Abstract
Aicardi-Goutières syndrome (AGS), Systemic Lupus Erythematosus (SLE), Familial Chilblain Lupus (FCL) and Retinal Vasculopathy and Cerebral Leukodystrophy (RVCL) {a new term encompassing three independently described conditions with a common etiology—Cerebroretinal Vasculopathy (CRV), Hereditary Vascular Retinopathy (HVR) and Hereditary Endotheliopathy, Retinopathy and Nephropathy (HERNS)}—have previously been regarded as distinct entities. However, recent genetic analysis has demonstrated that each of these diseases maps to chromosome 3p21 and can be caused by mutations in TREX1, the major human 3'–5' exonuclease. In this review, we discuss the putative functions of TREX1 in relationship to the clinical, genetic and functional characteristics of each of these conditions.
Keywords: TREX1, TREX2, DNase III, stroke, cerebrovascular disease
Introduction
Recently, mutations in the ubiquitously expressed human 3'–5' exonuclease TREX1 (DNase III) have been linked to four apparently independent diseases. In the case of Cerebroretinal Vasculopathy (CRV), Hereditary Endotheliopathy, Retinopathy and Nephropathy (HERNS) and Hereditary Vascular Retinopathy (HVR), a common etiology was first suspected based on clinical similarities and further supported when all showed evidence of linkage to a single locus on chromosome 3p21.1 We now know that these diseases consolidate to a single autosomal dominant inherited entity named Retinal Vasculopathy and Cerebral Leukodystrophy (RVCL) in which there are mutations affecting the carboxyl-terminus of TREX1.2 The other three diseases, Aicardi-Goutières syndrome (AGS), Systemic Lupus Erythematosus (SLE) and Familial Chilblain Lupus (FCL), share some clinical similarity but appear to be distinct clinical conditions. AGS, a severe, usually lethal disease, resembling an intrauterine viral infection has been associated with recessive mutations in TREX1 that impair its exonuclease activity.3 FCL is a rare, inherited form of lupus with prominent skin manifestations in which autosomal dominant mutations in TREX1 that decrease exonuclease activity have been described.4,5 Mutations in TREX1 have also been identified in ~3% of patients with SLE, a complex disease with diverse, systemic manifestations predominantly affecting women of child-bearing age.6 The goal of this review is to collate the existing information on these genetically related diseases with an emphasis on how the mutations in TREX1 lead to the disease state.
Nucleases Role in Cell Biology
Deoxyribonucleases (DNA nucleases) are essential to maintain genome stability and are involved in processes such as DNA replication, repair and recombination.7 These enzymes can be divided into two classes: endonucleases that hydrolyse the deoxyribose phosphodiester backbone within the DNA strand and exonucleases that hydrolyse the phosphodiester bonds at the DNA ends. Nucleases have selective affinity for single-stranded (ss) or double-stranded (ds) DNA. They differ in their mode of action (5'–3' or 3'–5' direction) and their main reaction products (5' mono- or dinucleotides and 3' mononucleotides).8
DNA replication during mitosis follows a complex sequence of events in which polymerases are responsible for the accurate duplication of the parental chromosomes. In mammalian cells, the estimated spontaneous mutation rate is 10−10–10−12 per cell division.9 This is, however, orders of magnitudes lower than the mutation rate of DNA polymerases such as Pol α and Pol β (10−4–10−5).10 They are not accurate enough to replicate our 3 billion base pair genome without deleterious consequences. Such a high mutation rate would be incompatible with life and is why some DNA polymerases (e.g., Pols γ, δ and ε) contain additional 3'–5' exonuclease activity. This “intrinsic” proofreading activity enables the polymerases to enhance the accuracy of DNA synthesis by removing incorrectly incorporated nucleotides before the replication process is reinitiated.
A second class of exonucleases is considered autonomous. These enzymes can hydrolyse their target sequences independently and may also assist DNA polymerases lacking this activity (e.g., Pol α) to increase their fidelity under normal conditions or in cases of genotoxic cell stress.9 TREX1 is the most abundant DNA 3'–5' exonuclease in mammalian cells.8,11
TREX1 Genetics and Structure
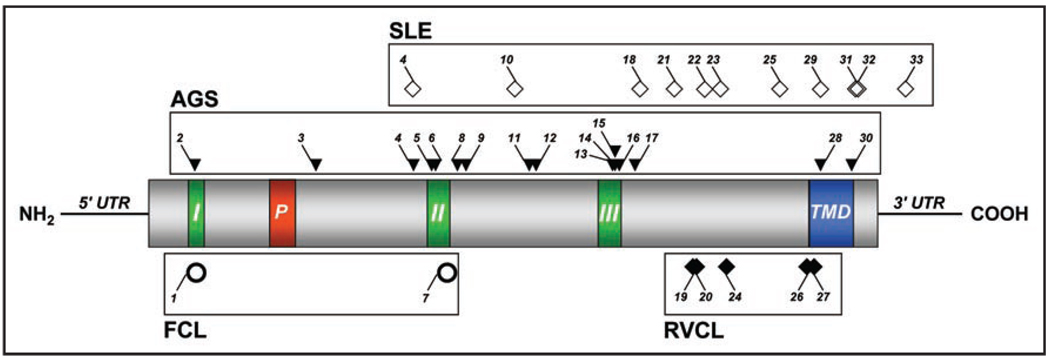
TREX1 (former DNase III, Three prime Repair EXonuclease) was identified in 1999 by Hoss et al.12 and Mazur and Perrino.13 The gene for TREX1 consists of a single exon and encodes a protein of 314 amino acids. Sequence homology places TREX1 in the DnaQ 3'–5' exonuclease family.14–16 The characteristic features of this family of exonucleases are three conserved sequence motifs, Exo I, Exo II and Exo III, which form the active site of the enzyme14–16 (Fig. 1). Recent crystal structures of murine Trex1 with DNA17 demonstrate a dimer with the active sites on opposing surfaces, allowing the potential for concurrent interaction with two 3' DNA ends. These structures demonstrate close similarity with another DnaQ 3'–5' exonuclease, the Escherichia coli DNA polymerase I. In addition to these three exonuclease motifs, TREX1 has a highly hydrophobic carboxyl-terminal region which is predicted to form a transmembrane helix.6,18 Deletion mutagenesis has demonstrated that this region is important in intracellular localization but has no role in the catalytic function.2,6 The TREX1 protein also contains a proline-rich sequence (PPII helix).17,18 This motif has been reported to play a crucial role in protein-protein interactions, specifically with Src homology 3, WW and EVH1 domains.19 The structure of TREX1 indicates that the PPII helix is surface exposed and available for protein interactions.17,18 This has been hypothesized to account for the interaction of TREX1 with the SET complex17 (see below).
Figure 1.
Schematic diagram of the TREX1 protein structure with sites of mutations associated with human disease. Numbers for each mutation correspond with those listed in Table 1. Regions I, II and III represent the exonuclease domains (Exo I–III). Region P represents the polyproline II helix (PPII). Region TMD represents the putative transmembrane domain.
