Abstract
The receptor for calcitonin gene-related peptide (CGRP) has been the target for the development of novel small molecule antagonists for the treatment of migraine. Two such antagonists, BIBN4096BS and MK-0974, have shown great promise in clinical trials and hence a deeper understanding of the mechanism of their interaction with the receptor is now required. The structure of the CGRP receptor is unusual since it is comprised of a hetero-oligomeric complex between the calcitonin receptor-like receptor (CRL) and an accessory protein (RAMP1). Both the CLR and RAMP1 components have extracellular domains which interact with each other and together form part of the peptide-binding site. It seems likely that the antagonist binding site will also be located on the extracellular domains and indeed Trp-74 of RAMP1 has been shown to form part of the binding site for BIBN4096BS. However, despite a chimeric study demonstrating the role of the N-terminal domain of CLR in antagonist binding, no specific residues have been identified. Here we carry out a mutagenic screen of the extreme N-terminal domain of CLR (residues 23–63) and identify a mutant, Met-42-Ala, which displays 48-fold lower affinity for BIBN4096BS and almost 900-fold lower affinity for MK-0974. In addition, we confirm that the Trp-74-Lys mutation at human RAMP1 reduces BIBN4096BS affinity by over 300-fold and show for the first time a similar effect for MK-0974 affinity. The data suggest that the non-peptide antagonists occupy a binding site close to the interface of the N-terminal domains of CLR and RAMP1.
Keywords: Antagonist, Receptor, GPCR, CGRP, Migraine, Pharmacology
Introduction
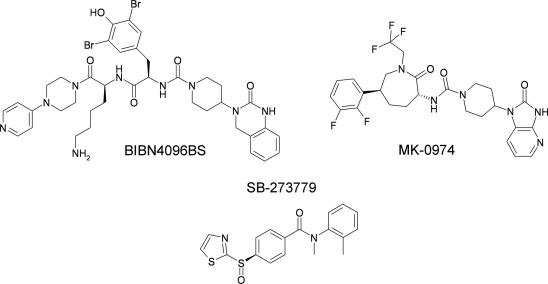
Calcitonin gene-related peptide (CGRP) is a 37-amino acid neuropeptide which has been implicated in the pathogenesis of migraine [1]. There have been concerted efforts within the pharmaceutical industry to develop novel small molecule antagonists of the CGRP receptor which have resulted in the development of two non-peptidic CGRP receptor antagonists (Fig. 1), BIBN4096BS (Olcegepant [2]) and MK-0974 (Telcagepant [3]). These compounds have been the subject of clinical trials that confirmed their clinical efficacy without the cardiovascular side effects commonly associated with triptans [4,5]. However, despite their clear therapeutic potential, relatively little is known regarding the molecular mechanisms underlying their binding to the CGRP receptor and their ability to antagonise CGRP action.
Fig. 1.
Chemical structures of three CGRP receptor antagonists: BIBN4096BS, MK-0974 and SB-273779.
The structure of the CGRP receptor (“CLR/RAMP1”) is unusual since it is comprised of a hetero-oligomeric complex between an accessory protein (RAMP1) and the calcitonin receptor-like receptor (CRL), a family B G protein-coupled receptor (GPCR). The CLR component shares the typical features of all family B GPCRs, namely an extracellular N-terminal domain of approximately 120 residues and a transmembrane domain consisting of seven transmembrane α-helices [6,7]. CLR is retained intracellularly in the absence of the RAMP partner which it requires for terminal glycosylation and translocation to the plasma membrane. The RAMP partner is not only essential for the successful transit of the CLR component to the cell surface but its particular type also determines the phenotype of the receptor itself [8]. Hence, while CRL/RAMP1 complex is a receptor for CGRP, heteromerisation of CLR with either the other RAMP types (RAMP2 or RAMP3) results in the formation of adrenomedullin receptors [8]. Interestingly, other family B GPCRs also interact with RAMPs although in many cases the effects upon their pharmacology are unexplored [9]. However, in the case of the calcitonin receptor (CTR), its pharmacology is switched from being a receptor for calcitonin in the absence of a RAMP to a receptor for amylin in the presence of a RAMP and, furthermore, the amylin receptor comprising CTR/RAMP1 also recognises CGRP with high potency [10].
