Abstract
Mammalian sperm require ATP for motility, and most of it is generated by glycolysis. The glycolytic enzymes segregate to the principal piece region of the flagellum, where some are bound tightly to a novel cytoskeletal structure defining this region, the fibrous sheath (FS), and others are easily extracted with detergents. One of the latter is the spermatogenic cell-specific variant isozyme of hexokinase type 1 (HK1S), characterized by an N-terminal 24-amino acid spermatogenic cell-specific region (SSR). Yeast two-hybrid screens carried out using the SSR as bait determined that HK1S is tethered to muscle-type phosphofructokinase (PFKM) in the principal piece region. This led to the identification of four testis-specific Pfkm splice variants, one that overlapped a variant reported previously (Pfkm_v1) and three that were novel (Pfkm_v2, Pfkm_v3, and Pfkm_v4). They differ from Pfkm transcripts found in somatic cells by encoding a novel 67-amino acid N-terminal extension, the testis-specific region (TSR), producing a spermatogenic cell-specific PFKM variant isozyme (PFKMS). An antiserum generated to the TSR demonstrated that PFKMS is present in the principal piece and is insoluble in 1% Triton X-100 detergent. In subsequent yeast two-hybrid screens, the TSR was found to interact with glutathione S-transferase mu class 5 (GSTM5), identified previously as a spermatogenic cell-specific component of the FS. These results demonstrated that HK1S is tethered in the principal piece region by PFKMS, which in turn is bound tightly to GSTM5 in the FS.
Keywords: flagellum, gamete biology, gametogenesis, glycolysis, multienzyme complex, sperm, spermatogenesis, testis
Spermatogenic cell-specific type 1 hexokinase is tethered to the fibrous sheath of the developing sperm by a testis-specific phosphofructokinase M isozyme that binds to glutathione S-transferase, mu class, 5.
INTRODUCTION
Glycolysis is the major source of ATP required for sperm flagellar activity, capacitation, and fertilization [1–3], and the enzymes for this process are localized in the principal piece region of the flagellum. This places the glycolytic enzymes and ATP production along most of the length of the flagellum in proximity to the dynein ATPase activity generating the flagellar beat. Some of these enzymes are known to be products of genes expressed specifically in spermatogenic cells, including lactate dehydrogenase C (LDHC) [4], phosphoglycerate kinase 2 (PGK2) [5, 6], glyceraldehyde 3-phosphate dehydrogenase S (GAPDHS) [7], and two aldolase A isozymes (ALDOART1 and ALDOART2) [8]. Others are products of variant transcripts, including spermatogenic cell-specific hexokinase type 1 (HK1S) [9, 10] and another aldolase A isozyme (ALDOA_V2) [8]. Furthermore, mammalian sperm contain atypical isozymes of phosphoglucose isomerase [11], triosephosphate isomerase [12], phosphoglycerate mutase [13], and enolase [14, 15], but it remains to be determined if they are products of genes or variant transcripts expressed specifically in spermatogenic cells.
The principal piece region accounts for about 70% of the length of the mouse sperm flagellum and is characterized by the presence of a fibrous sheath (FS). This novel cytoskeletal structure serves as a mechanoelastic structure involved in shaping the flagellar waveform [16], is composed of more than 20 proteins, and is a scaffold for glycolytic enzymes, signaling pathway components, and chemoprotective proteins [17]. Earlier studies indicated that some sperm glycolytic enzymes cofractionate with flagellar components [18], are difficult to extract [19], are associated with insoluble components [20], and are present in multienzyme complexes [20, 21]. More recent studies showed that GAPDHS [22, 23], lactate dehydrogenase A (LDHA), aldolase A [24], and pyruvate kinase (PK) [25] are tightly bound to the FS.
It remains to be determined how the glycolytic enzymes segregate to the principal piece region of the flagellum and are anchored in this region. Some of the glycolytic enzymes are highly resistant to extraction, including GAPDHS [23], ALDOART1, ALDOA_V2 [8], LDHA, and PK [24]. An unusual feature of GAPDHS is a novel proline-rich N-terminal 105-amino acid sequence not present in GAPDH [7], while ALDOART1 has a novel 55-amino acid N-terminal region, and ALDOA_V2 has a novel 54-amino acid N-terminal region [8]. It has been suggested that the novel N-terminal sequences are involved in delivering these enzymes to the principal piece and/or attaching them to the FS [8, 23, 24, 26].
While some of the glycolytic enzymes are highly resistant to extraction, others found in the principal piece region are readily solubilized by detergents, including HK1S, LDHC, and PGK2 [24]. While LDHC and PGK2 lack novel N-terminal features, HK1S has a novel N-terminal spermatogenic cell-specific region (SSR). In somatic cells, HK1 is tethered to porin (voltage-dependent anion channel) in the outer mitochondrial membrane by its N-terminal porin-binding domain [27, 28]. Replacement of the porin-binding domain by the SSR produces the spermatogenic cell-specific HK1S isozyme [9]. This led us to hypothesize that HK1S is localized to the principal piece by associating with other sperm components or multienzyme complexes in this region [9, 26]. This hypothesis was tested in the present study by using the SSR as bait in a yeast two-hybrid screen to search for HK1S-binding partners. We found that HK1S is tethered via its SSR to a previously unidentified spermatogenic cell-specific phosphofructokinase variant isozyme (PFKMS) confined to the principal piece. A subsequent yeast two-hybrid screen determined that PFKMS binds via its novel 67-amino acid N-terminal testis-specific region (TSR) to glutathione S-transferase mu class 5 (GSTM5), a highly insoluble spermatogenic cell-specific component of the FS identified previously by peptide sequencing [29]. This demonstrates that direct interactions occurring between two glycolytic enzymes and between a glycolytic enzyme and an FS protein, mediated by spermatogenic cell-specific peptide sequences, are responsible for anchoring HK1S and PKFMS to the FS.
MATERIALS AND METHODS
Materials and Animals
All reagents were purchased from Sigma (St. Louis, MO [http://www.sigma-aldrich.com]) unless indicated otherwise.
CD-1 mice used for protein experiments and immunohistochemistry were obtained from Charles River Laboratories (Raleigh, NC [http://www.criver.com]). All animal studies were approved by the National Institute of Environmental Health Sciences Institutional Animal Care and Use Committee and were performed according to U.S. Public Health Service guidelines.
Yeast Two-Hybrid Screening
Yeast two-hybrid screening was carried out as recommended by the supplier (BD Biosciences Clontech, San Jose, CA [http://www.clontech.com]). Yeast AH109 competent cells were prepared with the Frozen-EZ Yeast Transformation II kit (Zymo Research, Orange, CA [http://www.zymoresearch.com]). The cDNAs encoding the SSR of Hk1s cDNA and the TSR of Pfkm_v2 cDNA were subcloned into the yeast expression vector pKBTG7 (BD Biosciences Clontech) to generate pKBGT7-SSR and pKBGT7-TSR, respectively. Yeast AH109 cells were cotransformed with a mouse testis cDNA library (BD Biosciences Clontech) and pKBGT7-SSR or pKBGT7-TSR using the lithium acetate method [30].
Amplification of 5′ cDNA Ends
Rapid amplification of 5′ cDNA ends (5′ RACE) was performed using a Marathon-ready testis cDNA library and the BD Advantage 2 PCR kit (BD Biosciences Clontech) according to the supplier's recommended procedure. Adaptor primer 1 from the kit and primer (5′-CTG ATG TGC TCG CCA CCG TCC ACC A-3′) were used for PCR amplification. RACE products were ligated into the pGEM-T vector (Promega, Madison, WI [http://www.promega.com]) for sequencing.
Plasmids
The SSR, GSTM5 full length (FL), and GSTM5 mutants (GSTM5-N [amino acids 5–85] and GSTM5-C [amino acids 95–195]) were subcloned into the bacteria expression vector FLAG-CTC (Sigma). The TSR, PFKMS-FL, and PFKMS mutants (PFKMS-N [amino acids 16–383] and PFKMS-C [amino acids 403–685]) were subcloned into the pGEX-4T-1 vector (GE Healthcare Life Sciences; Piscataway, NY [http://www.gehealthcare.com]) for GST pull-down assays. In addition, HK1S, TSR, PFKMS-FL, and GSTM5-FL were subcloned into the pcDNA3.1(+)-His/Myc (Invitrogen, Carlsbad, CA [http://www.invitrogen.com]) vector for in vitro translation.
