Abstract
Riboswitches are RNA-based genetic control elements that function via a conformational transition mechanism when a specific target molecule binds to its binding pocket. To facilitate an atomic detail interpretation of experimental investigations on the role of the adenine ligand on the conformational properties and kinetics of folding of the add adenine riboswitch, molecular dynamics (MD) simulations were performed in both the presence and absence of the ligand. In the absence of ligand, structural deviations were observed in the J23 junction and the P1 stem. Destabilization of the P1 stem in the absence of ligand involves the loss of direct stabilizing interactions with the ligand, with additional contributions from the J23 junction region. The J23 junction of the riboswitch is found to be more flexible and the tertiary contacts among the junction regions are altered in the absence of the adenine ligand; results suggest that the adenine ligand associates and dissociates from the riboswitch in the vicinity of J23. Good agreement was obtained with the experimental data with the results indicating dynamic behavior of the adenine ligand on the nanosecond timescale to be associated with the dynamic behavior of hydrogen bonding with the riboswitch. Results also predict that direct interactions of the adenine ligand with U74 of the riboswitch are not essential for stable binding even though it is crucial for its recognition. The possibility of methodological artifacts and force field inaccuracies impacting the present observations was checked by additional MD simulations in the presence of 2,6-diaminopurine and in the crystal environment.
Keywords: adenine riboswitch, aptamer, molecular dynamics simulations, base pairing, binding energy
Introduction
A riboswitch is an untranslated part of selected mRNAs that forms a binding pocket to bind small molecules and control gene expression.1; 2; 3; 4; 5 It had previously been assumed that proteins are solely responsible for genetic control. However, recent studies have proved that the riboswitches also play a role in genetic control. Riboswitches assume different conformations in the presence of a specific metabolite thereby altering the expression level of a target mRNA and ultimately controlling cellular metabolism.1; 2; 3; 4; 5 Several riboswitches have been discovered that are involved in sensing the intracellular concentrations of metabolites such as adenine,6; 7 guanine,6; 8 thiamine pyrophosphate,9; 10; 11; 12 S-adenosylmethionine (SAM),13; 14 S-adenosynhomocysteine,15 glucosamine-6-phosphate,16, glycine,17 lysine,18 Mg2+,19 cyclic di-GMP,20 and coenzyme B12.21 In general, riboswitches are composed of two domains namely, an aptamer domain and an expression platform. The aptamer domain recognizes and directly binds small molecule ligands with high specificity. Binding of the ligand results in stabilization of a specific apatmer domain conformation leading to an alternate secondary structure for the expression platform, which suppresses or activates expression at the transcriptional or translational levels depending on the expression platform.5; 22; 23
The add adenine riboswitch is involved in gene expression via the activation of translation, whereas the xpt-pbuE guanine riboswitch controls gene expression through transcriptional termination.5; 6; 7; 22; 23 With the adenine riboswitch, the absence of adenine ligand leads to specific residues in the expression platform, the Shine-Dalgarno GAA sequences, becoming unavailable for translational activation due to base pairing. In the presence of adenine the aptamer fold is stabilized and the GAA sequences become available for interaction with ribosomal RNA and tRNA leading to the initiation of translation. On the contrary, the guanine riboswitch in the absence of the ligand facilitates transcription via stabilization of the antiterminator stem. The presence of the guanine ligand leads to the formation of the terminator stem thereby prohibiting transcription. Batey and coworkers have shown that purine riboswitches are capable of binding modified pyrimidines and purine analogs, which is accomplished by the inherent flexibility of riboswitches that allows optimal binding of these molecules by minor structural changes in the RNA.24; 25
The aptamer domains of the guanine riboswitch of xpt-pbuX in Bacillus subtilis and the adenine riboswitch from the add gene of Vibrio vulnificus bound to their respective ligands have been structurally characterized by X-ray crystallography.6; 26 These riboswitches are structurally very similar to each other consistent with their highly conserved sequences.5; 6 The secondary structure of the adenine riboswitch considered in the present study and a three dimensional (3D) representation of the structure are given in Figure 1. The adenine riboswitch consists of three stems (P1, P2 and P3), two hairpin loops (L2 and L3), and three junctions (J12, J23 and J31) that connect the three stems. The stems P2 and P3 are located approximately parallel to each other and are stabilized by tertiary interactions between the two loops L2 and L3. The adenine ligand forms a Watson-Crick (WC) type of interaction with U74 and is stabilized in the binding pocket by that interaction along with additional hydrogen bond and stacking interactions.6
Figure 1.
(a) The three dimensional structure of the adenine riboswitch (PDB ID: 1Y26) generated using VMD.60 The bound adenine base is depicted by a CPK representation. The stems (P1, P2 & P3), the hairpin loops (L2 and L3) that cap P2 and P3 stems, and the junctions (J12, J23 & J31) that connect the three stems are color coded. (b) The secondary structure of the riboswitch, color coded similar to the three dimensional structure. The regular base pair interactions in the stems, and tertiary interactions in the loops and junctions via WC base pairing are represented by dashed lines. The hydrogen bond and the stacking interactions of the riboswitch with the adenine ligand are denoted using bold dashed and bold solid lines. For the purpose of clarity, tertiary contacts via noncanonical base pair interactions are not shown.
As discussed above, riboswitches may assume two different conformations depending on whether the small molecule specific to the riboswitch is bound or unbound. Towards better understanding of both structural and dynamic changes associated with ligand binding several experimental studies have been done on the adenine and guanine riboswitches. Recently, Greenleaf et al. reported an ingenious experiment based on single molecule force measurements to investigate the folding pathway of the pbuE adenine riboswitch.27 A series of intermediates were observed during the unfolding of the riboswitch both in the absence and the presence of the adenine metabolite (Figure 2). Application of force to the folded riboswitch either in the bound or unbound state leads to the disruption of the P1 stem followed by the loss of tertiary contacts between L2 and L3 after which the stems P3 and P2 sequentially lose base pairing to yield the completely unfolded structure. A diagram of the proposed states of the riboswitch observed in the force experiment is presented in Figure 2. Recently, Lin and Thirumalai have explored the folding and unfolding events of the add adenine riboswitch using Langevin dynamics with a self-organized polymer model that yielded a picture of unfolding consistent with the experimental study, though in their model the P2 stem unfolds prior to the P3 stem.28 This difference was attributed to subtle differences in the add versus pbuE forms of the A switch.
Figure 2.
Adenine riboswitch unfolding pathway derived using single molecule force measurement studies reported by Greenleaf et al.27 ‘a’ is the fully folded adenine riboswitch and ‘e’ is its completely extended form. ‘b’, ‘c’ and ‘d’ are the hypothesized intermediate structures observed during the force induced unfolding. The P1 stem is disrupted as the first step to form ‘b’ followed by the loss of tertiary interactions between L2 and L3 resulting in ‘c’. After the formation of the intermediate ‘c’, the P3 stem unfolds to form ‘d’ followed by the unfolding of P2 stem leading to the unfolded structure, ‘e’. Different regions of the riboswitch are colored as in Figure 1.
Lemay et al. used fluorescence resonance energy transfer technique to study the folding of the pbuE adenine riboswitch.29; 30; 31 They reported a correlation between binding of the adenine ligand to the riboswitch and the extent of the loop-loop interaction. Eskandari and coworkers demonstrated that in spite of the structural homology of guanine and adenine sensing riboswitches, the ligand bound form of the pbuE adenine riboswitch exists in at least three globally distinct conformers in the bound state arising from the junction dynamics.32 A combined NMR and molecular dynamics study on the guanine riboswitch indicated that the interaction between the two loops in the unbound state is not favorable enough to stabilize the entire fold and a three step ligand induced folding of the riboswitch after it comes in contact with the target ligand was hypothesized.33 It also has been shown that the loop-loop interactions in the unbound riboswitch could also be stabilized by Mg2+ ions resulting in similar folds for both bound and unbound states.29; 34 However, it should be emphasized that despite their sequence similarities there appear to be “different core requirements” in the G versus the A switch such that observations made based on one switch are not necessarily applicable to the second.29 A recent study by Rieder et al. reported the folding mechanism of adenine sensing add riboswitch.35 They showed that the P2 and P3 stems along with L2 and L3 loops are largely preorganized in the absence of the ligand, but the L2-L3 tertiary interactions tighten during ligand binding. Recently, MD simulations on the aptamer domain of the guanine, adenine and SAM sensing riboswitches in the presence and absence of the ligand were reported.36; 37; 38; 39
In this study, the structure and dynamics of the adenine riboswitch both in the presence and absence of the adenine and DAP ligands are investigated via molecular dynamics (MD) simulations. While the structure of the bound state of the adenine riboswitch is available from X-ray crystal studies,6 the unbound state of the riboswitch is modeled in the present study starting from the bound crystallographic structure by deleting the coordinates of the adenine ligand. Accordingly, the present work addresses differences between the folded forms of the adenine riboswitch (a and b in Figure 2) in the presence and the absence of the adenine ligand. Atomic details of the structural features, flexibility patterns, and intramolecular interactions leading to the differential dynamic behavior of these two states are investigated, with emphasis on the interpretation of experimental data on the switch. Throughout the manuscript, the riboswitch in the presence of adenine will be referred to as the “holo” form and in the absence of the adenine ligand it will be referred as the “apo” form.
