Abstract
Second order nonlinear optical imaging of chiral crystals (SONICC) is explored for selective detection of integral membrane protein crystals grown in opaque and turbid environments. High turbidity is a hallmark of membrane protein crystallization due to the extensive use of detergent and/or lipids that often form various mesophases. Detection of crystals in such media by conventional optical methods (e.g., intrinsic UV fluorescence, birefringence, bright-field image analysis, etc.) is often complicated by optical scattering and by the small sizes of the crystals that routinely form. SONICC is shown to be well-suited for this application, by nature of its compatibility with imaging in scattering media and its high selectivity for protein crystals. Bright second harmonic generation (SHG) (up to 18 million counts/s) was observed from even relatively small crystals (5 micron) with a minimal background due to the surrounding lipid mesophase (~1 thousand counts/s). The low background nature of the resulting protein crystal images permitted the use of a relatively simple, particle counting analysis for preliminary scoring. Comparisons between a particle counting analysis of SONICC images and protocols based on the human expert analysis of conventional bright-field and birefringence images were performed.
Reliable detection of protein crystal formation is a critical step in automated high-throughput screening efforts. Many image analysis techniques have been applied to the classification of protein crystallization trials, such as Fisher linear discriminants1, self-organizing maps2, Bayes classifiers3, decision trees4, support vector machines5, Fourier descriptors5, and machine-learning approaches6. These techniques are primarily designed to classify droplets based on their likelihood of containing diffraction quality crystals, i.e. clear droplets and precipitates receive lower scores than those containing relatively large crystals. These techniques have been reported to have up to 85% classification accuracy, which has allowed the use data mining techniques designed to predict the likely precipitant conditions for novel proteins7. However, these techniques rely on bright-field images as inputs. Consequently, they are susceptible to analysis errors when detecting microcrystals (< 5 μm) and when imaging in highly scattering media.
These limitations are particularly problematic in crystallization trials of membrane proteins, as both of these complicating conditions routinely arise. Membrane proteins represent about 30 % of genome in most organisms and perform essential functions, including energy conversion, signal transduction, and chemical transport into and out of cells. Numerous diseases are associated with reduction or loss of function in membrane proteins, and many pathogens exploit membrane receptors to facilitate entrance inside the cells. Despite their importance, the number of membrane protein structures is far smaller than that of soluble proteins. Reliable crystal detection is one of the major stumbling blocks in obtaining initial crystal hits, which subsequently could be optimized into well diffracting membrane protein crystals. Therefore any technique capable of identifying promising crystallization conditions should facilitate quests for high-quality crystals. Detergents and lipids used to disperse the membrane protein are prone to phase separation, scattering light and rendering crystal detection algorithms based on the analysis of the bright-field inefficient.
Crystals grown in lipidic mesophases, such as lipidic cubic phase (LCP), sponge phase8,9 and bicelles10 are particularly challenging cases for reliable crystal detection. These methods take advantage of a more native membrane-like environment for obtaining protein crystals. Over the past few years, LCP crystallization provided a detailed structural view of the bacteriorhodpsin photocycle11 and yielded high-resolution structures of two human G protein-coupled receptors12,13. Although an LCP matrix is optically transparent and isotropic when prepared under controlled conditions14, it often develops numerous defects and voids upon contact with precipitant solution, obstructing detection of colorless protein crystals15. Additionally, LCP can transition to a highly turbid and birefringent lamellar or hexagonal phase as a result of interaction with precipitants or prolonged incubation16, making it extraordinarily difficult to detect colorless crystals.
Several approaches have been suggested to increase contrast for imaging and detection of protein crystals in such cases: crystal birefringence17,18, intrinsic protein fluorescence19, addition of fluorescent dyes20 and fluorescence of trace labeled protein molecules21, 22,22. Most of these methods have been added to the arsenal of modern commercial crystal imagers and are already being routinely used in structural laboratories. In this work, second-order nonlinear optical imaging of chiral crystals (SONICC) is explored as a complementary method for protein crystal detection in complex optically scattering matrices, including lipidic mesophases.
