Abstract
The application of metabolomics, the global analysis of metabolite levels, to the study of protozoan parasites has become an important tool for understanding the host/parasite relationship and holds promise for the development of direly needed therapeutics and improved diagnostics. Research advances over the past decade have opened the door for a systems biology approach to protozoan parasites with metabolomics providing a crucial readout of metabolic activity. In this review we highlight recent metabolomic approaches to protozoan parasites, including metabolite profiling, integration with genomics, transcription, and proteomic analysis, as well as the use of metabolic fingerprints for the diagnosis of parasitic infections.
Introduction
Enzymes are central to every aspect of biology and metabolite levels (as substrates, products and co-factors) are the crucial read-out of the combined enzyme and transporter activity within cells. Technological advances over the past decade now allow for the rapid and accurate measurement of the abundance levels of hundreds to thousands of metabolites simultaneously from complex mixtures. These techniques, collectively referred to as metabolomics, fill a crucial gap in the push towards the global characterization of cells as living system alongside their –omic siblings characterizing genes, transcripts, and proteins. Metabolomic analysis has been widely applied to study the systems biology of numerous model organisms across the tree of life, including Archeae (Trauger et al., 2008), Eubacteria, (Rabinowitz, 2007), fungi, (Brauer et al., 2006), plants, (Cho et al., 2008; Lisec et al., 2008), animals (Pedersen et al., 2008; Sun et al., 2007)), and human cell tissue culture (Khoo and Al-Rubeai, 2007). Application of metabolomics to the study of protozoan parasites, while still in its infancy, is rapidly emerging as a fertile approach to better understand the host/parasite interaction.
Protozoan parasites cause serious infections in humans and animals, including life-threatening diseases such as malaria (Plasmodium spp.), African sleeping sickness (Trypanosoma brucei), Chagas disease (Trypanosoma cruzi), and leishmaniasis (Leishmania spp.). While these diseases affect few in the developed world, they continue to threaten the health and livelihood of a large portion of humanity. To make matters worse, once efficacious drugs such as the anti-folates or 4-aminoquinolines targeting the malaria parasite have been rendered useless due to widespread parasite resistance against virtually all anti-malarial agents (Guerin et al., 2009).
From a systems biology perspective, studying parasitic disease is particularly enticing as it inherently involves the complex interplay of two interconnected biological systems with a net flow of energy and nutrients between the host and the parasite. In the process of adapting to their host niche, these organisms have evolved reduced metabolic capacity, often drastically so, while expanding mechanisms for avoiding host defense and utilizing metabolites from their hosts. Some, like the African trypanosome are so adept at avoiding the host defense mechanisms that they grow freely in the blood stream, where they are bathed in nutrients. Others such as the malaria parasite have adapted to life within host cells, which offers certain protections but can place additional barriers to nutrient acquisition. Many parasites also infect additional host species in order to complete their lifecycle, which can require a drastic remodeling of parasite metabolism to adapt to an entirely differently metabolic milieu, as in the mammal/arthropod transitions of vector-borne parasites (Aly et al., 2009).
The current paucity of effective preventative or therapeutic options is another strong motivation for the study of parasite metabolism. No effective vaccine exits for the prevention of protozoan parasitic disease in humans and the number of effective and affordable drugs, small to begin with, is progressively shrinking with rising incidence of resistance, making the development of novel anti-parasitic drugs imperative. Most drugs against infectious disease exploit differences in metabolism by differentially targeting pathogen enzymes that are absent or highly divergent from those of the host (Aguero et al., 2008). This strategy has been highly successful for evolutionarily distant bacterial pathogens but protozoan parasites, as fellow eukaryotes, present far fewer divergent drug targets due to a shared eukaryotic ancestry and their reduced metabolic capacity. Understanding parasite metabolic networks, including the role of novel organelles such as the apicomplexan apicoplast (the non-photosynthetic remnant of an algal symbiont) (Seeber et al., 2008) or the mitosomes/hydrogenosomes of low branching protozoa (Hackstein et al., 2006), will be crucial for developing effective new drugs.
The current state of research on protozoan parasites is primed for systems biology approaches, including global metabolite analysis. With few exceptions, one or more stages of the important protozoan pathogens of humans can be grown in culture to densities that allow isolation of significant amounts of DNA, RNA, proteins, and metabolites. Annotated draft sequences exist of genomes from all major groups of protozoan pathogens that infect humans (many accessible via www.EuPathDB.org). The available genomes and expressed sequence tags have opened the door for proteomics by allowing peptide mapping and for the design of DNA microarrays that have already been used extensively for transcriptome analysis of numerous parasites, including Plasmodium spp., T. gondii, and the kinetoplastids.
Integrating metabolomics with the analysis of the genomic, transcriptomic, and proteomic data already in place opens the door for systematic study of the host/parasite interaction that will lead to a deeper biological understanding and is likely to unveil additional targets for prevention and therapy. In this review we aim to outline the current status of using metabolomics in the study of protozoan parasites and highlight numerous promising applications.
Definitions and Methodology
Broadly defined, the metabolome is the collection of all small molecules (<1 kDa) of a given system and metabolomics is the characterization of this set by one or more techniques. The more narrowly defined term metabonomics refers to the quantitative measurement of metabolic responses over time within a system in response to an event (infection, drug treatment, starvation, etc.) (Holmes et al., 2008). In the case of a host/parasite system, metabolomics often involves the characterization and interplay of each organism's metabolome, or a characterization of the host cell's metabolome in the absence or presence of the parasite, known as the co-metabolome (Holmes et al., 2008).
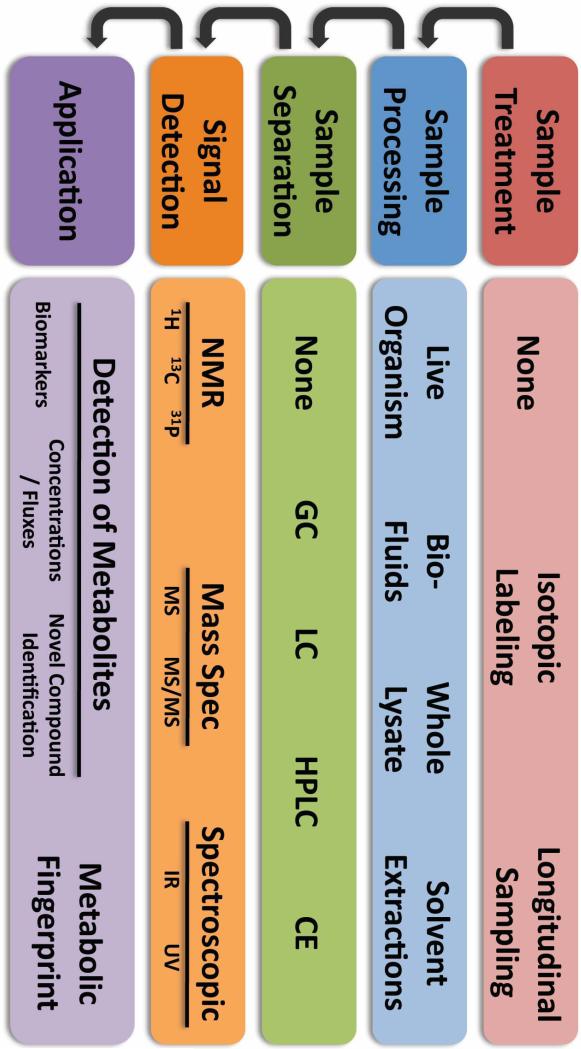
Historically, enzyme-based assays and tracing of radiolabeled nutrients were the major tools for assaying parasite metabolism (Sherman, 1998). Today, Nuclear Magnetic Resonance (NMR) spectroscopy and separation-linked single (MS) or tandem (MS/MS) mass spectrometry are the major tools in use for metabolomics. Their use in metabolomics has been expertly reviewed elsewhere (Bollard et al., 2005; Lenz and Wilson, 2007; Want et al., 2007). A cursory overview of some methodology options can be found in Figure 1 but a more detailed description and comparison of these technologies is outside the scope of this review.
Figure 1. Commonly-Used Methodologies in Metabolomics.