Results and Discussion
The present study was motivated by the folding model put forth based on single molecule force measurements used to probe the multistep folding pathway.27 In that study force was applied along the 5′ and 3′ ends of the fully folded pbuE riboswitch yielding a completely unfolded form which was then allowed to refold to different extents by altering the applied force. Based on the unfolding transitions, the authors hypothesized a sequential unfolding in the riboswitch from the fully folded conformation (Figure 2). The model sequentially involves (i) disruption of the P1 stem, (ii) loss of tertiary contacts, (iii) unfolding of the P3 stem, and (iv) unfolding of the P2 stem. The proposed free energy surfaces are exactly the same for the last three steps irrespective of whether adenine is bound or not, indicating that adenine dissociation coincides with unfolding of the P1 stem. Adenine binding was found to stabilize the riboswitch in the fully folded state by about 4 kcal/mol.27 Importantly, in the proposed model the order of folding of the different regions of the aptamer domain of the adenine riboswitch is the approximately the same as occurs during transcription. However, it should be emphasized that unfolding in that study was induced by force, which may impact the folding events as compared to those occurring in the absence of force.
To better understand structural and dynamic details of the riboswitch in the presence and the absence of the adenine ligand, MD simulations were performed in the present study on the holo pbuE riboswitch in the presence of adenine (H1), and 2,6-diaminopurine (DAP, H2). Two different models were used for the MD simulations of the apo structure by simply removing the adenine ligand. In one of the models, two water molecules were added to the void left by the ligand (A1) and in the other model the binding site was left empty (A2). These simulations were performed in solution while an additional holo riboswitch simulation was performed in the crystal environment (H3) to predict that impact of the crystal environment versus aqueous solution alone on the riboswitch. The two states being studied in the present work, therefore, correspond to state a in the presence of ligand and state b in the absence of adenine ligand (Figure 2), with information from the study allowing for an understanding of how the presence of ligand stabilizes the a state versus the b state.
Overall structures
To obtain details of the structural changes occurring upon going from state a to b as well as validate the computational model, root mean square differences (RMSD) were calculated for the apo and holo simulations with respect to the crystal structure (Table 1). Results in Table 1 also include RMSDs from the simulation of the holo switch in the crystal environment. RMSDs with respect to the experimental crystal structure were performed following alignment of the overall structures as well as for alignment of the individual regions of the riboswitch shown in Figure 1. As expected the holo MD simulations yielded structures more similar to the experimental structure with an overall RMSD of 3.8 Å or less (Table 1). The average overall RMSD from the MD simulation of the holo form in the crystal environment was 3.4 Å (Table 1), indicating the presence of the crystal environment to impact but not dominate the overall structure of the holo switch structure. In contrast, the overall RMSD values of the apo structures (A1 & A2) are larger (5.8 and 5.4 Å, respectively) indicating significant deviations from the crystal structure of the holo form.
Table 1.
Root mean square deviations (RMSD, Å) of the riboswitch in the presence and absence of adenine with respect to the crystallographic structure.
| H1a) | H1b) | H2a) | H2b) | H3a) | H3b) | A1a) | A1b) | A2a) | A2b) | |
|---|---|---|---|---|---|---|---|---|---|---|
| RNA | 3.83 ± 0.19 | 3.83 ± 0.19 | 2.85 ± 0.20 | 2.85 ± 0.20 | 3.43 ± 0.09 | 3.43 ± 0.09 | 5.78 ± 0.17 | 5.78 ± 0.17 | 5.37 ± 0.31 | 5.37 ± 0.31 |
| P1 | 4.40 ± 0.46 | 2.62 ± 0.14 | 3.46 ± 0.39 | 2.30 ± 0.15 | 3.29 ± 0.16 | 2.48 ± 0.11 | 7.07 ± 0.24 | 3.04 ± 0.10 | 6.71 ± 0.61 | 3.99 ± 0.14 |
| P2 | 3.27 ± 0.11 | 2.39 ± 0.08 | 2.88 ± 0.04 | 2.27 ± 0.02 | 3.27 ± 0.10 | 2.86 ± 0.08 | 4.23 ± 0.18 | 2.12 ± 0.02 | 3.58 ± 0.24 | 2.11 ± 0.22 |
| P3 | 3.12 ± 0.10 | 1.88 ± 0.04 | 2.83 ± 0.20 | 1.26 ± 0.03 | 2.45 ± 0.10 | 1.17 ± 0.03 | 3.04 ± 0.19 | 1.79 ± 0.01 | 3.37 ± 0.20 | 1.66 ± 0.04 |
| L2 | 3.09 ± 0.15 | 1.83 ± 0.08 | 2.66 ± 0.38 | 1.70 ± 0.35 | 3.67 ± 0.25 | 3.05 ± 0.26 | 2.92 ± 0.14 | 1.46 ± 0.04 | 6.07 ± 0.91 | 2.63 ± 0.26 |
| L3 | 4.97 ± 0.14 | 3.34 ± 0.23 | 2.54 ± 0.11 | 1.36 ± 0.08 | 3.36 ± 0.04 | 2.55 ± 0.07 | 4.49 ± 0.17 | 3.38 ± 0.10 | 6.24 ± 0.32 | 4.94 ± 0.20 |
| J12 | 2.84 ± 0.15 | 1.05 ± 0.00 | 1.66 ± 0.07 | 0.83 ± 0.01 | 2.13 ± 0.08 | 1.17 ± 0.09 | 4.06 ± 0.33 | 2.26 ± 0.14 | 3.16 ± 0.11 | 1.22 ± 0.02 |
| J23 | 3.70 ± 0.17 | 1.91 ± 0.03 | 2.05 ± 0.03 | 1.69 ± 0.05 | 5.10 ± 0.14 | 2.12 ± 0.12 | 10.31 ± 0.22 | 4.71 ± 0.06 | 6.06 ± 0.11 | 2.77 ± 0.08 |
| J31 | 4.64 ± 0.20 | 1.58 ± 0.13 | 2.13 ± 0.41 | 0.84 ± 0.08 | 2.96 ± 0.19 | 1.03 ± 0.04 | 4.72 ± 0.17 | 2.08 ± 0.22 | 4.87 ± 0.29 | 1.65 ± 0.04 |
| ADE | 2.38 ± 0.34 | 0.07 ± 0.00 | 1.50 ± 0.14 | 0.13 ± 0.00 | 2.78 ± 0.29 | 0.08 ± 0.00 | - | - | - | - |
The structures in the trajectories were aligned based on all the non-hydrogen atoms of the riboswitch
The alignment of the structures was based on the non-hydrogen atoms of the region of the riboswitch for which the RMSD were calculated.
For the individual regions of the structures, the RMSDs based on alignment with respect to regions themselves are significantly smaller especially with the holo structure in solution. For example, the RMSD values of P2 and P3 helices of the riboswitch are low in both the holo and apo structures. However, when aligning the structure to the entire RNA significantly larger RMSD values are obtained indicating a model where rigid body motions dominate structural alterations upon going from state a to b. The largest changes occur with the J23 loop (RMSD = 10.3 and 6.1 Å in A1 and A2, respectively) followed by the P1 helix (RMSD = 7.1 and 6.7 Å) of relative to the entire RNA. Such rigid body motions of the helicies with respect to each other and about the connecting J regions are consistent with previously reported studies32 and are consistent with the suggestion that changes in the stability of the riboswitch in the absence of the adenine ligand are associated with significant structural differences.27 In A2, major structural deviations are observed in L2 and L3 loops in addition to P1 and J23 regions. Investigation of the nature of the loss of the tertiary interactions indicated loss of hydrogen bonds between the two loops, as presented in Figure S2 of Supporting Information. Many of the interactions described by Serganov et al.6 are lost or significantly decreased, including the L2A33-L3A66 Hoogsteen and L2G38-L360 WC interactions. This indicates the impact of the starting structures on the resulting structures, and their dynamics (see Methods section).
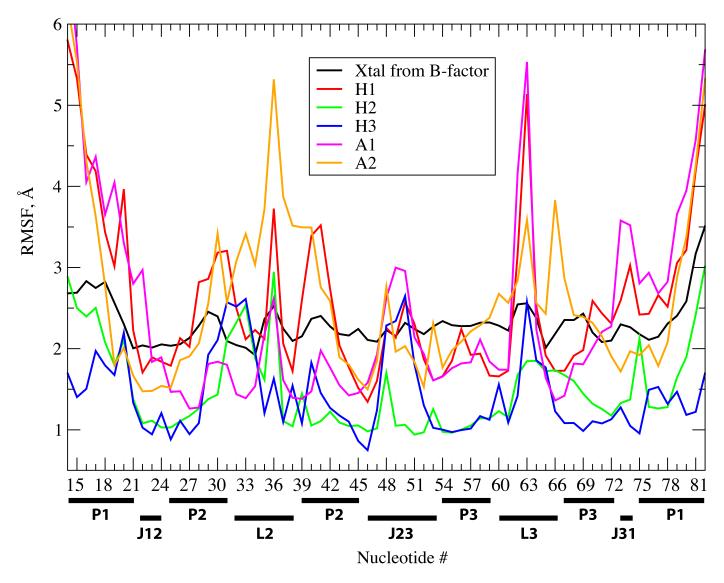
The flexibility of the riboswitch in the presence and absence of the ligand was assessed via the calculation of root mean square fluctuation (RMSF) values of each of the nucleotides (Figure 3). The figures also include the fluctuations calculated from the experimental X-ray temperature factors (eq. 1). In general, the RMS fluctuations of the apo forms from the MD simulation are qualitatively similar to those from the experimental crystal structure. For example, peaks are observed at both termini and in the vicinity of nucleotides 30, 40 and 63. More quantitative agreement of the experimental and simulation RMS fluctuations is not expected due to long-time conformational changes and lattice disorder contributions tending to favor larger fluctuations in the experimental values while the lack of crystal contacts will allow for increased flexibility in the solution simulations. Indeed, the systematically lower RMS fluctuations of the crystal simulation results as compared to experiment are due to lack of long time motions and lattice disorder in the former. Alternatively, the lack of crystal contacts leads to additional fraying at the termini of the RNA in the solution simulations, contributing to the high RMSF. Such events are common in DNA and RNA structures in solution; a recent NMR study on the pbuE A riboswitch did not observe signals for the terminal residues in the P1 helix, consistent with the present results.34
Figure 3.
