Abstract
Targeted protein degradation is largely performed by the ubiquitin-proteasome pathway, in which substrate proteins are marked by covalently attached ubiquitin chains that mediate recognition by the proteasome. It is currently unclear how the proteasome recognizes its substrates, as the only established ubiquitin receptor intrinsic to the proteasome is Rpn10/S5a1, which is not essential for ubiquitin-mediated protein degradation in budding yeast2. In the accompanying manuscript we report that Rpn133–7, a component of the 9-subunit proteasome base, functions as a ubiquitin receptor8, complementing its known role in docking deubiquitinating enzyme Uch37/UCHL54–6 to the proteasome. Here, we merge crystallography and NMR data to describe Rpn13’s ubiquitin binding mechanism. We determined the structure of Rpn13 alone and complexed with ubiquitin. The co-complex reveals a novel ubiquitin binding mode in which loops rather than secondary structural elements are used to capture ubiquitin. Further support for Rpn13’s role as a proteasomal ubiquitin receptor is demonstrated by its ability to bind ubiquitin and proteasome subunit Rpn2/S1 simultaneously. Finally, we provide a model structure of Rpn13 complexed to diubiquitin, which provides insights into how Rpn13’s role as a ubiquitin receptor is coupled to substrate deubiquitination by Uch37.
The structure of murine Rpn13 (mRpn13) (1–150) was determined at 1.7 Å resolution by X-ray crystallography, and found to contain a Pleckstrin Homology (PH) domain fold (Figure 1a and 1b) (Structure determination and refinement statistics are provided in the Supplement). In particular, whereas the first 21 N- and last 20 C-terminal amino acids are unstructured, residues 22–130 form a PH domain fold. This result was surprising, as primary sequence alignment did not identify Rpn13 to be homologous to previously characterized proteins. This finding coupled with its ubiquitin receptor properties8 prompted us to name the N-terminal domain of Rpn13 Pleckstrin-like receptor for ubiquitin (Pru).
Figure 1. Crystal structure of mRpn13 Pru reveals typical Pleckstrin homology (PH) fold.
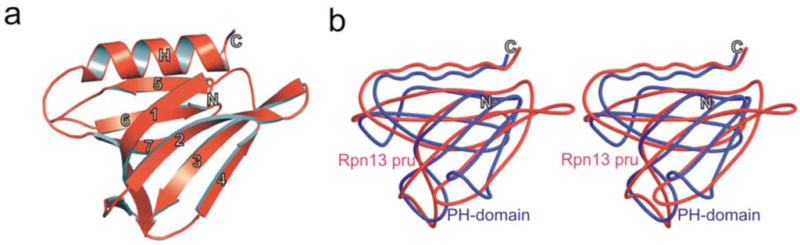
a, Ribbon representation of Rpn13 Pleckstrin-like receptor for ubiquitin (Rpn13 Pru). The PH fold consisting of a seven-stranded β-sandwich structure (1–7) capped by the C-terminal α-helix. b, Stereo representation of the structural alignment of Rpn13 Pru (red) and the PH domain (blue) from Pleckstrin (PDB accession code 1PLS).
Though very divergent at their sequence level, all PH domains have a common β-sandwich fold. The PH domain of Rpn13 is composed of a 4-stranded twisted antiparallel β-sheet (β1–4: residues 22–34, 45–52, 56–62, 71–74) that packs almost orthogonally against a second triple stranded β-sheet (β5–7: residues 80–85, 92–98, 103–110) (Supplementary Figure 1). Similar to other PH domains, Rpn13 Pru forms a hydrophobic core containing conserved hydrophobic residues (F26, V47, I49, F59, F82, Y94, L96, F107 and M109), which are located within β-sheets. One end of the β-sandwich is capped by a long C-terminal amphipathic α-helix (residues 117–128), which is stabilized by interactions between V124 and L128, whereas the other corner of the hydrophobic core is closed by three loops formed by residues located between strands S1/S2, S3/S4 and S6/S7 (Figure 1a and Supplementary Figure 1).