The diagram above outlines the major steps of metabolomic experiments along with some commonly-used options available for each step. Prior to sample processing biological materials of interest may be isotopically enriched to follow the metabolism of labeled compounds over time. Some technologies such as Nuclear Magnetic Resonance (NMR) spectroscopy allow for the monitoring of metabolites within live organisms or assaying whole lysate directly but often sample complexity is reduced by either limiting the material sampled to specific bio-fluids (e.g. urine, plasma) or performing solvent extractions (e.g. methanol/water, methanol/chloroform, perchlorate). Furthermore, mass spectrometry (MS) and spectroscopic (infrared (IR) or ultraviolet (UV)) approaches often utilize chromatography (gas (GC), liquid (LC), high-performance liquid (HPLC)) or capillary electrophoresis (CE) to limit the sample complexity entering the detector at any one time and to determine a characteristic retention time. Mass spectrometry utilizes the retention time for a given metabolite as well as the mass/charge ratio (MS), often in conjunction with the mass/charge ratio of fragmentation daughter ions in tandem MS (MS/MS), for identification. NMR exploits structurally-dependent changes in the magnetic resonance of suitable nuclei (e.g. 1H, 13C, 31P) for metabolite identification. Light spectroscopic approaches perform less well for the individual identification of metabolites but represent a more cost-effective approach to determine changes in the overall metabolic profile or fingerprint of a sample. Both NMR and MS approaches offer the possibility for not only the detection of dozens to hundreds of individual metabolites but recent advances also permit more accurate measurements of metabolite concentrations, which are critical for determining metabolite fluxes within a metabolic network.
Laying the Groundwork: Profiling Parasite Metabolomes
A number of pioneering studies published in the 1980's first applied NMR spectroscopy to parasite metabolism (expertly reviewed by S. N. Thompson (Thompson, 1991)). Many of these early studies focus on the profiling of major parasite metabolites. To avoid significant overlap with this comprehensive work, our review will focus only on subsequently published work.
Since 1990, 31P-NMR has continued to be used for profiling various aspects of parasite phosphate metabolism in Plasmodium spp., Toxoplasma gondii and Cryptosporidium parvum (Moreno et al., 2001), as well as the kinetoplastids (Mendoza et al., 2002). Similarly, the use of 13C-NMR for metabolite mapping has also continued, including the characterization of abundant metabolites in Entamoeba histolytica (Bakker-Grunwald et al., 1995) and P. falciparum infected erythrocytes (Lian et al., 2009), measuring glucose flux in P. falciparum and P. yoelii infected erythrocytes (Mehta et al., 2006), and measuring the suppression of glycolytic activity in uninfected erythrocytes by malaria conditioned medium (Mehta et al., 2006).
With increasing magnet strength and improved signal filtering the use of 1H-NMR for metabolite mapping has steadily grown and yielded several studies mapping the metabolome of kinetoplastid parasites including L. donovani (Gupta et al., 1999) and T. cruzi (Penin et al., 1998), as well as the lipidome of L. donovani (Adosraku et al., 1993). A recent metabolic analysis of P. falciparum trophozoite extracts compared several extraction conditions and identified over 50 metabolites with measured concentrations in the high nM/low μM range for 40 separate metabolites (Teng et al., 2009). A surprising finding of this study included the accumulation by the parasite of HEPES, a buffering agent used in culture medium, further highlighting the considerable differences between in vitro and in vivo growth conditions (reviewed in (LeRoux et al., 2009)). In a complementary study using an LC-MS/MS approach, Olszewski et al. sampledsynchronous P. falciparum blood-stage cultures at 8h intervals and assayed 89 known metabolites across the 48h intraerythrocytic cycle (Olszewski et al., 2009).
Kamleh and colleagues compared the metabolome of T. brucei procyclic forms fed either preferred carbon source glucose or proline, which is abundantly available in the Tsetse fly vector. Using hydrophilic interaction chromatography (HILIC) and electrospray ionization (ESI) MS over distinct 1000 M/Z peaks were resolved, including many phospholipids and lipophilic amino acids, which can be difficult to resolve using standard reverse phase HPLC. Several metabolites changed significantly in abundance following a switch in carbon source from glucose to proline, including several glutamine derivatives and phospholipids (Kamleh et al., 2008). Finally, an earlier study determined the uptake and catalysis of polyamines & thiols by T. cruzi trypomastigotes and amastigotes by HPLC/MALDI-TOF MS (Ariyanayagam et al., 2003).
Towards Parasite Systems Biology: Integrating -omics
While small molecule profiling provides an important baseline metabolic characterization of parasites, studying their metabolic response (and underlying regulation) due to environmental or genetic changes has the potential to yield greater insight into parasite biology by investigating how the interplay of genetics and environment affects changes in the metabolome and how changing metabolites feed back into enzymatic and gene expression level regulation.
Inferring Metabolic Networks: Pitfalls and Promises
The sequencing of parasite genomes has yielded unprecedented insights into how parasites have adapted to their various niches and have become increasingly dependent on their hosts by sacrificing the ability to synthesize nutrients de novo for less energy-intensive scavenging mechanisms. Identification of conserved metabolic enzyme pathways (Pinney et al., 2005) and nutrient transporters (Ginsburg and Stein, 2005; Martin et al., 2005) by comparative genomics has allowed the construction of metabolic networks for these organisms in silico (Chavali et al., 2008; Ma and Zeng, 2003). These networks have been a powerful tool in guiding parasite research by laying out parasites’ anabolic and catabolic potential (Pinney et al., 2005), highlighting potential drug targets (Cornish-Bowden and Cardenas, 2003; Pinney et al., 2007), and attempts to identify metabolic gaps and novel pathways (Huthmacher et al., 2008; Zamboni and Sauer, 2009).
A variety of interfaces and databases are available to browse the annotated genomes of protozoan parasites, with www.EuPathDB.org being the most comprehensive. The annotation depth and quality of parasite genomes varies considerably between species according to funding and human capital invested to date. Protein coding genes are generally automatically annotated by a variety of homology methods (Interproscan, OrthoMCL-DB) and assigned Gene Ontology function terms and Enzyme Commission numbers when possible. A number of automated and hand-annotated approaches have been used to build metabolic networks of protozoan parasites from functionally annotated genes, including the metaTiger KEGG interface, the ApiCyc and LeishCyc family of database, and the Malaria Parasite Metabolic Pathways (MPMP) database. In a recent review, Hagai Ginsburg outlines advantages and shortcomings of these approaches for building metabolic networks of the malaria parasite; lessons that are likely to apply generally to attempts of building metabolic pathways of other protozoan (Ginsburg, 2009). An inherent shortcoming of homology-based approaches for predicting metabolic networks is that they cannot predict the function of novel or highly divergent metabolic genes. Such “hypothetical” genes of unknown function represent a sizable fraction of predicted genes in the available parasite genomes (Carlton et al., 2008). Unidentified metabolic genes among them could alter the structure of metabolic networks considerably, depending on their activity.
Finding missing and essential links: Using metabolomics to improve metabolic networks
In silico metabolomic networks are particularly valuable for identifying metabolic gaps, “missing” enzymes based on otherwise complete pathways or orphan biochemical activities, and for highlighting differences in host and parasite metabolism that may be exploited for drug therapy. Apart from enzymatic evidence, various sources of experimental data have been used to improve the accuracy of metabolic networks based on genomic sequence including phylogenetic conservation of pathways (Chen and Vitkup, 2006), patterns of co-expression at the transcript (Brauer et al., 2005) or protein level (May et al., 2008), and, increasingly, metabolite presence and abundance (Brauer et al., 2006; Breitling et al., 2008; May et al., 2008). Recently, Mohanty et al. used a homology-based search for “missing enzymes” to assign putative function to several “hypothetical” genes and fill 14 of their 69 predicted metabolic gaps in P. falciparum (Mohanty and Srinivasan, 2009). Comprehensive profiling of parasite metabolomes is certain to reveal additional gaps through the identification of “orphan” metabolites or activities. Ultimately, the use of metabolomics for identification of novel previously uncharacterized metabolites will also set off the search of their synthesis pathways (Kalisiak et al., 2009).
Several studies have used metabolic network analysis to identify essential activities in protozoan parasites that may be promising targets for drug design. In an excellent example of this approach Chavali et al. mapped a metabolic network of Leishmania infantum that incorporated 560 genes, 1112 reactions and 1101 metabolites and 8 subcellular localizations across both the amastigote and trypomastigote life stages. Using this network the authors were able to predict minimal parasite nutritional requirements as well as metabolic reactions essential to parasite survival for both single and double knockout mutations (Chavali et al., 2008). In a similar approach, Roberts et al. used a combination of genomic and proteomic data to generate a compartmental model of T. cruzi epimastigote metabolism covering 215 genes, 162 reactions, and 158 metabolites. In addition to using this approach to predict essential enzymatic activities, they identified several enzymes critical for epimastigote survival previously thought to be non-essential as well as reactions previously thought essential that were dispensable. Taking their research a step further, these predictions were validated using RNA interference (Roberts et al., 2009). Using an in silico approach, Yeh et al. (Yeh et al., 2004) and Fatumo et al. (Fatumo et al., 2009) used metabolic network analysis to identify essential enzyme activities in Plasmodium falciparum. Analyses such as these are currently being combined with other considerations to prioritize parasite drug targets by the WHO/TDR in pathogens causing neglected diseases (Aguero et al., 2008).