RAMPs have a structure consisting of a single transmembrane segment and an extracellular domain comprising a three-helical bundle [11] which is envisaged to interact with the extracellular domain of CLR to form the peptide-binding site. However, the paucity of structural information describing the interaction between CLR and RAMP1 complicates the task of determining specific contact points between the receptor and its ligands. Nevertheless, a specific interaction between BIBN4096BS and the RAMP1 component of CLR/RAMP1 was identified following the observation that the antagonist exhibits approximately 200-fold species selectivity between primate and rat CGRP receptors [2]. The analysis of sequence differences between RAMP1 from these species, followed by associated site-directed mutagenesis studies, revealed that Trp-74 of human RAMP1 is key to the high-affinity binding of BIBN4096BS [12]. When mutated to lysine, as found in rat RAMP1, the antagonist’s affinity was decreased by two orders of magnitude [12]. MK-0974 also displays a marked species selectivity which is also RAMP1-dependent, although the specific residues involved in mediating this property have not been identified [3].
Since CLR/RAMP1 and CTR/RAMP1 share similar affinity for CGRP, yet are selective for BIBN4096BS [10,13] it suggests that CLR may also contribute directly to the binding of the antagonist. Indeed, Salvatore et al. [13] exploited this potential difference by means of a chimeric study of CLR and CTR, demonstrating that residues 37–63 of the N-terminus of CLR were required for high-affinity antagonist binding. In order to confirm the direct role of CLR in antagonist binding and to locate the non-peptide-binding site more precisely, we carried out a mutagenesis scan of the first 41 residues of the putative mature CLR sequence (residues 23–63).
Materials and methods
Constructs. Human CLR with an N-terminal hemagglutinin (HA) epitope tag (YPYDVPDYA) [8], was provided by Dr. Foord (GlaxoSmithKline, Stevenage, UK) and was sub-cloned into pcDNA3 (Invitrogen, Renfrew, UK) prior to mutagenesis. Introduction of the epitope did not effect the pharmacology of the receptor [8]. hRAMP1-pcDNA3.1 was purchased from UMR cDNA (Rolla, MO, USA).
Site-directed mutagenesis was carried out using the QuikChange protocol (Stratagene, La Jolla, CA), following the manufacturers instructions. These constructs were used to express the wild type, and mutant CLR and RAMP1 proteins in COS7 cells.
Cell culture. COS7 cells were cultured in Dulbecco’s Modified Eagle’s Medium (Sigma, Poole, UK) supplemented with 10% foetal calf serum (Lonza Wokingham Ltd., Wokingham, UK), 2 mM l-glutamine, 100 U/ml penicillin and 100 μg/ml streptomycin (Invitrogen). Cells were transfected with plasmid containing the cDNA encoding the receptors, using the Lipofectamine 2000 (Invitrogen) as follows: to two separate 15 ml falcon tubes, 1.6 ml of DMEM was added. To one tube, a further 30 μg of cDNA and 40 μl of Plus Reagent (Invitrogen) was introduced. Once 60 μl Lipofectamine 2000 (Invitrogen) had been transferred to the second tube, both were incubated at room temperature for 15 min. The contents of the two tubes were then incubated for a further 15 min. During this time, the COS7 cells were washed twice with phosphate-buffered saline (PBS). The transfection mixture was then added to the cells and placed back in the 37 °C incubator for 4 h. After addition of 15.5 ml of double serum CM10, the cells were returned to the incubator for 16 h. The next day, the media, along with transfection complexes were aspirated and replaced with 40 ml of CM10 and incubated for a further 24 h. The cells were then dissociated using TypLE Express (Invitrogen), centrifuged and resuspended in 1 ml Freezing Media (Sigma). After 16 h in a specialised freezing container at −80 °C, the cryovials were placed in a −140 °C freezer for long-term storage.
Ligands. All non-peptide ligands were synthesised and provided by GlaxoSmithKline. Human α-CGRP (herein referred to as CGRP) was purchased from Bachem (Saffron Walden, UK). The radioligand 125I-hCGRP was from PerkinElmer Life and Analytical Sciences (Waltham, MA).