RMS fluctuations averaged over each nucleotide computed for H1 (red line), H2 (green line), H3 (blue line), A1 (magenta line), and A2 (orange line) with the respect to the average structure along the MD trajectories. The fluctuation data (black line) calculated from the temperature factors (eq. 1) are also given for comparison. Different parts of the RNA (P1, P2, P3 and so on) are marked below the x-axis
The overall RMS fluctuations of the holo and apo forms are almost the same (2.59 for H1, and 2.47 and 2.64 Å for A1 and A2 respectively). However, the flexibility patterns of the holo and apo forms as a function of nucleotide differ significantly from each other. Most of nucleotides present in the junction regions (J12, J23 and J31) of the aptamer are more flexible in A1. Expectedly, the bases involved in binding the adenine ligand (A21, U22, U47, C50, A52, U74 and U75) except U51 are more flexible in its absence. This is consistent with the in-line probing experiments which showed that the ligand-binding region of the add A riboswitch to be more stable in the presence of ligand on long time scales.7 In contrast, the nucleotides involved in ligand binding (A21, U22, C50, U51, A52, U74 and U75) via hydrogen bonding and stacking interactions are less flexible in A2. Further analysis indicated that U47, A21, U51 and U74 forms a stable hydrogen bonded network resulting in a rigid conformation for the ligand binding regions (Figure S1 of supporting information). However, this is not in agreement with the NMR spectral study by Noeske et al, which did not observe any signals for U47, U51 and U74 precluding such a stable binding region in the absence of the adenine ligand.34 It should be noted that the nucleotides involved in the ligand binding exhibit significant RMSF values even in the Holo form (> 2 Å, Figure 3a). This is suggested to be related to the results from a recent fluorescence study of 2-aminopurine bound to the pbuE adenine riboswitch showing that the aptamer exhibits dynamic behavior on timescales of nanosecond or longer.32 However, such a behavior is not observed in the simulations involving DAP (H2) where the RMSF values of the nucleotides involved in ligand binding are comparatively smaller. More details of the nature of these fluctuations and their relationship to the fluorescence experiments are given below.
Dynamic cross correlation maps
Detailed analysis of the correlation of the movements of the nucleotides were investigated based on the cross correlation of the atomic fluctuations calculated using equation 2. The correlation coefficients were computed for all possible nucleotide pairs in both the holo and apo forms and are plotted in Figure 4, respectively. A value of 1 (dark blue) or −1 (dark red) for a given pair of nucleotides indicates strongly correlated movement between them, whereas a value of 0 indicates no correlation. Notably, correlated movements between the various regions of the riboswitch display different scenarios in the holo versus the apo forms. The movements of the bases constituting a base pair are expected to be strongly correlated. Positive correlations were observed for the base pairs in stems P2 (ie. P2a vs. P2b) and P3 in both the holo and apo forms. The correlations of the motions of the different parts of the riboswitch are similar in H1 and H2. However, the cross correlation coefficients corresponding to the P1 stem in the holo and apo forms are quite different from each other. The motions of each of the bases in P1 with its WC base pairing counterpart are fairly correlated in the holo form (Figure 4). However in the absence of the ligand, poor or no correlation is observed in both A1 and A2. For example, the nucleotides U17-A19 that are base paired to A79-U77, respectively, are not correlated in the apo form (Figure 4). This indicates a weaker P1 stem in the absence of adenine ligand and is consistent with the first step in the proposed unfolding of the pbuE adenine riboswitch being the disruption of the P1 stem27 leading to formation of the b state in Figure 2. The free energy barrier corresponding to the unfolding of P1 stem from the fully folded state was calculated to be lower in the absence of the adenine ligand (Figure 4E in ref. 27). Additionally the transition state for the disruption of P1 in the apo form occurs at a shorter extension length compared to that in the holo form. It is to be noted that the folding studies were done on a different adenine sensing riboswitch then studied presently, and the unfolding was achieved by force along the two termni of the RNA. The present results support the interpretation of the experimental data, as well as previously reported computational work,28 which indicated that lowered stability of the fully folded state in the absence of the ligand is due to weakened Watson-Crick base pairing between the two strands of P1.
Figure 4.
Computed dynamic cross correlation map for all the pairs of the nucleotides in the riboswitch. The maps for the (a) holo forms H1 and H2 are given in the upper and lower triangular regions respectively, and (b) apo forms, A1 and A2 in the upper and lower triangular regions respectively. Highly positive (C(i,j) close to 1) are given in dark blue, no correlations (C(i,j) close to 0) are given by white and highly negative (C(i,j) close to −1) are given in dark red. Grey colored lines are drawn to differentiate the various stems, loops and the junctions. P1a and P1b represent the two strands of the stem P1 and similar nomenclature are used for stems P2 and P3.
In addition to the differential behavior of the P1 stems, differences between the cross correlation maps of the holo and apo forms involve J23. Motions of the nucleotides in the P3 stem (P3a and P3b in Figure 4) are less correlated with the J23 junction in the apo form. Similarly, J23 motions are less correlated to the other junctions (J12 and J31) in the apo form. These differences are attributed to the movement of the J23 junction in A2 away from the adenine binding pocket in the absence of the ligand consistent with the RMSD data (Table 1). Major differences between A1 and A2 are observed in the correlation coefficients corresponding to L2 and L3. While they are highly correlated in A1, similar to that in the holo forms, their relative motions are less correlated with respect to each other in A2. The differential correlations between the various regions of the riboswitch are further understood based on the calculation of intramolecular interactions and are discussed in the following sections.
Adenine ligand binding
Interaction of the adenine ligand with the aptamer region of the riboswitch is essential for folding into the conformation that leads to the formation of the antiterminator strand which in turn leads to translational activation.5; 6; 22; 23 The crystal structure of the holo adenine riboswitch6 indicated that the adenine is stabilized in its binding pocket by the formation of direct hydrogen bonds from three bases and the 2′-hydroxyl group of a sugar group (Figure 5a). The probability distributions of the donor-acceptor distances of the hydrogen bonds formed between the riboswitch and the adenine ligand are given in Figure 5c. Results in Figure 5c include those for the holo simulations in solution and in the crystal environment. For the holo H1 solution simulation the histograms corresponding to the hydrogen bonds involving U22, U47 and U51 exhibit a sharp peak at about 3 Å indicating well-defined hydrogen bond interactions. However, additional distributions from 4-6 Å (except N3(U51)-N3) point to dynamic behavior of these hydrogen bonds. Interestingly, the probability distribution of the WC type base pairing hydrogen bonds between U74 and the adenine ligand in H1 have a zero population at hydrogen bonding distances. Examination of this base pairing as a function of simulation time revealed the hydrogen bonds to be lost at 3 ns and to not reform during the remainder of the trajectory. Thus, the holo solution simulation indicates the interactions between the ligand and the riboswitch to be highly dynamic, sampling multiple conformations. While such a result is consistent with fluorescence experiments indicating the present of multiple conformations of the holo riboswitch in solution being sampled on the nanosecond time scale,32 the result do raise a concern that methodological considerations may be leading to the loss of hydrogen bonding between the adenine ligand and the base U74 of the riboswitch. Such an observation was also reported by Sharma et al based on a 15 ns simulations.37 In a quantum mechanical study on the nonbonded interactions of adenine ligand with the neighboring nucleotides, Sharma et al showed that the total binding energy due to U74 is only about 25% of the total binding energy of the riboswitch with the adenine ligand.40 These observations suggest that hydrogen bonding of adenine with U74 may not be essential for binding, though the interaction is certainly key to ligand recognition.
Figure 5.
Image of the hydrogen bonding (a) and stacking interactions (b) of the nucleotides of the riboswitch with the adenine ligand. (c) Probability distributions of the donor-acceptor distances of the hydrogen bonds involved in riboswitch-ligand binding. Hydrogen bond distances from the crystal structure are depicted by green lines. Images generated using VMD.60
To test for possible limitations in the simulation methodology, including potential problems with the force field, two simulations of the holo switch were undertaken, one in presence of DAP (H2) and the other in its crystal environment (H3). H3 is expected to more rigorously reproduce the conditions in the crystallography experiment, allowing for discrimination of whether methodological versus environmental effects lead to the solution simulation results. As presented above, the presence of the crystal environment leads to lowered RMS differences and fluctuations, emphasizing the impact of the environment on the simulation results. Similarly, the presence of the crystal environment in the simulation leads to the adenine ligand-riboswitch hydrogen bonds to generally be well maintained as compared to the experimental structure (Figure 5C). Some heterogeneity in the switch-ligand interactions is still evident but in all cases well-defined hydrogen bond interactions are present in the simulation. This is particularly evident with U74, where the hydrogen bonds were lost in the holo solution simulation with adenine (H1). Similarly, in presence of DAP the all the hydrogen bonds including the two extra interactions involving the NH2 connected to C2 of DAP with U51 and U74 are well maintained. The results from these two simulations indicate that the loss of hydrogen bonding and conformational heterogeneity in the holo solution simulation of H1 may not be due to methodological artifacts, but rather associated with the inherent properties of the holo riboswitch bound to adenine in solution.
