Abstract
Breast milk transmission remains a major mode of infant HIV acquisition, yet anatomic and immunologic forces shaping virus quasispecies in milk are not well characterized. In this study, phylogenic analysis of envelope sequences of milk SIV variants revealed groups of nearly identical viruses, indicating local virus production. However, comparison of the patterns and rates of CTL escape of blood and milk virus demonstrated only subtle differences between the compartments. These findings suggest that a substantial fraction of milk viruses are produced by locally-infected cells, but are shaped by cellular immune pressures similar to that in the blood.
Findings
While transmission via breastfeeding remains a significant mode of infant HIV acquisition, the mechanisms of this transmission are not well understood. The level of both cell-free and cell-associated virus in milk have been linked to the risk of HIV infection of breastfeeding infants [1-3]. Therefore, viral variants in the milk are a likely source of transmitted virus.
Recent studies suggest that mucosal compartments are distinct from systemic compartments in their HIV/SIV-specific immune responses and virus quasispecies [4-8]. The immune response in milk may shape compartment-specific virus, as seen in other anatomic compartments such as semen and cervicovaginal fluid [6,9-11]. One study of the genetic diversity of milk virus suggested a difference in the dominant virus species in milk and peripheral blood [4]. A second study subsequently reported that the predominant virus in blood and milk were genetically similar, suggesting an equilibrium of virus between these compartments [12]. Thus, the production site of the virus transmitted via breastfeeding and the selection pressures that shape its genetic composition are not well understood.
It is well established that HIV/SIV-specific cellular immune responses are critical for control of systemic virus replication [13] and progression to AIDS [14]. While HIV/SIV-specific cellular immune responses have been identified in milk [15,16], the role of these responses in containing local virus replication is unknown. Immunodominant cytotoxic T lymphocyte (CTL) epitopes of SIV and HIV are under significant cellular immune pressure, leading to distinct mutations within these epitopes that facilitate virus escape from CTL recognition. Therefore, the rate at which these CTL escape mutations occur within a virus population may be indicative of the magnitude of the cellular immune pressure in that compartment. However, rates of CTL escape of mucosal and systemic virus quasispecies have not previously been compared. As the proportion of SIV-specific CD8+ T lymphocytes is two to three times higher in milk than in blood during acute SIV infection [15], a faster rate of CTL escape of the milk virus quasispecies than that in blood would indicate that this immune response in milk is acting on locally replicating virus.
The SIV-infected rhesus monkey model of HIV pathogenesis is an excellent model in which to study virus-specific immune responses and virus evolution in mucosal compartments, including breast milk [15]. In the present study, we sought to evaluate genetic similarities between virus quasispecies in blood and milk of chronically SIVmac251-infected rhesus monkeys through phylogenetic comparison of SIV envelope (env) sequences. We then compared the rate of CTL escape of two Mamu-A*01-restricted immunodominant epitopes (Tat TL8 and Gag p11C) in blood and milk virus. Animals were maintained in accordance with the guidelines of the "Guide for the Care and Use of Laboratory Animals" (National Research Council, National Academic Press, Washington, D.C., 1996).
