Abstract
Systemic lupus erythematosus (SLE) is an autoimmune disease characterized by the production of autoantibodies against a host of nuclear antigens. The pathogenesis of lupus is incompletely understood. Environmental factors may play a role via altering DNA methylation, a mechanism regulating gene expression. In lupus, genes including CD11a and CD70 are overexpressed in T cells as a result of promoter hypomethylation. T-cell DNA methyltransferase expression is regulated in part by the extracellular signal-regulated kinase (ERK) signaling pathway. In this study, we investigate the effects of decreased ERK pathway signaling in T cells using transgenic animals. We generated a transgenic mouse that inducibly expresses a dominant-negative MEK in T cells in the presence of doxycycline. We show that decreased ERK pathway signaling in T cells results in decreased expression of DNA methyltransferase 1 and overexpression of the methylation-sensitive genes CD11a and CD70, similar to T cells in human lupus. Our transgenic animal model also develops anti-dsDNA antibodies. Interestingly, microarray expression assays revealed overexpression of several interferon-regulated genes in the spleen similar to peripheral blood cells of lupus patients. This model supports the contention that ERK pathway signaling defects in T cells contribute to the development of autoimmunity.
Keywords: epigenetics, T cell, autoimmunity, interferon, methylation, lupus
Introduction
Genetic and environmental factors contribute to human lupus. Evidence for a genetic contribution comes from a higher concordance rate among monozygotic twins, familial aggregation, a number of known lupus-associated genetic polymorphisms and evidence for linkage at multiple loci across the human genome.1 Observations that the majority of lupus is sporadic, concordance in monozygotic twins is incomplete and drugs such as procainamide and hydralazine can trigger lupus-like autoimmunity indicate a role for exogenous agents.2,3 How environmental agents contribute to the development of autoimmunity is unclear. Recent evidence, though, suggests that the environment can modify T-cell gene expression through epigenetic mechanisms, resulting in autoreactive T cells capable of promoting autoimmunity. The epigenetic mechanism most strongly linked to lupus-like autoimmunity is DNA methylation.4
DNA methylation is the postsynthetic methylation of cytosine bases occurring in CG pairs. Promoters of active genes are typically hypomethylated, and methylation of promoter sequences renders the genes transcriptionally inactive.5 Methylation patterns are established during development by the de novo DNA methyltransferases Dnmt3a and 3b, then replicated during mitosis by the maintenance methyltransferase Dnmt1.6,7 Resting T cells express relatively little Dnmt1, but levels are upregulated during mitosis by signals through the extracellular signal-regulated kinase (ERK) and Jun N-terminal kinase pathways.8,9 Inhibiting DNA methyltransferase activity with inhibitors such as 5-azacytidine, or decreasing Dnmt1 levels with ERK pathway inhibitors, inhibits methylation of newly synthesized DNA, causing overexpression of genes suppressed by this mechanism.10
CD4+ T cells treated with DNA methylation inhibitors overexpress LFA-1 (CD11a/CD18) and become autoreactive, responding to self-class II MHC without the addition of exogenous antigen.11 The autoreactivity is due to demethylation and overexpression of the ITGAL (CD11a) gene,12,13 and T cells overexpressing LFA-1 by transfection display identical autoreactivity.14 Hypomethylated, autoreactive CD4+ cells also spontaneously kill autologous macrophages (Mø), causing release of antigenic nucleosomes through mechanisms including demethylation and subsequent overexpression of the PRF1 (perforin) gene.15-17 The hypomethylated cells also overstimulate autologous B-cell antibody production, due to demethylation and overexpression of the genes encoding CD70 (TNFSF7), and CD40L (CD40LG).18,19 Importantly, procainamide and hydralazine, which cause anti-nuclear antibodies in a majority of people and a lupus-like disease in a subset, are DNA methylation inhibitors.20 Procainamide is a competitive Dnmt1 inhibitor,21,22 whereas hydralazine inhibits ERK pathway signaling, thereby decreasing Dnmt1 expression.23 Adoptive transfer of CD4+ T cells treated with these drugs or 5-azacytidine causes anti-DNA antibodies and a lupuslike disease in mice.23-25 Although all strains studied develop anti-DNA antibodies, the clinical manifestations are strain dependent,24,25 suggesting a genetic requirement for full disease development similar to hydralazine-induced autoimmunity in people.26
Further, CD4+ T cells from patients with active lupus demethylate the same regulatory sequences, overexpress the same genes, and display the same autoreactive Mø killing and B cell overstimulation as cells demethylated in vitro with 5-azacytidine, procainamide and hydralazine.17,27,28 Together, these observations suggest that T-cell DNA demethylation may contribute to the development of idiopathic and some forms of drug-induced lupus.
T-cell DNA demethylation in idiopathic lupus may be due to decreased ERK pathway signaling. As noted above, inhibiting ERK pathway signaling decreases T-cell Dnmt1 expression, causing DNA demethylation and methylation-sensitive gene overexpression.8,23,28 Lupus T cells display similar decreases in ERK pathway signaling and Dnmt1 levels, and the degree of impairment is directly proportional to disease activity.29 Also, murine CD4+ T cells treated with hydralazine or the MEK inhibitor U0126 demethylate, overexpress methylation-sensitive T-cell genes and cause anti-DNA antibodies when injected into syngeneic mice.23
Based on these observations, we hypothesized that acquired ERK pathway signaling defects, occurring in mature T cells, may be fundamental to the development of lupus-like autoimmunity. To test this hypothesis, we generated transgenic mice with an inducible ERK pathway signaling defect. We report that inducing an ERK pathway signaling defect in mature T cells is sufficient to decrease Dnmt1 expression, cause overexpression of a methylation-sensitive T-cell genes, and induce interferon-regulated gene expression in a non-autoimmune prone mouse strain.
