1. Introduction
Understanding the function of biological macromolecules and their complexes at the physico-chemical level requires knowledge of both their structure and dynamics. Conventional biophysical techniques, such as crystallography and NMR, have yielded incredibly detailed structural information at the atomic level on highly populated static states.1 In the context of the energy landscape representation of macromolecules, highly populated states are located within the global free energy minimum region of a relatively rough free energy hypersurface.2–5 Much less is known, however, about lowly-populated, higher free energy states which cannot be trapped and are therefore invisible to conventional structural and biophysical techniques. Because such lowly-populated states are generally invisible does not mean that they are not important. Indeed, many biological processes, including macromolecular recognition and binding, enzyme catalysis, allostery and self-assembly, may proceed via intermediates involving fast but infrequent transitions between the free energy global minimum state and higher free energy states. Consider, for example, the process of recognition between two macromolecules. There are two opposing constraints governing this process, namely specificity and speed.6 If an interaction is too specific, the speed of recognition will likely be slow. If, on the other hand, the speed of interaction is too fast, specificity will likely be sacrificed. The dilemma, therefore, is how biological processes manage to achieve both speed and specificity. Extensive kinetic7–13 and theoretical work6,14–16 have suggested that one mechanism that can be used to enhance speed without sacrificing specificity is to reduce the dimensionality of the search process or more generally to incorporate a non-specific attractive potential.6 In the context of protein-DNA interactions where a transcription factor must be able to locate its specific DNA target site within a sea of non-specific DNA sites, non-specific DNA binding can be used to facilitate specific recognition via a combination of one-dimensional diffusion or sliding along the DNA coupled with jumping from one DNA molecule to another.17–26 Similarly for protein-protein interactions, non-specific encounter complexes can be rapidly formed and subsequently relax to the stereospecific complex via two-dimensional diffusion on the surface of the proteins coupled with electrostatic guiding.6,9,27–29 In this review, we will survey recent developments in paramagnetic relaxation enhancement (PRE), as measured by NMR spectroscopy, and show how this technique can provide unique information that permits one to both detect and visualize lowly-populated states of macromolecules and their complexes.
Macromolecular NMR spectroscopy has seen many developments over the last two decades. The advent of two-dimensional NMR led to the first three-dimensional structure determinations of small (<8–10 kDa) proteins and nucleic acids.30,31 The introduction of heteronuclear three- and four-dimensional NMR spectroscopy in the late 1980s and early 1990's32,33 extended the range of applicability of the method to systems in the 20–30 kDa range.34,35 Subsequently, the introduction of transverse relaxation optimized spectroscopy (TROSY)36–40 permitted structures of systems in the 40–80 kDa range to be determined41–46 and supra-molecular complexes in the 500 kDa range to be studied.47–50 Most NMR measurements, such as the nuclear Overhauser effect (NOE) which yields short (≤6 Å) interproton distance restraints that provide the mainstay of all NMR structure determinations, and three-bond scalar couplings that yield torsion angle restraints, are based on short-range local interactions. The power of the NOE is that a large number of short-range interproton distances between residues far apart in the linear sequence of amino acids are sufficiently constraining to yield the three-dimensional structure of a protein.51 Nevertheless, long-range information can still be very helpful. In this regard, the introduction of the routine use of residual dipolar couplings (RDCs) measured on weakly aligned macromolecules dissolved in dilute liquid crystalline media52,53 provides long-range orientational information (relative to an external alignment tensor) that has led to significant increases in coordinate accuracy.54–58 Long-range distance information in the 10–35 Å range, can also be obtained through the use of paramagnetic NMR.59–63
For investigations of dynamics by NMR, 2H-, 13C-, and 15N-relaxation measurements have provided a wealth of information on the pico- to nanosecond time scale. Recent advances in relaxation dispersion spectroscopy have yielded insights into processes on the micro- to millisecond time scale.64–67 Quantitative studies of slower exchange processes involving macromolecules can be carried out using techniques such as heteronuclear z-exchange spectroscopy25,68–71 and hydrogen exchange rate experiments.72,73
There are two NMR approaches that can be used to study lowly-populated states under equilibrium conditions, namely relaxation dispersion spectroscopy and paramagnetic relaxation enhancement. Relaxation dispersion spectroscopy provides detailed kinetic information on exchange processes between major species and lowly-populated states on the micro- to millisecond time scale, providing there are substantial chemical shift differences between the states. In the absence of substantial chemical shift differences between the states, exchange line broadening will not occur and no relaxation dispersion will be observed. In addition to kinetic rate constants, relaxation dispersion measurements yield chemical shifts for the minor species which can be used to obtain some qualitative structural insight into the nature of the minor species. More recently, combining relaxation dispersion with weak alignment has permitted the extraction, under favorable circumstances, of highly sensitive bond vector orientation information for the minor species in the form of RDCs and anisotropic chemical shifts.74,75
Over the last few years, paramagnetic NMR has emerged as an alternative, orthogonal, and powerful tool to investigate various dynamic processes involving macromolecules. In particular, the PRE arising from unpaired electrons with an isotropic g-tensor (such as a nitroxide spin-label or EDTA-Mn2+) has proved to be a particularly versatile tool that can provide information about structural aspects of the dynamic process owing to its <r−6> distance dependence between the paramagnetic center and the nucleus of interest. The PRE arises from magnetic dipolar interactions between a nucleus and the unpaired electrons of the paramagnetic center which result in an increase in nuclear relaxation rates. In contrast to the NOE where the effects are small and therefore limited to short range (< 6Å) interactions between protons, the PRE effect is very large owing to the large magnetic moment of an unpaired electron, permitting distances up to 35 Å (depending on the paramagnetic group) to be detected. The key for using the PRE to study lowly-populated states resides in the finding that the observed PRE rates in the fast exchange regime are population weighted averages of the PREs for the major and minor species.24 Thus, providing the paramagnetic center-proton distances are shorter in the minor species than the major one, the imprints of the minor species will be apparent in the observed PRE rates, thereby enabling structural information on the minor species to be extracted.
Despite the long history of the PRE (in fact, Solomon described the equations for the PRE in his famous paper on the NOE in 1955),76 application to biological macromolecules was limited for a long time to metal-binding proteins.60 General use of the PRE for non-metal binding proteins is dependent on the introduction of an extrinsic paramagnetic center via conjugation to a specific, solvent exposed, site.77 Further quantitative application of the PRE to structure determination requires the appropriate theoretical and computational framework for refinement directly against PRE relaxation rates taking into account the intrinsic flexibility of paramagnetic centers that are attached to the macromolecule via linkers with several rotatable bonds.78 The first applications of the PRE involved studies on spin-labeled lysozyme and bovine pancreatic trypsin inhibitor in the mid-1980s in which PRE effects were converted to approximate distance restraints,79,80 but it is only with recent biochemical and computational advances that the technique has come into more general use. Using this type of PRE data, macromolecular structures have been characterized for soluble proteins,81–84 protein-protein complexes,85–89 protein-oligosaccharide complexes,90–92 protein-nucleic acid complexes,78,93–96 and membrane proteins.97,98 Unfolded or partially unfolded states of proteins have also been investigated by PRE.99–109 A recent major advance in the field of dynamics of macromolecular interactions is the finding that in the fast exchange regime the intermolecular PRE can provide a powerful probe to detect and characterize lowly populated intermediates in macromolecular binding events, thereby providing structural information on encounter complexes that cannot be obtained by any other biophysical technique.24,27,28,110,111 The same principle can also be applied to other dynamic process such as nonspecific protein-DNA interactions,23,112 inter-domain motions113,114 and transient protein associations.115–117
The main focus of this review is on the PRE as a tool to investigate lowly-populated states of macromolecules and their complexes. In Section 2, we describe the theoretical aspects of the PRE and the effects of dynamics on the PRE. In this context, we explain how the PRE permits amplification of information for minor states in exchanging systems. The advantages of using the PRE on transverse magnetization arising from unpaired electrons with a isotropic g-tensor are also discussed. In Section 3, we describe experimental aspects of the PRE experiment including paramagnetic probes, pulse sequences and back-calculation of the PRE, including practical considerations. Finally, in Section 4, we review recent applications of the PRE to investigate various dynamic processes involving biological macromolecules.
2. Theoretical Aspects
2.1 Paramagnetic NMR parameters
2.1.1. Isotropic and anisotropic unpaired-electron systems
In paramagnetic systems there are three NMR experimental observables that yield long-range structural information: PREs, pseudo-contact shifts (PCS) and residual dipolar couplings (RDC). The PRE can be detected in any paramagnetic system, whereas PCS and RDCs can only be observed in systems with an anisotropic electron g-factor. Figure 1 summarizes the major long-range paramagnetic NMR observables in systems with isotropic and anisotropic electron g-factors. PCS and RDCs are dependent on the magnetic susceptibility tensor (usually referred to as the χ tensor). The electron g-tensor and χ-tensor are closely related. In general, if the electron g tensor is anisotropic, the χ tensor is also anisotropic. If the zero-field-splitting (ZFS) is negligible compared to the electron spin Zeeman energy, the relationship between the χ and g tensors is given by:118
| [1] |
where S is the electron spin quantum number; μ0, the permeability of free space; μB, the magnetic moment of the free electron; NA, Avogadro’s number; kB, the Boltzman constant; T, the temperature; and gkk, an element of the g-tensor (with k = x, y, or z). Nitroxide spin radicals, EDTA-Mn2+, and DTPA-Gd3+ are isotropic electron systems, whereas Fe3+, Dy3+ and many other paramagnetic metal ions represent anisotropic systems.
Figure 1.
Major long-range observables in paramagnetic NMR.
2.1.2 PRE through direct dipole-dipole interactions
The longitudinal (Γ1) and transverse (Γ2) PRE rates are conventionally described by the Solomon-Bloembergen (SB) equations:76,119
| [2] |
| [3] |
where g is the electron g-factor; γI , the proton gyromagnetic ratio; ωI/2π, the Larmor frequency of the proton; and JSB(ω), the generalized spectral density function for the reduced correlation function given by:
| [4] |
The correlation time τc is defined as where τr is the rotational correlation time of the macromolecule, and τs the effective electron relaxation time. The Solomon-Bloembergen theory makes the simplifying approximation that electron relaxation is not coupled to molecular tumbling. This is reasonable because the electron relaxation lifetime is comparable to or shorter than the rotational correlation time of a macromolecule. In the case of Mn2+ (S=5/2), electron relaxation is multi-exponential.120 Theoretical considerations, however, have shown that the SB equation with a single effective electron relaxation rate does not introduce any significant errors at high magnetic field strengths (> 10 Tesla).121
The Solomon-Bloembergen theory assumes that dipole-dipole interaction vectors are rigid in the molecular frame. Since PRE interaction vectors may be quite long (up to 35 Å for Mn2+), they are less susceptible to small fluctuations in atomic positions, making this assumption reasonable in many cases. However, this approximation will break down if the ensemble space sampled by the paramagnetic group is quite large. The extension of the Solomon-Bloembergen equations to deal with this problem will be described in Section 2.2.
2.1.3 PRE through Curie-spin relaxation
Curie-spin relaxation arises from dipole-dipole interaction between a nucleus and the time-averaged magnetization of the electrons. The Γ2 rate due to Curie-spin relaxation is given by:122
| [5] |
where k is the Boltzman constant, and T the temperature in Kelvin. (In some examples in the literature, the last two terms are neglected which is only valid when τs ≪ τr). From Eq. [5] it follows that the PRE rate for Curie-spin relaxation is approximately proportional to the square of the magnetic field. In the case of nitroxide spin radicals τc ≈ τr and therefore Curie-spin relaxation is negligible. For metal ions with an anisotropic g-tensor and a very short electron relaxation time (e.g., Fe3+, Dy3+), Curie-spin relaxation is the major component of the 1H-Γ2 rates for macromolecules. For isotropic metal ions such as Mn2+ and Gd3+, Curie-spin relaxation is almost negligible for medium-size macromolecules (~40 kDa). For example, in the case of the 20 kDa SRY/DNA- EDTA-Mn2+ complex, the contribution of Curie-spin relaxation to the overall Γ2 rates at 308 K is estimated to be only 2%, even at a 1H-frequency of 800 MHz.123 The relative contribution of Curie-spin relaxation depends on the size of the system. For a very large system with τrlarger than 50 ns, corresponding to a molecular weight > 100 kDa, the contribution could be larger than 20% at 800 MHz.
2.1.4. Pseudo-contact shifts
Pseudo-contact shifts (PCS) are observed only in paramagnetic systems with anisotropic unpaired electrons such as those in Dy3+, Tb3+, and Fe3+ ions. The magnitude of the PCS, δPCS, is given by:118
| [6] |
where r is the distance between the metal ion and nucleus; θ and ϕ, the angles between the metal-nucleus vector and the zz and xx principal axes of the magnetic susceptibility (χ) tensor, respectively; and Δχax and Δχrh are the axial and rhombic components, respectively, of the magnetic susceptibility tensor defined as:
| [7] |
| [8] |
It should be noted that the PCS displays a r−3 distance dependence, in contrast to the r−6 dependence for the PRE. As a result, the distance range for the experimentally detected PCS is relatively long. For example, the distance range for the PCS arising from Dy3+ can extend to ~40 Å, providing the principal axis of the χ tensor is fixed within the molecular frame, as is the case for metalloproteins. When an extrinsic metal ion is attached to a macromolecule using a chelator with a flexible linker, the magnitude of the PCS is significantly reduced owing to the fact that the principal axes of the χ tensor fluctuate within the frame of the macromolecule. Immobilization of the metal chelator, for example by using bidentate ligands that can be conjugated to two neighbouring sites simultaneously, will increase the observed PCS.124,125
There have been extensive studies in which PCS has been used to investigate dynamics in metal binding proteins.126–128 However, these PCS applications have not gained widespread popularity outside the area of metal binding proteins for two main reasons. First, the theoretical framework required to describe the relationship between PCS and dynamics has not yet been adequately established. Second, a practical problem arises from the presence of enantiomers for many metal chelators, such as EDTA, conjugated to macromolecules. Since the enantiomers exhibit slightly different χ tensors, the corresponding PCS's are also different, and as a result, multiple sets of cross-peaks appear.129–131 Metal chelators designed to solve this problem have been developed.124,125,129,132 With widespread use of these new compounds, PCS could become a widely used technique for investigations of dynamical processes.
2.1.5. Residual dipolar couplings
A molecule containing a paramagnetic center with an anisotropic g tensor will undergo partial alignment in the magnetic field giving rise to residual dipolar couplings (RDCs):118
| [9] |
where DAB is the residual dipolar coupling observed between nuclei A and B; rAB, the distance between nuclei A and B; θ, the angle between the A-B internuclear vector and the z axis of the χ tensor; ϕ, the angle between the projection of the A-B internuclear vector on the xy plane and the x axis of the χ tensor; B0, the static magnetic field strength; and h, Planck’s constant. Although Eqs. [6] and [9] are formally similar, they provide different, albeit complementary information, since the variables r, θ and #x003D5; are defined with respect to a paramagnetic center-nucleus vector in Eq. [6] and to an internuclear vector in Eq. [9].
For naturally occurring metal binding proteins where the metal ion(s) is at a fixed position in the molecular frame, the RDCs can be fairly large, especially at a high magnetic field. For example, in the case of the calbindin D9k with a Dy3+ ion bound at the C-terminal metal binding site, the metal-induced RDCs for backbone amide 15N-1H vectors spanned from −26 Hz to +19 Hz at 800 MHz and from −11 Hz to +8 Hz at 500 MHz.133
On the other hand, a Dy3+ ion bound to an extrinsic metal chelator attached to the macromolecule via a flexible linker will exhibit much smaller RDCs. This is because the flexibility of the linker reduces the magnitude of the molecular alignment of the macromolecule with respect to the magnetic field even though the alignment for the conjugated metal chelator may be sizable. In the case of the Trigger Factor protein with EDTA-Dy3+ conjugated to a cysteine residue, the largest observed |1DNH| RDC was only 8 Hz at 800 MHz.129
2.1.6 Advantages of paramagnetic systems with an isotropic g-tensor
As described above, paramagnetic systems with an anisotropic g-tensor can potentially provide three different NMR observables (PRE, PCS, and RDC) while systems with an isotropic g-tensor yield only the PRE. However, this does not necessarily mean that an anisotropic system is more useful in practice. In fact, there are a number of practical advantages to using an isotropic system, especially for dynamic investigations.
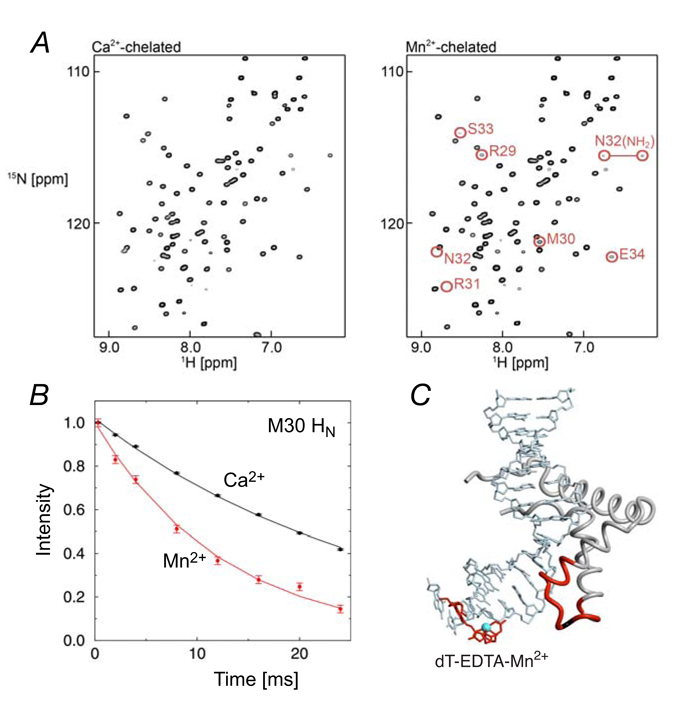
One advantage is that separate assignment of resonances in the paramagnetic state is unnecessary since there is no PCS for an isotropic g-tensor. Figure 2 shows an example for an SRY/DNA-EDTA complex.94 In this case, the paramagnetic and diamagnetic states consist of Mn2+ and Ca2+, respectively, chelated to an EDTA group that is covalently attached to the DNA. The chemical shifts for the diamagnetic and paramagnetic resonances are identical because the unpaired electrons of Mn2+ are isotropic. For an anisotropic electron system, however, resonances need to be assigned separately for the diamagnetic and paramagnetic states owing to the presence of PCS in the latter. From a practical perspective, resonance assignment for the paramagnetic state can be very challenging owing to lower sensitivity as a consequence of PRE. In favorable cases, z-exchange spectroscopy can be used for assignment purposes. In this approach, the paramagnetic and diamagnetic metal ions with the same or near identical affinities are mixed and the exchange cross-peaks are qualitatively used to correlate signals from the paramagnetic and diamagnetic states.134 For this method to work, however, the exchange rates for metal binding should range from approximately 0.1 to 100 s−1. Alternatively, resonance assignments for the paramagnetic state can be carried out using so-called ‘protonless’ NMR spectroscopy that employs 15N/13C heteronuclear correlation specroscopy in conjunction with direct 13C detection.135–142 Since the magnitudes of Γ2 for 13C and 15N nuclei are substantially smaller than that for 1H, this type of heteronuclear correlation spectroscopy permits assignment of 13C/15N resonances in the paramagnetic state. (Note, of course, that direct detection of 13C instead of 1H reduces the sensitivity of the experiments owing to the dependence of the signal-to-noise ratio on γ3/2, where γ is the gyromagnetic ratio of the detected nucleus.)
Figure 2.
(A) 1H-15N HSQC spectra recorded on SRY/DNA-EDTA complexes (Right, Mn2+-chelated; left, Ca2+-chelated).94 Note that the cross-peak locations for the diamagnetic (Ca2+) and paramagnetic (Mn2+) states are identical. (B) 1H transverse relaxation for Met-30 in the SRY/DNA-EDTA complexes (red, Mn2+-chelated; black, Ca2+-chelated). (C) Location of dT-EDTA-Mn2+ in the complex. Protein residues that exhibit a PRE 1HN-Γ2 rate larger than 15 s−1 are colored in red. Adapted from Iwahara et al.94 published in J. Am. Chem. Soc. (American Chemical Society) while the authors were US Government employees at the National Institutes of Health.