Groups of identical or nearly identical viruses in breast milk of chronically SIV-infected, lactating rhesus monkeys
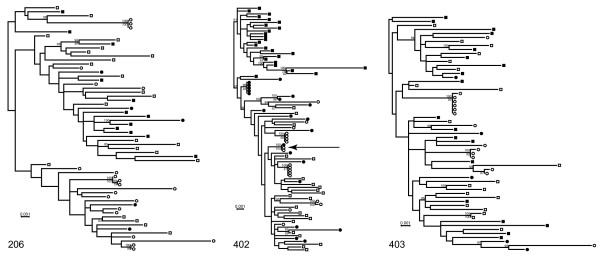
Cell-free and cell-associated SIV env amplicons were sequenced from plasma, peripheral blood mononuclear cells (PBMC), breast milk supernatant, and breast milk cells obtained from 3 chronically SIV-infected, lactating rhesus monkeys within a 7 day period between 1 and 1.5 years after SIVmac251 infection. SIV env cassettes containing the env open reading frame were amplified by single genome amplification (SGA) to reduce the possibility of PCR-induced recombination during amplification [17]. A nested PCR was performed on end-point diluted virus cDNA, extracted as previously described [15], with primers: outer forward (5' - GAAAGGCTGTAGATGTCTAGG - 3'), outer reverse (5' - CTCATCTGATACATTTACGGGG - 3'), inner forward (5' - GGGTAGTGGAGGTTCTGGAAG - 3'), inner reverse (5' - CCCTACCAAGTCATCATCTTC - 3') (GenBank D01065). Total SIV copy numbers measured by quantitative RT-PCR [15] ranged from 6.9 × 103 to 1.5 × 106 copies/ml (3.3 × 103 to 2.2 × 105 total copies measured) in breast milk and from 5.25 × 105 to 2.1 × 107 copies/ml (4.7 × 104 to 1.8 × 106 total copies measured) in plasma. These total SIV copy numbers were measured in the animals at the time of collection of the samples used for sequencing. Sequences were trimmed to the start codon of ENV and to the 3' region of env with unambiguous sequencing for each amplicon within the same monkey (length: 2397-2559 base pairs) and aligned using ClustalW version 2 [18]. PhyML version 3.0.1 [19] was used to infer the evolutionary model parameters and phylogenetic trees. For each analysis, a GTR+I+G model (general time reversible model with invariant sites and gamma-distributed site-to-site rate variation) was used. Bootstrap support was based on 100 resamplings. Bayesian posterior probabilities were calculated using MrBayes version 3.1.2 [20] with chains of 2.1 to 3 × 107 iterations and 25% burn in (Fig. 1).
Figure 1.
Identification of groups of identical or nearly identical envelope sequences in breast milk of lactating, chronically SIV-infected rhesus monkeys. Trees were inferred by maximum likelihood methods [19]. Cell-associated and cell-free blood virus quasispecies env sequences are indicated with closed and open squares, respectively, and cell-associated and cell-free milk virus quasispecies env sequences are indicated with closed and open circles, respectively. Bayesian posterior probabilities are shown above the branch and bootstrap support values are shown in italics below the branch. Sequences are shown for three chronically infected rhesus macaques: 206 (week 74 after SIV infection), 402 (week 67 after SIV infection), and 403 (week 49 after SIV infection).
Overall, the range of nucleotide sequence diversity of cell-free milk virus (monkey 206 = 1.03%, monkey 257 = 0.76%, monkey 403 = 0.75%) was similar to that in the cell-free blood virus (monkey 206 = 1.1%, monkey 257 = 0.85%, monkey 403 = 0.73%). Strikingly, groups of identical or nearly identical cell-free envs were identified in milk of all animals, while this was not observed for plasma-derived cell-free env sequences. These groups of envs were either identical or differed by a single nucleotide (0 - 0.04% nucleotide difference) and comprised one-third to three-quarters of the cell-free milk virus amplified in each animal (monkey 206 = 38.1%, monkey 257 = 76.5%, monkey 402 = 73.3%). Moreover, in animal 402, a single breast milk cell-associated virus variant had a sequence identical to that of a cluster of cell-free milk viruses, which suggests that the clonal amplification of cell-free milk virus variants is produced by a small number of productively-infected, resident cells. In fact, our findings are consistent with a recent report that actively replicating virus in breast milk evolves under drug pressure that is distinct from that in the plasma [21]. Thus, locally-produced virus is likely the major source of virus present in breast milk.
