Abstract
It is widely assumed that the key rate-limiting step in gene activation is the recruitment of RNA polymerase II (Pol II) to the core promoter2. Although there are well-documented examples where Pol II is recruited to a gene but stalls3–14, a general role for Pol II stalling in development has not been established. We have performed comprehensive Pol II ChIP-chip assays in Drosophila embryos and identified three distinct Pol II binding behaviors: active (uniform binding across the entire transcription unit), no binding, and stalled (binding at the transcription start site). The striking feature of the ~10% genes that are stalled is that they are highly enriched for developmental control genes that are repressed at the time of analysis or poised for activation during subsequent stages of development. We propose that Pol II stalling facilitates rapid temporal and spatial changes in gene activity during development.
Pol II stalling is probably best studied at heat shock genes in Drosophila, where Pol II engages in transcription but pauses immediately downstream of the transcriptional start site3,4,22. Upon activation by heat shock, Pol II is able to rapidly transcribe these genes. Regulation of Pol II activity after recruitment has also been described in bacteria23, yeast13 and mammalian cell lines3,6–12, and includes instances where Pol II is found in an inactive pre-initiation complex24,25. We will collectively refer to inactive Pol II near the transcription start site as stalled Pol II.
To determine at which genes Pol II stalling occurs during development, we analyzed global Pol II occupancy in whole Drosophila embryos. While this is one of the few systems where genomics approaches can easily be applied to developmental questions, interpretation is complicated by the occurrence of multiple tissues. To reduce the complexity, we used Toll10b embryos (2–4 hours after fertilization), a well characterized mutant that contains a homogenous population of mesodermal precursor cells at the expense of neuronal and ectodermal cells15–21. In Toll10b mutants, mesodermal genes are uniformly activated while genes required for the development of ectodermal and neural tissues are repressed throughout the embryo15–17. Previous whole-genome microarray experiments have identified the transcript levels of all genes in these mutants19,20. To distinguish between stalled and active Pol II, we used a mixture of antibodies that recognizes both the initiating and elongating forms of Pol II (see Methods), and performed whole-genome ChIP-chip assays as previously described21.
The results show that many genes known to be repressed in Toll10b embryos display strikingly high levels of Pol II near the transcription start site (Fig.1A–D). In some cases the prominent Pol II peak is tightly restricted to the promoter region (e.g. at the tup gene in Fig. 1A), while at other genes Pol II is also found at low levels throughout the transcription unit (e.g. the sog and brk genes Fig. 1C,D). This is consistent with previous evidence that some genes such as sog are transiently activated but then repressed at later stages26, while other genes such as tup are never activated in Toll10b mutants19,20.
Fig. 1. Different classes of Pol II binding profiles.
ChIP-chip assays were performed with 2–4 hr Toll10b embryos using antibodies that recognize both the initiating and elongating form of Pol II. The enrichment ratios of Pol II are shown on the y-axis. (A–D) display the binding patterns across genes that are repressed in Toll10b embryos. All four genes display high levels of Pol II near the transcription start sites. At some genes such as tup (A), Pol II is tightly restricted to this region, whereas at other genes including sog (C) and brk (D), Pol II is also detected at lower levels throughout the transcription unit. (E,F) Pol II is uniformly distributed across the transcription units of genes that are actively transcribed. The Heartbroken (Hbr, also called stumps or Dof—downstream of FGF) locus (E) is specifically activated in mesodermal precursor cells, while RpL3 (F) is a ribosomal gene. (G,H) No Pol II binding is found at many genes that are inactive during embryogenesis. The eyeless (ey) gene (G) is expressed during eye development at larval stages but not in the early embryo. Likewise, the torso (tor) gene (H) is only active during oogenesis but not in the early embryo.
The Pol II profile of repressed genes is clearly distinct from those of active genes (Fig. 1E, F). For example, the Hbr gene, which encodes an FGF receptor specifically expressed in mesodermal precursors (Fig. 1E), and ribosomal genes such as RpL3 (Fig.1F), show uniformly high levels of Pol II throughout the transcription unit. Furthermore, genes that are silent in the early embryo simply lack Pol II binding altogether (Fig. 1G, H). Thus, there appears to be three distinct classes of genes: those with Pol II distributed throughout the transcription unit, those genes with preferential enrichment of Pol II at the transcription site, and genes that lack Pol II binding altogether.