Another advantage of using a paramagnetic system with an isotropic g-tensor is that the analysis of the PRE is much more straightforward. For an anisotropic system, the PRE Γ2 rates are difficult to interpret quantitatively owing to two factors. First, Γ2 for an anisotropic system arises principally from Curie-spin relaxation, for which cross-correlations with other relaxation mechanisms are significant.143,144 While cross-correlation could potentially provide an additional source of information for structural studies on a static system,143,146 the effect simply complicates the study of dynamic systems. Second, the exchange contributions arising from conformational dynamics or molecular interactions to the transverse relaxation rate, R2, can be substantially different for the diamagnetic and paramagnetic states owing to the presence of PCS. In such a situation, the PRE Γ2 rate cannot simply be measured as a difference between the R2 rates for the diamagnetic and paramagnetic states.112 Although the PRE Γ1 rate can be measured for both anisotropic and isotropic systems, cross-relaxation and hydrogen exchange with water molecules can, in general, significantly reduce the accuracy of the Γ 1 measurement. For an isotropic system, on the other hand, the contribution of Curie-spin relaxation to the PRE Γ2 rate is negligible and the PRE Γ2 rate is dominated by direct dipole-dipole interactions between a nucleus and the unpaired electrons of the paramagnetic center. In this case, the PRE Γ2 relaxation does not exhibit cross-correlation with other relaxation mechanisms because of cancellation by rapid electron relaxation. (Note that Curie-spin relaxation is caused by time-averaged magnetization and is independent of electron relaxation). In addition, since there is no PCS for an isotropic system, the exchange contributions to R2 are identical for the diamagnetic and paramagnetic states and are therefore cancelled out when Γ2 is measured as are the transverse relaxation rates in the paramagnetic and diamagnetic states, respectively.
2.2 Effect of fast dynamics on the PRE arising from a paramagnetic probe conjugated to a macromolecule
Artificially introduced paramagnetic groups are generally attached to the macromolecule of interest through linkers that have several rotatable bonds. Hence the paramagnetic centers are intrinsically flexible. In addition, the observed 1H nuclei may be located in mobile regions of the macromolecule. Here, we describe the effect of fast dynamics in the picosecond to nanosecond timescale on the PRE for paramagnetic systems with an isotropic g-tensor.
2.2.1 Model-free extension of the Solomon-Bloemenbergen equations
A ‘model-free’ formalism147 can be readily incorporated into the Solomon-Bloembergen theory to evaluate the influence of internal motions on the PRE.78 If the internal motions are not coupled with overall tumbling of the molecule, the correlation function, CI(t), for internal motion of the interaction vector within the molecular frame can be approximated by:
| [11] |
where S2 is the square of the generalized order parameter (not to be confused with the electron spin quantum number S), and τi the correlation time for internal motion. Although the ‘model-free’ formalism is commonly used for fixed-length interaction vectors such as 15N-1H or 13C-1H bond vectors, it can also be applied to variable length vectors such as those for homonuclear 1H-1H dipolar interactions.148,149 In this case, the order parameter is defined as:149
| [12] |
where Y2m (Ω) are second-order spherical harmonics, and Ωmol are Euler angles in the molecular frame. As Brüschweiler et al.148 demonstrated for 1H-1H dipolar interactions, it is useful to approximate the order parameter S2 by decomposition into its radial and angular components:
| [13] |
where the angular (S2PRE,angular) and radial (S2PRE,radial) order parameters are defined as:
| [14] |
| [15] |
It is assumed that the effective electron relaxation rate is not influenced by the correlation times for either overall or internal motions of the interaction vector. For a transition metal ion, electron relaxation primarily arises from modulation of the zero-field splitting tensor (as a consequence of collisions with solvent molecules) and is therefore governed by a very short lifetime between collisions τυ (~5 ps for water at physiological temperature).120 Since internal motion faster than τυ would affect the effective electron relaxation rate , we focus on cases where the correlation time τi for internal motion is significantly longer than τυ. Under these conditions, incorporation of the correlation function CI(t) for internal motions into the correlation function for the PRE transforms JSB(ω) in the SB equation (Eq. [4]) into:
| [16] |
where τt is the total correlation time defined as . When the original SB equations (Eqs. [2–4]) are expressed in the form:
| [17] |
incorporation of the model-free formalism transforms Eq. [17] into:
| [18] |
For simplicity, we refer to the SB equations incorporating the model-free formalism as the SBMF equations. The SBMF equations reduce to the SB equations under conditions where the internal motion is either very slow (and eventually, τt≈ τc) or highly restricted in space (S2 ≈ 1).
The SB (Eq. [17]) and SBMF (Eq. [18]) equations are valid for systems with isotropic tumbling. For non-globular systems exhibiting significant diffusion anisotropy, the PRE also depends on the angles between the principal axes of the diffusion tensor and the electron-nucleus interaction vector. The effect of anisotropic tumbling can be readily incorporated according to previous literature.150,151 However, the impact of anisotropic tumbling is not significant unless the rotational correlation time dominates the overall correlation time.
2.2.2. Effect of fast internal motions on PRE 1H-Γ1 and Γ2
The impact of motional effects on 1H-Γ1 and 1H-Γ2 rates are very different. Owing to the large distances involved, the variation in the values of the order parameters for PRE interaction vectors is rather small. However, the internal motion correlation time τi is likely to have a wide range of values depending on the internal dynamics of individual 1H nuclei in the macromolecule (e.g. sidechain versus backbone). The contribution of τi can be evaluated using the second term of the SBMF equation (Eq. [18]). The dependence of 1H-Γ1 and 1H-Γ2 rates on τi are shown in Figure 3. In the case where the order parameters S2 for the PRE interaction vectors are as small as 0.5, the 1H-Γ1 rate exhibits a strong dependence on τi (in particular for τi < 1/ωH); indeed the value of Γ1 can be up to five times larger than that of Γ1 in the absence of internal motion, depending on the value of τi (Figure 3A). The 1H-Γ2 rate, on the other hand, is significantly less sensitive to the internal correlation time τi (Figure 3B). Thus, for structure analysis using 1H-PRE data arising from flexible paramagnetic groups, the analysis of 1H-Γ1 requires accurate information on the values of τi for the individual PRE interaction vectors, whereas a simple estimation of τi is sufficient for analysis of 1H-Γ2 data. Consequently, 1H-Γ2 data provide far more useful structural restraints since they are not affected by the motional effects within the macromolecule. In addition, proton T1 relaxation is not a single-exponential process owing to cross-relaxation (and water-exchange in the case of amide protons), and hence the observed 1H-Γ1 values will also be perturbed by such effects.
Figure 3.
Influence of the correlation time τi for internal motion on (A) 1H-Γ1 and (B) 1H-Γ2 rates at 500 MHz. The vertical axis displays the Γ/Γ0 ratio, where Γ is calculated with the full SBMF equation (Eq. 12) and Γ0 with only the first term of the SBMF equation. Γ0 corresponds to Γ when τi→0. Curves, calculated with the correlation time set to 3 ns, are shown with three different values of the order parameter S2 for PRE interaction vectors: S2MnH = 0.9 (solid line), 0.7 (dashed line) and 0.5 (long-short dashed line). The dependence of the 1H-Γ1 and 1H-Γ2 rates on τi are field-dependent and the maximum of the 1H-Γ1/1H-Γ10 curve is located around τi ~ 1/ωI Adapted from Iwahara et al.78 published in J. Am. Chem. Soc. (American Chemical Society) while the authors were US Government employees at the National Institutes of Health.
2.3. Effect of slow dynamics on the PRE
In Section 2.2, we described the effect of fast dynamics (~ 10−11−10−8 s timescale) on the PRE where the impact of dynamics is at the level of the auto-correlation function. Dynamics more relevant to macromolecular interactions, however, typically occur on slower timescales. Here we describe the effects of slower dynamics (~10−6−10−1 s timescale) on the PRE. The principle of amplification of information relating to low population states in an exchanging system will also be discussed.
2.3.1. PRE Γ2 rates in an exchanging system
The effect of slower dynamics on the PRE can be simulated using the McConnell equations,152 which represent the most fundamental description of the effects of chemical exchange in NMR. The matrix form of the McConnell equations for transverse magnetization is given by:
| [19] |
where m is a vector containing the transverse magnetizations of the exchanging states; R, a relaxation matrix; K, a kinetic matrix; and W, a chemical shift matrix. In the case of two-state exchange, A⇔B, the vector and matrices in Eq. [19] are given as follows:
| [20] |
| [21] |
| [22] |
| [23] |
where M+ is a transverse magnetization represented by a complex variable Mx+iMy; R2, a transverse relaxation rate; kAB and kBA, kinetic rate constants for the A→B and B→A processes, respectively; and Ω/2π, the chemical shift frequency. In the paramagnetic state, the relaxation matrix contains both the intrinsic R2 and the PRE Γ2 rates, while in the diamagnetic state the relaxation matrix contains only the intrinsic R2 rates. Expansion of Eqs. [20–23] for a three-state (e.g., see Led and Grant153) or general N-state exchange is straightforward.
Using the McConnell equations, one can simulate apparent PRE rates for exchanging systems. Numerical solutions of the McConnell equations can be readily obtained using either the exponential matrix formalism or by numerical integration. Simulations require initial conditions determined by the equilibrium populations of individual states. If the number of states exceeds two, it is convenient to consider the equilibrium populations in terms of a partition function, Q. For example, the partition function Q for a consecutive three-state exchange A⇔B⇔C (with species A as a reference) system is given by:
| [24] |
and the populations of states A, B and C are calculated to be 1/Q, (kAB/kBA)/Q and (kAB/kBA)(kBC/kCB)/Q, respectively. Solutions of the McConnell equations provide time-domain data (i.e. free induction decay) and subsequent Fourier transformation yields an NMR spectrum for the exchanging system. Lineshape fitting against the simulated spectrum permits analysis of apparent relaxation rates. Two simulations with and without the PRE Γ2 rate in the R matrix provide apparent relaxation rates for paramagnetic and diamagnetic states, and the apparent PRE rate can be calculated as in exactly the same manner as in an experimental measurement. Strictly speaking, this corresponds to in the case without a refocusing pulse. Effects of refocusing pulses can be considered in the context of the McConnell equations as described by Allerhand.154 However, simulations of apparent PRE rates with and without refocusing pulses result in virtually identical Γ2 values. This is due to the fact that the exchange contributions to the R2 rates for the paramagnetic and diamagnetic states are virtually identical and are therefore cancelled in the calculation of Γ2.
2.3.2. PRE-based amplification of information relating to minor states
The PRE provides a unique technique for obtaining structural information about low population states in an exchanging system.24 The underlying basis of this phenomenon can be ascertained from simulations based on the McConnell equations. Figure 4 illustrates the impact on the transverse PRE 1H-Γ2 rate observed on the resonance of a major species A (population pA, 99%) with a paramagnetic center-proton distance of rA = 30 Å on the presence of a minor species B (population pB, 1%) with a corresponding distance of rB = 8 Å. Due to the r−6 dependence of the PRE, the PRE rates span a very broad range. For this particular case, with a molecular weight of ~30 kDa, the PRE 1H-Γ2 rate arising from Mn2+ for species A (Γ2,A) is only 2 s−1, whereas that for species B (Γ2,B) is 5.6 × 103 s−1. Figure 5 shows the relationship between kex and Γ2app for this system. The apparent value of Γ2(Γ2app) observed for the resonance of the major species A is highly dependent on the exchange rate kex between the major and minor species. If kex is slow (< 50 s−1), the presence of the minor species B has no effect and the value of Γ2app is the same as that expected for the major species A. For larger kex, however, Γ2app is highly influenced by the minor species B. When kex >> |Γ2,B - Γ2,A| (the fast exchange regime on the Γ2 relaxation time scale), Γ2app is the weighted population average of the Γ2 rates for the two species:
| [25] |
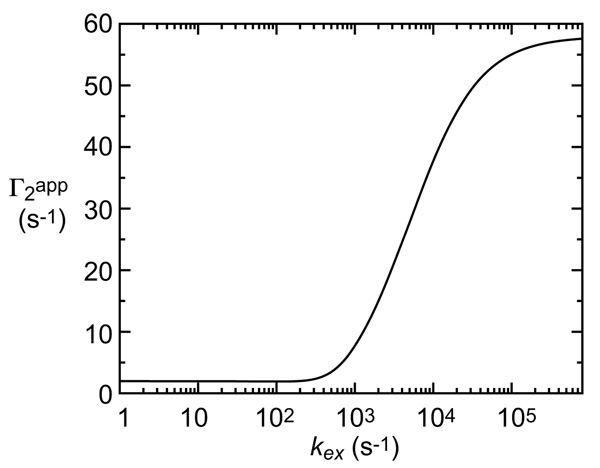
It should be noted that even with pB set to as low as 0.01, Γ2app is dominated by the term arising from species B under these conditions and has a value ~30-fold larger than Γ2,A because Γ2,B is so much larger than Γ2,A. As a result one can infer the presence of the minor species and, in addition, obtain some structural information on it because the PRE is a distance-dependent quantity.
Figure 4.
PRE-based amplification of information on low population states. (A) Two site exchange system comprising a major species A (99%) and a minor species B (1%). (B) Lineshape simulation of the resonance of species A illustrating the effect of exchange on the apparent PRE Γ2app rate. When the exchange rate kex is fast, the apparent PRE Γ2app rate can be dominated by the invisible minor state even though its population is as small as 1%. In these simulations, the intrinsic relaxation rates for species A and B were set to 50 s−1 for the diamagnetic state. The resonances arising from species A and B are located at −50 and 50 Hz, respectively. The chemical shifts for the diamagnetic and paramagnetic are assumed to be identical since a paramagnetic system with an isotropic electron g-tensor such as Mn2+ does not generate a pseudo-contact shift. Adapted from Iwahara et al.24 published in Nature (Nature Publishing Group) while the authors were US Government employees at the National Institutes of Health.
Figure 5.
Relationship between exchange rate kex and apparent PRE Γ2app rate for the two-state exchange system shown in Figure 4.
Although amplification of information relating to the minor state represents a fascinating phenomenon, it is important to be aware that quantitative analysis is only possible under conditions where kex >> |Γ2,B - Γ2,A|. If kex does not satisfy this inequality, information on kex and the chemical shift difference |ΩA - ΩB| between the corresponding resonances of the two species are required as shown in Figure 6. Further, when |ΩA - ΩB| << kex<< |Γ2,B - Γ2,A| holds because of a very large Γ2,B (i.e. a short rB distance), the apparent PRE Γ2app becomes virtually independent of rB.
Figure 6.
Relationships between chemical shift difference |ΩA−ΩB| and apparent PRE Γ2app rate for the two-state exchange system in Figure 4.
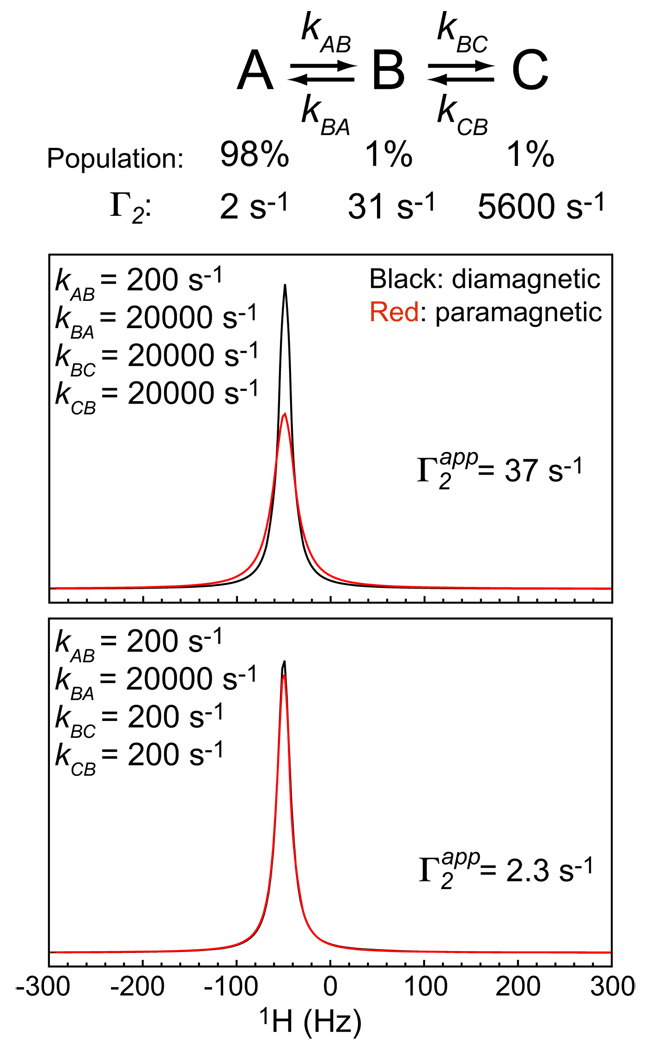
Apparent PRE Γ2app rates in exchanging systems comprising more than two states follow a similar pattern to that for a two-state exchange system. Figure 7 shows simulations for a consecutive three-state exchange system A ⇔B⇔C. Values of the PRE Γ2 rates for A, B, and C were set to 2, 31, and 5.6 × 103 s−1, respectively, corresponding to electron-1H distances of rA = 30 Å, rB = 19 Å, and rC = 8 Å, respectively. In these simulations, two sets of kinetic rate constants were used: in the first set kAB = 200 s−1 and kBA=kBC=kCB=20000 s−1 (Figure 7; top); in the second set kAB = kBC=kCB= 200 s−1 and kBA =20000 s−1 (Figure 7; bottom). For both cases, the populations of A, B, and C are 98%, 1%, and 1%, respectively (See the partition function given by Eq. [24]). However, the apparent Γ2app rate is highly affected by the presence of state C only for the former set of rate constants, as is easily deduced from the simulations for a two-state exchange system.
Figure 7.
PRE simulations for a consecutive three-state exchange system.
3. Experimental analysis of 1H PRE data
3.1. Paramagnetic probes
Chemical probes used for paramagnetic NMR can be divided into two classes: (1) nitroxide stable radicals >N-O•, and (2) metal chelators (such as EDTA, DTPA and metal-binding peptides) that bind paramagnetic metal ions with very high affinity. These two classes of paramagnetic probes can be covalently attached to both proteins and nucleic acids, and used for the observation of intramolecular or intermolecular PREs (Figures 8A and 8B). Free probes, in the form of paramagnetic co-solute molecules, are also useful to identify solvent accessible regions of the molecular surface of a macromolecule (Figure 8C). In this section, we describe paramagnetic probes that have gained widespread use in paramagnetic NMR for structural and dynamic investigations of macromolecular systems.
Figure 8.
Three types of PRE. (A) Intramolecular PREs arising from the paramagnetic group within the same molecule. (B) Intermolecular PREs arising from the paramagnetic group located on the interaction partner. (C) Solvent PREs arising from random collisions between a macromolecule and paramagnetic co-solute molecules.
3.1.1. Paramagnetic probes attached to proteins
A solvent-exposed cysteine residue, which can be readily introduced by site-directed mutagenesis, is most commonly used as the conjugation site for site-specific incorporation of a paramagnetic probe on a protein. Figure 9 shows several different types of paramagnetic probes designed for cysteine modification. Compounds 1–4 are nitroxide stable spin radicals,81,82,102,114,155,156 while compounds 5–7 are derivatives of metal chelators.84,124,129,157–159 Most of the nitroxide-derivatives shown in Figure 9 were originally developed for EPR.
Figure 9.
Paramagnetic probes for protein labeling at a surface exposed cysteine residue. (1) MTSSL,81,82 (2) Iodoacetamido-PROXYL,102 (3) Iodoacetamido-TEMPO, (4) Maleimide-TEMPO,114,155 (5) S-(2-pyridylthio)cysteaminyl-EDTA,84,157,158 (6) MTS-EDTA,129 and (7) CLaNP-5.2.124,159 These compounds are conjugated to a cysteine thiol group (Compounds 2–4 can also react with other types of amino acids, depending on reaction conditions). Gray-colored groups are replaced with a cysteine Sγ atom after conjugation. For maleimide-TEMPO (compound 4), the C=C double bond becomes a single bond and the cysteine Sγ atom is bonded to one of the carbon atoms. Compound 7 is conjugated to two cysteine residues in close spatial proximity on the surface of the protein. For the metal chelators (compounds 5–7), atoms involved in metal coordination are indicated by an asterisk.
Conjugation of a paramagnetic probe based on disulfide chemistry81,82,84 is probably the most commonly used. This type of reaction is thiol-specific and virtually no conjugation reactions other than on cysteine residues are found. Although this reaction is convenient, the final product is not very stable owing to disulfide-exchange reactions causing gradual accumulation of cross-linked protein dimer in the NMR sample. For example, in the case of a 6-month-old sample comprising a cysteaminyl-EDTA-Mn2+ conjugate of 15N-labeled HPr (E32C), the dimer population was found to be as high as 30%.123 Considering that a relatively small percentage of diamagnetic contamination can affect the PRE measurements (see section 3.2.4), NMR experiments on this type of conjugate should be carried out within a week or two of sample preparation.
In terms of sample stability, paramagnetic probes conjugated with iodo- or bromoacetamide-derivatives102,156 are advantageous since they irreversibly form a stable C–S bond. A drawback is their lower reaction specificity. These reagents can also react with histidine, methionine and lysine residues at a slower rate. Careful optimization of the reaction conditions to avoid undesired conjugations is therefore necessary, as described by Gillespie and Shortle.102 Although cysteine modification by the maleimide-derivative is popular in biochemistry, Kosen pointed out the instability of the maleimide conjugate and recommended the use of the other compounds.77
The presence of multiple cysteine residues in a protein makes selective incorporation of a paramagnetic group difficult, especially if such cysteine residues are functionally important and cannot therefore be mutated to serine or alanine. Direct incorporation of a spin-labeled amino-acid in solid-phase peptide synthesis is feasible.160–165 The method is potentially very powerful in that it permits the site-specific incorporation of nitroxide stable radicals at desired positions even if there are cysteine residues at different locations. However, application of this method is currently limited to polypeptides shorter than about 50 residues. Incorporation of a reactive keto group using the method developed by Schultz and co-workers166 may provide an alternative approach to site-directed incorporation of paramagnetic groups for PRE studies.