Time-resolved fluorescence experiments indicate the presence of relaxation events occurring on the sub-nanosecond to nanosecond time range in the pbuE riboswitch.32 Autocorrelation functions of the hydrogen bond distances were therefore calculated and are presented in Figure 6. These correlation functions were then modeled as multiple exponential decays, with two- or three-exponential decays giving satisfactory fits in all cases; results of the fitting are included in the individual panels in Figure 6. As is evident the relaxation behavior of the hydrogen bond interactions is occurring on sub-nanosecond to nanosecond time scales. The experimental study observed three relaxation times, and suggested these to be due to at least three different conformations sampled by the riboswitch, that differ in structure both locally at the binding pocket, and more globally. The sub-ns to ns hydrogen bonding dynamics observed here most likely contribute to the local dynamics and is related to the fluorescence relaxation times observed experimentally, while the larger structural changes may occur at timescales longer than the present simulations. Further, more rapid local fluctuations in hydrogen bonding may contribute to the slower interconversion between conformational states indicated in the fluorescence experiments. These possibilites along with the use of different ligands and different riboswitches limit more quantitative comparison of the experimental and calculated results.
Figure 6.
Autocorrelation functions of selected distances between riboswitch and adening ligand atoms in H1. Experimental autocorrelation functions are in red and fits of the decays to multiple exponential functions are in green. Included in each panel are the decays times in ns from the fits and the final squared correlation coefficient. Distances studies include (A) O2’(U22)-N7, (B) O2(U47)-N9, (C) O4(U51)-N9, (D) N3(U51)-N3, (E) N3(U74)-N1 and (F) O4(U74)-N6.
To better understand the energetic contributions to adenine binding the overall, hydrogen bonding, and stacking interaction energies where calculated (Table 2). The total interaction energies of −47 −68, and −52 kcal/mol between the ligand and the riboswitch obtained from the simulations of H1, H2 and H3, respectively, indicates that the adenine is bound strongly in the binding pocket in the solution simulation in spite of the loss of WC type base pair interactions with U74 in H1. DAP is found to interact more favorably with the riboswitch compared to adenine, which is in good agreement with experimental studies showing DAP to have higher affinity.7; 8 Stabilization of the ligand in its binding pocket is dominated by the junctions J23 and J12, and the P1 stem. In the H2 and H3 simulations, J31 has favorable interactions (−13.1 and −9.8 kcal/mol) with the ligand mainly due to the WC type base pair interactions involving U74. There are also significant stacking interactions involving the A21-U75 base pair, which is located on the top of the P1 stem (Figure 1). The presence of significant interactions involving A21-U75 is consistent with their high level of conservation and with results from a study by Batey and coworkers.24 That study, on the B. subtilis xpt-pbuX guanine riboswitch (GR) and a variant of GR where C74 has been converted to a uracil (termed GRA), making the riboswitch adenine specific, showed that mutation of the A21-U75 to both the U21-A75 and G21-C75 pairs lead to a significant decrease in binding of adenine to GRA. The two stems P2 and P3, and the two loops L2 and L3 do not have direct contact with the bound adenine, which explains the lack of favorable interaction between them (Table 2). The interaction energy of the three bases (U47, U51 & U74) and the sugar group of U22 (Figure 5a) with the adenine ligand are favorable (−20.6, −34.1 and −26.3 kcal/mol) confirming the well-defined hydrogen bonds between these bases and the ligand (Figure 5c). Consistent with the probability distribution, there is no significant contribution to the favorable adenine ligand binding from U74 in the solution simulation of H1. However, U74 has been shown to be essential for the recognition of adenine 7; 8 and hence crucial in the genetic control by the expression platform.6; 41
Table 2.
The interaction energies (kcal/mol) of the adenine or DAP ligand with the riboswitch and its various regions, and the stacking interaction energies of the ligand with select bases of the riboswitch.
| Interaction Energy | H-bonding Interaction Energy | Stacking Interaction Energy | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| H1 | H2 | H3 | H1 | H2 | H3 | H1 | H2 | H3 | |||
| P1-lig | −9.1 ± 0.4 | −10.0 ± 0.1 | −4.6 ± 1.4 | U22-lig | −7.7 ± 0.4 | −4.2 ± 0.1 | −5.0 ± 1.1 | A21-lig | −8.0 ± 0.2 | −7.9 ± 1.2 | −1.7 ± 0.4 |
| P2-lig | 0.8 ± 0.2 | −0.3 ± 0.3 | 0.2 ± 0.3 | U47-lig | −4.2 ± 0.3 | −5.1 ± 0.1 | −2.8 ± 0.5 | U22-lig | −5.2 ± 0.2 | −6.6 ± 0.2 | −6.3 ± 0.2 |
| P3-lig | 2.7 ± 0.2 | 1.4 ± 0.1 | 2.3 ± 0.1 | U51-lig | −8.6 ± 0.5 | −12.2 ± 0.2 | −7.6 ± 1.7 | C50-lig | −0.5 ± 0.1 | −1.6 ± 0.4 | −0.8 ± 0.2 |
| L1-lig | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.0 ± 0.0 | U74-lig | −0.1 ± 0.0 | −12.6 ± 0.4 | −10.8 ± 0.2 | A52-lig | −4.2 ± 0.2 | −5.0 ± 0.1 | −4.5 ± 0.4 |
| L2-lig | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.0 ± 0.0 | Total | −20.6 ± 0.6 | −34.1 ± 0.2 | −26.3 ± 0.5 | U75-lig | −0.7 ± 0.1 | −3.2 ± 0.9 | −3.4 ± 0.6 |
| J12-lig | −13.0 ± 0.3 | −11.5 ± 0.2 | −11.7 ± 1.0 | Total | −18.7 ± 0.4 | −24.2 ± 0.9 | −16.7 ± 0.6 | ||||
| J23-lig | −29.3 ± 0.2 | −34.7 ± 0.6 | −28.3 ± 2.0 | ||||||||
| J31-lig | 1.2 ± 0.1 | −13.1 ± 0.5 | −9.8 ± 0.3 | ||||||||
| RNA-lig | −46.6 ± 0.5 | −68.2 ± 0.5 | −51.8 ± 0.6 | ||||||||
For the calculation of hydrogen bonding interaction energy of U22, the sugar and phosphate atoms were considered; stacking interaction was computed by considering the uracil base.
As presented above, a simulation of the holo switch in the crystal environment and in the presence of DAP indicate that the lack of interactions between U74 and the adenine ligand is not a methodological artifact. This suggests that the lack of direct interactions of adenine with U74 may be due to conformational substates being sampled in the simulation that are not observable in experiments performed to date. The simulation (H1) presented here is suggested to have sampled conformations corresponding to the partially unfolded states observed based on fluorescence studies.32 However, NMR experiments on the pbuE A riboswitch have observed an interaction of the adenine ligand with U74 in solution.42 Thus, the loss of WC type interaction between U74 and adenine ligand in the holo simuluation (H1) may still be an artifact of the simulation. While artifacts cannot be excluded, the results are consistent with recent studies by Batey and coworkers on the mechanism of discrimination of purine bases by the guanine riboswitch (GR) and its C74U mutation (GRA).24; 43 In those studies, which included multiple crystal structures of different ligands bound to the two switches, it was evident that of the nucleotides comprising the purine binding site U or C74 undergoes the largest conformational variations. Thus, U74 can sample multiple conformations, though the extent of those variations may be overestimated in the present study. While requiring additional investigation, these observations suggests a model where U74 may be crucial for initial substrate recognition but may not be essential for stable binding of the ligand in the folded state.
Additional considerations with respect to riboswitch-adenine ligand interactions are differences in the experimentally determined energetics of binding of guanine to the G switch and adenine and DAP to the A switch and how those energetics may effect specificity. First, guanine is bound to the guanine riboswitch approximately 2 orders of magnitude tighter that of adenine to the adenine riboswitch. Similarly, the binding affinity for adenine is approximately 2 orders of magnitude less favorable than that of the adenine analog DAP bound to the adenine riboswitch.7; 8 In addition, the on-rate of adenine is about 3 times higher than that of DAP for the adenine riboswitch such that the off-rate for adenine must be significantly higher than that of DAP.44 These results indicate that adenine’s interaction with the riboswitch is not ideal (e.g. less than maximal interactions with the RNA are occurring) consistent with the possibility that adenine’s action in regulating translational activation may be kinetically controlled such that a high binding affinity is not required. This would be consistent with direct hydrogen bonding between U74 and the adenine ligand not occurring in all conformational substates of the riboswitch.7; 8 In addition, the binding of guanine to the adenine riboswitch is only two orders of magnitude less favorable than that of adenine; 30 μM versus 300 nM, respectively,8; 26 which corresponds to a free energy difference of approximately 2.7 kcal/mol based on the van’t Hoff relationship.45 As guanine would present different hydrogen bonding groups to U74, which could involve GU wobble base pairs,46 this further suggests that the highly specific interactions between the ligand and U74 of adenine riboswitch are not dominating the overall binding affinity.