Results
Generation of transgenic mice
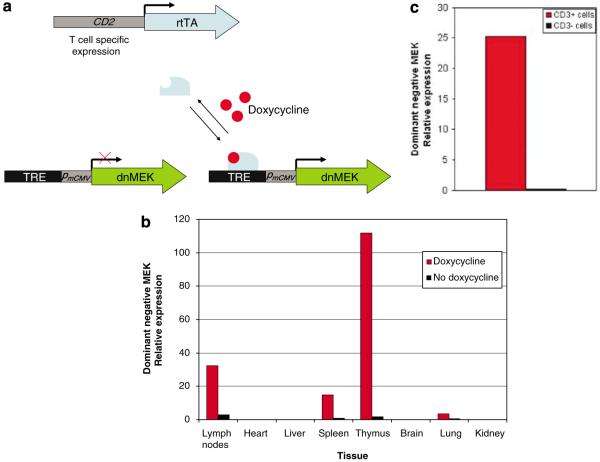
MEK is a protein kinase, which upon phosphorylation by Raf, results in ERK phosphorylation and therefore activation of the ERK signaling pathway. We generated a double-transgenic mouse (dnMEK/CD2-rtTA) that inducibly expresses a dominant-negative MEK selectively in T cells in the presence of doxycycline. The dnMEK mutant, kindly donated by Dr Kun-Liang Guan, contains triple mutations in the regulatory and kinase domains of wild-type MEK1 (substitutions of serines 218, 222, and lysine 97 by alanine), that block Raf-1-mediated MEK phosphorylation. Expression of this construct leads to ~60% reduction in ERK phosphorylation in Swiss 3T3 cells and does not impair cell survival.30 This mutant was cloned into pTRE-2, containing a tetracycline response element, then injected into C57BL/6 × (C57BL/6 × SJL) F1 mouse eggs and implanted into psuedopregnant females. Mice with the transgene were backcrossed onto a C57BL/6 background 6–10 times, then bred with a BL/6 transgenic strain containing the reverse tetracycline transactivator under the control of a CD2 promoter (CD2-rtTA). The double-transgenic mice (dnMEK/CD2-rtTA) would be predicted to express the dnMEK when doxycycline is added to their drinking water (Figure 1a).
Figure 1.
Conditional expression of a dnMEK in T cells. (a) A transgenic mouse strain was generated by subcloning a dnMEK cDNA downstream of a Tet-responsive promoter. This promoter contains the Tet response element (TRE), consisting of seven copies of the 42-bp tet operator sequence (tetO). Breeding with another transgenic mouse containing the reverse tetracycline-regulated transactivator (rtTA) under the control of the CD2 promoter yielded a double-transgenic mouse (dnMEK/CD2-rtTA) capable of T-cell specific expression of the dnMEK. When doxycycline is provided, rtTA binds to the tetO in the Tet-responsive promoter and activates the transcription of the dnMEK. (b) A representative of two independent experiments showing the expression of dominant-negative MEK (dnMEK) in various tissues in dnMEK/CD2-rtTA double-transgenic mice assessed by real-time reverse transcription (RT)–PCR. The expression of the transgene is restricted to the lymph nodes, spleen and thymus and is induced by doxycycline (2 mg ml−1 in drinking water). (c) dnMEK expression is restricted to CD3+ splenocytes. CD3+ and CD3− splenocytes were isolated and incubated for 24 h with doxycycline at a concentration of 1 μg ml−1. RNA was isolated and dnMEK expression was assessed by real-time RT-PCR and normalized to GAPDH (representative of two experiments).
‘Leakiness’ and inducibility of dnMEK expression in the dnMEK/CD2-rtTA mice was tested in various tissues including heart, lung, kidney, brain, thymus and spleen by real-time RT-PCR and primers specific for the dnMEK in mice with and without doxycycline. As expected, significant expression of dnMEK mRNA was detected in the lymph nodes, spleen and thymus in the presence of doxycycline but not in its absence (Figure 1b). Further-more, the dnMEK was expressed only in CD3+ T cells in the presence of doxycycline (Figure 1c). To test whether the expression of dnMEK decreases ERK signaling in T cells, CD3+ cells isolated from spleens of dnMEK/CD2-rtTA double-transgenic mice were incubated with doxycycline, without doxycycline or with the MEK inhibitor PD 98059 for 24 h. CD3+ cells incubated with doxycycline demonstrated reduced ERK signaling as measured by phosphorylated ERK (active MAPK), upon stimulation with phorbol myristate acetate (Figure 2).
Figure 2.
(a) A representative western blot demonstrating extracellular signal-regulated kinase (ERK) signaling as measured by phosphorylated ERK 1 and 2 (p-ERK 1/2) compared to total ERK (t-ERK 1/2). CD3+ cells isolated from spleens of double-transgenic mice and wild-type mice were incubated with doxycycline (1 μg ml−1), without doxycycline or with the MEK inhibitor PD 98059 (50 μm) for 24 h. CD3+ cells from double-transgenic mice incubated with doxycycline demonstrated reduced ERK signaling upon stimulation with phorbol myristate acetate. (b) A relative reduction in p-ERK in double-transgenic mice with versus without doxycycline (0.52 ± 0.08 versus 1 ± 0.00 (mean ± s.e.m.), P = 0.0032, n = 3). No reduction in p-ERK was observed in wild-type mice with doxycycline (n = 2).
DnMEK/CD2-rtTA double-transgenic mice demonstrate reduced Dnmt1 expression and the overexpression of T-cell methylation-sensitive genes
To test whether decreased ERK signaling in T cells decreases Dnmt1 expression, double-transgenic mice were given doxycycline (2 mg/ml) in their drinking water for 1 week, then treated and untreated mice were killed and the spleens extracted. RNA was isolated from the whole spleen, and Dnmt1 mRNA levels were measured by real-time reverse transcription (RT)-PCR. Dnmt1 transcripts were reduced by ~50% with doxycycline treatment (Figure 3a, P = 0.02). This confirms earlier reports that ERK signaling pathway is involved in regulating Dnmt1 expression.29 We also measured Dnmt1 expression in vitro in stimulated T cells from dnMEK/CD2-rtTA mice as compared to CD2-rtTA mice treated with doxycycline (2 μg ml−1). There was a significant reduction in Dnmt1 mRNA levels in stimulated T cells from dnMEK/CD2-rtTA (0.40 ± 0.05 versus 1.36 ± 0.28 (mean ± s.e.m.), P = 0.028, n = 3) (Figure 3b).
Figure 3.