A short amino-acid sequence that binds to a paramagnetic metal ion can be genetically introduced. The ATCUN motif is a short N-terminal sequence (NH2-X1-X2-His) that binds paramagnetic Cu2+ with high affinity. In the case of the Cu2+ ion bound to the ATCUN motif, the electron g-tensor is almost isotropic, and therefore Curie-spin relaxation and pseudo-contact shifts are negligible. PRE 1H-Γ2 rates arising from ATCUN-Cu2+ can be used for quantitative analysis. The HHP motif (NH2-His-His-Pro) placed at the N-terminus of a protein binds Ni2+ and results in the formation of a dimer in which two molecules of the protein are bound to a single Ni2+ ion.167 The lanthanide binding motif (Tyr-Val-Asp-Thr-Asn-Asn-Asp-Gly-Ala-Tyr-Glu-Gly-Asp-Glu-Leu) is a 15-residue sequence that specifically binds lanthanide ions and can be incorporated either genetically168 or chemically.125
3.1.2. Paramagnetic probes attached to nucleic acids
For chemically synthesized nucleic acids the incorporation of a paramagnetic group is straightforward. Phosphoramidite derivatives of nucleotides bearing a paramagnetic group have been developed and they can be incorporated at any desired position of the DNA or RNA during the course of solid-phase synthesis. Just as for proteins, either a nitroxide spin radical or a metal-chelator for incorporation of a paramagnetic metal ion can be covalently attached to nucleic acids.
EDTA-derivatized nucleotides (Figure 10) have been used most frequently in PRE studies of protein-DNA interactions.23,24,78,94,112,169 The phosphoramidite for dT-EDTA is commercially available and dT-EDTA can be incorporated at any desired position using a normal DNA synthesizer. Synthesis of a similar EDTA-derivative of the cytosine nucleotide (dC-EDTA) has also been carried out.170 Although dT-EDTA was originally developed for sequence specific DNA cleavage via the Fenton reaction with Fe2+ chelated to the EDTA group,171) cleavage does not occur with either Mn2+ and Ca2+.94 Since divalent ions bind DNA with a dissociation constant Kdiss in the 10−4−10−3 M range,172–174 it is important to remove divalent ions at undesired locations by extensive washing.24,94
Figure 10.
EDTA-derivatized DNA bases.94,170,171 These modified bases can be incorporated at any desired position by solid-phase DNA synthesis.
Various types of nitroxide-conjugated nucleotides have been developed mostly for EPR (see review by Keyes and Bobst175). Some of these derivaives may not be suitable for NMR purposes because of requirements of larger amounts, higher conjugation efficiency and higher chemical stability. To date, the nitroxide-labeled bases represented by compounds 3 and 4 in Figure 11 have been successfully used for NMR investigations.95,176,177
Figure 11.
3.1.3. Paramagnetic co-solutes for probing molecular surfaces
Small paramagnetic compounds added to macromolecular solutions as co-solute molecules provide a unique means of obtaining information on the molecular surface of macromolecules. Random collisions of a macromolecule with the paramagnetic co-solute at relatively high concentration cause sizeable PREs for 1H nuclei of the macromolecule depending on their depth from the molecular surface (Figure 8C). For simplicity and clarification, we refer to such PREs as solvent PREs.
Three paramagnetic co-solute molecules have gained popularity: TEMPOL,178,179 O2,180,181 and Gd-diethylenetriamine pentacetic acid-bismethylamide (Gd-DTPA-BMA)23,182 (Figure 12). Since these are neutral molecules with no net charge, their spatial distribution with respect to a macromolecule in solution is generally assumed to be uniform. This approximation appears to be valid for O2 and Gd-DTPA-BMA and the resulting PRE patterns can be predicted reasonably well from the macromolecular structure (see Section 3.4.2). Hydrophobic patches on macromolecular surfaces, however, tend to bias the spatial distribution of TEMPOL via preferential hydrophobic interactions. The solvent PRE is also useful for identifying binding interfaces in macromolecular complexes.23,181 For the latter application, the solvent PREs for the free and complex states are compared. Since the binding interface is exposed in the free state but buried in the complex, the solvent PRE rates for the binding interface decrease upon complex formation.
Figure 12.
Co-solute paramagnetic probes for investigations of molecular surface.
In some circumstances, probes that exhibit a biased distribution of collision sites on macromolecules can also be useful. For example, use of a positively charged paramagnetic probe (e.g. charged derivatives of TEMPOL) can permit the identification of negatively charged patches on the molecular surface.183,184 Another example is the use of spin-labeled lipids to identify lipid-contacting regions of membrane proteins.185,186
3.2. Pulse sequences for the measurement of PRE 1H-Γ2 rates
The PRE rate Γ2 is measured as the difference in transverse relaxation rates between the paramagnetic (R2,para) and diamagnetic (R2,dia) states of the macromolecule:
| [25] |
This subtraction cancels out relaxation mechanisms common to both states such that the only remaining relaxation mechanism arises from electron-nucleus interactions (i.e. the PRE effect). For the case of systems with an isotropic electron g-tensor, chemical exchange contributions to the transverse relaxation rate R2, are also canceled. This is not the case, however, for systems with an anisotropic electron g-tensor. In this instance the exchanging species may experience different pseudo-contact shifts, and therefore the exchange contributions can be significantly different in the paramagnetic and diamagnetic states.
In many papers in the literature, the ratio Ipara/Idia of the signal intensities for the paramagnetic (Ipara) and diamagnetic (Idia) states is inappropriately referred to as the PRE. Such a ratio is not physically meaningful, because it depends not only on the PRE 1H-Γ2 rate, but also on the corresponding PRE 1H-Γ1 rate, the delays for coherence transfers, and the type of data processing. For quantitative PRE investigations of macromolecules, measurement of actual PRE 1H-Γ2 rates is required and for systems with an isotropic electron g-tensor such measurements are straightforward.
3.2.1. Γ2 measurement for 15N-attached 1H-nuclei
Figure 13 shows the pulse sequence used to measure 1H-Γ2 rates on 1H-nuclei directly bonded to 15N.83,123 The period for the 1H transverse relaxation measurement is incorporated in the first INEPT scheme. During the 1H transverse period of T + 2τa, 15N 180 pulses swap the slow and fast relaxing components (represented by Hy − 2HyNz and Hy + 2HyNz, respectively) and the contributions from the two components are identical, making the overall decay a single-exponential process with an average relaxation rate. The observed relaxation rate corresponds to 1/T2 for 1H in-phase terms, since the transverse relaxation rates for Hy and 2HxNz terms are expected to be virtually identical for macromolecules. (Note that T1 relaxation of N2 is much slower than 1H–T2 relaxation). Identical experiments are performed for the paramagnetic and diamagnetic samples to obtain Γ2 (cf. Eq. [25]). This pulse sequence can readily be modified to a transverse relaxation optimized (TROSY) version. In this instance, Boltzmann 15N-magnetization should be destroyed so that all observables are modulated by 1H transverse relaxation during the time interval T.
Figure 13.
Pulse sequence for the measurement of PRE Γ2 rates on HN protons. Details are given in Iwahara et al.123 published in J. Magn. Reson (Elsevier) while the authors were US Government employees at the National Institutes of Health.
A two-time-point measurement provides a simple means of obtaining Γ2 rates and their corresponding errors without making use of any fitting procedures. In this approach, 1H-Γ2 rates are determined from two time points (T = 0 and ΔT) for transverse relaxation as follows:
| [26] |
where Idia and Ipara are the peak intensities for the diamagnetic and paramagnetic states, respectively. It should be noted that effects of homonuclear 3JHNHα-modulation during the delay T are cancelled out by using identical times and taking ratios for the two states. The errors in Γ2 can be propagated from Eq. [26] based on partial derivatives with respect to peak intensities, and are given by:
| [27] |
where σdia and σpara are the standard deviations of the noise in the spectra recorded for the diamagnetic and paramagnetic states, respectively. Figure 14 shows a comparison of PRE 1H-Γ2 rates determined from two- versus eight-time points for the same overall measurement time and it can be seen that agreement is excellent.
Figure 14.
Comparison of 1H-Γ2 rates obtained from a simple two-time point measurement versus a conventional eight-time point measurement. Adapted from Iwahara et al.123 published in J. Magn. Reson (Elsevier) while the authors were US Government employees at the National Institutes of Health.
3.2.2. Γ2 measurement for 13C-attached 1H-nuclei
Figure 15 shows the pulse sequence used to measure PRE 1H-Γ2 rates for 1Hα protons directly bonded to 13C.78. This experiment is recorded as a 2D 1H–15N correlation experiment with the initial excitation on 1Hα nuclei, followed by coherence transfers to 1HN through 13Cα, 13C and 15N nuclei. A scheme for 1H-Γ2 relaxation is incorporated in the first INEPT transfer step. Since CSA-DD cross relaxation is negligible for Hα, the scheme is simpler than that for the Γ2 measurement on HN. 2D spectra with different T-delays are recorded in an interleaved manner to determine PRE Γ2 rates.
Figure 15.
Pulses sequences for the measurement of PRE Γ2 rates on (A) 1Ha directly bonded to 13C and (B) all other 13C-attached protons. Details are given in supporting information of Iwahara et al.78 published in J. Am. Chem. Soc. (American Chemical Society) while the authors were US Government employees at the National Institutes of Health.
The same scheme can also be incorporated into a 1H-13C HSQC experiment to measure PRE 1H-Γ2 for other 13C-attached protons (Figure 15). Although the T2 process for methyl CH3 protons is bi-exponential due to cross-correlation,187 a two-time point measurement using Eq. [26] still gives reasonably accurate Γ2 values as long as the PRE affects both components of the bi-exponential decay equally. Indeed, the PRE measured on methyl protons in the SRY/DNA-EDTA-Mn2+ complex using this method exhibited excellent agreement with those calculated from the structure.78 In favorable cases, even two methyl groups in the same Leu or Val residue can be stereospecifically distinguished based on the measured 1H-Γ2 rates and the three-dimensional structure.
3.3. Practical considerations for Γ2 measurements
3.3.1. Choice of time points
In the context of a two-time point measurement of Γ2, the time-points should be chosen to minimize the errors in the measured Γ2 rates. Figure 16 illustrates the theoretical relationship between ΔT and the error in Γ2, obtained using Eq. [27] and the following equation:
| [28] |
where τ represents the overall 1H transverse relaxation period for the coherence transfers (set to 9.9 ms). Eq. [28] assumes a Lorentzian line-shape in the 1H-dimension, equal recovery levels during the repetition delay, and the same concentration and number of scans for both the diamagnetic and paramagnetic samples. Under these conditions, the error σ(Γ2) is minimal when Ta = 0 and Tb ~ 1.15/(R2dia+ Γ2). As can be seen from Figure 16, the larger the value of Γ2, the narrower the optimal range of ΔT. Therefore, the second time-point should be set to be optimal for a relatively large Γ2 value expected for the system under study. For example, if the range of expected Γ2 rates is 0 to 75 s−1, a second time point Tb at ~1.15(R2,dia+ 50) seconds represents a reasonable choice. (However, if diamagnetic contamination in the paramagnetic sample is greater than ~3%, smaller values are required for accurate measurement of intramolecular Γ2 rates; see below).
Figure 16.
Error in PRE Γ2 as function of ΔT in two-time point measurement. Adapted from Iwahara et al.123 published in J. Magn. Reson. (Elsevier) while the authors were US Government employees at the National Institutes of Health.
3.3.2. Undesired solvent PRE effects at high sample concentration
For PRE analysis, the sample concentration should be relatively low (~0.2–0.5 mM) to avoid contributions from additional undesired PREs arising from random "elastic" collisions between a molecule and the paramagnetic group of another molecule. This effect corresponds to the ‘solvent PRE’ discussed in Section 3.1.3. Whether or not this effect is significant can be easily ascertained by carrying out PRE measurements at different sample concentrations since the magnitude of the solvent PRE is directly proportional to the sample concentration. For example, solvent PRE effects were observed for some residues of ubiquitin tagged with ATCUN-Cu2+ at its N-terminus at high sample concentration (~2 mM) but were insignificant at lower concentrations (< 0.6 mM).83
Due to the requirement of low sample concentration, the measurement of PRE Γ2 rates can be somewhat time-consuming in the absence of a cryogenic probe. Total measurement time (including data acquisition for both diamagnetic and paramagnetic samples) on a cryoprobe required to obtain reasonably precise data with errors less than ~10% for Γ2 ~50 s−1 is about two days for a ~0.3 mM non-deuterated sample comprising a system of ~20 kDa in size.
3.3.3. Field dependence of PRE 1H-Γ2
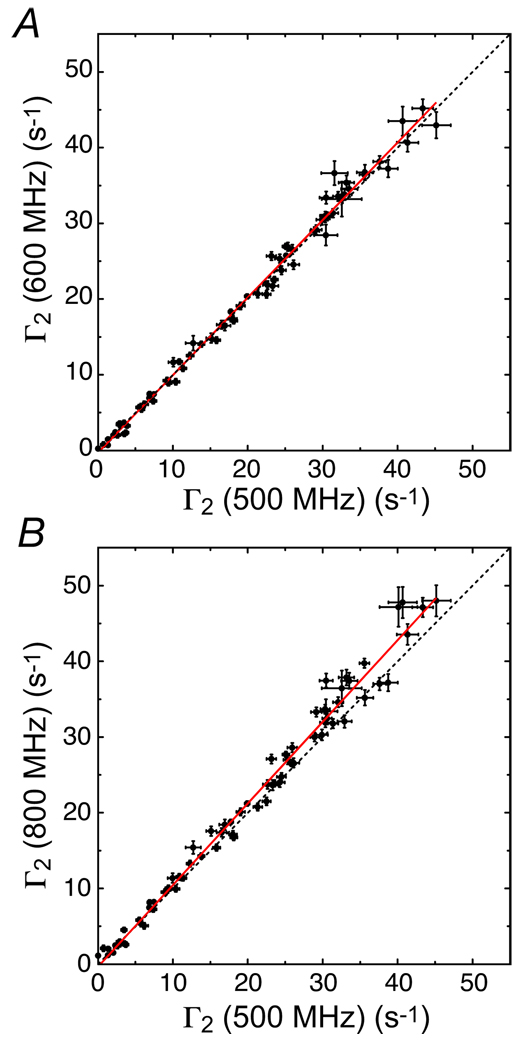
Since Γ2 for a macromolecule is dominated by the value of the spectral density function at zero frequency, 1H-Γ2 rates measured at high magnetic field (B0 > 10 Tesla) should be dependent on B0 if (a) the PRE correlation time τc is field-dependent, or (b) the contribution from Curie-spin relaxation (whose rate is proportional to B02) is non-negligible.
In the case of SRY/DNA-EDTA-Mn2+ complex the field-dependence of 1H-τ2 is very weak (Figure 17). The relative contribution of Curie-spin relaxation depends upon the size of the system. For a very large system with a rotational correlation time τr larger than 50 ns (corresponding to a molecular weight in excess of 100 kDa), the contribution could be larger than 20% at 800 MHz. In the case of a nitroxide spin label, the electron spin relaxation time τs (>10−7 s) is much longer than τr,188,189, and the PRE correlation time τc, defined as , is virtually identical to τr, resulting in a field-independent 1H-Γ2. For a macromolecular system with conjugated EDTA-Mn2+, for which τs is comparable to τr,78,94,130, 1H-Γ2 could be field-dependent because of the field-dependence of τs.
Figure 17.
Field-dependence of 1H-Γ2 rates arising from dT-EDTA-Mn2+ in the SRY/DNA-EDTA-Mn2+ complex. Adapted from Iwahara et al.123 published in J. Magn. Reson. (Elsevier) while the authors were US Government employees at the National Institutes of Health.
3.2.4. Effect of diamagnetic impurities on PRE 1H-Γ2
In practice, no matter how carefully a paramagnetic sample may be prepared, contamination by trace amounts (~1–5%) of the corresponding diamagnetic species is almost impossible to avoid owing to several factors: (a) incomplete conjugation of the extrinsic paramagnetic group and insufficient purification of the conjugated species; (b) the presence of diamagnetic impurities in the paramagnetic stock solution (e.g. trace amounts of diamagnetic metals such as Zn2+ and Ca2+ in stock solutions of Mn2+ for the EDTA-Mn2+ system; the reduced species for a nitroxide spin label); (c) chemical instability of the conjugated states (discussed in a later section). Thus, it is important to ascertain the impact of diamagnetic contamination on the measured 1H-Γ2 rates. In fact, the effect of diamagnetic impurities on PRE measurements can be significant but fortunately can be reduced with careful setting of experimental conditions as shown below.123
Under conditions where the chemical shifts for the diamagnetic and paramagnetic states are identical and they do not exchange with each other, the signal intensity for the paramagnetic sample containing a trace amount of the diamagnetic species with population pd is given by:
| [29] |
Figure 18 displays the theoretical relationship between the true value of 1H-Γ2 and the apparent value (Γ2app) that would be obtained from a two-time point measurement for two values of ΔT and diamagnetic contaminations ranging from 0 to 5%. Γ2app is always smaller than the true Γ2 value. For the case with R2,dia = 50 s−1, Γ2 = 60 s−1 and pd = 2%, the apparent value of Γ2 from the two-time point measurement with ΔT = 18 ms is 52 s−1. For pd = 5%, Γ2app is reduced to 44 s−1. Note that the percentage error in Γ2 is much larger than pd. This is due to the fact that the PRE significantly reduces the contribution of the first term in Eq. [29]. The deviation from the true value of Γ2 can be reduced by using a relatively small value of ΔT, thereby reducing the relative contribution from the second term in Eq. [29]. For example, the apparent values of Γ2 obtained with ΔT = 6 ms are 55 s−1 for pd = 2% and 48 s−1 for pd = 5%.
Figure 18.
Theoretical effect of diamagnetic impurities on the measurement of 1H-Γ2 rates. Adapted from Iwahara et al.123 published in J. Magn. Reson (Elsevier) while the authors were US Government employees at the National Institutes of Health.
Figure 19 shows correlations between experimental 1HN-Γ2 data measured with ΔT = 4 ms and 40 ms on 15N-HPr(E32C) conjugated with EDTA-Mn2+ at Cys32. As expected from the above considerations, the measured Γ2 values derived from the experiment with ΔT = 40 ms are systematically smaller than those obtained with ΔT = 4 ms, indicating the presence of a diamagnetic impurity. From this correlation, pd was estimated to be 3%. The same population for the diamagnetic impurity can also be obtained from the intensities of residual peaks that should be completely broadened beyond detection, yet appear with low intensity because of the diamagnetic contamination (Figure 19b).
Figure 19.
Experimentally observed effect of diamagnetic impurities on 1H-Γ2 rate measurements. Adapted from Iwahara et al.123 published in J. Magn. Reson (Elsevier) while the authors were US Government employees at the National Institutes of Health.
In the case of intermolecular PRE measurements on a complex where dissociation and association processes are in the fast exchange regime on the relaxation time scale, the observed Γ2 is simply scaled down by a factor of (1 - pd) and the effect of a diamagnetic impurity is much weaker than for the intramolecular case considered above.
3.4. PRE back-calculation from three-dimensional structure
In this section, we review the computational methods required to back-calculate PRE rates from macromolecular structures. While the back-calculation is straightforward for a static system, motional effects need to be taken into consideration for a dynamic system.
3.4.1. PRE arising from paramagnetic probes attached to macromolecules
Two averaging techniques have been proposed for back-calculating NMR parameters in systems with flexibility: time-averaging of restrained molecular dynamics trajectories190–193, and ensemble-averaging.194–200 To date, the latter approach has been adopted for PRE back-calculation.78,100,105
The computational method we have developed to back-calculate 1H-PREs makes use of a multiple conformer representation for the paramagnetic group.78 This approach has been implemented as the ‘prePot’ pseudo-energy term in the macromolecular structure determination software package Xplor-NIH201,202 The target function for the PRE restraints, EPRE, is defined as:
| [30] |
where kPRE is a force constant; wi is a weighting factor that is defined for each restraint; Γ2obs(i) and Γ2calc(i) (cf. Eq. 18 in Section 2.2.1) are the observed and calculated PRE rates, respectively; and the summation is over individual 1H-Γ2 PRE data. To account for the finite conformational space occupied by a flexible paramagnetic group, N different conformers are used to represent the paramagnetic group in the calculations, while the remainder of the molecule is represented by a single structure. Individual conformers are treated independently under the restraints of the 1H-PRE target function and can overlap with each other since interactions between the conformers are excluded from the non-bonded interaction energy term. The N conformers represent a discrete jump model where fractions of individual sites and the transition probability over a long period of time are uniform and equal to 1/N. Since members of the ensemble can overlap structurally, this model is also valid for a case with fewer sites and uneven distributions providing N is large enough. To obtain the ensemble average of Γ2, the ensemble-averaged quantity <r−6> for the electron-proton distance r must be calculated as follows:
| [31] |
where np is the number of equivalent protons (i.e. 1 for NH or CH; 2 for equivalent aromatic protons that are degenerate owing to fast ring flipping; 3 for methyl protons). This quantity is used to back-calculate the PRE Γ2 rate based on either the SB (Eq. [17]) or SBMF (Eq. [18]) equations. The direct application of the SBMF equation requires the explicit use of order parameters S2 for the PRE interaction vectors as defined by Eq. [13]. These order parameters are calculated directly from the N-site discrete jump model of the paramagnetic group used in the simulated annealing calculations, assuming that positional fluctuations of the analyzed 1H nucleus are small compared to the paramagnetic center-1H distance and hence do not contribute to S2 for the 1H-PRE interaction vector. For the N-state jump model, the order parameters can be calculated using Eq. [13] together with the equations:
| [32] |
| [33] |
Owing to increased complexity, the use of the SBMF equation in the PRE back-calculation is computationally more expensive. Figure 20 displays correlations between experimental Γ2 data obtained for the SRY-DNA complex with the dT-EDTA-Mn2+ paramagnetic label located at three separate sites (one at a time) and the calculated Γ2 rates using a three-conformer ensemble for the paramagnetic labels. Quantitative agreement between observed (Γ2obs) and calculated (Γ2calc) rates is given by the PRE Q-factor calculated as:
| [34] |
For the SRY-DNA complex, the overall Q-factor for a three-member ensemble representation for the paramagnetic labels is 0.20 for backbone protons (HN and Hα) and 0.26 for side chain protons.
