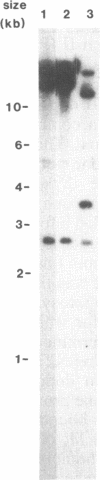
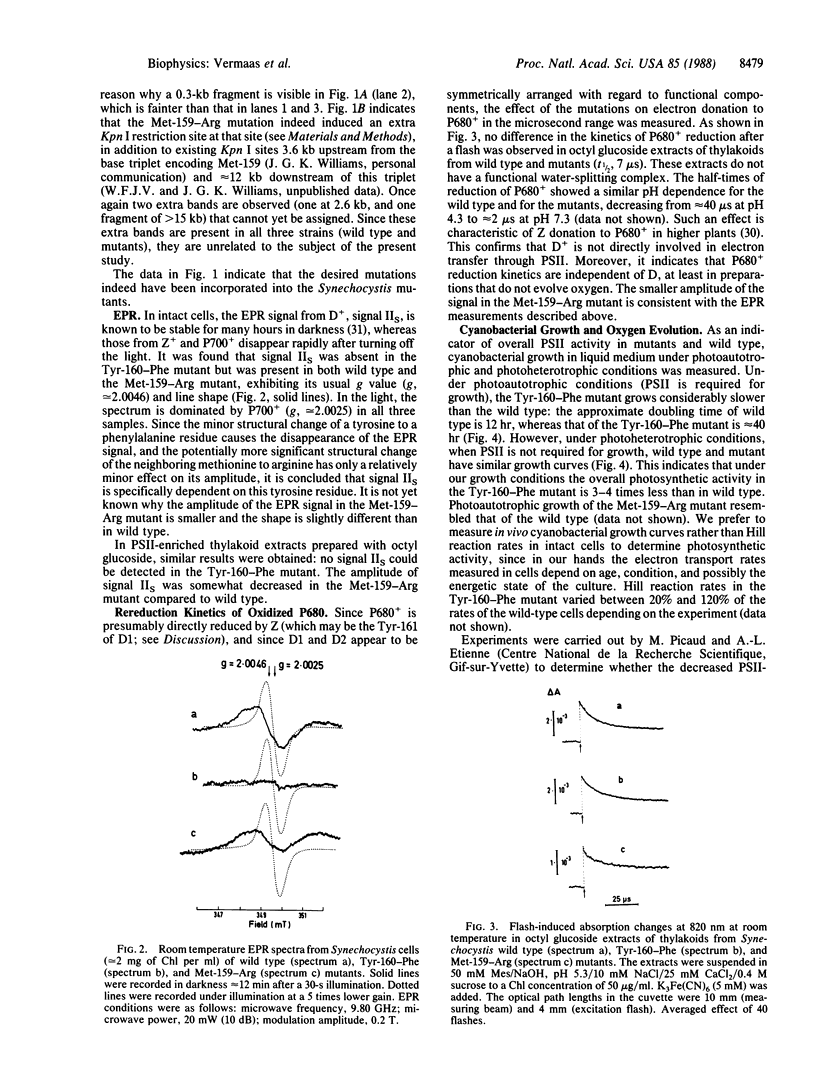
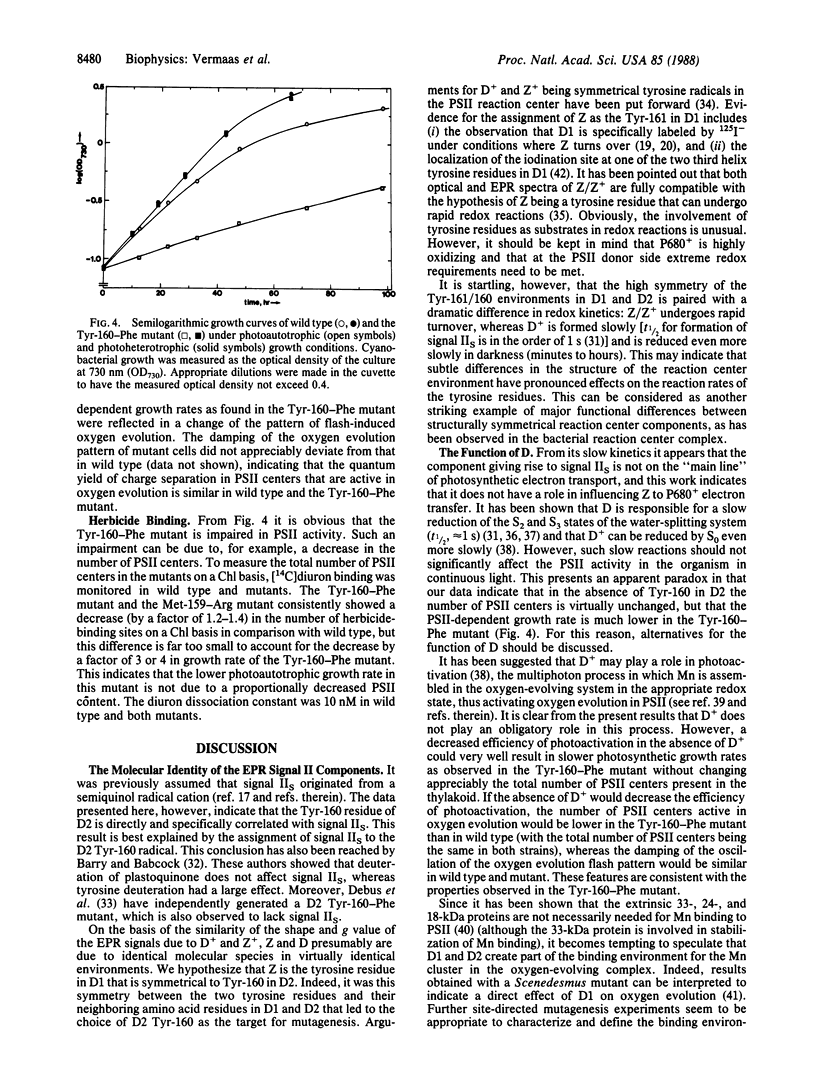
Abstract
The chemical nature of electron donor(s) in photosystem II in photosynthetic membranes was analyzed by site-directed mutagenesis of the gene encoding the protein D2 of the photosystem II reaction center. Mutation of the Tyr-160 residue of the D2 protein into phenylalanine results in the disappearance of the electron paramagnetic resonance signal IIS originating from D+, the oxidized form of the slow photosystem II electron donor D. Signal IIS is still present if a neighboring residue in D2, Met-159, is mutated into arginine. Both mutants have normal rereduction kinetics of the oxidized primary electron donor, P680+, in octyl glucoside-extracted thylakoids, indicating that D is not directly involved in P680+ reduction. However, overall photosystem II activity appears to be impaired in the Tyr-160-Phe mutant: photosystem II-dependent growth of this mutant is slowed down by a factor of 3-4, whereas photoheterotrophic growth rates in wild type and mutant are essentially identical. Binding studies of diuron, a photosystem II herbicide, show that there is no appreciable decrease in the number of photosystem II centers in the Tyr-160-Phe mutant. The decrease in photosystem II activity in this mutant may be interpreted to indicate a role of D in photoactivation, rather than one as an important redox intermediate in the photosynthetic electron-transport chain.