TREX1 Functions
Elucidation of the definitive in vivo function of TREX1 has proved problematic. TREX1 is an autonomous non-processive 3'–5' DNA-specific exonuclease with a preference for ssDNA or mispaired 3' termini.12,13,20,21 Further analysis has suggested that, unusual for an exonuclease, TREX1 has a significant preference for particular DNA sequences and that this correlates with exonuclease activity.18 This exonuclease function, in addition to slight homology with known editing enzymes, suggests that it may serve a DNA-editing role in DNA replication or gap filling during DNA repair. However, this has not been borne out by an increase in spontaneous mutation rate or a higher cancer incidence in the Trex1 knockout mouse as would have been predicted if TREX1 served an obligatory role of editing mismatched 3' termini generated during DNA repair or DNA replication. Instead, the Trex1 knockout mouse displayed an autoimmune-like inflammatory myocarditis leading to a dilated cardiomyopathy and a dramatically reduced lifespan.22
Another role for TREX1 has been suggested by its association with the SET complex.23 This protein complex is involved in granzyme A-mediated cell death, a caspase-independent pathway which involves ssDNA damage. The killer lymphocyte associated protease, granzyme A, causes mitochondrial damage and superoxide generation that induces nuclear translocation of the SET complex. It then cleaves the NM23-H1 inhibitor, SET, freeing NM23-H1 to make a ssDNA cut (introduces a DNA nick) that is then extended by TREX1. Cells with silenced TREX1 are relatively resistant to apoptotic cell death but remain sensitive to the caspase-activating granzyme B.23
A role for TREX1 in cell homeostasis has recently been described by Yang and colleagues.24 They demonstrated that TREX1 deficiency results in constitutive activation of the ATM-dependent DNA damage checkpoint. This results in impaired G1/S transition in Trex1-deficient cells. Additionally, a 60–65 bp ssDNA species accumulates in the cytoplasm. Importantly, similar phenotypic observations were obtained from both Trex1-deficient mouse cells as well as from patient cells homozygous for a non-functional mutant form of the protein. These roles of TREX1 have been postulated to be critical in controlling autoimmunity24,25 and will be discussed later.
TREX1 versus TREX2
TREX1 has a homologue, TREX2, which has ~40% amino acid sequence identity with TREX1.13,26 TREX2 is also an autonomous DNA 3'–5' exonuclease,13,26 important for cell proliferation.26 TREX2 lacks the ~75 amino acid carboxyl-terminal hydrophobic domain found in TREX1. This region is responsible for intracellular localization and contains the non-repetitive proline-rich region which plays a crucial role in protein-protein interactions. TREX2 contains a conserved DNA binding loop positioned adjacent to the active site that has a sequence distinct from the corresponding loop in the TREX1 enzyme. These differences suggest non-overlapping physiological roles for these proteins.
Aicardi-Goutières Syndrome
In 1984, the autosomal recessive Aicardi-Goutières syndrome (AGS)(OMIM 225750) was first described in eight children from five unrelated families who developed progressive encephalopathy of early onset, brain atrophy, demyelination, basal ganglia calcifications and chronic cerebrospinal fluid (CSF) lymphocytosis.27 These clinical findings mimic those observed with intrauterine infections but evaluation for an infectious etiology was negative. Affected children typically present before 4 months of age with failure to progress in motor and social skills while one-third of cases present later, between 4 and 12 months of age, with loss of previously acquired motor and mental skills.28 Neurological manifestations also include spasticity and an acquired microcephaly. Extra-neurological features observed in a subset of patients include hepatosplenomegaly, anemia, thrombocytopenia, elevated liver transaminases and chilblains (ulcerating lesions on fingers, toes or ear lobes).28 Due to the similarity between AGS and intrauterine infection, levels of interferon α (IFNα) were measured and found to be elevated in the CSF of affected infants.29
Two other familial diseases, the microcephaly and intracranial calcification syndrome (MICS)30,31 and Cree Indian encephalitis,32 which were initially described as separate disorders, have considerable overlap with AGS. All three are inherited as autosomal recessive diseases, can have increased levels of IFNα in the CSF, and manifest various extra-neurological findings such as hepatosplenomegaly, thrombocytopenia, elevated liver transaminases and chilblains.33,34 Furthermore, Cree encephalitis has been found to be allelic with AGS.3,34 Thus, AGS, MICS and Cree encephalitis appear to represent the same disorder.
Several cases in the literature have also reported an overlap between AGS and infantile systemic lupus erythematosus (SLE).35–37 In addition to findings compatible with the diagnosis of AGS, the affected children had autoantibodies typically found in lupus with antigenic specificity for cardiolipin, ssDNA, dsDNA and RNA-protein complexes. Notably, neuro-lupus has also been associated with increased levels of IFNα in the CSF despite being a non-infectious disorder.38
The recent elucidation of the genetic basis for AGS provides a rationale for its clinical diversity. TREX1 mutations were first demonstrated to cause AGS by Crow et al. (AGS1, OMIM 225750).3 Functional characterization of only a few AGS associated TREX1 mutations have been performed3,5,13 (see Table 1); however, in the recessive mutations examined, a defect in exonuclease activity was demonstrated. This is further supported by Yang et al., who have demonstrated the presence of ssDNA in Trex1-null cells, findings also seen in AGS patient cells carrying homozygous mutations in TREX1.24
Table 1.
TREX1 mutations found in human disease
| # | Nucleotide change | Amino acid change | Exonuclease Function | Intracellular Localization | Disease | Segregation | Reference |
|---|---|---|---|---|---|---|---|
| 1 | 520→A | D18N | Decreased | NT | FCL | Het | 43 |
| 2 | 58_59insG | E20fs | NT | NT | AGS | Hom | 3 |
| 3 | 212_2l3dupTG | A72fs | NT | NT | AGS | Hom | 42 |
| 4 | 341G→A | R114H | Decreased | Perinuclear | AGS | Hom | 2, 3 |
| 4 | 341G→A | R114H | Decreased | Perinuclear | SLE | Het | 2, 6 |
| 5 | 365T→C | V122A | NT | NT | AGS | Hom | 42 |
| 6 | 366_368dupGGC | Al24ins | NT | NT | AGS | Hom | 42 |
| 7 | 375dupT | G126fs | Decreased | NT | FCL | Het | 5 |
| 8 | 393_408dup | E137fs | NT | NT | AGS | Hom | 42 |
| 9 | 397delC | L133fs | NT | NT | AGS | Hom | 42 |
| 10 | 473C→T | A158V | NT | NT | SLE | Het | 6 |
| 11 | 490C→T | R164X | NT | NT | AGS | Hom | 3 |
| 12 | 500delG | S167fs | NT | NT | AGS | Hom | 42 |
| 13 | 598G→A | D200N | Normal | NT | AGS | Het | 5 |
| 14 | 600_60linsGAT | D201ins | Decreased | NT | AGS | Comp het, R114H | 3, 17 |
| 15 | 602T→A | V201D | Decreased | NT | AGS | Hom | 3, 17 |
| 16 | 609_662dup | A203ins | NT | NT | AGS | Hom | 42 |
| 17 | 625_628dupCAGT | W210fs | NT | NT | AGS | Hom | 42 |
| 18 | 634delC | P212fs | NT | Cytoplasm | SLE | Het | 6 |
| 19 | 703_704insG | V235fs | Normal | Nucleus/Cytoplasm | RVCL | Het | 2 |
| 20 | 706_707insA | T236fs | Normal | Nucleus/Cytoplasm | RVCL | Het | 2 |
| 21 | 679G→A | G227S | NT | NT | SLE | Comp het, A247P? | 6 |
| 22 | 720G→C | R240S | NT | NT | SLE | Het | 6 |
| 23 | 739G→C | A247P | NT | NT | SLE | Comp het, G227S? | 6 |
| 24 | 742_745dupGTCA | T249fs | NT | Nucleus/Cytoplasm | RVCL | Het | 2 |
| 25 | 812_8l3insAA | P272fs | Normal | Nucleus | SLE | Het | 6 |
| 26 | 850_851insA | R284fs | NT | NT | RVCL | Het | 2 |
| 27 | 857_858insG | R287fs | NT | Nucleus/Cytoplasm | RVCL | Het | 2 |
| 28 | 868_885del | P290del | NT | NT | AGS | Hom | 42 |
| 29 | 869C→T | P290L | NT | NT | SLE | Het | 6 |
| 30 | 907A→C | T303P | NT | NT | AGS | Hom | 42 |
| 31 | 914A→G | Y305C | NT | NT | SLE | Het | 6 |
| 32 | 917G→C | G306A | NT | NT | SLE | Het | 6 |
| 33 | 979delC | 3' UTR | NT | NT | SLE | Het | 6 |
Numbers correspond to those shown in Figure 1. NT: Not tested.