Additional stabilization of the adenine ligand at equilibrium comes from stacking interactions. The adenine ligand is stacked between the A52-U22 base pair and the A21-U75-C50 triple in the binding pocket (Figure 5b). The stacking energies of these bases with the adenine base were calculated and given in Table 2. The total stacking interaction energy contributed by the five bases is −19 kcal/mol in H1 (−24 and −17 kcal/mol from H2 and H3, respectively) further emphasizing the strong contribution of this term to binding. After the first 3.8 ns of the H1 solution simulation, A21 undergoes partial base opening and moved towards C50 resulting in more favorable stacking interaction with the adenine base. The stacking interaction energy when A21 was base paired to U75 is −3.4 kcal/mol (up to 3.8 ns) and the favorable energy increases to −7.7 kcal/mol in the partially base open state (3.8-40 ns). Such movement of A21 was not observed in the crystal simulation, H3, which is also reflected by the less favorable stacking energy (−1.7 kcal/mol), though the stacking energy of U75 (−3.4 kcal/mol) is more favorable in this simulation. The presence of strong stacking interactions between the riboswitch and its ligand is consistent with the observed fluorescence quenching of 2-aminopurine32 as well as with the results of Batey and coworkers on the guanine riboswitch, as discussed above.24
Intramolecular interactions
Energetics of the intramolecular interactions responsible for the structural and dynamic results discussed above are analyzed in this section, focusing on base pairing in the stems and the tertiary contacts (Tables 3 & 4). Maintenance of the P2 and P3 helices, as evidenced by the RMSD analysis, (Table 1) is seen in the average base pair energies in P2 and P3 being very similar in both the apo and holo forms of the riboswitch (Table 3), with the P3 stem more stable in H2 and H3. Recent studies in the pbuE riboswitch showed that the P2 and P3 stems are preorganized even in the absence of ligand,29; 33 consistent with the present observation. Concerning the P1 stem, the interaction energies involving base pairing revealed a destabilizing effect in the apo form (Table 3). The interaction energies are more favorable by about 3 to 4 kcal/mol per base pair in the presence of ligand. The decrease in the favorable interaction in stem P1 further explains the reduced correlation between the motions of its bases and supports the disruption of the P1 stem during unfolding of the riboswitch (Table 1 and Figure 2). Interestingly, the base pair near the ligand-binding region (ie. A21-U75) is more stable in the absence of the ligand compared to the holo systems especially in the two solution simulations. Additionally, the terminal base pairs in the P1 stem are found to be relatively weak in the absence of the ligand compared to the holo simulations suggesting that the presence of ligand may stabilize these base pairs. Another interesting observation is the highly favorable interaction energy of the C18-G78 base pair, which is consistent with the suggestion that this pair is a “structural keystone” by Block and coworkers.27
Table 3.
Base pair interaction energies (kcal/mol) of the three stems of the riboswitch P1, P2 and P3 calculated in the presence and the absence of adenine or DAP. The average interaction energies for each of the stems are also given.
| H1 | H2 | H3 | A1 | A2 | |
|---|---|---|---|---|---|
| P1 Stem | |||||
| G14-U82 | −9.9 ± 0.6 | −9.5 ± 0.5 | −9.5 ± 0.5 | −2.0 ± 0.9 | −4.0 ± 1.1 |
| C15-G81 | −18.6 ± 1.5 | −20.5 ± 1.0 | −21.5 ± 0.3 | −1.6 ± 1.5 | −13.4 ± 1.1 |
| U16-A80 | −11.3 ± 0.1 | −11.5 ± 0.0 | −11.4 ± 0.0 | −9.8 ± 0.8 | −11.3 ± 0.0 |
| U17-A79 | −8.6 ± 1.6 | −1.9 ± 0.2 | −5.2 ± 1.2 | −3.0 ± 1.8 | −6.7 ± 1.4 |
| C18-G78 | −23.2 ± 0.2 | −22.0 ± 0.9 | −16.6 ± 2.6 | −22.5 ± 0.1 | −12.0 ± 1.0 |
| A19-U77 | −6.1 ± 1.6 | −10.6 ± 0.1 | −8.8 ± 1.3 | −4.1 ± 1.5 | −3.7 ± 0.1 |
| U20-A76 | −0.5 ± 0.2 | −1.2 ± 0.7 | −4.9 ± 1.8 | −5.3 ± 0.7 | −0.2 ± 0.0 |
| A21-U75 | −3.3 ± 0.4 | −6.0 ± 1.1 | −11.2 ± 0.1 | −6.9 ± 1.0 | −6.9 ± 0.1 |
| Average | −10.2 | −10.4 | −11.1 | −6.9 | −7.3 |
| P2 Stem | |||||
|---|---|---|---|---|---|
| U25-A45 | −11.4 ± 0.1 | −11.5 ± 0.0 | −11.4 ± 0.1 | −11.6 ± 0.0 | −11.5 ± 0.0 |
| C26-G44 | −20.7 ± 1.5 | −24.2 ± 0.0 | −22.3 ± 1.3 | −24.0 ± 0.1 | −22.1 ± 1.0 |
| C27-G43 | −23.9 ± 0.1 | −24.1 ± 0.0 | −23.9 ± 0.1 | −24.1 ± 0.0 | −23.8 ± 0.1 |
| U28-G42 | −8.1 ± 1.0 | 1.7 ± 0.2 | −11.5 ± 0.2 | −12.1 ± 0.0 | −8.0 ± 1.8 |
| A29-U41 | −3.7 ± 1.1 | −4.4 ± 0.1 | −2.9 ± 0.6 | −2.5 ± 0.1 | −2.9 ± 0.9 |
| A30-U40 | −2.7 ± 0.6 | −11.0 ± 0.2 | −0.6 ± 0.2 | −10.9 ± 0.1 | −3.4 ± 1.6 |
| U31-U39 | −1.4 ± 0.6 | −5.2 ± 2.3 | 0.0 ± 0.0 | −0.1 ± 0.0 | −1.5 ± 0.8 |
| Average | −10.3 | −11.2 | −10.4 | −12.2 | −10.5 |
| P3 Stem | |||||
|---|---|---|---|---|---|
| C54-G72 | −23.2 ± 0.4 | −24.2 ± 0.1 | −23.1 ± 0.3 | −24.2 ± 0.0 | −24.0 ± 0.0 |
| A55-U71 | −10.6 ± 0.3 | −11.6 ± 0.0 | −11.6 ± 0.0 | −11.5 ± 0.0 | −11.3 ± 0.1 |
| A56-U70 | −4.0 ± 1.4 | −11.6 ± 0.1 | −11.5 ± 0.0 | −11.5 ± 0.0 | −11.4 ± 0.1 |
| G57-C69 | −23.5 ± 0.1 | −23.5 ± 0.7 | −24.0 ± 0.0 | −23.5 ± 0.1 | −22.9 ± 0.1 |
| A58-U68 | −10.9 ± 0.4 | −11.3 ± 0.2 | −11.6 ± 0.1 | −2.2 ± 0.1 | −11.4 ± 0.1 |
| G59-C67 | −22.3 ± 0.4 | −23.7 ± 0.2 | −22.7 ± 0.4 | −23.5 ± 0.1 | −10.5 ± 2.1 |
| Average | −15.8 | −17.6 | −17.4 | −16.1 | −15.2 |
Table 4.
Interaction energies (kcal/mol) between the bases of the different regions of the riboswitch in the presence and absence of the adenine or DAP ligand.
| H1 | H2 | H3 | A1 | A2 | |
|---|---|---|---|---|---|
| P1-P2 | 0.1 ± 0.0 | 0.3 ± 0.0 | 0.2 ± 0.0 | 0.2 ± 0.0 | 0.2 ± 0.0 |
| P1-P3 | −0.5 ± 0.0 | −0.5 ± 0.0 | −0.5 ± 0.0 | −0.5 ± 0.0 | −0.5 ± 0.0 |
| P1-L2 | −0.1 ± 0.0 | −0.1 ± 0.0 | −0.1 ± 0.0 | −0.1 ± 0.0 | 0.0 ± 0.0 |
| P1-L3 | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.1 ± 0.0 | 0.1 ± 0.0 |
| P1-J12 | −1.1 ± 0.1 | −0.7 ± 0.0 | −1.4 ± 0.1 | −0.2 ± 0.1 | −6.6 ± 0.1 |
| P1-J23 | −4.3 ± 0.4 | −19.9 ± 1.0 | −2.1 ± 0.3 | −0.2 ± 0.0 | −14.5 ± 0.2 |
| P1-J31 | 0.2 ± 0.1 | −2.6 ± 0.6 | −2.6 ± 0.1 | −3.1 ± 0.6 | −3.3 ± 0.3 |
| P2-P3 | 1.5 ± 0.4 | 2.1 ± 0.1 | 3.2 ± 0.2 | 3.3 ± 0.1 | 0.4 ± 0.3 |
| P2-L2 | −7.2 ± 0.7 | −6.3 ± 0.8 | −6.3 ± 1.0 | −3.9 ± 0.1 | −11.4 ± 1.9 |
| P2-L3 | −2.4 ± 0.6 | −0.8 ± 0.0 | −0.7 ± 0.1 | −2.1 ± 0.3 | −1.0 ± 0.3 |
| P2-J12 | −7.4 ± 0.2 | −6.9 ± 0.2 | −7.0 ± 0.4 | −3.9 ± 0.3 | −7.9 ± 0.1 |
| P2-J23 | −11.6 ± 0.3 | −7.5 ± 0.2 | −14.9 ± 0.7 | −15.8 ± 0.5 | −19.2 ± 2.2 |
| P2-J31 | 0.1 ± 0.0 | 0.1 ± 0.0 | 0.2 ± 0.0 | 0.0 ± 0.0 | 0.2 ± 0.0 |
| P3-L2 | −2.2 ± 0.3 | −1.7 ± 0.2 | −4.8 ± 0.3 | −5.2 ± 0.2 | 0.4 ± 0.3 |
| P3-L3 | −10.9 ± 0.2 | −11.1 ± 0.1 | −12.7 ± 0.4 | −16.8 ± 0.2 | −7.5 ± 1.0 |
| P3-J12 | −12.2 ± 0.1 | −10.8 ± 0.3 | −12.6 ± 0.2 | −10.8 ± 0.1 | −12.5 ± 0.2 |
| P3-J23 | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.3 ± 0.1 | −0.1 ± 0.0 | −0.6 ± 0.1 |
| P3-J31 | −3.7 ± 0.3 | −1.4 ± 0.1 | −1.3 ± 0.0 | −2.5 ± 0.6 | −1.5 ± 0.0 |
| L2-L3 | −105.0 ± 1.7 | −96.7 ± 5.0 | −88.9 ± 5.0 | −105.4 ± 0.4 | −63.4 ± 2.3 |
| L2-J12 | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.0 ± 0.0 |
| L2-J23 | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.0 ± 0.0 |
| L2-J31 | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.0 ± 0.0 |
| L3-J12 | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.0 ± 0.0 |
| L3-J23 | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.1 ± 0.0 |
| L3-J31 | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.0 ± 0.0 |
| J12-J23 | −29.3 ± 0.1 | −26.3 ± 0.1 | −29.8 ± 0.2 | −15.7 ± 1.1 | −29.0 ± 0.2 |
| J12-J31 | −9.0 ± 0.4 | −8.4 ± 0.1 | −7.1 ± 0.6 | −7.4 ± 0.5 | −7.1 ± 0.1 |
| J23-J31 | −0.4 ± 0.1 | −3.9 ± 0.3 | −3.1 ± 0.5 | 0.0 ± 0.0 | −9.9 ± 0.3 |
Table 4 lists the interaction energies between the bases of all possible pairs of the three stems, three junctions and two loops. Most of the interaction energies involving the holo systems are similar to each other except P1-J23 which is more favorable in the presence of the DAP ligand. Between the two apo MD simulations, significant differences are observed. In A1, the largest difference is seen in the interaction energies between J12 and J23, being less favorable in the apo form compared to the holo forms. In addition, less favorable interactions are observed between P1-J23, P2-L2, P2-J12, P3-J12, J12-J23, while more favorable interactions occur for P2-J23 and P3-L3 in A1. These changes, along with the RMSD analysis discussed above, indicate that the absence of the adenine ligand leads to alterations of J23 and P1, changes that are due to direct interactions of these regions with the ligand (Figure 1). These changes lead to increased interactions of P2 with J23. This interaction appears to lead to lowered P2-L2 interactions and increased P3-L3 interactions, changes that impact the orientation of the P2 and P3 helicies consistent with the RMSD data (Table 1). Thus, the intramolecular interaction energies reveal two significant differences between the holo systems, and A1, destabilization of the P1 stem (Table 3) and a decrease in favorable interactions involving J23 with J12 (Table 4). These differences then impact other region-region interactions resulting in rigid body motions of these regions with respect to each other as observed in the RMSD analysis.