Radioligand binding. Ice-cold distilled water (1.5 ml) was added to a vial of frozen COS7 cells which was then mixed and centrifuged in a bench-top centrifuge for 5 min at 13,000g. The pelleted cells were resuspended thoroughly in ice-cold distilled water and centrifuged as previously described. The pellet was then resuspended in 10 ml ice-cold distilled water and left on ice for 10 min prior to centrifugation (13,000g for 30 min). The cells were then resuspended in ice-cold PBS and centrifuged for 5 min. After three additional PBS washes, the crude membrane pellet was resuspended in 1 ml of HEPES binding buffer (HBB; 20 mM HEPES, 100 mM NaCl, 1 mM EDTA, 3 mM MgSO4, pH 7.5) supplemented with 50 μg/ml bacitracin and forced through a 23G needle. Aliquots (0.1 ml) were snap-frozen in liquid nitrogen and stored at −70 °C.
Membranes were slowly thawed on ice before diluting to a concentration that gave total radioligand binding of <10% total counts added. In a reaction volume of 200 μl, 75 pM (∼60,000 cpm) 125I-hCGRP, various concentrations of unlabelled hCGRP competitor ligand (1 μM to 1 pM) and COS7 membranes expressing the receptor of interest were combined. All ligand and membranes were diluted to appropriate concentration in HBB supplemented with 0.3% NFM and 5050 μg/ml bacitracin. Assays were carried out for 1 h in MultiScreen 96-well Filtration Plates (polyvinylidene fluoride filters, 0.45 μm pore size, Millipore, Bedford, MA) pre-soaked for 1 h in 1% non-fat milk/PBS. After the incubation, membrane-associated radioligand was harvested by transferring the assay mixture to the filtration plate housed in a vacuum manifold. The wells of the filtration plate were washed three times with 0.2 ml ice-cold PBS before harvesting the filter discs. Filter-bound radioactivity was measured in a gamma counter (RiaStar 5405 counter; PerkinElmer Life and Analytical Sciences, Waltham, MA). Total radioligand bound was <10% and non-specific binding was ∼1% of total counts added.
cAMP assays. COS7 cells transiently expressing the relevant receptors were thawed at 37 °C for 30 min, then washed in PBS and counted with an automated cell culture analyser (Cedex AS20, Innovatis). The cells were resuspended in stimulation buffer: HBSS, 5 mM HEPES, 0.1% BSA, 500 μM IBMX (all Sigma), pH 7.4 at a concentration of 1 × 106 cells per ml. Cell number had been optimised from previous experimentation in order that raw data fell within a range determined by a standard cAMP concentration curve. Antagonist solutions were prepared in DMSO (Sigma) and 0.1 μl was added to each well of a white 384-well low volume OptiPlate (Greiner) to give final concentrations ranging from 1 μM to 10 pM. Cells (5 μl) were added to each well followed by 15 min incubation at room temperature. At this point 0.1 μl CGRP at final concentrations ranging from 1 μM to 1 pM was added to each well using a Hummingbird (Digilab Genomic Solutions) followed by 5 μl of LANCE Stimulation buffer (PerkinElmer) containing 0.01 (v/v) Alexa Fluor® 647-labelled antibody (resulting in 5000 cells per well and 0.005 (v/v) Alexa Fluor®). The plates were incubated at room temperature for 30 min.
The detection mix was prepared by diluting Eu-W8044 labelled streptavidin (PerkinElmer) and biotin cAMP (PerkinElmer) in Detection buffer 2250 and 750-fold, respectively. This mixture was incubated at room temperature for 30 min to allow complex formation. Detection mix was (10 μl) added to each well (which lyses the cells) and incubated for 2 h at room temperature. The acceptor fluorescence signal was then read at 665 nm on a ViewLux instrument (PerkinElmer).
Data analysis. Curves in the figures represent one of at least three independent experiments for which each point is the mean of triplicate values with SEM displayed as error bars. All curves were fitted using non-linear regression with the aid of GraphPad PRISM 5 software (San Diego, CA). Binding data were normalised to the maximal specific binding within each data set and IC50 values were calculated with a single site binding model. Response curves were normalised to the maximal binding with CGRP alone. Antagonist data were analyzed to generate pA2 values [14]. Values in the tables represent the mean with SEM calculated from at least three independent experiments.
Results and discussion
While it is known that BIBN4096BS interacts with the CGRP receptor via RAMP1 [12], its high selectivity for the CGRP receptor (CLR/RAMP1) over the amylin receptor (CTR/RAMP1) implicates the CLR component in antagonist binding [10,13]. In this study we have carried out a mutagenesis scan of the first 41 residues of the putative mature CLR sequence (residues 23–63) in order to identify potential binding sites for non-peptidic antagonists.