Many properties of metabolic networks cannot be fully determined based on interconnectivity and co-expression alone but depend critically on the reaction rates of each step (Sauer, 2006). Understanding metabolite flux through the network can be probed by feeding isotopically-labeled nutrients and following label-incorporation into down-stream metabolites by successive metabolomic analyses (Zamboni et al., 2009). Isotopic labeling can reveal non-intuitive network behavior and network redundancy (Blank et al., 2005), as well as highlight potential drug targets (Munger et al., 2006) and mechanisms of action (Kwon et al., 2008) that are not apparent based on interconnectivity alone. With the exception of work on the E. histolytica glycolysis pathway (Moreno-Sanchez et al., 2008), very little work has been done on metabolic flux analysis in protozoan parasites. The application to host-parasites systems is likely to be an important tool for rational drug design of novel anti-parasitic compounds and understanding the mechanisms of action of existing drugs such as artemisinin, which remains contentious (Eastman and Fidock, 2009).
These studies illustrate how interpreting parasite metabolism in light of the genome alone can be misleading and it is important to remember that in silico metabolic networks only reveal the metabolic potential of these organisms. The actualization of this potential is restricted by numerous levels of regulation, including life-cycle specific gene-expression, subcellular protein localization, enzyme concentration and post-translational regulation, metabolite transport, as well as substrate, product, and cofactor abundance. Global transcriptional and proteomic analyses have helped narrow these questions by revealing which parts of the networks are co-expressed but are also unable to speak directly to metabolic activity. Hence integrating mechanisms of regulation with the actual read-out of metabolic activity, metabolite levels and fluxes, is crucial for a systematic understanding of parasite metabolism and rational identification of drug targets. Of course, obligate intracellular parasites pose a unique challenge to flux analysis since two interconnected metabolic networks are at work – that from the host and that of the parasite. Therefore, defining the source of the labeled metabolites is more difficult. Furthermore, nutrients solely obtained from the host cell will be difficult to label for the study of parasite metabolism, making flux analysis virtually impossible for these nutrients.
Break Something and Measure what Happens: Applications to Reverse Genetics
Combining metabolomics with reverse genetics (genetic or chemical knock-outs/downs) is very appealing particularly for targets with potential enzymatic activity as it allows for the rapid characterization of hundreds of metabolic phenotypes simultaneously. For this reason, it has been one of the areas in the study of parasites where metabolomics has taken a solid foothold. In the Apicomplexa, due to their haploid genomes, knockout parasite lines are particularly well-suited for metabolomic studies. The susceptibility of T. brucei to RNA interference (Wang et al., 2000) enables a powerful combination of reverse genetics and metabolomics among the kinetoplasts.
Van Weelden et al. used a combination of HPLC-UV/conductivity detection, 14C radiolabeled glucose, and 13C-NMR to measure glycolysis and TCA cycle intermediates, as well as ATP/ADP/AMP/P ilevels in a T. brucei wild-type strain and two independent aconitase knockout strains to validate the dispensability of the TCA cycle for energy generation in T. brucei procyclic forms (van Weelden et al., 2003). In a second metabolomic study of T. brucei Coustou et al. used 13C-NMR to compare the utilization of D-[1-13C]-glucose (preferred) or L-[4-13C]-proline (highly available in the insect host) as the primary carbon source for wild-type procyclic forms. Combining this approach with RNAi knockdown mutants the authors were able to develop a detailed metabolic model of proline utilization in the presence and absence of glucose (Coustou et al., 2008).
Studies in Leishmania have used metabolomics to study the critical role of fructose-1,6-bisphosphatase in gluconeogenesis for L. major survival in macrophages by illustrating the parasite's inability to utilize labeled fatty acids as gluconeogenic substrates by 13C-NMR and demonstrating the loss of virulence in fructose-1,6-bisphosphatase knockout parasites (Naderer et al., 2006). De Souza et al. used the effect of glucose transporter and phosphomannose isomerase ablation on the metabolome L. Mexicana to validate improved noise filtering using a new peak clustering algorithms by comparing metabolite extracts form wild-type strains and knockout mutants using GC-MS (De Souza et al., 2006).
In an apicomplexan example, Olszweski et al. used LC-MS of growth medium from cultures of synchronized malaria parasites to link arginine depletion and ornithine enrichment to the parasite arginase by demonstrating stable arginine and ornithine levels in ex vivo cultures of arginase knockout P. berghei parasites (Olszewski et al., 2009).
Currently, there are numerous genetically altered parasite lines available (www.MR4.org, www.ATCC.org, www.pberghei.eu), but one clear metabolic example of interest stems from recent studies that have generated type II fatty acid synthesis (FAS II) knockout mutants in Plasmodium spp. (Vaughan et al., 2009; Yu et al., 2008) and Toxoplasma gondii (Mazumdar et al., 2006). These papers demonstrate genetically that FAS II is essential only during late stage liver development in Plasmodium but necessary for tachyzoite growth in Toxoplasma. It will be interesting to characterize the metabolic phenotypes downstream of these mutations under both lethal and non-lethal conditions using metabolomic approaches for both parasites.
From Metabolites to Genes: Applications to Forward Genetics
The application of quantitative trait locus analysis to quantitative measurements of global metabolite levels (mQTL) has emerged as a powerful tool dissecting the genetic basis of metabolism in plants (Keurentjes et al., 2006). Assaying 84 metabolites (50 of known chemical structure) by GC-MS in shoots of 429 genotyped inbred A. thaliana lines, Lisec et al. were able to identify 157 metabolite specific QTLs, mapping to enzymes in the appropriate metabolic pathway for 67% of QTLs. The number of contributing loci varied from just one for each of 42 metabolites to six QTLs contributing to tyrosine metabolism. Furthermore, the study revealed the presence of mQTL hot spots, loci that affect the levels of many different metabolites (Lisec et al., 2008).
Several sets of genotyped F1 lines are available from crosses in both P. falciparum (Hayton et al., 2008; Walliker et al., 1987; Wellems et al., 1990) and T. gondii (Sibley and Boothroyd, 1992; Su et al., 2002). Using QTL analysis, these crosses have been used successfully to map the genetic basis of a number of phenotypes, including resistance to a variety of drugs (Ferdig et al., 2004; Fidock et al., 2000; Mu et al., 2003; Nair et al., 2008; Yuan et al., 2009), host cell binding (Hayton et al., 2008), and virulence (Saeij et al., 2006; Taylor et al., 2006). Given a sufficient difference in metabolic phenotypes of the parental strains, metabolomic analyses of daughter lines from these crosses could be used to map the genetic basis of a variety of phenotypes, including starvation responses, compensatory mechanism in response to manipulation of metabolic genes, metabolic effects of drug treatment, potentially identifying unintended, secondary drug targets.
Controlling the Machinery: Metabolomics and Gene Regulation
Understanding the interplay of an organism's environment and regulation of gene expression was among the first problems of molecular genetics (Jacob and Monod, 1961). What started out as a simple system of less than a dozen components controlling lactose utilization by E. coli has grown into the study of systems biology monitoring gene expression across on organism's genome and measuring hundreds of metabolites within the organism and the environment.
Correlating genetic responses to environmental stimuli has yielded a deeper, systematic understanding of the biology of a great variety of organisms, such as environmental adaptation by extremophiles (Trauger et al., 2008), optimizing bacterial biofuel production (Yang et al., 2009), understanding yeast metabolic cycle changes in response to environmental disturbances (Gasch et al., 2000), and temperature stress in fruit flies (Pedersen et al., 2008). But combining the study of gene expression at either the transcript or protein level with metabolomic approaches has been rare in the study of host-parasite interaction.
In order to compare the response of mice either resistant or susceptible to cerebral malaria, Rae et al. measured gene transcript levels of 21 metabolic and immune response genes expressed in the brain as well as determining brain metabolite levels using 13C- and 31P-NMR, but were unable to correlate changes in gene expression and metabolite levels in mice susceptible to cerebral malaria (Rae et al., 2004). This could possibly be due to the fact that correlation of gene expression and metabolite changes occur in a more localized fashion and are lost when integrated over a whole organ as diverse as the brain.
Two P. falciparum studies have correlated changes in malaria gene expression with metabolite measurements over the intraerythrocytic developmental cycle using global transcription microarray and LC-MS/MS analysis. Olszewski et al. monitored the levels of over 90 metabolites in synchronized P. falciparum culture at 8h intervals, sampling both the media and infected red blood cells while simultaneously extracting total RNA for microarray analysis. Several metabolites correlated strongly with transcript levels of enzymes involved in their synthesis and/or catabolism, including 5-methylthioinosine and α-ketoglutarate (Olszewski et al., 2009).
