Abstract
Natural killer (NK) cells are innate immune effector cells that make up ~10–15% of the peripheral blood lymphocytes in humans and are primarily involved in immunosurveillance to eliminate transformed and virally-infected cells. They were originally defined by their ability to spontaneously eliminate rare cells lacking expression of class I major histocompatibility complex (MHC-I) self molecules, which is commonly referred to as “missing self” recognition. The molecular basis for missing self recognition emerges from the expression of MHC-I-specific inhibitory receptors on the NK cell surface that tolerize NK cells toward normal MHC-I-expressing cells. By lacking inhibitory receptor ligands, tumor cells or virus-infected cells that have down-modulated surface MHC-I expression become susceptible to attack by NK cells. Killer cell Ig-like receptors (KIR; CD158) constitute a family of MHC-I binding receptors that play major roles in regulating the activation thresholds of NK cells and some T cells in humans. Here, we review the multiple levels of KIR diversity that contribute to the generation of a highly varied NK cell repertoire and explain how this diversity can influence susceptibility to a variety of diseases, including cancer. We further describe strategies by which KIR can be manipulated therapeutically to treat cancer, through the exploitation of KIR/MHC-I ligand mismatch to potentiate hematopoietic stem cell transplantation and the use of KIR blockade to enhance tumor cell killing.
Introduction
Natural killer (NK) cells are lymphocytes of the innate immune system that play important roles to protect from viral infections and the development of cancer.1, 2 In humans, NK cells constitute 10–15% of peripheral blood lymphocytes and are considered large granular lymphocytes due to their expression of dense intracellular cytolytic granules.3, 4 Upon encountering certain abnormal tumor or virally-infected cells, NK cells are spontaneously activated to release the contents of these granules, namely perforin and granzymes, toward the target cell.3, 5 Perforin and granzymes initiate apoptosis of the target cell.6, 7 Cytotoxicity requires the one-on-one recognition of and the adhesion to abnormal cells.8, 9 Therefore, NK cells are thought to be particularly important in eliminating single-cell tumors, especially leukemias, lymphomas and metastasizing tumor cells.10 Although originally named for their capacity to elicit cytotoxicity, NK cells are also a potent source of cytokines and chemokines, especially interferon (IFN)-γ, tumor necrosis factor (TNF)-α, and GM-CSF.11 In addition to direct effects on the tumor and virally-infected cells, these cytokines can promote the differentiation, activation and/or recruitment of other immune cells.12–14
Tumor cell recognition by NK cells is based upon their unique capacity to identify and attack cells that express diminished levels of cell surface major histocompatibility class I (MHC-I) molecules,15, 16 which are normally expressed on virtually every cell of the body. Many abnormal tumor or virally-infected cells have been shown to down-modulate MHC-I, which allows them to escape detection by cytolytic T cells.17, 18 The down-regulation of MHC-I (loss of self molecules) makes abnormal cells sensitive to NK cell cytotoxicity, however, and this process has been termed “missing self” recognition. When this “missing self” concept was first described in the late 1980s,19 it was difficult to comprehend how a lymphocyte could recognize the loss of a cell surface marker, since T and B cells were known to become activated in response to the gain of foreign molecules in the body.20, 21 It has since become clear that NK cell responsiveness is controlled by a balance of signals generated from cell surface activating and inhibitory receptors, and that the MHC-I-binding inhibitory receptors are vital to tolerizing NK cells toward normal cells through detection of these self molecules.22, 23
Controlling NK cell responsiveness
The major NK cell activating receptors are the natural cytotoxicity receptors (NCR: NKp30, NKp44, and NKp46), the Fc receptor CD16, NKG2D, and activating killer cell Ig-like receptors (KIR).24, 25 The ligands for NCR have only recently begun to be identified,26–28 while NKG2D recognizes the non-classical MHC-I molecules, MICA/MICB and ULBPs,29, 30 and activating KIR seem to recognize classical MHC-I molecules.31 On the other hand, CD16 binds the Fc portion of IgG antibodies to initiate antibody-dependent cellular cytotoxicity (ADCC)32 and provides NK cells with the ability to recognize and kill target cells coated with antibodies. In fact, CD16 has been shown to contribute to the anti-tumor properties of rituximab and herceptin antibodies in the treatment of B cell lymphoma and breast cancer, respectively.33, 34 Activation can be augmented further by the co-engagement of a variety of co-receptors (e.g. 2B4, CD2, LFA-1, and DNAM-1).4 All of these aforementioned activating receptors promote both cytotoxicity and cytokine production responses through stimulating intracellular protein tyrosine kinase cascades (Figure 1).23
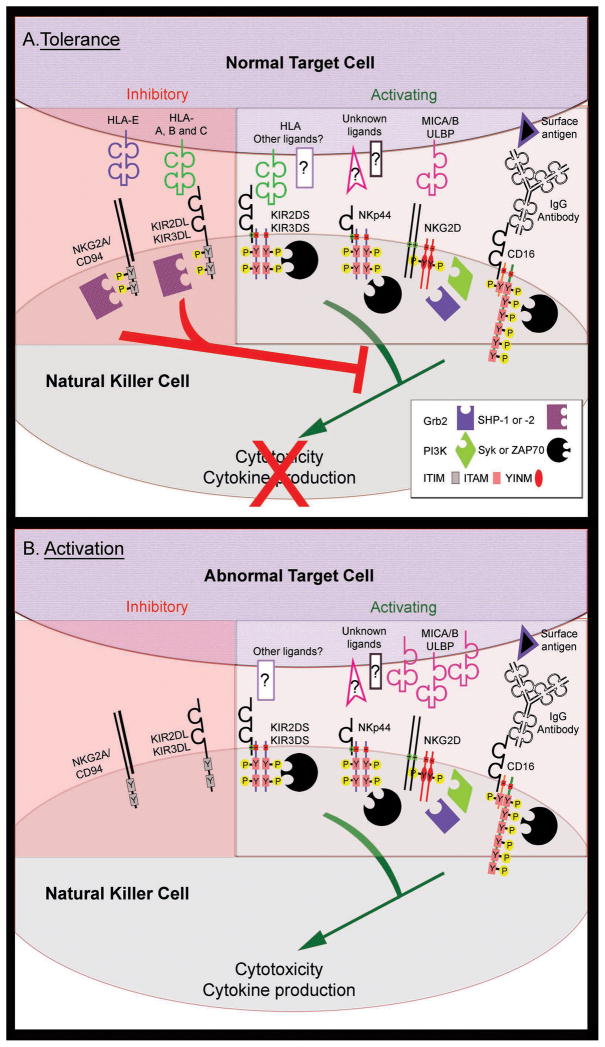
Figure 1. NK cell activity is controlled by the balance of inhibitory and activating receptors.