However, we did not observe phylogenetic evidence of compartmentalization of blood and milk virus quasispecies, as the blood and milk viral variants were interspersed in the phylogenetic tree of sequences derived from each monkey. While the milk viruses did not cluster in the phylogenetic trees, all animals exhibited significant compartmentalization between blood and milk cell-free virus quasispecies as determined by the Slatkin-Maddison compartmentalization test of the minimum possible number of inter-compartment migration events compared to the distribution of migration events in 1000 randomized trees [22,23] implemented by HYPHY software [24] (monkey 206: p = 0.001; monkey 402: p = 0.003; monkey 403: p = 0.001). When repeated identical and nearly identical sequences were removed from the data set subjected to the Slatkin-Maddison analysis, significant compartmentalization between blood and breast milk was only evident in animal 206 (monkey 206: p = 0.006; monkey 402: p = 0.795; monkey 403: p = 0.505). Therefore, whether breast milk and blood represent distinct anatomic compartments for SIV in the setting of chronic infection remains an open question.
Rate of CTL escape at immunodominant epitopes Tat TL8 and Gag p11C in breast milk virus
As a robust SIV-specific CD8+ T lymphocyte response occurs in breast milk during acute SIV infection, we investigated whether CTL escape of two Mamu-A*01-restricted immunodominant epitopes Tat TL8 and Gag p11C occurred at a different rate in milk and blood virus quasispecies. The Tat TL8 epitope was amplified by SGA with primers described above (6 to 31 amplicons per compartment). The Gag p11C epitope was analyzed by SGA with primers: outer forward (5' - GTCTGCGTCATCTGGTGCATTC - 3'), outer reverse (5' - TGTTTGTTCTGCTCTTAAGCTTTTGTAG - 3'), inner forward (5' - CAAAACAGATAGTGCAGAGACACCTAGTG - 3'), inner reverse (5' - GAAATGGCTCTTTTGGTCCTT - 3'). Virus quasispecies with at least one nonsynonymous mutation within an immunodominant epitope was defined as a CTL escape variant. CTL escape at the Tat TL8 epitope occurs rapidly in the blood so that all virus quasispecies have a mutated Tat TL8 epitope by day 28 of infection [25], whereas CTL escape at the Gag p11C epitope occurs much later due to structural constraints on the virus at that epitope [26,27].
CTL escape occurred in milk virus at both the Tat TL8 and Gag p11C epitope with patterns of epitope position mutations similar to those that occurred in the blood virus (data not shown). Although there were subtle differences in the rate of CTL escape of the milk and blood virus populations, these did not achieve statistical significance. At day 18 after infection, a lower proportion of virus remained wild type at the Tat TL8 epitope in blood (median = 34.3%; range = 12.5% - 90.9%) than in breast milk (median = 67.6%, range = 31% - 100%) of all monkeys (Table 1). However, CTL escape at the Tat TL8 epitope occurred in all virus variants from both compartments by day 28 after infection in all monkeys.
Table 1.
CTL escape of breast milk and blood virus quasispecies at the Tat TL8 epitope during acute SIV infection.
| Days after SIV infection | |||||
|---|---|---|---|---|---|
| Animal # | Day 14 | Day 18 | Day 21 | Day 28 | |
| 206 | blood | 100%a (17/17)b | 14.3% (3/21) | 0% (0/26) | NDc |
| milk | 100% (30/30) | 87.5% (28/32) | 17.4% (4/23) | 0% (0/17) | |
| 257 | blood | 100% (29/29) | 90.9% (20/22) | 38.5% (10/26) | 0% (0/20) |
| milk | 100% (21/21) | 100% (30/30) | 54.5% (12/22) | 0% (0/31) | |
| 402 | blood | 100% (28/28) | 12.5% (3/24) | 7.7% (2/26) | ND |
| milk | 100% (18/18) | 31.0% (9/29) | 16.7% (1/6) | 0% (0/32) | |
| 403 | blood | 100% (29/29) | 54.2% (13/24) | 9.4% (3/32) | ND |
| milk | 100% (23/23) | 47.6% (10/21) | 17.4% (4/23) | 0% (0/31) | |
a % reported is the proportion of virus quasispecies that did not have an amino acid substitution within the Tat TL8 epitope (remained wild type).
b In parentheses is the number of virus quasispecies that remained wild type at the Tat TL8 epitope/number of virus quasispecies sequenced.
c ND = not done
We used the Spearman's rank correlation test to determine if the proportion of virus variants with CTL escape of Tat TL8 at day 18 after SIV infection was associated with the proportion of Tat TL8-specific CD8+ T lymphocytes or the magnitude of the milk virus load [15]. The proportion of virus with evidence of CTL escape in the milk virus population most strongly correlated with the peak milk virus load (R2 = 0.986, p = 0.083). Therefore, the slightly slower rate of CTL escape mutation of the Tat TL8 epitope in milk is likely a result of a lower rate of virus replication in the breast milk compartment.