Taking a broad systems approach van Brummelen et al. characterized the global effect of inhibiting the bi-functional enzyme PfAdoMetDC/ODC on the Plasmodium falciparum intraerythrocytic developmental cycle by following global transcript, protein, and metabolite abundance, using DNA microarrays, 2D PAGE-MALDI-MS/MS, and LC-MS/MS respectively (van Brummelen et al., 2009). PfAdoMetDC/ODC inhibition resulted in growth arrest and compensatory changes at the transcript and protein level including down regulation of several enzymes in the polyamine synthesis pathway downstream of PfAdoMetDC/ODC while ornithine decarboxylase, which generates the PfAdoMetDC/ODC substrate ornithine, increased along with lysine decarboxylase, which produces the polyamine cadaverine independently of PfAdoMetDC/ODC. As expected metabolite analysis revealed a significant drop in parasite polyamine levels, specifically spermidine and putrescine, with a concomitant rise in the upstream metabolite S-adenosylmethionine.
These studies mark the first steps towards parasite systems biology and illustrate how parasite metabolite levels fluctuate in response to changes in parasite gene expression at the transcriptional and protein level and how metabolite concentrations can feed back to regulate expression of metabolic genes.
It takes two to tango: Metabolomics of host/parasite systems
By its very nature, parasite metabolism cannot be understood without integrating it with that of the host. Metabolomic profiling of host fluids and tissues and changes in response to infection with protozoan parasites is critical for understanding parasite pathogenesis and shows great promise for use in diagnostics.
Biomarkers of infection
A major promise of the application of metabolomics to the study of parasites is the identification of diagnostic biomarkers. Standard diagnostic methods for parasitic infections rely heavily on microscopy, which is time-intensive and requires highly trained personnel as well as specialized equipment. Metabolomic identification of infection biomarkers is an important step towards the development of rapid and cost-effective diagnostics (Holmes et al., 2008). Successful identification of infection biomarkers has been shown using a variety of experimental systems, including in the plasma of lymphocytic choriomeningitis virus infected mice (Wikoff et al., 2009b), the cerebral spinal fluid of simian immunodeficiency virus infected rhesus macaques (Wikoff et al., 2008), and sera of Hepatitis B virus infected humans (Yang et al., 2006).
Furthermore, sample acquisition for diagnostic purposes can be intrusive and requires sterile equipment not always available, making the use of biomarkers from non-intrusive samples (saliva/urine) particularly appealing. This approach has already been applied successfully in the diagnosis of a number of non-parasite disease states (reviewed in Nordstrom and Lewensohn, 2009), including early diagnosis of various cancers, markers of renal failure, and response to a variety of drugs. The development of metabolic biomarkers of parasite infection for diagnostic purposes remains in its infancy but promising work has emerged over the past few years.
Recently, several studies have used both NMR (Li et al., 2009; Nishina et al., 2004; Saric et al., 2009; Wang et al., 2004) and UV spectroscopy (Angulo et al., 2009) for metabolomic analysis of host samples (plasma, urine, or stool) to identify biomarkers of infection with a variety of parasitic helminths. These approaches were able to differentiate infected from uninfected animals, often early in the course of infection, using either abundance changes in the metabolomic profile or metabolites unique to infected animals. Similarly, metabolite analysis of histological samples was able to demonstrate tissue specific responses to infection. A shared thread of these studies was significant changes in metabolites produced by the intestinal flora following infection with intestinal and extra-intestinal helminths alike, indicating that changes in the composition of the intestinal flora can have significant effects on the metabolite profile of the host (Wikoff et al., 2009a; Yap et al., 2008) and is a common outcome of infection.
Markers of Protozoan Infection
Few metabolomic studies have attempted to develop biomarkers of protozoan infections. Using 1H-NMR and working in the P. berghei mouse model Li et al. found drastic decreases in plasma glucose levels and a concomitant rise of lactate and pyruvate as the result of anaerobic glycolysis by the parasite, as well as a drop in glycerophosphoryl choline, which act as a source of choline for the parasite. Analysis of urine from infected mice again found evidence of changes in the intestinal flora and notably excretion of pipecolic acid and two unknown metabolites absent from the urine of uninfected mice, though many of these changes only became apparent when parasitemia exceeded 15% of circulating erythrocytes (Li et al., 2008).
Using 13C- or 31P-NMR and the rodent malaria P. berghei model, Rae et al. compared brain glucose metabolism & bioenergetics in response to infection with the ANKA strain, which causes cerebral malaria, to infection with K173 strain, which doesn't. In mice with cerebral malaria there was a notable reduction in brain metabolic activity along with a shift in D-[1-13C]-Glucose utilization from the TCA cycle to (1) anaerobic metabolism, (2) the glutamate/glutamine cycle, and (3) the GABA shunt. The increased utilization of the glutamate/glutamine cycle preceded the increases in lactate and alanine suggesting that increased gluteminergic metabolism is not the result of brain hypoxia but due to other metabolic disturbances. Mice infected with K173 strain on the other hand displayed evidence of increased (~2-fold) cerebral metabolic rate with elevated glucose utilization and relatively uniform incorporation into metabolites. Energy metabolism as monitored by 31P-NMR revealed a significant increase in inorganic phosphate and phosphocreatine consistent with increased anaerobic metabolism and cerebral hypoxia in mice with cerebral malaria (Rae et al., 2004).
A third study compared the effects of P. berghei ANKA infection in mice susceptible (CBA/J) or resistant (BALB/c) to cerebral malaria via in vivo NMR and 1H/31P-NMR metabolite profiling of blood and brain extracts. Cerebral malaria-susceptible mice showed brain anomalies absent from resistant mice at similar parasitemia levels, including an unexpected elevation of cerebral blood flow. Measuring brain metabolite levels revealed a notable drop in choline-derived compounds levels, important for membrane and myelin homeostasis (Penet et al., 2007). Furthermore, this study found increases in brain glutamine and alanine levels in susceptible mice, consistent with increased blood ammonia levels as well as the decreased levels of brain myo-inositol, which are thought to compensate for increased intracellular osmolarity due to glutamine accumulation. These changes are compatible with hepatic encephalopathy and were supported by significantly increased blood bilirubin levels, significant changes liver enzyme function, and an overall increase of hepatic tissue pathology in susceptible mice (Penet et al., 2007).
Tracking the urine and plasma metabolome over 33 days by 1H-NMR was able to differentiate mice infected by African trypanosomes from uninfected animals starting one day post-infection with differences becoming more pronounced as the infection progressed (Wang et al., 2008). Notable changes in urine were seen for several markers including 3-carboxy-2-methyl-3-oxopropanamine as a unique marker of infection. Analysis of plasma metabolite levels was able to correlate increases in lactate, oxaloacetate, and glucose as well as decreases in glutamine and lysine with infection. Starting on day 7 there were also notable changes in plasma markers. The identification of several unique urinary metabolites present early in disease progression is promising as treatment options for African trypanosomiasis are considerably better while the parasite still remains confined to the bloodstream. Once the parasite crosses the blood-brain barrier, prospects of survival dim considerably. Identification of urine or plasma biomarkers of central nervous system invasion by the parasite would mark a major advance the diagnostic standard of microscopic inspection of cerebral spinal fluid.
So far metabolomic biomarker studies of parasite infections have performed well in differentiating infected from uninfected animals and in highlighting the effect of infection on the intestinal flora. However, these approaches remain far from being useful in a diagnostic context, as many of these biomarkers appear to simply be indicators of infection rather than being pathogen specific (Koulman et al., 2009). Blinded studies using several pathogens and the same host will be needed to validate the use of metabolomics in producing correct differential diagnoses. For instance, Changes in the composition of the host's intestinal microflora can result in major changes of blood and urine metabolites (Wikoff et al., 2009a; Yap et al., 2008) and appears to be a common side effect in parasitic infections. The recurrence of cometabolites (e.g. trimethylamine, hippurate) produced by the intestinal flora as prominent biomarkers in these model systems using controlled nutritious and isogenic hosts foreshadows the large potential for variability where the diversity of host genetics, intestinal flora composition, and nutritional status is considerably higher.
One successful application of metabolomics to medical parasitology has been in the diagnosis and monitoring of large hydatid cysts in the brains and peritoneal cavity and are caused by cestodes in the genus Echinococcus. These cysts can be diagnosed and monitored by MRI and display characteristically high end product metabolites of the parasite's anaerobic glycolysis, which decrease with successful treatment (Seckin et al., 2008). Such metabolite-based diagnostics hold great promise for the future and will ultimately enable rapid assessment of patient treatment in the field.