NK cell activity resulting from the interaction of a NK cell with a normal, MHC-I bearing target (A, tolerance) and an abnormal, tumor cell that has lost MHC-I expression (B, activation). Engagement of activating receptors (KIR, NKG2D, CD16 and the NCRs; only NKp44 is shown) with ligands on target cells stimulates NK cell cytotoxicity and the production of cytokines through transmembrane charge-based association with homo- or heterodimers of accessory molecules. The accessory molecules recruit signaling effector molecules (Syk or ZAP-70, phosphatidylinositol 3-kinase (PI3K), or Grb2) to tyrosine phosphorylated (Y-P) cytoplasmic motifs (ITAM or YINM), which mediate downstream activation signaling. Some activating receptor ligands are upregulated after cellular stress, cancerous transformation or viral infection (e.g. the NKG2D ligands, MICA/B and ULBP), further increasing NK cell activity. Normal cells are protected from NK cell-dependent cytotoxicity through the engagement of inhibitory receptors with MHC-I molecules (HLA-A, -B, -C and –E) on the normal target cell surface. Upon engagement with MHC-I, inhibitory KIR (KIR2DL and KIR3DL) and NKG2A/CD94 receptors become phosphorylated on tyrosine residues within the cytoplasmic ITIM sequences. ITIM phosphorylation leads to the recruitment of SH2 domain-containing phosphatases SHP-1 and/or SHP-2, which dominantly suppress the membrane-proximal tyrosine phosphorylation events to block activation signaling.
In contrast, two main types of NK cell inhibitory receptors that recognize MHC-I molecules provide the molecular basis for “missing self” recognition. These are the inhibitory KIR and the heterodimeric NKG2A/CD94 receptor.35, 36 Inhibitory KIR recognize subsets of the classical MHC-I molecules (human leukocyte antigens (HLA) -A, -B, and –C), while NKG2A/CD94 detects the non-classical MHC-I molecule, HLA-E.37 Engagement of inhibitory KIR and CD94/NKG2A with the ubiquitous MHC-I molecules on the surface of most cells establishes NK cell tolerance toward normal cells. Upon interaction with MHC-I ligands on the target cells, KIR and NKG2A/CD94 recruit protein tyrosine phosphatases to the plasma membrane, which counteract activating receptor signals to inhibit cytotoxicity and cytokine production (Figure 1).38–41 The balance of signals from activating and inhibitory receptors can be significantly influenced by changes in surface expression levels of ligands on the target cells, which can alter the overall activation threshold of NK cells. In this way, MHC-I-deficient cells lack inhibitory receptor ligands and become targets of NK cell-mediated attack.
KIR family members and their signaling functions
The KIR (also known as CD158) are a family of receptors encoded by 14 polymorphic genes [KIR2DL1–5, KIR3DL1–3, KIR2DS1–5, KIR3DS1],35 seven of which are inhibitory and seven of which are activating (Table I). Although the current review focuses primarily on the impacts of KIR expression on NK cell function, it is important to note that KIR are also expressed on subsets of T cells, including invariant NKT cells, and can thereby also directly influence their function.42 The KIR nomenclature is based upon structural features of the extracellular domain (2D versus 3D; referring to the number of extracellular Ig-like domains) and for the length of the cytoplasmic tail (L, long versus S, short). KIR function can be predicted from the length of the cytoplasmic domain, where L receptors are generally inhibitory and all S receptors are activating.43, 44 The only exception to this rule is KIR2DL4, which is an unique activating receptor that stimulates potent cytokine production, but minimal cytotoxicity.45, 46 Inhibitory receptors contain one or two immunoreceptor tyrosine-based inhibitory motifs [ITIM; (I/V)xYxx(L/V)], which are necessary and sufficient for inhibitory KIR function.40, 47 When inhibitory KIR engage with MHC-I on target cells, the ITIM sequences are phosphorylated by Src family protein tyrosine kinases, which creates specific docking sites for the SHP-1 and SHP-2 protein tyrosine phosphatases (Figure 1).23, 48 Recruitment of SHP-1/2 leads to the dominant suppression of activating receptor signals transduced via protein tyrosine kinases.38, 39, 47 On the other hand, activating KIR lack ITIMs, but alternatively possess a charged transmembrane residue, which facilitates physical association with the transmembrane accessory proteins DAP12 or FcεRI–γ (Figure 1).23, 48, 49 DAP12 and FcεRI–γ deliver activating signals through immunoreceptor tyrosine-based activation motifs [ITAM; Yxx(L/I/V)x6–8Yxx(L/I/V)] in their cytoplasmic domains, which are phosphorylated by Src family kinases and recruit Syk/ZAP-70 family protein tyrosine kinases to mediate downstream activation signaling.23 Consistent with activating KIR, NCR and CD16 associate with ITAM-containing accessory proteins (DAP12, FcεRI-γ and TCR-ζ) and promote activation through the recruitment of Syk/ZAP-70. Alternatively, NKG2D associates with the accessory protein DAP10 to promote activation via recruitment of phosphatidylinositol 3-kinase and Grb2.50
Table I.