Despite much effort, we were unable to crystallize the Rpn13 Pru:ubiquitin complex; however, we were able to determine the structure of this complex by molecular docking, based on the crystal structure of mRpn13 Pru and intermolecular NOEs and chemical shift perturbation data derived by using NMR. The topology of the complexed structure was readily defined by twelve unambiguous intermolecular NOE interactions between human Rpn13 (hRpn13) Pru and ubiquitin (Figure 2a). We were able to use the hRpn13:ubiquitin NOEs in conjunction with the mRpn13 crystal structure because all of the amino acids exhibiting intermolecular NOEs are strictly conserved between murine and human Rpn13. Importantly, the mRpn13 Pru:ubiquitin structure reveals a novel ubiquitin binding mode in which residues of the S2–S3, S4–S5, and S6–S7 loops capture ubiquitin (Figure 2b). At the core of the contact surface, respective hydrogen bonds are formed between sidechain oxygens of D78 and D79 in hRpn13 and Nε2 and Nδ1 of H68 in ubiquitin. Moreover, F76 engages in hydrophobic interactions with I44, Q49, and V70 of ubiquitin (Supplementary Figure 2). These contacts are enabled by the strictly conserved P77, which causes the S4–S5 loop to turn. The intermolecular NOE data for this complex were fully satisfied without Rpn13 Pru or ubiquitin structural rearrangements, and the r.m.s. deviations between the free and complexed state of Rpn13 Pru and ubiquitin are 0.91 and 0.75 Å for backbone atoms and 1.20 and 1.15 Å for all non-hydrogen atoms, respectively.
Figure 2. Structure of Rpn13 Pru:ubiquitin complex defines a novel ubiquitin-binding motif.
a, Representative NOE interactions identified between Rpn13 Pru and ubiquitin. Each panel contains a selected region of an 15N dispersed NOESY experiment recorded on 15N, 13C and 70% 2H-labeled hRpn13 Pru mixed with equimolar quantities of unlabeled ubiquitin. All of the resonances displayed in this panel were unambiguously assigned as intermolecular NOE interactions with ubiquitin. Ubiquitin and Rpn13 Pru assignments are provided at the top and bottom of the expanded regions, respectively.
b, Stereo representation of the mRpn13 Pru:ubiquitin complex oriented with ubiquitin at the top. At the interaction surface, secondary structural elements of ubiquitin and Rpn13 Pru are displayed in green and blue, respectively. Residues at the contact surface with intermolecular NOEs are in yellow (ubiquitin) or red (Rpn13 Pru) whereas those suggested to be at the contact surface only by the NMR titration experiments are displayed in purple (ubiquitin) or cyan (Rpn13 Pru). M31, C88, and E111, which shift upon hRpn2 (797–953) addition are displayed in dark green.
c, Specific amino acid substitutions were made within the S2–S3, S4–S5, and S6–S7 loops of mRpn13 Pru by in vitro mutagenesis and the protein products expressed as GST-fusions and used in GST pull down assays to highlight the importance of these loops for tetraubiquitin binding.
d, Ubiquitin (blue), hRpn2 (797–953) (green), or ubiquitin and hRpn2 (797–953) (red) were added to hRpn13 Pru domain, which was monitored by 1H, 15N HSQC experiments. Comparison with the spectrum acquired on the protein alone (black) indicates that S55, F76 and D78 bind ubiquitin in a manner that is independent of hRpn2 (797–953), whereas M31, C88 and E111 bind hRpn2 in a ubiquitin -independent manner.