Membrane preparations from COS7 cells co-expressing RAMP1-WT with either CLR-WT or the N-terminal mutations of CLR were first screened for their ability to bind 125I-CGRP in the presence and absence of 1 μM unlabelled CGRP (data not shown). When co-expressed with RAMP1-WT, four of the 41 mutant CLR variants (I41A, A44L, C48A and Y49A) consistently resulted in no detectable 125I-CGRP binding. Since this could indicate either loss of CGRP affinity, absence of receptor expression or severe indirect structural disruption of the receptor structure, these mutants were not analysed further in this study. The remaining 37 mutant CLR variants resulted in levels of specific 125I-CGRP binding that were comparable to CLR-WT, indicating the expression of intact CGRP receptors and enabling further more detailed pharmacological analysis.
LANCE cAMP assays were used to carry out concentration response curves for CGRP at COS7 cells co-expressing RAMP1-WT with either CLR-WT or the 37 CLR variants. The mutations had no significant effect upon CGRP potency (Table 1) indicating that these residues do not play an important role in binding to the peptide agonist. The ability of BIBN4096BS to competitively antagonise CGRP-induced receptor activation was analysed using six different concentrations of the non-peptidic antagonist in order to right-shift the CGRP concentration response curve and hence determine pA2 values. Of the 37 mutant receptors, only CLR-M42A resulted in a significant loss of BIBN4096BS affinity (48-fold, p < 0.05), with pA2 values of 8.95 ± 0.09 at the mutant compared with 10.63 ± 0.08 at CLR-WT (Table 1).
Table 1.
Pharmacological properties of CLR variants co-expressed with wild type RAMP1-WT.
| CLR variant | CGRP EC50 | BIBN4096BS pA2 |
|---|---|---|
| CLR-WT | 9.64 ± 0.06 | 10.63 ± 0.08 |
| CLR-E23A | 9.55 ± 0.04 | 10.41 ± 0.12 |
| CLR-L24A | 9.54 ± 0.07 | 10.26 ± 0.11 |
| CLR-E25A | 9.47 ± 0.10 | 10.24 ± 0.14 |
| CLR-E26A | 9.43 ± 0.13 | 10.35 ± 0.14 |
| CLR-S27A | 9.52 ± 0.10 | 10.38 ± 0.09 |
| CLR-P28A | 9.60 ± 0.08 | 10.46 ± 0.16 |
| CLR-E29A | 9.70 ± 0.07 | 10.42 ± 0.11 |
| CLR-D30A | 9.55 ± 0.09 | 10.36 ± 0.16 |
| CLR-S31A | 9.69 ± 0.09 | 10.22 ± 0.17 |
| CLR-I32A | 9.54 ± 0.08 | 10.38 ± 0.13 |
| CLR-Q33A | 9.16 ± 0.11 | 10.13 ± 0.13 |
| CLR-L34A | 9.55 ± 0.08 | 10.30 ± 0.11 |
| CLR-G35A | 9.47 ± 0.07 | 10.41 ± 0.11 |
| CLR-V36A | 9.69 ± 0.15 | 10.16 ± 0.13 |
| CLR-T37A | 9.34 ± 0.13 | 10.73 ± 0.06 |
| CLR-R38A | 9.48 ± 0.08 | 10.33 ± 0.13 |
| CLR-N39A | 9.59 ± 0.07 | 10.43 ± 0.17 |
| CLR-K40A | 9.44 ± 0.13 | 10.50 ± 0.13 |
| CLR-M42A | 9.49 ± 0.10 | 8.95 ± 0.09⁎⁎ |
| CLR-T43A | 9.52 ± 0.06 | 10.05 ± 0.10 |
| CLR-Q45A | 9.57 ± 0.12 | 10.13 ± 0.14 |
| CLR-Y46A | 9.54 ± 0.11 | 10.36 ± 0.15 |
| CLR-E47A | 9.49 ± 0.08 | 10.51 ± 0.14 |
| CLR-Q50A | 9.51 ± 0.08 | 10.19 ± 0.16 |
| CLR-K51A | 9.68 ± 0.06 | 10.47 ± 0.21 |
| CLR-I52A | 9.47 ± 0.06 | 10.28 ± 0.11 |
| CLR-M53A | 9.67 ± 0.16 | 10.31 ± 0.20 |
| CLR-Q54A | 9.57 ± 0.14 | 10.43 ± 0.12 |
| CLR-D55A | 9.51 ± 0.11 | 10.42 ± 0.11 |
| CLR-P56A | 9.48 ± 0.10 | 10.39 ± 0.16 |
| CLR-I57A | 9.61 ± 0.16 | 10.42 ± 0.13 |
| CLR-Q58A | 9.69 ± 0.13 | 10.46 ± 0.15 |
| CLR-Q59A | 9.88 ± 0.11 | 10.07 ± 0.13 |
| CLR-A60L | 9.62 ± 0.16 | 10.46 ± 0.14 |
| CLR-E61A | 9.40 ± 0.08 | 10.45 ± 0.17 |
| CLR-G62A | 9.76 ± 0.05 | 10.58 ± 0.18 |
| CLR-V63A | 9.50 ± 0.10 | 10.64 ± 0.15 |
p < 0.01 relative to pA2 of BIBN4096BS at WT receptor.