Keywords: photosynthesis, protein engineering, oxygen evolution, electron paramagnetic resonance, electron transport
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Allen J. P., Feher G., Yeates T. O., Rees D. C., Deisenhofer J., Michel H., Huber R. Structural homology of reaction centers from Rhodopseudomonas sphaeroides and Rhodopseudomonas viridis as determined by x-ray diffraction. Proc Natl Acad Sci U S A. 1986 Nov;83(22):8589–8593. doi: 10.1073/pnas.83.22.8589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babcock G. T., Sauer K. Electron paramagnetic resonance signal II in spinach chloroplasts. I. Kinetic analysis for untreated chloroplasts. Biochim Biophys Acta. 1973 Dec 14;325(3):483–503. doi: 10.1016/0005-2728(73)90209-0. [DOI] [PubMed] [Google Scholar]
- Barry B. A., Babcock G. T. Tyrosine radicals are involved in the photosynthetic oxygen-evolving system. Proc Natl Acad Sci U S A. 1987 Oct;84(20):7099–7103. doi: 10.1073/pnas.84.20.7099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C. H., Tiede D., Tang J., Smith U., Norris J., Schiffer M. Structure of Rhodopseudomonas sphaeroides R-26 reaction center. FEBS Lett. 1986 Sep 1;205(1):82–86. doi: 10.1016/0014-5793(86)80870-5. [DOI] [PubMed] [Google Scholar]
- Conjeaud H., Mathis P. The effects of pH on the reductions kinetics of P-680 in Tris-treated chloroplasts. Biochim Biophys Acta. 1980 May 9;590(3):353–359. doi: 10.1016/0005-2728(80)90206-6. [DOI] [PubMed] [Google Scholar]
- Debus R. J., Barry B. A., Babcock G. T., McIntosh L. Site-directed mutagenesis identifies a tyrosine radical involved in the photosynthetic oxygen-evolving system. Proc Natl Acad Sci U S A. 1988 Jan;85(2):427–430. doi: 10.1073/pnas.85.2.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel H., Epp O., Deisenhofer J. Pigment-protein interactions in the photosynthetic reaction centre from Rhodopseudomonas viridis. EMBO J. 1986 Oct;5(10):2445–2451. doi: 10.1002/j.1460-2075.1986.tb04520.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanba O., Satoh K. Isolation of a photosystem II reaction center consisting of D-1 and D-2 polypeptides and cytochrome b-559. Proc Natl Acad Sci U S A. 1987 Jan;84(1):109–112. doi: 10.1073/pnas.84.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oka A., Sugisaki H., Takanami M. Nucleotide sequence of the kanamycin resistance transposon Tn903. J Mol Biol. 1981 Apr 5;147(2):217–226. doi: 10.1016/0022-2836(81)90438-1. [DOI] [PubMed] [Google Scholar]
- Rutherford A. W. How close is the analogy between the reaction centre of Photosystem II and that of purple bacteria? Biochem Soc Trans. 1986 Feb;14(1):15–17. doi: 10.1042/bst0140015. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar H. K., Viitanen P. V., Padan E., Trumble W. R., Poonian M. S., McComas W., Kaback H. R. Oligonucleotide-directed site-specific mutagenesis of the lac permease of Escherichia coli. Methods Enzymol. 1986;125:214–230. doi: 10.1016/s0076-6879(86)25019-3. [DOI] [PubMed] [Google Scholar]
- Velthuys B. R., Visser J. W. The reactivation of EPR signal II in chloroplasts treated with reduced dichlorophenol-indophenol: evidence against a dark equilibrium between two oxidation states of the oxygen evolving system. FEBS Lett. 1975 Jul 15;55(1):109–112. doi: 10.1016/0014-5793(75)80971-9. [DOI] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- Yeates T. O., Komiya H., Rees D. C., Allen J. P., Feher G. Structure of the reaction center from Rhodobacter sphaeroides R-26: membrane-protein interactions. Proc Natl Acad Sci U S A. 1987 Sep;84(18):6438–6442. doi: 10.1073/pnas.84.18.6438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoller M. J., Smith M. Oligonucleotide-directed mutagenesis of DNA fragments cloned into M13 vectors. Methods Enzymol. 1983;100:468–500. doi: 10.1016/0076-6879(83)00074-9. [DOI] [PubMed] [Google Scholar]