Conclusions
Global metabolite analysis is a powerful tool for understanding the biology of protozoan parasites and host/parasite interactions. In this review we have presented a wide variety of applications for metabolomics to protozoan parasites, including work towards integrating global metabolite levels with genomic, transcriptional, and proteomic analysis in search of systematic understanding of parasite metabolism and gene regulation. Given the dire need for better anti-parasitic drugs and diagnostics, metabolomics is becoming a valuable tool in defining markers of infection and identifying potential drug targets. Additionally, characterizing the effect of novel drugs on the host and parasite metabolite levels promises to identify their mechanism of action and guide further refinement. A number of fascinating questions in parasite biology are likely to benefit tremendously through metabolomic analysis. Of great interest is the dissection of potentially non-canonical biochemical pathways and or parasite-specific metabolism. One such example is deciphering the actual function of the tricarboxylic acid (TCA) cycle of blood stage malaria, as it is known to not be a major source of energy or reducing power in these parasites (Mogi and Kita, 2009; Seeber et al., 2008). Probing the TCA cycle using metabolite flux using 13C-enriched nutrients is likely to yield novel insights into how these parasites utilize TCA cycle enzymes and their contribution to central carbon metabolism.
Recent work by Blume et. al. showed that TgGT1, the major glucose transporter at the parasite plasma membrane of T. gondii, is non-essential for parasite growth in vitro but results in growth and motility defects that can partially ameliorated by increasing media glutamine levels (Blume et al., 2009). Metabolomic characterization of this substrate switch could provide insight into the robustness of T. gondii metabolism and its ability to adapt to changing nutritional environments.
Other promising applications include understanding how parasite metabolic requirements vary upon differentiating from one life stage to the next or following transmission to new host species. For example, as Toxoplasma parasites differentiate into slow-growing bradyzoites, they become increasingly resistant to many drugs effective against the fast-growing tachyzoite stage, yet the underlying metabolic changes responsible for this resistance are still poorly understood. Similarly, when malaria parasites differentiate into sexual stage gametocytes they become increasingly resistant to many drugs effective against the other blood stages. A further area of interest would be the use of metabolomics to explore the host response to parasite infection both in terms of how parasites manage to siphon off energy and metabolites and how the host can modify its metabolism to combat the infection, as in the case of tryptophan restriction in mice to control growth of Toxoplasma (Daubener et al., 2001). With the recent advances in methodologies for measuring complex mixtures of cellular biomolecules, metabolomic studies of parasites hold great promise for the improved understanding and control of protozoan parasites.
Acknowledgements
We thank Bryson D. Bennett and Kellen L. Olszewski for valuable insight and comments on the manuscript. This work was funded by the Burroughs Wellcome Fund and an NIH Director's New Innovators award (1DP2OD001315-01) with support from the Center for Quantitative Biology (P50 GM071508). BFCK is a Howard Hughes Medical Institute Fellow of the Damon Runyon Cancer Research Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Note: We apologize to the numerous authors whose work we were unable to cite due to space limitations.
Reference List
- Adosraku RK, Anderson MM, Anderson GJ, Choi G, Croft SL, Yardley V, Phillipson JD, Gibbons WA. Proton NMR lipid profile of Leishmania donovani promastigotes. Mol. Biochem. Parasitol. 1993;62:251–262. doi: 10.1016/0166-6851(93)90114-d. [DOI] [PubMed] [Google Scholar]
- Aguero F, Al-Lazikani B, Aslett M, Berriman M, Buckner FS, Campbell RK, Carmona S, Carruthers IM, Chan AW, Chen F, et al. Genomic-scale prioritization of drug targets: the TDR Targets database. Nat. Rev. Drug Discov. 2008;7:900–907. doi: 10.1038/nrd2684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aly AS, Vaughan AM, Kappe SH. Malaria parasite development in the mosquito and infection of the mammalian host. Annu. Rev. Microbiol. 2009;63:195–221. doi: 10.1146/annurev.micro.091208.073403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angulo S, Garcia-Perez I, Legido-Quigley C, Barbas C. The autocorrelation matrix probing biochemical relationships after metabolic fingerprinting with CE. Electrophoresis. 2009;30:1221–1227. doi: 10.1002/elps.200800554. [DOI] [PubMed] [Google Scholar]
- Ariyanayagam MR, Oza SL, Mehlert A, Fairlamb AH. Bis(glutathionyl)spermine and other novel trypanothione analogues in Trypanosoma cruzi. J. Biol. Chem. 2003;278:27612–27619. doi: 10.1074/jbc.M302750200. [DOI] [PubMed] [Google Scholar]
- Bakker-Grunwald T, Martin JB, Klein G. Characterization of glycogen and amino acid pool of Entamoeba histolytica by 13C-NMR spectroscopy. J. Eukaryot. Microbiol. 1995;42:346–349. doi: 10.1111/j.1550-7408.1995.tb01592.x. [DOI] [PubMed] [Google Scholar]
- Blank LM, Kuepfer L, Sauer U. Large-scale 13C-flux analysis reveals mechanistic principles of metabolic network robustness to null mutations in yeast. Genome Biol. 2005;6:R49. doi: 10.1186/gb-2005-6-6-r49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blume M, Rodriguez-Contreras D, Landfear S, Fleige T, Soldati-Favre D, Lucius R, Gupta N. Host-derived glucose and its transporter in the obligate intracellular pathogen Toxoplasma gondii are dispensable by glutaminolysis. Proc. Natl. Acad. Sci. U. S. A. 2009;106:12998–13003. doi: 10.1073/pnas.0903831106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollard ME, Stanley EG, Lindon JC, Nicholson JK, Holmes E. NMR-based metabonomic approaches for evaluating physiological influences on biofluid composition. NMR Biomed. 2005;18:143–162. doi: 10.1002/nbm.935. [DOI] [PubMed] [Google Scholar]
- Brauer MJ, Saldanha AJ, Dolinski K, Botstein D. Homeostatic adjustment and metabolic remodeling in glucose-limited yeast cultures. Mol. Biol. Cell. 2005;16:2503–2517. doi: 10.1091/mbc.E04-11-0968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brauer MJ, Yuan J, Bennett BD, Lu W, Kimball E, Botstein D, Rabinowitz JD. Conservation of the metabolomic response to starvation across two divergent microbes. Proc. Natl. Acad. Sci. U. S. A. 2006;103:19302–19307. doi: 10.1073/pnas.0609508103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitling R, Vitkup D, Barrett MP. New surveyor tools for charting microbial metabolic maps. Nat. Rev. Microbiol. 2008;6:156–161. doi: 10.1038/nrmicro1797. [DOI] [PubMed] [Google Scholar]
- Carlton JM, Adams JH, Silva JC, Bidwell SL, Lorenzi H, Caler E, Crabtree J, Angiuoli SV, Merino EF, Amedeo P, et al. Comparative genomics of the neglected human malaria parasite Plasmodium vivax. Nature. 2008;455:757–763. doi: 10.1038/nature07327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavali AK, Whittemore JD, Eddy JA, Williams KT, Papin JA. Systems analysis of metabolism in the pathogenic trypanosomatid Leishmania major. Mol. Syst. Biol. 2008;4:177. doi: 10.1038/msb.2008.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Vitkup D. Predicting genes for orphan metabolic activities using phylogenetic profiles. Genome Biol. 2006;7:R17. doi: 10.1186/gb-2006-7-2-r17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho K, Shibato J, Agrawal GK, Jung YH, Kubo A, Jwa NS, Tamogami S, Satoh K, Kikuchi S, Higashi T, et al. Integrated transcriptomics, proteomics, and metabolomics analyses to survey ozone responses in the leaves of rice seedling. J. Proteome Res. 2008;7:2980–2998. doi: 10.1021/pr800128q. [DOI] [PubMed] [Google Scholar]