Killer cell Ig-like Receptor (KIR) family members
| Gene name | Alternate names | Recognition motif on ligands | Common alleles of ligands |
|---|---|---|---|
| KIR2DL1 | CD158a, nkat1 | HLA-C2 | C2: Cw2, Cw4, Cw5, Cw6 |
| KIR2DL2 | CD158b1, nkat6 | HLA-C1 > HLA-C2 | C1: Cw1, Cw3, Cw7, Cw8 |
| KIR2DL3 | CD158b2, nkat2 | HLA-C1 > HLA-C2 | C1: Cw1, Cw3, Cw7, Cw8 |
| KIR2DL5A* | CD158f | Unknown | |
| KIR2DL5B* | KIR2DL5.2 | Unknown | |
| KIR3DL1 | NKB1, nkat3 | HLA-Bw4 and some HLA-A | B08, B27, B57, A24 |
| KIR3DL2 | CD158k, nkat4 | Certain HLA-A allotypes | A3, A11 |
| KIR3DL3 | CD158z | Unknown | |
| KIR2DL4 | CD158d | HLA-G | |
| KIR2DS1 | CD158h | HLA-C2A | C2: Cw2, Cw4, Cw5, Cw6 |
| KIR2DS2 | CD158j, nkat5 | HLA-C1A | C1: Cw1, Cw3, Cw7, Cw8 |
| KIR2DS3 | nkat7 | HLA-C1A | C1: Cw1, Cw3, Cw7, Cw8 |
| KIR2DS4 | CD158i, nkat8 | disease peptide?; HLA-CA | Cw3, Cw4B |
| KIR2DS5 | CD158g, nkat9 | Unknown | |
| KIR3DS1 | CD158e2, nkat10 | HLA-Bw4? |
KIR ligands
Individual KIR recognize distinct subsets of the classical human MHC-I molecules, HLA-A, -B, and -C.4 HLA are encoded by a collection of genes on a separate chromosome (6p21.3) from the KIR genes (19q13.4) and are thereby inherited independently.51 KIR have evolved rapidly in higher mammals, especially primates, to become a highly polymorphic family of receptors with the capacity to detect common elements found on broad subsets of the even more polymorphic HLA molecules.52, 53 Similar to the T cell receptor (TCR), KIR bind to HLA across the peptide binding groove of MHC-I; however, unlike the TCR, KIR only contact the C-terminal end of the peptide presented on MHC-I.54 Nonetheless, distinct peptides have been shown to diminish KIR binding to MHC-I, suggesting that certain tumor- or virus-derived peptides may diminish inhibitory KIR recognition to lower the NK cell activation threshold.55–57
Together the different inhibitory KIR have the capacities to recognize 100% of the known HLA-C allotypes (which can be divided into C1 and C2 subgroups) and subsets of HLA-B and HLA-A allotypes. Ligand binding specificity of KIR is determined by specific sequence elements in the HLA molecules (Table I and Figure 2A). KIR2DL1 binds HLA-C2 allotypes, which characteristically have a lysine at position 80.58–60 KIR2DL2/KIR2DL3, which segregate as alleles of the same gene, bind HLA-C1 allotypes that contain an asparagine at position 80.21, 61, 62 KIR2DL2 is reported to have higher affinity for some HLA-C1 allotypes, and as a consequence, has been shown to function as a stronger inhibitory receptor than KIR2DL3 in several contexts.63, 64 Some reports suggest (either with direct binding or functional assays) that both KIR2DL2/3 can also bind some HLA-C2 allotypes and a few unconventional HLA-B allotypes.63, 65 Therefore, KIR2DL1–3 together are able to inhibit NK cytotoxicity against cells expressing any HLA-C allotype. KIR3DL1 recognizes a specific motif termed ‘Bw4’66 that is found in roughly 40% of the known HLA-B allotypes and some HLA–A allotypes. The remaining HLA-B allotypes are typified by a Bw6 motif, which is not recognized by KIR3DL1.67–69 HLA-Bw4 motifs can be further divided into allotypes with an isoleucine or threonine at position 80, which exhibit higher and lower affinity for KIR3DL1, respectively.67, 68, 70 Therefore, KIR3DL1 will only inhibit NK cell cytotoxicity against target cells expressing a discrete subset of HLA-B and HLA-A allotypes. KIR3DL2 is only known to recognize HLA-A3 and HLA-A11 allotypes, and these interactions can be modulated by certain presented peptides.55, 71, 72 To date, the ligands of KIR2DL5 and KIR3DL3 are unknown and remain to be identified.
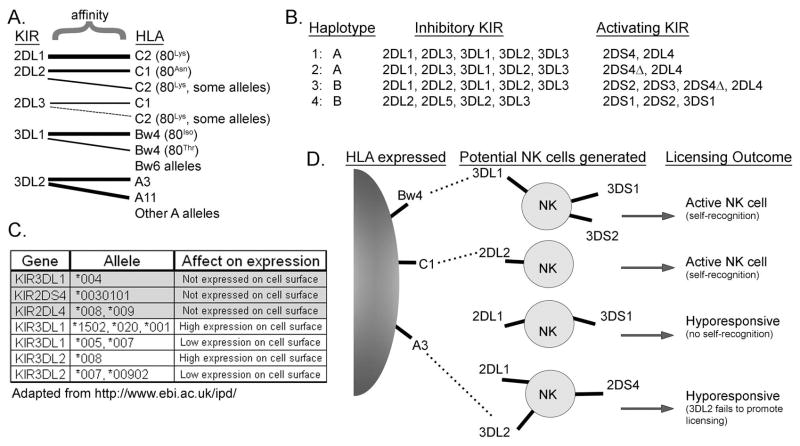
Figure 2. Multifactorial diversity of KIR inheritance and expression.