Figure 20.
Correlation between observed and calculated PRE 1H-Γ2 rates for the NMR structure of the SRY-DNA complex. The experimental data comprise PREs collected for the EDTA-Mn2+ paramagnetic label at three different sites, one at a time. Each paramagnetic labels was represented by a three-conformer ensemble.78 Adapted from Iwahara et al.94 published in J. Am. Chem. Soc. (American Chemical Society) while the authors were US Government employees at the National Institutes of Health.
The introduction of a multiple ensemble representation for the paramagnetic labels results in a substantial decrease in both the working (i.e. PRE data included in the refinement) and free PRE Q-factors (i.e. PRE data excluded from refinement and used for complete cross-validation), indicating that the back-calculation method results in improved coordinate accuracy without overfitting of the data. Independent confirmation that the ensemble representation of the paramagnetic label results in a substantial improvement in coordinate accuracy is provided by cross-validation against RDCs.78 Indeed, if the paramagnetic label is represented by only a single species, coordinate accuracy decreases substantially.78
The method of PRE back-calculation presented in this section can also be applied to an exchanging system, as discussed later on in Section 4.2, providing that every process involved is in fast exchange on the PRE timescale.
3.4.2. Otting-LeMaster approach for predicting solvent PREs
The PRE arising from paramagnetic co-solute molecules such as those displayed in Figure 12 depends on many factors, including translational diffusion constants, electron relaxation time and rotational correlation time. Hwang and Freed proposed a general theory to describe the solvent PRE effect in which pair-correlation functions related to a potential of averaged forces between the molecules are considered.203,204 Despite its generality, application of the Hwang-Freed theory to macromolecules is extremely complicated and consequently of no practical use. Although the hard-sphere model that provides the simplest pair-correlation functions was found to be effective for systems involving small molecules, the model was found to be too simple for macromolecules and predictions based on the model are in poor agreement with the experimental data.
An empirical approach to predict solvent PREs from three-dimensional structure was proposed independently by two groups.180,182 We refer to this approach as the Otting-LeMaster approach. In this method, the coordinates of the macromolecule are embedded in a cubic lattice with grid spacing < 1.0 Å (Figure 21). For each hydrogen atom of the macromolecule, the following quantity is calculated:
| [35] |
where rgrid-H is a distance between a 1H-nucleus and a grid point; δgrid is 0 for grid points that overlap with the macromolecule and 1 for the others. All grid points within the van der Waals radius plus the molecular radius of the paramagnetic co-solute from any atom in the macromolecule are regarded as overlapped. Summation is over the entire lattice. A solvent PRE is predicted to be aAOL, for which a is a scaling factor common to all protons in the macromolecule. Since the scaling factor a includes the effects of the concentration of co-solute, correlation-times, etc… and can therefore not be determined explicitly, its value is obtained by numerical optimization against the experimental data. Profiles of solvent PREs predicted with this empirical approach have been found to be in a reasonably good agreement with experimental observations. LeMaster’s group applied this approach to solvent PRE 1H-Γ1 data arising from O2 for the backbone amide 1HN protons in rubredoxin proteins.180 Otting’s group applied the method to solvent PRE 1H-Γ1 data arising from Gd-DTPA-BMA for 13C-attached 1H nuclei in ubiquitin.182 The same approach was also found to be useful for the prediction of solvent PRE 1H-Γ2 data arising from Gd-DTPA-BMA for specific- and non-specific protein-DNA complexes (Figure 22).23
Figure 21.
Otting-LeMaster180,182 approach for predicting solvent PREs from structure. For the prediction, distances from a 1H nucleus to grid points that can be occupied by co-solute molecules are calculated and used in Eq. [35].
Figure 22.
Comparison of observed and calculated solvent PRE profiles for the HoxD9 homeodomain free in solution and bound to DNA in specific and non-specific complexes. (A) Experimental solvent PRE profiles for free HoxD9 (black) and HoxD9 bound specifically (red) and non-specifically (green) to DNA. (B) Predicted solvent PREs ratios for free to specifically bound HoxD9 calculated using the Otting-LeMaster approach.23 (C) Experimentally observed solvent PRE ratios for free to non-specifically bound HoxD9. (D) Experimentally observed solvent PRE ratios for free to specifically bound HoxD9. Adapated from Iwahara et al.23 published in Proc. Natl. Acad. Sci. U. S. A. (National Academy of Sciences) while the authors were US Government employees at the National Institutes of Health.
Since the Otting-LeMaster approach permits back-calculation of solvent PREs from the structure, solvent PRE data may potentially be used as restraints in NMR structure calculations. Such restraints would be unique in that they constrain depths of observed 1H nuclei from the molecular surface. Currently, however, two problems need to be overcome to make this practical. First, back-calculation of solvent PREs using this approach is computationally too expensive owing to the requirement for three-dimensional integration. Second, the partial derivatives of the back-calculated solvent PREs with respect to the atomic coordinates, which are essential to build a pseudo-energy potential for structure calculation, are difficult to compute.
4. Applications
In this section we discuss a series of PRE applications aimed at studying species involved in rapid dynamic processes and interconversions at equilibrium. These include the study of non-specific protein-DNA interactions, investigations into the search process whereby a specific DNA-binding protein locates its target site, the detection and characterization of encounter complexes in both specific and non-specific protein-protein interactions, the probing of large scale interdomain motions involving lowly-sampled states, and finally investigations aimed at probing the conformational space sampled by denatured or intrinsically disordered proteins.
4.1. Investigations of the target search process in protein-DNA interactions
4.1.1. Non-specific protein-DNA interactions
Non-specific protein-DNA interactions play a crucial role in increasing the efficiency whereby specific DNA binding proteins locate their cognate DNA binding site within a sea of non-specific DNA sites. Non-specific protein-DNA interactions, however, are particularly difficult to study since they are intrinsically dynamic. That is to say these interactions involve both one-dimensional diffusion of the protein along the DNA and hopping of the protein from one DNA molecule or segment to another.9,20,205 As a consequence, characterizing non-specific interactions of a protein with DNA in terms of both structure and dynamics is highly challenging since the experimental observables represent an ensemble average of many rapidly exchanging states with the protein moving between multiple overlapping sites on the DNA. One way around this problem is to design an experimental system in which the protein is constrained to only a single location on the DNA, for example by using a length of DNA that is exactly equal to the length of the DNA binding site and/or by making use of cross-linking.206–210 However, the dynamic nature of the non-specific interactions is lost, and, further, the biological relevance is questionable since it is hard to exclude that such structures may represent partially specific complexes or are heavily influenced by extrinsic effects imposed for example, by crystal packing forces or cross-linking agents. An alternative approach is to directly study non-specific interactions in a naturally dynamic environment by means of the PRE.
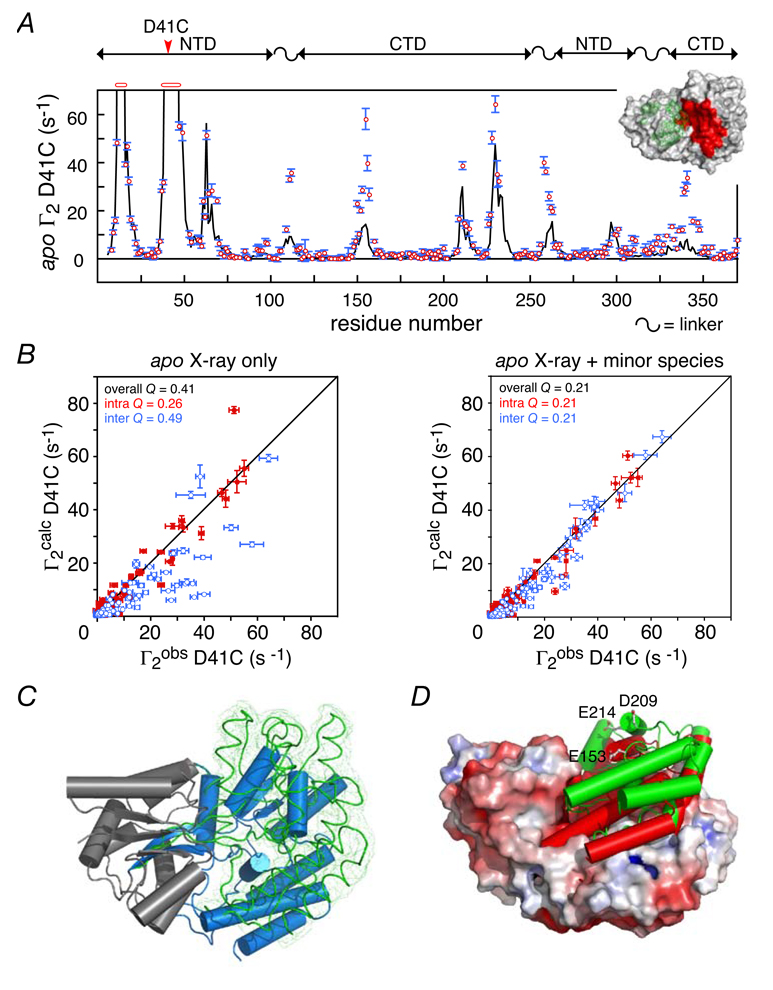
The first non-specific protein-DNA interaction we studied involved the A box of the classical HMG-box protein, HMGB-1. 112 HMGB-1A is an architectural protein that binds non-specifically in the minor groove of DNA causing the DNA to bend.211,212 The DNA binding domain of HMGB-1A is highly homologous to that of the male sex-determining factor SRY, another member of the HMG-box family.213,214 In contrast to HMGB-1A, SRY binds sequence specifically to DNA with an affinity that is approximately 2 orders of magnitude higher (KD ~ 20 nM versus 0.9 µM). The 1H–15N correlation spectrum of the HMGB-1A/DNA complex with a 14-bp DNA duplex reveals a single set of well-resolved narrow cross-peaks (Figure 23A). From such a spectrum it would be impossible to ascertain whether HMGB-1A was binding to only a single site on the DNA or to multiple sites with rapid exchange between the available binding modes (Figure 23B). Intermolecular PRE data recorded on the complexes with paramagnetic labels (dT-EDTA-Mn2+) positioned at either end of the DNA resolve the situation unambiguously (Figure 24). The intermolecular PRE profiles obtained with SRY are fully consistent with a single specific complex and, as expected, the PRE profiles observed for the two paramagnetic labels are completely different (Figure 24A, lower left-hand panel). In the case of the HMGB-1A/DNA complex, however, the intermolecular PRE profiles obtained with the two paramagnetic labels are virtually identical and, moreover, the magnitude of the PREs is considerably larger than that observed for SRY (Figure 24A, upper left-hand panel). The observed PRE profiles for the HMGB-1A/DNA complex are characteristic of a protein binding to multiple DNA binding sites in two possible orientations differing by a 180° rotation (cf. Figure 24 A, upper, right-hand panels).
Figure 23.
NMR characterization of the HMGB-1A/DNA non-specific complex. (A) 1H-15N correlation spectrum of U-[15N/13C]-HMGB-1A in the presence of a 14-bp DNA duplex (natural isotopic abundance) at a ratio of 1:1.5 protein to DNA to ensure that all protein is bound and only a single molecule of protein is bound per 14-bp DNA duplex. (B) The 1H-15N correlation spectrum does not permit one to distinguish between a single binding mode (top) and multiple binding modes in rapid exchange with one another (bottom). Adapted from Iwahara et al.112 published in J. Am. Chem. Soc. (American Chemical Society) while the authors were US Government employees at the National Institutes of Health.
Figure 24.
PRE characterization of HMGB-1A binding non-specifically to DNA. (A) Comparison of the intermolecular PRE profiles observed for the non-specific HMGB-1A/DNA complex (top) and the specific SRY/DNA complex (bottom) with two DNA duplexes bearing the dT-EDTA-Mn2+ paramagnetic label at the two ends of the DNA. A diagrammatic depiction of the states giving rise to the observed PREs is shown on the right-hand side of the figure. (B) Semi-quantitative analysis of the intermolecular PRE data used to estimate the distribution and occupancy of HMGB-1A binding sites along a 14-bp DNA duplex. There are a total of 13 potential intercalation sites for Phe-37 and HMGB-1A can bind in two orientations related by a 180° rotation (top left panel) giving rise to a total of 26 states. Minimization of the χ2 function shown in Eq. [36] results in a PRE Q-factor of 0.36 (top right panel) with optimized distributions for the two orientations shown in the bottom two panels. Adapted from Iwahara et al. 112 published in J. Am. Chem. Soc. (American Chemical Society) while the authors were US Government employees at the National Institutes of Health.
The intermolecular PRE data for the HMGB-1A complex can be further analyzed using a semiquantitative approach to obtain an estimate of the populations of the individual binding sites.112 With N discrete binding sites in rapid exchange, the ensemble average <Γ2> rate is given by a population weighted average of the Γ2 rates for the individual states:
| [36] |
where ρk and Γ2k are the population and Γ2 rate for state k, respectively. The populations ρk can be obtained by back-calculating the Γ2 rates from structural models of the 26 possible states, 13 in each of the two possible orientations (Figure 25B), and minimizing the χ2 function:
| [37] |
where i is the residue number, Γ2,obs(i) the observed value of Γ2 for residue i; Γ2,calck(i) the calculated value of Γ2 for binding state k; and σ(i) the experimental error for Γ2,obs(i). The structural models were generated from the crystal structure of the HMGB-1A/cisplatin-modified DNA complex.212 Despite the relatively crude nature of the model, the overall Q-factor after minimization is 0.36 with a correlation coefficient of 0.71. The occupancy of HMGB-1A molecules in orientations 1 and 2 is essentially equal, with maximal occupancy for sites 5–8 in orientation 1 and sites 6–9 in orientation 2. The site of intercalation of Phe37 is located at the edge of the DNA binding site, and full coverage of the DNA binding site (~7 bp) is obtained at intercalation sites 5–13 in orientation 1 and intercalation sites 1–9 in orientation 2 (Figure 24B). Thus one would expect reduced occupancy of sites 1–4 in orientation 1 and sites 10–13 in orientation 2, as a consequence of a progressive decrease in protein-DNA contacts. However, the reduced populations for sites 9–13 in orientation 1 and sites 1–5 in orientation 2 presumably reflect a subtle degree of sequence preference since the distribution is not the same as that expected from simple statistical considerations if the interaction energy at a given site was only dependent on base-pair coverage.
Figure 25.
Intermolecular PREs observed for the specific HoxD9/DNA complex in low and high salt corresponding to slow and fast exchange regimes on the PRE relaxation time scale, respectively, for the interconversion between the specific complex and lowly-populated (<1%) non-specific complexes. (A) 24-bp DNA duplex with the specific site centrally located (boxed) and the location of the 4 paramagnetic labels (one at a time) indicated. (B) Diagrammatic representation of the specific complex (left) and the target search process whereby the specific complex is located (right). (C ) and (D) Correlation between observed and calculated PREs for all 4 sites at low (20 mM NaCl) and high (160 mM) NaCl), respectively. The calculated rates are obtained from a model derived from the crystal structure of the Antennapedia homeodomain/DNA complex. (E) and (F) Intermolecular PRE profiles observed at low (20 mM NaCl) and high (100 and 160 mM NaCl), respectively. The PRE data are mapped on the structural model of the specific complex, with the color coding depicting the observed PRE rates. Adapted from Iwahara et al.24 published in Nature (Nature Publishing Group) while the authors were US Government employees at the National Institutes of Health.
The PRE has also been used to characterize diffusion and hopping of the homeodomain HoxD9 on non-specific DNA.23 As in the case of HMGB-1A, very similar intermolecular PRE profiles with large PREs are observed with the paramagnetic label positioned at either end of the DNA duplex. Interestingly, the RDCs measured on HoxD9 bound specifically and non-specifically to DNA in a dilute liquid crystalline medium of phage pf1 are virtually identical with a correlation coefficient of 0.99 and a pairwise rms difference of only 2.3 Hz. Further, the measured RDCs for both the specific and non-specific complexes are in excellent agreement with those calculated from the crystal structure of the Antennapedia homeodomain-DNA complex215 with RDC R-factors56 of 13.7 and 15.2%, respectively. These data indicate that (i) the binding orientation of HoxD9 with respect to the long axis of the DNA at each non-specific site is effectively identical to that for the specific complex; (ii) end-effects that could potentially result in alternative binding modes are insignificant; (iii) alignment in the pf1 medium is dominated by the negative charges on the DNA phosphate backbone and is minimally sensitive to protein location on the DNA; and (iv) the DNA is essentially straight.23 (In the case of the HMGB-1A/DNA complex where HMGB-1A induces a ~50° bend in the DNA, the alignment would be dependent upon protein location since this determines the site of the bend on the DNA, and therefore the overall shape of the DNA).
4.1.2. Locating the cognate target site in specific protein-DNA interactions
A sequence specific DNA binding protein must be able to locate its specific target site within a sea of non-specific sites. Non-specific DNA binding can facilitate this process by permitting both intramolecular one-dimensional sliding on the DNA coupled with intermolecular translocation involving hopping from one DNA molecule to another or from one non-specific DNA site to another.9,20,205 Intermolecular PRE measurements provide a direct means for investigating these processes.
The HoxD9 homeodomain binds specifically to DNA with a KDiss of ~1.5 nM in 100 mM salt compared to a KDiss of 0.2–0.4 µM for non-specific binding.23,24 Intermolecular PRE data were collected for the HoxD9/DNA complex with the specific DNA site located in the center of a 24-bp DNA duplex and paramagnetic labels (dT-EDTA-Mn2+) placed at four different sites (Figure 25A).24 At low salt (20 mM NaCl) the intermolecular PRE data are completely consistent with a model derived from the crystal structure of the Antennapedia homeodomain/DNA complex215 with a combined Q-factor for all 4 paramagnetic labels of 0.26 (Figure 25C). Thus, large magnitude PREs are only observed for residues in close proximity to the label in the specific complex (Figure 25E). As the salt concentration is raised (100 and 160 mM NaCl), however, the intermolecular PRE data become completely inconsistent with the structure with a Q-factor of 0.66 (Figure 25D) and large intermolecular PREs are observed on regions of the protein far away from the paramagnetic label in the specific complex (Figure 25F). Moreover, the PRE data observed with the paramagnetic label at sites 1 (base pair 2) and 4 (base pair 23) are very similar, and likewise for the PRE data observed with the paramagnetic label at sites 2 (base pair 5) and 3 (base pair 18) (Figure 25F). In all cases, however, the residues of HoxD9 that exhibit large intermolecular PREs are located close to or at the DNA binding interface and can therefore readily come close to the paramagnetic labels. The 1H–15N correlation spectrum of HoxD9 is minimally perturbed by salt and the RDCs for HoxD9 at low and high salt are highly correlated. The observed intermolecular PREs at high salt must therefore reflect the footprint of a stochastic search process in which there is rapid exchange between HoxD9 bound to non-specific sites at low occupancy and HoxD9 bound to the specific site. The occupancy of the minor species can be estimated to be less than 1% based on the observed KDiss values for specific and non-specific DNA binding of HoxD9.