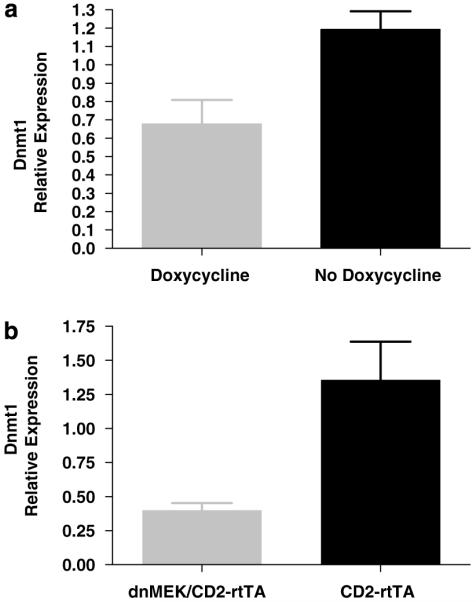
(a) Dnmt1 expression in the spleen of dnMEK/CD2-rtTA mice. Double-transgenic mice given doxycycline and thus expressing the dnMEK gene show decreased Dnmt1 expression as compared to double-transgenic mice without doxycycline (0.678 ± 0.132 versus 1.194 ± 0.098 (mean ± s.e.m.); P = 0.02, n = 4). (b) Reduced Dnmt1 expression in vitro in doxycycline-treated T cells from dnMEK/CD2-rtTA mice as compared to CD2-rtTA controls. Dnmt1 mRNA levels were determined by real-time RT-PCR and normalized to GAPDH (n = 3, P = 0.028).
To assess if reduced Dnmt1 expression could cause overexpression of methylation-sensitive genes in vivo, double-transgenic mice were given doxycycline (4 mg/ml) for 2 weeks, then killed, and transcripts of the methylation-sensitive genes CD11a and CD70 were quantitated in the spleen by real-time RT–PCR. Both genes were overexpressed in doxycycline-treated double-transgenic mice, as predicted (Figure 4). The expression of CD11a has previously been reported to be methylation-sensitive in both mouse and human T cells.15,23,31 To confirm that the expression of CD70 is also methylation-sensitive in mouse T cells, wild-type mouse spleen cells were stimulated with Con-A overnight, and then incubated with or without the DNA methylation inhibitor 5-azacytidine (5 μm) in interleukin 2 (IL-2; 50 U ml−1) containing culture media for an additional 72 h. T cells were isolated and the expression of CD70 measured by real-time RT-PCR. We found that CD70 expression increased with 5-azacytidine treatment in wild-type mouse T cells (1.18 ± 0.26 to 5.12 ± 0.84 (mean ± s.e.m.), P = 0.011, n = 3) confirming that CD70 is a methylation-sensitive gene in mouse T cells.
Figure 4.
Expression of methylation-sensitive genes in dnMEK/CD2-rtTA mice. CD11a and CD70 expression were determined in the spleen by real-time reverse transcription (RT)–PCR and normalized to GAPDH. (a) Increased expression of the methylation-sensitive gene Itgal (CD11a) in double-transgenic mice with as compared to without doxycycline (P = 0.036, n = 7). (b) Increased expression of the methylation-sensitive gene Tnfsf7 (CD70) in double-transgenic mice with as compared to without doxycycline (P = 0.032, n = 10).
We next measured cytokine production in vitro in stimulated T cells from dnMEK/CD2-rtTA mice as compared to CD2-rtTA mice treated with doxycycline (2 μg ml−1) using a bead-based immunofluorescence assay (Luminex Inc., Austin, TX, USA). Multiplex cytokine reagents were purchased from Invitrogen (Carlsbad, CA, USA). Following red blood cell lysis, splenocytes (3 × 106 cells per ml) were stimulated with Con-A overnight and cytokine levels were measured in the supernatants. We found no difference in the production of the cytokines measured including IFN-γ, IL-12, IL-6, GM-CSF and TNF-α.
Defective ERK signaling in T cells induces anti-dsDNA antibody production and differential expression of interferon-regulated genes
We then compared the development of anti-dsDNA antibodies in three dnMEK/CD2-rtTA mice receiving doxycycline (4 mg/ml) and three not, administered continuously over 12 weeks. As tetracyclines can cause a lupus-like disease in some people,32 six wild-type mice were also given doxycycline as a control. In addition, anti-dsDNA antibodies were measured in seven wild-type mice without doxycycline treatment. Figure 5a shows the kinetics of the anti-dsDNA response in a representative double-transgenic mouse receiving doxycycline, and a double-transgenic mouse not receiving the drug. Anti-ds DNA antibodies appear between 4 and 8 weeks, and increase through 12 weeks. Figure 5b shows the mean anti-dsDNA titers in the four groups, measured after 12 weeks of doxycycline or control drinking water. Only the double-transgenic mice given doxycycline were positive for anti-dsDNA antibodies (Figure 5).
Figure 5.
Development of anti-dsDNA antibodies in dnMEK/CD2-rtTA mice. (a) An example showing serial anti-dsDNA antibody titer in two littermate mice; double transgenic with and without doxycycline. (b) Anti-dsDNA antibody titer at 12 weeks following doxycycline treatment. Sera were collected from double-transgenic mice as well as wild-type mice with and without doxycycline. Double-transgenic mice with, but not without, doxycycline develop anti-dsDNA antibodies specific for lupus (0.319 ± 0.039 versus 0.156 ± 0.040 (mean absorbance ± s.e.m.); P = 0.04). The cutoff for positive result in this assay was 0.253 (dotted line).
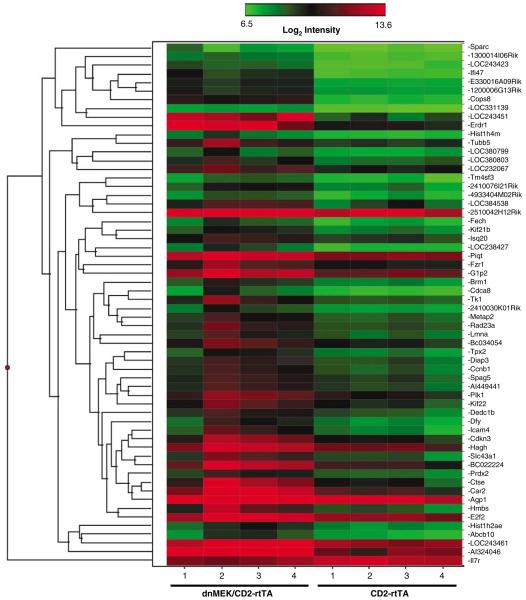
Expression microarray experiments were performed to detect differential gene expression in the spleens of four dnMEK/CD2-rtTA mice and four CD2-rtTA controls after two weeks of doxycycline treatment (4 mg/ml). We identified 60 genes that were differentially expressed by at least twofold between the dnMEK/CD2-rtTA mice and CD2-rtTA mice (Figure 6). Of interest, several differentially expressed genes are interferon-regulated genes (Figure 7a). Isg15 (G1p2) and Isg20 are part of the ‘interferon signature’ genes described in lupus patients and scleroderma patients respectively.33,34 We validated the microarray results by confirming the expression of four transcripts differentially expressed in dnMEK/CD2-rtTA mice using real-time RT-PCR (Tk1, Isg15, Isg20 and Il7r; Figure 7b).