A. Inhibitory KIR have separate and sometimes overlapping affinity for distinct HLA molecules. KIR2DL1 has strong affinity for HLA-C2, while KIR2DL2 and KIR2DL3 recognize HLA-C1 with strong affinity and HLA-C2 weakly. KIR3DL1 recognizes Bw4 motifs, but not Bw6 motifs in HLA-B and some -A allotypes, while KIR3DL2 is only known to recognize HLA-A3 and –A11. The ligand interactions are indicated, with wider lines signifying a stronger affinity, while the HLA groups lacking lines (e.g. Bw6 and other A alleles) are not recognized by any KIR. B. KIR genes are inherited in gene arrays named haplotypes of which two major subtypes exist (designated haplotypes A and B). The A haplotypes encode mainly inhibitory KIR (KIR2DL/KIR3DL) with only 1–2 activating KIR (KIR2DL4 and/or KIR2DS4), while the more diverse B haplotypes encode additional activating KIR (KIR2DS/KIR3DS). Representative examples for both haplotypes from four hypothetical human donors are shown with the inherited inhibitory and activating KIR listed. Due to a 22 bp deletion leading to a premature stop codon in a large fraction of KIR2DS4 genes (denoted KIR2DS4), many individuals with A and some B haplotypes fail to express functional KIR2DS4 protein. C. Minor KIR allelic polymorphism can affect both the presence of KIR on the cell surface and their level of surface expression. Many of these minor polymorphisms vary at only one or a few amino acids and it is currently unclear whether some variations also alter HLA recognition. Examples of some KIR2DL4, KIR2DS4, KIR3DL1 and KIR3DL2 alleles that affect expression are shown. D. The variegated expression of KIR during NK cell differentiation will generate a pool of NK cells that is heterogeneous for KIR expression and NK cell activity. In this example, a person lacking expression of HLA-C2 would generate a hyporesponsive NK cell if KIR2DL1 was the only inhibitory receptor expressed on an individual NK cell (no self-recognition). In contrast, NK cells that express inhibitory KIR for which ligand is present (KIR2DL2, KIR3DL1), would become fully functional (self-recognition). Adding to the complexity, Fauriat et al. showed that KIR3DL2 was unable to license NK cells,92 indicating that an NK cell only expressing KIR3DL2 as a self-recognizing receptor would also be hyporesponsive. Due to their low affinity, interactions between activating (“S”) KIR and HLA are not indicated.
Given the extensive homology between the extracellular domains of activating and inhibitory KIR (~99%), numerous studies have reported that activating KIR recognize the same HLA molecules as their inhibitory counterparts, although with significantly weaker affinities (see Table I).58, 64, 73–75 This binding specificity is uncertain, however, due to the lower affinities and inconsistencies between some reports. For example, two studies have implicated both HLA-C1 and –C2 as potential ligands for KIR2DS4.76, 77 It is possible that the activating KIR-HLA affinities are enhanced by specific peptides presented in the HLA molecules, as has been shown for KIR2DS1 interactions with Epstein-Barr virus-infected cells.75 Alternatively, entirely distinct ligands for activating KIR may exist, since KIR2DS4 has been shown to recognize a non-MHC-I polypeptide on the surface of melanoma cells.78
KIR diversity
Unlike HLA, in which the majority of diversity stems from sequence polymorphism within each of the inherited genes (two copies of HLA-A, -B, and –C are inherited, one from each parent), KIR diversity is multifactorial (Figure 2).79 This multifactorial nature of KIR diversity is based upon: 1) inheritance of diverse allelic combinations of the 14 available KIR genes (haplotypes) by individuals in the human population, 2) the existence of minor allelic sequence polymorphisms within each KIR gene, and 3) variegated expression of distinct KIR molecules on the surfaces of individual NK cells in the peripheral blood.44
The KIR genes are located in a ~150 kb segment within the leukocyte receptor complex on chromosome 19q13.4. They are tandemly arrayed into haplotypes that vary widely in the number of KIR genes present and the distribution of activating and inhibitory alleles.44 The most common human haplotype, designated haplotype A, encodes for mostly inhibitory receptors, including KIR3DL3, KIR2DL2/3, KIR2DL1, KIR3DL1, and KIR3DL2, and the activating KIR2DS4 and KIR2DL4 (Figure 2B).35 Importantly, KIR2DS4 has been found to be non-functional in 80% of European Americans, due to a small deletion,43, 80 and a high proportion of inherited KIR2DL4 alleles are not expressed on the cell surface.81 Therefore, many A haplotypes lack expression of any activating KIR. In contrast, B haplotypes are much more diverse in the total number of encoded genes and number of activating KIR, since the B haplotype is broadly defined as possessing at least one more activating KIR2DS/KIR3DS gene in addition to KIR2DS4 (Figure 2B).35, 43 The distribution of A and B KIR haplotypes varies widely between distinct ethnic groups, being roughly equal in individuals of European descent, while the A haplotype dominates in Korean and Japanese populations, but is overshadowed by the B haplotype in Australian Aborigines.80, 82–84 HLA ligand distribution is also stratified by ethnic group. For instance, 92% of Japanese express the KIR2DL2/3 ligand, HLA-C1, as compared to 66% of Caucasians.85 Therefore, distinct ethnic groups can potentially express vastly different combinations of KIR and HLA ligands.