Additional hydrophobic contacts exist, as Rpn13’s sidechain methyl group of A100 and the Cα group of G101 partially bury ubiquitin’s F45, which is solvent-exposed in the free protein. Similarly, Rpn13’s strictly conserved F98 located on S6 also becomes less solvent accessible through interactions with A46 and G47 of ubiquitin. Calculation of the electrostatic potential of mRpn13’s ubiquitin binding surface indicates that a hydrophobic region containing L56 and F76 is available to interact with ubiquitin’s L8, I44 and V70 (Supplementary Figure 3). Complementary electrostatic interactions between Rpn13 Pru and ubiquitin also stabilize the complex, including interactions of D78 and D79 of Rpn13 Pru with ubiquitin’s H68, as well as D53 and D54 with ubiquitin’s R42 and R72.
In total, the contact surface of Rpn13 Pru and ubiquitin comprises 1256 Å2, which is large for ubiquitin receptors. Thus, the relatively high affinity of hRpn13 binding to monoubiquitin8 is explained by the enlarged contact surface as compared to that of EAP45 GLUE (1000 Å2 in total) 9. Published values for the total buried surface of Cue2-1cue and Dsk2UBA upon ubiquitin binding are even smaller: 960 and 800 Å2, respectively 10, 11.
To analyze the significance of specific interactions identified in the mRpn13:ubiquitin complex, we made several amino acid substitutions including L56A, I75R, F76R, D79N, and F98R. The ubiquitin binding competency of the resulting amino acid substituted proteins was tested by using GST-4xUb (created by the in frame expression of GST and four ubiquitin sequences) in pull down assays (Figure 2c). These experiments validate our mRpn13:ubiquitin structure and provide strong evidence for the importance of the S2–S3, S4–S5, and S6–S7 loops in ubiquitin binding by mRpn13. In particular, the single amino acid substitutions L56A, I75R, F76R, and F98R abrogate ubiquitin binding and a strong reduction is observed for the D79N mutation. We tested how three of these mutations affect mRpn13 structural integrity. In particular, NMR experiments were performed on mRpn13 Pru with the L56A, F76R or D79N mutations incorporated and compared to the wild-type mRpn13 Pru domain. These comparisons demonstrated that the loss of ubiquitin binding was not caused by loss of structural integrity (Supplementary Figure 4). Altogether, our results demonstrate that ubiquitin binding is defined by key interactions with residues within the S2–S3, S4–S5, and S6–S7 loops.
To function as a proteasomal ubiquitin receptor, Rpn13 must bind ubiquitin and proteasome components simultaneously. In both yeast and mammals, Rpn13 binds to Rpn2 via its Pru domain5, 12, 13. Although the Pru domain also binds ubiquitin, we found that Rpn2 binding does not disturb the Rpn13 loops that bind ubiquitin. By using a nested set of N-terminal deletions in human Rpn2 (hRpn2), we determined a fragment spanning 797–953 to bind mRpn13 (Supplementary Figure 5). The addition of this fragment (hRpn2 (797–953)) to hRpn13 did not affect residues at the ubiquitin contact surface, which shift only upon ubiquitin addition, as demonstrated for S55, F76, and D78 (Figure 2d). By contrast, M31, C88, and E111, which are unaffected by ubiquitin, shift after hRpn2 (797–953) addition. Furthermore, when both Rpn2 and ubiquitin were added, S55, F76, and D78 contact ubiquitin while M31, C88, and E111 contact Rpn2 (Figure 2d), indicating that the two binding surfaces are largely independent. M31, C88, and E111 are conserved in mRpn13 and map to S1, the S5–S6 loop, and the region linking S7 to H1, respectively. These elements are clustered in a region that is opposite to the ubiquitin binding loops of Rpn13.