Figure 6.
Hierarchical Clustering of microarray data for 60 genes that are differentially expressed between dnMEK/CD2-rtTA mice and CD2-rtTA mice (n = 4). Both mice groups received doxycycline. Red indicates higher gene expression levels and green indicates lower expression levels.
Figure 7.
(a) Interferon-regulated genes differentially expressed between dnMEK/CD2-rtTA and CD2-rtTA mice. (b) The differential expression of four representative genes was confirmed by real-time reverse transcription (RT)–PCR and normalized to GAPDH.
Using Ingenuity Pathway Analysis software (Ingenuity Systems Inc., Redwood City, CA, USA), two interesting biological networks were identified that incorporated differentially expressed genes in dnMEK/CD2-rtTA mice (Figure 8). Network ‘A’ is particularly interesting, as it reveals the central role for interferon-γ in the regulation of several differentially expressed genes. Network ‘B’ identifies a central role for both β-estradiol and TGFβ1 in some of the differentially expressed genes in dnMEK/CD2-rtTA mice.
Figure 8.

Networks of genes that are deferentially expressed in the dnMEK/CD2-rtTA mice compared to CD2-rtTA controls. Networks were identified using the Ingenuity software. (a) Cell death and immunological disease network, and (b) Connective tissue development network. Key: Solid lines indicate direct interaction, dotted lines indicate indirect interaction. An arrow from (a) to (b) indicates that (a) acts on (b), a line without an arrowhead indicates binding only and a line with a small vertical line at the end from (a) to (b) indicates (a) inhibits (b). Red indicates genes that are upregulated, green indicates genes that are downregulated and white indicates genes that are not user specified but incorporated into the network through relationships with other genes. Node shapes are: square, cytokine; diamond (vertical), enzyme; diamond (horizontal), peptidase; dotted rectangular (vertical), ion channel; solid rectangular (vertical), G-protein coupled receptor; triangle, kinase; oval (horizontal), transcription regulator; oval (vertical), transmembrane receptor; trapezoid, transporter; circle, other.
Discussion
This is a novel mouse model demonstrating that induction of a T-cell ERK pathway signaling defect results in changes consistent with human lupus. This indicates an important role for defective T-cell ERK signaling in the pathogenesis of lupus. The production of anti-dsDNA antibodies in this animal model can be induced by inhibiting T-cell ERK signaling. Indeed, disease activity in lupus patients correlates well with both the degree of T-cell ERK signaling defect as well as the anti-dsDNA antibody titer.29,35 Inducing a T-cell ERK signaling defect, resulted in reduced expression of Dnmt1, supporting an important role for ERK signaling in regulating Dnmt1 expression.29 As expected, the expression of T-cell methylation-sensitive genes was increased in the spleen when ERK signaling defect was induced. The overexpression of these methylation-sensitive genes such as CD11a, and CD70 is also observed in patients with active lupus as well as in human T cells treated with DNA methylation inhibitors.15,18
Microarray expression experiments performed on spleen RNA from mice with T-cell ERK signaling defect revealed differential expression of 60 genes. Interestingly, eight genes that are differentially expressed are interferon-regulated genes (Figure 7). The increased level and activity of interferon-α is well described in lupus patients.36 Indeed, a number of interferon-regulated genes are differentially expressed in peripheral blood mononuclear cells from lupus patients, constituting what is referred to as the ‘interferon signature’ in lupus.33,37 Recent data implies a correlation between high interferon-α activity and anti-dsDNA antibody production in lupus patients suggesting that the interferon pathway could be involved in the pathogenesis of lupus.38 In lupus mouse models, however, a more prominent role for interferon-γ rather than interferon-α has been reported.39,40 A prominent role for interferon-γ is also seen in our mouse model as suggested by the differentially expressed gene profiles (Figure 8). In the MRL/lpr lupus-prone mouse, 24 interferon-regulated genes are differentially expressed in the spleen relative to MRL+/+ controls.41 The finding that interferon-γ expression progressively increased in the spleens of MRL/lpr mice with time, whereas the levels of interferon-α and interferon-β remained stable, suggest an ‘interferon-γ signature’ in the MRL/lpr lupus-prone mouse.41 Interestingly, MRL/lpr mice also have T-cell DNA methylation defect as demonstrated by reduced T-cell Dnmt1 expression, and hypomethylation with overexpression of methylation-sensitive genes such as CD70,42 similar to what we observed in the dnMEK/CD2-rtTA mouse model.
The mouse model described herein reproduces several abnormalities that exist in lupus patients. These include a defective Dnmt1 expression, overexpression of CD11a and CD70, the production of anti-dsDNA antibodies and the overexpression of interferon-regulated genes. However, the lack of a lupus clinical phenotype indicates the importance of genetics in lupus susceptibility. Our model suggests that episodic environmental exposures that decrease ERK signaling in T cells can trigger autoimmunity. However, genetic susceptibility is crucial for developing clinical pathology. In human lupus these environmental agents can be certain drugs such as hydralazine and procainamide, in the case of druginduced lupus, or environmental triggers that are yet to be identified, in the case of idiopathic lupus. In either case, genetic susceptibility is necessary but not sufficient to cause disease in the vast majority of lupus patients.