In addition to the diversity of KIR gene haplotypes, minor polymorphisms within KIR and HLA genes can significantly influence their ligand affinities and levels of cell surface expression (Figure 2C). For example, distinct alleles of KIR3DL1, one of the most polymorphic KIR genes, can be expressed at widely varying levels on the cell surface.85, 86 Some KIR alleles encode receptors that are not expressed on the cell surface at all (e.g. KIR3DL1*004, KIR2DL4*008, KIR3DS1*049N); however, it is currently unclear whether these intracellular forms still provide some function.45, 87, 88 In addition, several reports have found that residues outside the HLA-Bw4 motif can influence both the binding affinity and the inhibitory capacity of KIR3DL1.70, 89, 90 Similar to these influences on HLA-Bw4, residues outside the HLA-C1 and HLA-C2 motifs have been shown to influence the binding affinities for KIR2DL2/3 and KIR2DL1, respectively.63 Therefore, allelic polymorphisms can influence both the affinities of KIR for HLA, as well as the levels of KIR or HLA surface expression, and the full impacts of this diversity have not yet been fully defined.
Variation in KIR expression within the peripheral NK cell pool
In addition to the diversity at the genomic level, KIRs are expressed in a variegated pattern on distinct NK cells, thereby creating a repertoire of NK cells expressing distinct combinations of KIRs within the peripheral blood (Figure 2D).79, 91 During NK cell development, multiple KIR are randomly expressed on the NK cell surface. Since KIR and HLA are independently inherited on distinct chromosomes, some inhibitory KIR within the repertoire can be expressed without concomitant expression of their cognate ligands. Once initiated during NK cell development, KIR expression is stably maintained throughout the lifetime of an NK cell and in the resulting daughter cells.92–94 This stochastic expression generates NK cells either lacking or expressing one or multiple KIR on individual NK cells in the repertoire.92 During NK cell maturation, the NKG2A/CD94 dimer is expressed prior to KIR and can be co-expressed more readily in NK cells expressing fewer inhibitory KIR.92
Additional factors influence the composition of KIR expression on NK cells during development. Fischer et al. showed that 2DL3 is expressed prior to 2DL1 during NK cell differentiation, and that ordered expression impacted upon the overall frequency of KIR expression (i.e., the proportion of 2DL3+ NK cells was greater than the 2DL1+ fraction).95 Gene copy number can also influence the incidence of overall KIR surface expression, since each allele is independently regulated. Therefore, people who inherit two alleles of a particular KIR will have a greater proportion of their NK cell pool expressing that specific KIR as compared to those with only one allele.85, 96
KIR/HLA inheritance and susceptibility to diseases including cancer
Since the interactions of KIR with cognate HLA ligands can dramatically influence overall responsiveness of NK, T or NKT cells expressing these receptors, they have the potential to influence both the innate and adaptive immune responses.97, 98 Furthermore, since both KIR and HLA ligands are encoded by highly polymorphic genes, it is perhaps not surprising that susceptibility to pathological conditions can be influenced by the combined inheritance of distinct KIR/HLA gene combinations (referred to as the KIR/HLA compound genotypes).98 A wide array of studies have reported associations between distinct KIR/HLA compound genotypes with susceptibility or resistance to viral infections (Hepatitis C, HIV), autoimmune or chronic inflammatory diseases (vasculitis, psoriasis, diabetes, idiopathic bronchiectasis, birdshot chorioretinopathy), and conditions affecting the outcome of pregnancy (spontaneous abortion and pre-eclampsia).31, 43, 96 In addition, several studies have described KIR/HLA compound genotypes that associate with susceptibilities to certain cancers (melanoma, leukemia, cervical neoplasia, Hodgkin’s lymphoma).99–103
Coordinate inheritance of certain combinations of KIR and HLA genes (or specific alleles) have been shown to correlate with disease susceptibility in ways that suggest a variety of possible mechanistic influences.31, 43, 96 Many studies found that a less inhibitory KIR/HLA compound genotype (such as homozygosity of low affinity KIR2DL3 with cognate HLA-C1 ligand) was positively correlated with susceptibility to inflammatory disorders. These studies suggest that a lower threshold of activation (due to weak inhibition) leads to hyper-responsive NK cells and T cells, which may promote an overactive immune system. In contrast, a less inhibitory KIR/HLA compound genotype was shown to be protective against some viral infections. These data suggest that NK cells and cytolytic T cells exhibiting weaker inhibitory KIR interactions are more able to clear the infection. However, the Carrington lab has found that the activating KIR3DS1/Bw4 and inhibitory KIR3DL1/Bw4 receptor/ligand combinations were both protective in HIV/AIDS (as measured by incidence of progression to AIDS and viral load),104 suggesting that KIR/HLA combinations can have multiple influences on immune responsiveness, which are difficult to explain without further mechanistic insight.
Self-recognizing KIR/HLA interactions can impact NK cell responsiveness in at least two ways. First, the combined potency of interactions can shift the balance between activating and inhibitory signals, whereby a strongly interacting inhibitory KIR/HLA compound genotype would lead to less-responsive NK cells. In this way, KIR act as rheostats to regulate the NK cell activation threshold. In this context, a strong inhibitory KIR/HLA compound genotype may be deleterious when fighting infection or cancer, due to a decreased NK cell response to the presence of the virus or tumor cells. Second, the potency of self-recognizing KIR/HLA interactions can also impact upon development of the total pool of responsive, mature NK cells in a process referred to as NK cell education or licensing. During NK cell development, self-recognition of MHC-I by inhibitory receptors promotes the terminal maturation and functional competence of NK cells.94 In contrast, NK cells lacking expression of self-recognizing inhibitory receptors (KIR or NKG2A) fail to mature normally and become hyporesponsive. Therefore, KIR/HLA interactions, particularly self-recognizing inhibitory KIR/HLA interactions, have the potential to impact NK cell responsiveness: 1) during NK cell maturation where they promote the generation of functional pools of NK cells and 2) by controlling the activation threshold of the mature NK cells. These two mechanisms by which KIR/HLA interactions influence responsiveness of the mature NK cell pool have the potential to be contradictory. Therefore, it may be difficult to predict how a particular KIR/HLA compound genotype may impact the immune response in vivo.