The intermolecular PRE profiles observed for the HoxD9/DNA complex at high salt can be attributed to two processes: intramolecular sliding and intermolecular translocation. From 15Nz-exchange spectroscopy the latter is known to occur rapidly between specific sites, as well as nonspecific sites, located on different DNA molecules without necessitating the dissociation of HoxD9 from its specific DNA site into free solution, a process that is very slow (kdiss < 0.01 s−1).23,25 The apparent rate of intermolecular translocation is dependent on the concentration of free DNA as well as salt concentration. To ascertain the relative contributions of intra- and intermolecular translocation PRE measurements were carried out on two samples comprising equimolar concentrations of specific and non-specific DNA duplexes with a ratio of protein:DNAspecific:DNAnon-specific of 1:1.5:1.5.24 In sample 1, the non-specific duplex bears the paramagnetic label so that any observed PRE must involve rapid exchange via intermolecular translocation between the major specific complex (that is observed in the NMR spectrum) and lowly-populated complexes on the non-specific DNA duplex (Figure 26A). In sample 2, the specific DNA duplex has the paramagnetic label and therefore the intermolecular PREs observed on the resonances of the specific complex can arise from both intra- and intermolecular translocation events. The PRE profiles observed for the two samples are very similar (Figure 26B). However, the PREs for residues 24–33 (loop between helices 1 and 2 and N-terminus of helix 2) and 41–42 (N-terminus of helix 3) are 30–100% larger for sample 2 than those for sample 1, whereas the PREs for the N-terminal arm (that binds in the minor groove of the DNA) is essentially the same for the two samples (Figure 26B, bottom panel). The data from sample 1 indicate that intermolecular translocation is a major contributor to the observed PREs, while the larger effects for selected regions seen in sample 2 are directly attributable to intramolecular translocation which gives rise to bias since the orientation of HoxD9 on the specific site will be favored as the protein slides along the DNA (Figure 26C). The alternative orientation (180° relative to that at the specific site) requires at least one intermolecular translocation event from the DNA bearing the specific site to non-specific DNA sites located on another DNA molecule (either the specific duplex, in which case only one translocation event is needed or the non-specific one in which case two translocation events are required from the specific to the non-specific duplex and back to the specific duplex).
Figure 26.
Assessing the contributions of intramolecular sliding and intermolecular translocation for the HoxD9/DNA complex. (A) PRE data were collected on two samples comprising an equimolar concentration of specific and non-specific DNA duplexes with the paramagnetic label attached to the end of the non-specific duplex in sample 1 and to the end of the specific duplex in sample 2. The location of the specific site and the paramagnetic label are indicated in blue and red, respectively. (B) PRE profiles observed for samples 1 (blue) and 2 (red) are shown in the top panel, and the ratio of the observed PRE rates for the two samples is shown in the bottom panel. (C) Schematic representation of sliding along the DNA starting from the specific site with HoxD9 colored according to the Γ2(sample 2)/Γ2(sample 1) ratio. Adapted from Iwahara et al.24 published in Nature (Nature Publishing Group) while the authors were US Government employees at the National Institutes of Health.
4.2. Encounter complexes in protein-protein association
Transient kinetic studies on many protein-protein interactions suggest that the initial step involves the formation of a pre-equilibrium encounter complex that subsequently relaxes to the stereospecific complex. This picture is further supported by the results of both mutagenesis studies and Brownian dynamics simulations which have shown that the association rate constant can be modulated by charge alterations on the protein surface distal from the specific interaction site.10–12,14–16,216 These data suggest that, by analogy with protein-DNA interactions, non-specific encounter complexes may accelerate the formation of the stereospecific complex either via a reduction in the dimensionality of the search process, in this instance from three dimensions to two, or by the presence of a non-specific attractive potential.6,217 The presence of an ensemble of encounter complexes in rapid exchange with the stereospecific complex (Figure 27) can be probed under equilibrium conditions by means of intermolecular PRE measurements, providing the stereospecific complex is relatively weak (i.e. KDiss in the micromolar range or higher).
Figure 27.
Schematic representation of protein-protein association with the initial binding event involving the formation of an ensemble of encounter complexes that subsequently relax to the stereospecific complex. The two proteins are displayed in red and blue with the respective interaction surfaces for the stereospecific complex shown in white and yellow. Adapted from Suh et al.28 published in J. Am. Chem. Soc. (American Chemical Society) while the authors were US Government employees at the National Institutes of Health.
4.2.1. Protein-protein complexes from the bacterial phosphotransferase system
The protein-protein complexes of the bacterial phosphotransferase system (PTS) are relatively weak and in fast exchange with equilibrium dissociation constants (Kdiss) spanning the micromolar to millimolar range.218 These complexes direct phosphoryl transfer from one protein to the next along the PTS pathway that is coupled to sugar transport across the membrane and sugar phosphorylation. We carried out intermolecular PRE measurements on three complexes involving the interaction of the protein HPr with its upstream partner, the N-terminal domain of enzyme I (EIN) (Kdiss ~ 10 µM) and two downstream partners, enzymes IIAMannitol (IIAMtl) and IIAMannose (IIAMan) (Kdiss = 30–50 µM).27 In all three cases structures of the stereospecific complex had been determined by NMR on the basis of extensive NOE and RDC data.42,44,219 These structures were fully consistent with the formation of a pentacoordinate phosphoryl transition state with trigonal bipyramidal geometry without necessitating any significant displacement in the backbone coordinates of the residues immediately adjacent to the active site histidines. To investigate the presence of non-specific encounter complexes HPr was labeled with EDTA-Mn2+ at three sites (E5C, E25C and E32C).27
The intramolecular PRE data for HPr within the EIN-HPr complex are fully consistent with the structure of HPr and the PRE Q-factor for all three paramagnetic sites combined is only 0.18 (Figure 28A). However, agreement between the observed intermolecular PRE Γ2 rates measured on 15N-labeled EIN and those calculated from the structure of the stereospecific complex show much poorer agreement with a Q-factor of 0.61 (Figure 28B). The regions of agreement and discrepancy are clearly visualized in the PRE profiles (Figure 28C). For all three paramagnetic labels, there are clearly regions where there is good qualitative agreement between observed and calculated Γ2 rates. However, in each case, there are also regions where large PRE Γ2 rates are observed but small PRE Γ2 rates are predicted (Figure 28C). These discrepancies can be attributed to the presence of a small population of nonspecific encounter complexes in rapid exchange with the stereospecific complex.27
Figure 28.
PREs observed for the EIN-HPR complex with EDTA-Mn2+ conjugated to an engineered surface cysteine residue at three separate sites (E5C, E25C and E32C). (A) Correlation between observed and calculated intermolecular Γ2 rates for HPr within the EIN-HPr complex. (B) Correlation between observed and calculated intermolecular Γ2 rates measured on EIN arising from paramagnetically labeled HPr. (C) Intermolecular PRE profiles observed for the three paramagnetically labeled sites, with the experimental Γ2 rates shown as red circles, and the expected Γ2 rates calculated from the structure of the stereospecific complex42 denoted by the black line. The insets depict ribbon diagrams of the complex showing the paramagnetic label (with the distribution of Mn2+ displayed as an ensemble of three red balls) on HPr and the location on EIN of intermolecular PREs that display significant discrepancies between observed and calculated values (with color coding of the difference ΔΓ2 between observed and calculated Γ2 rates shown at the top of the figure). Adapted from Tang et al.27 published in Nature (Nature Publishing Group) while the authors were US Government employees at the National Institutes of Health.
A semi-quantitative description of the encounter complex ensemble can be obtained by rigid body simulated annealing refinement202,220 against the PRE data using a representation comprising the stereospecific complex (whose structure is known and fixed) with population p in rapid exchange with an ensemble of encounter complexes comprising N states with population (1 - p) (Figure 29A).27 These calculations minimize the difference between observed and calculated Γ2 rates for all three paramagnetic sites simultaneously. The target function comprises the PRE potential, a quartic van der Waals repulsion term to prevent atomic overlap between EIN and HPr, and a very weak radius of gyration restraint to ensure that each member of the ensemble makes at least some intermolecular contacts. The paramagnetic groups on HPr are represented by a three-conformer ensemble to account for their intrinsic flexibility. The coordinates of the EDTA-Mn2+ moieties (with atomic overlap of EDTA-Mn2+ groups permitted since these represent a distribution of states) were optimized by simulated annealing in torsion angle space against the intramolecular PRE data for HPr within the context of the EIN-HPr complex as described in Section 3.4.1. The intramolecular PRE Q-factor was 0.18 (Figure 28A). Calculations were carried out either by keeping EIN fixed and allowing an ensemble of HPr molecules to rotate and translate, or by the converse (HPr fixed and ensemble of EIN molecules) with essentially identical results. Complete cross-validation indicates that the encounter complex ensemble is best represented by an ensemble size of N = 10–20 at a population of ~10% (Figures 29B and C), yielding an overall intermolecular PRE Q-factor of 0.21 (Figure 29D).
Figure 29.
Semi-quantitative depiction of the non-specific encounter complex ensemble of EIN-HPr. (A) The representation used for rigid body ensemble refinement comprises an equilibrium mixture of the stereospecific complex (whose structure is known and fixed) and a non-specific encounter complex ensemble of size N in rapid exchange with one another. The overall calculated Γ2 rate is a population weighted average of the Γ2 rates of the species present in solution. (B) Dependence of the working (Qe and Qee) and complete cross-validated (Qfree) PRE Q-factors on ensemble size N at a population of 10% for the encounter complex ensemble. (Qe is the average Q-factor for all 100 calculated ensembles, and Qee is the ensemble of ensembles average Q-factor.) Complete cross-validation shows that the optimal size of N lies between 10 and 20. (C) Dependence of the working Q factors on population of the encounter complex ensemble for an ensemble size of N = 20. (D) Correlation between observed and calculated intermolecular Γ2 rates for N = 20 and a population of 10% for the encounter complex ensemble. (E) Two views depicting the atomic probability distribution of the encounter complex ensemble of HPr (green mesh plotted at a threshold of 20% maximum and calculated from 100 independent calculations of ensemble size N = 20 at a population of 10%) on the molecular surface of EIN (color coded by electrostatic potential, ± 8 kT) with the location of HPr in the stereospecific complex shown as a blue ribbon. Adapted from Tang et al.27 published in Nature (Nature Publishing Group) while the authors were US Government employees at the National Institutes of Health.
A simple, concise and informative manner to visualize the encounter complex ensemble is afforded by a reweighted atomic probability density map (Figure 29E) in which the contribution of each atom is rescaled depending on the distribution of its positions in the ensemble.221 The reweighted atomic density ρ at point q is defined as:
| [38] |
where Ns is the number of structures in the ensemble, Na the number of atoms in each structure, qkl the position of atom l in structure k, ρa(q) a normalized distribution for a single atom; and
| [39] |
By this means, for a given atom, the relative probability of a given position is preserved but the contribution of one atom relative to another is reweighted such that the maxima of each contribution is constant, thereby yielding an isosurface that provides a comprehensive representation of the most likely structures. In the case of the EIN-HPr complex, the probability map for the encounter complex ensemble is constructed from 100 independent calculations, each of ensemble size N = 20; that is a total of Ns = 2000 structures.
Two features of the atomic probability map for the EIN-HPr encounter complex ensemble stand out (Figure 29E). First, there is a qualitative correlation between the encounter complex ensemble distribution and the electrostatic surface potential, with the positively charged surface of HPr populating the largely negative surface of EIN. Second, the occupancy of the encounter complexes in the vicinity of the stereospecific complex is low, indicating that once HPr locates this region formation of the stereospecific complex occurs with high probability. Thus, binding and specific recognition occurs within the context of a rough, funnel-shaped energy landscape29,222 possessing many local minima comprising the sites occupied by the encounter complexes and a global minimum occupied by the stereospecific complex. The characteristics of the non-specific encounter complexes are distinct from the stereospecific complex. The latter possesses a well-defined array of complementary van der Waals and electrostatic interactions with the latter being critical in determining the orientation of binding. In the case of the non-specific encounter complex ensemble, the buried accessible surface area is on average an order of magnitude smaller and the gap index is much larger indicating that the interface is less well packed. In addition, the average fraction of charged residues at the interfaces of the non-specific encounter complexes appears to be somewhat increased relative to that at the interface of the stereospecific complex.
The salt dependence of the PREs from 0 to 0.5 M NaCl reveals that the magnitude of PREs attributable to the stereospecific complex display only a small dependence on salt concentration. This can be fully accounted for by the dependence of the overall Kdiss on salt concentration, as determined by isothermal titration calorimetry, resulting in a small decrease in the population of bound EIN as the salt concentration is increased (Figure 30). The magnitude of the intermolecular PREs originating from the ensemble of non-specific encounter complexes, however, is much more sensitive to salt concentration, implying that the population of non-specific encounter complexes is significantly more affected by ionic strength than that of the stereospecific complex (Figure 30).28 These results demonstrate the importance of electrostatic interactions in the formation of the ensemble of nonspecific encounter complexes, and are fully consistent with Debye-Hückel theory since the less compact interfacial packing for the non-specific encounter complexes relative to the stereospecific complex permits more effective screening of intermolecular electrostatic interactions by ions in solution.28
Figure 30.
Differential salt dependence of intermolecular PREs attributable to the stereospecific EIN-HPR complex and the non-specific encounter complex ensemble. (A) Correlation of intermolecular Γ2 rates arising from the stereospecific complex (left-hand panels) and the ensemble of non-specific encounter complexes (right-hand panels) at 0.15 (red), 0.3 (green) and 0.5 (blue) M NaCl versus the corresponding Γ2 rates at 0M NaCl for EIN-HPr(E5C) and EIN-HPr(E25C) paramagnetically labeled with EDTA-Mn2+. The residues for which the observed Γ2 rates are in agreement with those back-calculated on the basis of the stereospecific complex are attributed to the stereospecific complex (residues 115–125 for the EIN/E5C–HPr complex, and residues 40–80 and 127–135 for the EIN/E25C–HPr complex, while the remainder are considered to originate primarily from the ensemble of non-specific encounter complexes (residues 58–91 for the EIN/E5C–HPr complex, and residues 89–92 and 107–123 for the EIN/E25C–HPr complex) (cf. Figure 28C).28. (B) Ionic strength dependence of the Kdiss for the EIN-HPr complex determined by isothermal titration calorimetry (left-hand panel) and dependence of the slopes of the Γ2(NaCl) versus Γ2(0 M NaCl) correlations (from panel A) versus the normalized bound population pbound(NaCl)/pbound (0 M NaCl) of stereospecific complex. Adapted from Suh et al.28 published in J. Am. Chem. Soc. (American Chemical Society) while the authors were US Government employees at the National Institutes of Health.
Similar results were also observed for the IIAMtl-HPr and IIAMan-HPr complexes (Figure 31).27 (Note that IIAMtl and IIAMan bear no similarity in terms of primary, secondary, tertiary or quaternary structure). With the paramagnetic label (E5C–EDTA-Mn2+) placed on the opposite side of HPr's binding site for IIAMtl and IIAMan, there are regions on the surface of IIAMtl and IIAMan in both complexes that exhibit intermolecular Γ2 rates that are much larger than those back-calculated from the respective structures of the stereospecific complexes (Figures 31A and E). The largest discrepancies involve regions on IIAMtl and IIAMan rich in acidic residues (Figures 31B and C and Figures 31 F and G), the distribution of which correlates qualitatively with the negative electrostatic potential on the surface of these proteins (Figures 31D and H), suggesting that long-range electrostatic interactions play an important role in the formation of these particular encounter complexes.
Figure 31.
Observed and calculated intermolecular PREs for the IIAMtl-HPr (panels A-D) and IIAMan-HPr (panels E-H) complexes with HPr paramagnetically labeled with EDTA-Mn2+ at position E5C. A comparison of the observed PRE profiles (red circles) with those back-calculated from the structures of the stereospecific complexes (black lines) are shown in (A) and (E). Structures of the two stereospecific complexes color coded according to the difference, ΔΓ2, between observed and calculated intermolecular Γ2 rates are shown in (B) and (F). Residues of IIAMtl (C) and IIAMan (G) that display a large ΔΓ2 are colored in cyan on a molecular surface representation, with HPr shown as a green ribbon; corresponding electrostatic potential isosurfaces (± 5 kT) are shown in (D) and (H), respectively, using the same views. Adapted from Tang et al.27 published in Nature (Nature Publishing Group) while the authors were US Government employees at the National Institutes of Health.
Recently, replica exchange Monte Carlo simulations using a coarse-grained energy function have been used to study the interactions of the EIN-HPr, IIAMtl-HPr and IIAMan-HPr complexes from a theoretical perspective.29 The simulations recover the structures of the stereospecific complexes together with an ensemble of non-specific encounter complexes at an occupancy of ~10%, and reproduce the overall experimental binding affinities well. The picture that emerges from both the experimental and theoretical work is one in which the overall funnel-shaped energy landscape of complex formation is dominated by the stereospecific complex, a small number of structured nonspecific encounter complexes and a diffuse cloud of loosely bound encounter complexes connecting the stereospecific and nonspecific binding sites with each other and the unbound state.29
4.2.2. Encounter complexes in protein-protein electron transfer
In the phosphoryl transfer complexes of the PTS discussed in the previous section the configuration of the stereospecific complex is dictated by rigid constraints imposed by chemistry with only small variations in the distance between the two active site histidines depending on whether the phosphoryl transfer reaction proceeds via an associative or dissociative mechanism.42,44,219 In the case of electron transfer reactions, however, which proceed principally via electron tunneling modulated by the intervening bonding networks, such highly strict geometric requirements are not present, and hence a multitude of closely related configurations can still potentially support electron transfer.223
The crystal structure of the complex between cytochrome c peroxidase (CcP) and cytochrome c (Cc) provides a static picture of an electron transfer complex.224 Brownian dynamics simulations, however, suggest a multitude of complexes, including several discrete and energetically accessible minima.14 While the most stable minimum corresponds to the crystal structure, other minima have smaller metal-metal separation.223 Since the CCp-Cc complex is weak, the PRE provides an ideal approach to answer these questions experimentally.110 Using a very qualitative interpretation of the PRE data in which the PREs were converted to approximate distance restraints with bounds of ±4 Å, Volkov et al.110 showed that most of the PRE restraints were consistent with a single structure that is close to the crystal structure (backbone rms difference of 2.2 Å). However, a number of non-randomly distributed PRE restraints were consistently violated, indicating that the single docked structure is insufficient to explain all the experimental PRE data. Volkov et al.110 were able to show that the dominant protein-protein orientation, corresponding approximately to the crystal structure, is occupied >70% of the lifetime of the complex, with the remainder of the time being spent in an ensemble of encounter complexes. The authors were further able to deduce that the conformational space explored by Cc on the surface of CcP in the encounter complexes constitutes about half the molecular surface of CcP with the crystal structure configuration located at approximately the center of this surface.
In the context of electron transfer, the results on the Cc-CcP complex are not only relevant to the mechanism of protein complex formation but also to the modulation of electron transfer rates.110 The observation that the dominant protein-protein orientation in solution corresponds approximately to that observed in the crystal structure is consistent with the finding that electron transfer rates are similar in solution and in the crystal.225,226 However, forward and backward electron transfer rates are differentially modulated by dynamic processes, and forward electron transfer rates appear to be conformationally gated implying that the most stable binding configuration is not necessarily the most electron transfer active.226
The relevance of encounter complexes to electron transfer is illustrated even more dramatically in the case of the adrenoxin (Adx) and Cc.111 Adx and Cc associate to form a very weak and short-lived complex that is electron transfer active. In this instance the PRE data indicate that the Adx-Cc complex is in fact a pure encounter complex ensemble.
4.2.3 Ultra-weak self-association
Ultra-weak macromolecular self-association (Kdiss > 10 mM) is beyond the limit of detection of conventional biophysical techniques such as sedimentation velocity and therefore very difficult to study. Such weak interactions, however, likely play a critical role in the initial nucleation events involved in spontaneous self-assembly of higher-order architectures, including viral capsid formation, crystallization and amyloid fibril formation.
As an example of ultra-weak self-association we investigated the self-association of HPr using the PRE.116 No evidence of self-association could be detected by sedimentation velocity providing an upper limit of 1–2% for any high order soluble species. Intermolecular PRE measurements were carried out using a 1:1 mixture of 15N-labeled HPr and HPr paramagnetically labeled with EDTA-Mn2+ at three positions (one at a time), E5C, E25C and E32C. Large intermolecular PREs were observed with the paramagnetic label at E5C and E32C, compared to negligible effects with the label at E25C, thereby unequivocally demonstrating the presence of very weak self-association (Figure 32A). Self-association could be completely eliminated by the addition of EIN indicating that the EIN-HPr interface overlaps with that for self-association of HPr. The PREs are also modulated differentially by ionic strength, as well as by a surface charge mutation (S46D), revealing the presence of several clusters of self-associated species. Using similar computational methodology to that employed to explore the distribution of nonspecific encounter complexes for the interaction of EIN and HPr (Section 4.2.1), it could be shown that the self-associated state was optimally represented by an ensemble of species of size N = 4 at an overall occupancy of ~1% (Figure 31B), corresponding to a Kdiss ≥ 15 mM. From the atomic probability distribution representing the self-associated species one can deduce that the ensemble of self-associated states is driven by both electrostatic and hydrophobic interactions (Figure 32C).
Figure 32.
Ultra-weak self association of HPr. (A) Intermolecular PRE profiles with EDTA-Mn2+ at positions E5C (top) and E32C (bottom panel). The location of the intermolecular PREs is shown in the insets. (B) Correlation between observed and calculated PREs with the self-associated state represented by an ensemble of size N = 4 at a population of 1%. (C) Reweighted atomic probability density maps (green mesh, plotted at threshold of 20% maximum) showing the distribution of 15N-labeled HPr on the surface of paramagnetically labeled HPr (left panel) and of paramagnetically-labeled HPr on the surface of 15N-labeled HPr (right panel with residues experiencing large intermolecular PREs colored in red, green, blue and yellow using the same color scheme as in (A)). Adapted from Tang et al.116 published in J. Am. Chem. Soc. (American Chemical Society) while the authors were US Government employees at the National Institutes of Health.