As predicted by previous studies of CTL escape at the Gag p11C epitope, CTL escape at the Gag p11C epitope occurred at amino acid position 2 during chronic infection in both blood and milk virus populations (between weeks 17 and 46 after SIV infection) [27]. Interestingly, CTL escape appears to have occurred more quickly at Gag p11C in milk than in blood of 2 of 4 monkeys included in this study. The majority of the milk virus quasispecies was mutated at Gag p11C by week 37 in monkey 206 and week 41 in monkey 257, whereas the majority of blood virus quasispecies remained wild type at Gag p11C until week 45 and 46, respectively (Table 2) We were unable to detect more rapid CTL escape at the Gag p11C epitope in the milk virus population in one rapid progressor monkey who died less than one year after SIV inoculation (animal 403) and one monkey that was not lactating at the time of Gag p11C CTL escape (animal 402). In addition, there was no detectable correlation between the proportion Gag p11C-specific CD8+ T lymphocytes or milk virus load and the rate of Gag p11C CTL escape in milk using nonparametric linear regression analysis.
Table 2.
CTL escape at the Gag p11c epitope in breast milk and blood virus quasispecies during acute SIV infection.
| Weeks after SIV infection | ||||||||
|---|---|---|---|---|---|---|---|---|
| Animal # | Week 33 | Week 35 | Week 36 | Week 37 | Week 38 | Week 40 | Week 45 | |
| 206 | blood | 100%a (14/14)b |
65.4% (17/26) |
59.4% (19/32) |
61.5% (16/26) |
41.7% (10/24) |
30.4% (7/23) |
8.3% (2/24) |
| milk | 80% (4/5) |
29.0% (9/31) |
83.3% (35/42) |
3.1% (1/32) |
NDc | ND | ND | |
| Week 37 | Week 39 | Week 41 | Week 42 | Week 45 | Week 46 | |||
| 257 | blood | 100% (16/16) |
ND | ND | 100% (28/28) |
21.2% (7/33) |
0% (0/17) |
|
| milk | ND | 50% (5/10) |
0% (0/5) |
ND | 14.3% (1/7) |
6.7% (1/15) |
||
| Week 33 | Week 35 | Week 36 | Week 37 | |||||
| 402 | blood | 0% (0/22) |
0% (0/35) |
0% (0/27) |
0% (0/19) |
|||
| milk | 0% (0/22) |
12.5% (1/8) |
0% (0/15) |
0% (0/5) |
||||
| Week 15 | Week 16 | Week 17 | ||||||
| 403 | blood | 73.1% (19/26) |
28.0% (7/25) |
0% (0/26) |
||||
| milk | 100% (15/15) |
18.5% (5/27) |
4.3% (1/23) |
|||||
a % reported is the proportion of virus quasispecies that did not have an amino acid substitution within the Gag p11C epitope (remained wild type).
b In parentheses is the number of virus quasispecies that remained wild type at the Tat TL8 epitope/number of virus quasispecies sequenced.
c ND = not done or unable to amplify virus variants
In summary, CTL escape of milk virus occurred with a pattern and rate similar to that seen in blood virus, suggesting that similar cellular immune pressures shape virus quasispecies evolution in these compartments. While SIV-specific CD8+ T lymphocyte responses during acute SIV infection appear in milk concurrently with the reduction of milk virus load from peak [15], we were not able to demonstrate that the high proportion of SIV-specific CD8+ T lymphocytes observed in milk drives earlier CTL escape of milk virus. However, the small number of evaluated monkeys and restricted sampling schedule make it difficult to demonstrate distinct rates of CTL escape in blood and milk virus. Although a limited number of animals were included in this study, clonal amplification of virus in milk and not in plasma of all animals studied suggests that replication of breast milk virus occurs locally.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
SP designed and coordinated the study, performed sequence data analysis, and drafted the manuscript. HK, AW, LM performed single genome amplification and sequencing. KG and AC participated in study design and supervised and performed nonhuman primate procedures. GL created the phylogenetic tree and performed phylogenetic sequence analysis. BH performed phylogenetic sequence analysis and assisted in data interpretation and manuscript writing. NL participated in the design and coordination of the study and assisted in the drafting of the manuscript.