Although the search for impacts of KIR/HLA on disease is burgeoning, caution is warranted when interpreting immune functions from some of these correlative studies. For instance, interpretation of many of the published studies is confounded by an array of issues: 1) limited sample size (thus lacking strong statistical significance), 2) a lack of an appropriate control group (e.g. matched for age and ethnicity, since age can affect disease onset and the distribution of KIR and HLA can be vastly different between ethnic groups), 3) a failure in some studies to remove KIR alleles that are not expressed on the cell surface from the analysis, 4) a failure to consider KIR in the context of its ligand(s) and 5) a lack of full understanding of all of the impacts of distinct KIR polymorphisms on HLA ligand recognition. Moreover, different diseases are influenced in distinct ways by the immune system and the KIR/HLA relationship could potentially impact a disease at multiple stages (e.g. initiation versus progression of disease versus age of onset); therefore, it becomes difficult to predict common themes to explain how KIR/HLA compound genotypes impact upon the susceptibility to diverse diseases.
Influences of KIR/HLA combinations in hematopoietic stem cell-based cancer therapy
Hematopoietic stem cell transplantion (HSCT) is commonly used to treat hematologic malignancies, where eradication of tumor cells and reconstitution of the recipient’s immune system with transplanted cells is required.105 In contrast to other forms of tissue transplantation, partial HLA allogenicity in HSCT can provide beneficial graft-versus-tumor effects to treat hematologic malignancies, although graft-versus-host disease (GVHD) can be a negative consequence. In recent years, allogenicity between donor inhibitory KIR and recipient HLA ligands has been shown to further influence HSCT to treat such malignancies.
Given the tremendous diversity of KIR and HLA genes between individuals and the variegated expression of KIR on individual NK cells during development, it is likely that some donor-derived NK cells transplanted in HSCT would be ‘alloreactive’ toward the tissues of a recipient lacking appropriate HLA ligand (referred to as a KIR/HLA ligand mismatch).105–107 Therefore, during HSCT, a mismatch between the KIR expressed by the transplant donor and the HLA ligands expressed by the transplant recipient would result in an alloreactive NK cell pool that would be reactive toward recipient tissues, and hence tumor cells. KIR-HLA ligand mismatch has been shown to be particularly beneficial in “haploidentical” HSCT, in which the stem cells share only partial match in HLA haplotype with that of the recipient. Parents or siblings are generally suitable haploidentical donors that are usually readily available.
In 2002, Ruggeri et al. showed that the presence of alloreactive NK cells significantly improved the therapeutic benefit of haploidentical HSCT in treating patients with acute myeloid leukemia (AML) by promoting the graft-versus-tumor response, increasing survival and decreasing relapse.108 Interestingly, they also found that NK cell alloreactivity decreased the incidence of the most grievous side effect of HSCT, GVHD, which is mediated by allogeneic donor-derived T cells. Consistent with an earlier report, the authors further provided evidence that the reduced GVHD was due to depletion of residual antigen presenting cells in the transplant recipient.108, 109 It is important to note that allogeneic NK cells do not seem to directly mediate GVHD responses to normal tissues in HSCT recipients, possibly due to minimal expression of activating receptor ligands on normal cells.
Unfortunately, initial attempts by other groups failed to replicate the beneficial effect of KIR-HLA mismatch on therapeutic outcome of HSCT.110–112 These initial follow-up studies, however, utilized significantly different HSCT treatment protocols, as compared to that of Ruggeri et al. Subsequent studies107, 113, 114 have found that incorporation of certain treatment parameters is critical to achieve beneficial impacts of KIR/HLA ligand mismatch in haploidentical HSCT therapies: 1) aggressive myeloablative and immunosuppressive pre-conditioning of the recipient, such as full body irradiation and chemotherapy, to preempt graft rejection and decrease cancer burden, 2) extensive T-cell depletion of the HSC prior to engraftment to prevent GVHD, and 3) infusion of very high numbers of HSC.
Another factor that may significantly benefit the outcome of haploidentical HSCT is the presence of a large proportion of NK cells in the peripheral blood of the donor that would be alloreactive toward the recipient (fraction of NK cells lacking inhibitory KIR that recognize the recipient’s HLA).65, 114 It has been shown that the maturing NK cell pool established within the HSCT recipient takes on a KIR expression profile that becomes similar to that of the donor.115, 116 Therefore, a larger fraction of allogeneic NK cells in the donor to recipient direction would presumably be particularly beneficial. Fauriat et al. (2008) recently found that the alloreactive fraction of the NK cell pool varies widely (0 to 62%) among potential donors, showing that not all individuals would be appropriate donors for HSCT.92, 106 The direct infusion of mature allogeneic NK cells is also being performed by some groups, and has been shown to be safe and provide some effective anti-leukemic responses.117 Future development of this technique also shows therapeutic promise.
In addition to the potential benefits in haploidentical HSCT, KIR-HLA relationships can also influence the outcomes of other HSCT conditions. For example, Hsu et al. showed increased disease-free survival in patients with AML and myelodysplastic syndrome that had received HLA-identical HSCT with inhibitory KIR/HLA ligand mismatch.118 Similarly, Leung et al. showed survival benefit with inhibitory KIR/HLA ligand mismatch after autologous HSCT (re-infusing the patients’s HSC after intervening chemotherapy) to treat various lymphomas and solid tumors.119 In contrast, a recent report from Cooley et al. found that AML patients receiving T cell-replete HSCT did not benefit from inhibitory KIR-HLA ligand mismatch.120 That study, however, showed significantly improved relapse-free survival when the donor possessed at least one B haplotype of KIR (which contain more activating KIR genes than haplotype A), indicating that introducing more activating KIR on the transplanted NK cells can also be beneficial under these conditions.120
In summary, many reports have described beneficial impacts of KIR-mediated NK cell allogenicity in HSCT therapies to treat AML and other cancers. The influences of allogenicity due to KIR/HLA ligand mismatch are complex and mechanistic foundations are currently unclear, however. Future work will need to focus on optimizing transplantation conditions and improving our molecular understanding of which KIR/HLA combinations provide the most benefit.