Second harmonic generation (SHG), or the frequency doubling of light, underpins SONICC measurements23. The unique symmetry requirements of SHG result in destructive interference and no coherent signal from randomly oriented assemblies of proteins (e.g., proteins in solution or in amorphous aggregates), but produce bulk-allowed SHG from all crystals of homochiral molecules, such as proteins. However, most salt crystals adopt centrosymmetric, SHG-inactive structures. Similarly, amorphous materials (liquids, solutions, glasses, and disordered aggregates) are symmetry-forbidden to produced coherent SHG. Because SHG scales with the square of the intensity, only the unattenuated light that reaches the focal plane significantly contributes to the detected signal when using the most common beam-scanning confocal configurations. Furthermore, any SHG that is detected can be assumed to have arisen from within the focal volume, such that image resolution is not substantially degraded even in highly scattering media. In the present study, SONICC images in turbid lipidic cubic phase crystallization trials are shown to be compatible with relatively simple analysis methods by using standard particle counting methods to determine the average size and SHG signal of the crystals in the image. These values can be used to order rank crystallization trials on their likelihood of containing high quality crystals and compared to conventional scoring methods based on expert visual inspection of bright-field and birefringence images.
Experimental/Computational Methods
The LCP crystallization trials were imaged by bright-field, birefringence, UV-fluorescence, fluorescence of trace-labeled protein, as well as by SONICC. Bright-field, birefringence and UV-fluorescence images were taken using an automated incubator/imager (RockImager 1000, Formulatrix). The bright-field and birefringence images were acquired through a Navitar 12x variable zoom lens with 1.5x lens attachment using a Qimaging MicroPublisher 5.0 RTV CCD color camera at 2×2 binning (1280×960 pixels). A transillumination from a cold white LED light source conditioned with a variable numerical aperture Koehler-type condenser was used. The condenser aperture was adjusted to provide a maximum contrast for the bright-field images and was fully opened for acquiring birefringence images. A linear polarizer and an analyzer were aligned parallel for the bright-field and crossed at 90° for the birefringence images. Typically 5–6 slices, depending on the zoom level, were taken across the depth of the drops and combined into one extended focus image. The UV-fluorescence images were acquired through a 20x UV-objective (NA 0.4). The fluorescence images of Cy3 labeled protein were acquired by a Zeiss AxioImager A1 microscope using an HBO 100 epi-illumination with 543/22 nm excitation and 575–640 nm emission filters, EC-plan 10x objective (NA 0.3) and a Photometrics CoolSNAP HQ2 cooled (−30°C) CCD monochrome camera (1392×1040 pixels). SONICC images were acquired using a custom system performing beam scanning with a resonant vibrating mirror (~8 kHz) to direct the beam along the fast scan axis, and a computerized stage (Prior Optiscan II) for sample-scanning along the slow axis. The incident beam was generated from a Spectra-Physics Mai Tai, 100 fs, 80 Mhz, 100 to 250 mW average power at 800 nm during imaging, focused onto the sample using a 10x objective (0.3 N.A.). The depth of field and beam waist were measured to be ~60 μm and ~2 μm, respectively, which were both greater than theoretically achievable given the N.A. of the objective due to under filling the back of the objective. This depth of field was smaller than the thickness of the LCP samples (~140 μm). The plane through the middle of the droplet was used in all cases. The SHG was collected in epi and transmission (Burle, XP2920PC), with dichroic mirrors used to reject the incident beam and narrow band-pass filters (Chroma) centered around 400 nm. SONICC images included in this work were generated from the sum of the epi and transmitted beams, although qualitatively comparable images were obtained for each independent measurement. The final image sizes and acquisition times varied, but were approximately 140 pixels on the fast scan axis and 280 pixels on the slow axis, corresponding to a field of view of 560 μm × 560 μm. SONICC images were acquired at rates of ~1 min per frame and were resized to facilitate overlay. The relatively long acquisition times were limited primarily by the software used for instrument automation. These exposures are well-below those expected to induce detectable changes in the crystal quality as assessed by X-ray diffraction (manuscript in preparation). The minimum crystal size detectable by SONICC is expected to vary considerably depending on the symmetry of the crystal, the nature of the protein, and the arrangement of the protein within the unit cell. However, based on previous studies of soluble proteins, it is reasonable to expect routine detection limits corresponding to protein crystals equivalent in volume to approximately 100 nm × 100 nm × 100 nm under low (10x) magnification23.