To further characterize these three groups, we developed a principled method that classifies genes based on their Pol II enrichment profiles (Fig. 2, Supplemental Materials). Similar to an analysis performed in E.coli23, we calculated the ratio between Pol II enrichment at the transcription start site versus internal regions of the transcription unit (Fig. 2). We were able to assign 76% of the protein coding genes (10,220 of 13,448 genes) into one of three classes (Fig. 2B). At least 27% of all genes display an active Pol II profile in which Pol II is detected uniformly throughout the transcription unit. At least 12% of all genes (1,614 of 13,448) show disproportionate accumulation of Pol II near the transcription start site. Among this group, Pol II is tightly restricted to the transcription start site at 62% of genes. At the remaining 38% of genes, Pol II is also detected within the transcription unit, presumably because these genes -like sog – are expressed at low levels in at least a subset of cells during the timeframe of the analysis (2–4 hrs after fertilization). Finally, 37% of all genes lack Pol II binding altogether.
Fig. 2. Whole-genome analysis of Pol II binding.
(A) Genes were assigned to either one of three classes - stalled Pol II, active Pol II or no Pol II, and - based on their Stalling Index (box). The Stalling Index is the ratio between the maximum enrichment near the transcription start site (maxTSS) (± 300 bp) and the median enrichment of the probes distributed across the transcription unit (mediantranscript) (excluding the first 600 bp). Stalling Index values of >4 qualified as “stalled Pol II”, whereas Stalling Index values <2 qualified as “uniform (active) Pol II”. If no probe within the TSS region was significantly bound, the gene was assigned to the “no Pol II” category. (B) Over 76% of all protein-coding genes could be assigned to one of three categories based on Stalling Index values: 12% have stalled Pol II, 23% display the active form of Pol II and 37% of genes have no Pol II. Among the genes with stalled Pol II, 62% have Pol II tightly restricted to the transcription start site.
Several lines of evidence confirm that the ~1,600 genes with disproportionate enrichment of Pol II at the transcription start site represent a form of stalled Pol II (Fig. 3). First, all heat shock genes, which are the classical example of Pol II stalling4,22 fall into this class (Fig. 3A). Second, the Pol II peaks map an average of ~50 bp downstream of the transcription start site, consistent with the location of stalled Pol II at heat shock genes4,5,22(Fig. 3B). Since this is an average profile, it is possible that a fraction of Pol II occupancy comes from inactive pre-initiation complexes. However, the majority of detected Pol II signal appears to come from Pol II that is stalled downstream of the transcription start site. Third, Pol II stalling at these genes is consistent with comprehensive expression analysis using whole-genome tiling arrays20. Genes with Pol II tightly restricted to the transcription start site are either silent or only weakly expressed in Toll10b mutants (Fig. 3C). In contrast, genes with similar levels of Pol II binding, but uniform distribution throughout the transcription unit, are expressed at significant levels in these mutants (Fig. 3C). Finally, we used permanganate footprint assays as an independent method to confirm stalled Pol II at selected genes5,14. For example, the rho gene displays clear permanganate sensitivity downstream of the transcription start site (+36 bp), consistent with the Pol II stalling profile seen in Toll10b mutants (Fig. 3D; see Fig. 1B).
Fig. 3. Confirmation of the class of genes with Pol II stalling.
(A) The group of stalled genes contains all known heat shock genes where Pol II stalling has been well documented. (B) Metagene analysis shows that the average peak of “stalled Pol” II is ~ 50 bp downstream of transcription (arrow). The profile for “no Pol II” and “active Pol II” is shown as comparison. (C) Analysis of the transcript levels confirms that genes with the stalled Pol II profile (tightly restricted to the transcription site) are either silent or expressed at low levels. Genes that show Pol II enrichment at comparable levels throughout the transcription unit (active Pol II) are expressed at significantly higher levels. Genes with no Pol II are not expressed. The transcript levels are represented as box and whiskers plot of the fold-ratios (measured by whole-genome tiling arrays20). The box represents the 25th and 75th percentile with the median as red bar. The whiskers refer to the first and 99th percentile. The scale on the y-axis is a log scale. (D) A permanganate footprint assay of the rho gene confirms stalled Pol II downstream of the transcription start site. Genomic sequences of A+G are shown as marker (lane 1). In comparison with purified genomic DNA, which was either not treated (lane 2) or treated (lane 3) with KMnO4, a prominent hypersensitive T residue is detected in Toll10b mutant embryos (lane 4), implying the existence of a transcription bubble at the region around +37 in vivo. The bottom panel shows actual sequences from +26 to +47 of the rho locus (relative to TTS as +1).