Preparation of Recombinant TSR
Recombinant protein containing the TSR was produced for use as an immunogen. The TSR-encoding region of the Pfkm_v2 cDNA was subcloned with pET-21b vector (EMD Chemicals, Inc., Gibbstown, NJ [http://www.emdbiosciences.com]) and expressed in Escherichia coli BL21(DE3) (Stratagene, Cedar Creek, TX [http://www.stratagene.com]), and exponential growth was induced with 0.5 mM isopropyl-d-thiogalactoside. The recombinant protein was purified using Ni-NTA beads (EMD Chemicals, Inc.) and the MagneHis Protein purification system (Promega) as recommended by the suppliers.
Purification of Antiserum to TSR
Serum was collected from New Zealand white rabbits following immunization with the recombinant TSR peptide (Covance, Denver, PA [http://www.covance.com]). Proteins were precipitated from the serum by saturating with ammonium sulfate at 4°C for 2 h and were collected by centrifugation at 24 553 × g for 30 min at 4°C. The pellet was resuspended in PBS and dialyzed against 10 mM Tris (pH 7.5) overnight at 4°C. The dialysate was affinity purified using recombinant TSR linked to CNBr-4B beads (GE Healthcare Life Sciences) as recommended by the supplier. This is referred to hereafter as TSR antiserum.
Northern Blot Analysis
Total RNA was extracted from testes of mice aged 10–30 days and from tissues (brain, heart, kidney, liver, spleen, lung, skeletal muscle, thymus, ovary, and epididymis) of adult mice using Trizol reagent (Invitrogen), separated by electrophoresis on 1.0% agarose gels containing 0.66 M formaldehyde, and transferred to Hybond N nylon membranes (GE Healthcare Life Sciences) in 10× saline-sodium citrate (SSC). The probes were labeled with [32P] using a random prime labeling kit (Roche Applied Sciences, Indianapolis, IN [http://roche.com]) with the 320-base pair (bp) cDNA encoding the TSR from Pfkms_v2 as template. Hybridization was performed in Hybrisol I (Millipore Corporation, Billerica, MA [http://www.millipore.com]) at 42°C overnight. The membranes were washed with 2× SSC containing 0.1% SDS at room temperature for 15 min and then in 0.1× SSC containing 0.1% SDS at 50°C for 15 min, and they were subjected to autoradiography.
Pull-Down Assays
Recombinant GST-PFKMS-FL, GST-PFKMS mutants (GSTM5-N and GSTM5-C), GST-phosphofructokinase (PFK), and GST proteins were incubated with recombinant FLAG-tagged SSR protein, and GST pull-down assays were carried out using the μMACS GST-tag isolation kit (Miltenyi Biotech, Auburn, CA [http://www.miltenyibiotec.com]). Elutes were analyzed by immunoblot analysis with antibodies to GST (RGST-45A-Z; Immunology Consultants Laboratory, Inc., Newberg, OR [http://www.icllab.com]) and to FLAG (F7425; Sigma). FLAG pull-down assays were performed as recommended by the supplier (Sigma). Recombinant GST-TSR protein was incubated with FLAG vector only protein or with recombinant FLAG-tagged GSTM5-FL (FLAG-GSTM5-FL), the N-terminal portion of GSTM5 (FLAG-GSTM5-N), or the C-terminal portion of GSTM5 (FLAG-GSTM5-C) protein for 2 h at 4°C. FLAG-M2-agarose (A2220; Sigma) was added to the protein mixtures, and after incubation overnight at 4°C, the agarose beads were washed with Tris-buffered saline (TBS), mixed with 1× sample buffer, and heated at 95°C for 5 min. The materials released were separated by SDS-PAGE and immunoblotted with antibody to GST.
In Vitro Translation
Using plasmids already described, in vitro translation of the SSR of HK1S and the TSR of PFKMS and GSTM5 was carried out in the TNT coupled reticulocyte lysate system (Promega) according to the supplier's instructions. Translation was performed for 2 h at 30°C in a volume of 25 μl containing 20 μCi of [35S] methionine (GE Healthcare Life Sciences). After incubation, the labeled proteins were separated from unincorporated [35S] methionine using MicroSpin G-25 columns (GE Healthcare Life Sciences).
Immunoprecipitation
Testes were homogenized in a lysis buffer containing 20 mM Tris-HCl (pH 8.0), 150 mM NaCl, 1 mM ethylenediaminetetraacetic acid (EDTA), 0.1% SDS, 0.5% NP-40, 1% Triton X-100, and protease inhibitor cocktail (Roche Applied Sciences). Homogenates were incubated for 30 min on ice and then centrifuged at 15 800 × g for 30 min, and the supernatant was used for immunoprecipitation studies. The in vitro-translated [35S]-labeled proteins were incubated with testis lysate for 2 h at 4°C. Ten micrograms of antibody was added and incubated for 2 h at 4°C, and then Immunopure immobilized protein G beads (Pierce Biotechnology, Rockford, IL [http://piercenet.com]) were added and incubated for 2 h at 4°C on a rotating platform. The beads were washed with TBS, mixed with 1× sample buffer, and heated for 5 min at 86°C. After centrifugation at 5000 × g for 3 min at room temperature, the supernatants were separated by SDS-PAGE, and the gels were immersed in Amplify (GE Healthcare Life Sciences) for 30 min, dried, and exposed to Hyperfilm MP (GE Healthcare Life Sciences) at −70°C.
Sample Preparation Procedures
Skeletal muscle and testes were homogenized in lysis buffer (20 mM PBS, 1% Triton X-100, 0.2 mM sodium orthovanadate, and 0.1 mM PMSF), incubated for 30 min on ice, and centrifuged at 15 800 × g for 30 min, and the soluble fraction (supernatant) was collected. Sperm used in most studies were isolated from the cauda epididymis, incubated in M2 medium (Sigma) for 5 min at room temperature, pelleted in Eppendorf tubes by centrifugation at 7500 × g for 3 min, and washed in PBS. To prepare sperm samples for SDS-PAGE, the pellet was suspended in PBS containing 0.1% Triton X-100 (PBSTX) and an equal volume of 2× sample buffer (4% SDS, 100 mM Tris-HCl [pH 6.8], 20% glycerol, 2% 2-mercaptoethanol, and 0.001% bromophenol blue).
For comparative solubility studies, the sperm pellet was suspended in PBS containing 1% Triton X-100 or urea (1 M, 2 M, 3 M, or 4 M) for 2 h on ice or in PBS containing 6 M urea for 2 h at room temperature. After centrifugation at 12 334 × g for 15 min at 4°C, the supernatants were suspended in 2× sample buffer. For sequential solubility studies, sperm were treated using a procedure modified from one reported previously [31]. Sperm were suspended in NP-40 lysis buffer (1% NP-40,10 mM Hepes [pH 7.5], 50 mM NaCl, 10 mM EDTA, 1% NP-40, 0.2 mM PMFS, and 10 μg/ml of aprotinin) for 1 h on ice, centrifuged at 500 × g for 10 min at 4°C, and the supernatant was collected. The pellet was washed with 50 mM Tris-HCl (pH 8.2), resuspended in urea lysis buffer (6 M urea, 50 mM Tris-HCl [pH 8.0], 10 mM EDTA, 6 M urea, 0.2 mM dithiothreitol [DTT], and 10 μg/ml of aprotinin) for 2 h on ice, and centrifuged at 12 000 × g for 20 min at 4°C, and the supernatant was collected. Pellets (6 M urea-insoluble fraction) were suspended in 1× sample buffer. For studies of quiescent sperm, sperm were collected from the caput and cauda epididymis directly into 1.5-ml tubes without dilution as described previously [32], suspended in PBSTX, and centrifuged at 4528 × g for 5 min 4°C, and the supernatants and pellets were collected. For activated sperm, sperm were collected as already described. Samples of quiescent and activated sperm from caput and cauda epididymis were suspended in 2× sample buffer containing 2% 2-mercaptoethanol (reducing conditions) or without 2-mercaptoethanol (nonreducing conditions).