The study on the unfolding of the pbuE adenine riboswitch under force showed that the first step is the disruption of the P1 stem and the energetic requirement is lower in the apo form compared to the holo form, which is explained by the reduced interactions between the base pairs of P1.27 Additionally, in the force measuring optical tweezer experiment, the corresponding transition state occurred at a shorter extension length in the apo form compared to the holo form, which is supported by the current results that the P1 stem forms weaker base pairing interactions in the former. The interaction energies also allow for hypotheses to be developed for an atomic-detailed mechanism leading to the significant structural perturbations in the P1 stem and the J23 junction in the absence of adenine ligand. Obviously, with P1 the loss of the direct, favorable interactions of the terminal base pair in the stem (Figure 1 and Table 2) with the adenine ligand leads to destabilization. In addition, it appears that interactions with the joint regions may also play a role. Both J12 and J23 have favorable interactions with the ligand (Table 2), which leads to significant structural changes in these regions in the apo form (Table 1). This leads to J23 having significantly less favorable interactions with P1 in the holo form than in the apo form (Table 4). Such differential interactions are hypothesized to contribute to the destabilization of the P1 stem, thereby facilitating unfolding as observed experimentally.29; 32 Visual examination of the trajectories from the apo simulation indicate structural transition of the J23 junction from a closed to an open state in the absence of the adenine ligand (Figure 7). Also, U74, which plays an integral role in recognizing the adenine ligand, becomes more accessible to the solvent environment. The direction of opening of the J23 junction also indicates that the adenine ligand may preferably bind to the switch entering the pocket from the side of J23. Recently, in NMR experiments of the pbuE apo riboswitch imino resonances were not observed for U47, U51 and U74, nucleotides involved in direct interactions with the adenine ligand.34 In addition, resonances were also not observed for U20, U22, G45, G46, U49, U71, G72 and U75. Those results suggest that the central region of the riboswitch that interacts with the adenine ligand, including regions J12, J23 and J31 (Figure 1) are unstructured, in agreement with the present results. On the other hand, major differences are found in the interaction energies between the two loops L2 and L3 of A2, which is also supported by the decreased correlation of the movement between these two loops (Figure 4). Experimental studies have shown that binding of the ligand to the riboswitch leads to tightening and stabilization of the tertiary interactions between L2 and L3.29; 33; 35; 47 Hence, the two models of the apo forms of the riboswitch, A1 and A2, developed in this study represent two extremes of the experimental regimen, with the results obtained from the individual simulations allowing for interpretation of the available experimental data.
Figure 7.
The snapshots of the three dimensional structures obtained from the apo simulations (A1) illustrating the opening of the J23 junction, and its movement away from the adenine regions. The initial and final (40 ns) structures are also given for comparison. The color codes used are the same as in Figure 1.
Summary
MD simulations were performed to investigate the structural differences between the holo and apo forms of the adenine sensing add riboswitch. Preliminary analysis based on RMS deviations with respect to the available X-ray crystal structure of the holo form of the adenine riboswitch show good agreement with the simulated structure in the presence of the crystal environment, supporting the quality of the computational methodology. Comparison of the holo and apo MD simulations revealed significant structural changes in the P1 and J23 regions of the riboswitch. While the overall fluctuations of the riboswitch are very similar in the apo and holo forms, the flexibility patterns of the various regions are unique. The adenine-binding pocket is found to be flexible in the holo form, with fluctuations occurring on the nanosecond and subnanosecond time scales associated with changes in hydrogen bonding between the adenine ligand and the riboswitch. It is suggested that these fluctuations may be occurring in one or more of the conformations identified based on time-resolved fluorescence experiments32 and may contribute to transitions between those conformations. The present results along with the those experiments suggests that the adenine binding region maintains significant flexibility in the holo form on short, ns time scales; however on longer timescales, as evidenced by in-line probing experiments,7 larger scale dynamics are decreased in the holo form.
The WC base pair interaction between U74 and the bound adenine observed in the crystal structure6 as indicated to exist in solution based on NMR experiments is lost in the holo form solution MD simulation after the initial 3 ns. However, the interaction energies between the riboswitch and the bound adenine are still highly favorable and stable binding is predicted to be not affected by the loss of adenine ligand-U74 interactions. It is therefore suggested that U74 is important in the kinetic recognition of the target adenine base but may not be essential for stabilizing the base in the binding pocket. Interaction energies and the correlation coefficients indicate that the adenine ligand is important for stabilizing the P1 stem. Contributing to this destabilization is opening of the J23 junction in the apo form, which is associated with a decrease in the tertiary interactions involving P1 and with J12. The atomic details of this conformational transition and the position of U74 indicate that the adenine ligand may associate and dissociate with the switch along the side of the riboswitch where J23 is located.
As discussed above, the loss of the U74-adenine ligand WC interactions is not consistent with crystallographic and NMR data. To check that the lack of U74-adenine ligand interactions was not a methodological artifact, a MD simulation of the holo form in the crystal environment was performed in which the WC interaction between U74 and the adenine ligand was retained throughout the 40 ns simulation. In addition, hydrogen bonding of the ligand and U74 was maintained throughout a 20 ns simulation of the holo switch in which DAP was the ligand. While these results are supportive of the quality of the applied methodology, artifacts associated with the simulations cannot be excluded. However, the simulation results are in agreement with a range of experimental data such that the presented observations should be of utility for the interpretation of experimental studies as well as stimulate new studies.
Methods
The CHARMM 48 and NAMD49 programs were used to run the MD simulations reported in this study using the CHARMM27 all atom nucleic acid force field50; 51 and the modified TIP3P water model.52 The coordinates of the add adenine sensing riboswitch were from the X-ray crystal structure published by Serganov et al. (PDB accession code: 1Y26).6 Hydrogen atoms were added to the riboswitch in standard orientations and to the water molecules found in the crystal structure using the Hbuild utility in CHARMM. A total of five MD simulations were performed, which will be referred to as H1, H2, H3, A1 and A2 in the reminder of the manuscript of which the first three are holo systems, and the last two are apo systems. H1: The riboswitch was solvated in a truncated octahedron TIP3P water box and the solvent molecules within 1.9 Å of the non-hydrogen atoms of the crystallographic structure were deleted. The system was then electrically neutralized by placing Mg2+ ions at random positions in the solvent environment; the resultant concentration of Mg2+ was 107 mM. H2: The adenine ligand in the original crystal structure was replaced by DAP, and solvated as performed for H1 followed by random placement of Mg2+ ions. The H2 simulation was done using the NAMD molecular dynamics simulation package for 20 ns. H3: MD simulations were also performed on the holo form of the riboswitch in the crystal environment on a single asymmetric unit of the P21212 space group.6 The asymmetric unit was solvated and Mg2+ ions were added in random positions in regions of the crystal occupied by solvent to yield an electronically neutral system. The crystallographic water molecules were included in the simulation, and additional water molecules were added to fill the void between the asymmetric units. The symmetry related molecules in the unit cell of the riboswitch were included via the CRYSTAL module in CHARMM. A1: For the apo riboswitch MD simulation the adenine ligand in the riboswitch was deleted from the crystal structure and solvation was done using the procedure given above. A2: The adenine ligand was simply deleted from the starting structure used for H1, and the resulting configuration was used as the starting structure for A2 simulation. The primary difference between the starting structures of A1 and A2 is the presence of two water molecules in place of the adenine ligand in the former, and void in the latter. In all the simulations, the Mg2+ ions and the water coordinates from the original crystal structure were retained.