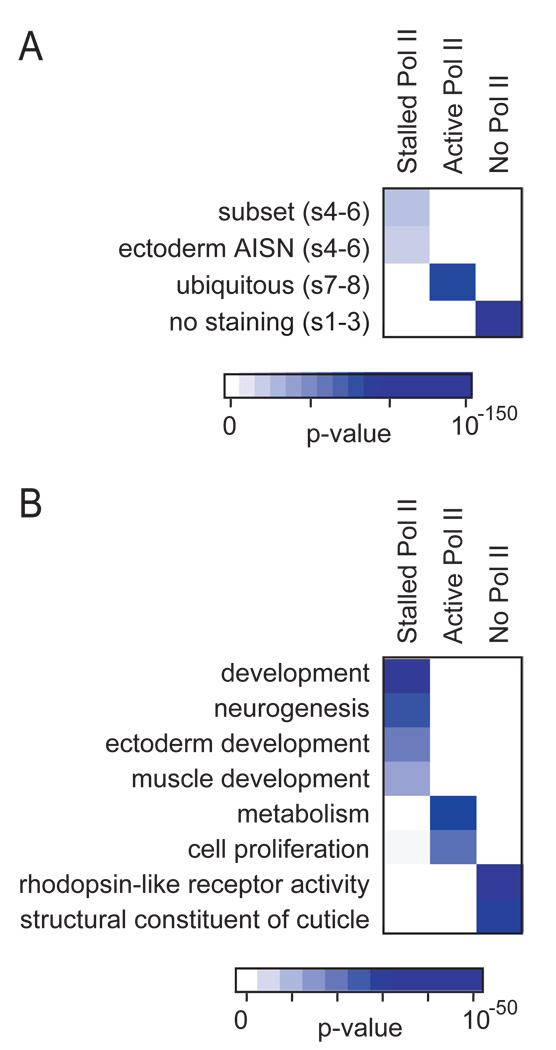
There are significant differences in the expression and functions of genes in the active, stalled or no Pol II classes based on in situ expression patterns (ImaGO)27 and functional annotations (GO)28 (Fig. 4). The set of genes with stalled Pol II is highly enriched for developmentally regulated genes, particularly those expressed in ectodermal and neuronal precursor cells (Fig. 4A). Consistent with the in situ hybridization data, genes with stalled Pol II are highly enriched for functions in development, including neurogenesis, ectoderm development and muscle differentiation (Fig. 4B). Many of these genes encode sequence-specific transcription factors (Hox, T-box, bHLH, zinc fingers, and HMG) and components of cell signaling pathways (FGF, Wnt, Notch, EGF, TGFβ, JNK, and TNF) (see Supplemental Materials).
Fig. 4. Functional analysis of the three classes of genes.
Representative categories of enrichment among genes with stalled Pol II, active Pol II or no Pol II for gene sets are shown for the (A) ImaGO database, which contains the in situ expression patterns of a substantial fraction of all protein coding genes in the Drosophila genome and (B) the gene sets from the Biological Process categories in the Gene Ontology (GO) database. The scale bars below indicate the significance of each test (hypergeometric distribution). Genes containing stalled Pol II (green) are significantly enriched for genes expressed in a subset of cells and in those of developing ectoderm (AISN = anlage in statu nascendi) at the time of the analysis (120–240 min). The stage of the enriched categories is indicated in parenthesis (s1–3 = ~0–100 min, s4–6 = ~100–200 min, s7–8 = ~200–250 min). The genes expressed as subset (s4–6) are largely identical to those of the ectoderm AISN (s4–6) category because mesoderm, neurectoderm and dorsal ectoderm are specified at that stage. Functional analysis confirms that genes with stalled Pol II are enriched for genes with roles in development, in particular those required for neurogenesis, ectoderm differentiation, and muscle development. Genes displaying active Pol II (red) are enriched for genes that display ubiquitous expression in developing embryos. They are enriched for functions that mediate cell proliferation and metabolic functions such as protein and nucleotide metabolism. Genes lacking Pol II (grey) tend to be inactive during embryogenesis, and deployed at later stages of the life cycle such as for cuticle function and vision.