26S proteasomes exhibit high affinity for ubiquitinated substrates. Ubiquitin chains linked via isopeptide bonds between K48 and the C-terminal glycine of neighboring ubiquitin molecules are known to trigger proteasomal degradation of the labeled protein14, 15. We found that Rpn13 Pru binds K48-linked tetraubiquitin in a manner comparable to that of monoubiquitin. More specifically, tetraubiquitin and monoubiquitin caused chemical shift changes to the same hRpn13 residues (Supplementary Figure 6a), including L56, F76, and F98, and shifted them almost identically (Figure 3a). Only two residues in hRpn13 exhibit changes that are specific to K48-linked tetraubiquitin, namely L73 and R104 (Figure 3a and Supplementary Figure 6a, red). These residues and the sidechain atoms of neighboring K103 are proximal to each other and in the mRpn13:monoubiquitin structure they are directed towards K48’s sidechain atoms. This arrangement is congruent with Rpn13 binding the proximal subunit of diubiquitin, namely that which forms an isopeptide using K48 (Figure 3b). We tested this model further by monitoring the sidechain atoms of the proximal or distal subunit of diubiquitin upon hRpn13 Pru addition. More specifically, unlabeled hRpn13 Pru was added to diubiquitin with either its proximal or distal subunit 13C labeled, and the effect recorded by 1H, 13C HMQC experiments. Significant resonance shifting characteristic of hRpn13 binding was observed for the I44 γ1, I44 δ1, and A46 methyl groups of the proximal subunit (Figure 3c, left panel, Supplementary Figure 7a). By contrast, resonances of the distal subunits exhibited only minor perturbations, most likely due to loss of intramolecular interactions with the proximal subunit. These data provide strong evidence that the major contacts formed between hRpn13 Pru and diubiquitin involve residues of the proximal subunit, at least when these two proteins are at equimolar concentration. Further evidence of this binding mode is provided by analytical ultracentrifugation data, which revealed 1:1 stoichiometry between hRpn13 Pru and diubiquitin (Supplementary Figure 7b).
Figure 3.
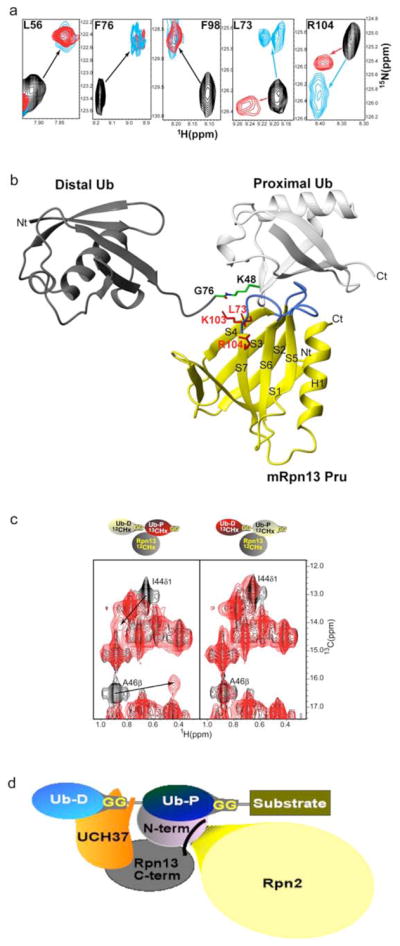
a, Comparison of spectra acquired with monoubiquitin versus K48-linked tetraubiquitin reveal identical affects for Rpn13’s L56, F76 and F98 but differences for L73 and R104. Monoubiquitin (red) or tetraubiquitin (cyan) was added to hRpn13, which was monitored by 1H, 15N HSQC experiments. The spectrum of free hRpn13 is indicated in black, and the molar ratio of monoubiquitin (red) or tetraubiquitin (cyan) to hRpn13 was 1:1 in the represented spectra.
b, Computer-generated model of the mRpn13 Pru:diubiquitin complex. White and grey ribbon diagrams display the proximal and distal ubiquitin, respectively, whereas a balls-and-sticks representation is used for the K48–G76 isopeptide bond linkage. Rpn13 Pru is colored in yellow and loops recognizing ubiquitin in blue, whereas L73, K103 and R104 are displayed in red. Diubiquitin was created by Insight II software based on atomic coordinates for the mRpn13 Pru:ubiquitin complex and monoubiquitin (PDB entry 1D3Z). In this model, the distal subunit of diubiquitin is positioned arbitrary, as its only constraints prohibit steric clashes with other atoms.