Contributor Information
Sallie R Permar, Email: sallie.permar@childrens.harvard.edu.
Helen H Kang, Email: hhk2001@med.cornell.edu.
Andrew B Wilks, Email: awilk1@bidmc.harvard.edu.
Linh V Mach, Email: lmach@bidmc.harvard.edu.
Angela Carville, Email: angela.carville@hms.harvard.edu.
Keith G Mansfield, Email: keith.mansfield@hms.harvard.edu.
Gerald H Learn, Email: jlearn@uab.edu.
Beatrice H Hahn, Email: bhahn@uab.edu.
Norman L Letvin, Email: nletvin@bidmc.harvard.edu.
Acknowledgements
This work was supported by the Center for HIV/AIDS Vaccine Immunology (SP and NL; AI067854), the Pediatric Infectious Disease Society/St. Jude Children's Hospital Basic Science Research Award (SP), and NIH K08AI087992 (SP).
We are grateful to Jesus Gonzalez-Salazar, Maria Salazar, Jeff Anderson, Aravind Basavapathruni, James Whitney, George Shaw, and Bette Korber for assistance with sequence analysis and data interpretation.
References
- John GC, Nduati RW, Mbori-Ngacha DA, Richardson BA, Panteleeff D, Mwatha A, Overbaugh J, Bwayo J, Ndinya-Achola JO, Kreiss JK. Correlates of mother-to-child human immunodeficiency virus type 1 (HIV-1) transmission: association with maternal plasma HIV-1 RNA load, genital HIV-1 DNA shedding, and breast infections. J Infect Dis. 2001;183:206–212. doi: 10.1086/317918. [DOI] [PubMed] [Google Scholar]
- Rousseau CM, Nduati RW, Richardson BA, Steele MS, John-Stewart GC, Mbori-Ngacha DA, Kreiss JK, Overbaugh J. Longitudinal analysis of human immunodeficiency virus type 1 RNA in breast milk and of its relationship to infant infection and maternal disease. J Infect Dis. 2003;187:741–747. doi: 10.1086/374273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semba RD, Kumwenda N, Hoover DR, Taha TE, Quinn TC, Mtimavalye L, Biggar RJ, Broadhead R, Miotti PG, Sokoll LJ, Hoeven L van der, Chiphangwi JD. Human immunodeficiency virus load in breast milk, mastitis, and mother-to-child transmission of human immunodeficiency virus type 1. J Infect Dis. 1999;180:93–98. doi: 10.1086/314854. [DOI] [PubMed] [Google Scholar]
- Becquart P, Chomont N, Roques P, Ayouba A, Kazatchkine MD, Belec L, Hocini H. Compartmentalization of HIV-1 between breast milk and blood of HIV-infected mothers. Virology. 2002;300:109–117. doi: 10.1006/viro.2002.1537. [DOI] [PubMed] [Google Scholar]
- Hel Z, Nacsa J, Kelsall B, Tsai WP, Letvin N, Parks RW, Tryniszewska E, Picker L, Lewis MG, Edghill-Smith Y, Moniuszko M, Pal R, Stevceva L, Altman JD, Allen TM, Watkins D, Torres JV, Berzofsky JA, Belyakov IM, Strober W, Franchini G. Impairment of Gag-specific CD8(+) T-cell function in mucosal and systemic compartments of simian immunodeficiency virus mac251- and simian-human immunodeficiency virus KU2-infected macaques. J Virol. 2001;75:11483–11495. doi: 10.1128/JVI.75.23.11483-11495.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiessling AA, Fitzgerald LM, Zhang D, Chhay H, Brettler D, Eyre RC, Steinberg J, McGowan K, Byrn RA. Human immunodeficiency virus in semen arises from a genetically distinct virus reservoir. AIDS Res Hum Retroviruses. 1998;14(Suppl 1):S33–41. [PubMed] [Google Scholar]