In contrast, the set of genes with uniform Pol II binding is highly enriched for ubiquitously expressed genes (Fig. 4A), which function mostly in metabolism and cell proliferation (Fig. 4B). The set of genes that lacks Pol II binding is highly enriched in genes that show no staining in whole-embryo in situ hybridizations, confirming that they are not expressed during early embryogenesis (Fig. 4A). Many of these genes have functions in adult cells such as cuticle proteins or proteins required for vision (Fig. 4B).
Pol II stalling could reflect two nonexclusive developmental functions. It could be indicative of active transcriptional repression, or prepare genes for activation at later stages of embryogenesis. The second model is particularly attractive since Pol II stalling has already been shown to prepare heat shock genes for rapid activation4,22. We found evidence for both models.
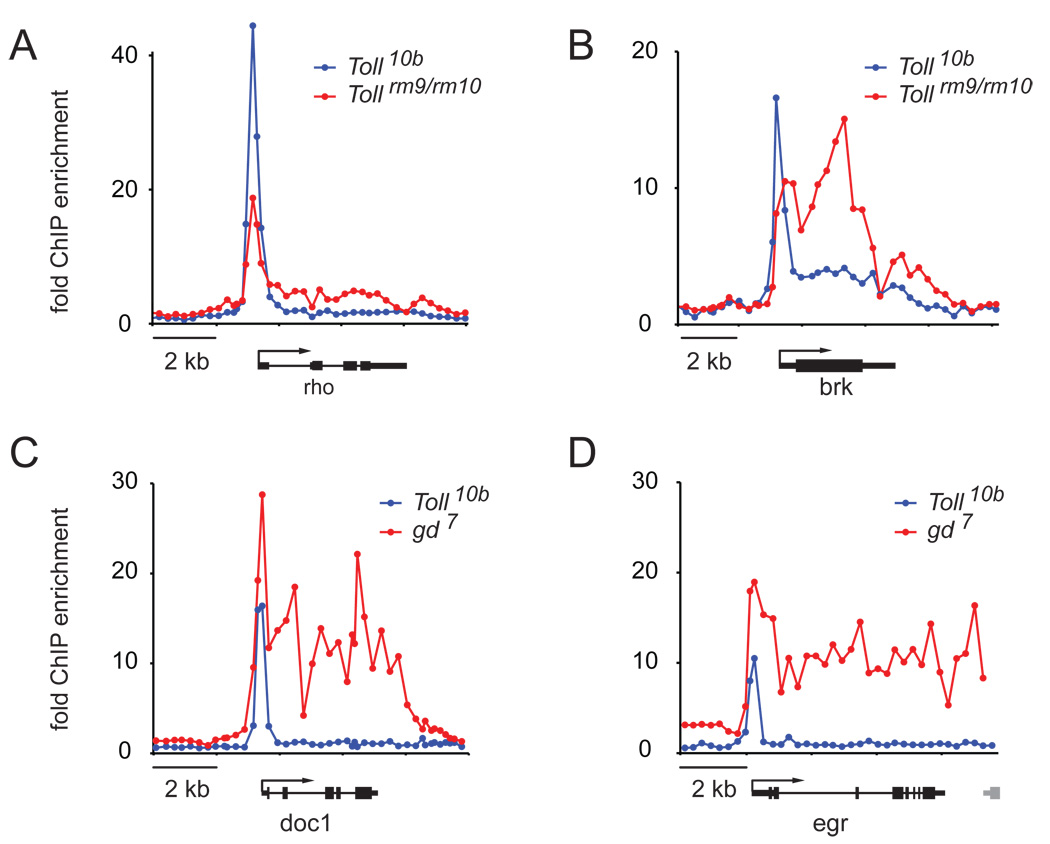
Pol II stalling is particularly prevalent among genes expressed in the neuroectoderm and dorsal ectoderm, which are repressed in Toll10b embryos. To test whether Pol II stalling is specific for repressed genes, we examined the Pol II profile of repressed genes in mutant embryos in which these genes are active. For this, we used two well defined mutants, Toll rm9/rm10 and gd7 (2–4 h), in which cells adopt neurectodermal and dorsal fate, respectively15,19,20,29. Indeed, at these genes, Pol II is redistributed into the transcription unit in these mutants (Fig. 5) and some genes now display the active Pol II profile (Fig. 5B–D). These results demonstrate that Pol II stalling is associated with cell-type specific repression and thus is subject to dynamic changes during development.