Although AGS is classically inherited in an autosomal recessive manner, there is an isolated report of an individual with an autosomal dominant form of the disease caused by a TREX1 mutation (AGS5; OMIM 610905).5 This individual possessed a D200N mutation which, on functional analysis, demonstrated close to normal levels of exonuclease activity. Although the mechanism of action remains to be established, it is proposed that this mutant alters the specificity of TREX1 or interferes with protein-protein interactions.
In addition to mutations in TREX1, mutations in three other genes [RNASEH2A (AGS4, OMIM 606034); RNASEH2B (AGS2, OMIM 610326); RNASEH2C (AGS3, OMIM 610330)] have been reported to cause AGS.39 RNase H2 is the principal source of ribonuclease activity in the cell,40,41 however, the mechanism through which a reduction in ribonuclease activity leads to disease remains speculative. Increased amounts of RNA-DNA hybrids stimulating an innate immune response with overproduction of IFNα have been hypothesized. In addition to these four causative genes, additional genes responsible for AGS are suggested by a cohort of affected individuals in whom mutations have not been identified.42
Analysis of a large cohort of AGS has revealed genotype:phenotype correlations in the disease.42 For instance, individuals with TREX1 mutations tend to present at birth while individuals with mutations in RNASEH2B present later. AGS caused by RNASEH2B mutations also seems to have a milder phenotype with a lower mortality and relatively preserved intellectual function.42
Familial Chilblain Lupus
Familial chilblain lupus (FCL, OMIM 610448) is a rare cutaneous form of SLE. It is an autosomal dominant disease in which affected members present in early childhood with painful bluish-red inflammatory cutaneous lesions, typically on fingers, toes, ear helices, nose and cheeks. These lesions worsen with cold or wet exposure. They usually heal without scarring but may ulcerate leading to atrophic and hypopigmented skin and, in severe cases, to destruction of interphalangeal joints and distal toes.4,5,43 Some affected individuals also have antinuclear antibodies and immune complexes.4,5,43 Progression to SLE is documented in 18% of individuals with chilblain lupus44 but has not yet been described in individuals with FCL.
A SNP-based genome-wide linkage analysis mapped FCL to chromosome 3p and a subsequent haplotype analysis narrowed the locus to 3p21-14, an area that includes the gene for TREX1.4 Although FCL is not associated with neurological pathology, there is some overlap with AGS in which some affected individuals develop chilblains and autoantibodies. Hence, TREX1 was considered a plausible candidate gene and sequencing of affected individuals revealed several mutations.
Rice et al.5 reported a family with compound heterozygous mutations in three siblings (c.375dupT/F17S), though the disease segregated with only the c.375dupT mutation which was assumed to be inherited from the unstudied affected father. The c.375dupT mutation resulted in a truncated protein missing the last 188 amino acids, which would be predicted to be functionally significant. The mother carrying the F17S mutation was unaffected. This change is assumed to be a rare polymorphism. Exonuclease assays on lymphoblastoid cell lines derived from the affected individuals demonstrate decreased enzymatic activity.5
Lee-Kirsch et al.43 described a heterozygous mutation (D18N) in a family with FCL. Functional analysis of this mutation revealed a loss of exonuclease function. Further analysis demonstrated reduced sensitivity to granzyme A-mediated cell death in patient-derived lymphoblastoid cell lines.
Retinal Vasculopathy and Cerebral Leukodystrophy
Cerebroretinal Vasculopathy (CRV) is an inherited disorder first described by Grand et al., in 1988.45 It begins in middle age with predominant central nervous system, especially retinal, involvement. Study of eight patients spanning three generations in the initial pedigree showed 100% penetrance with an autosomal dominant mode of inheritance. The disease manifestations begin during the fourth or fifth decade and there is 100% mortality over a 5 to 10 year period secondary to progressive neurological decline. Typical ophthalmologic findings on retinal fluorescein angiograms are capillary dropouts, particularly in the macular region, leading to loss of central vision, prominent juxta-foveolar capillary obliteration and telangiectasias. Neurological manifestations commonly observed were transient ischemic attacks and strokes with motor and sensory loss, cognitive dysfunction, headaches, personality disorders, depression and anxiety. CT scans often show mass lesions with displacement of the surrounding structures and central contrast enhancement commonly in the frontoparietal region. Histopathology demonstrates coagulative necrosis secondary to an obliterative vasculopathy and minimal inflammatory infiltrate (“as if the brain had been radiated”). Autopsies demonstrate involvement of pons, cerebellum and basal ganglia in addition to the frontoparietal region.45 The CRV family has now been followed for over two decades (Atkinson JP, unpublished data). Hepatic and renal findings are not as clinically prominent as those seen in the nervous system but noteworthy from a clinical point of view in about one-third. Elevation of liver alkaline phosphatase is common and at autopsy, nodular regenerative hyperplasia is found. Renal dysfunction of a glomerular origin with proteinuria and elevation of creatinine is also observed. Renal histopathology is most suggestive of accelerated arteriolonephrosclerosis. Small vessel type gastrointestinal bleeding is also seen. Taken together, these data implicate small vessel vasculopathy leading to premature infarction and necrosis of the tissue.
A smaller family of Ashkenazi Jewish ancestry was next reported, in which the affected individuals had evidence of retinal vasculopathy on fluorescein angiograms and periventricular white matter lesions on brain MRI.46 No follow-up on this family was possible. However, in the original report45 there was also a patient of Ashkenazi Jewish origin with probable CRV whose disease has now been confirmed genetically.2 In 1999 and 2000, Weil et al.,47 and Niedermayer et al.,48 respectively reported two other families thought to have CRV.
In 1990, Storimans et al. published a preliminary report describing Hereditary Vascular Retinopathy (HVR), a syndrome of retinal vasculopathy, migraines and Raynaud’s phenomena in a Dutch kindred.49 It was further described by Terwindt et al., in 1998.50 As initially reported, these patients did not appear to have pseudotumors, renal dysfunction or shortened life expectancy similar to that seen in CRV patients. Furthermore, the visual acuity in these patients was largely preserved due to predominant peripheral retinal involvement. 50 However, further follow-up indicates that the clinical course is similar to the CRV and HERNS kindreds (see below; from Arn MJM van den Maagdenberg to JPA, personal communication).