Both the apo and holo systems were subjected to a 500-step adopted basis Newton-Raphson minimization with mass weighted harmonic restraints of 5.0 kcal/mol/Å2 on the non-hydrogen atoms of the riboswitch. In all the minimizations and the subsequent MD runs, periodic boundary conditions were applied using the CRYSTAL module53 in CHARMM. The solvent molecules and the ions were equilibrated using a 100 ps MD simulation in the NPT ensemble maintaining the harmonic restraints used for the minimization. The electrostatic interactions were treated using the particle mesh Ewald method54; 55 in all the minimizations and the MD simulations the electrostatic real-space and the Lennard-Jones (LJ) interactions were truncated at 12 Å with a LJ force switch smoothing function from 8 to 12 Å. Nonbond interaction lists were updated heuristically. Covalent bonds involving hydrogen atoms were constrained using the SHAKE algorithm, which allowed the use of a 2 fs integration time step during the simulations. Production simulations of both systems were carried out for 40 ns in the NPT ensemble with the Leapfrog integrator. Hoover chains56 were used for temperature control and the Langevin piston method57 was employed to maintain the pressure of 1 atm with a piston mass of 600 amu. The MD simulation of H2 in presence of DAP was done using NAMD program following the system preparation protocol as described above.
Coordinates from the MD simulations were saved every 5 ps during the production simulations for analysis. All the analyses that are discussed in the paper were performed over the last 35 ns of the MD simulations unless specified (last 15 ns in the case of H2). The rotational and translational motions of the RNA in the trajectory were removed by superimposing the non-hydrogen atoms of the riboswitch excluding the terminal base pairs on to the X-ray crystal structure. Root mean square differences (RMSD) were calculated following least squares fitting of all the non-hydrogen atoms in the RNA and following least squares fitting of the non-hydrogen atoms in the individual segments being analyzed. The experimental atomic fluctuations of the bound state of the riboswitch were estimated from the temperature factors in the X-ray crystal data using equation 1. The RMSD values signifies the extent of deviation of the structures obtained using the MD simulations with respect to the reference structure, in this case the X-ray crystal structure. On the other hand, RMSF values presented here gives the fluctuation of each of the nucleotides in comparison to the average structure from the trajectories.
| Eq (1) |
The dynamic features of the holo and apo forms of the riboswitch and the extent of correlation of the motions of the different regions were assessed via the calculation of cross correlation coefficients, C(i,j) given by equation 2.58; 59 In the equation, Δri and Δrj are the displacement vectors for atoms i and j, respectively, and the angle brackets denotes the ensemble average. In the present study, the correlation coefficients were averaged over each nucleotide and the resultant correlation coefficients are presented in the form of a two dimensional graph.
| Eq (2) |
For the calculation of the interaction energies, the INTER command in CHARMM program was applied using infinite nonbond cutoffs. The interaction energies thus calculated include both electrostatic and van der Waals contributions. The RMS deviations and the energies were averaged over 5 ns blocks during the last 35 or 15 ns of the simulations and the resulting average values were used for the calculation of the reported averages and standard errors of the mean. Dynamic analysis of riboswitch-adenine ligand hydrogen-bond interactions was performed by calculating the autocorrelation functions of selected atom-atom distances. The resulting correlation functions were then fit with 2 and 3 exponential functions using the program MATLAB from which the relaxation times were extracted.
Supplementary Material
Acknowledgements
Dr. E. Prabhu Raman, University of Maryland Baltimore, is acknowledged for his help with the MD simulation involving DAP. NIH (GM 51501) is thanked for financial support. We acknowledge the Department of Defense High Performance Computing for their generous allocation of computer time.
Abbreviations used
- MD
molecular dynamics
- RMSD
root mean square deviation
- RMSF
root mean square fluctuation
- WC
Watson-Crick
- NMR
nuclear magnetic resonance
- DAP
2,6-diaminopurine
- LJ
Lennard-Jones
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Breaker RR. Complex riboswitches. Science. 2008;319:1795–1797. doi: 10.1126/science.1152621. [DOI] [PubMed] [Google Scholar]
- 2.Edwards TE, Klein DJ, Ferre-D’Amare AR. Riboswitches: small-molecule recognition by gene regulatory RNAs. Current Opinion in Structural Biology. 2007;17:273–279. doi: 10.1016/j.sbi.2007.05.004. [DOI] [PubMed] [Google Scholar]
- 3.Coppins RL, Hall KB, Groisman EA. The intricate world of riboswitches. Current Opinion in Microbiology. 2007;10:176–181. doi: 10.1016/j.mib.2007.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Serganov A, Patel DJ. Ribozymes, riboswitches and beyond: regulation of gene expression without proteins. Nature Reviews Genetics. 2007;8:776–790. doi: 10.1038/nrg2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim JN, Breaker RR. Purine sensing by riboswitches. Biology of the Cell. 2008;100:1–11. doi: 10.1042/BC20070088. [DOI] [PubMed] [Google Scholar]
- 6.Serganov A, Yuan YR, Pikovskaya O, Polonskaia A, Malinina L, Phan AT, Hobartner C, Micura R, Breaker RR, Patel DJ. Structural basis for discriminative regulation of gene expression by adenine- and guanine-sensing mRNAs. Chemistry & Biology. 2004;11:1729–1741. doi: 10.1016/j.chembiol.2004.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mandal M, Breaker RR. Adenine riboswitches and gene activation by disruption of a transcription terminator. Nature Structural & Molecular Biology. 2004;11:29–35. doi: 10.1038/nsmb710. [DOI] [PubMed] [Google Scholar]
- 8.Mandal M, Boese B, Barrick JE, Winkler WC, Breaker RR. Riboswitches control fundamental biochemical pathways in Bacillus subtilis and other bacteria. Cell. 2003;113:577–586. doi: 10.1016/s0092-8674(03)00391-x. [DOI] [PubMed] [Google Scholar]
- 9.Miranda-Rios J, Navarro M, Soberon M. A conserved RNA structure (thi box) is involved in regulation of thiamin biosynthetic gene expression in bacteria. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:9736–9741. doi: 10.1073/pnas.161168098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Edwards TE, Ferre-D’Amare AR. Crystal structures of the thi-box riboswitch bound to thiamine pyrophosphate analogs reveal adaptive RNA-small molecule recognition. Structure. 2006;14:1459–1468. doi: 10.1016/j.str.2006.07.008. [DOI] [PubMed] [Google Scholar]
- 11.Serganov A, Polonskaia A, Phan AT, Breaker RR, Patel DJ. Structural basis for gene regulation by a thiamine pyrophosphate-sensing riboswitch. Nature. 2006;441:1167–1171. doi: 10.1038/nature04740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thore S, Leibundgut M, Ban NN. Structure of the eukaryotic thiamine pyrophosphate riboswitch with its regulatory ligand. Science. 2006;312:1208–1211. doi: 10.1126/science.1128451. [DOI] [PubMed] [Google Scholar]
- 13.Winkler WC, Nahvi A, Sudarsan N, Barrick JE, Breaker RR. An mRNA structure that controls gene expression by binding S-adenosylmethionine. Nature Structural Biology. 2003;10:701–707. doi: 10.1038/nsb967. [DOI] [PubMed] [Google Scholar]
- 14.Fuchs RT, Grundy FJ, Henkin TM. The S-MK box is a new SAM-binding RNA for translational regulation of SAM synthetase. Nature Structural & Molecular Biology. 2006;13:226–233. doi: 10.1038/nsmb1059. [DOI] [PubMed] [Google Scholar]
- 15.Wang JX, Lee ER, Morales DR, Lim J, Breaker RR. Riboswitches that Sense S-adenosylhomocysteine and Activate Genes Involved in Coenzyme Recycling. 2008;29:691. doi: 10.1016/j.molcel.2008.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Winkler WC, Nahvi A, Roth A, Collins JA, Breaker RR. Control of gene expression by a natural metabolite-responsive ribozyme. Nature. 2004;428:281–286. doi: 10.1038/nature02362. [DOI] [PubMed] [Google Scholar]
- 17.Mandal M, Lee M, Barrick JE, Weinberg Z, Emilsson GM, Ruzzo WL, Breaker RR. A glycine-dependent riboswitch that uses cooperative binding to control gene expression (vol 306, pg, 275, 2004) Science. 2004;306:1477–1477. doi: 10.1126/science.1100829. [DOI] [PubMed] [Google Scholar]
- 18.Grundy FJ, Lehman SC, Henkin TM. The L box regulon: Lysine sensing by leader RNAs of bacterial lysine biosynthesis genes. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:12057–12062. doi: 10.1073/pnas.2133705100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cromie MJ, Shi Y, Latifi T, Groisman EA. An RNA sensor for intracellular Mg2+ Cell. 2006;125:71–84. doi: 10.1016/j.cell.2006.01.043. [DOI] [PubMed] [Google Scholar]