c, hRpn13 Pru interacts with the I44 δ1 and A46 methyl groups of the proximal subunit. 1H, 13C HMQC spectra were acquired on samples containing either no (black) or equimolar unlabeled hRpn13 (1–150) (red) mixed with diubiquitin in which its proximal (left) or distal (right) subunit is 13C labeled. The shifted resonances are labeled.
d, Model for how Rpn13 participates in Uch37 deconjugation of ubiquitinated substrates. Rpn13’s C-terminal domain (grey) binds Uch37 (orange) as its N-terminal domain binds the polyubiquitin chain and Rpn2/S1 (yellow). In this model, Uch37 binds to the distal subunit (light blue) of the chain while Rpn13 binds the proximal subunit (dark blue).
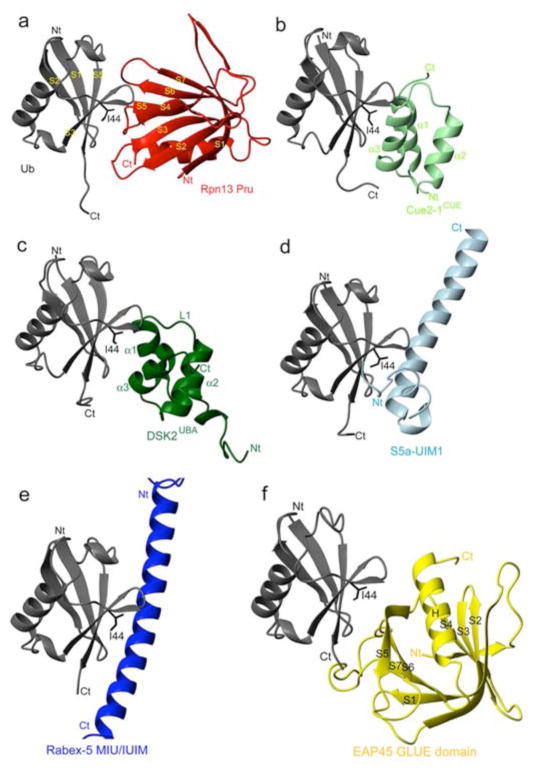
In conclusion, we reveal that the ubiquitin-binding region of proteasome subunit Rpn13 adopts a PH domain fold and solve its structure complexed with ubiquitin to unveil a new ubiquitin-binding mode. PH domains are present in a remarkably large number of proteins16, but Rpn13 Pru is the first example of a PH domain structure within the 26S proteasome. Rpn13, like many other ubiquitin receptors, binds to the L8, I44, and V70 hydrophobic pocket of ubiquitin (Figure 2b). However, it is the first to bind this region using exclusively loops (Figure 4a). Most of the ubiquitin receptors characterized to date use α-helices to bind this surface of ubiquitin, including the UBA, CUE, UIM, DUIM, MIU and GAT binding motifs. Among them, the UBA and CUE domains are structurally homologous with a common 3-helical bundle architecture. Cue2–1cue binds ubiquitin through the α1 and α3 helices (Figure 4b)10, whereas the Dsk2UBA uses the loop between α1 and α2, as well as the C-terminal part of α3 (Figure 4c)11. Structural characterization of the UIMs demonstrated that a single α-helix is sufficient for binding this region of ubiquitin17–19. The UIM helix includes a conserved alanine neighbored by a bulky hydrophobic residue, each of which packs against ubiquitin’s I44 as demonstrated in the S5a UIM1:ubiquitin complex (Figure 4d)19. Rabex-5 MIU/IUIM20, 21 and the pol η UBZ domain22 similarly bind this region in ubiquitin through a single α-helix, but in the reverse orientation (Figure 4e). The GLUE domain of ESCRT-II EAP45, which exhibits a split pleckstrin-homology topology, is the only previously known ubiquitin-binding PH domain23. However, it binds ubiquitin in a different manner: the I44-containing surface of ubiquitin is contacted by residues within secondary structural elements including the EAP45 C-terminal helix corresponding to H1 in Rpn1323 (Figure 4f). Moreover, although the longer S6–S7 loop of EAP45 is involved in binding ubiquitin, the S2–S3 and S4–S5 loops are not; instead, contacts are formed by residues from S5 and S6.