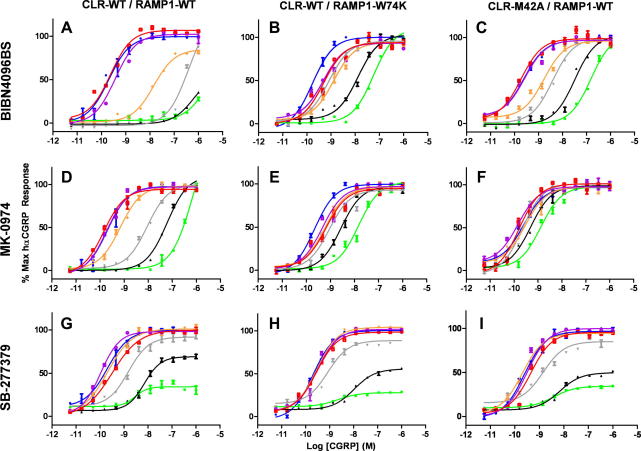
Whole COS7 cells and membrane preparations co-expressing RAMP1-WT with either CLR-WT or CLR-M42A were analysed in more detail in order to determine the role of Met-42 (Table 2, Fig. 2). While the Met-42-Ala mutation had no significant effect upon CGRP affinity or potency, a very substantial reduction in MK-0974 affinity was observed (891-fold; Fig. 2F).
Table 2.
Pharmacological properties of wild type CGRP receptor and mutants of RAMP1 and CLR.
| Receptor | CGRP | CGRP | BIBN4096BS | MK-0974 | SB-273779 |
|---|---|---|---|---|---|
| pIC50 | pEC50 | pA2 | pA2 | pA2 | |
| CGRP-WT | 8.60 ± 0.04 | 9.64 ± 0.06 | 10.63 ± 0.08 | 9.74 ± 0.09 | 7.10 ± 0.14 |
| CLR-M42A/RAMP1-WT | 8.36 ± 0.10 | 9.49 ± 0.10 | 8.95 ± 0.09∗ | 6.79 ± 0.06∗∗ | 7.08 ± 0.14 |
| CLR-WT/RAMP1-W74K | 8.33 ± 0.12 | 9.35 ± 0.15 | 8.13 ± 0.11∗ | 7.29 ± 0.09∗ | 7.11 ± 0.06 |
∗p < 0.05 and ∗∗p < 0.01, relative to same antagonist at WT receptor.
Fig. 2.
Concentration–response curves for CGRP at the either wild type CGRP receptor “CLR-WT/RAMP1-WT” (A,D,G), “CLR-WT/RAMP1-W74K” (B,E,H) or “CLR-M42A/RAMP1-WT” (C,F,I). The effect on the concentration–response curves of various concentrations of the antagonists BIBN4096BS (A–C) and MK-0974 (D–F) is shown: no antagonist (blue), 10 pM (red), 100 pM (purple), 1 nM (orange), 10 nM (grey), 100 nM (black), 1 μM (green). The concentration range used for SB-273779 (G–I) was 100-fold higher for each color. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this paper.)
In addition, the known BIBN4096BS binding residue on RAMP1, Trp-74, was mutated to Lys, its equivalent in rat [12] and co-expressed with CLR-WT (Table 2, Fig. 2). As with the co-expression of CLR-M42A/RAMP1-WT, the CLR-WT/RAMP1-W74K combination resulted in normal CGRP affinity and efficacy but displayed reduced affinity for both BIBN4096BS (316-fold, Fig. 2B) and MK-0974 (282-fold, Fig. 2E). Although it was known that MK-0974 displayed species selectivity, this is the first confirmation that, like BIBN4096BS, the selectivity is due to the Trp/Lys change at residue 74 of RAMP1.