Immunoblot Analysis
Proteins extracted from sperm and skeletal muscle were separated by SDS-PAGE on 4%–20% gradient ready gels or 10% ready gels (Bio-Rad Laboratories, Hercules, CA [http://www.bio-rad.com]) and were transferred from the gels to Immobilon-P PVDF membranes (Millipore Corporation). The membranes were immunostained with TSR antiserum and antisera to SSR [10], PFKM (100-1156; Rockland Immunochemicals, Inc., Gilbertsville, PA [http://www.rockland-inc.com]), α-tubulin (T5168; Sigma), β-actin (A5441; Sigma), or GSTM5 (gift of Dr. Irving Listowsky, Albert Einstein College of Medicine, Bronx, NY) [33] and detected using ECL reagents (GE Healthcare Life Sciences) as recommended by the supplier.
Immunohistochemistry
Testes were fixed in Bouin solution, dehydrated, and embedded in paraffin, and 6-μm sections were prepared using standard procedures. Slides were deparaffinized and immunostained with TSR antiserum and an Elite ABC kit for rabbit IgG (Vector Laboratories, Burlingame, CA [http://www.vectorlabs.com]), and the peroxidase-labeled secondary antibody was detected using 3,3′-diaminobenzidine tetrahydrocholoride (Sigma) as the chromogen.
Sperm were isolated from the cauda epididymis as already described, fixed in 2% paraformaldehyde in PBS for 15 min at room temperature, suspended in 50 mM glycine in PBS for 30 min to block free aldehyde groups, and applied to slides as described previously [34]. The slides were immersed in ice-cold methanol for 1 min to permeabilize the sperm, immunostained with TSR antiserum and antiserum to rabbit IgG conjugated to fluorescein isothiocyanate (MP Biomedicals, Irvine, CA [http://mpbio.com]), and observed using an Axioplan microscope (Carl Zeiss, Thornewood, NJ [http://www.zeiss.com/micro]), and images were recorded using a QImaging camera and software (QImaging, Tucson, AZ [http://www.qimaging.com]).
PFK Enzymatic Assay
Sperm isolated from the cauda epididymis as already described were permeabilized for 30 min on ice in PBS containing 1% Triton X-100, 0.2 mM sodium orthovanadate, and 0.1 mM PMSF. The sample was centrifuged at 12 000 × g for 5 min at 4°C, the supernatant was collected, and the pellet was resuspended in PBST. Enzymatic activity was measured in the pellet and supernatant of sperm from the cauda epididymis as described previously [35]. In brief, the sample (30 μl) was assayed at 25°C in 1 ml of assay buffer (pH 8.2) containing 75 mM glycelglycine, 75 mM disodium β-glycerophosphate, 3 mM Na2-EDTA, 18 mM MgCl2, 9 mM ammonium sulfate, 10 mg/ml of NADH, 3 mM DTT, 30 mM ATP, 30 mM fructose-6-phosphate, 18 U/ml of triosephosphate isomerase, 18 U/ml of α-glycerophosphate dehydrogenase, and 36 U/ml of aldolase. Enzymatic activity was determined using a UV spectrophotometer (Amersham Pharmacia Biotech, Piscataway, NJ) at 340 nm to measure the decrease in NADH. After 3 min of preincubation, the reaction was initiated by adding 1–3 μg of sample. The negative control included assay buffer without fructose-6-phosphate. The protein concentration of the pellet and supernatant was measured with Bradford reagent (Bio-Rad Laboratories). These assay results are reported as the average of four separate experiments, each performed in duplicate. Data are expressed as the mean ± SD of the four experiments.
Quantitative Real-Time RT-PCR
Total RNA was extracted using Trizol reagent from testes of mice aged 10–30 days old. The cDNA for quantitative real-time RT-PCR (qPCR) was synthesized with reverse transcriptase (RT; Applied Biosystems, Foster City, CA). The qPCR analyses were performed using SYBR Green PCR Master Mix reagents (Applied Biosystems) and the ABI PRISM 7900HT Sequence Detection System (Applied Biosystems) according to the manufacturer's protocols. The cDNA served as a template in a 50-μl reaction mixture and was processed using an initial denaturation step at 95°C for 10 min, followed by 45 amplification cycles (95°C for 10 sec and 60°C for 1 min) and one dissociation stage cycle (95°C for 15 sec, 60°C for 15 sec, and 95°C for 15 sec). The reactions were carried out using primer for upstream exon -s2 (5′-GAAGCTTCAACCTCCAACA-3′ and 5′-AGTGTCAGGAATATACAG-3′) and for Pfkm exon 1 (5′-GAGTGGATCATGACCCAT-3′ and 5′-GCATCTCCACCAGAGGTCA-3′). The relative steady-state transcript levels were calculated using cycle time (Ct) values and the following equation: Relative Quantity = 2−ΔΔCt. The expression levels were normalized using primer for Gapdh (5′-TGCACCACCAACTGCTTAG-3′ and 5′-GATGCAGGGATGATGTTC-3′) as an internal control for each sample. The relative ratios (folds) of transcript levels in each sample were calculated using the Day 10 value as 1. The qPCR reactions were performed in triplicate with each of the samples.
Conventional RT-PCR
To confirm the presence of the Pfkm variants in mouse testis, total RNA was extracted from testis using Trizol reagent according to manufacturer's instructions (Invitrogen). Total RNA (1 μg) was treated with RNase-free DNase I and was reverse transcribed using oligo dT primers. The PCR was carried out using forward primers in the upstream exons -s1 (5′-ACCTGGCTCCTC CCCGTGTT-3′), -s2 (5′-GAAGCTTCAACCTCCAACA-3′), -s3 (5′-GAAGTAGCCATGCGTAGAGA-3′), -s4 (5′-CCATGTTGACCTTTGACACT-3′), -s5 (5′-GACGTGGAGGGAGCGT-3′), and a reverse primer located in exon 1 (5′-GCATCTCCACCAGAGGTCA-3′). The 20-μl reaction mixture was processed using an initial denaturation step at 95°C for 2 min, followed by 50 amplification cycles (95°C for 30 sec, 54°C for 30 sec, and 72°C for 20 sec) and one step at 72°C for 7 min. Products were separated on 2% agarose gels.
RESULTS
HK1S Is Tethered to the FS by PFKM
The localization of HK1S mainly in the principal piece region of the flagellum led to the hypothesis that HK1S is tethered through its SSR to proteins associated with the FS [10, 26]. This was tested in the present study using the SSR as bait in a yeast two-hybrid screen. Of the seven cDNA clones isolated and characterized, one encoded a 246-bp sequence with high identity to amino acids 321–401 in mouse muscle-type PFK (Pfkm [GenBank accession number BC005526.1]). The identification of PFKM as a binding partner of HK1S was confirmed using pull-down assays (described herein).
Identification of Testis-Specific Splice Variants of Pfkm
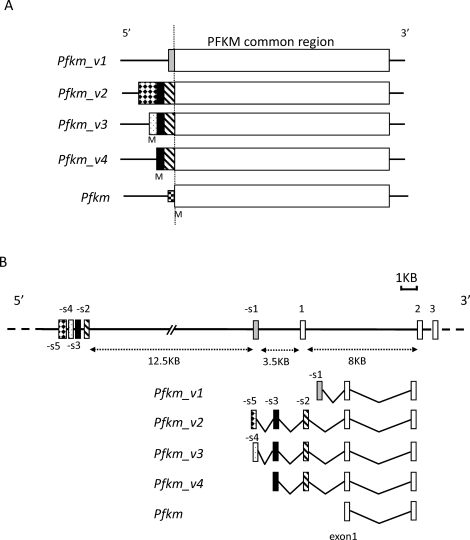
The use of 5′ RACE and a mouse testis cDNA library to further characterize the Pfkm transcript giving rise to the yeast prey clone led to the unexpected identification of four Pfkm variant transcripts (Pfkm_v1, Pfkm_v2, Pfkm_v3, and Pfkm_v4). They are distinguished from Pfkm transcripts in somatic cells by the presence of additional sequences on their 5′ ends that are identical to regions approximately 3.5–16 kilobase (kb) upstream of the Pfkm translation start site on chromosome 15. These sequences are encoded by five exons and are referred to provisionally as spermatogenic cell exons -s1 to -s5 (Fig. 1, A and B).
FIG. 1.