In 1997, Jen et al., described a Chinese American family with 11 affected members spanning three generations who manifested a CRV-like illness.51 This group named the disease Hereditary Endotheliopathy with Retinopathy, Nephropathy and Stroke (HERNS). Ultrastructural studies showed distinctive multilaminated vascular basement membrane in the brain and other tissues, including the kidney, gastrointestinal tract and skin. Genetic analysis ruled out linkage to the cerebral autosomal dominant arteriopathy with subcortical infarcts and leukoencephalopathy (CADASIL) locus on chromosome 19.51
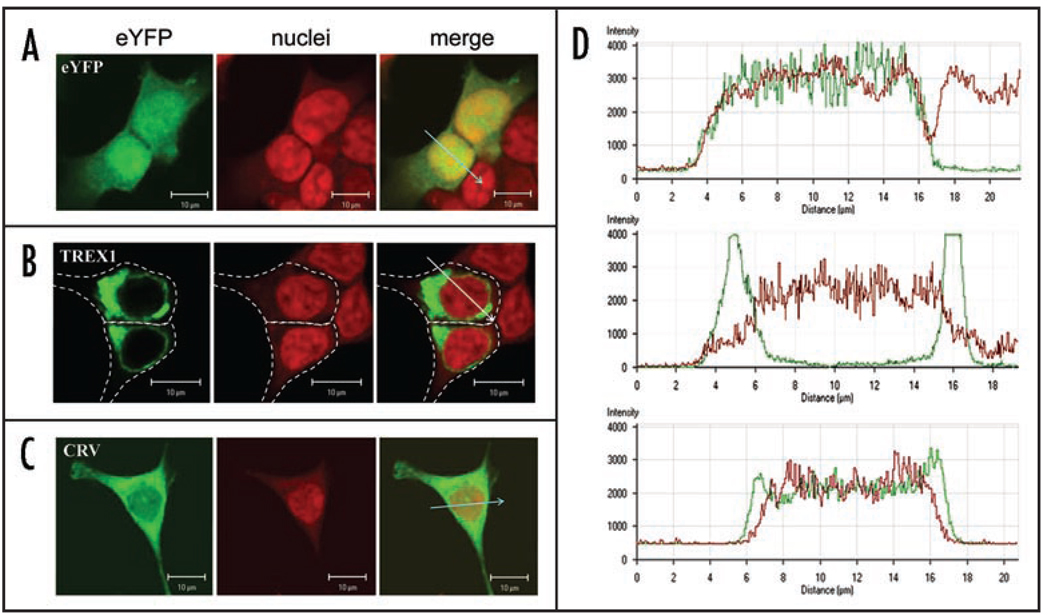
A whole genome screen in the extended Dutch family found probable linkage to 3p21.1-p21.3.1 Genetic analysis of patients from the CRV and HERNS kindreds demonstrated linkage to the same region. In 2005, Cohn et al., reported a family diagnosed with HERNS and prominent peripheral retinal involvement who also mapped to 3p21.1-p21.3.52 In 2007, CRV, HERNS and HVR were grouped together as Retinal Vasculopathy with Cerebral Leukodystrophy (RVCL, OMIM 192315) and the causative gene mutations were identified as carboxylterminus frame shifts in TREX1.2 These mutant TREX1 forms (lacking their native carboxyl-termini), no longer localize to their usual perinuclear site.2 Instead, they are now apparently capable of freely diffusing throughout the cell (Fig. 2C and corresponding intensity profiles D). Their distribution profile is indistinguishable from that of the fluorescent protein alone (Fig. 2A). Importantly, the exonuclease activity of these proteins is fully preserved. In contrast, fluorescent protein-tagged wild type TREX1 or TREX1 with deficient enzymatic activity are concentrated in the expected perinuclear space and are not detectable in the cytoplasm or nucleus (Fig. 2B).
Figure 2.
Functional consequences of TREX1 mutations associated with RVCL. Confocal microscopy of HEK293T cells showing transiently expressed yellow fluorescent protein (eYFP)-tagged TREX1 proteins (green), TOPRO3 staining of nuclei (red), and the overlay for eYFP alone (A), wild-type TREX1 (B), and the CRV mutant form of TREX1 (C) as well as the corresponding intensity profile for each across region of drawn arrow (D).
A recent report by Winkler et al., details a familial disease they call Hereditary Systemic Angiopathy (HSA) which bears a remarkable resemblance to RVCL.53 The affected individuals present in the fourth to fifth decade with visual disturbances, migraine-like headaches, and neurological symptoms including seizures, motor paresis and cognitive decline. As the disease progresses, some develop renal and/or hepatic dysfunction. Furthermore, the pathologic findings correlate with those seen in RVCL. Although the authors had included CRV, HVR and HERNS in their differential diagnosis as separate entities, it is now clear that all three represent the spectrum encompassing RVCL and HSA would also seem to fall within that spectrum.
Systemic Lupus Erythematosus and Sjogren’s Syndrome
Systemic Lupus Erythematosus (SLE) (OMIM152700) and Sjögren’s syndrome (SS) (OMIM 270150) are prototypes of autoimmune diseases because of the generation of a wide array of autoantibodies. SLE is clinically very heterogeneous and linkage studies and candidate gene studies have suggested many genes are involved in its pathogenesis (Table 1).54,55
As with AGS, SLE and SS are notable for the generation of antinuclear antibodies and an IFNα activation signature. Rice et al. reported that at least one parent of a patient with AGS had SLE.5 As a result of these similarities and the autoimmune phenotype of the Trex1 knockout mouse, Lee-Kirsch et al. sequenced the coding regions of TREX1 in lupus cohorts from the United Kingdom, Germany and Finland and discovered mutations in ~3% of individuals with SLE.6 Subsequent work also demonstrated TREX1 mutations in individuals with SS.6
Mutations in SLE were seen throughout the TREX1 gene. Although functional analysis of the missense changes was not performed, at least one of the mutants described in SLE had been previously reported in AGS, R114H. Functional assessment of this mutant demonstrated decreased exonuclease activity, establishing that at least some of the SLE mutants affect enzymatic function. 36 Other missense mutants, however, lie outside the catalytic domain and their functional significance remains unclear.6
Two frameshift mutations were also observed in SLE. Analogous to the mutations reported in RVCL, these mutations would not be predicted to disrupt enzymatic function. Functional analysis of one of these mutants, D272fs, failed to show any major enzymatic deficiency; however, as with all mutations seen in RVCL, the frameshift mutations in SLE also altered subcellular localization. Interestingly, the two mutants observed in SLE had different intracellular distributions: the P212fs mutant distributed throughout the cytoplasm in endosomal vesicles while the D272fs mutant was almost exclusively localized within the nucleus, possibly in association with subnuclear organelles.6
In a recent study by Hur et al., although no conclusive association between TREX1 polymorphisms and SLE was demonstrated, certain TREX1 polymorphisms were protective against the development of autoantibodies.56
Discussion
This review has described four independent conditions found to have a common underlying etiology through mutations in the major mammalian autonomous 3'–5' exonuclease TREX1. Although mutations in TREX1 are connected to all four conditions, there are differences with respect to the functional changes which allow some genotype:phenotype correlations to be defined (Table 2).
Table 2.