Hence, both BIBN4096BS and MK-0974 bind to the CGRP receptor in a similar location involving both the RAMP1 (via Trp-74) and CLR (via Met-42). Interestingly, although the two molecules appear to share a similar binding site, the interaction with the CLR component is more important for MK-0974 than BIBN4096BS. While the mutation of Trp-74 in RAMP1 resulted in a similar reduction in affinity for both ligands, MK-0974 was almost 20-fold more sensitive to the Met-42 mutation in CLR.
The affinity of the unrelated non-peptidic antagonist SB-273779 (Fig. 1) was not affected by either the CRL-M42A or RAMP1-W74K mutations (Fig. 2G–I). SB-273779 has irreversible binding characteristics the CGRP receptor [15] and is non-selective between CGRP and AMY1 receptors (data not shown). Combined with major differences in structure, it is highly likely that SB-273779 binds through a different series of amino acids compared with BIBN4096BS and MK-0974.
The unaltered affinity of both CGRP and SB-273779 demonstrates that the mutations do not disrupt the structure of the receptor and suggests that the reduced affinity of BIBN4096BS and MK-0974 at the mutated receptors results from the disruption of direct ligand interaction sites mediated by Trp-74 of RAMP1 and Met-42 of CLR. Furthermore, it suggests that Trp-74 of RAMP1 and Met-42 of CLR may reside close to the RAMP1/CLR interface. The crystal structure of the extracellular domain of RAMP1 [11] demonstrates that Trp-74 is solvent exposed and on the outside surface of the structure. Likewise, analyses of the crystal structures of related family B GPCR N-terminal domains [16–18] demonstrate that the equivalent residues to Met-42 of CLR are also on the outside surface of the domain. Although there are still too many degrees of freedom to accurately model the CLR–RAMP1 interface, it is clear that a domain–domain interaction which involves both residues can be easily accommodated. There is already evidence within the literature which supports the importance of the extreme N-terminus of CLR in association of the two constituent proteins of the CGRP receptor. For example, in a chimeric study of the parathyroid hormone-1 receptor, it was shown that residues 23–60 from CLR were sufficient for full expression and oligomerisation of RAMP1 with PTH1 receptor.
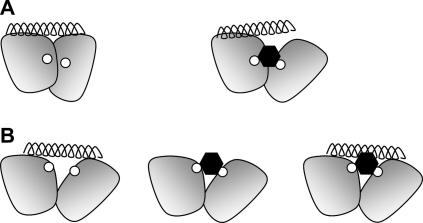
In principle an antagonist could work by directly intervening in the conventional association of CLR with RAMP1, thereby disrupting the peptide-binding site. However, since this complex forms in the endoplasmic reticulum and is maintained even after endocytosis, it suggests it is highly stable and unlikely to be readily disrupted by an antagonist [19,20]. Nevertheless, a more modest alteration of the CLR:RAMP1 interface could be envisaged that results in an allosteric modification of the peptide-binding site (Fig. 3A), with compounds such as BIBN4096BS and MK-0974 acting as a lever between the N-termini of RAMP1 and CLR, breaking nearby interactions which are critical for binding of the peptidic ligand. Indeed, there is evidence that BIBN4096BS acts allosterically [10] and it is interesting that neither of the mutations that have defined the two known contact sites for these non-peptide antagonists disrupt CGRP binding. However, this does not rule out some degree of overlap at other places in the binding sites and it is possible that the peptide and non-peptide ligands occupy over-lapping space while utilising different residues to obtain high affinity (Fig. 3B). Hence, it is possible that the non-peptides antagonise CGRP action by either an indirect allosteric action (Fig. 3A) or by competing for a partially over-lapping binding site (Fig. 3B).
Fig. 3.
Two models for the mechanism of action of the antagonists BIBN4096BS and MK-0974. The N-terminal domains of CLR and RAMP1 are depicted by the two shaded shapes, the antagonist-binding residues Met-42 and Trp-74 are each shown as small white circles, the antagonist as a black hexagon and the peptide ligand as a helix. Both A(left) and B(left) depict how the peptide can interact with both domains but not with the antagonist-binding residues. In A(right) the antagonist binding at the CLR:RAMP1 interface causes an allosteric effect which alters the peptide-binding site. In B the antagonist binding (centre) results in the occupation of some of the space required for the binding of the peptide, leading to competitive binding (right).