Therapeutic manipulation of KIR to treat cancer
Since inhibitory KIR play prominent roles in regulating NK cell activation, therapeutic strategies designed to diminish KIR function may be able to potentiate NK cell activity in treating cancer and viral infections.121 This could be particularly relevant in treating patients with tumors that still express HLA molecules, and therefore are not appropriate targets for attack by NK cells. In addition to their relevance to inhibitory KIR function, SHP-1 and SHP-2 phosphatases are common signaling effectors for a wide array of inhibitory receptors in a variety of tissues, and therefore, pharmacological inhibition of these phosphatases would not provide enough therapeutic specificity to enhance only NK cell function in vivo. Alternatively, specific therapeutic blockade of ligand recognition by inhibitory KIR offers intriguing potential to specifically lower the NK cell activation threshold.
A recent report by Binyamin et al. used an in vitro autologous primary NK cell/transformed target cell model system to demonstrate that antibody-mediated blockade of KIR significantly boosted CD16-dependent ADCC responses.122 Importantly, inhibitory KIR blockade alone did not significantly increase killing of autologous transformed cells in these in vitro studies, indicating that additional self-tolerance mechanisms are in place to prevent attack of cells not targeted by anti-tumor antibody. Similarly increased tumor cell killing and lack of self-reactivity was recently demonstrated by antibody-mediated KIR blockade using an in vivo KIR/HLA transgenic mouse model.123 Furthermore, using their in vitro autologous target cell system, Binyamin et al. showed that KIR blockade could better potentiate ADCC responses by NK cells from donors homozygous for the low affinity allele of CD16 (FcγRIIIA 158F).122 This is important, since follicular lymphoma patients homozygous for the low affinity allele of FcγRIIIA (158F vs. 158V) exhibit reduced clinical response rates to rituximab therapy.34, 124, 125 Another recent report has described a newly developed human antibody that blocks KIR2DL1–3 and showed impressive capacity to improve natural cytotoxicity and ADCC responses toward AML blasts in humanized mouse studies.126 Together, these reports provide preclinical evidence that KIR blockade may be a therapeutically viable option to boost NK cell mediated cytotoxicity responses toward tumors in cancer patients.
Conclusions
In conclusion, our understanding of the diversity of KIR/HLA interactions is improving, but new findings are continually revealing increasing levels of complexity. It is becoming clear, however, that the interactions between these highly polymorphic receptor and ligand repertoires can alter immune function to influence susceptibility to many diseases including cancer and the effectiveness of HSCT to treat leukemias, especially AML. Further study is warranted to better define the influences of distinct polymorphic variants of KIR and HLA on surface expression and receptor/ligand affinities, as well as to define the remaining ligands for KIR. In addition, more studies are needed to better understand the mechanistic basis of how certain KIR/HLA interactions are influencing NK cell maturation and function in certain individuals to affect their health. Also, it is important to consider that some of the impacts of KIR may be mediated through effects on subsets of T cells. Finally, additional work is needed to establish how certain KIR/HLA combinations can be selected to optimize the outcome of hematopoietic stem cell transplantation to treat cancer.
Acknowledgments
We thank Drs. Jennifer Rhodes (FCCC) and Wesley Rose (Arcadia University, Glenside, PA) for constructive criticism during manuscript preparation. This work was supported by R01 grants CA-083859 and CA-100226 (K.S.C), training grant CA009035-32 (A.K.P) and partially by Centers of Research Excellence grant CA06927 (FCCC) from the National Institutes of Health. The research was also supported in part by an appropriation from the Commonwealth of Pennsylvania. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the National Cancer Institute.
References
- 1.Biron CA. Activation and function of natural killer cell responses during viral infections. Curr Opin Immunol. 1997;9:24–34. doi: 10.1016/s0952-7915(97)80155-0. [DOI] [PubMed] [Google Scholar]
- 2.Trinchieri G. Biology of natural killer cells. Adv Immunol. 1989;47:187–376. doi: 10.1016/S0065-2776(08)60664-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Caligiuri MA. Human natural killer cells. Blood. 2008;112:461–9. doi: 10.1182/blood-2007-09-077438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu J, Lanier LL. Natural killer cells and cancer. Adv Cancer Res. 2003;90:127–56. doi: 10.1016/s0065-230x(03)90004-2. [DOI] [PubMed] [Google Scholar]
- 5.Smyth MJ, Cretney E, Kelly JM, Westwood JA, Street SE, Yagita H, et al. Activation of NK cell cytotoxicity. Mol Immunol. 2005;42:501–10. doi: 10.1016/j.molimm.2004.07.034. [DOI] [PubMed] [Google Scholar]
- 6.Lieberman J. The ABCs of granule-mediated cytotoxicity: new weapons in the arsenal. Nat Rev Immunol. 2003;3:361–70. doi: 10.1038/nri1083. [DOI] [PubMed] [Google Scholar]