Figure 4. Structural comparison of ubiquitin receptors complexed with ubiquitin.
a–f, Complex structures of ubiquitin and specific receptors displayed with ubiquitin in the same orientation (grey) and ubiquitin’s I44 shown in black sticks. Each receptor has unique color coding: a, Rpn13 Pru (red); b, Cue2–1CUE (PDB code 1OTR; light green); c, Dsk2UBA (PDB code 1WR1; dark green); d, S5a-UIM1 (PDB code 1YX5; light blue); e, Rabex-5 MIU/IUIM (PDB code 2FIF; dark blue); f, EAP45 GLUE domain (PDB code 2HTH; yellow). All structures are compared by a best-fit superposition of bound ubiquitin (grey). In c, L1 denotes the loop connecting α1 and α2.
In addition to its unique monoubiquitin binding mode, we have demonstrated that Rpn13 has a novel preference for diubiquitin elements within K48-linked chains8 and that it most likely interacts directly with the isopeptide bond within a ubiquitin chain. This ubiquitin binding mode is consistent with Rpn13’s functional relationship with Uch37, which it adds to the RP’s collection of chain processing enzymes4–6. For diubiquitin, hRpn13 binding to the proximal subunit would leave the distal one available to interact with Uch37 (Figure 3d). Evidence exists for Uch37 binding to the distal subunit of polyubiquitin; it is reported to be incapable of disassembling ubiquitin chains in which the distal ubiquitin contains the L8A and I44A mutations24 and dismantles chains by removing one ubiquitin moiety at a time from the distal end25. Uch37’s distal end deconjugation of ubiquitin chains25 complements that of Ubp6 and Rpn11, as Ubp6 can deconjugate multiple ubiquitin’s in a single cleavage event26 and Rpn11 performs “en bloc” deubiquitination from the proximal end27, 28. Deubiquitinating activities, particularly that of Ubp6, are antagonized by another RP component, the chain elongation factor Hul529. With so many receptors and chain-processing enzymes within the RP, the detailed pathway by which a substrate is degraded may be subject to many stochastic variations. Whether this unanticipated design promotes high substrate flux through the proteasome is unclear, but it seems well suited to allow the cell to fine-tune proteasome activity.
Methods
Expression, purification, crystallization and structure determination of mRpn13 Pru
mRpn13 Pru was over-expressed in E. coli strain BL21(DE3) RIL (Stratagene) and purified by GST-affinity chromatography using a PreScission Protease cleavage site followed by size exclusion chromatography. Rpn13 Pru was crystallized by the hanging drop vapor diffusion method and frozen in a stream of liquid nitrogen during X-ray exposure. Single anomalous dispersion (SAD) methods were performed using synchrotron radiation at the BW6 beamline at the DESY-centre in Hamburg, Germany. Native data were collected to 1.7 Å resolution (Supplementary Table 1). Details about recombinant DNA modifications, expression and purification of mutant forms of Rpn13 Pru, data processing, phase determination, model building and structural refinement are described in Supplementary Information.
NMR spectroscopy
Chemical shift assignments and spectra were acquired as described in Supplementary Information. mRpn13 Pru:ubiquitin and mRpn13 Pru:diubiquitin complexes were generated as described in Supplementary Information.
Supplementary Material
Acknowledgments
Help of G. Bourenkov (DESY, BW6, Hamburg, Germany) during synchrotron data collection is gratefully acknowledged and we thank J. Lary, J. Cole, and the National Analytical Ultracentrifugation Facility of the University of Connecticut for performing the sedimentation experiments. NMR data were acquired in the UMN BMBB NMR facility and the data processed in the MSI BSCL. This work was supported by the National Institutes of Health CA097004 (KW), GM43601 (DF), and GM008700-Chemistry Biology Interface Training Grant (LR), Deutsche Forschungsgemeinschaft (DI 931/3-1) and the Cluster of Excellence “Macromolecular Complexes” of the Goethe University Frankfurt (EXC115) to ID, and Deutsche Forschungsgemeinschaft SFB740/TP B4 (MG).