Preparation of crystallization trials
LCP crystallization trials were performed with two integral membrane proteins: a human β2-adrenergic receptor fused to lysozyme from T4 phage (β2AR-T4L); and a human A2A adenosine receptor fused to lysozyme from T4 phage (A2AR-T4L) following published protocols and conditions15,12,13. These trials were a screen of condition around the published, optimized condition. The A2AR-T4L was trace-labeled (<0.1% labeling efficiency) with Cy3 succinimidyl ester (GE Healthcare) as described in Cherezov, et al., 2008. All trials were setup in 96-well glass sandwich plates24 by an in meso crystallization robot25 using 50 nL of protein laden LCP and 1 μL of precipitant solution per well. The glass sandwich plates were assembled from a 1 mm-thick glass base slide (Paul Marienfeld GmbH), a 140 μm-thick double sticky tape (9500PC, 3M) spacer with 96 holes (cut and punched by Saunders). The trials were covered and sealed with a 0.2 mm-thick glass coverslip (Paul Marienfeld GmbH) after setting up crystallization trials. Since the coverslip had significantly lower transmission to UV light (<0.1% at 280 nm) than the base glass slide (~10% at 280 nm), the UV imaging was carried out through the base glass slide. All other types of imaging were performed with plates oriented in such way that the coverslip was facing the objective.
Manual scoring of trials based on image inspection
Protocols for manually scoring crystallization trials in lipidic cubic phase have been described in detail previously15, and were used without modification. In brief, each trial was assigned a numerical score from 0 to 9 by a human expert based on several visual identifiers. Score 0 was given to drops in which LCP transformed into a birefringent phase, either lamellar or hexagonal, as a result of effects of precipitant solutions16 or of slow lipid transesterification and hydrolysis, frequently occurring at high pH26. Both the lamellar and inverted hexagonal phases do not have 3D continuity, which is required for protein diffusion, and thus do not support growth of 3D crystals. Protein crystals may still form before the drop completely transforms into one of these phases. However, high turbidity and birefringence, characteristic to both of them, preclude crystal detection by conventional optical imaging techniques. Score 1 was assigned to drops in which lipidic mesophase was completely dissolved by the precipitant. Protein typically precipitates in such conditions. Scores from 2 (clear LCP drop) to 9 (3D crystal) were arranged in the order of increasing desired outcome of the trials. By this convention, drops with scores of 5 (crystallites or spherulites), 6 (microcrystals) and above were considered as a positive results amenable to further optimization.
Automated scoring based on particle counting in SONICC images
The particle counting analysis of SONICC images was performed in ImageJ27. Specifically, a threshold of 100,000 counts per second was set and the number of pixels and integrated counts were calculated for each contiguous area of pixels above this threshold for each SONICC image. The average crystal size was calculated by taking the total number of contiguous pixels above the threshold and dividing by the number of regions. To generate a score for each image, the average crystal size was multiplied by the integrated intensity. The log of that result was then rescaled to fall within a 0–9 scoring range.
Results and Discussion
In meso crystallization trials were prepared with 3 different integral membrane proteins using grid screens around published successful conditions. 41 representative drops were selected for comparison of imaging techniques using bright-field, birefringence, UV fluorescence, labeled protein fluorescence and SONICC. Representative images summarizing the commonly observed outcomes are shown in Figure 1, including well-formed crystals easily detectable by all methods and several challenging cases. Line traces are shown in Figure 2 quantitatively comparing intrinsic UV fluorescence and SONICC for two trials that are qualitatively similar by conventional optical methods, but distinctly different by SONICC.
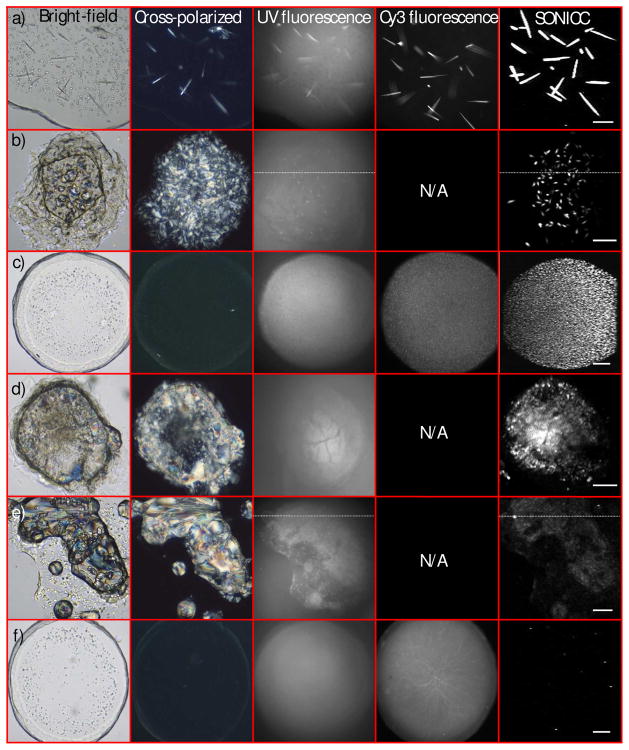
Figure 1.