Materials and methods
Generation and breeding of transgenic mice
A human dominant negative MEK1 cDNA (hereafter referred to as dnMEK)30 was excised from the parent vector pCMV-dnMEK with the restriction enzyme BamHI (Roche, Basel, Switzerland) and subcloned into the multiple cloning site of pTRE-2 vector (Clontech Laboratories, Palo Alto, CA, USA) at the BamHI restriction site via T4 DNA ligase (Promega, Madison, WI, USA). The orientation and integrity of the dnMEK insert was determined by direct sequencing. The resulting transgene, TRE2-dnMEK, was excised from the vector by the restriction enzymes NotI and HindIII (Roche) and was purified prior to microinjection into C57BL/6 × (C57BL/6 × SJL) F1 mouse eggs implanted into psuedopregnant females by personnel at the University of Michigan Animal Model Core. Founders were identified by PCR on tail biopsy DNA, and the founders backcrossed onto a C57BL/6 background. Double-transgenic mice were generated by crossing TRE2-dnMEK transgenic mice with CD2-rtTA mice. CD2-rtTA mice express the reverse tetracycline transactivator (rtTA) under the control of a CD2 promoter allowing for lymphoid specific expression.43
Screening for the double-transgenic mice (dnMEK/CD2-rtTA) was by PCR on tail-biopsy DNA. The following genotyping primers were used: dnMEK forward, 5′-ACGCCATCCACGCTGTTTTG-3′; reverse, 5′-TTCGCTGCTGCTCATCAAGC-3′; rtTA forward, 5′-GTGATTAACAGCGCATTA-3′; reverse, 5′-ATCAATTCAAGGCCGAAT-3′.
In order to generate wild-type littermate controls, double-transgenic mice were bred with wild-type C57BL/6 mice. Mice were maintained in specific pathogen-free environment and all our protocols and procedures were approved by the Institutional Animal Care and Use Committee at the Oklahoma Medical Research Foundation. Animals were given doxycycline in the drinking water at a concentration of 2 or 4 mg per ml in different experiments. Doxycycline solution was made in 5% sucrose to cover the bitter taste of doxycycline, and in all in-vivo experiments, double-transgenic control mice were given 5% sucrose without doxycycline.
RNA isolation
Both Trizol (Invitrogen, Carlsbad, CA, USA) and RNeasy kits (Qiagen, Valencia, CA, USA) were used to isolate tissue RNA. Briefly, tissues are first homogenized in Trizol, chloroform added and then the aqueous layer mixed with an equal volume of 70% ethanol and loaded onto the RNeasy columns. The remainder of the RNA isolation was according to the RNeasy kit manufacturer’s instructions. DNA digestion was performed using Turbo DNA-free (Ambion Inc., Austin, TX, USA) following the manufacturer’s instructions.
Real-time RT-PCR
One-step real-time RT-PCR was performed using the QuantiTect SYBR Green RT-PCR kit (Qiagen) and the Rotor-Gene 3000 real-time thermocycler (Corbett Research, Australia). A total of 150 ng of RNA was used for each reaction. The following amplification conditions were used: 30 min at 50 °C, 15 min at 95 °C, 54 cycles of 15 s at 94 °C and 20 s at 56 °C and 30 s at 72 °C. Quantitation was performed by comparison with internal standards prepared by serial dilutions. Various transcript expression levels were always normalized for the housekeeping gene GAPDH. The following primers were used: mouse GAPDH forward, 5′-CAACGACCCCTTCATTGACCTC-3′; reverse, 5′-GCCTCACCCCATTTGATGTTAGTG-3′; mouse CD11a forward, 5′-CAGATTGAAGATGGGGTTGTCG-3′; reverse, 5′-CGGGACGATTTTGTAACATAGGTC-3′; mouse CD70 forward, 5′-TGGCTGTGGGCATCTGCTC-3′; reverse, 5′-ACATCTCCGTGGACCAGGTATG-3′; mouse Dnmt1 forward, 5′-GGAAGGCTACCTGGCTAAAGTCAAG-3′; reverse, 5′-ACTGAAAGGGTGTCACTGTCCGAC-3′, mouse Tk1 forward, 5′-GCAGATTCAGGTGATTCTCGGG-3′; reverse, 5′-GCATACTTGATGACCAGGCACTTG-3′; mouse Isg15 forward, 5′-ACGGTCTTACCCTTTCCAGTC-3′; reverse, 5′-CCCCTTTCGTTCCTCACCAG-3′; mouse Isg20 forward, 5′-GTCACGCCTCAGCACATGGT-3′; reverse, 5′-CCACCAGCTTGCCTTTCAGAA-3′; mouse Il7r forward, 5′TATGTGGGGCTCTTTTACGAGT-3′; reverse, 5′-GCCTCGGCTTTAACTATTGTGT-3′. All primers were purchased from Integrated DNA Technologies Inc. (Coralville, IA, USA).
Expression microarray
RNA
RNA was isolated from the spleens using RNeasy kit (Qiagen) following the manufacturer’s protocol. RNA concentration was determined with a Nanodrop scanning spectrophotometer (Nanodrop Technologies, Wilmington, DE, USA), and then qualitatively assessed for degradation using the ratio of 28:18 rRNA using a capillary gel electrophoresis system (Agilent 2100 Bioanalyzer, Agilent Technologies, Santa Clara, CA, USA).
Labeling and hybridization
Biotinylated amplified RNA was produced from 200 ng total RNA per sample using a modification of the Eberwine protocol,44 as described in the Illumina TotalPrep RNA Amplification Kit from Ambion Inc. Briefly, RNA was reverse transcribed with oligo(dT) primer containing a T7 promoter. RNA containing biotin-UTP ribonucleotides is amplified by in-vitro transcription to generate anti-sense RNA. This RNA is hybridized overnight at 58 °C to Sentrix Mouse-6 v1.1 Expression BeadChip microarrays obtained from Illumina Corp. (San Diego, CA, USA). The arrays have 46 657 genes, splice variants and control beads. These arrays contain 50-mer oligonucleotides coupled to beads that are mounted on glass slides. Each bead has approximately 20- to 30-fold redundancy per microarray. Microarrays are washed under high stringency, labeled with streptavidin-Cy3, and scanned with an Illumina BeadStation 500 scanner.