4.2.4 Transient events involved in N-terminal auto-processing of HIV-1 protease
Mature HIV-1 protease is a dimer in which residues from each subunit contribute to the catalytic active site.227 HIV-1 protease plays a critical role in viral maturation by processing the Gag and Gag-Pol polyproteins into mature structural and functional proteins, including itself. The full-length transframe region (TFR)-protease precursor is monomeric but undergoes maturation by intramolecular N-terminal cleavage of a putative precursor dimer.228–230 This cleavage event is associated with the appearance of mature-like catalytic activity. Cleavage at the C-terminal end of the protease occurs intermolecularly and involves an active protease dimer.231 How then does N-terminal cleavage of the HIV-1 protease precursor occur when the N- and C-termini of the mature protease are part of an intersubunit β-sheet located distal from the active site? To address this question we carried out PRE measurements on a mini-precursor protease construct, SFNFPRD25N, comprising a four-residue N-terminal extension derived from the TFR and a D25N mutation to abolish catalytic activity.117 The four-residue N-terminal extension is sufficient to result in a monomeric species whose secondary and tertiary structures are the same as those of the mature protease, with the exception of the N- and C-termini which are disordered and no longer form an intersubunit β-sheet. The experimental design is similar to that employed in the study of HPr self-association. The mini-precursor was spin-labeled at three sites (T12C, E34C and V82C, one at a time) and significant intermolecular PRE effects were observed with the nitroxide label at two of the three sites (T12C and V82C) (Figure 33A). These data permit one to reconstruct the probability distribution of very weakly self-associated states (Figure 33C). The mini-precursor forms highly transient, lowly populated (3–5%) dimeric encounter complexes (Kdiss = 3–5 mM) that involve the mature dimer interface but occupy a wide range of subunit orientations relative to the mature dimer. The occupancy of the mature dimer orientation is extremely low, thereby accounting for the very low enzymatic activity of the precursor. Additional PRE measurements with the nitroxide label positioned close to the N-terminus of the mini-precursor, reveals that the N-terminal extension makes both transient intra- and intersubunits contacts with the substrate binding site and cleft (Figures 33B and D). Thus, the N-terminal extension is available for auto-cleavage when the correct dimmer orientation is sampled within the encounter complex ensemble of mini-precursor self-associated species (Figure 33D).
Figure 33.
PRE investigation of transient events involved in N-terminal auto-processing of the SFNFPRD25N HIV-1 protease mini-precursor. (A) Intermolecular PRE profiles observed with nitroxide labels at positions T12C and V82C located at the periphery of the substrate binding cleft and dimer interface of the mature protease. The observed Γ2 rates are shown as red circles, the calculated Γ2 rates (derived from ensemble rigid body simulated annealing calculations) for a N = 4 representation for the ensemble of self self-associated states at a population of 5% are shown as black lines (PRE Q-factor = 0.22); and the Γ2 rates back-calculated from the structure of the mature protease at populations of 1 and 2% are shown as blue and green lines, respectively. (B) PRE profiles observed with the nitroxide spin label attached at the N-terminus of SFNFPRD25N with intermolecular PREs (obtained from a mixed sample of 15N-labeled precursor and nitroxide-labeled precursor) shown in red and the sum of intra- and intermolecular PREs (obtained from a sample comprising dual 15N- and nitroxide labeled mini-precursor) in blue. (C) Atomic probability density map (grey mesh plotted at a threshold of 20% of maximum) showing the distribution of the spin-labeled (T12C, E34C and V82C) subunit relative to the isotopically labeled subunit (red ribbon) in the SFNFPRD25N encounter complexes, with the location of the second subunit in the mature protease shown as a blue ribbon. (D) Inter- and intramolecular PREs with Γ2 rates > 10 s−1 color-coded in red and blue, respectively, onto the molecular surface of the mature protease dimer originating from the N-terminal spin-label. Adapted from Tang et al.117 published in Nature (Nature Publishing Group) while the authors were US Government employees at the National
4.3 Transient domain-domain interactions in proteins
Large-scale domain rearrangements are known to play a critical role in ligand binding, recognition, catalysis and regulation. Such motions include transitions between alternative states (e.g. closed versus open conformations) as well as transitions from intrinsically disordered states to ordered ones upon binding.
4.3.1. Interdomain dynamics in maltose-binding protein
Maltose binding protein (MBP) belongs to a large class of periplasmic proteins in which ligand binding is associated with large domain reorientation from an open apo state to a closed holo state.232 The ligand binding site consists of residues from the N- and C-terminal domains and is fully accessible in the apo state; in the holo state the ligand is buried by the two domains. For this reason, MBP and related proteins, have been compared to a Venus Fly Trap. The general question that arises for proteins such as MBP is whether the apo state exists as a single species in which the holo conformation is energetically inaccessible in the absence of ligand and interdomain rearrangement is induced by ligand binding, or whether the predominantly open apo state coexists in rapid equilibrium with alternative lowly-populated species that approximate to the closed holo state.
In the case of MBP, domain reorientation upon ligand binding involves a ~35° rigid body domain reorientation between the N- (NTD) and C-(CTD) domains, involving hinge bending within the linker region connecting the two domains.233,234 RDCs measured on the apo and holo states of MBP are fully consistent with the respective crystals structures (with RDC R-factors of ~13–14%).235 Since RDCs are exquisitely sensitive to bond vector orientation,58 it follows that the predominant conformation of the apo and holo states in solution corresponds to the open and closed states observed in the apo and holo crystal structures, respectively. However, RDCs represent a linear weighted average of the species present in solution and therefore will not be sensitive to the presence of lowly-populated states.114 Thermodynamic and mutagenesis data further indicate that the upper limit for the occupancy of the holo conformation in the absence of ligand is only 0.002%.236
PRE measurements on MBP were carried out by placing a nitroxide label (one at a time) on the surface of the NTD (D41C) and CTD (S211C).114 In the presence of ligand, the PRE data from both sites are fully consistent with the holo crystal structure. In the absence of ligand, however, a different picture emerges: although the PRE data obtained for S211C are compatible with the apo crystal structure, the PRE profile for D41C reveals major discrepancies between the observed and calculated interdomain PREs observed on the CTD. Specifically, the magnitude of the PREs originating from D41C and observed within the CTD are considerably larger than expected from the apo crystal structure (Figure 34A) with an interdomain PRE Q-factor of 0.49 (Figure 34B). The PRE profiles for MBP in the absence of ligand cannot be accounted for by a simple mixture of apo and holo conformations. Further, although the PRE data can be fit to a single alternative conformation, this conformation is inconsistent with the RDC data (RDC R-factor of 24±3% compared to 14% for the apo crystal structure). One can therefore conclude that apo MBP in solution exists as a mixture of at least two species, with the predominant one corresponding to the apo crystal structure. Simulated annealing refinement against the PRE data using a two-member ensemble comprising a major open state (corresponding to the apo crystal structure) and a minor species in a 95%:5% mixture satisfies the experimental PRE data with Q-factors of 0.21 and 0.24 for the D41C (Figure 34B) and S211C data, respectively, and is fully consistent with both RDC and small angle X-ray scattering data.114 In this instance representation of the minor species by a larger ensemble of states does not result in any significant improvement in agreement with the experimental PRE data and yields the same physical and structural picture.
Figure 34.
Dynamic interconversion between major open and minor, partially-closed states of apo MBP. (A) Comparison of the observed PRE profile for apo MBP spin-labeled at position D41C (red circles) with that back-calculated on the basis of the apo crystal structure (open state). (B) Correlation between observed and calculated Γ2 rates for the D41C data obtained with the apo crystal structure alone (left panel) and upon inclusion of a minor, partially-closed species at an occupancy of 5% (right panel) derived by conjoined rigid body/torsion angle simulated annealing refinement. (C) Comparison of the major open (blue cylinder) and minor partially-cosed (green smoothed backbone trace with reweighted atomic probability map shown as a green mesh) forms of the CTD of apo MBP with the NTD of the two species superimposed (grey ribbon). (D) Comparison of the relative orientation of the CTD in the minor partially-closed state of apo MBP (green cylinder) and the holo crystal structure (closed state) of MBP (red cylinders) with the crystal structure (open state) of apo MBP shown as a molecular surface color-coded according to electrostatic potential. Adapted from Tang et al.114 published in Nature (Nature Publishing Group) while the authors were US Government employees at the National Institutes of Health.
The lowly-populated species calculated from the PRE data represents a partially-closed state (Figure 34C) that is distinct from the holo conformation.114 The transition between the major open and minor partially-closed apo species involves a hinge body rotation of 33°. The minor partially-closed apo state and the holo conformation differ by an 18° rotation as well as a 6 Å translation (Figure 34D). The latter relieves electrostatic repulsion in the holo conformation between negatively charged residues that line the substrate binding cleft, thereby rendering the holo conformation energetically inaccessible in the absence of sugar substrate.236 In the presence of sugar, an array of intermolecular hydrogen bonds between these residues and the substrate stabilize the holo conformation. In the absence of sugar, translation of the CTD out of the sugar-binding pocket in the partially-closed apo state not only reduces electrostatic repulsion between the NTD and CTD but also leaves the sugar binding surface on the CTD exposed and therefore accessible for binding. The presence of a partially-closed apo state in rapid equilibrium with the predominant open state may facilitate the transition to the holo conformation upon sugar binding. Further, since the predominant structural fluctuations in apo MBP do not involve the energetically disfavored holo conformation but a related partially-closed conformation, one could regard the conformational transition to the holo conformation upon sugar binding as an example of induced fit.
4.3.2. Conformational space sampled by denatured and intrinsically disordered proteins
Denatured and intrinsically disordered proteins occupy an ensemble of non-native conformational states.237 One might expect that these states constitute a statistical random coil, but this need not necessarily be the case owing to the presence of residual structure or local non-native collapse around hydrophobic clusters.238 By comparing the observed PREs with those expected from a statistical random coil, one can deduce the existence of residual structure or compaction. PRE measurements have been carried out on a wide range of partially unfolded, unfolded and intrinsically unstructured proteins.99–109
The expected profile for a random coil, calculated using the mean field approximation to an unperturbed wormlike chain, predicts that the PRE should extend to ~15 residues from the site of a nitroxide spin label.108 Depending on the location of the spin label, this behavior can be reproduced. In other cases, however, PRE effects have been observed to extend well beyond the 15 residue limit and in addition, may not be evenly distributed. In general the PRE data on unfolded and intrinsically disordered proteins have provided definitive evidence of multiple distinct hydrophobic clusters which include both non-native interactions and in some instances residual native contacts.99–108 For chemically-denatured N-PGK,109 on the other hand, significant PREs, distributed almost uniformly throughout the sequence, were observed from multiple spin labels, suggesting the existence of a collapsed state with no coherent three-dimensional structure and a radius approximately 20% larger than that of the folded state.
5. Concluding remarks
Although the PRE was first described in the 1950's76 and early 1960's,119 and has been extensively used in the study of metalloproteins,59,60,118 it is only relatively recently that the appropriate theoretical and computational framework has been established to permit direct quantitative refinement against PRE relaxation rates for systems in which an extrinsic paramagnetic center (covalently attached to the macromolecule of interest via a linker with several rotatable bonds) samples a wide range of conformational space.78 Quantitative refinement against PRE relaxation rates not only affords a powerful tool in three-dimensional solution structure determination, but also permits structural information on lowly-populated states to be derived under conditions where the minor and major species are in fast exchange on the PRE relaxation time scale.
From the perspective of structure determination, the PRE permits a large number of long-range distance interactions, up to 35 Å in the case of Mn2+, between the paramagnetic center and the protons of the macromolecule(s) to be probed. In contrast, the NOE which forms the backbone of all NMR structure determination, is restricted to short-range interproton distances less than about 6 Å.30–32,51 It is not simply the ability to sample long-range distances that makes the PRE so useful. The unambiguous identification of a PRE effect is entirely straightforward since the assignment of cross-peaks in 2D or 3D correlation spectra are already known from through-bond scalar triple resonance experiments. This is in stark contrast to the assignment of NOE cross-peaks where extensive spectral overlap can render unambiguous identification to a single interproton interaction difficult even in four-dimensional spectra.
While the NOE cannot provide information concerning long-range order, since the effects are limited to 6 Å, it is evident that a large number of such short-range contacts, even if interpreted in the most generous and qualitative manner, are in fact highly constraining from a structural perspective when dealing with globular proteins.51 Qualitative interpretation of PRE data, however, is much less useful in this regard since a distance restraint of 20±5 Å or 20±10Å is not particularly conformationally restrictive. This is why direct refinement against the PRE rates is so important from the perspective of structure determination, since it is only by this means that the full power of the PRE can be truly exploited.78
The PRE is not the only biophysical technique that can probe long-range distance interactions. Fluorescence resonance energy transfer (FRET)239 and double electron-electron resonance (DEER)240,241 allow separations of 10–100 Å and 20–60 Å to be studied, and in contrast to solution NMR methods are not limited by molecular size. However, both FRET and DEER require the introduction of two probes, a fluorescence donor and acceptor for the former and two nitroxide labels for the later, and more importantly, only a single pairwise distance can be measured per sample, with each distance requiring a new double chromophore or double spin-labeled sample. The PRE, of course, can probe hundreds of distances simultaneously in a single sample, thereby permitting one to derive not only three-dimensional structural information, but also to detect and visualize lowly-populated species.
As with any labeling method, the introduction of extrinsic PRE labels can potentially alter the interaction energy, structure or kinetics of the interacting molecules. For this reason care should be taken in choosing the sites of labeling. The absence of significant structural perturbations involving the high-populated state of the labeled molecule and the complex can be easily assessed by NMR spectroscopy using chemical shifts as an indicator. The absence of any significant chemical shift perturbations (e.g. in a 1H–15N correlation spectrum) outside the immediate vicinity of the labeled residue indicates that any potential structural perturbations are too small to be characterized by NMR. Likewise, residual dipolar couplings can be readily used to verify the absence of any significant pertubations in backbone structure. Overall interactions energies can be derived by titration experiments (e.g. using NMR, fluorescence or isothermal titration calorimetry to determine overall equilibrium constants). In the examples discussed here involving protein-protein and protein-DNA interactions, as well as multidomain proteins, no significant structural or energetic perturbations were detected upon introduction of the paramagnetic labels.23–25,27,78,94,112,114,116,117
The application of the PRE for structure determination of complexes or multi-domain proteins is restricted to systems in slow exchange on the PRE relaxation time scale. Under these conditions, the observed intermolecular or interdomain PREs reflect a single state, and direct refinement against the PRE rates using a single structure representation of the macromolecule can proceed in the same way as using the intramolecular PREs for structure determination/refinement of a single protein.78
Perhaps the most exciting aspects of the PRE are the recent findings that under fast exchanging conditions, intermolecular or interdomain PREs can provide the footprint of low population species providing a significant number of paramagnetic center-proton distances are shorter in the minor species than the major one.24 This PRE footprint permits one to detect, visualize and characterize lowly-populated states of macromolecules and their complexes under equilibrium conditions, shedding information on states that are effectively invisible to other biophysical and structural techniques. These states are of considerable interest since they play a crucial role in a wide range of dynamical processes including molecular recognition and binding, allostery, induced-fit and self-assembly. In this review we have shown how the PRE in the fast exchange regime can be used to demonstrate sliding and hopping of transcription factors on DNA;24 to identify and structurally characterize non-specific encounter complexes in protein-protein association;27,28,110,111 to detect ultra-weak self-association,116 a process that is vital, for example, for N-terminal auto-processing of the monomeric HIV-1 protease precursor;117 to probe large scale interdomain motions in multidomain proteins involving rare transitions;114 and to characterize ensembles of unfolded and intrinsically disordered states of proteins including the sampling of non-native partially collapsed states. 99–109
Acknowledgments
We thank Attila Szabo and Chun Tang for useful discussions. This work was supported by the Intramural Program of the National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health (to G.M.C.), the AIDS Targeted Antiviral Program of the Office of the Director of the National Institutes of Health (to G.M.C.), and by grant H-1683 from the Welch Foundation (to J.I.).
Biographies

Marius Clore is Chief of the Protein NMR Section at the NIH (1988-present). He received his MD and PhD degrees at University College Hospital Medical School and the MRC National Institute for Medical Research in London, and was joint Head of the Biological NMR group at the Max Planck Institute for Biochemistry in Munich (1984–1988). His major research interests lie in the application of NMR spectroscopy to study the structure and dynamics of macromolecular complexes.

Junji Iwahara is currently an Assistant Professor in the Department of Biochemistry and Molecular Biology at the University of Texas Medical Branch (2007-present). He received his PhD degree at the University of Tokyo and was a Senior Research Fellow in the Laboratory of Chemical Physics at the NIH (2002–2007). His current interests lie in the use of NMR spectroscopy to study highly dynamic processes such as protein translocation on DNA.