Comparison of SONICC and conventional optical methods for protein crystal detection, from six representative outcomes of crystallization trials (a–f). Bright-field and birefringence (cross-polarized) images were obtained using white-light illumination, with UV and Cy3 fluorescence excited with ~280 nm and ~543 nm light, respectively. All SONICC images were acquired with 800 nm incident light with detection at 400 nm. Corresponding scores for these trials are summarized in Table I. The following proteins were used in these images: cy3-labeled A2AR-T4L bound to ZM241385 (a,c,f), and β2AR-T4L bound to timolol (b), clenbuterol (d) and alprenolol (e). 100 micron scale bar.
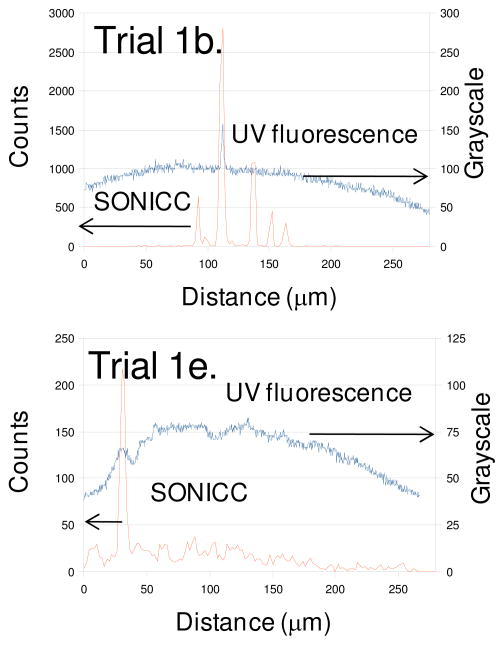
Figure 2.
Line traces corresponding to the dashed lines in Figure 1b and 1e, comparing UV fluorescence and SONICC for detection of unlabelled protein crystals.
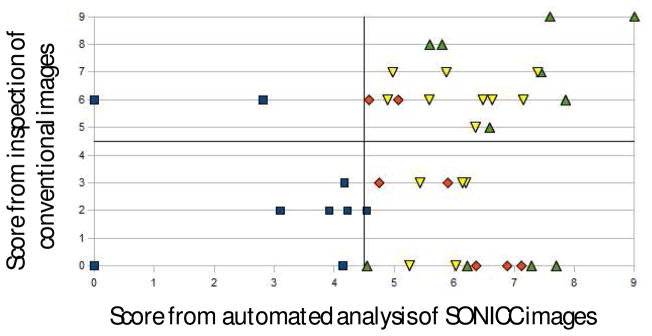
Two sets of scores were assigned to the 41 different crystallization trials based on manual inspection of bright-field and birefringence images and automated particle counting analysis of SONICC images. The particle counting analysis and visual inspection scores for the images in Figure 1 are summarized in Table I. These trials illustrate the breadth of SONICC image types and scores encountered in this study. The SONICC images were classed into 4 categories based in visual inspection of the SONICC images, large (larger than ~15 μm) crystals (1a), small (between 2 μm and 15 μm) crystals (1b and 1c), crystal showers (1d) (less than 2 μm crystal space less than 2 μm apart) and negligible crystallization (1e and 1f) (minimal, if any crystalline material). Both scoring methods and SONICC image classification are summarized in Figure 3.
Table I.
Comparison of scores for the crystallization trials shown in Figure 1.
| a | b | c | d | e | f | |
|---|---|---|---|---|---|---|
| Visual inspection | 9 | 0 | 6 | 0 | 0 | 2 |
| Average area (μm2) | 975 | 216 | 18 | 215 | 44 | 42 |
| Total SHG (counts) | 4.96×106 | 1.20×106 | 1.21×105 | 3.28×105 | 2020 | 3270 |
| Automated Score | 9.00 | 7.70 | 5.58 | 7.12 | 4.15 | 4.22 |
| SONICC image classification | Large crystals | Large crystals | Small crystals | Crystal shower | Negligible crystals | Negligible crystals |
Figure 3.