Analysis
Illumina BeadChip arrays (Illumina Inc., San Diego, CA, USA) were scanned with the Illumina Bead Array Reader and bead intensities were quantitated with the Illumina BeadStudio software. Raw values were quantile-normalized using MATLAB software (The MathWorks Inc., Natick, MA, USA) and the resulting normalized values were imported into BRB ArrayTools (Biometric Research Branch, National Cancer Institute) where they were then log transformed. Genes were filtered using the ‘Log Expression Variation Filter’ to screen out genes that are not likely to be informative, based on the variance of each gene across the arrays. In this case, the filter was set to exclude genes that fell below the fiftieth percentile of gene variance. We identified genes that were differentially expressed between the two classes by using a multivariate permutation test.45 We used the multivariate permutation test to provide a median false discovery rate (FDR) of 10% (80% confidence). The test statistics used were random variance t-statistics for each gene.46 Although t-statistics were used, the multivariate permutation test is nonparametric and does not require the assumption of Gaussian distributions. Data were exported to Excel (Microsoft, Redmond, WA, USA) where averages of the classes were used to calculate expression ratios. Genes that were differentially expressed (<10% FDR) and simultaneously had ratios twofold or larger were used in further analyses. Pathways were explored by placing the only genes that passed our significance/ratio threshold into Ingenuity Pathways Analysis (Ingenuity Systems, www.ingenuity.com).
Cell separation and culture
CD3+ lymphocytes were isolated from the spleens of double-transgenic mice via magnetic bead separation using indirect labeling (Miltenyi Biotec, Auburn, CA, USA) following the manufacturer’s protocol. Cell purity was routinely checked by flow cytometry. Where indicated, cells were cultured in RPMI 1640 with 10% fetal calf serum and were incubated with doxycycline, without doxycycline or with the MEK inhibitor PD 98059 (50 μm) for 24 h. Phorbol myristate acetate (50 ng ml−1) was added and cells were harvested after 15 min. Where indicated, mouse spleen cells were incubated with con-A (5 μg ml−1) overnight for T-cell stimulation.
Protein isolation and western blot
CD3+ T cells were harvested, centrifuged and resuspended in radioimmunoprecipitation assay buffer (50 mm Tris-HCL, pH 7.4, 150 mm NaCl, 0.25% deoxycholic acid, 1% NP-40, 1 mm EDTA, 100 μg ml−1 phenylmethylsulfonyl fluoride, 100 μm sodium orthovanadate, 1 mm dithiothreitol and a protease inhibitor cocktail (Roche)). The suspension was rotated at 4 °C for 30 min, insoluble material removed by centrifugation at 16 000 g for 30 min and the supernatant saved as whole cell lysate. The amount of cellular protein present in the whole cell lysate was measured using the bicinchoninic acid (BCA) protein assay (Pierce, Rockford, IL, USA). Isolated proteins (20 μg) were diluted in Laemmli loading buffer and denatured by boiling for 5 min followed by electrophoresis in 10–12% SDS–polyacrylamide gels. The fractionated proteins were then electrophoretically transferred to nitrocellulose membranes (Schleicher and Schuell, Keene, NH, USA) and stained with Ponceau S (Sigma-Aldrich, St Louis, MO, USA) to verify equal amounts of protein between lanes prior to western blot analyses. Membranes were blocked for 2 h in Tris-buffered saline (TBS) containing 0.1% Tween (Sigma-Aldrich) and 5% nonfat dry milk (Bio-Rad Laboratories, Hercules, CA, USA). After a 16-h incubation with a rabbit polyclonal anti-active MAPK (1:5000; Promega) in TBS, 0.1% Tween, 5% nonfat dry milk, blots were washed three times with TBS containing 0.1% Tween and incubated with a horseradish peroxidase-linked anti-rabbit antibody (1:2000; Cell Signaling Technologies, Danvers, MA, USA) for 1 h. After three washes with TBS–0.1% Tween the membranes were treated with ECL chemiluminiscence detection system (Amersham, Piscataway, NJ, USA), exposed to X-ray film (Kodak, Rochester, NY, USA) and developed to visualize the labeled protein bands. Molecular mass was estimated by comparison of sample bands with prestained molecular mass marker (Bio-Rad Laboratories). Blots were stripped and reblotted with a rabbit anti-ERK antibody (1:5000; Promega). Values were normalized to total ERK.
Statistical analysis
For continuous variables, differences between groups were tested using Student’s t-test. P-values of <0.05 were considered statistically significant.
Acknowledgements
We are indebted to Dr Kun-Liang Guan for donating the dominant-negative MEK1 mutant, and to Dr Rose Zamoyska and Dr Avery August for providing the CD2-rtTA mice. We are thankful to Dr John Attwood for assistance and discussion. We are also thankful to Maggie Van Keuren and Dr Thomas L Saunders at the University of Michigan Transgenic Animal Model Core for generating the TRE2-dnMEK transgenic mouse and for very helpful discussion. We are indebted to the Microarray Research Facility at OMRF and Dr Mark Barton Frank, Dr Michael Centola, Dr R Craig Cadwell and Dr Jonathan D Wren for analyzing the microarray data. The microarray core receives funding from the NIH grants number P20RR016478, P20RR020143, P20RR017703, P20RR15577, and U19AI062629. This publication was made possible by NIH Grant Number P20RR015577 from the National Center for Research Resources (AHS), PHS grants AR42525, AG25877, and ES015214, and a Merit grant from the Department of Veterans Affairs (BR).
Footnotes
Conflict of interest
None.