Clinical, pathological and laboratory features of diseases associated with TREX1 mutations
| Features | Retinal Vasculopathy with Cerebral Leukodystrophy (RVCL) |
Systemic Lupus Erythematosus (SLE) | Aicardi-Goutières Syndrome (AGS) | Familial Chilblain Lupus (FCL) |
|---|---|---|---|---|
| Inheritance | Autosomal Dominant | Polygenic Rare monogenic forms | Autosomal Recessive* | Autosomal Dominant |
| Genes | TREX1 | TREX1, DNASE1, HLA, FCGR2A/B, BLK, FCGR3A/B, CIQ, CIR, CIS, MBL, CRP, CR2, C2, C4, IRF5, TYK2, PTPN22, PDCDI, CTLA4, TLR5, TNFSF4, BANK1, ITGAM, KIAA1542 PXK, STAT4 | TREX1 (ACSI) RNASEH2A (AGS4) RNASEH2B (AGS2) RNASEH2C (AGS3) |
TREX1 |
| Onset | Between 30–50 years | Usually between 15–40 years | Usually by 4 months | Early childhood |
| Mortality | Usually within 10 years from time of onset | 5–20% 10 year mortality | Usually by 10 years of age | Non-lethal |
|
Neurological Manifestations |
||||
| Clinical | Strokes, pseudotumors, seizures, migraine-like headaches, motor/sensory/cerebellar deficits, personality changes, decrease in mental acuity | Strokes, seizures, psychosis, mood disorders, aseptic meningitis, transverse myelitis, mononeuritis multiplex, peripheral neuropathy, cognitive dysfunction | Failure to progress developmentally (or deterioration) in motor and social skills, peripheral spasticity, dystonic posturing, truncal hypotonia, seizures, acquired microcephaly | None reported |
| Radiological | Scattered areas of increased signal in deep white matter, enhancing lesions (often irregular) with mass effect and edema progressing to a multifocal process | Small white matter lesions, focal areas of infarction | Basal ganglia calcification, white matter hypodensities, progressive brain atrophy | N/A |
| Pathological | Localized areas of coagulative necrosis with fibrinoid necrosis of the walls of the vessels (resembling obliterative vasculopathy), multilayering of the basement membrane of capillaries | Immune-complex vasculitis (often with perivascular lymphocytic infiltrate without destruction of vessel wall), thrombotic occlusion of the blood vessels (APLS), necrotizing vasculitis with fibrinoid necrosis & neutrophilic infiltration (rare), accelerated atherosclerosis | Brain atrophy, wide-spread demyelination, calcification in white matter & basal ganglia, multiple small infarcts, small vessel proliferation with thickened adventitial and medial walls, astrocytosis | N/A |
|
Ophthalmologic Manifestations |
Predominant macular involvement, microaneurysms and telangiectasia, capillary dropout, progressive visual defects | Keratoconjunctivitis sicca, iritis, episcleritis, keratitis, retinal vasculitis, choroidopathy papillitis, ischemic optic neuropathy, retrobulbar optic neuritis | Reduced/absent vision in some, abnormal eye movements, optic atrophy/pale papillae, congenital glaucoma | None reported |
|
Other organ involvement |
Renal impairment, proteinuria, hematuria, micronodular cirrhosis, GI bleeding, anemia, Raynaud's phenomenon | Cutaneous rash, photosensitivity, oral/nasal ulcers, pleuritis pericarditis, GI dysmotility, pancreatitis, hepatitis, nephritis, pneumonitis, pulmonary hypertension & hemorrhage, arthritis, myositis, antiphospholipid antibody syndrome, Raynaud's phenomenon, cytopenias | Chilblains, anemia, thrombocytopenia, hepatosplenomegaly, intermittent sterile pyrexias, hypothyroidism, insulin dependent diabetes mellitus, scoliosis, cardiomegaly | Chilblains, large joint arthralgias |
|
Laboratory Abnormalities |
||||
| Autoantibodies | +/− | ++++ | +/− | + |
| Interferon-α | Unknown | ↑ | ↑ | Unknown |
| CSF | Elevated protein | Pleocytosis, elevated protein including IgG and oligoclonal bands | Chronic lymphocytosis | N/A |
One autosomal dominant case reported;4 ANA, antinuclear antibodies; APLS, antiphospholipid; CSF, cerebrospinal fluid; GI, gastrointestinal.
Homozygous TREX1 mutations cause the typical autosomal recessive form of AGS. Where functional analysis has been performed, these mutations result in decreased exonuclease function.3,5,17 Only one documented case of autosomal dominant AGS has been described in the literature. Although the reported mutation does not alter exonuclease activity in vitro, it may be non-functional in vivo.5
FCL is associated with heterozygous mutations in TREX1. In all individuals examined these mutations result in reduced exonuclease activity.5,43 Some heterozygous parents of children with AGS have been reported to present with chilblains following cold exposure.5 Although a much milder condition than AGS, FCL has sufficient phenotypic overlap (chilblain-like lesions and antinuclear antibodies) to suggest that the difference in these diseases may be a gene dosage effect. Therefore it appears that a partial loss of exonuclease activity is sufficient to cause FCL
RVCL is also associated with heterozygous mutations in TREX1. In contrast to the heterozygous mutations characterizing FCL, all the mutations described are in the carboxyl-terminus of TREX1 and disrupt the predicted transmembrane domain.6,18 They do not diminish the enzymatic function of TREX1 but alter its intracellular localization.2 We speculate that the phenotype seen in RVCL is due to loss of the carboxyl-terminus which results in dissemination of TREX1 throughout the cell. We hypothesize that this leads to a detrimental gain-of-function phenotype. Alternatively, the mutations could just result in insufficient quantities of TREX1 in the correct location to fulfill its physiological role. How the latter explanation would fit with the phenotypic differences seen between RVCL and FCL is unclear.
Heterozygous mutations in TREX1 have also been identified in a small number of SLE patients. Unlike the mutations in FCL which disrupt exonuclease function and RVCL which disrupt intracellular localization, the mutations observed in SLE are diverse. Some mutations disrupt exonuclease activity, others result in altered intracellular localization, and many are of unknown significance. SLE is a complex clinical disease and heterozygous mutations in TREX1 appear to account for only ~3% of cases.6 Detailed phenotypic examination of these individuals may result in a clearer understanding of the differences among RVCL, AGS and FCL.
These conditions also provide a window into a better understanding of the in vivo roles of TREX1. In the 10 years since TREX1 was discovered, there has been much speculation on the cellular function of this enzyme. Based on its involvement in these human diseases and from knockout animal studies, it seems that TREX1 does not have a requisite role in DNA repair. In contrast, it appears to be involved in the regulation of immunity through several non-mutually exclusive pathways.
Yang et al., have recently demonstrated that TREX1-deficient cells accumulate ~60 bp ssDNA species in the cytoplasm.24 Such DNA intermediates are not exclusively generated during DNA replication. For example, DNA viruses and retroviruses are additional sources of DNA species that could accumulate if a degrading enzyme is lacking. In the absence of TREX1, the DNA intermediates required for the viral replication cycle could accumulate and become immunostimulatory. A similar scenario applies to active endogeneous retroviruses (and retrotransposons) residing in the human genome.25
Cell surveillance for viral DNA and RNA is in the form of Toll-like receptors, some of which reside in the cytoplasm.57 Under normal physiological conditions host DNA is sequestered from these receptors in the nucleus or mitochondria. It is possible that the cytoplasmic ssDNA plays a pathogenic role by mimicking viral DNA and stimulating these receptors. This will result in the production of antiviral cytokines including IFNα.
It is intriguing that both SLE and AGS are associated with high levels of IFNα. SLE has long been associated with high serum IFNα levels58 and these levels correlate with disease activity and severity.58–60 Evidence that the raised IFNα levels may be important in disease pathogenesis comes from individuals treated with IFNα for malignancies and chronic hepatitis C who have been seen to develop autoimmune diseases including SLE.61,62 Mouse models of SLE also lend weight to a pathogenic role for IFNα. Administration of IFNα, either exogenously,63,64 through an adenovirus vector,65 or by injection of IFNα-inducing agents66 in mouse models of lupus have demonstrated increased severity of disease. Additionally, some lupus-prone mice lacking the Type I interferon receptor have a milder disease phenotype.67,68 A role for IFNα in the pathogenesis of AGS is suggested by the reproduction of the neuropathology in transgenic mice with astrocytes chronically producing IFNα who develop a progressive inflammatory encephalopathy, calcifications and neurodegeneration.69
Although no studies have examined IFNα in RVCL, several pieces of evidence may suggest such an association. Although distinct from the retinopathy of RVCL, there is a retinopathy associated with interferon that is characterized by cotton wool spots, retinal hemorrhages and microaneurysms.70–74 Migraine and Raynaud’s phenomenon are also side-effects of IFNα treatment and are seen in RVCL. If the pathogenic role of IFNα in RVCL can be confirmed, then this would at last provide some hope for a condition which currently has a grim prognosis. Chloroquine and glucocorticoids have been used in SLE and inhibit IFNα production75 and the IFN signature76 respectively. Humanized monoclonal anti-IFNα antibodies77 and soluble IFNα receptors78 may also become therapeutic options. However, further investigation is required to confirm a pathogenic role for IFNα in RVCL as IFNα may not be the only link between TREX1 mutations and immunity.