Comparison of manual and automated scoring from different sets of images. The graph is divided into four regions based on the minimum score that was considered a positive in each case. Color coding and shapes indicate manual categorization based on inspection of the SONICC images: large crystals (green up-triangles), small crystals (yellow down-triangles), crystal showers (red diamonds), and minimal or no crystallization (blue squares).
Discussion
A clear positive by all imaging techniques used in this study is shown in Figure 1a. In this trial, there was little interference from the LCP in the bright-field image and the crystals were large enough to allow easy identification in the birefringence image. Since birefringence imaging depends sensitively on crystal orientation, crystals with the principal polarizability axes oriented coparallel with one of the two polarizers will not be detected, consistent with images in Figure 1a. Crystals were also clearly evident in both the UV and Cy3 fluorescence images, although with a relatively high background in the UV fluorescence image. The signal to background ratios for the Cy3 fluorescence and UV fluorescence were approximately 5/1 and 1.5/1, respectively. By comparison, SONICC images of the same crystallization trial produced a signal to background ratio exceeding 12,000/1. The substantial improvement in contrast by SONICC is attributed primarily to the absence of a significant background from - proteins randomly oriented in LCP, which collectively interfere destructively to produce no coherent background SHG. In contrast, disordered proteins in LCP still contributed to the background in absorption and fluorescence measurements.
Although such relatively large, well-formed crystals can be easily detected using a number of optical methods, LCP droplets are routinely more complicated to score (e.g., Figure 1b-1f). For example, some cocktails of precipitants induce phase changes in the lipid framework from LCP into birefringent mesophases, typically lamellar or hexagonal. Definitive identification of crystals grown under these conditions (Figures 1b, 1d and 1e) is particularly challenging by conventional imaging approaches. For the sets of images in 1b and 1e, protein crystals were not apparent from inspection of the bright-field and birefringence images due to the high inhomogeneity within the lipid matrix. Intrinsic UV fluorescence improved the contrast for areas of high protein concentration, which appeared in both trials. However, contrast for protein crystals still suffered from the high background, presumably arising from a combination of diffuse out-of-plane fluorescence and scatter of the short-wavelength emission by the turbid lipid matrix. Furthermore, the UV fluorescence only indicates regions of high local protein concentration, but cannot easily distinguish between small crystals and amorphous aggregates.
The line traces included in Figure 2 allow quantitative comparison of the SONICC and UV fluorescence images acquired within birefringent mesophases, one of which contained numerous well-formed protein crystals as determined by SONICC (Figure 1b) and one of which did not (Figure 1e). The conventional images (bright-field, birefringence, and UV fluorescence) for the two trials were qualitatively similar, and both received similar scores in the manual analysis based on image inspection. However, SONICC clearly indicates the presence of six well-formed crystals bisecting the line trace, while the intrinsic fluorescence misses all but one crystal. The contrast in the UV fluorescence image in Figure 1e can potentially be attributed to amorphous protein aggregation rather than ordered crystal formation based on comparison with the SONICC images. With the exception of one localized region producing relatively weak SHG (presumably from an isolated poorly-ordered microcrystal), a low baseline SHG was observed throughout the droplet. The line trace was chosen to include this spurious region to emphasize the notable difference in SHG intensity when compared with that from the well-formed crystals in Figure 1b.
The scores from the automated analysis of the SONICC images (abscissa in Figure 3) were in general agreement with the outcomes assessed by visual inspection of the corresponding SONICC images (indicated by the color/shape coding in Figure 3). These results for the trials selected for Figure 1 are compiled in Table I. The manual and automated scores for well-formed crystals such as shown in Figure 1a and 1b were in good agreement, both suggesting relatively large crystal sizes (>20 μm) and high brightness. Representative images containing numerous well-ordered small crystals as determined by SONICC are shown in Figure 1c and 1d. The microcrystals had greater separation in Figure 1c, producing a small size score by automated analysis. In contrast, the low separation between microcrystals in Figure 1d resulted in an overestimation of the average crystal size in the automated approach. Finally, the low automated score and the absence of significant crystal formation in Figures 1e and 1f are mutually consistent.