- Cornish-Bowden A, Cardenas ML. Metabolic analysis in drug design. C. R. Biol. 2003;326:509–515. doi: 10.1016/s1631-0691(03)00117-3. [DOI] [PubMed] [Google Scholar]
- Coustou V, Biran M, Breton M, Guegan F, Riviere L, Plazolles N, Nolan D, Barrett MP, Franconi JM, Bringaud F. Glucose-induced remodeling of intermediary and energy metabolism in procyclic Trypanosoma brucei. J. Biol. Chem. 2008;283:16342–16354. doi: 10.1074/jbc.M709592200. [DOI] [PubMed] [Google Scholar]
- Daubener W, Spors B, Hucke C, Adam R, Stins M, Kim KS, Schroten H. Restriction of Toxoplasma gondii growth in human brain microvascular endothelial cells by activation of indoleamine 2,3-dioxygenase. Infect. Immun. 2001;69:6527–6531. doi: 10.1128/IAI.69.10.6527-6531.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Souza DP, Saunders EC, McConville MJ, Likic VA. Progressive peak clustering in GC-MS Metabolomic experiments applied to Leishmania parasites. Bioinformatics. 2006;22:1391–1396. doi: 10.1093/bioinformatics/btl085. [DOI] [PubMed] [Google Scholar]
- Eastman RT, Fidock DA. Artemisinin-based combination therapies: a vital tool in efforts to eliminate malaria. Nat. Rev. Microbiol. 2009 doi: 10.1038/nrmicro2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatumo S, Plaimas K, Mallm JP, Schramm G, Adebiyi E, Oswald M, Eils R, Konig R. Estimating novel potential drug targets of Plasmodium falciparum by analysing the metabolic network of knock-out strains in silico. Infect. Genet. Evol. 2009;9:351–358. doi: 10.1016/j.meegid.2008.01.007. [DOI] [PubMed] [Google Scholar]
- Ferdig MT, Cooper RA, Mu J, Deng B, Joy DA, Su XZ, Wellems TE. Dissecting the loci of low-level quinine resistance in malaria parasites. Mol. Microbiol. 2004;52:985–997. doi: 10.1111/j.1365-2958.2004.04035.x. [DOI] [PubMed] [Google Scholar]
- Fidock DA, Nomura T, Talley AK, Cooper RA, Dzekunov SM, Ferdig MT, Ursos LM, Sidhu AB, Naude B, Deitsch KW, et al. Mutations in the P. falciparum digestive vacuole transmembrane protein PfCRT and evidence for their role in chloroquine resistance. Mol. Cell. 2000;6:861–871. doi: 10.1016/s1097-2765(05)00077-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasch AP, Spellman PT, Kao CM, Carmel-Harel O, Eisen MB, Storz G, Botstein D, Brown PO. Genomic expression programs in the response of yeast cells to environmental changes. Mol Biol Cell. 2000;11:4241–57. doi: 10.1091/mbc.11.12.4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginsburg H. Caveat emptor: limitations of the automated reconstruction of metabolic pathways in Plasmodium. Trends Parasitol. 2009;25:37–43. doi: 10.1016/j.pt.2008.08.012. [DOI] [PubMed] [Google Scholar]
- Ginsburg H, Stein WD. How many functional transport pathways does Plasmodium falciparum induce in the membrane of its host erythrocyte? Trends Parasitol. 2005;21:118–121. doi: 10.1016/j.pt.2005.01.004. [DOI] [PubMed] [Google Scholar]
- Guerin PJ, Bates SJ, Sibley CH. Global resistance surveillance: ensuring antimalarial efficacy in the future. Curr. Opin. Infect. Dis. 2009;22:593–600. doi: 10.1097/QCO.0b013e328332c4a7. [DOI] [PubMed] [Google Scholar]
- Gupta N, Goyal N, Singha UK, Bhakuni V, Roy R, Rastogi AK. Characterization of intracellular metabolites of axenic amastigotes of Leishmania donovani by 1H NMR spectroscopy. Acta Trop. 1999;73:121–133. doi: 10.1016/s0001-706x(99)00020-0. [DOI] [PubMed] [Google Scholar]
- Hackstein JH, Tjaden J, Huynen M. Mitochondria, hydrogenosomes and mitosomes: products of evolutionary tinkering! Curr. Genet. 2006;50:225–245. doi: 10.1007/s00294-006-0088-8. [DOI] [PubMed] [Google Scholar]
- Hayton K, Gaur D, Liu A, Takahashi J, Henschen B, Singh S, Lambert L, Furuya T, Bouttenot R, Doll M, et al. Erythrocyte binding protein PfRH5 polymorphisms determine species-specific pathways of Plasmodium falciparum invasion. Cell. Host Microbe. 2008;4:40–51. doi: 10.1016/j.chom.2008.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes E, Wilson ID, Nicholson JK. Metabolic phenotyping in health and disease. Cell. 2008;134:714–717. doi: 10.1016/j.cell.2008.08.026. [DOI] [PubMed] [Google Scholar]
- Huthmacher C, Gille C, Holzhutter HG. A computational analysis of protein interactions in metabolic networks reveals novel enzyme pairs potentially involved in metabolic channeling. J. Theor. Biol. 2008;252:456–464. doi: 10.1016/j.jtbi.2007.09.042. [DOI] [PubMed] [Google Scholar]
- Jacob F, Monod J. Genetic regulatory mechanisms in the synthesis of proteins. J. Mol. Biol. 1961;3:318–356. doi: 10.1016/s0022-2836(61)80072-7. [DOI] [PubMed] [Google Scholar]
- Kalisiak J, Trauger SA, Kalisiak E, Morita H, Fokin VV, Adams MW, Sharpless KB, Siuzdak G. Identification of a new endogenous metabolite and the characterization of its protein interactions through an immobilization approach. J. Am. Chem. Soc. 2009;131:378–386. doi: 10.1021/ja808172n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamleh A, Barrett MP, Wildridge D, Burchmore RJ, Scheltema RA, Watson DG. Metabolomic profiling using Orbitrap Fourier transform mass spectrometry with hydrophilic interaction chromatography: a method with wide applicability to analysis of biomolecules. Rapid Commun. Mass Spectrom. 2008;22:1912–1918. doi: 10.1002/rcm.3564. [DOI] [PubMed] [Google Scholar]
- Keurentjes JJ, Fu J, de Vos CH, Lommen A, Hall RD, Bino RJ, van der Plas LH, Jansen RC, Vreugdenhil D, Koornneef M. The genetics of plant metabolism. Nat Genet. 2006;38:842–9. doi: 10.1038/ng1815. [DOI] [PubMed] [Google Scholar]
- Khoo SH, Al-Rubeai M. Metabolomics as a complementary tool in cell culture. Biotechnol. Appl. Biochem. 2007;47:71–84. doi: 10.1042/BA20060221. [DOI] [PubMed] [Google Scholar]
- Koulman A, Lane GA, Harrison SJ, Volmer DA. From differentiating metabolites to biomarkers. Anal. Bioanal Chem. 2009;394:663–670. doi: 10.1007/s00216-009-2690-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon YK, Lu W, Melamud E, Khanam N, Bognar A, Rabinowitz JD. A domino effect in antifolate drug action in Escherichia coli. Nat. Chem. Biol. 2008;4:602–608. doi: 10.1038/nchembio.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenz EM, Wilson ID. Analytical strategies in metabonomics. J. Proteome Res. 2007;6:443–458. doi: 10.1021/pr0605217. [DOI] [PubMed] [Google Scholar]
- LeRoux M, Lakshmanan V, Daily JP. Plasmodium falciparum biology: analysis of in vitro versus in vivo growth conditions. Trends Parasitol. 2009;25:474–481. doi: 10.1016/j.pt.2009.07.005. [DOI] [PubMed] [Google Scholar]
- Li JV, Holmes E, Saric J, Keiser J, Dirnhofer S, Utzinger J, Wang Y. Metabolic profiling of a Schistosoma mansoni infection in mouse tissues using magic angle spinning-nuclear magnetic resonance spectroscopy. Int. J. Parasitol. 2009;39:547–558. doi: 10.1016/j.ijpara.2008.10.010. [DOI] [PubMed] [Google Scholar]
- Li JV, Wang Y, Saric J, Nicholson JK, Dirnhofer S, Singer BH, Tanner M, Wittlin S, Holmes E, Utzinger J. Global metabolic responses of NMRI mice to an experimental Plasmodium berghei infection. J. Proteome Res. 2008;7:3948–3956. doi: 10.1021/pr800209d. [DOI] [PubMed] [Google Scholar]
- Lian LY, Al-Helal M, Roslaini AM, Fisher N, Bray PG, Ward SA, Biagini GA. Glycerol: an unexpected major metabolite of energy metabolism by the human malaria parasite. Malar J. 2009;8:38. doi: 10.1186/1475-2875-8-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisec J, Meyer RC, Steinfath M, Redestig H, Becher M, Witucka-Wall H, Fiehn O, Torjek O, Selbig J, Altmann T, Willmitzer L. Identification of metabolic and biomass QTL in Arabidopsis thaliana in a parallel analysis of RIL and IL populations. Plant J. 2008;53:960–972. doi: 10.1111/j.1365-313X.2007.03383.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H, Zeng AP. Reconstruction of metabolic networks from genome data and analysis of their global structure for various organisms. Bioinformatics. 2003;19:270–277. doi: 10.1093/bioinformatics/19.2.270. [DOI] [PubMed] [Google Scholar]