Fig. 5. Pol II profile comparison of genes in the repressed vs. active state.
Genes are shown that are repressed in Toll10b embryos but are active in either Tollrm9/Tollrm10 embryos (A, B) or gd7 embryos (C,D) at 2–4 h after fertilization, were examined by Pol II ChIP assays. The enrichment ratios of Pol II are shown on the y-axis: active state (red) and repressed state (blue). The results show that the degree of Pol II stalling is dependent on the gene’s activity, with some genes showing a complete switch between the stalled and active form of Pol II.
Previous studies have shown that the repression of a large set of genes in Toll10b embryos depends on Snail, a well-studied repressor that is constitutively expressed in Toll10b embryos but not in Toll rm9/rm10 and gd7 embryos16,17,19–21,30. We found a statistically significant association between repression by Snail and Pol II stalling (Supplemental Materials). For example, among the 139 genes that are occupied by Snail21 and display reduced expression in the Toll10b mutant19, 54% exhibit stalled Pol II, while only 19% of all genes with reduced expression display Pol II stalling (p < 10−23, Supplemental Materials). This suggests that Pol II stalling in Toll10b embryos may be regulated by Snail. A role for developmental repressors in regulating Pol II stalling is also consistent with a recent study of Drosophila segmentation14.
Multiple lines of evidence suggest that Pol II stalling also occurs at genes that are poised for activation in older embryos. Genes with stalled Pol II are highly over-represented among genes that are rapidly induced within 12 hours after the time-frame of our analysis (p < 10−27, Supplemental Materials). Moreover, genes with stalled Pol II are enriched for genes expressed in the derivatives of the mesoderm precursors present in Toll10b mutants, such as the developing heart and muscle cells (p < 10−15, Supplemental Materials). These genes, such as Drop and bagpipe, are not yet activated at the timeframe of the analysis31,32, but nonetheless show high levels of Pol II near the transcription start site (Supplemental Materials).
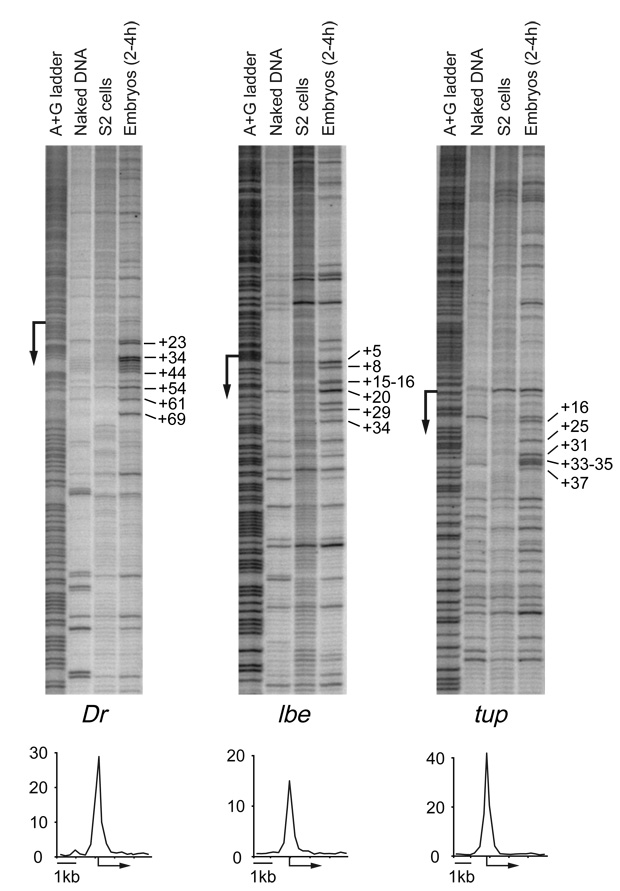
To confirm that muscle genes indeed show stalled Pol II before activation, we performed permanganate assays on wild-type Drosophila embryos at 2–4 hours after fertilization (Fig. 6). Drop and ladybird show a clear permanganate footprint downstream of transcription. These footprints were specific to the early embryo stage, since S2 cells, a cell line derived from older embryos, did not show a permanganate footprint under the same conditions (Fig. 6). These results confirm that Pol II stalling is dynamically regulated and suggest that one of its functions is to prepare genes for activation.