Conclusions
In summary, we have identified the first interaction site on CLR that interacts with the non-peptide antagonists BIBN4096BS and MK-0974. In addition, we have shown that the high affinity of MK-0974, like that of BIBN4096BS, is dependent upon Trp-74 in RAMP1. The location of these interaction sites highlight the domain–domain interface between the extracellular domains of CLR and RAMP1 and suggest a mechanism to explain the binding and antagonism of BIBN4096BS and MK-0974.
Acknowledgments
P.M. was funded through a BBSRC CASE award in collaboration with GlaxoSmithKline. J.B. was funded by a studentship awarded by the British Heart Foundation (FS/05/054).
References
- 1.Brain S.D., Poyner D.R., Hill R.G. CGRP receptors: a headache to study, but will antagonists prove therapeutic in migraine? Trends Pharmacol. Sci. 2002;23:51–53. doi: 10.1016/s0165-6147(02)01945-4. [DOI] [PubMed] [Google Scholar]
- 2.Doods H., Hallermayer G., Wu D.M., Entzeroth M., Rudolf K., Engel W., Eberlein W. Pharmacological profile of BIBN4096BS, the first selective small molecule CGRP antagonist. Br. J. Pharmacol. 2000;129:420–423. doi: 10.1038/sj.bjp.0703110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Salvatore C.A., Hershey J.C., Corcoran H.A., Fay J.F., Johnston V.K., Moore E.L., Mosser S.D., Burgey C.S., Paone D.V., Shaw A.W., Graham S.L., Vacca J.P., Williams T.M., Koblan K.S., Kane S.A. Pharmacological characterization of MK-0974, a potent and orally active calcitonin gene-related peptide receptor antagonist for the treatment of migraine. J. Pharmacol. Exp. Ther. 2008;324:416–421. doi: 10.1124/jpet.107.130344. [DOI] [PubMed] [Google Scholar]
- 4.Olesen J., Diener H.C., Husstedt I.W., Goadsby P.J., Hall D., Meier U., Pollentier S., Lesko L.M. Calcitonin gene-related peptide receptor antagonist BIBN 4096 BS for the acute treatment of migraine. N. Engl. J. Med. 2004;350:1104–1110. doi: 10.1056/NEJMoa030505. [DOI] [PubMed] [Google Scholar]
- 5.Ho T.W., Ferrari M.D., Dodick D.W., Galet V., Kost J., Fan X., Leibensperger H., Froman S., Assaid C., Lines C., Koppen H., Winner P.K. Efficacy and tolerability of MK-0974 (telcagepant), a new oral antagonist of calcitonin gene-related peptide receptor, compared with zolmitriptan for acute migraine: a randomised, placebo-controlled, parallel-treatment trial. Lancet. 2008;372:2115–2123. doi: 10.1016/S0140-6736(08)61626-8. [DOI] [PubMed] [Google Scholar]
- 6.Fluhmann B., Muff R., Hunziker W., Fischer J.A., Born W. A human orphan calcitonin receptor-like structure. Biochem. Biophys. Res. Commun. 1995;206:341–347. doi: 10.1006/bbrc.1995.1047. [DOI] [PubMed] [Google Scholar]
- 7.Njuki F., Nicholl C.G., Howard A., Mak J.C., Barnes P.J., Girgis S.I., Legon S. A new calcitonin-receptor-like sequence in rat pulmonary blood vessels. Clin. Sci. (Lond.) 1993;85:385–388. doi: 10.1042/cs0850385. [DOI] [PubMed] [Google Scholar]
- 8.McLatchie L.M., Fraser N.J., Main M.J., Wise A., Brown J., Thompson N., Solari R., Lee M.G., Foord S.M. RAMPs regulate the transport and ligand specificity of the calcitonin-receptor-like receptor. Nature. 1998;393:333–339. doi: 10.1038/30666. [DOI] [PubMed] [Google Scholar]
- 9.Christopoulos A., Christopoulos G., Morfis M., Udawela M., Laburthe M., Couvineau A., Kuwasako K., Tilakaratne N., Sexton P.M. Novel receptor partners and function of receptor activity-modifying proteins. J. Biol. Chem. 2003;278:3293–3297. doi: 10.1074/jbc.C200629200. [DOI] [PubMed] [Google Scholar]