Footnotes
ACCESSION NUMBERS
Coordinates and structure factors of mRpn13 Pru and mRpn13 Pru:ubiquitin have been deposited in the Protein Data Bank with accession number 2R2Y and 2Z59, respectively.
References
- 1.Deveraux Q, Ustrell V, Pickart C, Rechsteiner M. A 26 S protease subunit that binds ubiquitin conjugates. J Biol Chem. 1994;269:7059–61. [PubMed] [Google Scholar]
- 2.Fu H, et al. Multiubiquitin chain binding and protein degradation are mediated by distinct domains within the 26 S proteasome subunit Mcb1. J Biol Chem. 1998;273:1970–81. doi: 10.1074/jbc.273.4.1970. [DOI] [PubMed] [Google Scholar]
- 3.Jorgensen JP, et al. Adrm1, a putative cell adhesion regulating protein, is a novel proteasome-associated factor. J Mol Biol. 2006;360:1043–52. doi: 10.1016/j.jmb.2006.06.011. [DOI] [PubMed] [Google Scholar]
- 4.Yao T, et al. Proteasome recruitment and activation of the Uch37 deubiquitinating enzyme by Adrm1. Nat Cell Biol. 2006;8:994–1002. doi: 10.1038/ncb1460. [DOI] [PubMed] [Google Scholar]
- 5.Hamazaki J, et al. A novel proteasome interacting protein recruits the deubiquitinating enzyme UCH37 to 26S proteasomes. Embo J. 2006;25:4524–36. doi: 10.1038/sj.emboj.7601338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Qiu XB, et al. hRpn13/ADRM1/GP110 is a novel proteasome subunit that binds the deubiquitinating enzyme, UCH37. Embo J. 2006;25:5742–53. doi: 10.1038/sj.emboj.7601450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Verma R, et al. Proteasomal proteomics: identification of nucleotide-sensitive proteasome-interacting proteins by mass spectrometric analysis of affinity-purified proteasomes. Mol Biol Cell. 2000;11:3425–39. doi: 10.1091/mbc.11.10.3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Husnjak K, et al. Proteasome subunit Rpn13 is a novel ubiquitin receptor. doi: 10.1038/nature06926. (accompanying manuscript) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hirano S, et al. Structural basis of ubiquitin recognition by mammalian Eap45 GLUE domain. Nat Struct Mol Biol. 2006;13:1031–2. doi: 10.1038/nsmb1163. [DOI] [PubMed] [Google Scholar]
- 10.Kang RS, et al. Solution structure of a CUE-ubiquitin complex reveals a conserved mode of ubiquitin binding. Cell. 2003;113:621–30. doi: 10.1016/s0092-8674(03)00362-3. [DOI] [PubMed] [Google Scholar]
- 11.Ohno A, et al. Structure of the UBA domain of Dsk2p in complex with ubiquitin molecular determinants for ubiquitin recognition. Structure. 2005;13:521–32. doi: 10.1016/j.str.2005.01.011. [DOI] [PubMed] [Google Scholar]
- 12.Ito T, et al. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc Natl Acad Sci U S A. 2001;98:4569–74. doi: 10.1073/pnas.061034498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gandhi TK, et al. Analysis of the human protein interactome and comparison with yeast, worm and fly interaction datasets. Nat Genet. 2006;38:285–93. doi: 10.1038/ng1747. [DOI] [PubMed] [Google Scholar]
- 14.Chau V, et al. A multiubiquitin chain is confined to specific lysine in a targeted short-lived protein. Science. 1989;243:1576–83. doi: 10.1126/science.2538923. [DOI] [PubMed] [Google Scholar]