Yang et al. also demonstrated that Trex1-deficient cells had constitutive activation of the ATM-dependent DNA-damage checkpoint resulting in impaired G1/S transition.24 This has been hypothesized to impair T-lymphocyte development which may reduce their ability to regulate self tolerance.25 TREX1 has also been shown to be involved in granzyme A-mediated cell death. Granzyme A is released by cytotoxic T-cells and NK-cells. It has been demonstrated that granzyme A-mediated cell death is impaired in FCL43 which may lead to the retention of autoreactive lymphocytes resulting in disease.
Thus, the last two years have seen the remarkable discovery of mutations in one gene, TREX1, which are responsible for four distinct clinical diseases. They have, however, areas of clinical and genetic overlap which point to a common pathological mechanism. Further definition of the physiological and pathological role of TREX1 will hopefully lead to treatment advances for all these conditions.
Abbreviations
- AGS
aicardi-goutières syndrome
- SLE
systemic lupus erythematosus
- FCL
familial chilblain lupus
- RVCL
retinal vasculopathy and cerebral leukodystrophy
- CRV
cerebroretinal vasculopathy
- HVR
hereditary vascular retinopathy
- HERNS
hereditary endotheliopathy, retinopathy and nephropathy
- HSA
hereditary systemic angiopathy
References
- 1.Ophoff RA, DeYoung J, Service SK, et al. Hereditary vascular retinopathy, cerebroretinal vasculopathy and hereditary endotheliopathy with retinopathy, nephropathy and stroke map to a single locus on chromosome 3p21.1-p21.3. Am J Hum Genet. 2001;69:447–453. doi: 10.1086/321975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Richards A, van den Maagdenberg AM, Jen JC, et al. Truncations in the carboxyl-terminus of human 3'–5' DNA exonuclear TREX1 cause autosomal dominant retinal vasculopathy with cerebral leukodystrophy. Nat Genet. 2007;39:1068–1070. doi: 10.1038/ng2082. [DOI] [PubMed] [Google Scholar]
- 3.Crow YJ, Hayward BE, Parmar R, et al. Mutations in the gene encoding the 3’–5’ DNA exonuclease TREX1 cause Aicardi-Goutieres syndrome at the AGS1 locus. Nat Genet. 2006;38:917–920. doi: 10.1038/ng1845. [DOI] [PubMed] [Google Scholar]
- 4.Lee-Kirsch MA, Gong M, Schulz H, et al. Familial chilblain lupus, a monogenic form of cutaneous lupus erythematosus, maps to chromosome 3p. Am J Hum Genet. 2006;79:731–737. doi: 10.1086/507848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rice G, Newman WG, Dean J, et al. Heterozygous mutations in TREX1 cause familial Chilblain lupus and dominant Aicardi-Goutieres syndrome. Am J Hum Genet. 2007;80:811–815. doi: 10.1086/513443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee-Kirsch MA, Gong M, Chowdhury D, et al. Mutations in the gene encoding the 3'–5' DNA exonuclease TREX1 are associated with systemic lupus erythematosus. Nat Genet. 2007;39:1065–1067. doi: 10.1038/ng2091. [DOI] [PubMed] [Google Scholar]
- 7.Hubscher U, Maga G, Spadari S. Eukaryotic DNA polymerases. Annu Rev Biochem. 2002;71:133–163. doi: 10.1146/annurev.biochem.71.090501.150041. [DOI] [PubMed] [Google Scholar]
- 8.Lindahl T, Gally JA, Edelman GM. Properties of deoxyribonuclease 3 from mammalian tissues. J Biol Chem. 1969;244:5014–5019. [PubMed] [Google Scholar]
- 9.Shevelev IV, Hubscher U. The 3'–5' exonucleases. Nat Rev Mol Cell Biol. 2002;3:364–376. doi: 10.1038/nrm804. [DOI] [PubMed] [Google Scholar]
- 10.Drake JW, Charlesworth B, Charlesworth D, Crow JF. Rates of spontaneous mutation. Genetics. 1998;148:1667–1686. doi: 10.1093/genetics/148.4.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lindahl T. Excision of pyrimidine dimers from ultraviolet-irradiated DNA by exonucleases from mammalian cells. Eur J Biochem. 1971;18:407–414. doi: 10.1111/j.1432-1033.1971.tb01257.x. [DOI] [PubMed] [Google Scholar]
- 12.Hoss M, Robins P, Naven TJ, Pappin DJ, Sgouros J, Lindahl T. A human DNA editing enzyme homologous to the Escherichia coli DnaQ/MutD protein. EMBO J. 1999;18:3868–3875. doi: 10.1093/emboj/18.13.3868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mazur DJ, Perrino FW. Identification and expression of the TREX1 and TREX2 cDNA sequences encoding mammalian 3’→5’ exonucleases. J Biol Chem. 1999;274:19655–19660. doi: 10.1074/jbc.274.28.19655. [DOI] [PubMed] [Google Scholar]
- 14.Barnes MH, Spacciapoli P, Li DH, Brown NC. The 3'–5' exonuclease site of DNA polymerase III from gram-positive bacteria: definition of a novel motif structure. Gene. 1995;165:45–50. doi: 10.1016/0378-1119(95)00530-j. [DOI] [PubMed] [Google Scholar]
- 15.Strauss BS, Sagher D, Acharya S. Role of proofreading and mismatch repair in maintaining the stability of nucleotide repeats in DNA. Nucleic Acids Res. 1997;25:806–813. doi: 10.1093/nar/25.4.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Taft-Benz SA, Schaaper RM. Mutational analysis of the 3'→5' proofreading exonuclease of Escherichia coli DNA polymerase III. Nucleic Acids Res. 1998;26:4005–4011. doi: 10.1093/nar/26.17.4005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.de Silva U, Choudhury S, Bailey SL, Harvey S, Perrino FW, Hollis T. The crystal structure of TREX1 explains the 3' nucleotide specificity and reveals a polyproline II helix for protein partnering. J Biol Chem. 2007;282:10537–10543. doi: 10.1074/jbc.M700039200. [DOI] [PubMed] [Google Scholar]
- 18.Brucet M, Querol-Audi J, Serra M, et al. Structure of the dimeric exonuclease TREX1 in complex with DNA displays a proline-rich binding site for WW domains. J Biol Chem. 2007 doi: 10.1074/jbc.M700236200. [DOI] [PubMed] [Google Scholar]
- 19.Zarrinpar A, Bhattacharyya RP, Lim WA. The structure and function of proline recognition domains. Sci STKE. 2003;2003:8. doi: 10.1126/stke.2003.179.re8. [DOI] [PubMed] [Google Scholar]
- 20.Bebenek K, Matsuda T, Masutani C, Hanaoka F, Kunkel TA. Proofreading of DNA polymerase eta-dependent replication errors. J Biol Chem. 2001;276:2317–2320. doi: 10.1074/jbc.C000690200. [DOI] [PubMed] [Google Scholar]
- 21.Mazur DJ, Perrino FW. Structure and expression of the TREX1 and TREX2 3’→5’ exonuclease genes. J Biol Chem. 2001;276:14718–14727. doi: 10.1074/jbc.M010051200. [DOI] [PubMed] [Google Scholar]
- 22.Morita M, Stamp G, Robins P, et al. Gene-targeted mice lacking the Trex1 (DNase III) 3’→5’ DNA exonuclease develop inflammatory myocarditis. Mol Cell Biol. 2004;24:6719–6727. doi: 10.1128/MCB.24.15.6719-6727.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chowdhury D, Beresford PJ, Zhu P, et al. The exonuclease TREX1 is in the SET complex and acts in concert with NM23-H1 to degrade DNA during granzyme A-mediated cell death. Mol Cell. 2006;23:133–142. doi: 10.1016/j.molcel.2006.06.005. [DOI] [PubMed] [Google Scholar]