A) The Pfkm_v1, Pfkm_v2, Pfkm_v3, and Pfkm_v4 transcripts are derived from different combinations of exons, and their 5′ ends (upstream of Pfkm exon 1) differ in length. M indicates the putative initiation methionine. B) The genomic structure of Pfkm, indicating the location of the exons giving rise to the variant transcripts. The rectangles show the location of the spermatogenic cell expressed exons (-s1 to -s5) and exons (1–3) transcribed in somatic and germ cells. The Pfkm_v1 transcript uses exon -s1, and Pfkm_v2, Pfkm_v3, and Pfkm_v4 use exons -s2 and -s3. In addition, Pfkm_v2 uses exon -s5, and Pfkm_v3 uses exon -s4.
The Pfkm_v1 transcript incorporates exon -s1, which contains a Pfkm variant sequence reported previously [36] (TE-PFK-M [GenBank accession number D21867]) plus an additional 60 nucleotides on the 5′ end (GenBank accession number GQ428207). All or part of the exon -s1 sequence is present in more than 60 expressed sequence tags (ESTs), many of which map to UniGene Mm.272582 containing sequences related to Pfkm. However, we were unable to identify a likely ATG start codon upstream of exon -s1, and a BLASTP search failed to identify significant similarities between the hypothetical 39- residue peptide sequence encoded by the exon and any mouse proteins, suggesting that exon -s1 and thus Pfkm_v1 are not translated.
The 5′ ends of Pfkm_v2, Pfkm_v3, and Pfkm_v4 differed (Fig. 1A), but all contained an identical sequence originating from exon -s2 and exon -s3 (GenBank accession numbers GQ428206 and GQ428205) that encodes a 67-amino acid sequence not present in the typical PFKM isozyme. This sequence is referred to hereafter as the TSR and is the hallmark of the spermatogenic cell-specific PFKM isozyme, provisionally named PFKMS (Fig. 2). Putative ATG translation start codons were identified for exon -s3 and exon -s4 (GenBank accession number GQ428204) but not for exon -s5 (GenBank accession number GQ428203), suggesting that it is not translated. The PCR results suggested that transcript levels for Pfkm_v1, Pfkm_v2, and Pfkm_v3 were lower than Pfkm_v4 transcript levels in adult mouse testis (Supplemental Fig. S1 available at www.biolreprod.org). Using the BLAST program to query the National Center for Biotechnology Information databases, we found that the sequence of exon -s2 was contained in an EST from newborn mouse brain (CB523377) that included exon 1 of Pfkm, but there were no significant matches with other RNA or EST sequences. In addition, part of exon -s5 and all of exon -s4, exon -s3, and exon -s2 were identical to regions in a sequence from newborn mouse thymus that was purportedly from a cDNA clone (AK138788) but contained intronic sequences. The four Pfkm variants also contained the Pfkm coding sequence.
FIG. 2.
Amino acid sequences of the ubiquitous (PFKM) and testis-specific (PFKMS) PFK isozymes. The sequences in the dashed-line box are present in the ubiquitous PFKM in somatic cells, and the PFKM translation initiation codon (M) is underlined. The sequences in the solid-line box are present in the TSR of PFKMS. The PFKMS initiation codons (M) are underlined and bold.
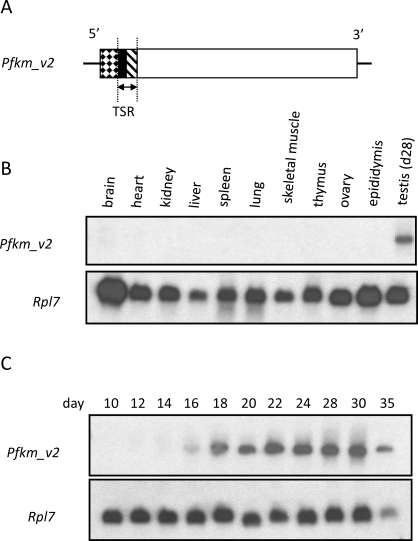
Northern blot analysis was used to determine if the variants were expressed in other tissues. When a portion of the sequence encoding the TSR (Fig. 3A) was used to probe RNA from 11 tissues, an approximate 4.4-kb transcript was detected only in testis (Fig. 3B). When this probe was used for Northern blot analysis with RNA from testes of mice aged 10–35 days, transcripts were first seen on Postnatal Day 16 (Fig. 3C), corresponding to the age when midpachytene spermatocytes are present in the synchronous first wave of spermatogenesis.
FIG. 3.
Northern blot analyses using a probe of the TSR (arrows) from Pfkm_v2 (A) and RNA from various tissues of CD-1 mice (B) or whole testes of juvenile mice 10–35 days old (C). The Rpl7 transcript was used as an internal control.
Sequences encoding peptides similar to the TSR of mouse PFKMS are present upstream of the genes for PFKM in human, chimpanzee, rhesus monkey, dog, rat, and horse (Supplemental Fig. S2). The first 28 amino acids of the TSR-like region in human, chimpanzee, rhesus monkey, dog, rat, and horse are approximately 85% similar to the TSR of mouse. The 68-amino acid sequences of the TSR-like region are highly similar among human, chimpanzee, and rhesus monkey and are 61% similar to the TSR of mouse. Sequences similar to those encoding the mouse TSR were not found in cat, sheep, pig, or cow.
Expression during spermatogenesis of transcripts encoding the TSR was examined by qPCR using cDNA from testes of mice aged 10–30 days (Supplemental Fig. S1A). The level of TSR-containing (exon -s2) sequences increased substantially between Days 14 and 18 and continued to increase to age 30 days (relative to the level at Day 10). While there also was an increase in the level of transcripts encoding a region present in both PFKM and PFKMS (exon 1), the relative increase was substantially less than for TSR-specific sequences. This suggests that the level of transcripts for PFKM in the testis is relatively constant after Day 10, and the increase that does occur is due to transcripts for PFKMS. Transcripts for each of the Pfkm transcript variants were detected by RT-PCR in total RNA from mouse testis using primers in exons -s1 to -s5 and in exon 1 (Supplemental Fig. S1B).
SSR of HK1S Does Not Interact with TSR of PFKMS
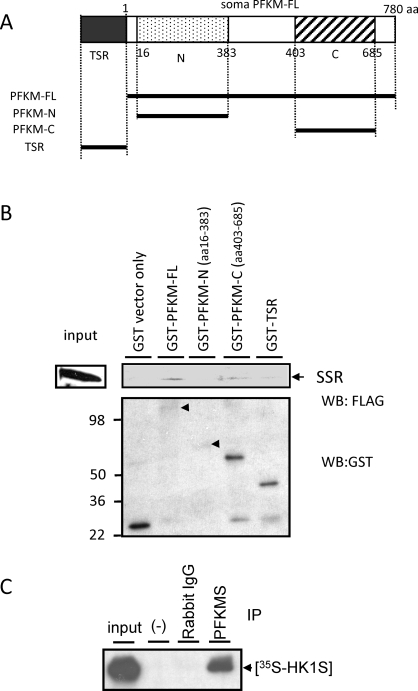
Pull-down assays were used to rule out that the SSR did not also interact directly with the TSR of PFKMS. We first established that the SSR bound to PFKM-FL tagged with GST but not with GST alone (Fig. 4B). Pull-down assays then were used to localize the SSR-binding region on PFKMS. The SSR bound to GST-tagged PFKM-FL and the C-terminal region of PFKMS (PFKM-C) but not to the GST-tagged N-terminal region of PFKMS (PFKM-N) or to the TSR (Fig. 4B). Direct interaction between in vitro-translated [35S]-HK1S and PFKMS was confirmed with a coimmunoprecipitation assay using TSR antiserum (Fig. 4C). These results strongly suggested that the SSR of HK1S interacts with the C-terminal portion of PFKMS but not with the TSR.
FIG. 4.
The SSR of HK1S interacts with PFKM in a GST pull-down and coimmunoprecipitation assays. A) Diagram of recombinant proteins containing either TSR of PFKMS fused to GST (GST-TSR) or all or part of PFKM fused to GST, including full length (GST-PFKM-FL), the N-terminal region, and the C-terminal region (GST-PFKM-N and GST-PFKM-C). B) GST-PFKM-FL, GST-PFKM-N, and GST-PFKM-C proteins were immobilized on glutathione-sepharose resin and incubated with FLAG-tagged SSR protein, and the recombinant proteins that interacted were eluted from the resin. The eluted recombinant proteins were separated by SDS-PAGE, and then Western blot analysis was performed with antibodies to FLAG (upper panel) or GST (lower panel). C) In vitro-translated [35S]-HK1S added to testis lysate coimmunoprecipitated with PFKMS when TSR antiserum and protein G beads were added but not when TSR antiserum was deleted (-) or rabbit IgG was added instead of TSR antiserum.