Crystallization conditions were selected such that the majority of the 41 trials included in the comparison were expected to produce crystals. Consistent with this expectation, 80% of the trials were observed to contain optimized or optimizable protein crystals as determined by SONICC and 49% were observed by visual inspection. The higher number of trials containing crystals by SONICC is due to a combination of two effects. First, SONICC readily identifies promising crystallization conditions in highly turbid, birefringent mesophases as in Figures 1b and 1c providing positive hits for trials that appear as false-negatives by manual inspection of conventional optical images (those receiving a visual inspection score of zero in the lower right of Figure 3). Second, showers of numerous small microcrystals produce comparatively large scores in the particle counting analysis of the SONICC images, but are often rated low by visual inspection. For example, Figure 3 shows a case in which a trial scored 3 by visual inspection and almost 6 by particle counting analysis. A few large crystals were observed amidst a shower of smaller microcrystals in the SONICC image, but were obscured in the conventional images. Similarly, six of the ten trials that received moderate scores by analysis of the SONICC images but zero-scores by visual inspection corresponded to crystal showers. Although crystal showers are themselves poorly suited for diffraction analysis, discrimination between microcrystals and amorphous aggregates can potentially provide a point of reference for subsequent optimization. The two imaging and scoring approaches also differ significantly in the two trials receiving moderate scores of 6 by visual inspection, but low scores by SONICC (upper-left region). These two trials generated no significant SHG. It is suggested that these trials represent false-positives as determined by visual inspection.
Overall, automated scoring of SONICC images and manual scoring of traditional techniques agree qualitatively on 24 of the trials (corresponding to the top right and bottom left quadrants in Figure 3) and disagree on 2 in the top left quadrant, and 15 in the bottom right quadrant. In the case of the top left quadrant, the disagreement is attributed to false positives from visual inspection, arising from the intrinsic difficulties in reliably scoring trials in turbid mesophases. The large numbers of apparent false-negatives arising from visual inspection in the lower right quadrant are particularly noteworthy. These false-negatives are attributed in part to the inability of conventional imaging to easily discriminate between showers of well-ordered microcrsytals (<2–3 μm) and amorphous aggregates of comparable size. However, 9 of the 15 of the apparent false-negatives by visual inspection contained small or large crystals based on inspection of the corresponding SONICC images. Of these same trials, 6 received scores of 0 by visual inspection.
Summary and Conclusion
SONICC imaging was found to compare favorably with conventional optical imaging approaches for protein crystal detection, particularly in scattering and inhomogeneous environment that generally frustrate reliable crystal detection by conventional means. A general comparison of SONICC and other common optical methods for protein crystal detection are summarized in Table II. In these crystallization trials in LCP, SONICC resulted in images of well-formed crystals with signal to background ratios >12,000/1, compared to 1.5/1 for UV fluorescence and 5/1 for fluorescence from protein crystals incorporating Cy3 labels. Manual scores of crystallization trials based on established protocols for image interpretation were compared with manual and automated analysis of SONICC images. SONICC agreed well with inspection of conventional images in most instances, but also indicated apparent identification by SONICC of both false positives and numerous false negatives generated by conventional imaging methods. Approximately 1/3 of the small or large crystals as determined by SONICC were considered negatives based on inspection of conventional images. SONICC imaging is directly compatible with a broad range of protein crystallization methods, suggesting promise as a general technique for aiding in automated crystallization screenings.
Table II.
Summary of the strengths and limitations of several different optical methods for assessment of protein crystal trials.
| Pros | Cons | |
|---|---|---|
| Bright-field | Provides good edge contrast for relatively large crystals in transparent media. | False positives and negatives are common, particularly in inhomogeneous media. Difficult to interpret for automated crystal scoring. Cannot discriminate between protein crystals and salt crystals. |
| Birefringence | Provides contrast for crystals. | Detection depends on crystal orientation and space group. Inapplicable in birefringent mesophases. Cannot discriminate between protein crystals and salt crystals. Inapplicable in highly scattering media. |
| UV fluorescence | Can discriminate between protein and salt crystals. | Exposes crystals to UV light. Requires tryptophan. Contrast can be reduced by a high background and/or autoquenching within the crystal. Challenging in highly scattering media. |
| Labeled fluorescence | Provides excellent contrast. Can discriminate between protein and salt crystals. | Requires labeling of the protein, which introduces complexity in sample preparation and can potentially negatively impact crystal quality. Challenging in highly scattering media. |
| SONICC | Substantially improved contrast relative to fluorescence-based methods through the suppression of contributions from disordered proteins. Ability to detect <2μm crystals and discriminate between crystal showers and amorphous aggregation. Can discriminate between protein and salt crystals. Low susceptibility to complications from optical scatter. | Requires an ultrafast laser source and beam-scanning for illumination. |
Acknowledgments
The authors acknowledge Ellen Chien, Kirk Allin and Tam Trinh for their help with protein expression, Mark Griffith and Michael Hanson for providing protein samples. This work was supported in part by the NIH Roadmap Initiative grant P50 GM073197, Protein Structure Initiative grant U54 GM074961, NSF-MRI 0722558, and NSF-CHE 0640549.