- 24.Yang YG, Lindahl T, Barnes DE. Trex1 Exonuclease Degrades ssDNA to Prevent Chronic Checkpoint Activation and Autoimmune Disease. Cell. 2007;131:873–886. doi: 10.1016/j.cell.2007.10.017. [DOI] [PubMed] [Google Scholar]
- 25.Coscoy L, Raulet DH. DNA mismanagement leads to immune system oversight. Cell. 2007;131:836–838. doi: 10.1016/j.cell.2007.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen MJ, Ma SM, Dumitrache LC, Hasty P. Biochemical and cellular characteristics of the 3'→5' exonuclease TREX2. Nucleic Acids Res. 2007;35:2682–2694. doi: 10.1093/nar/gkm151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Aicardi J, Goutieres F. A progressive familial encephalopathy in infancy with calcifications of the basal ganglia and chronic cerebrospinal fluid lymphocytosis. Ann Neurol. 1984;15:49–54. doi: 10.1002/ana.410150109. [DOI] [PubMed] [Google Scholar]
- 28.Goutieres F. Aicardi-Goutieres syndrome. Brain Dev. 2005;27:201–206. doi: 10.1016/j.braindev.2003.12.011. [DOI] [PubMed] [Google Scholar]
- 29.Goutieres F, Aicardi J, Barth PG, Lebon P. Aicardi-Goutieres syndrome: an update and results of interferon-alpha studies. Ann Neurol. 1998;44:900–907. doi: 10.1002/ana.410440608. [DOI] [PubMed] [Google Scholar]
- 30.Burn J, Wickramasinghe HT, Harding B, Baraitser M. A syndrome with intracranial calcification and microcephaly in two sibs, resembling intrauterine infection. Clin Genet. 1986;30:112–116. doi: 10.1111/j.1399-0004.1986.tb00578.x. [DOI] [PubMed] [Google Scholar]
- 31.Reardon W, Hockey A, Silberstein P, et al. Autosomal recessive congenital intrauterine infecction-like syndrome of microcephaly, intracranial calcification and CNS disease. American Journal Medical Genetics. 1994;52:58–65. doi: 10.1002/ajmg.1320520112. [DOI] [PubMed] [Google Scholar]
- 32.Black DN, Watters GV, Andermann E, et al. Encephalitis among Cree children in northern Quebec. J Med Genet. 1988;40:183–187. doi: 10.1002/ana.410240402. [DOI] [PubMed] [Google Scholar]
- 33.Sanchis A, Cervero L, Bataller A, et al. Genetic syndromes mimic congenital infections. J Pediatr. 2005;146:701–705. doi: 10.1016/j.jpeds.2005.01.033. [DOI] [PubMed] [Google Scholar]
- 34.Crow YJ, Black DN, Ali M, et al. Cree encephalitis is allelic with Aicardi-Goutieres syndrome: implications for the pathogenesis of disorders of interferon alpha metabolism. J Med Genet. 2003;40:183–187. doi: 10.1136/jmg.40.3.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dale RC, Tang SP, Heckmatt JZ, Tatnall FM. Familial systemic lupus erythematosus and congenital infection-like syndrome. Neuropediatrics. 2000;31:155–158. doi: 10.1055/s-2000-7492. [DOI] [PubMed] [Google Scholar]
- 36.Rasmussen M, Skullerud K, Bakke SJ, Lebon P, Jahnsen FL. Cerebral thrombotic microangiopathy and antiphospholipid antibodies in Aicardi-Goutieres syndrome—report of two sisters. Neuropediatrics. 2005;36:40–44. doi: 10.1055/s-2004-830532. [DOI] [PubMed] [Google Scholar]
- 37.De Laet C, Goyens P, Christophe C, Ferster A, Mascart F, Dan B. Phenotypic overlap between infantile systemic lupus erythematosus and Aicardi-Goutieres syndrome. Neuropediatrics. 2005;36:399–402. doi: 10.1055/s-2005-873058. [DOI] [PubMed] [Google Scholar]
- 38.Lebon P, Lenoir GR, Fischer A, Lagrue A. Synthesis of intrathecal interferon in systemic lupus erythematosus with neurological complications. Br Med J (Clin Res Ed) 1983;287:1165–1167. doi: 10.1136/bmj.287.6400.1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Crow YJ, Leitch A, Hayward BE, et al. Mutations in genes encoding ribonuclease H2 subunits cause Aicardi-Goutières syndrome and mimic congenital viral brain infection. Nat Genet. 2006;38:910–916. doi: 10.1038/ng1842. [DOI] [PubMed] [Google Scholar]
- 40.Frank P, Braunshofer-Reiter C, Wintersberger U, Grimm R, Busen W. Cloning of the cDNA encoding the large subunit of human RNase HI, a homologue of the prokaryotic RNase HII. Proc Natl Acad Sci USA. 1998;95:12872–12877. doi: 10.1073/pnas.95.22.12872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Eder PS, Walder JA. Ribonuclease H from K562 human erythroleukemia cells. Purification, characterization and substrate specificity. J Biol Chem. 1991;266:6472–6479. [PubMed] [Google Scholar]
- 42.Rice G, Patrick T, Parmar R, et al. Clinical and molecular phenotype of Aicardi-Goutieres syndrome. American Journal of Human Genetics. 2007;81:713–725. doi: 10.1086/521373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee-Kirsch MA, Chowdhury D, Harvey S, et al. A mutation in TREX1 that impairs susceptibility to granzyme A-mediated cell death underlies familial chilblain lupus. J Mol Med. 2007;85:531–537. doi: 10.1007/s00109-007-0199-9. [DOI] [PubMed] [Google Scholar]
- 44.Millard LG, Rowell NR. Chilblain lupus erythematosus (Hutchinson). A clinical and laboratory study of 17 patients. Br J Dermatol. 1978;98:497–506. doi: 10.1111/j.1365-2133.1978.tb01935.x. [DOI] [PubMed] [Google Scholar]
- 45.Grand MG, Kaine J, Fulling K, et al. Cerebroretinal vasculopathy: A new hereditary syndrome. Ophthalmology. 1988;95:649–659. doi: 10.1016/s0161-6420(88)33131-3. [DOI] [PubMed] [Google Scholar]
- 46.Gutmann DH, Fischbeck KH, Sergott RC. Hereditary retinal vasculopathy with cerebral white matter lesions. Am J Med Genet. 1989;34:217–220. doi: 10.1002/ajmg.1320340217. [DOI] [PubMed] [Google Scholar]
- 47.Weil S, Reifenberger G, Dudel C, Yousry TA, Schriever S, Noachtar S. Cerebroretinal vasculopathy mimicking a brain tumor: a case of a rare hereditary syndrome. Neurology. 1999;53:629–631. doi: 10.1212/wnl.53.3.629. [DOI] [PubMed] [Google Scholar]
- 48.Niedermayer I, Graf N, Schmidbauer J, Reiche W. Cerebroretinal vasculopathy mimicking a brain tumor. Neurology. 2000;54:1878–1879. doi: 10.1212/wnl.54.9.1878-a. [DOI] [PubMed] [Google Scholar]
- 49.Storimans CW, Oosterhuis JA, van Schooneveld MJ, Bos PJ, Maaswinkel-Mooy PD. Familial vascular retinopathy. A preliminary report. Doc Ophthalmol. 1990;75:259–261. doi: 10.1007/BF00164839. [DOI] [PubMed] [Google Scholar]
- 50.Terwindt GM, Haan J, Ophoff RA, et al. Clinical and genetic analysis of a large Dutch family with autosomal dominant vascular retinopathy, migraine and Raynaud’s phenomenon. Brain. 1998;121:303–316. doi: 10.1093/brain/121.2.303. [DOI] [PubMed] [Google Scholar]
- 51.Jen J, Cohen AH, Yue Q, et al. Hereditary endotheliopathy with retinopathy, nephropathy and stroke (HERNS) Neurology. 1997;49:1322–1330. doi: 10.1212/wnl.49.5.1322. [DOI] [PubMed] [Google Scholar]