- Mora JR, Bono MR, Manjunath N, Weninger W, Cavanagh LL, Rosemblatt M, Von Andrian UH. Selective imprinting of gut-homing T cells by Peyer's patch dendritic cells. Nature. 2003;424:88–93. doi: 10.1038/nature01726. [DOI] [PubMed] [Google Scholar]
- Sullivan ST, Mandava U, Evans-Strickfaden T, Lennox JL, Ellerbrock TV, Hart CE. Diversity, divergence, and evolution of cell-free human immunodeficiency virus type 1 in vaginal secretions and blood of chronically infected women: associations with immune status. J Virol. 2005;79:9799–9809. doi: 10.1128/JVI.79.15.9799-9809.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delwart EL, Mullins JI, Gupta P, Learn GH Jr, Holodniy M, Katzenstein D, Walker BD, Singh MK. Human immunodeficiency virus type 1 populations in blood and semen. J Virol. 1998;72:617–623. doi: 10.1128/jvi.72.1.617-623.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellerbrock TV, Lennox JL, Clancy KA, Schinazi RF, Wright TC, Pratt-Palmore M, Evans-Strickfaden T, Schnell C, Pai R, Conley LJ, Parrish-Kohler EE, Bush TJ, Tatti K, Hart CE. Cellular replication of human immunodeficiency virus type 1 occurs in vaginal secretions. J Infect Dis. 2001;184:28–36. doi: 10.1086/321000. [DOI] [PubMed] [Google Scholar]
- Overbaugh J, Anderson RJ, Ndinya-Achola JO, Kreiss JK. Distinct but related human immunodeficiency virus type 1 variant populations in genital secretions and blood. AIDS Res Hum Retroviruses. 1996;12:107–115. doi: 10.1089/aid.1996.12.107. [DOI] [PubMed] [Google Scholar]
- Henderson GJ, Hoffman NG, Ping LH, Fiscus SA, Hoffman IF, Kitrinos KM, Banda T, Martinson FE, Kazembe PN, Chilongozi DA, Cohen MS, Swanstrom R. HIV-1 populations in blood and breast milk are similar. Virology. 2004;330:295–303. doi: 10.1016/j.virol.2004.09.004. [DOI] [PubMed] [Google Scholar]
- Schmitz JE, Kuroda MJ, Santra S, Sasseville VG, Simon MA, Lifton MA, Racz P, Tenner-Racz K, Dalesandro M, Scallon BJ, Ghrayeb J, Forman MA, Montefiori DC, Rieber EP, Letvin NL, Reimann KA. Control of viremia in simian immunodeficiency virus infection by CD8+ lymphocytes. Science. 1999;283:857–860. doi: 10.1126/science.283.5403.857. [DOI] [PubMed] [Google Scholar]
- Ramduth D, Chetty P, Mngquandaniso NC, Nene N, Harlow JD, Honeyborne I, Ntumba N, Gappoo S, Henry C, Jeena P, Addo MM, Altfeld M, Brander C, Day C, Coovadia H, Kiepiela P, Goulder P, Walker B. Differential immunogenicity of HIV-1 clade C proteins in eliciting CD8+ and CD4+ cell responses. J Infect Dis. 2005;192:1588–1596. doi: 10.1086/496894. [DOI] [PubMed] [Google Scholar]
- Permar SR, Kang HH, Carville A, Mansfield KG, Gelman RS, Rao SS, Whitney JB, Letvin NL. Potent simian immunodeficiency virus-specific cellular immune responses in the breast milk of simian immunodeficiency virus-infected, lactating rhesus monkeys. J Immunol. 2008;181:3643–3650. doi: 10.4049/jimmunol.181.5.3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabbaj S, Edwards BH, Ghosh MK, Semrau K, Cheelo S, Thea DM, Kuhn L, Ritter GD, Mulligan MJ, Goepfert PA, Aldrovandi GM. Human immunodeficiency virus-specific CD8(+) T cells in human breast milk. J Virol. 