- 20.Sudarsan N, Lee ER, Weinberg Z, Moy RH, Kim JN, Link KH, Breaker RR. Riboswitches in eubacteria sense the second messenger cyclic di-GMP. Science’s STKE. 2008;321:411. doi: 10.1126/science.1159519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nahvi A, Sudarsan N, Ebert MS, Zou X, Brown KL, Breaker RR. Genetic control by a metabolite binding mRNA. Chemistry & Biology. 2002;9:1043–1049. doi: 10.1016/s1074-5521(02)00224-7. [DOI] [PubMed] [Google Scholar]
- 22.Grundy FJ, Henkin TM. From ribosome to riboswitch: Control of gene expression in bacteria by RNA structural rearrangements. Critical Reviews in Biochemistry and Molecular Biology. 2006;41:329–338. doi: 10.1080/10409230600914294. [DOI] [PubMed] [Google Scholar]
- 23.Nudler E, Mironov a. S. The riboswitch control of bacterial metabolism. Trends in Biochemical Sciences. 2004;29:11–17. doi: 10.1016/j.tibs.2003.11.004. [DOI] [PubMed] [Google Scholar]
- 24.Gilbert SD, Reyes FE, Edwards AL, Batey RT. Adaptive ligand binding by the purine riboswitch in the recognition of guanine and adenine analogs. Structure. 2009;17:857–68. doi: 10.1016/j.str.2009.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gilbert SD, Mediatore SJ, Batey RT. Modified pyrimidines specifically bind the purine riboswitch. Journal of the American Chemical Society. 2006;128:14214. doi: 10.1021/ja063645t. [DOI] [PubMed] [Google Scholar]
- 26.Batey RT, Gilbert SD, Montange RK. Structure of a natural guanine-responsive riboswitch complexed with the metabolite hypoxanthine. Nature. 2004;432:411–415. doi: 10.1038/nature03037. [DOI] [PubMed] [Google Scholar]
- 27.Greenleaf WJ, Frieda KL, Foster DAN, Woodside MT, Block SM. Direct observation of hierarchical folding in single riboswitch aptamers. Science. 2008;319:630–633. doi: 10.1126/science.1151298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lin J-C, Thirumalai D. Relative stability of helices determines the fording landscape of adenine riboswitch aptamers. J. Am. Chem. Soc. 2008;130:14080–14081. doi: 10.1021/ja8063638. [DOI] [PubMed] [Google Scholar]
- 29.Lemay JF, Penedo JC, Tremblay R, Lilley DMJ, Lafontaine DA. Folding of the adenine riboswitch. Chemistry & Biology. 2006;13:857–868. doi: 10.1016/j.chembiol.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 30.Gilbert SD, Batey RT. Riboswitches: Fold and function. Chemistry & Biology. 2006;13:805–807. doi: 10.1016/j.chembiol.2006.08.002. [DOI] [PubMed] [Google Scholar]
- 31.Fernandez-Luna MT, Miranda-Rios J. Riboswitch folding - One at a time and step by step. Rna Biology. 2008;5:20–23. doi: 10.4161/rna.5.1.5974. [DOI] [PubMed] [Google Scholar]
- 32.Eskandari S, Prychyna O, Leung J, Avdic D, O’Neill MA. Ligand-directed dynamics of adenine riboswitch conformers. Journal of the American Chemical Society. 2007;129:11308. doi: 10.1021/ja073159l. + [DOI] [PubMed] [Google Scholar]
- 33.Buck J, Furtig B, Noeske J, Wohnert J, Schwalbe H. Time-resolved NMR methods resolving ligand-induced RNA folding at atomic resolution. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:15699–15704. doi: 10.1073/pnas.0703182104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Noeske J, Schwalbe H, Wohnert J. Metal-ion binding and metal-ion induced folding of the adenine-sensing riboswitch aptamer domain. Nucleic Acids Research. 2007;35:5262–5273. doi: 10.1093/nar/gkm565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rieder R, Lang K, Graber D, Micura R. Ligand-induced folding of the adenosine deaminase A-riboswitch and implications on riboswitch translational control. ChemBioChem. 2007;8:896–902. doi: 10.1002/cbic.200700057. [DOI] [PubMed] [Google Scholar]
- 36.Villa A, Wohnert J, Stock G. Molecular dynamics simulation study of the binding of purine bases to the aptamer domain of the guanine sensing riboswitch. Nucleic Acids Research. 2009 doi: 10.1093/nar/gkp486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sharma M, Bulusu G, Mitra A. MD simulations of ligand-bound and ligand-free aptamer: Molecular level insights into the binding and switching mechanism of the add A-riboswitch. RNA. 2009;15:1673. doi: 10.1261/rna.1675809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huang W, Kim J, Jha S, Aboul-ela F. A mechanism for S-adenosyl methionine assisted formation of a riboswitch conformation: a small molecule with a strong arm. Nucl. Acids Res. 2009:gkp664. doi: 10.1093/nar/gkp664. %R 10.1093/nar/gkp664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Whitford PC, Schug A, Saunders J, Hennelly SP, Onuchic JN, Sanbonmatsu KY. Nonlocal helix formation is key to understanding S-Adenosylmethionine-1 riboswitch function. Biophysical Journal. 2009;96:7–9. doi: 10.1016/j.bpj.2008.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sharma P, Sharma S, Chawla M, Mitra A. Modeling the noncovalent interactions at the metabolite binding site in purine riboswitches. Journal of Molecular Modeling. 2009;15:633–649. doi: 10.1007/s00894-008-0384-y. [DOI] [PubMed] [Google Scholar]
- 41.Gilbert SD, Stoddard CD, Wise SJ, Batey RT. Thermodynamic and kinetic characterization of ligand binding to the purine riboswitch aptamer domain (vol 359, pg 754, 2006) Journal of Molecular Biology. 2006;363:624–624. doi: 10.1016/j.jmb.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 42.Noeske J, Richter C, Grundl MA, Nasiri HR, Schwalbe H, Wohnert J. An intramolecular base triple as the basis of ligand specificity and affinity in the guanine- and adenine-sensing riboswitch RNAs. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:1372–1377. doi: 10.1073/pnas.0406347102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gilbert SD, Mediatore SJ, Batey RT. Modified pyrimidines specifically bind the purine riboswitch. J Am Chem Soc. 2006;128:14214–5. doi: 10.1021/ja063645t. [DOI] [PubMed] [Google Scholar]
- 44.Wickiser JK, Cheah MT, Breaker RR, Crothers DD. The kinetics of ligand binding by an adenine-sensing riboswitch. Biochemistry. 2005;44:13404–13414. doi: 10.1021/bi051008u. [DOI] [PubMed] [Google Scholar]
- 45.Atkins PW. Physical Chemistry. 6th edit W.H. Freeman & Co.; 1997. [Google Scholar]
- 46.Chen X, McDowell JA, Kierzek R, Krugh TR, Turner DH. Nuclear magnetic resonance spectroscopy and molecular modeling reveal that different hydrogen bonding patterns are possible for G.U pairs: one hydrogen bond for each G.U pair in r(GGCGUGCC)2 and two for each G.U pair in r(GAGUGCUC)2. Biochemistry. 2000;39:8970–8982. [PubMed] [Google Scholar]
- 47.Prychyna O, Dahabieh MS, Chao J, O’Neill MA. Sequence-dependent folding and unfolding of ligand-bound purine riboswitches. Biopolymers. 2009;91 doi: 10.1002/bip.21283. [DOI] [PubMed] [Google Scholar]
- 48.Brooks BR, Brooks CL, III, Mackerell AD, Jr., Nilsson L, Petrella RJ, Roux B, Won Y, Archontis G, Bartels C, Boresch S, Caflisch A, Caves L, Cui Q, Dinner AR, Feig M, Fischer S, Gao J, Hodoscek M, Im W, Kuczera K, Lazaridis T, Ma J, Ovchinnikov V, Paci E, Pastor RW, Post CB, Pu JZ, Schaefer M, Tidor B, Venable RM, Woodcock HL, Wu X, Yang W, York DM, Karplus M. CHARMM: The biomolecular simulation program. Journal of Computational Chemistry. 2009;30:1545–1614. doi: 10.1002/jcc.21287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kale L, Schulten K. Scalable molecular dynamics with NAMD. Journal of Computational Chemistry. 2005;26:1781. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Foloppe N, MacKerell AD., Jr. All-atom empirical force field for nucleic acids: 1) Parameter optimization based on small molecule and condensed phase macromolecular target data. J. Comp. Chem. 2000;21:86–104. [Google Scholar]
- 51.MacKerell AD, Jr., Banavali NK. All-atom empirical force field for nucleic acids: 2) Application to solution MD simulations of DNA. J. Comp. Chem. 2000;21:105–120. [Google Scholar]
- 52.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of Simple Potential Functions for Simulating Liquid Water. Journal of Chemical Physics. 1983;79:926–935. [Google Scholar]
- 53.Field MJ, Karplus M. CRYSTAL: Program for Crystal Calculations in CHARMM. Harvard University; Cambridge, MA: 1992. [Google Scholar]
- 54.Darden T, Perera L, Li LP, Pedersen L. New tricks for modelers from the crystallography toolkit: the particle mesh Ewald algorithm and its use in nucleic acid simulations. Structure with Folding & Design. 1999;7:R55–R60. doi: 10.1016/s0969-2126(99)80033-1. [DOI] [PubMed] [Google Scholar]
- 55.Essmann U, Perera L, Berkowitz ML, Darden TA, Lee H, Pedersen LG. A smooth particle mesh ewald method. J Chem Phys. 1995;103:8577–8593. [Google Scholar]
- 56.Hoover WG. Canonical dynamics: equilibrium phase-space distributions. Phy. Rev. 1985;A 31:1695–1697. doi: 10.1103/physreva.31.1695. [DOI] [PubMed] [Google Scholar]
- 57.Feller SE, Zhang Y, Pastor RW, Brooks RW. Constant Pressure Molecular Dynamics Simulation: The Langevin Piston Method. J. Chem. Phys. 1995;103:4613–4621. [Google Scholar]
- 58.Brooks CLK, M., Pettitt BM. Proteins: A theoretical perspective of dynamics, structure, and thermodynamics. John Wiley & Sons; New York: 1988. [Google Scholar]
- 59.McCcammon JAH, C. S. Dynamics of proteins and nucleic acids. Cambridge University Press; Cambridge: 1986. [Google Scholar]
- 60.Humphrey W, Dalke A, Schulten K. VMD: Visual Molecular Dynamics. J. Molec. Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.