- Martin RE, Henry RI, Abbey JL, Clements JD, Kirk K. The ‘permeome’ of the malaria parasite: an overview of the membrane transport proteins of Plasmodium falciparum. Genome Biol. 2005;6:R26. doi: 10.1186/gb-2005-6-3-r26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May P, Wienkoop S, Kempa S, Usadel B, Christian N, Rupprecht J, Weiss J, Recuenco-Munoz L, Ebenhoh O, Weckwerth W, Walther D. Metabolomics- and proteomics-assisted genome annotation and analysis of the draft metabolic network of Chlamydomonas reinhardtii. Genetics. 2008;179:157–166. doi: 10.1534/genetics.108.088336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazumdar J, Wilson EH, Masek K, Hunter CA, Striepen B. Apicoplast fatty acid synthesis is essential for organelle biogenesis and parasite survival in Toxoplasma gondii. Proc. Natl. Acad. Sci. U. S. A. 2006;103:13192–13197. doi: 10.1073/pnas.0603391103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta M, Sonawat HM, Sharma S. Glycolysis in Plasmodium falciparum results in modulation of host enzyme activities. J Vector Borne Dis. 2006;43:95–103. [PubMed] [Google Scholar]
- Mendoza M, Mijares A, Rojas H, Rodriguez JP, Urbina JA, DiPolo R. Physiological and morphological evidences for the presence acidocalcisomes in Trypanosoma evansi: single cell fluorescence and 31P NMR studies. Mol. Biochem. Parasitol. 2002;125:23–33. doi: 10.1016/s0166-6851(02)00166-4. [DOI] [PubMed] [Google Scholar]
- Mogi T, Kita K. Diversity in mitochondrial metabolic pathways in parasitic protists Plasmodium and Cryptosporidium. Parasitol. Int. 2009 doi: 10.1016/j.parint.2010.04.005. [DOI] [PubMed] [Google Scholar]
- Mohanty S, Srinivasan N. Identification of “missing” metabolic proteins of Plasmodium falciparum: a bioinformatics approach. Protein Pept. Lett. 2009;16:961–968. doi: 10.2174/092986609788923257. [DOI] [PubMed] [Google Scholar]
- Moreno B, Bailey BN, Luo S, Martin MB, Kuhlenschmidt M, Moreno SN, Docampo R, Oldfield E. (31)P NMR of apicomplexans and the effects of risedronate on Cryptosporidium parvum growth. Biochem. Biophys. Res. Commun. 2001;284:632–637. doi: 10.1006/bbrc.2001.5009. [DOI] [PubMed] [Google Scholar]
- Moreno-Sanchez R, Encalada R, Marin-Hernandez A, Saavedra E. Experimental validation of metabolic pathway modeling. FEBS J. 2008;275:3454–3469. doi: 10.1111/j.1742-4658.2008.06492.x. [DOI] [PubMed] [Google Scholar]
- Mu J, Ferdig MT, Feng X, Joy DA, Duan J, Furuya T, Subramanian G, Aravind L, Cooper RA, Wootton JC, Xiong M, Su XZ. Multiple transporters associated with malaria parasite responses to chloroquine and quinine. Mol Microbiol. 2003;49:977–89. doi: 10.1046/j.1365-2958.2003.03627.x. [DOI] [PubMed] [Google Scholar]
- Munger M, Bajad SU, Coller HA, Shenk T, Rabinowitz JD. Dynamics of the Cellular Metabolome during Human Cytomegalovirus Infection. PLoS Pathogens. 2006;2:E132. doi: 10.1371/journal.ppat.0020132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naderer T, Ellis MA, Sernee MF, De Souza DP, Curtis J, Handman E, McConville MJ. Virulence of Leishmania major in macrophages and mice requires the gluconeogenic enzyme fructose-1,6-bisphosphatase. Proc. Natl. Acad. Sci. U. S. A. 2006;103:5502–5507. doi: 10.1073/pnas.0509196103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair S, Miller B, Barends M, Jaidee A, Patel J, Mayxay M, Newton P, Nosten F, Ferdig MT, Anderson TJ. Adaptive copy number evolution in malaria parasites. PLoS Genet. 2008;4:e1000243. doi: 10.1371/journal.pgen.1000243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishina M, Suzuki M, Matsushita K. Trichinella spiralis: activity of the cerebral pyruvate recycling pathway of the host (mouse) in hypoglycemia induced by the infection. Exp. Parasitol. 2004;106:62–65. doi: 10.1016/j.exppara.2004.01.007. [DOI] [PubMed] [Google Scholar]
- Nordstrom A, Lewensohn R. Metabolomics: Moving to the Clinic. J. Neuroimmune Pharmacol. 2009 doi: 10.1007/s11481-009-9156-4. [DOI] [PubMed] [Google Scholar]
- Olszewski KL, Morrisey JM, Wilinski D, Burns JM, Vaidya AB, Rabinowitz JD, Llinas M. Host-parasite interactions revealed by Plasmodium falciparum metabolomics. Cell. Host Microbe. 2009;5:191–199. doi: 10.1016/j.chom.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen KS, Kristensen TN, Loeschcke V, Petersen BO, Duus JO, Nielsen NC, Malmendal A. Metabolomic signatures of inbreeding at benign and stressful temperatures in Drosophila melanogaster. Genetics. 2008;180:1233–1243. doi: 10.1534/genetics.108.089144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penet MF, Kober F, Confort-Gouny S, Le Fur Y, Dalmasso C, Coltel N, Liprandi A, Gulian JM, Grau GE, Cozzone PJ, Viola A. Magnetic resonance spectroscopy reveals an impaired brain metabolic profile in mice resistant to cerebral malaria infected with Plasmodium berghei ANKA. J. Biol. Chem. 2007;282:14505–14514. doi: 10.1074/jbc.M608035200. [DOI] [PubMed] [Google Scholar]
- Penin P, Sanchez-Moreno M, de Diego JA. Proton nuclear magnetic resonance analysis of metabolic end products of the Bolivia strain of Trypanosoma cruzi and three of its clones. Comp. Biochem. Physiol. A. Mol. Integr. Physiol. 1998;120:571–574. doi: 10.1016/s1095-6433(98)10076-4. [DOI] [PubMed] [Google Scholar]
- Pinney JW, Papp B, Hyland C, Wambua L, Westhead DR, McConkey GA. Metabolic reconstruction and analysis for parasite genomes. Trends Parasitol. 2007;23:548–554. doi: 10.1016/j.pt.2007.08.013. [DOI] [PubMed] [Google Scholar]
- Pinney JW, Shirley MW, McConkey GA, Westhead DR. metaSHARK: software for automated metabolic network prediction from DNA sequence and its application to the genomes of Plasmodium falciparum and Eimeria tenella. Nucleic Acids Res. 2005;33:1399–1409. doi: 10.1093/nar/gki285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabinowitz JD. Cellular metabolomics of Escherchia coli. Expert Rev. Proteomics. 2007;4:187–198. doi: 10.1586/14789450.4.2.187. [DOI] [PubMed] [Google Scholar]
- Rae C, McQuillan JA, Parekh SB, Bubb WA, Weiser S, Balcar VJ, Hansen AM, Ball HJ, Hunt NH. Brain gene expression, metabolism, and bioenergetics: interrelationships in murine models of cerebral and noncerebral malaria. FASEB J. 2004;18:499–510. doi: 10.1096/fj.03-0543com. [DOI] [PubMed] [Google Scholar]
- Roberts SB, Robichaux JL, Chavali AK, Manque PA, Lee V, Lara AM, Papin JA, Buck GA. Proteomic and network analysis characterize stage-specific metabolism in Trypanosoma cruzi. BMC Syst. Biol. 2009;3:52. doi: 10.1186/1752-0509-3-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeij JP, Boyle JP, Coller S, Taylor S, Sibley LD, Brooke-Powell ET, Ajioka JW, Boothroyd JC. Polymorphic secreted kinases are key virulence factors in toxoplasmosis. Science (New York, N.Y.) 2006;314:1780–1783. doi: 10.1126/science.1133690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saric J, Li JV, Wang Y, Keiser J, Veselkov K, Dirnhofer S, Yap IK, Nicholson JK, Holmes E, Utzinger J. Panorganismal metabolic response modeling of an experimental Echinostoma caproni infection in the mouse. J. Proteome Res. 2009;8:3899–3911. doi: 10.1021/pr900185s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer U. Metabolic networks in motion: 13C-based flux analysis. Mol. Syst. Biol. 2006;2:62. doi: 10.1038/msb4100109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seckin H, Yagmurlu B, Yigitkanli K, Kars HZ. Metabolic changes during successful medical therapy for brain hydatid cyst: case report. Surg. Neurol. 2008;70:186–189. doi: 10.1016/j.surneu.2007.05.047. [DOI] [PubMed] [Google Scholar]