References
- 1.Sestak AL, Nath SK, Sawalha AH, Harley JB. Current status of lupus genetics. Arthritis Res Ther. 2007;9:210. doi: 10.1186/ar2176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sarzi-Puttini P, Atzeni F, Iaccarino L, Doria A. Environment and systemic lupus erythematosus: an overview. Autoimmunity. 2005;38:465–472. doi: 10.1080/08916930500285394. [DOI] [PubMed] [Google Scholar]
- 3.James JA, Kaufman KM, Farris AD, Taylor-Albert E, Lehman TJ, Harley JB. An increased prevalence of Epstein-Barr virus infection in young patients suggests a possible etiology for systemic lupus erythematosus. J Clin Invest. 1997;100:3019–3026. doi: 10.1172/JCI119856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sawalha AH, Richardson BC. DNA methylation in the pathogenesis of systemic lupus erythematosus. Curr Pharmacogen. 2005;3:73–78. [Google Scholar]
- 5.Richardson B, Yung R. Role of DNA methylation in the regulation of cell function. J Lab Clin Med. 1999;134:333–340. doi: 10.1016/s0022-2143(99)90147-6. [DOI] [PubMed] [Google Scholar]
- 6.Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- 7.Bird AP. Use of restriction enzymes to study eukaryotic DNA methylation: II. The symmetry of methylated sites supports semi-conservative copying of the methylation pattern. J Mol Biol. 1978;118:49–60. doi: 10.1016/0022-2836(78)90243-7. [DOI] [PubMed] [Google Scholar]
- 8.Deng C, Yang J, Scott J, Hanash S, Richardson BC. Role of the ras-MAPK signaling pathway in the DNA methyltransferase response to DNA hypomethylation. Biol Chem. 1998;379:1113–1120. doi: 10.1515/bchm.1998.379.8-9.1113. [DOI] [PubMed] [Google Scholar]
- 9.MacLeod AR, Rouleau J, Szyf M. Regulation of DNA methylation by the Ras signaling pathway. J Biol Chem. 1995;270:11327–11337. doi: 10.1074/jbc.270.19.11327. [DOI] [PubMed] [Google Scholar]
- 10.Lu R, Wang X, Chen ZF, Sun DF, Tian XQ, Fang JY. Inhibition of the extracellular signal-regulated kinase/mitogen-activated protein kinase pathway decreases DNA methylation in colon cancer cells. J Biol Chem. 2007;282:12249–12259. doi: 10.1074/jbc.M608525200. [DOI] [PubMed] [Google Scholar]
- 11.Richardson B. Effect of an inhibitor of DNA methylation on T cells. II. 5-Azacytidine induces self-reactivity in antigen-specific T4+ cells. Hum Immunol. 1986;17:456–470. doi: 10.1016/0198-8859(86)90304-6. [DOI] [PubMed] [Google Scholar]
- 12.Richardson B, Powers D, Hooper F, Yung RL, O’Rourke K. Lymphocyte function-associated antigen 1 overexpression and T cell autoreactivity. Arthritis Rheum. 1994;37:1363–1372. doi: 10.1002/art.1780370915. [DOI] [PubMed] [Google Scholar]
- 13.Lu Q, Ray D, Gutsch D, Richardson B. Effect of DNA methylation and chromatin structure on ITGAL expression. Blood. 2002;99:4503–4508. doi: 10.1182/blood.v99.12.4503. [DOI] [PubMed] [Google Scholar]
- 14.Yung R, Powers D, Johnson K, Amento E, Carr D, Laing T, et al. Mechanisms of drug-induced lupus. II. T cells overexpressing lymphocyte function-associated antigen 1 become autoreactive and cause a lupus like disease in syngeneic mice. J Clin Invest. 1996;97:2866–2871. doi: 10.1172/JCI118743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Richardson BC, Strahler JR, Pivirotto TS, Quddus J, Bayliss GE, Gross LA, et al. Phenotypic and functional similarities between 5-azacytidine-treated T cells and a T cell subset in patients with active systemic lupus erythematosus. Arthritis Rheum. 1992;35:647–662. doi: 10.1002/art.1780350608. [DOI] [PubMed] [Google Scholar]
- 16.Yung R, Kaplan MJ, Ray D, Schneider K, Mo R-R, Johnson K, et al. Autoreactive murine Th1 and Th2 cells kill syngeneic macrophages and induce autoantibodies. Lupus. 2001;10:539–546. doi: 10.1191/096120301701549660. [DOI] [PubMed] [Google Scholar]
- 17.Kaplan MJ, Lu Q, Wu A, Attwood J, Richardson B. Demethylation of promoter regulatory elements contributes to perforin overexpression in CD4+ lupus T cells. J Immunol. 2004;172:3652–3661. doi: 10.4049/jimmunol.172.6.3652. [DOI] [PubMed] [Google Scholar]
- 18.Oelke K, Lu Q, Richardson D, Wu A, Deng C, Hanash S, et al. Overexpression of CD70 and overstimulation of IgG synthesis by lupus T cells and T cells treated with DNA methylation inhibitors. Arthritis Rheum. 2004;50:1850–1860. doi: 10.1002/art.20255. [DOI] [PubMed] [Google Scholar]
- 19.Lu Q, Wu A, Tesmer L, Ray D, Yousif N, Richardson B. Demethylation of CD40LG on the inactive X in T cells from women with lupus. J Immunol. 2007;179:6352–6358. doi: 10.4049/jimmunol.179.9.6352. [DOI] [PubMed] [Google Scholar]
- 20.Cornacchia E, Golbus J, Maybaum J, Strahler J, Hanash S, Richardson B. Hydralazine and procainamide inhibit T cell DNA methylation and induce autoreactivity. J Immunol. 1988;140:2197–2200. [PubMed] [Google Scholar]
- 21.Scheinbart LS, Johnson MA, Gross LA, Edelstein SR, Richardson BC. Procainamide inhibits DNA methyltransferase in a human T cell line. J Rheumatol. 1991;18:530–534. [PubMed] [Google Scholar]
- 22.Lee BH, Yegnasubramanian S, Lin X, Nelson WG. Procainamide is a specific inhibitor of DNA methyltransferase 1. J Biol Chem. 2005;280:40749–40756. doi: 10.1074/jbc.M505593200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Deng C, Lu Q, Zhang Z, Rao T, Attwood J, Yung R, et al. Hydralazine may induce autoimmunity by inhibiting extracellular signal-regulated kinase pathway signaling. Arthritis Rheum. 2003;48:746–756. doi: 10.1002/art.10833. [DOI] [PubMed] [Google Scholar]
- 24.Yung RL, Quddus J, Chrisp CE, Johnson KJ, Richardson BC. Mechanism of drug-induced lupus. I. Cloned Th2 cells modified with DNA methylation inhibitors in vitro cause autoimmunity in vivo. J Immunol. 1995;154:3025–3035. [PubMed] [Google Scholar]