- 52.Cohn AC, Kotschet K, Veitch A, Delatycki MB, McCombe MF. Novel ophthalmological features in hereditary endotheliopathy with retinopathy, nephropathy and stroke syndrome. Clinical & Experimental Ophthalmology. 2005;33:181–183. doi: 10.1111/j.1442-9071.2005.00976.x. [DOI] [PubMed] [Google Scholar]
- 53.Winkler DT, Lyrer P, Probst A, et al. Hereditary Systemic Angiopathy (HSA) with cerebral calcifications, retinopathy, progressive nephropathy and hepatopathy. J Neurol. 2008;255:77–88. doi: 10.1007/s00415-008-0675-3. [DOI] [PubMed] [Google Scholar]
- 54.Rahman A, Isenberg DA. Systemic lupus erythematosus. N Engl J Med. 2008;358:929–939. doi: 10.1056/NEJMra071297. [DOI] [PubMed] [Google Scholar]
- 55.Crow MK. Collaborations, genetic associations and lupus erythematosus. N Engl J Med. 2008;358:956–961. doi: 10.1056/NEJMe0800096. [DOI] [PubMed] [Google Scholar]
- 56.Hur JW, Sung YK, Shin HD, Park BL, Cheong HS, Bae SC. TREX1 polymorphisms associated with autoantibodies in patients with systemic lupus erythematosus. Rheumatol Int. 2007 doi: 10.1007/s00296-007-0509-0. [DOI] [PubMed] [Google Scholar]
- 57.Unterholzner L, Bowie AG. The interplay between viruses and innate immune signaling: recent insights and therapeutic opportunities. Biochem Pharmacol. 2008;75:589–602. doi: 10.1016/j.bcp.2007.07.043. [DOI] [PubMed] [Google Scholar]
- 58.Hooks JJ, Moutsopoulos HM, Geis SA, Stahl NI, Decker JL, Notkins AL. Immune interferon in the circulation of patients with autoimmune disease. N Engl J Med. 1979;301:5–8. doi: 10.1056/NEJM197907053010102. [DOI] [PubMed] [Google Scholar]
- 59.Bengtsson AA, Sturfelt G, Truedsson L, et al. Activation of type I interferon system in systemic lupus erythematosus correlates with disease activity but not with antiretroviral antibodies. Lupus. 2000;9:664–671. doi: 10.1191/096120300674499064. [DOI] [PubMed] [Google Scholar]
- 60.Dall’era MC, Cardarelli PM, Preston BT, Witte A, Davis JC., Jr Type I interferon correlates with serological and clinical manifestations of SLE. Ann Rheum Dis. 2005;64:1692–1697. doi: 10.1136/ard.2004.033753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gota C, Calabrese L. Induction of clinical autoimmune disease by therapeutic interferon-alpha. Autoimmunity. 2003;36:511–518. doi: 10.1080/08916930310001605873. [DOI] [PubMed] [Google Scholar]
- 62.Ronnblom LE, Alm GV, Oberg KE. Autoimmunity after alpha-interferon therapy for malignant carcinoid tumors. Ann Intern Med. 1991;115:178–183. doi: 10.7326/0003-4819-115-3-178. [DOI] [PubMed] [Google Scholar]
- 63.Heremans H, Billiau A, Colombatti A, Hilgers J, de Somer P. Interferon treatment of NZB mice: accelerated progression of autoimmune disease. Infect Immun. 1978;21:925–930. doi: 10.1128/iai.21.3.925-930.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Adam C, Thoua Y, Ronco P, Verroust P, Tovey M, Morel-Maroger L. The effect of exogenous interferon: acceleration of autoimmune and renal diseases in (NZB/W) F1 mice. Clin Exp Immunol. 1980;40:373–382. [PMC free article] [PubMed] [Google Scholar]
- 65.Mathian A, Weinberg A, Gallegos M, Banchereau J, Koutouzov S. IFNalpha induces early lethal lupus in preautoimmune (New Zealand Black × New Zealand White) F1 but not in BALB/c mice. J Immunol. 2005;174:2499–2506. doi: 10.4049/jimmunol.174.5.2499. [DOI] [PubMed] [Google Scholar]
- 66.Walker SE. Accelerated mortality in young NZB/NZW mice treated with the interferon inducer tilorone hydrochloride. Clin Immunol Immunopathol. 1977;8:204–212. doi: 10.1016/0090-1229(77)90110-6. [DOI] [PubMed] [Google Scholar]
- 67.Santiago-Raber ML, Baccala R, Haraldsson KM, et al. Type-I interferon receptor deficiency reduces lupus-like disease in NZB mice. J Exp Med. 2003;197:777–788. doi: 10.1084/jem.20021996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Braun D, Geraldes P, Demengeot J. Type I Interferon controls the onset and severity of autoimmune manifestations in lpr mice. J Autoimmun. 2003;20:15–25. doi: 10.1016/s0896-8411(02)00109-9. [DOI] [PubMed] [Google Scholar]
- 69.Akwa Y, Hassett DE, Eloranta ML, et al. Transgenic expression of IFNalpha in the central nervous system of mice protects against lethal neurotropic viral infection but induces inflammation and neurodegeneration. J Immunol. 1998;161:5016–5026. [PubMed] [Google Scholar]
- 70.Guyer DR, Tiedeman J, Yannuzzi LA, et al. Interferon-associated retinopathy. Arch Ophthalmol. 1993;111:350–356. doi: 10.1001/archopht.1993.01090030068041. [DOI] [PubMed] [Google Scholar]
- 71.Sugano S, Yanagimoto M, Suzuki T, et al. Retinal complications with elevated circulating plasma C5a associated with interferon-alpha therapy for chronic active hepatitis C. Am J Gastroenterol. 1994;89:2054–2056. [PubMed] [Google Scholar]
- 72.Hayasaka S, Fujii M, Yamamoto Y, Noda S, Kurome H, Sasaki M. Retinopathy and sub-conjunctival haemorrhage in patients with chronic viral hepatitis receiving interferon alfa. Br J Ophthalmol. 1995;79:150–152. doi: 10.1136/bjo.79.2.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Soushi S, Kobayashi F, Obazawa H, et al. [Evaluation of risk factors of interferon-associated retinopathy in patients with type C chronic active hepatitis] Nippon Ganka Gakkai Zasshi. 1996;100:69–76. [PubMed] [Google Scholar]
- 74.Kawano T, Shigehira M, Uto H, et al. Retinal complications during interferon therapy for chronic hepatitis C. Am J Gastroenterol. 1996;91:309–313. [PubMed] [Google Scholar]
- 75.Lebon P. Inhibition of herpes simplex virus type 1-induced interferon synthesis by monoclonal antibodies against viral glycoprotein D and by lysosomotropic drugs. J Gen Virol. 1985;66:2781–2786. doi: 10.1099/0022-1317-66-12-2781. [DOI] [PubMed] [Google Scholar]
- 76.Bennett L, Palucka AK, Arce E, et al. Interferon and granulopoiesis signatures in systemic lupus erythematosus blood. J Exp Med. 2003;197:711–723. doi: 10.1084/jem.20021553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chuntharapai A, Lai J, Huang X, et al. Characterization and humanization of a monoclonal antibody that neutralizes human leukocyte interferon: a candidate therapeutic for IDDM and SLE. Cytokine. 2001;15:250–260. doi: 10.1006/cyto.2001.0934. [DOI] [PubMed] [Google Scholar]
- 78.Han CS, Chen Y, Ezashi T, Roberts RM. Antiviral activities of the soluble extracellular domains of type I interferon receptors. Proc Natl Acad Sci USA. 2001;98:6138–6143. doi: 10.1073/pnas.111139598. [DOI] [PMC free article] [PubMed] [Google Scholar]