Expression of PFKMS
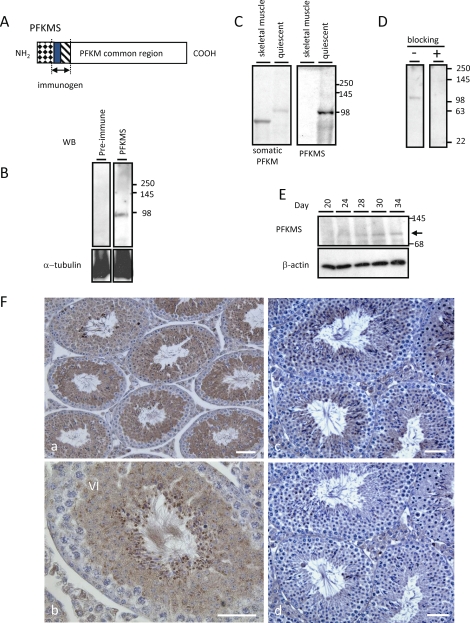
The TSR antiserum (Fig. 5A) was used to determine the developmental expression and localization of PFKMS in mouse sperm and testis. The antiserum bound to a protein of approximately 98 kDa on immunoblots of sperm (Fig. 5B) but not to a protein in muscle (Fig. 5C). In contrast, an antiserum to PFKM (100-1156; Rockland Immunochemicals, Inc.) bound to PFKM from skeletal muscle and to PFKMS from sperm because of their shared sequences. As expected, the molecular mass of PFKMS was greater than that of PFKM because of the additional 67 amino acids on the N-terminus (Fig. 1A). Furthermore, the sperm protein was not recognized by the preimmune serum (Fig. 5B) or by TSR antiserum preabsorbed with recombinant TSR peptide (Fig. 5D). On immunoblots of extracts of testes from mice aged 20–34 days probed with TSR antiserum, PFKMS protein was first seen at Day 20 (Fig. 5E), indicating that translation is delayed until approximately 4 days after Pfkm variant transcripts are detected. On sections of adult mouse testis (Fig. 5F [a and c]) subjected to immunohistochemistry with TSR antiserum, the PFKMS protein first was detected in round spermatids (steps 5 and 6), consistent with immunoblot analysis results, and was present in the cytoplasm and flagellum throughout the remainder of spermiogenesis. Immunostaining was blocked by preabsorption of the antiserum with recombinant TSR peptide (Fig. 5F [d]).
FIG. 5.
Western blot analysis and immunohistochemistry using TSR antiserum. A) The region of PFKMS recognized by TSR antiserum is encoded by exons -s3 and -s2. B) A band of approximately 98 kDa was detected in sperm with antiserum but not with preimmune serum. C) PFKMS was detected with antiserum in extracts of quiescent sperm but not in skeletal muscle. However, proteins were detected in both quiescent sperm and skeletal muscle with an antibody recognizing the region common to PFKM and PFKMS. D) Detection of PFKMS in sperm was blocked when antiserum was preabsorbed with recombinant TSR protein but not when it was preabsorbed with preimmune serum. E) PFKMS was detected faintly by antiserum in lysates of Day 20 testis and more clearly in lysates of testes at later stages of neonatal testis development. F) Immunohistochemistry on sections of adult testis. PFKMS was first detected in the cytoplasm of round spermatids (steps 5 and 6) (a and b) and subsequently in the flagellum of elongated spermatids extending into the seminiferous tubule lumen. The immunostaining seen in a section of testis (c) was not seen in a parallel section (d) when antiserum was preabsorbed with recombinant TSR protein. Bar = 25 μm.
PFKMS Is Anchored in the Principal Piece of the Sperm Flagellum
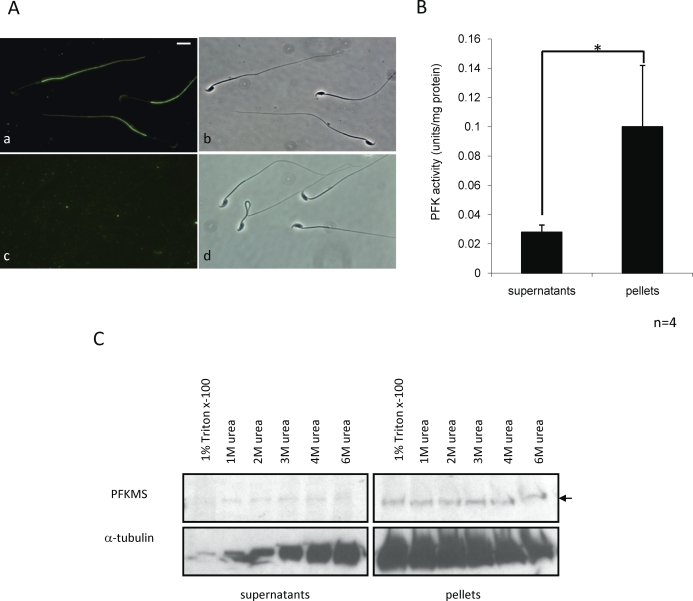
Indirect immunofluorescence microscopy with TSR antiserum and cauda epididymal sperm established that PFKMS was localized to the principal piece region of the sperm flagellum (Fig. 6A [a and b]). The immunostaining was eliminated when the antiserum was preabsorbed with recombinant TSR peptide (Fig. 6A [c and d]). To determine if PFKMS is anchored in this region, a PFK activity assay was carried out using sperm extracted with 1% Triton X-100 in PBS. The PFK activity in the pellet fraction was approximately 4-fold higher than that in the supernatant fraction (Fig. 6B). In addition, immunoblot analysis was performed using sperm extracted with 1% Triton X-100 in PBS or in 1 M, 2 M, 3 M, 4 M, or 6 M urea in PBS (Fig. 6C). PFKMS was detected mainly in the pellet fractions in all samples, with low amounts present in the supernatant fractions in urea extracts and none detected in the 1% Triton X-100 extract. These results suggested that PFKMS is bound strongly to one or more other proteins in the FS.
FIG. 6.
Immunolocalization of PFKMS, PFK activity, and solubility of PFKMS in sperm. A) PFKMS was detected by TSR antiserum in the principal piece region of the sperm flagellum (a) but was not detected when antiserum was preabsorbed with recombinant TSR protein (c). Phase-contrast images (b and d) are shown of the sperm immunostained with unabsorbed and preabsorbed TSR antiserum (a and c). Bar = 8 μm. B) PFK activity was measured in supernatants and pellets of sperm lysed in PBS containing 1% Triton X-100. PFK activity in the pellets was significantly higher than that in the supernatants (*P < 0.05). Data are expressed as the mean ± SD of three separate assays, each performed in duplicate. C) PFKMS was detected by TSR antiserum in the supernatants and pellets following lysis of sperm in various concentrations of urea and 1% Triton X-100/PBS, with the exception of supernatants of sperm lysed only with 1% Triton X-100.
TSR Interacts with GSTM5
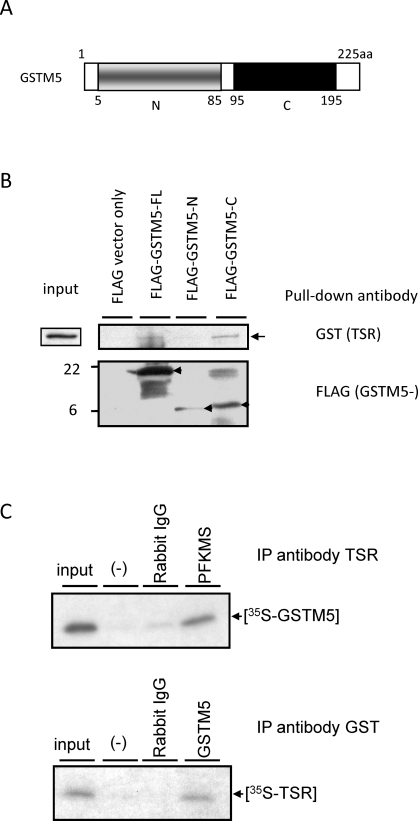
Yeast two-hybrid screens with the TSR (Fig. 3A) as bait were carried out to determine what protein anchors PFKMS to the FS. Four cDNA clones were isolated and characterized, two for Gstm5 cDNAs and two of which appeared to contain genomic sequences. The Gstm5 cDNAs were identical to a 570-bp region of the mouse Gstm5 cDNA sequence reported previously [29] (GenBank accession number NM_010360.1). To confirm this interaction, a pull-down assay was performed with GST-tagged TSR, FLAG-GSTM5-FL, FLAG-GSTM5-N, and FLAG-GSTM5-C (Fig. 7B). Interaction with GST-tagged TSR occurred for all three, but the amount of GSTM5-C and GSTM5-N that bound appeared to be greater than the amount of GSTM5-FL. Interaction between PFKMS and GSTM5 was confirmed by coimmunoprecipitation with TSR antiserum and GSTM5 antiserum (Fig. 7C). These results strongly suggested that PFKMS is anchored through its TSR to GSTM5 in the FS.