References
- 1.Branden C, Tooze J. Introduction to Protein Structure. 2nd ed. New York: Garland Publishing Inc; 1998. [Google Scholar]
- 2.Bryngelson JD, Onuchic JN, Socci ND, Wolynes PG. Proteins. 1995;21:167. doi: 10.1002/prot.340210302. [DOI] [PubMed] [Google Scholar]
- 3.Onuchic JN, Luthey-Schulten Z, Wolynes PG. Annu. Rev. Phys. Chem. 1997;48:545. doi: 10.1146/annurev.physchem.48.1.545. [DOI] [PubMed] [Google Scholar]
- 4.Miyashita O, Onuchic JN, Wolynes PG. Proc. Natl. Acad. Sci. U. S. A. 2003;100:12570. doi: 10.1073/pnas.2135471100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Miyashita O, Wolynes PG, Onuchic JN. J. Phys. Chem. B. 2005;109:1959. doi: 10.1021/jp046736q. [DOI] [PubMed] [Google Scholar]
- 6.Zhou HX, Szabo A. Phys. Rev. Lett. 2004;93:178101. doi: 10.1103/PhysRevLett.93.178101. [DOI] [PubMed] [Google Scholar]
- 7.Berg OG, von Hippel PH. Annu Rev Biophys Biophys Chem. 1985;14:131. doi: 10.1146/annurev.bb.14.060185.001023. [DOI] [PubMed] [Google Scholar]
- 8.Berg OG, von Hippel PH. J. Mol. Biol. 1987;193:723. doi: 10.1016/0022-2836(87)90354-8. [DOI] [PubMed] [Google Scholar]
- 9.von Hippel PH, Berg OG. J. Biol. Chem. 1989;264:675. [PubMed] [Google Scholar]
- 10.Schreiber G, Fersht AR. Nature Struct. Biol. 1996;3:427. doi: 10.1038/nsb0596-427. [DOI] [PubMed] [Google Scholar]
- 11.Vijayakumar M, Wong KY, Schreiber G, Fersht AR, Szabo A, Zhou HX. J. Mol. Biol. 1998;278:1015. doi: 10.1006/jmbi.1998.1747. [DOI] [PubMed] [Google Scholar]
- 12.Selzer T, Schreiber G. Proteins. 2001;45:190. doi: 10.1002/prot.1139. [DOI] [PubMed] [Google Scholar]
- 13.Schreiber G. Curr. Opin. Struct. Biol. 2002;12:41. doi: 10.1016/s0959-440x(02)00287-7. [DOI] [PubMed] [Google Scholar]
- 14.Northrup SH, Boles JO, Reynolds JC. Science. 1988;241:67. doi: 10.1126/science.2838904. [DOI] [PubMed] [Google Scholar]
- 15.Gabdoulline RR, Wade RC. Curr. Opin. Struct. Biol. 2002;12:204. doi: 10.1016/s0959-440x(02)00311-1. [DOI] [PubMed] [Google Scholar]
- 16.Spaar A, Dammer C, Gabdoulline RR, Wade RC, Helms V. Biophys. J. 2006;90:1913. doi: 10.1529/biophysj.105.075507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Berg OG, Winter RB, von Hippel PH. Biochemistry. 1981;20:6929. doi: 10.1021/bi00527a028. [DOI] [PubMed] [Google Scholar]
- 18.Winter RB, Berg OG, von Hippel PH. Biochemistry. 1981;20:6961. doi: 10.1021/bi00527a030. [DOI] [PubMed] [Google Scholar]
- 19.Halford SE, Szczelkun MD. Eur. Biophys. J. 2002;31:257. doi: 10.1007/s00249-002-0224-4. [DOI] [PubMed] [Google Scholar]
- 20.Halford SE, Marko JF. Nucleic Acids Res. 2004;32:3040. doi: 10.1093/nar/gkh624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hu T, Shklovskii BI. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 2006;74:021903. doi: 10.1103/PhysRevE.74.021903. [DOI] [PubMed] [Google Scholar]
- 22.Hu T, Shklovskii BI. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 2007;76:051909. doi: 10.1103/PhysRevE.76.051909. [DOI] [PubMed] [Google Scholar]
- 23.Iwahara J, Zweckstetter M, Clore GM. Proc. Natl. Acad. Sci. U. S. A. 2006;103:15062. doi: 10.1073/pnas.0605868103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Iwahara J, Clore GM. Nature. 2006;440:1227. doi: 10.1038/nature04673. [DOI] [PubMed] [Google Scholar]
- 25.Iwahara J, Clore GM. J. Am. Chem. Soc. 2006;128:404. doi: 10.1021/ja056786o. [DOI] [PubMed] [Google Scholar]
- 26.Doucleff M, Clore GM. Proc. Natl. Acad. Sci. U. S. A. 2008;105:13871. doi: 10.1073/pnas.0805050105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tang C, Iwahara J, Clore GM. Nature. 2006;444:383. doi: 10.1038/nature05201. [DOI] [PubMed] [Google Scholar]
- 28.Suh JY, Tang C, Clore GM. J. Am. Chem. Soc. 2007;129:12954. doi: 10.1021/ja0760978. [DOI] [PubMed] [Google Scholar]
- 29.Kim YC, Tang C, Clore GM, Hummer G. Proc. Natl. Acad. Sci. U. S. A. 2008;105:12855. doi: 10.1073/pnas.0802460105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wüthrich K. NMR of Proteins and Nucleic Acids. New York: John Wiley & Sons; 1986. [Google Scholar]
- 31.Clore GM, Gronenborn AM. Crit. Rev. Biochem. Mol. Biol. 1989;24:479. doi: 10.3109/10409238909086962. [DOI] [PubMed] [Google Scholar]
- 32.Clore GM, Gronenborn AM. Science. 1991;252:1390. doi: 10.1126/science.2047852. [DOI] [PubMed] [Google Scholar]
- 33.Bax A, Grzesiek S. Acc. Chem. Res. 1993;26:131. [Google Scholar]
- 34.Clore GM, Wingfield PT, Gronenborn AM. Biochemistry. 1991;30:2315. doi: 10.1021/bi00223a005. [DOI] [PubMed] [Google Scholar]
- 35.Garrett DS, Seok YJ, Liao DI, Peterkofsky A, Gronenborn AM, Clore GM. Biochemistry. 1997;36:2517. doi: 10.1021/bi962924y. [DOI] [PubMed] [Google Scholar]
- 36.Salzmann M, Pervushin K, Wider G, Senn H, Wüthrich K. Proc. Natl. Acad. Sci. U. S. A. 1998;95:13585. doi: 10.1073/pnas.95.23.13585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pervushin KQ. Rev. Biophys. 2000;33:161. doi: 10.1017/s0033583500003619. [DOI] [PubMed] [Google Scholar]
- 38.Riek R, Pervushin K, Wüthrich K. Trends Biochem. Sci. 2000;25:462. doi: 10.1016/s0968-0004(00)01665-0. [DOI] [PubMed] [Google Scholar]
- 39.Tugarinov V, Muhandiram R, Ayed A, Kay LE. J. Am. Chem. Soc. 2002;124:10025. doi: 10.1021/ja0205636. [DOI] [PubMed] [Google Scholar]
- 40.Tugarinov V, Sprangers R, Kay LE. J. Am. Chem. Soc. 2004;126:4921. doi: 10.1021/ja039732s. [DOI] [PubMed] [Google Scholar]
- 41.Caffrey M, Cai M, Kaufman J, Stahl SJ, Wingfield PT, Covell DG, Gronenborn AM, Clore GM. EMBO J. 1998;17:4572. doi: 10.1093/emboj/17.16.4572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Garrett DS, Seok YJ, Peterkofsky A, Gronenborn AM, Clore GM. Nature Struct. Biol. 1999;6:166. doi: 10.1038/5854. [DOI] [PubMed] [Google Scholar]
- 43.Williams DC, Jr., Cai M, Clore GM. J. Biol. Chem. 2004;279:1449. doi: 10.1074/jbc.M309790200. [DOI] [PubMed] [Google Scholar]
- 44.Williams DC, Jr., Cai M, Suh JY, Peterkofsky A, Clore GM. J. Biol. Chem. 2005;280:20775. doi: 10.1074/jbc.M501986200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tugarinov V, Choy WY, Orekhov VY, Kay LE. Proc. Natl. Acad. Sci. U. S. A. 2005;102:622. doi: 10.1073/pnas.0407792102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hu J, Hu K, Williams DC, Jr, Komlosh ME, Cai M, Clore GM. J. Biol. Chem. 2008;283:11024. doi: 10.1074/jbc.M800312200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fiaux J, Bertelsen EB, Horwich AL, Wüthrich K. Nature. 2002;418:207. doi: 10.1038/nature00860. [DOI] [PubMed] [Google Scholar]
- 48.Horst R, Bertelsen EB, Fiaux J, Wider G, Horwich AL, Wüthrich K. Proc. Natl. Acad. Sci. U. S. A. 2005;102:12748. doi: 10.1073/pnas.0505642102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sprangers R, Gribun A, Hwang PM, Houry WA, Kay LE. Proc. Natl. Acad. Sci. U. S. A. 2005;102:16678. doi: 10.1073/pnas.0507370102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sprangers R, Kay LE. Nature. 2007;445:618. doi: 10.1038/nature05512. [DOI] [PubMed] [Google Scholar]
- 51.Clore GM, Gronenborn AM. Proc. Natl. Acad. Sci. U. S. A. 1998;95:5891. doi: 10.1073/pnas.95.11.5891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tjandra N, Bax A. Science. 1997;278:1111. doi: 10.1126/science.278.5340.1111. [DOI] [PubMed] [Google Scholar]
- 53.Clore GM, Starich MR, Gronenborn AM. J. Am. Chem. Soc. 1998;120:10571. [Google Scholar]
- 54.Tjandra N, Omichinski JG, Gronenborn AM, Clore GM, Bax A. Nature Struct. Biol. 1997;4:732. doi: 10.1038/nsb0997-732. [DOI] [PubMed] [Google Scholar]
- 55.Clore GM, Starich MR, Bewley CA, Cai M, Kuszewski J. J. Am. Chem. Soc. 1999;121:6513. [Google Scholar]
- 56.Clore GM, Garrett DS. J. Am. Chem. Soc. 1999;121:9008. [Google Scholar]
- 57.Clore GM. Proc. Natl. Acad. Sci. U. S. A. 2000;97:9021. doi: 10.1073/pnas.97.16.9021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bax A, Kontaxis G, Tjandra N. Methods Enzymol. 2001;339:127. doi: 10.1016/s0076-6879(01)39313-8. [DOI] [PubMed] [Google Scholar]
- 59.Bertini I, Luchinat C, Aime S. NMR of Paramagnetic Substances. Amsterdam: Elsevier; 1996. [Google Scholar]
- 60.Bertini I, Luchinat C, Piccioli M. Methods Enzymol. 2001;339:314. doi: 10.1016/s0076-6879(01)39320-5. [DOI] [PubMed] [Google Scholar]
- 61.Clore GM, Tang C, Iwahara J. Curr. Opin. Struct. Biol. 2007;17:603. doi: 10.1016/j.sbi.2007.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Clore GM. Mol. Biosyst. 2008;4:1058. doi: 10.1039/b810232e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pintacuda G, John M, Su XC, Otting G. Acc. Chem. Res. 2007;40:206. doi: 10.1021/ar050087z. [DOI] [PubMed] [Google Scholar]
- 64.Palmer AG, Kroenke CD, Loria JP. Methods Enzymol. 2001;339:204. doi: 10.1016/s0076-6879(01)39315-1. [DOI] [PubMed] [Google Scholar]
- 65.Mittermaier A, Kay LE. Science. 2006;312:224. doi: 10.1126/science.1124964. [DOI] [PubMed] [Google Scholar]
- 66.Korzhnev DM, Kay LE. Acc. Chem. Res. 2008;41:442. doi: 10.1021/ar700189y. [DOI] [PubMed] [Google Scholar]
- 67.Loria JP, Berlow RB, Watt ED. Acc. Chem. Res. 2008;41:214. doi: 10.1021/ar700132n. [DOI] [PubMed] [Google Scholar]
- 68.Montelione GT, Wagner G. J. Am. Chem. Soc. 1989;111:3096. [Google Scholar]
- 69.Wider G, Neri D, Wüthrich K. J. Biomol. NMR. 1991;1:93. [Google Scholar]
- 70.Farrow NA, Zhang O, Forman-Kay JD, Kay LE. J. Biomol. NMR. 1994;4:727. doi: 10.1007/BF00404280. [DOI] [PubMed] [Google Scholar]
- 71.Sahu D, Clore GM, Iwahara J. J. Am. Chem. Soc. 2007;129:13232. doi: 10.1021/ja074604f. [DOI] [PubMed] [Google Scholar]
- 72.Bai Y. Chem. Rev. 2006;106:1757. doi: 10.1021/cr040432i. [DOI] [PubMed] [Google Scholar]
- 73.Krishna MM, Hoang L, Lin Y, Englander SW. Methods. 2004;34:51. doi: 10.1016/j.ymeth.2004.03.005. [DOI] [PubMed] [Google Scholar]
- 74.Vallurupalli P, Hansen DF, Kay LE. Proc. Natl. Acad. Sci. U. S. A. 2008;105:11766. doi: 10.1073/pnas.0804221105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Vallurupalli P, Hansen DF, Kay LE. J. Am. Chem. Soc. 2008;130:2734. doi: 10.1021/ja710817g. [DOI] [PubMed] [Google Scholar]
- 76.Solomon I. Phys. Rev. 1955;99:559. [Google Scholar]
- 77.Kosen PA. Methods Enzymol. 1989;177:86. doi: 10.1016/0076-6879(89)77007-5. [DOI] [PubMed] [Google Scholar]
- 78.Iwahara J, Schwieters CD, Clore GM. J. Am. Chem. Soc. 2004;126:5879. doi: 10.1021/ja031580d. [DOI] [PubMed] [Google Scholar]
- 79.Schmidt PG, Kuntz ID. Biochemistry. 1984;23:4261. doi: 10.1021/bi00313a038. [DOI] [PubMed] [Google Scholar]
- 80.Kosen PA, Scheek RM, Naderi H, Basus VJ, Manogaran S, Schmidt PG, Oppenheimer NJ, Kuntz ID. Biochemistry. 1986;25:2356. doi: 10.1021/bi00357a009. [DOI] [PubMed] [Google Scholar]
- 81.Battiste JL, Wagner G. Biochemistry. 2000;39:5355. doi: 10.1021/bi000060h. [DOI] [PubMed] [Google Scholar]
- 82.Gaponenko V, Howarth JW, Columbus L, Gasmi-Seabrook G, Yuan J, Hubbell WL, Rosevear PR. Protein Sci. 2000;9:302. doi: 10.1110/ps.9.2.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Donaldson LW, Skrynnikov NR, Choy WY, Muhandiram DR, Sarkar B, Forman-Kay JD, Kay LE. J. Am. Chem. Soc. 2001;123:9843. doi: 10.1021/ja011241p. [DOI] [PubMed] [Google Scholar]
- 84.Dvoretsky A, Gaponenko V, Rosevear PR. FEBS Lett. 2002;528:189. doi: 10.1016/s0014-5793(02)03297-0. [DOI] [PubMed] [Google Scholar]
- 85.Card PB, Erbel PJ, Gardner KH. J. Mol. Biol. 2005;353:664. doi: 10.1016/j.jmb.2005.08.043. [DOI] [PubMed] [Google Scholar]
- 86.Gross JD, Moerke NJ, von der Haar T, Lugovskoy AA, Sachs AB, McCarthy JE, Wagner G. Cell. 2003;115:739. doi: 10.1016/s0092-8674(03)00975-9. [DOI] [PubMed] [Google Scholar]
- 87.Mahoney NM, Rastogi VK, Cahill SM, Girvin ME, Almo SC. J. Am. Chem. Soc. 2000;122:7851. [Google Scholar]
- 88.Mal TK, Ikura M, Kay LE. J. Am. Chem. Soc. 2002;124:14002. doi: 10.1021/ja028109p. [DOI] [PubMed] [Google Scholar]
- 89.Rumpel S, Becker S, Zweckstetter MJ. Biomol. NMR. 2008;40:1. doi: 10.1007/s10858-007-9204-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Jain NU, Venot A, Umemoto K, Leffler H, Prestegard JH. Protein Sci. 2001;10:2393. doi: 10.1110/ps.17401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Johnson PE, Brun E, MacKenzie LF, Withers SG, McIntosh LP. J. Mol. Biol. 1999;287:609. doi: 10.1006/jmbi.1999.2627. [DOI] [PubMed] [Google Scholar]
- 92.Macnaughtan MA, Kamar M, Alvarez-Manilla G, Venot A, Glushka J, Pierce JM, Prestegard JH. J. Mol. Biol. 2007;366:1266. doi: 10.1016/j.jmb.2006.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Varani L, Gunderson SI, Mattaj IW, Kay LE, Neuhaus D, Varani G. Nature Struct. Biol. 2000;7:329. doi: 10.1038/74101. [DOI] [PubMed] [Google Scholar]
- 94.Iwahara J, Anderson DE, Murphy EC, Clore GM. J. Am. Chem. Soc. 2003;125:6634. doi: 10.1021/ja034488q. [DOI] [PubMed] [Google Scholar]
- 95.Ramos A, Varani GJ. Am. Chem. Soc. 1998;120:10992. [Google Scholar]
- 96.Ueda T, Kato A, Ogawa Y, Torizawa T, Kuramitsu S, Iwai S, Terasawa H, Shimada I. J. Biol. Chem. 2004;279:52574. doi: 10.1074/jbc.M409942200. [DOI] [PubMed] [Google Scholar]
- 97.Roosild TP, Greenwald J, Vega M, Castronovo S, Riek R, Choe S. Science. 2005;307:1317. doi: 10.1126/science.1106392. [DOI] [PubMed] [Google Scholar]
- 98.Liang BY, Bushweller JH, Tamm LK. J. Am. Chem. Soc. 2006;128:4389. doi: 10.1021/ja0574825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Bertoncini CW, Jung YS, Fernandez CO, Hoyer W, Griesinger C, Jovin TM, Zweckstetter M. Proc. Natl. Acad. Sci. U. S. A. 2005;102:1430. doi: 10.1073/pnas.0407146102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Dedmon MM, Lindorff-Larsen K, Christodoulou J, Vendruscolo M, Dobson CM. J. Am. Chem. Soc. 2005;127:476. doi: 10.1021/ja044834j. [DOI] [PubMed] [Google Scholar]
- 101.Gillespie JR, Shortle D. J. Mol. Biol. 1997;268:170. doi: 10.1006/jmbi.1997.0953. [DOI] [PubMed] [Google Scholar]
- 102.Gillespie JR, Shortle D. J. Mol. Biol. 1997;268:158. doi: 10.1006/jmbi.1997.0954. [DOI] [PubMed] [Google Scholar]
- 103.Kristjansdottir S, Lindorff-Larsen K, Fieber W, Dobson CM, Vendruscolo M, Poulsen FM. J. Mol. Biol. 2005;347:1053. doi: 10.1016/j.jmb.2005.01.009. [DOI] [PubMed] [Google Scholar]
- 104.Lietzow MA, Jamin M, Jane Dyson HJ, Wright PE. J. Mol. Biol. 2002;322:655. doi: 10.1016/s0022-2836(02)00847-1. [DOI] [PubMed] [Google Scholar]
- 105.Marsh JA, Neale C, Jack FE, Choy WY, Lee AY, Crowhurst KA, Forman-Kay JD. J. Mol. Biol. 2007;367:1494. doi: 10.1016/j.jmb.2007.01.038. [DOI] [PubMed] [Google Scholar]
- 106.Song J, Guo LW, Muradov H, Artemyev NO, Ruoho AE, Markley JL. Proc. Natl. Acad. Sci. U. S. A. 2008;105:1505. doi: 10.1073/pnas.0709558105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Vise P, Baral B, Stancik A, Lowry DF, Daughdrill GW. Proteins. 2007;67:526. doi: 10.1002/prot.21364. [DOI] [PubMed] [Google Scholar]
- 108.Felitsky DJ, Lietzow MA, Dyson HJ, Wright PE. Proc. Natl. Acad. Sci. U. S. A. 2008;105:6278. doi: 10.1073/pnas.0710641105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Cliff MJ, Craven CJ, Marston JP, Hounslow AM, Clarke AR, Waltho JP. J. Mol. Biol. 2009;385:266. doi: 10.1016/j.jmb.2008.10.004. [DOI] [PubMed] [Google Scholar]
- 110.Volkov AN, Worrall JA, Holtzmann E, Ubbink M. Proc. Natl. Acad. Sci. U. S. A. 2006;103:18945. doi: 10.1073/pnas.0603551103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Xu X, Reinle W, Hannemann F, Konarev PV, Svergun DI, Bernhardt R, Ubbink M. J. Am. Chem. Soc. 2008;130:6395. doi: 10.1021/ja7101357. [DOI] [PubMed] [Google Scholar]
- 112.Iwahara J, Schwieters CD, Clore GM. J. Am. Chem. Soc. 2004;126:12800. doi: 10.1021/ja046246b. [DOI] [PubMed] [Google Scholar]
- 113.Baker KA, Hilty C, Peti W, Prince A, Pfaffinger PJ, Wider G, Wüthrich K, Choe S. Biochemistry. 2006;45:1663. doi: 10.1021/bi0516430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Tang C, Schwieters CD, Clore GM. Nature. 2007;449:1078. doi: 10.1038/nature06232. [DOI] [PubMed] [Google Scholar]
- 115.Hansen DF, Hass MA, Christensen HM, Ulstrup J, Led JJ. J. Am. Chem. Soc. 2003;125:6858. doi: 10.1021/ja034013z. [DOI] [PubMed] [Google Scholar]
- 116.Tang C, Ghirlando R, Clore GM. J. Am. Chem. Soc. 2008;130:4048. doi: 10.1021/ja710493m. [DOI] [PubMed] [Google Scholar]
- 117.Tang C, Louis JM, Aniana A, Suh JY, Clore GM. Nature. 2008;455:693. doi: 10.1038/nature07342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Bertini I, Luchinat C, Parigi G. Prog. Nucl. Magn. Reson. Spect. 2002;40:249. [Google Scholar]
- 119.Bloembergen N, Morgan LO. J. Chem. Phys. 1961;34:842. [Google Scholar]
- 120.Rubinstein M, Baram A, Luz Z. Mol. Phys. 1971;20:67. [Google Scholar]
- 121.Westlund PO. Mol. Phys. 1995;85:1165. [Google Scholar]
- 122.Guéron M. J. Magn. Reson. 1975;19:58. [Google Scholar]
- 123.Iwahara J, Tang C, Clore GM. J. Magn. Reson. 2007;184:185. doi: 10.1016/j.jmr.2006.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Keizers PH, Desreux JF, Overhand M, Ubbink M. J. Am. Chem. Soc. 2007;129:9292. doi: 10.1021/ja0725201. [DOI] [PubMed] [Google Scholar]