Fig. 6. Pol II stalling at genes prior to activation.
In vivo permanganate footprints were performed on the genes Drop (Dr), ladybird (lbe) and tail-up (tup) using early wild-type embryos (2–4 h) and S2 cells (107 cells), a cell line derived from older embryos. Positions of A+G (lane 1) and the transcription start site (TSS, arrow) are shown for orientation. T residues sensitive to KMnO4 treatment are shown for naked DNA as control (lane 2), S2 cells (lane 3) and early embryos (lane 4). Hypersensitive T residues in the early embryo sample (selected positions marked on the right) indicate an open transcription bubble. The footprints found at the muscle regulating genes Dr and lbe are similar to the one of tup, a gene known to be repressed at that stage. These results show that Pol II is found in a stalled form at the Dr and lbe genes prior to activation during embryonic development.
Our genome-wide analysis revealed that genes in Drosophila embryos are found in three distinct dynamic states: active, stalled or no Pol II. Stalled Pol II is particularly associated with developmental genes that are repressed and poised for activation. We propose that Pol II stalling allows genes to rapidly respond to developmental signals and thus facilitates the dynamic temporal and spatial expression patterns of developmental control genes.
Materials and Methods
Drosophila stock
Toll10b is a dominant gain-of-function mutation of the maternal gene Toll15. Embryos were collected from Toll10b/+ females obtained directly from the balanced stock (Toll10b/TM3 Sb Ser and Toll10b/OR60). Toll rm9 and Toll rm10 are recessive Toll mutations15 and embryos were collected from Toll rm9 /Toll rm9 females. gd7 embryos were collected from gd7/gd7 females29. Wild-type embryos were white11,18.
ChIP-chip assays
The chromatin immunoprecipitation (ChIP) experiments coupled to micrroarrays (chip) were performed as described21. The protocol can be downloaded from http://web.wi.mit.edu/young/pol2/
Antibodies
The antibodies used were 8WG16 and H14 (see Supplemental Materials for further information).
Arrays
We used Drosophila whole-genome tiling arrays printed by Agilent as described21. Probes of 60mers span the entire eukaryotic portion of the Drosophila melanogaster genome. While the spacing of these probes is ~280 bp on average, an additional probe is present between the two probes that flank each known TSS. Thus, the resolution around transcriptional start sites is ~ 140 bp. The data are available from ArrayExpress and our web site http://web.wi.mit.edu/young/pol2/.
Data analysis
We used the Rosetta error model to control for noise at probes, thus a probe required a p-value < 0.001. We did not use our previous algorithms for detecting bound probes and then assigning genes. Rather, we calculated parameters indicating Pol II enrichment directly for each gene (see Supplementary Materials for a detailed description). A combination of Pol II enrichment at the start site and median enrichment across the gene were used to classify Pol II as either absent, stalled or active.
Statistics
For the identification of gene sets that are over-represented in the three classes of genes, we used the hypergeometric distribution test (see Supplemental Materials for further information).
Permanganate footprint assays
Transcription bubble assays with KMnO4 were performed as described previously5,14. Embryos were collected 2–4-h AED (after egg deposition), dechorionated and partially homogenized before treatment with KMnO4. Embryos were treated with 20 mM or 40 mM KMnO4 for 60s on ice. The transcription start sites of the examined genes were identified and confirmed using ESTs in Flybase and previous expression analysis using tiling arrays20. The linker primers and gene-specific primers used for ligation-mediated PCR are listed in Supplemental Materials.