FIG. 7.
The TSR of PFKMS interacts with GSTM5 in pull-down and coimmunoprecipitation assays. A) Diagram of recombinant proteins with the FLAG sequence fused to GSTM5-FL or truncated GSTM5 sequences (GSTM5-N and GSTM5-C). B) GST-TSR was pulled down with recombinant FLAG-GSTM5-FL and GSTM5-C proteins by a FLAG antibody linked to agarose beads but not by vector only. C) Upper panel: [35S]-GSTM5 was coimmunoprecipitated with PFKMS by the antibody to TSR but not by rabbit IgG or when primary antibody was deleted (-). Lower panel: [35S]-TSR of PFKMS was coimmunoprecipitated with GSTM5 by an antibody to GSTM5 but not by rabbit IgG or when primary antibody was deleted (-).
Differences in the Solubility of HK1S, PFKMS, and GSTM5
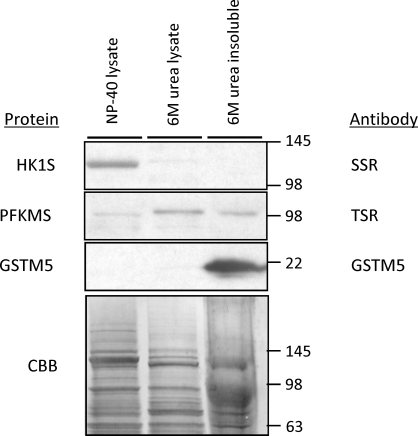
Results of the preceding studies suggested that binding between the SSR of HK1S and PFKMS is weaker than the binding between the TSR of PFKMS and GSTM5. This was confirmed by performing immunoblot analysis with lysates of sperm treated sequentially with NP-40 and 6 M urea (Fig. 8). HK1S was abundant only in the NP-40 sperm lysate supernatants, and PFKMS was present mainly in the 6 M urea lysate supernatant, while GSTM5, a highly insoluble component of the FS [24, 29], was present only in the pellet remaining after extraction with 6 M urea.
FIG. 8.
Differences in solubility of HK1S, PFKMS, and GSTM5. Most of the HK1S (detected with SSR antibody), some PFKMS (detected with TSR antibody), but no GSTM5 (detected with GSTM5 antibody) was extracted from sperm by 1% NP-40. The majority of PFKMS was solubilized by 6 M urea, but some of the PFKMS and all of the GSTM5 protein were present in the 6 M urea-insoluble sperm fraction. The lower panel is an image of a gel with these samples stained with Coomassie brilliant blue (CBB) before transfer for Western blot analysis. Values are in kilodaltons.
PFKM Dimerization and PFK Activity
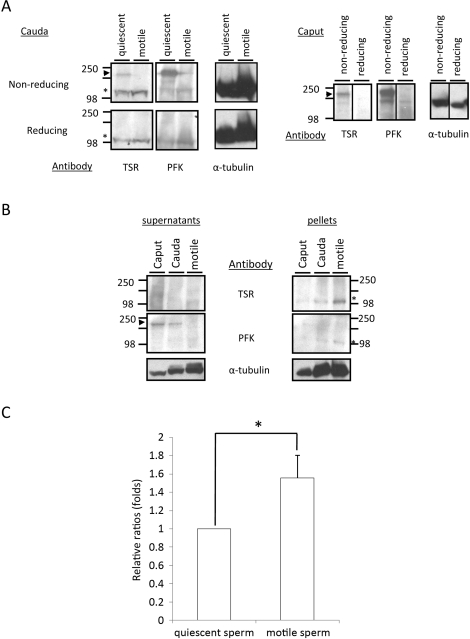
We found previously that hexokinase enzymatic activity was lower in sperm immediately after isolation from the epididymis (quiescent) than at 5 min after dilution in M2 medium (motile) [32]. HK1S was present mainly as a dimer in quiescent sperm and as a monomer in motile sperm, and the conversion of dimer to monomer was associated with a reduction of disulfide bonds [32]. Immunoblot analysis and PFK enzymatic activity measurements were used to determine if PFKMS undergoes a similar change in activity and subunit association. A dimer-sized (approximately 200 kDa) band was detected with the PFKM antibody and the TSR antibody in quiescent sperm from the caput and cauda epididymis separated by PAGE under nonreducing conditions but not in motile sperm separated under reducing conditions (Fig. 9A [arrowheads]). However, a monomer-sized band (approximately 100 kDa) was detected with both antibodies in quiescent and motile sperm separated under nonreducing and reducing conditions (Fig. 9A [asterisks]). It is unclear why loss of the dimer-sized band recognized by antibodies in caput sperm extracts or under nonreducing conditions is not accompanied by a concomitant increase in the intensity of the monomer-sized band in cauda sperm extracts or under reducing conditions. We speculate that cleavage of disulfide bonds during epididymal maturation, as occurs with HK1S [32], and cleavage of disulfide bonds under reducing gel conditions cause a loss of shape-dependent epitopes recognized by the antibodies. However, there are other possible explanations such as selective proteolysis or the loss of cytoplasmic droplets during epididymal maturation, and additional studies will be required to understand these changes.
FIG. 9.
Conversion of PFKMS from dimer to monomer correlates with change in PFK activity. A) Western blot analysis with TSR antiserum on extracts of quiescent and motile cauda sperm (left panels) and extracts of caput sperm (right panels) separated by PAGE under nonreducing and reducing conditions. PFKMS dimers (arrowhead) were detected with TSR and PFKM antisera in extracts of quiescent sperm on nonreducing gels, and monomers (asterisks) were detected in extracts of quiescent and motile sperm by both antisera on nonreducing and reducing gels. Studies were repeated four times, and representative examples are shown. B) Western blot analysis with TSR and PFKM antisera on supernatants (left panel) and pellets (right panel) of lysates of quiescent caput and cauda sperm and motile cauda sperm on nonreducing gels. PFKMS dimers (arrowhead) were detected in the supernatants of quiescent caput and cauda sperm by the PFKM antiserum, and monomers (asterisks) were detected by TSR antiserum in quiescent and motile cauda sperm. C) PFK enzymatic activities in the pellets from lysates of quiescent and motile sperm, expressed as relative folds. Values represent the mean ± SD (n = 4), *P < 0.05.
Quiescent caput and cauda sperm and motile cauda sperm were lysed in PBS containing 0.5% SDS, 0.5% NP-40, and 1% Triton X-100, and lysates and pellets were subjected to PAGE under nonreducing conditions, followed by immunoblot analysis. A dimer-sized band (approximately 200 kDa) was detected with the PFKM antibody in supernatants of quiescent caput and cauda sperm (Fig. 9B [arrowhead]), and monomer-sized bands (approximately 100 kDa) were detected with TSR antiserum and PFKM antibody in the pellets of quiescent caput and cauda sperm and activated cauda sperm (Fig. 9B [asterisks]). In addition, PFK activity in the pellets of motile sperm was higher than that in the pellets of quiescent sperm (Fig. 9C). This suggests that PFKMS might either undergo changes in activity similar to that seen with HK1S for the dimer to monomer conversion occurring concurrent with change of sperm from quiescent to actively motile, or PFKMS activity may increase due to greater availability of substrate that results from the increased activity of HK1S upstream in the glycolytic pathway. Additional studies will be required to address these possibilities.