Contributor Information
David J. Kissick, Department of Chemistry, Purdue University, West Lafayette IN, 47907
Ellen J. Gualtieri, Department of Chemistry, Purdue University, West Lafayette IN, 47907
Garth J. Simpson, Email: gsimpson@purdue.edu, Department of Chemistry, Purdue University, West Lafayette IN, 47907
Vadim Cherezov, Email: vcherezo@scripps.edu, Department of Molecular Biology, The Scripps Research Institute, La Jolla CA 92037.
References
- 1.Cumbaa CA, Lauricella A, Fehrman N, Veatch C, Collins R, Luft J, DeTitta G, Jurisica I. Automatic classification of sub-microlitre protein-crystallization trials in 1536-well plates. Acta Crystallographica Section D-Biological Crystallography. 2003;59:1619–1627. doi: 10.1107/s0907444903015130. [DOI] [PubMed] [Google Scholar]
- 2.Spraggon G, Lesley SA, Kreusch A, Priestle JP. Computational analysis of crystallization trials. Acta Crystallographica Section D-Biological Crystallography. 2002;58:1915–1923. doi: 10.1107/s0907444902016840. [DOI] [PubMed] [Google Scholar]
- 3.Wilson J. Towards the automated evaluation of crystallization trials. Acta Crystallographica Section D-Biological Crystallography. 2002;58:1907–1914. doi: 10.1107/s0907444902016633. [DOI] [PubMed] [Google Scholar]
- 4.Bern M, Goldberg D, Stevens RC, Kuhn P. Automatic classification of protein crystallization images using a curve-tracking algorithm. Journal of Applied Crystallography. 2004;37:279–287. doi: 10.1107/s0021889804001761. [DOI] [Google Scholar]; Zhu XQ, Sun SH, Bern M. 26th Annual International Conference of the IEEE-Engineering-in-Medicine-and-Biology-Society; San Francisco, CA. 2004. pp. 1628–1631. ieee. [DOI] [PubMed] [Google Scholar]
- 5.Walker CG, Foadi J, Wilson J. Classification of protein crystallization images using Fourier descriptors. Journal of Applied Crystallography. 2007;40:418–426. doi: 10.1107/s0021889807011156. [DOI] [Google Scholar]
- 6.Liu R, Freund Y, Spraggon G. Image-based crystal detection: a machine-learning approach. Acta Crystallographica Section D-Biological Crystallography. 2008;64:1187–1195. doi: 10.1107/s090744490802982x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cumbaa C, Jurisica I. Journal of Strucural and Functional Genomics. 2005;6:195–202. doi: 10.1007/s10969-005-5243-9. [DOI] [PubMed] [Google Scholar]
- 8.Cherezov V, Clogston J, Papiz MZ, Caffrey M. Room to move: Crystallizing membrane proteins in swollen lipidic mesophases. Journal of Molecular Biology. 2006;357:1605–1618. doi: 10.1016/j.jmb.2006.01.049. [DOI] [PubMed] [Google Scholar]
- 9.Wadsten P, Wohri AB, Snijder A, Katona G, Gardiner AT, Cogdell RJ, Neutze R, Engstrom S. Lipidic sponge phase crystallization of membrane proteins. Journal of Molecular Biology. 2006;364:44–53. doi: 10.1016/j.jmb.2006.06.043. [DOI] [PubMed] [Google Scholar]
- 10.Faham S, Bowie JU. Bicelle crystallization A new method for crystallizing membrane proteins yields a monomeric bacteriorhodopsin structure. Journal of Molecular Biology. 2002;316:1–6. doi: 10.1006/jmbi.2001.5295. [DOI] [PubMed] [Google Scholar]
- 11.Landau EM, Pebay-Peyroula E, Neutze R. 2nd Nobel Symposium on Membrane Proteins: Structure Function and Assembly (126) Vol. 555. Friiberghs Herrgard; Sweden: 2003. pp. 51–56. [Google Scholar]