Supplementary Material
Acknowledgment
We would like to thank Robert Zinzen for collecting the Toll10b, Tollrm9/rm10 and gd7 embryos, Tom Volkert and Jennifer Love for microarray experimental support, and members of the Young lab for critical reading of the manuscript. This research was supported in part by the Intramural Research Program of the NIH, National Institute of Environmental Health Sciences (K.A.), by NIH grants HG002668 and GM069676 to R.A.Y., GM46638 to M.L., a grant by the Moore Foundation and a postdoctoral fellowship by Schering (A.S.). R.A.Y. consults for Agilent Technologies.
Footnotes
Author Contributions
J.Z. and M.L. designed the experiment. J.Z. designed the arrays and carried out the experiments and analysis. A.S., M.K. and J.Z. analyzed expression data and functional categories, J.-W. H., S. N. and K. A. carried out the permanganate footprint assays, J.Z., M.L. and R.A.Y. prepared the manuscript.
References
- 1.Levine M, Davidson EH. Gene regulatory networks for development. Proc Natl Acad Sci U S A. 2005;102:4936–4942. doi: 10.1073/pnas.0408031102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ptashne M. Regulation of transcription: from lambda to eukaryotes. Trends Biochem Sci. 2005;30:275–279. doi: 10.1016/j.tibs.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 3.Conaway JW, Shilatifard A, Dvir A, Conaway RC. Control of elongation by RNA polymerase II. Trends Biochem Sci. 2000;25:375–380. doi: 10.1016/s0968-0004(00)01615-7. [DOI] [PubMed] [Google Scholar]
- 4.Saunders A, Core LJ, Lis JT. Breaking barriers to transcription elongation. Nat Rev Mol Cell Biol. 2006;7:557–567. doi: 10.1038/nrm1981. [DOI] [PubMed] [Google Scholar]
- 5.Gilmour DS, Lis JT. RNA polymerase II interacts with the promoter region of the noninduced hsp70 gene in Drosophila melanogaster cells. Mol Cell Biol. 1986;6:3984–3989. doi: 10.1128/mcb.6.11.3984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bender TP, Thompson CB, Kuehl WM. Differential expression of c-myb mRNA in murine B lymphomas by a block to transcription elongation. Science. 1987;237:1473–1476. doi: 10.1126/science.3498214. [DOI] [PubMed] [Google Scholar]
- 7.Strobl LJ, Eick D. Hold back of RNA polymerase II at the transcription start site mediates down-regulation of c-myc in vivo. Embo J. 1992;11:3307–3314. doi: 10.1002/j.1460-2075.1992.tb05409.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Krumm A, Meulia T, Brunvand M, Groudine M. The block to transcriptional elongation within the human c-myc gene is determined in the promoter-proximal region. Genes Dev. 1992;6:2201–2213. doi: 10.1101/gad.6.11.2201. [DOI] [PubMed] [Google Scholar]
- 9.Laspia MF, Wendel P, Mathews MB. HIV-1 Tat overcomes inefficient transcriptional elongation in vitro. J Mol Biol. 1993;232:732–746. doi: 10.1006/jmbi.1993.1427. [DOI] [PubMed] [Google Scholar]
- 10.Cheng C, Sharp PA. RNA polymerase II accumulation in the promoter-proximal region of the dihydrofolate reductase and gamma-actin genes. Mol Cell Biol. 2003;23:1961–1967. doi: 10.1128/MCB.23.6.1961-1967.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aiyar SE, et al. Attenuation of estrogen receptor alpha-mediated transcription through estrogen-stimulated recruitment of a negative elongation factor. Genes Dev. 2004;18:2134–2146. doi: 10.1101/gad.1214104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aida M, et al. Transcriptional pausing caused by NELF plays a dual role in regulating immediate-early expression of the junB gene. Mol Cell Biol. 2006;26:6094–6104. doi: 10.1128/MCB.02366-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Radonjic M, et al. Genome-wide analyses reveal RNA polymerase II located upstream of genes poised for rapid response upon S. cerevisiae stationary phase exit. Mol Cell. 2005;18:171–183. doi: 10.1016/j.molcel.2005.03.010. [DOI] [PubMed] [Google Scholar]