DISCUSSION
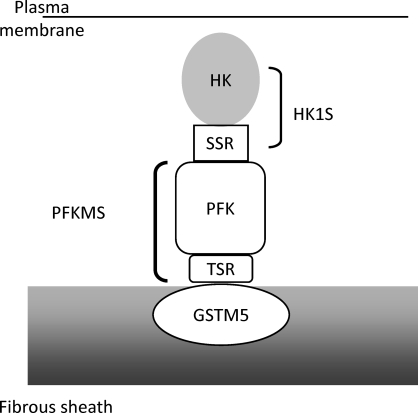
Earlier studies found that mammalian sperm are capable of glycolysis [1, 2] and that some glycolytic enzymes are tightly bound to insoluble sperm components [18–21]. More recent studies established that several sperm glycolytic enzymes are products of novel genes or variant transcripts expressed specifically in testis [4–10]. However, it was unknown how the glycolytic enzymes localize to the principal piece region of the flagellum or bind to the FS. It was hypothesized that the unique N-terminal regions of GAPDHS [23, 26], HK1S [10, 26], and ALDOART1 and ALDOA_V2 [8] are involved in their localization or binding to the FS. The present study tested this hypothesis and found that the HK1S isozyme is tethered through its N-terminal 24-amino acid SSR to a novel PFKMS isozyme, which in turn is anchored through its 67-amino acid TSR to GSTM5, an insoluble component of the FS (Fig. 10).
FIG. 10.
Proposed model of the HK1S, PFKMS, and GSTM5 complex in the FS of sperm. HK1S binds to the FS by tethering through its SSR to PFKMS, which anchors through its TSR to GSTM5, a component of the FS.
The GAPDHS [23], ALDOART1, ALDOA_V2 [8], LDHA, and PK [24] glycolytic enzymes in mouse sperm were shown to resist solubilization by vigorous extraction procedures and to remain tightly bound to the isolated FS. In contrast, LDHC, PGK2, enolase, and HK1S were solubilized from mouse sperm by treatment with mild detergents [24]. Similar observations have been reported for GAPDH in bull and boar sperm [18, 22], ALDO in bull sperm [18, 21], and PK in boar sperm [25]. Nevertheless, there appear to be species differences in the solubility of sperm glycolytic enzymes. Unlike the mouse, lactate dehydrogenase activity remained in the tail fraction of bull sperm [18] and in hypotonically treated boar sperm [37], but less than one fourth of the hexokinase activity in bull and ram sperm was released by homogenization [19]. In addition, while the majority of PFK activity was insoluble in bull and ram sperm [19], it was soluble in boar sperm [37].
Most of the studies that identified genes and transcripts for spermatogenic cell-specific glycolytic isozymes in sperm were in the mouse. While variant transcripts were identified for HK1S in human [38] and spermatogenic cell-specific genes were identified for PGK2 in human [6], GAPDHS in rat [39] and human [40], and LDHC in human [41], it remains to be determined how many of the sperm glycolytic enzymes in most species are products of spermatogenic cell-specific transcripts and genes. However, orthologues of mouse GSTM5 are present in human and rat sperm FS [42, 43], and TSR-like sequences might anchor PFKMS to the GSTM5 orthologues in sperm of other species.
An unexpected finding in this study was the presence in testis of four Pfkm variant transcripts that differed on their 5′ ends from each other and from the typical Pfkm transcript. An antiserum generated against the TSR encoded by two of the exons (exons -s3 and -s2) confirmed that a novel PFKMS isoform containing the TSR is synthesized in spermatids and localizes to the principal piece region of the sperm flagellum. Because the two exons are present in the sequences of three variants (Pfkm_v2, Pfkm_v3, and Pfkm_v4), it is uncertain which is responsible for the PFKMS protein. However, we were unable to identify a potential translation start site for Pfkm_v2 and consider it less likely to give rise to PFKMS than Pfkm_v3 and Pfkm_v4, which have putative translation start codons. Because transcript levels for Pfkm_v4 appear to be more abundant than for Pfkm_v3 (Supplemental Fig. S1), we suspect that it is the primary source of PFKMS.
We previously reported the following findings [32]: 1) HK1S activity was lower in quiescent sperm immediately after isolation from the epididymis than in actively motile sperm; 2) approximately two thirds of the HK1S activity in mouse sperm was in the pellet fraction of immotile sperm, while less than one third of the activity was in the pellet fraction of motile sperm; 3) the majority of HK1S was present as dimers in quiescent sperm and as monomers in actively motile sperm; and 4) the dimer to monomer conversion involved reduction of disulfide bonds. In the present study, we report the following findings: 1) PFK activity was lower in immotile sperm than in motile sperm, 2) most of the PFK enzymatic activity remained in the detergent-insoluble pellet fraction and was extracted only partially with 6 M urea, 3) PFKMS was present in dimers in extracts of quiescent sperm and in monomers in extracts of actively motile sperm analyzed under nonreducing conditions, and 4) PFKMS was present as monomers under reducing conditions. Thus, enzymatic activity of both HK1S and PFKMS is lower in quiescent than in actively motile sperm, and the change in activity correlates with a dimer to monomer conversion. However, HK1S is solubilized more easily than PFKMS, and previous studies [24, 29] demonstrated that GSTM5 remains present in the FS fraction after a rigorous extraction procedure. These results indicate that a hierarchy of binding affinities exists from low to high between HK1S and PFKMS, between PFKMS and GSTM5, and between GSTM5 and other FS components. However, it remains to be determined how the glycolytic enzymes are transported to the principal piece region of the flagellum and how the others are held in this region. While some of the enzymes have novel N-terminal regions that might be involved in these processes (e.g., GAPDHS, ALDOART1, and ALDOART2), some do not (e.g., PGK2 and LDHC), and the mechanisms involved in the segregation of most enzymes to this region remain to be identified.
Acknowledgments
The authors thank Eugenia H. Goulding for valuable technical support, Clyde Roger and Linwood Koonce for help with animals, other members of the Gamete Biology Group for constructive suggestions, and Tracy Clement and Oleg Alekseev for helpful comments on the manuscript.
Footnotes
Supported by the Intramural Research Program of the NIH, National Institute of Environmental Health Sciences.
REFERENCES
- Mukai C, Okuno M.Glycolysis plays a major role for adenosine triphosphate supplementation in mouse sperm flagellar movement. Biol Reprod 2004; 71: 540–547. [DOI] [PubMed] [Google Scholar]
- Miki K, Qu W, Goulding EH, Willis WD, Bunch DO, Strader LF, Perreault SD, Eddy EM, O'Brien DA.Glyceraldehyde 3-phosphate dehydrogenase-S, a sperm-specific glycolytic enzyme, is required for sperm motility and male fertility. Proc Natl Acad Sci U S A 2004; 101: 16501–16506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odet F, Duan C, Willis WD, Goulding EH, Kung A, Eddy EM, Goldberg E.Expression of the gene for mouse lactate dehydrogenase C (Ldhc) is required for male fertility. Biol Reprod 2008; 79: 26–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg E.Lactate dehydrogenase-X from mouse testes and spermatozoa. Methods Enzymol 1975; 41: 318–323. [DOI] [PubMed] [Google Scholar]
- Boer PH, Adra CN, Lau YF, McBurney MW.The testis-specific phosphoglycerate kinase gene pgk-2 is a recruited retroposon. Mol Cell Biol 1987; 7: 3107–3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarrey JR, Thomas K.Human testis-specific PGK gene lacks introns and possesses characteristics of a processed gene. Nature 1987; 326: 501–505. [DOI] [PubMed] [Google Scholar]
- Welch JE, Schatte EC, O'Brien DA, Eddy M.Expression of a glyceraldehydes 3-phosphate dehydrogenase gene specific to mouse spermatogenic cells. Biol Reprod 1992; 46: 869–878. [DOI] [PubMed] [Google Scholar]