- Seeber F, Limenitakis J, Soldati-Favre D. Apicomplexan mitochondrial metabolism: a story of gains, losses and retentions. Trends Parasitol. 2008;24:468–478. doi: 10.1016/j.pt.2008.07.004. [DOI] [PubMed] [Google Scholar]
- Sherman I. Malaria: Parasite Biology, Pathogenesis, and Protection. American Society for Microbiology; Washington, DC: 1998. Carbohydrate Metabolism of Asexual Stages. pp. 123–143. [Google Scholar]
- Sibley LD, Boothroyd JC. Construction of a molecular karyotype for Toxoplasma gondii. Mol. Biochem. Parasitol. 1992;51:291–300. doi: 10.1016/0166-6851(92)90079-y. [DOI] [PubMed] [Google Scholar]
- Su C, Howe DK, Dubey JP, Ajioka JW, Sibley LD. Identification of quantitative trait loci controlling acute virulence in Toxoplasma gondii. Proc. Natl. Acad. Sci. U. S. A. 2002;99:10753–10758. doi: 10.1073/pnas.172117099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun G, Yang K, Zhao Z, Guan S, Han X, Gross RW. Shotgun metabolomics approach for the analysis of negatively charged water-soluble cellular metabolites from mouse heart tissue. Anal. Chem. 2007;79:6629–6640. doi: 10.1021/ac070843+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor S, Barragan A, Su C, Fux B, Fentress SJ, Tang K, Beatty WL, Hajj HE, Jerome M, Behnke MS, et al. A secreted serine-threonine kinase determines virulence in the eukaryotic pathogen Toxoplasma gondii. Science (New York, N.Y.) 2006;314:1776–1780. doi: 10.1126/science.1133643. [DOI] [PubMed] [Google Scholar]
- Teng R, Junankar PR, Bubb WA, Rae C, Mercier P, Kirk K. Metabolite profiling of the intraerythrocytic malaria parasite Plasmodium falciparum by (1)H NMR spectroscopy. NMR Biomed. 2009;22:292–302. doi: 10.1002/nbm.1323. [DOI] [PubMed] [Google Scholar]
- Thompson SN. Applications of nuclear magnetic resonance in parasitology. J. Parasitol. 1991;77:1–20. [PubMed] [Google Scholar]
- Trauger SA, Kalisak E, Kalisiak J, Morita H, Weinberg MV, Menon AL, Poole FL, 2nd, Adams MW, Siuzdak G. Correlating the transcriptome, proteome, and metabolome in the environmental adaptation of a hyperthermophile. J. Proteome Res. 2008;7:1027–1035. doi: 10.1021/pr700609j. [DOI] [PubMed] [Google Scholar]
- van Brummelen AC, Olszewski KL, Wilinski D, Llinas M, Louw AI, Birkholtz LM. Co-inhibition of Plasmodium falciparum S-adenosylmethionine decarboxylase/ornithine decarboxylase reveals perturbation-specific compensatory mechanisms by transcriptome, proteome, and metabolome analyses. J. Biol. Chem. 2009;284:4635–4646. doi: 10.1074/jbc.M807085200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Weelden SW, Fast B, Vogt A, van der Meer P, Saas J, van Hellemond JJ, Tielens AG, Boshart M. Procyclic Trypanosoma brucei do not use Krebs cycle activity for energy generation. J. Biol. Chem. 2003;278:12854–12863. doi: 10.1074/jbc.M213190200. [DOI] [PubMed] [Google Scholar]
- Vaughan AM, O'Neill MT, Tarun AS, Camargo N, Phuong TM, Aly AS, Cowman AF, Kappe SH. Type II fatty acid synthesis is essential only for malaria parasite late liver stage development. Cell. Microbiol. 2009;11:506–520. doi: 10.1111/j.1462-5822.2008.01270.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walliker D, Quakyi IA, Wellems TE, McCutchan TF, Szarfman A, London WT, Corcoran LM, Burkot TR, Carter R. Genetic analysis of the human malaria parasite Plasmodium falciparum. Science. 1987;236:1661–6. doi: 10.1126/science.3299700. [DOI] [PubMed] [Google Scholar]
- Wang Y, Holmes E, Nicholson JK, Cloarec O, Chollet J, Tanner M, Singer BH, Utzinger J. Metabonomic investigations in mice infected with Schistosoma mansoni: an approach for biomarker identification. Proc. Natl. Acad. Sci. U. S. A. 2004;101:12676–12681. doi: 10.1073/pnas.0404878101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Utzinger J, Saric J, Li JV, Burckhardt J, Dirnhofer S, Nicholson JK, Singer BH, Brun R, Holmes E. Global metabolic responses of mice to Trypanosoma brucei brucei infection. Proc. Natl. Acad. Sci. U. S. A. 2008;105:6127–6132. doi: 10.1073/pnas.0801777105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Morris JC, Drew ME, Englund PT. Inhibition of Trypanosoma brucei gene expression by RNA interference using an integratable vector with opposing T7 promoters. J. Biol. Chem. 2000;275:40174–40179. doi: 10.1074/jbc.M008405200. [DOI] [PubMed] [Google Scholar]
- Want EJ, Nordstrom A, Morita H, Siuzdak G. From exogenous to endogenous: the inevitable imprint of mass spectrometry in metabolomics. J. Proteome Res. 2007;6:459–468. doi: 10.1021/pr060505+. [DOI] [PubMed] [Google Scholar]
- Wellems TE, Panton LJ, Gluzman IY, do Rosario VE, Gwadz RW, Walker-Jonah A, Krogstad DJ. Chloroquine resistance not linked to mdr-like genes in a Plasmodium falciparum cross. Nature. 1990;345:253–5. doi: 10.1038/345253a0. [DOI] [PubMed] [Google Scholar]
- Wikoff WR, Anfora AT, Liu J, Schultz PG, Lesley SA, Peters EC, Siuzdak G. Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites. Proc. Natl. Acad. Sci. U. S. A. 2009a;106:3698–3703. doi: 10.1073/pnas.0812874106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wikoff WR, Kalisak E, Trauger S, Manchester M, Siuzdak G. Response and recovery in the plasma metabolome tracks the acute LCMV-induced immune response. J. Proteome Res. 2009b;8:3578–3587. doi: 10.1021/pr900275p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wikoff WR, Pendyala G, Siuzdak G, Fox HS. Metabolomic analysis of the cerebrospinal fluid reveals changes in phospholipase expression in the CNS of SIV-infected macaques. J. Clin. Invest. 2008;118:2661–2669. doi: 10.1172/JCI34138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Zhao X, Liu X, Wang C, Gao P, Wang J, Li L, Gu J, Yang S, Xu G. High performance liquid chromatography-mass spectrometry for metabonomics: potential biomarkers for acute deterioration of liver function in chronic hepatitis B. J. Proteome Res. 2006;5:554–561. doi: 10.1021/pr050364w. [DOI] [PubMed] [Google Scholar]
- Yang S, Tschaplinski TJ, Engle NL, Carroll SL, Martin SL, Davison BH, Palumbo AV, Rodriguez M, Jr, Brown SD. Transcriptomic and metabolomic profiling of Zymomonas mobilis during aerobic and anaerobic fermentations. BMC Genomics. 2009;10:34. doi: 10.1186/1471-2164-10-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap IK, Li JV, Saric J, Martin FP, Davies H, Wang Y, Wilson ID, Nicholson JK, Utzinger J, Marchesi JR, Holmes E. Metabonomic and microbiological analysis of the dynamic effect of vancomycin-induced gut microbiota modification in the mouse. J. Proteome Res. 2008;7:3718–3728. doi: 10.1021/pr700864x. [DOI] [PubMed] [Google Scholar]
- Yeh I, Hanekamp T, Tsoka S, Karp PD, Altman RB. Computational analysis of Plasmodium falciparum metabolism: organizing genomic information to facilitate drug discovery. Genome Res. 2004;14:917–24. doi: 10.1101/gr.2050304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu M, Kumar TR, Nkrumah LJ, Coppi A, Retzlaff S, Li CD, Kelly BJ, Moura PA, Lakshmanan V, Freundlich JS, et al. The fatty acid biosynthesis enzyme FabI plays a key role in the development of liver-stage malarial parasites. Cell. Host Microbe. 2008;4:567–578. doi: 10.1016/j.chom.2008.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J, Johnson RL, Huang R, Wichterman J, Jiang H, Hayton K, Fidock DA, Wellems TE, Inglese J, Austin CP, Su XZ. Genetic mapping of targets mediating differential chemical phenotypes in Plasmodium falciparum. Nat. Chem. Biol. 2009;5:765–771. doi: 10.1038/nchembio.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamboni N, Fendt SM, Ruhl M, Sauer U. (13)C-based metabolic flux analysis. Nat. Protoc. 2009;4:878–892. doi: 10.1038/nprot.2009.58. [DOI] [PubMed] [Google Scholar]
- Zamboni N, Sauer U. Novel biological insights through metabolomics and 13C-flux analysis. Curr. Opin. Microbiol. 2009;12:553–558. doi: 10.1016/j.mib.2009.08.003. [DOI] [PubMed] [Google Scholar]