- 14.Wang X, Lee C, Gilmour DS, Gergen JP. Transcription elongation controls cell fate specification in the Drosophila embryo. Genes Dev. 2007;21:1031–1036. doi: 10.1101/gad.1521207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schneider DS, Hudson KL, Lin TY, Anderson KV. Dominant and recessive mutations define functional domains of Toll, a transmembrane protein required for dorsal-ventral polarity in the Drosophila embryo. Genes Dev. 1991;5:797–807. doi: 10.1101/gad.5.5.797. [DOI] [PubMed] [Google Scholar]
- 16.Leptin M. twist and snail as positive and negative regulators during Drosophila mesoderm development. Genes Dev. 1991;5:1568–1576. doi: 10.1101/gad.5.9.1568. [DOI] [PubMed] [Google Scholar]
- 17.Kosman D, Ip YT, Levine M, Arora K. Establishment of the mesoderm-neuroectoderm boundary in the Drosophila embryo. Science. 1991;254:118–122. doi: 10.1126/science.1925551. [DOI] [PubMed] [Google Scholar]
- 18.Furlong EE, Andersen EC, Null B, White KP, Scott MP. Patterns of gene expression during Drosophila mesoderm development. Science. 2001;293:1629–1633. doi: 10.1126/science.1062660. [DOI] [PubMed] [Google Scholar]
- 19.Stathopoulos A, Van Drenth M, Erives A, Markstein M, Levine M. Whole-genome analysis of dorsal-ventral patterning in the Drosophila embryo. Cell. 2002;111:687–701. doi: 10.1016/s0092-8674(02)01087-5. [DOI] [PubMed] [Google Scholar]
- 20.Biemar F, et al. Comprehensive identification of Drosophila dorsal-ventral patterning genes using a whole-genome tiling array. Proc Natl Acad Sci U S A. 2006;103:12763–12768. doi: 10.1073/pnas.0604484103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zeitlinger J, et al. Whole-genome ChIP-chip analysis of Dorsal, Twist, and Snail suggests integration of diverse patterning processes in the Drosophila embryo. Genes Dev. 2007;21:385–390. doi: 10.1101/gad.1509607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lis J, Wu C. Protein traffic on the heat shock promoter: parking, stalling, and trucking along. Cell. 1993;74:1–4. doi: 10.1016/0092-8674(93)90286-y. [DOI] [PubMed] [Google Scholar]
- 23.Reppas NB, Wade JT, Church GM, Struhl K. The transition between transcriptional initiation and elongation in E. coli is highly variable and often rate limiting. Mol Cell. 2006;24:747–757. doi: 10.1016/j.molcel.2006.10.030. [DOI] [PubMed] [Google Scholar]
- 24.Agalioti T, et al. Ordered recruitment of chromatin modifying and general transcription factors to the IFN-beta promoter. Cell. 2000;103:667–678. doi: 10.1016/s0092-8674(00)00169-0. [DOI] [PubMed] [Google Scholar]
- 25.Soutoglou E, Talianidis I. Coordination of PIC assembly and chromatin remodeling during differentiation-induced gene activation. Science. 2002;295:1901–1904. doi: 10.1126/science.1068356. [DOI] [PubMed] [Google Scholar]
- 26.Markstein M, Markstein P, Markstein V, Levine MS. Genome-wide analysis of clustered Dorsal binding sites identifies putative target genes in the Drosophila embryo. Proc Natl Acad Sci U S A. 2002;99:763–768. doi: 10.1073/pnas.012591199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tomancak P, et al. Systematic determination of patterns of gene expression during Drosophila embryogenesis. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-12-research0088. research0088.1-0088.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ashburner M, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang AM, Rusch J, Levine M. An anteroposterior Dorsal gradient in the Drosophila embryo. Genes Dev. 1997;11:1963–1973. doi: 10.1101/gad.11.15.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Boulay JL, Dennefeld C, Alberga A. The Drosophila developmental gene snail encodes a protein with nucleic acid binding fingers. Nature. 1987;330:395–398. doi: 10.1038/330395a0. [DOI] [PubMed] [Google Scholar]
- 31.Sandmann T, et al. A temporal map of transcription factor activity: mef2 directly regulates target genes at all stages of muscle development. Dev Cell. 2006;10:797–807. doi: 10.1016/j.devcel.2006.04.009. [DOI] [PubMed] [Google Scholar]
- 32.Furlong EE. Integrating transcriptional and signalling networks during muscle development. Curr Opin Genet Dev. 2004;14:343–350. doi: 10.1016/j.gde.2004.06.011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.