- 12.Cherezov V, Rosenbaum DM, Hanson MA, Rasmussen SGF, Thian FS, Kobilka TS, Choi HJ, Kuhn P, Weis WI, Kobilka BK, Stevens RC. High-resolution crystal structure of an engineered human beta(2)-adrenergic G protein-coupled receptor. Science. 2007;318:1258–1265. doi: 10.1126/science.1150577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jaakola VP, Griffith MT, Hanson MA, Cherezov V, Chien EYT, Lane JR, Ijzerman AP, Stevens RC. The 2.6 Angstrom Crystal Structure of a Human A(2A) Adenosine Receptor Bound to an Antagonist. Science. 2008;322:1211–1217. doi: 10.1126/science.1164772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Caffrey M. A lipid’s eye view of membrane protein crystallization in mesophases. Current Opinion in Structural Biology. 2000;10:486–497. doi: 10.1016/s0959-440x(00)00119-6. [DOI] [PubMed] [Google Scholar]
- 15.Caffrey M, Cherezov V. Crystallizing membrane proteins using lipidic mesophases. Nature Protocols. 2009;4:706–731. doi: 10.1038/nprot.2009.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cherezov V, Fersi H, Caffrey M. Crystallization screens: Compatibility with the lipidic cubic phase for in meso crystallization of membrane proteins. Biophysical Journal. 2001;81:225–242. doi: 10.1016/S0006-3495(01)75694-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Echalier A, Glazer RL, Fulop V, Geday MA. Assessing crystallization droplets using birefringence. Acta Crystallographica Section D-Biological Crystallography. 2004;60:696–702. doi: 10.1107/s0907444904003154. [DOI] [PubMed] [Google Scholar]
- 18.Nollert P. Microscope detection options for colorless protein crystals grown in lipidic cubic phases. Journal of Applied Crystallography. 2003;36:1295–1296. doi: 10.1107/s0021889803013724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Judge RA, Swift K, Gonzalez C. An ultraviolet fluorescence-based method for identifying and distinguishing protein crystals. Acta Crystallographica Section D-Biological Crystallography. 2005;61:60–66. doi: 10.1107/s0907444904026538. [DOI] [PubMed] [Google Scholar]
- 20.Groves MR, Muller IB, Kreplin X, Muller-Dieckmann J. A method for the general identification of protein crystals in crystallization experiments using a noncovalent fluorescent dye. Acta Crystallographica Section D-Biological Crystallography. 2007;63:526–535. doi: 10.1107/s0907444906056137. [DOI] [PubMed] [Google Scholar]
- 21.Forsythe E, Achari A, Pusey ML. Trace fluorescent labeling for high-throughput crystallography. Acta Crystallographica Section D-Biological Crystallography. 2006;62:339–346. doi: 10.1107/s0907444906000813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fry EH, Qin W, Fleck EN, JRA, Chiu ML. The Open Strucutal Biology Journal. 2009;3:11–15. [Google Scholar]
- 23.Wampler RD, Kissick DJ, Dehen CJ, Gualtieri EJ, Grey JL, Wang HF, Thompson DH, Cheng JX, Simpson GJ. Selective Detection of Protein Crystals by Second Harmonic Microscopy. Journal of the American Chemical Society. 2008;130:14076-+. doi: 10.1021/ja805983b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cherezov V, Caffrey M. Nano-volume plates with excellent optical properties for fast, inexpensive crystallization screening of membrane proteins. Journal of Applied Crystallography. 2003;36:1372–1377. doi: 10.1107/s002188980301906x. [DOI] [Google Scholar]
- 25.Cherezov V, Peddi A, Muthusubramaniam L, Zheng YF, Caffrey M. A robotic system for crystallizing membrane and soluble proteins in lipidic mesophases. Acta Crystallographica Section D-Biological Crystallography. 2004;60:1795–1807. doi: 10.1107/s0907444904019109. [DOI] [PubMed] [Google Scholar]
- 26.Murgia S, Caboi F, Monduzzi M, Ljusberg-Wahren H, Nylander T. In: Conference on Lipid and Polymer-Lipid Systems. Nylander T, Lindman B, editors. Vol. 120. Chia Laguna; Italy: 2000. pp. 41–46. [Google Scholar]
- 27.Abramoff MD, Magelhaes PJ, Ram SJ. Biophotonics International. 2004;11:36–42. [Google Scholar]