- 15.Finley D, et al. Inhibition of proteolysis and cell cycle progression in a multiubiquitination-deficient yeast mutant. Mol Cell Biol. 1994;14:5501–9. doi: 10.1128/mcb.14.8.5501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lemmon MA. Pleckstrin homology domains: not just for phosphoinositides. Biochem Soc Trans. 2004;32:707–11. doi: 10.1042/BST0320707. [DOI] [PubMed] [Google Scholar]
- 17.Fisher RD, et al. Structure and ubiquitin binding of the ubiquitin-interacting motif. J Biol Chem. 2003;278:28976–84. doi: 10.1074/jbc.M302596200. [DOI] [PubMed] [Google Scholar]
- 18.Swanson KA, Kang RS, Stamenova SD, Hicke L, Radhakrishnan I. Solution structure of Vps27 UIM-ubiquitin complex important for endosomal sorting and receptor downregulation. Embo J. 2003;22:4597–606. doi: 10.1093/emboj/cdg471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang Q, Young P, Walters KJ. Structure of S5a bound to monoubiquitin provides a model for polyubiquitin recognition. J Mol Biol. 2005;348:727–39. doi: 10.1016/j.jmb.2005.03.007. [DOI] [PubMed] [Google Scholar]
- 20.Lee S, et al. Structural basis for ubiquitin recognition and autoubiquitination by Rabex-5. Nat Struct Mol Biol. 2006;13:264–71. doi: 10.1038/nsmb1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Penengo L, et al. Crystal structure of the ubiquitin binding domains of rabex-5 reveals two modes of interaction with ubiquitin. Cell. 2006;124:1183–95. doi: 10.1016/j.cell.2006.02.020. [DOI] [PubMed] [Google Scholar]
- 22.Bomar MG, Pai MT, Tzeng SR, Li SS, Zhou P. Structure of the ubiquitin-binding zinc finger domain of human DNA Y-polymerase eta. EMBO Rep. 2007;8:247–51. doi: 10.1038/sj.embor.7400901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Alam SL, et al. Structural basis for ubiquitin recognition by the human ESCRT-II EAP45 GLUE domain. Nat Struct Mol Biol. 2006;13:1029–30. doi: 10.1038/nsmb1160. [DOI] [PubMed] [Google Scholar]
- 24.Lam YA, DeMartino GN, Pickart CM, Cohen RE. Specificity of the ubiquitin isopeptidase in the PA700 regulatory complex of 26 S proteasomes. J Biol Chem. 1997;272:28438–46. doi: 10.1074/jbc.272.45.28438. [DOI] [PubMed] [Google Scholar]
- 25.Lam YA, Xu W, DeMartino GN, Cohen RE. Editing of ubiquitin conjugates by an isopeptidase in the 26S proteasome. Nature. 1997;385:737–40. doi: 10.1038/385737a0. [DOI] [PubMed] [Google Scholar]
- 26.Hanna J, et al. Deubiquitinating enzyme Ubp6 functions noncatalytically to delay proteasomal degradation. Cell. 2006;127:99–111. doi: 10.1016/j.cell.2006.07.038. [DOI] [PubMed] [Google Scholar]
- 27.Verma R, et al. Role of Rpn11 metalloprotease in deubiquitination and degradation by the 26S proteasome. Science. 2002;298:611–5. doi: 10.1126/science.1075898. [DOI] [PubMed] [Google Scholar]
- 28.Yao T, Cohen RE. A cryptic protease couples deubiquitination and degradation by the proteasome. Nature. 2002;419:403–7. doi: 10.1038/nature01071. [DOI] [PubMed] [Google Scholar]
- 29.Crosas B, et al. Ubiquitin chains are remodeled at the proteasome by opposing ubiquitin ligase and deubiquitinating activities. Cell. 2006;127:1401–13. doi: 10.1016/j.cell.2006.09.051. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.