- Vemuganti SA, Bell TA, Scarlett CO, Parker CE, de Villena FP, O'Brien DA.Three male germline-specific aldolase A isozymes are generated by alternative splicing and retrotransposition. Dev Biol 2007; 309: 18–31. [DOI] [PubMed] [Google Scholar]
- Mori C, Welch JE, Fulcher KD, O'Brien DA, Eddy EM.Unique hexokinase messenger ribonucleic acids lacking the porin-binding domain are developmentally expressed in mouse spermatogenic cells. Biol Reprod 1993; 49: 191–203. [DOI] [PubMed] [Google Scholar]
- Mori C, Nakamura N, Welch JE, Gotoh H, Goulding EH, Fujioka M, Eddy EM.Mouse spermatogenic cell-specific type 1 hexokinase (mHk1-s) transcripts are expressed by alternative splicing from the mHk1 gene and the HK1-S protein is localized mainly in the sperm tail. Mol Reprod Dev 1998; 49: 374–385. [DOI] [PubMed] [Google Scholar]
- Buehr M, McLaren A.Interlitter variation in progeny of chimaeric male mice. J Reprod Fertil 1984; 72: 213–221. [DOI] [PubMed] [Google Scholar]
- Russell DL, Kim KH.Expression of triosephosphate isomerase transcripts in rat testis: evidence for retinol regulation and a novel germ cell transcript. Biol Reprod 1996; 55: 11–18. [DOI] [PubMed] [Google Scholar]
- Fundele R, Illmensee K, Jägerbauer EM, Fehlau M, Krietsch WK.Sequential expression of maternally inherited phosphoglycerate kinase-1 in the early mouse embryo. Differentiation 1987; 35: 31–36. [DOI] [PubMed] [Google Scholar]
- Edwards YH, Grootegoed JA.A sperm-specific enolase. J Reprod Fertil 1983; 68: 305–310. [DOI] [PubMed] [Google Scholar]
- Gitlits VM, Toh BH, Loveland KL, Sentry JW.The glycolytic enzyme enolase is present in sperm tail and displays nucleotide-dependent association with microtubules. Eur J Cell Biol 2000; 79: 104–111. [DOI] [PubMed] [Google Scholar]
- Fawcett DW.The mammalian spermatozoon. Dev Biol 1975; 44: 394–436. [DOI] [PubMed] [Google Scholar]
- Eddy EM.The scaffold role of the fibrous sheath. Soc Reprod Fertil Suppl 2007; 65: 45–62. [PubMed] [Google Scholar]
- Mohri H, Mohri T, Eranster L.Isolation and enzymic properties of the midpiece of bull spermatozoa. Exp Cell Res 1965; 38: 217–246. [DOI] [PubMed] [Google Scholar]
- Harrison RA.Glycolytic enzymes in mammalian spermatozoa: activities and stabilities of hexokinase and phosphofructokinase in various fractions from sperm homogenates. Biochem J 1971; 124: 741–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storey BT, Kayne FJ.Energy metabolism of spermatozoa, VII: interactions between lactate, pyruvate and malate as oxidative substrates for rabbit sperm mitochondria. Biol Reprod 1978; 18: 527–536. [DOI] [PubMed] [Google Scholar]
- Gillis BA, Tamblyn TM.Association of bovine sperm aldolase with sperm subcellular components. Biol Reprod 1984; 31: 25–35. [DOI] [PubMed] [Google Scholar]
- Westhoff D, Kamp G.Glyceraldehyde 3-phosphate dehydrogenase is bound to the fibrous sheath of mammalian spermatozoa. J Cell Sci 1997; 110: 1821–1829. [DOI] [PubMed] [Google Scholar]
- Bunch DO, Welch JE, Magyar PL, Eddy EM, O'Brien DA.Glyceraldehyde 3-phosphate dehydrogenase-S protein distribution during mouse spermatogenesis. Biol Reprod 1998; 58: 834–841. [DOI] [PubMed] [Google Scholar]
- Krisfalusi M, Miki K, Magyar PL, O'Brien DA.Multiple glycolytic enzymes are tightly bound to the fibrous sheath of mouse spermatozoa. Biol Reprod 2006; 75: 270–278. [DOI] [PubMed] [Google Scholar]
- Feiden S, Stypa H, Wolfrum U, Wegener G, Kamp G.A novel pyruvate kinase (PK-S) from boar spermatozoa is localized at the fibrous sheath and the acrosome. Reproduction 2007; 134: 81–95. [DOI] [PubMed] [Google Scholar]
- Eddy EM, Welch JE, Mori C, Fulcher KD, O'Brien DA.Role and regulation of spermatogenic cell-specific gene expression: enzymes of glycolysis. Bartke A.Function of Somatic Cells in the Testis New York:Springer-Verlag;1994: 362–372. [Google Scholar]
- Adams V, Griffin L, Towbin J, Gelb B, Worley K, McCabe ER.Porin interaction with hexokinase and glycerol kinase: metabolic microcompartmentation at the outer mitochondrial membrane. Biochem Med Metab Biol 1991; 45: 271–291. [DOI] [PubMed] [Google Scholar]
- Smith AD, Wilson JE.Effect of ligand binding on the tryptic digestion pattern of rat brain hexokinase: relationship of ligand-induced conformational changes to catalytic and regulatory functions. Arch Biochem Biophys 1991; 291: 59–68. [DOI] [PubMed] [Google Scholar]
- Fulcher KD, Welch JE, Klapper DG, O'Brien DA, Eddy EM.Identification of a unique mu-class glutathione S-transferase in mouse spermatogenic cells. Mol Reprod Dev 1995; 42: 415–424. [DOI] [PubMed] [Google Scholar]
- Gietz RD, Schiestl RH.Transforming yeast with DNA. Methods Mol Cell Biol 1995; 5: 255–269. [Google Scholar]
- Fenderson BA, Toshimori K, Muller CH, Lane TF, Eddy EM.Identification of a protein in the fibrous sheath of the sperm flagellum. Biol Reprod 1988; 38: 345–357. [DOI] [PubMed] [Google Scholar]
- Nakamura N, Miranda-Vizuete A, Miki K, Mori C, Eddy EM.Cleavage of disulfide bonds in mouse spermatogenic cell-specific type 1 hexokinase isozyme is associated with increased hexokinase activity and initiation of sperm motility. Biol Reprod 2008; 79: 537–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe JD, Nieves E, Listowsky I.Subunit diversity and tissue distribution of human glutathione S-transferases: interpretations based on electrospray ionization-MS and peptide sequence-specific antisera. Biochem J 1997; 325: 481–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saling PM, Irons GP, Waibel R.Mouse sperm antigens that participate in fertilization, I: inhibition of sperm fusion with the egg plasma membrane using monoclonal antibodies. Biol Reprod 1985; 33: 515–526. [DOI] [PubMed] [Google Scholar]
- Kemp RG.Phosphofructokinase from rabbit skeletal muscle. Methods Enzymol 1975; 62: 71–77. [DOI] [PubMed] [Google Scholar]
- Yamada S, Nakajima H, Kuehn MR.Novel testis- and embryo-specific isoforms of the phosphofructokinase-1 muscle type gene. Biochem Biophys Res Commun 2004; 316: 580–587. [DOI] [PubMed] [Google Scholar]
- Jones AR, Piccolo F.Glycolytic enzyme activity in hypotonically treated boar spermatozoa. Reprod Fertil Dev 1999; 11: 409–413. [DOI] [PubMed] [Google Scholar]
- Mori C, Nakamura N, Welch JE, Shiota K, Eddy EM.Testis-specific expression of mRNAs for a unique human type 1 hexokinase lacking the porin-binding domain. Mol Reprod Dev 1996; 44: 14–22. [DOI] [PubMed] [Google Scholar]
- Welch JE, Barbee RR, Magyar PL, Bunch DO, O'Brien DA.Expression of the spermatogenic cell-specific glyceraldehyde 3-phosphate dehydrogenase (GAPDS) in rat testis. Mol Reprod Dev 2006; 73: 1052–1060. [DOI] [PubMed] [Google Scholar]
- Welch JE, Brown PL, O'Brien DA, Magyar PL, Bunch DO, Mori C, Eddy EM.Human glyceraldehyde 3-phosphate dehydrogenase-2 gene is expressed specifically in spermatogenic cells. J Androl 2000; 21: 328–339. [PubMed] [Google Scholar]
- Cooker LA, Brooke CD, Kumari M, Hofmann MC, Millan JL, Goldberg E.Genomic structure and promoter activity of the human testis lactate dehydrogenase gene. Biol Reprod 1993; 48: 1309–1319. [DOI] [PubMed] [Google Scholar]
- Rowe JD, Patskovsky Y, Patskovska LN, Novikova E, Listowsky I.Rationale for reclassification of a distinct subdivision of mammalian class mu-glutathione S transferases that are primarily expressed in testis. J Biol Chem 1998; 273: 9593–9601. [DOI] [PubMed] [Google Scholar]
- Rowe JD, Tchaikovskaya T, Shintani N, Listowsky I.Selective expression of a glutathione S-transferase subclass during spermatogenesis. J Androl 1998; 19: 558–567. [PubMed] [Google Scholar]