- 25.Quddus J, Johnson KJ, Gavalchin J, Amento EP, Chrisp CE, Yung RL, et al. Treating activated CD4+ T cells with either of two distinct DNA methyltransferase inhibitors, 5-azacytidine or procainamide, is sufficient to cause a lupus-like disease in syngeneic mice. J Clin Invest. 1993;92:38–53. doi: 10.1172/JCI116576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mansilla-Tinoco R, Harland SJ, Ryan PJ, Bernstein RM, Dollery CT, Hughes GR, et al. Hydralazine, antinuclear antibodies, and the lupus syndrome. Br Med J (Clin Res Ed) 1982;284:936–939. doi: 10.1136/bmj.284.6320.936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lu Q, Kaplan M, Ray D, Ray D, Zacharek S, Gutsch D, et al. Demethylation of ITGAL (CD11a) regulatory sequences in systemic lupus erythematosus. Arthritis Rheum. 2002;46:1282–1291. doi: 10.1002/art.10234. [DOI] [PubMed] [Google Scholar]
- 28.Lu Q, Wu A, Richardson BC. Demethylation of the same promoter sequence increases CD70 expression in lupus T cells and T cells treated with lupus-inducing drugs. J Immunol. 2005;174:6212–6219. doi: 10.4049/jimmunol.174.10.6212. [DOI] [PubMed] [Google Scholar]
- 29.Deng C, Kaplan MJ, Yang J, Ray D, Zhang Z, McCune WJ, et al. Decreased Ras-mitogen-activated protein kinase signaling may cause DNA hypomethylation in T lymphocytes from lupus patients. Arthritis Rheum. 2001;44:397–407. doi: 10.1002/1529-0131(200102)44:2<397::AID-ANR59>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 30.Syu LJ, Guan KL, Saltiel AR. Inducible expression of a mutant form of MEK1 in Swiss 3T3 cells. J Cell Biochem. 1997;67:367–377. [PubMed] [Google Scholar]
- 31.Yung R, Chang S, Hemati N, Johnson K, Richardson B. Mechanisms of drug-induced lupus. IV. Comparison of procainamide and hydralazine with analogs in vitro and in vivo. Arthritis Rheum. 1997;40:1436–1443. doi: 10.1002/art.1780400811. [DOI] [PubMed] [Google Scholar]
- 32.Gordon MM, Porter D. Minocycline induced lupus: case series in the West of Scotland. J Rheumatol. 2001;28:1004–1006. [PubMed] [Google Scholar]
- 33.Bennett L, Palucka AK, Arce E, Cantrell V, Borvak J, Banchereau J, et al. Interferon and granulopoiesis signatures in systemic lupus erythematosus blood. J Exp Med. 2003;197:711–723. doi: 10.1084/jem.20021553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tan FK, Zhou X, Mayes MD, Gourh P, Guo X, Marcum C, et al. Signatures of differentially regulated interferon gene expression and vasculotrophism in the peripheral blood cells of systemic sclerosis patients. Rheumatology (Oxford) 2006;45:694–702. doi: 10.1093/rheumatology/kei244. [DOI] [PubMed] [Google Scholar]
- 35.Swaak AJ, Groenwold J, Aarden LA, Statius van Eps LW, Feltkamp EW. Prognostic value of anti-dsDNA in SLE. Ann Rheum Dis. 1982;41:388–395. doi: 10.1136/ard.41.4.388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Crow MK, Kirou KA. Interferon-alpha in systemic lupus erythematosus. Curr Opin Rheumatol. 2004;16:541–547. doi: 10.1097/01.bor.0000135453.70424.1b. [DOI] [PubMed] [Google Scholar]
- 37.Baechler EC, Batliwalla FM, Karypis G, Gaffney PM, Ortmann WA, Espe KJ, et al. Interferon-inducible gene expression signature in peripheral blood cells of patients with severe lupus. Proc Natl Acad Sci USA. 2003;100:2610–2615. doi: 10.1073/pnas.0337679100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Niewold TB, Hua J, Lehman TJ, Harley JB, Crow MK. High serum IFN-alpha activity is a heritable risk factor for systemic lupus erythematosus. Genes Immun. 2007;8:492–502. doi: 10.1038/sj.gene.6364408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lawson BR, Prud’homme GJ, Chang Y, Gardner HA, Kuan J, Kono DH, et al. Treatment of murine lupus with cDNA encoding IFN-gammaR/Fc. J Clin Invest. 2000;106:207–215. doi: 10.1172/JCI10167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Richards HB, Satoh M, Jennette JC, Croker BP, Yoshida H, Reeves WH. Interferon-gamma is required for lupus nephritis in mice treated with the hydrocarbon oil pristane. Kidney Int. 2001;60:2173–2180. doi: 10.1046/j.1523-1755.2001.00045.x. [DOI] [PubMed] [Google Scholar]
- 41.Liu J, Karypis G, Hippen KL, Vegoe AL, Ruiz P, Gilkeson GS, et al. Genomic view of systemic autoimmunity in MRLlpr mice. Genes Immun. 2006;7:156–168. doi: 10.1038/sj.gene.6364286. [DOI] [PubMed] [Google Scholar]
- 42.Sawalha AH, Jeffries M. Defective DNA methylation and CD70 overexpression in CD4+ T cells in MRL/lpr lupus-prone mice. Eur J Immunol. 2007;37:1407–1413. doi: 10.1002/eji.200636872. [DOI] [PubMed] [Google Scholar]
- 43.Legname G, Seddon B, Lovatt M, Tomlinson P, Sarner N, Tolaini M, et al. Inducible expression of a p56Lck transgene reveals a central role for Lck in the differentiation of CD4 SP thymocytes. Immunity. 2000;12:537–546. doi: 10.1016/s1074-7613(00)80205-8. [DOI] [PubMed] [Google Scholar]
- 44.Van Gelder RN, von Zastrow ME, Yool A, Dement WC, Barchas JD, Eberwine JH. Amplified RNA synthesized from limited quantities of heterogeneous cDNA. Proc Natl Acad Sci USA. 1990;87:1663–1667. doi: 10.1073/pnas.87.5.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Simon R, Korn E, McShane L, Radmacher M, Wright G, Zhao Y. Design and Analysis of DNA Microarray Investigations. Springer-Verlag; New York: 2003. [Google Scholar]
- 46.Wright GW, Simon RM. A random variance model for detection of differential gene expression in small microarray experiments. Bioinformatics. 2003;19:2448–2455. doi: 10.1093/bioinformatics/btg345. [DOI] [PubMed] [Google Scholar]