- 10.Hay D.L., Christopoulos G., Christopoulos A., Sexton P.M. Determinants of 1-piperidinecarboxamide, N-[2-[[5-amino-l-[[4-(4-pyridinyl)-l-piperazinyl]carbonyl]pentyl]amino]-1-[(3,5-dibromo-4-hydroxyphenyl)methyl]-2-oxoethyl]-4-(1,4-dihydro-2-oxo-3(2H)-quinazolinyl) (BIBN4096BS) affinity for calcitonin gene-related peptide and amylin receptors – the role of receptor activity modifying protein 1. Mol. Pharmacol. 2006;70:1984–1991. doi: 10.1124/mol.106.027953. [DOI] [PubMed] [Google Scholar]
- 11.Kusano S., Kukimoto-Niino M., Akasaka R., Toyama M., Terada T., Shirouzu M., Shindo T., Yokoyama S. Crystal structure of the human receptor activity-modifying protein 1 extracellular domain. Protein Sci. 2008;17:1907–1914. doi: 10.1110/ps.036012.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mallee J.J., Salvatore C.A., LeBourdelles B., Oliver K.R., Longmore J., Koblan K.S., Kane S.A. Receptor activity-modifying protein 1 determines the species selectivity of non-peptide CGRP receptor antagonists. J. Biol. Chem. 2002;277:14294–14298. doi: 10.1074/jbc.M109661200. [DOI] [PubMed] [Google Scholar]
- 13.Salvatore C.A., Mallee J.J., Bell I.M., Zartman C.B., Williams T.M., Koblan K.S., Kane S.A. Identification and pharmacological characterization of domains involved in binding of CGRP receptor antagonists to the calcitonin-like receptor. Biochemistry. 2006;45:1881–1887. doi: 10.1021/bi052044w. [DOI] [PubMed] [Google Scholar]
- 14.Schild H.O. pA, a new scale for the measurement of drug antagonism. Br. J. Pharmacol. Chemother. 1947;2:89–206. doi: 10.1111/j.1476-5381.1947.tb00336.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aiyar N., Daines R.A., Disa J., Chambers P.A., Sauermelch V.F., Quiniou M.J., Khandoudi N., Gout B., Douglas S.A., Willette R.N. Pharmacology of SB-273779, a nonpeptide calcitonin gene-related peptide 1 receptor antagonist. J. Pharmacol. Exp. Ther. 2001;296:768–775. [PubMed] [Google Scholar]
- 16.Parthier C., Kleinschmidt M., Neumann P., Rudolph R., Manhart S., Schlenzig D., Fanghanel J., Rahfeld J.U., Demuth H.U., Stubbs M.T. Crystal structure of the incretin-bound extracellular domain of a G protein-coupled receptor. Proc. Natl. Acad. Sci. USA. 2007;104:13942–13947. doi: 10.1073/pnas.0706404104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Runge S., Thogersen H., Madsen K., Lau J., Rudolph R. Crystal structure of the ligand-bound glucagon-like peptide-1 receptor extracellular domain. J. Biol. Chem. 2008;283:11340–11347. doi: 10.1074/jbc.M708740200. [DOI] [PubMed] [Google Scholar]
- 18.Pioszak A.A., Xu H.E. Molecular recognition of parathyroid hormone by its G protein-coupled receptor. Proc. Natl. Acad. Sci. USA. 2008;105:5034–5039. doi: 10.1073/pnas.0801027105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hilairet S., Belanger C., Bertrand J., Laperriere A., Foord S.M., Bouvier M. Agonist-promoted internalization of a ternary complex between calcitonin receptor-like receptor, receptor activity-modifying protein 1 (RAMP1), and beta-arrestin. J. Biol. Chem. 2001;276:42182–42190. doi: 10.1074/jbc.M107323200. [DOI] [PubMed] [Google Scholar]
- 20.Hilairet S., Foord S.M., Marshall F.H., Bouvier M. Protein–protein interaction and not glycosylation determines the binding selectivity of heterodimers between the calcitonin receptor-like receptor and the receptor activity-modifying proteins. J. Biol. Chem. 2001;276:29575–29581. doi: 10.1074/jbc.M102722200. [DOI] [PubMed] [Google Scholar]