2002;76:7365–7373. doi: 10.1128/JVI.76.15.7365-7373.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salazar-Gonzalez JF, Bailes E, Pham KT, Salazar MG, Guffey MB, Keele BF, Derdeyn CA, Farmer P, Hunter E, Allen S, Manigart O, Mulenga J, Anderson JA, Swanstrom R, Haynes BF, Athreya GS, Korber BT, Sharp PM, Shaw GM, Hahn BH. Deciphering human immunodeficiency virus type 1 transmission and early envelope diversification by single-genome amplification and sequencing. J Virol. 2008;82:3952–3970. doi: 10.1128/JVI.02660-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Clustal W and Clustal X version 2. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Guindon S, Delsuc F, Dufayard JF, Gascuel O. Estimating maximum likelihood phylogenies with PhyML. Methods Mol Biol. 2009;537:113–137. doi: 10.1007/978-1-59745-251-9_6. full_text. [DOI] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Andreotti M, Galluzzo CM, Guidotti G, Germano P, Altan AD, Pirillo MF, Marazzi MC, Vella S, Palombi L, Giuliano M. Comparison of HIV Type 1 Sequences from Plasma, Cell-Free Breast Milk, and Cell-Associated Breast Milk Viral Populations in Treated and Untreated Women in Mozambique. AIDS Res Hum Retroviruses. 2009;25:707–711. doi: 10.1089/aid.2008.0276. [DOI] [PubMed] [Google Scholar]
- Pillai SK, Pond SL, Liu Y, Good BM, Strain MC, Ellis RJ, Letendre S, Smith DM, Gunthard HF, Grant I, Marcotte TD, McCutchan JA, Richman DD, Wong JK. Genetic attributes of cerebrospinal fluid-derived HIV-1 env. Brain. 2006;129:1872–1883. doi: 10.1093/brain/awl136. [DOI] [PubMed] [Google Scholar]
- Slatkin M, Maddison WP. A cladistic measure of gene flow inferred from the phylogenies of alleles. Genetics. 1989;123:603–613. doi: 10.1093/genetics/123.3.603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pond SL, Frost SD, Muse SV. HyPhy: hypothesis testing using phylogenies. Bioinformatics. 2005;21:676–679. doi: 10.1093/bioinformatics/bti079. [DOI] [PubMed] [Google Scholar]
- Allen TM, O'Connor DH, Jing P, Dzuris JL, Mothe BR, Vogel TU, Dunphy E, Liebl ME, Emerson C, Wilson N, Kunstman KJ, Wang X, Allison DB, Hughes AL, Desrosiers RC, Altman JD, Wolinsky SM, Sette A, Watkins DI. Tat-specific cytotoxic T lymphocytes select for SIV escape variants during resolution of primary viraemia. Nature. 2000;407:386–390. doi: 10.1038/35036559. [DOI] [PubMed] [Google Scholar]
- Peyerl FW, Barouch DH, Yeh WW, Bazick HS, Kunstman J, Kunstman KJ, Wolinsky SM, Letvin NL. Simian-human immunodeficiency virus escape from cytotoxic T-lymphocyte recognition at a structurally constrained epitope. J Virol. 2003;77:12572–12578. doi: 10.1128/JVI.77.23.12572-12578.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh WW, Cale EM, Jaru-Ampornpan P, Lord CI, Peyerl FW, Letvin NL. Compensatory substitutions restore normal core assembly in simian immunodeficiency virus isolates with Gag epitope cytotoxic T-lymphocyte escape mutations. J Virol. 2006;80:8168–8177. doi: 10.1128/JVI.00068-06. [DOI] [PMC free article] [PubMed] [Google Scholar]