- 125.Su XC, McAndrew K, Huber T, Otting G. J. Am. Chem. Soc. 2008;130:1681. doi: 10.1021/ja076564l. [DOI] [PubMed] [Google Scholar]
- 126.Bertini I, Del Bianco C, Gelis I, Katsaros N, Luchinat C, Parigi G, Peana M, Provenzani A, Zoroddu MA. Proc. Natl. Acad. Sci. U. S. A. 2004;101:6841. doi: 10.1073/pnas.0308641101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Bertini I, Gupta YK, Luchinat C, Parigi G, Peana M, Sgheri L, Yuan J. J. Am. Chem. Soc. 2007;129:12786. doi: 10.1021/ja0726613. [DOI] [PubMed] [Google Scholar]
- 128.Hulsker R, Baranova MV, Bullerjahn GS, Ubbink MJ. Am. Chem. Soc. 2008;130:1985. doi: 10.1021/ja077453p. [DOI] [PubMed] [Google Scholar]
- 129.Ikegami T, Verdier L, Sakhaii P, Grimme S, Pescatore B, Saxena K, Fiebig KM, Griesinger C. J. Biomol. NMR. 2004;29:339. doi: 10.1023/B:JNMR.0000032611.72827.de. [DOI] [PubMed] [Google Scholar]
- 130.Pintacuda G, Moshref A, Leonchiks A, Sharipo A, Otting G. J. Biomol. NMR. 2004;29:351. doi: 10.1023/B:JNMR.0000032610.17058.fe. [DOI] [PubMed] [Google Scholar]
- 131.Vlasie MD, Comuzzi C, van den Nieuwendijk AM, Prudencio M, Overhand M, Ubbink M. Chemistry. 2007;13:1715. doi: 10.1002/chem.200600916. [DOI] [PubMed] [Google Scholar]
- 132.Su XC, Man B, Beeren S, Liang H, Simonsen S, Schmitz C, Huber T, Messerle BA, Otting G. J. Am. Chem. Soc. 2008;130:10486. doi: 10.1021/ja803741f. [DOI] [PubMed] [Google Scholar]
- 133.Barbieri R, Bertini I, Cavallaro G, Lee YM, Luchinat C, Rosato A. J. Am. Chem. Soc. 2002;124:5581. doi: 10.1021/ja025528d. [DOI] [PubMed] [Google Scholar]
- 134.John M, Headlam MJ, Dixon NE, Otting G. J. Biomol. NMR. 2007;37:43. doi: 10.1007/s10858-006-9098-6. [DOI] [PubMed] [Google Scholar]
- 135.Babini E, Bertini I, Capozzi F, Felli IC, Lelli M, Luchinat C. J. Am. Chem. Soc. 2004;126:10496. doi: 10.1021/ja047573m. [DOI] [PubMed] [Google Scholar]
- 136.Balayssac S, Bertini I, Luchinat C, Parigi G, Piccioli M. J. Am. Chem. Soc. 2006;128:15042. doi: 10.1021/ja0645436. [DOI] [PubMed] [Google Scholar]
- 137.Bermel W, Bertini I, Felli IC, Kummerle R, Pierattelli R. J. Am. Chem. Soc. 2003;125:16423. doi: 10.1021/ja037676p. [DOI] [PubMed] [Google Scholar]
- 138.Bermel W, Bertini I, Felli IC, Kummerle R, Pierattelli R. J. Magn. Reson. 2006;178:56. doi: 10.1016/j.jmr.2005.08.011. [DOI] [PubMed] [Google Scholar]
- 139.Bertini I, Felli IC, Gonnelli L, Pierattelli R, Spyranti Z, Spyroulias GA. J. Biomol. NMR. 2006;36:111. doi: 10.1007/s10858-006-9068-z. [DOI] [PubMed] [Google Scholar]
- 140.Bertini I, Jimenez B, Piccioli M. J. Magn. Reson. 2005;174:125. doi: 10.1016/j.jmr.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 141.Bertini I, Jimenez B, Pierattelli R, Wedd AG, Xiao Z. Proteins. 2008;70:1196. doi: 10.1002/prot.21577. [DOI] [PubMed] [Google Scholar]
- 142.Caillet-Saguy C, Delepierre M, Lecroisey A, Bertini I, Piccioli M, Turano P. J. Am. Chem. Soc. 2006;128:150. doi: 10.1021/ja054902h. [DOI] [PubMed] [Google Scholar]
- 143.Boisbouvier J, Gans P, Blackledge M, Brutscher B, Marion D. J. Am. Chem. Soc. 1999;121:7700. [Google Scholar]
- 144.Pintacuda G, Kaikkonen A, Otting G. J. Magn. Reson. 2004;171:233. doi: 10.1016/j.jmr.2004.08.019. [DOI] [PubMed] [Google Scholar]
- 145.Madhu PK, Grandori R, Hohenthanner K, Mandal PK, Muller N. J. Biomol. NMR. 2001;20:31. doi: 10.1023/a:1011292410478. [DOI] [PubMed] [Google Scholar]
- 146.Hus JC, Marion D, Blackledge M. J. Mol. Biol. 2000;298:927. doi: 10.1006/jmbi.2000.3714. [DOI] [PubMed] [Google Scholar]
- 147.Lipari G, Szabo A. J. Am. Chem. Soc. 1982;104:4546. [Google Scholar]
- 148.Brüschweiler R, Roux B, Blackledge M, Griesinger C, Karplus M, Ernst RR. J. Am. Chem. Soc. 1992;114:2289. [Google Scholar]
- 149.Olejniczak ET, Dobson CM, Karplus M, Levy RM. J. Am. Chem. Soc. 1984;106:1923. [Google Scholar]
- 150.Benetis N, Kowalewski J. J. Magn. Reson. 1985;65:13. [Google Scholar]
- 151.Woessner DE. J. Chem. Phys. 1962;37:647. [Google Scholar]
- 152.McConnell HM. J. Chem. Phys. 1958;28:430. [Google Scholar]
- 153.Led JJ, Grant DM. J. Am. Chem. Soc. 1977;99:5845. doi: 10.1021/ja00460a001. [DOI] [PubMed] [Google Scholar]
- 154.Allerhand A, Thiele E. J. Chem. Phys. 1966;45:902. [Google Scholar]
- 155.Griffith OH, McConnell HM. Proc. Natl. Acad. Sci. U. S. A. 1966;55:8. doi: 10.1073/pnas.55.1.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Ogawa S, McConnell HM. Proc. Natl. Acad. Sci. U. S. A. 1967;58:19. doi: 10.1073/pnas.58.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Ebright YW, Chen Y, Pendergrast PS, Ebright RH. Biochemistry. 1992;31:10664. doi: 10.1021/bi00159a004. [DOI] [PubMed] [Google Scholar]
- 158.Ermacora MR, Delfino JM, Cuenoud B, Schepartz A, Fox RO. Proc. Natl. Acad. Sci. U. S. A. 1992;89:6383. doi: 10.1073/pnas.89.14.6383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Keizers PH, Saragliadis A, Hiruma Y, Overhand M, Ubbink M. J. Am. Chem. Soc. 2008;130:14802. doi: 10.1021/ja8054832. [DOI] [PubMed] [Google Scholar]
- 160.Hanson P, Millhauser G, Formaggio F, Crisma M, Toniolo C. J. Am. Chem. Soc. 1996;118:7618. [Google Scholar]
- 161.Milov AD, Tsvetkov YD, Gorbunova EY, Mustaeva LG, Ovchinnikova TV, Raao J. Biopolymers. 2002;64:328. doi: 10.1002/bip.10208. [DOI] [PubMed] [Google Scholar]
- 162.Monaco V, Formaggio F, Crisma M, Toniolo C, Hanson P, Millhauser G, George C, Deschamps JR, Flippen-Anderson JL. Bioorg. Med. Chem. 1999;7:119. doi: 10.1016/s0968-0896(98)00220-x. [DOI] [PubMed] [Google Scholar]
- 163.Monaco V, Formaggio F, Crisma M, Toniolo C, Hanson P, Millhauser GL. Biopolymers. 1999;50:239. doi: 10.1002/(SICI)1097-0282(199909)50:3<239::AID-BIP2>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 164.Shenkarev ZO, Paramonov AS, Balashova TA, Yakimenko ZA, Baru MB, Mustaeva LG, Raap J, Ovchinnikova TV, Arseniev AS. Biochem. Biophys. Res. Commun. 2004;325:1099. doi: 10.1016/j.bbrc.2004.10.115. [DOI] [PubMed] [Google Scholar]
- 165.Lindfors HE, de Koning PE, Drijfhout JW, Venezia B, Ubbink M. J Biomol NMR. 2008;41:157. doi: 10.1007/s10858-008-9248-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Wang L, Zhang Z, Brock A, Schultz PG. Proc. Natl. Acad. Sci. U. S. A. 2003;100:56. doi: 10.1073/pnas.0234824100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Jensen MR, Lauritzen C, Dahl SW, Pedersen J, Led JJ. J. Biomol. NMR. 2004;29:175. doi: 10.1023/B:JNMR.0000019251.09648.c4. [DOI] [PubMed] [Google Scholar]
- 168.Wöhnert J, Franz KJ, Nitz M, Imperiali B, Schwalbe H. J.Am. Chem. Soc. 2003;125:13338. doi: 10.1021/ja036022d. [DOI] [PubMed] [Google Scholar]
- 169.Chirivino E, Giordano C, Faini S, Cellai L, Fragai M. Chem. Med. Chem. 2007;2:1153. doi: 10.1002/cmdc.200600311. [DOI] [PubMed] [Google Scholar]
- 170.Giordano C, Del Bianco C, Faini S, Napoli A, Sindona G, Cellai L. Synthesis-Stuttgart. 2004:1835. [Google Scholar]
- 171.Dreyer GB, Dervan PB. Proc. Natl. Acad. Sci. U. S. A. 1985;82:968. doi: 10.1073/pnas.82.4.968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Braunlin WH, Drakenberg T, Nordenskiold L. Biopolymers. 1987;26:1047. doi: 10.1002/bip.360260705. [DOI] [PubMed] [Google Scholar]
- 173.Froystein NA, Davis JT, Reid BR, Sletten E. Acta Chem. Scand. 1993;47:649. doi: 10.3891/acta.chem.scand.47-0649. [DOI] [PubMed] [Google Scholar]
- 174.Rai P, Cole TD, Wemmer DE, Linn S. J. Mol. Biol. 2001;312:1089. doi: 10.1006/jmbi.2001.5010. [DOI] [PubMed] [Google Scholar]
- 175.Keyes RS, Bobst AM. In: In Biological Magnetic Resonance. Berliner LJ, editor. Vol. 14. New York: Plenum Press; 1998. [Google Scholar]
- 176.Cai S, Zhu L, Zhang Z, Chen Y. Biochemistry. 2007;46:4943. doi: 10.1021/bi061738h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Qin PZ, Hideg K, Feigon J, Hubbell WL. Biochemistry. 2003;42:6772. doi: 10.1021/bi027222p. [DOI] [PubMed] [Google Scholar]
- 178.Fesik SW, Gemmecker G, Olejniczak ET, Petros AM. J. Am. Chem. Soc. 1991;113:7080. [Google Scholar]
- 179.Petros AM, Mueller L, Kopple KD. Biochemistry. 1990;29:10041. doi: 10.1021/bi00495a005. [DOI] [PubMed] [Google Scholar]
- 180.Hernandez G, Teng CL, Bryant RG, LeMaster DM. J. Am. Chem. Soc. 2002;124:4463. doi: 10.1021/ja017340k. [DOI] [PubMed] [Google Scholar]
- 181.Sakakura M, Noba S, Luchette PA, Shimada I, Prosser RS. J. Am. Chem. Soc. 2005;127:5826. doi: 10.1021/ja047825j. [DOI] [PubMed] [Google Scholar]
- 182.Pintacuda G, Otting G. J. Am. Chem. Soc. 2002;124:372. doi: 10.1021/ja016985h. [DOI] [PubMed] [Google Scholar]
- 183.Likhtenshtein GI, Adin I, Novoselsky A, Shames A, Vaisbuch I, Glaser R. Biophys.J. 1999;77:443. doi: 10.1016/S0006-3495(99)76902-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Teng CL, Bryant RG. J Magn Reson. 2006;179:199. doi: 10.1016/j.jmr.2005.12.001. [DOI] [PubMed] [Google Scholar]
- 185.Hilty C, Wider G, Fernandez C, Wüthrich K. ChemBioChem. 2004;5:467. doi: 10.1002/cbic.200300815. [DOI] [PubMed] [Google Scholar]
- 186.Kutateladze TG, Capelluto DG, Ferguson CG, Cheever ML, Kutateladze AG, Prestwich GD, Overduin M. J.Biol. Chem. 2004;279:3050. doi: 10.1074/jbc.M309007200. [DOI] [PubMed] [Google Scholar]
- 187.Ollerenshaw JE, Lidar DA, Kay LE. Phys. Rev. Lett. 2003;91:217904. doi: 10.1103/PhysRevLett.91.217904. [DOI] [PubMed] [Google Scholar]
- 188.Thomas DD, Dalton LR, Hyde JS. J. Chem. Phys. 1976;65:3006. [Google Scholar]
- 189.Wien RW, Morrisett JD, McConnell HM. Biochemistry. 1972;11:3707. doi: 10.1021/bi00770a008. [DOI] [PubMed] [Google Scholar]
- 190.Pearlman DA, Kollman PA. J. Mol. Biol. 1991;220:457. doi: 10.1016/0022-2836(91)90024-z. [DOI] [PubMed] [Google Scholar]
- 191.Torda AE, Brunne RM, Huber T, Kessler H, Vangunsteren WF. J. Biomol. NMR. 1993;3:55. doi: 10.1007/BF00242475. [DOI] [PubMed] [Google Scholar]
- 192.Torda AE, Scheek RM, Vangunsteren WF. Chem. Phys. Lett. 1989;157:289. [Google Scholar]
- 193.Torda AE, Scheek RM, Vangunsteren WF. J. Mol. Biol. 1990;214:223. doi: 10.1016/0022-2836(90)90157-H. [DOI] [PubMed] [Google Scholar]
- 194.Bonvin AM, Brunger AT. J. Mol. Biol. 1995;250:80. doi: 10.1006/jmbi.1995.0360. [DOI] [PubMed] [Google Scholar]
- 195.Bonvin AM, Vis H, Breg JN, Burgering MJ, Boelens R, Kaptein R. J.Mol. Biol. 1994;236:328. doi: 10.1006/jmbi.1994.1138. [DOI] [PubMed] [Google Scholar]
- 196.Brüschweiler R, Blackledge M, Ernst RR. J. Biomol. NMR. 1991;1:3. doi: 10.1007/BF01874565. [DOI] [PubMed] [Google Scholar]
- 197.Clore GM, Schwieters CD. J. Am. Chem. Soc. 2004;126:2923. doi: 10.1021/ja0386804. [DOI] [PubMed] [Google Scholar]
- 198.Clore GM, Schwieters CD. Biochemistry. 2004;43:10678. doi: 10.1021/bi049357w. [DOI] [PubMed] [Google Scholar]
- 199.Clore GM, Schwieters CD. J. Mol. Biol. 2006;355:879. doi: 10.1016/j.jmb.2005.11.042. [DOI] [PubMed] [Google Scholar]
- 200.Schwieters CD, Clore GM. Biochemistry. 2007;46:1152. doi: 10.1021/bi061943x. [DOI] [PubMed] [Google Scholar]
- 201.Schwieters CD, Kuszewski JJ, Tjandra N, Clore GM. J.Magn. Reson. 2003;160:65. doi: 10.1016/s1090-7807(02)00014-9. [DOI] [PubMed] [Google Scholar]
- 202.Schwieters CD, Kuszewski JJ, Clore GM. Prog. Nucl. Magn. Reson. Spect. 2006;48:47. [Google Scholar]
- 203.Freed JH. J. Chem. Phys. 1978;68:4034. [Google Scholar]
- 204.Hwang LP, Freed JH. J. Chem. Phys. 1975;63:4017. [Google Scholar]
- 205.Givaty O, Levy Y. J. Mol. Biol. 2009;385:1087. doi: 10.1016/j.jmb.2008.11.016. [DOI] [PubMed] [Google Scholar]
- 206.Winkler FK, Banner DW, Oefner C, Tsernoglou D, Brown RS, Heathman SP, Bryan RK, Martin PD, Petratos K, Wilson KS. EMBO J. 1993;12:1781. doi: 10.2210/pdb4rve/pdb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Banerjee A, Santos WL, Verdine GL. Science. 2006;311:1153. doi: 10.1126/science.1120288. [DOI] [PubMed] [Google Scholar]
- 208.Kalodimos CG, Biris N, Bonvin AM, Levandoski MM, Guennuegues M, Boelens R, Kaptein R. Science. 2004;305:386. doi: 10.1126/science.1097064. [DOI] [PubMed] [Google Scholar]
- 209.Viadiu H, Aggarwal AK. Mol. Cell. 2000;5:889. doi: 10.1016/s1097-2765(00)80329-9. [DOI] [PubMed] [Google Scholar]
- 210.Gewirth DT, Sigler PB. Nature Struct. Biol. 1995;2:386. doi: 10.1038/nsb0595-386. [DOI] [PubMed] [Google Scholar]
- 211.Agresti A, Bianchi ME. Curr. Opin. Genet. Dev. 2003;13:170. doi: 10.1016/s0959-437x(03)00023-6. [DOI] [PubMed] [Google Scholar]
- 212.Ohndorf UM, Rould MA, He Q, Pabo CO, Lippard SJ. Nature. 1999;399:708. doi: 10.1038/21460. [DOI] [PubMed] [Google Scholar]
- 213.Werner MH, Huth JR, Gronenborn AM, Clore GM. Cell. 1995;81:705. doi: 10.1016/0092-8674(95)90532-4. [DOI] [PubMed] [Google Scholar]
- 214.Murphy EC, Zhurkin VB, Louis JM, Cornilescu G, Clore GM. J. Mol. Biol. 2001;312:481. doi: 10.1006/jmbi.2001.4977. [DOI] [PubMed] [Google Scholar]
- 215.Fraenkel E, Pabo CO. Nature Struct. Biol. 1998;5:692. doi: 10.1038/1382. [DOI] [PubMed] [Google Scholar]
- 216.Selzer T, Albeck S, Schreiber G. Nature Struct. Biol. 2000;7:537. doi: 10.1038/76744. [DOI] [PubMed] [Google Scholar]
- 217.Adams G, Delbruck M. In: Structural Chemistry and Molecular Biology. Rich A, Davidson N, editors. San Francisco: Freeman & Co.; 1968. [Google Scholar]
- 218.Deutscher J, Francke C, Postma PW. Microbiol. Mol. Biol. Rev. 2006;70:939. doi: 10.1128/MMBR.00024-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Cornilescu G, Lee BR, Cornilescu CC, Wang G, Peterkofsky A, Clore GM. J. Biol. Chem. 2002;277:42289. doi: 10.1074/jbc.M207314200. [DOI] [PubMed] [Google Scholar]
- 220.Schwieters CD, Clore GM. J. Magn. Reson. 2001;152:288. doi: 10.1006/jmre.2001.2413. [DOI] [PubMed] [Google Scholar]
- 221.Schwieters CD, Clore GM. J. Biomol. NMR. 2002;23:221. doi: 10.1023/a:1019875223132. [DOI] [PubMed] [Google Scholar]
- 222.Levy Y, Wolynes PG, Onuchic JN. Proc. Natl. Acad. Sci. U. S. A. 2004;101:511. doi: 10.1073/pnas.2534828100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Nocek JM, Zhou JS, De Forest S, Priyadarshy S, Beratan DN, Onuchic JN, Hoffman BM. Chem. Rev. 1996;96:2459. doi: 10.1021/cr9500444. [DOI] [PubMed] [Google Scholar]
- 224.Pelletier H, Kraut J. Science. 1992;258:1748. doi: 10.1126/science.1334573. [DOI] [PubMed] [Google Scholar]
- 225.Kang SA, Marjavaara PJ, Crane BR. J. Am. Chem. Soc. 2004;126:10836. doi: 10.1021/ja049230u. [DOI] [PubMed] [Google Scholar]
- 226.Hoffman BM, Celis LM, Cull DA, Patel AD, Seifert JL, Wheeler KE, Wang J, Yao J, Kurnikov IV, Nocek JM. Proc. Natl. Acad. Sci. U. S. A. 2005;102:3564. doi: 10.1073/pnas.0408767102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Wlodawer A, Vondrasek J. Annu. Rev. Biophys. Biomol. Struct. 1998;27:249. doi: 10.1146/annurev.biophys.27.1.249. [DOI] [PubMed] [Google Scholar]
- 228.Louis JM, Nashed NT, Parris KD, Kimmel AR, Jerina DM. Proc. Natl. Acad. Sci. U. S. A. 1994;91:7970. doi: 10.1073/pnas.91.17.7970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Louis JM, Wondrak EM, Kimmel AR, Wingfield PT, Nashed NT. J. Biol. Chem. 1999;274:23437. doi: 10.1074/jbc.274.33.23437. [DOI] [PubMed] [Google Scholar]
- 230.Louis JM, Weber IT, Tozser J, Clore GM, Gronenborn AM. Adv. Pharmacol. 2000;49:111. doi: 10.1016/s1054-3589(00)49025-3. [DOI] [PubMed] [Google Scholar]
- 231.Wondrak EM, Nashed NT, Haber MT, Jerina DM, Louis JM. J. Biol. Chem. 1996;271:4477. doi: 10.1074/jbc.271.8.4477. [DOI] [PubMed] [Google Scholar]
- 232.Tam R, Saier MH., Jr Microbiol. Rev. 1993;57:320. doi: 10.1128/mr.57.2.320-346.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Sharff AJ, Rodseth LE, Spurlino JC, Quiocho FA. Biochemistry. 1992;31:10657. doi: 10.1021/bi00159a003. [DOI] [PubMed] [Google Scholar]
- 234.Quiocho FA, Spurlino JC, Rodseth LE. Structure. 1997;5:997. doi: 10.1016/s0969-2126(97)00253-0. [DOI] [PubMed] [Google Scholar]
- 235.Evenas J, Tugarinov V, Skrynnikov NR, Goto NK, Muhandiram R, Kay LE. J. Mol. Biol. 2001;309:961. doi: 10.1006/jmbi.2001.4695. [DOI] [PubMed] [Google Scholar]
- 236.Millet O, Hudson RP, Kay LE. Proc. Natl. Acad. Sci. U. S. A. 2003;100:12700. doi: 10.1073/pnas.2134311100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Dyson HJ, Wright PE. Nature Rev. Mol. Cell. Biol. 2005;6:197. doi: 10.1038/nrm1589. [DOI] [PubMed] [Google Scholar]
- 238.Neri D, Billeter M, Wider G, Wüthrich K. Science. 1992;257:1559. doi: 10.1126/science.1523410. [DOI] [PubMed] [Google Scholar]
- 239.Hillisch A, Lorenz M, Diekmann S. Curr. Opin. Struct. Biol. 2001;11:201. doi: 10.1016/s0959-440x(00)00190-1. [DOI] [PubMed] [Google Scholar]
- 240.Altenbach C, Kusnetzow AK, Ernst OP, Hofmann KP, Hubbell WL. Proc. Natl. Acad. Sci. U. S. A. 2008;105:7439. doi: 10.1073/pnas.0802515105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Klug CS, Feix JB. Methods Cell. Biol. 2008;84:617. doi: 10.1016/S0091-679X(07)84020-9. [DOI] [PubMed] [Google Scholar]