- 7.Shresta S, Pham CT, Thomas DA, Graubert TA, Ley TJ. How do cytotoxic lymphocytes kill their targets? Curr Opin Immunol. 1998;10:581–7. doi: 10.1016/s0952-7915(98)80227-6. [DOI] [PubMed] [Google Scholar]
- 8.Patarroyo M, Prieto J, Rincon J, Timonen T, Lundberg C, Lindbom L, et al. Leukocyte-cell adhesion: a molecular process fundamental in leukocyte physiology. Immunol Rev. 1990;114:67–108. doi: 10.1111/j.1600-065x.1990.tb00562.x. [DOI] [PubMed] [Google Scholar]
- 9.Robertson MJ, Caligiuri MA, Manley TJ, Levine H, Ritz J. Human natural killer cell adhesion molecules. Differential expression after activation and participation in cytolysis. J Immunol. 1990;145:3194–201. [PubMed] [Google Scholar]
- 10.Ruggeri L, Mancusi A, Perruccio K, Burchielli E, Martelli MF, Velardi A. Natural killer cell alloreactivity for leukemia therapy. J Immunother. 2005;28:175–82. doi: 10.1097/01.cji.0000161395.88959.1f. [DOI] [PubMed] [Google Scholar]
- 11.Perussia B. The Cytokine Profile of Resting and Activated NK Cells. Methods. 1996;9:370–8. doi: 10.1006/meth.1996.0042. [DOI] [PubMed] [Google Scholar]
- 12.Degli-Esposti MA, Smyth MJ. Close encounters of different kinds: dendritic cells and NK cells take centre stage. Nat Rev Immunol. 2005;5:112–24. doi: 10.1038/nri1549. [DOI] [PubMed] [Google Scholar]
- 13.Di Santo JP. Natural killer cells: diversity in search of a niche. Nat Immunol. 2008;9:473–5. doi: 10.1038/ni.f.201. [DOI] [PubMed] [Google Scholar]
- 14.French AR, Yokoyama WM. Natural killer cells and viral infections. Curr Opin Immunol. 2003;15:45–51. doi: 10.1016/s095279150200002x. [DOI] [PubMed] [Google Scholar]
- 15.Storkus WJ, Alexander J, Payne JA, Dawson JR, Cresswell P. Reversal of natural killing susceptibility in target cells expressing transfected class I HLA genes. Proc Natl Acad Sci U S A. 1989;86:2361–4. doi: 10.1073/pnas.86.7.2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Storkus WJ, Howell DN, Salter RD, Dawson JR, Cresswell P. NK susceptibility varies inversely with target cell class I HLA antigen expression. J Immunol. 1987;138:1657–9. [PubMed] [Google Scholar]
- 17.Collins KL, Baltimore D. HIV’s evasion of the cellular immune response. Immunol Rev. 1999;168:65–74. doi: 10.1111/j.1600-065x.1999.tb01283.x. [DOI] [PubMed] [Google Scholar]
- 18.Restifo NP, Kawakami Y, Marincola F, Shamamian P, Taggarse A, Esquivel F, et al. Molecular mechanisms used by tumors to escape immune recognition: immunogenetherapy and the cell biology of major histocompatibility complex class I. J Immunother Emphasis Tumor Immunol. 1993;14:182–90. doi: 10.1097/00002371-199310000-00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Karre K, Ljunggren HG, Piontek G, Kiessling R. Selective rejection of H-2-deficient lymphoma variants suggests alternative immune defence strategy. Nature. 1986;319:675–8. doi: 10.1038/319675a0. [DOI] [PubMed] [Google Scholar]
- 20.Dal Porto JM, Gauld SB, Merrell KT, Mills D, Pugh-Bernard AE, Cambier J. B cell antigen receptor signaling 101. Mol Immunol. 2004;41:599–613. doi: 10.1016/j.molimm.2004.04.008. [DOI] [PubMed] [Google Scholar]
- 21.Smith-Garvin JE, Koretzky GA, Jordan MS. T cell activation. Annu Rev Immunol. 2009;27:591–619. doi: 10.1146/annurev.immunol.021908.132706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Joncker NT, Raulet DH. Regulation of NK cell responsiveness to achieve self-tolerance and maximal responses to diseased target cells. Immunol Rev. 2008;224:85–97. doi: 10.1111/j.1600-065X.2008.00658.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.MacFarlane AWt, Campbell KS. Signal transduction in natural killer cells. Curr Top Microbiol Immunol. 2006;298:23–57. doi: 10.1007/3-540-27743-9_2. [DOI] [PubMed] [Google Scholar]
- 24.Lanier LL. NK cell receptors. Annu Rev Immunol. 1998;16:359–93. doi: 10.1146/annurev.immunol.16.1.359. [DOI] [PubMed] [Google Scholar]
- 25.Moretta A, Bottino C, Vitale M, Pende D, Cantoni C, Mingari MC, et al. Activating receptors and coreceptors involved in human natural killer cell-mediated cytolysis. Annu Rev Immunol. 2001;19:197–223. doi: 10.1146/annurev.immunol.19.1.197. [DOI] [PubMed] [Google Scholar]
- 26.Pogge von Strandmann E, Simhadri VR, von Tresckow B, Sasse S, Reiners KS, Hansen HP, et al. Human leukocyte antigen-B-associated transcript 3 is released from tumor cells and engages the NKp30 receptor on natural killer cells. Immunity. 2007;27:965–74. doi: 10.1016/j.immuni.2007.10.010. [DOI] [PubMed] [Google Scholar]
- 27.Brandt CS, Baratin M, Yi EC, Kennedy J, Gao Z, Fox B, et al. The B7 family member B7-H6 is a tumor cell ligand for the activating natural killer cell receptor NKp30 in humans. J Exp Med. 2009;206:1495–503. doi: 10.1084/jem.20090681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hecht ML, Rosental B, Horlacher T, Hershkovitz O, De Paz JL, Noti C, et al. Natural cytotoxicity receptors NKp30, NKp44 and NKp46 bind to different heparan sulfate/heparin sequences. J Proteome Res. 2009;8:712–20. doi: 10.1021/pr800747c. [DOI] [PubMed] [Google Scholar]
- 29.Diefenbach A, Raulet DH. The innate immune response to tumors and its role in the induction of T-cell immunity. Immunol Rev. 2002;188:9–21. doi: 10.1034/j.1600-065x.2002.18802.x. [DOI] [PubMed] [Google Scholar]
- 30.Steinle A, Li P, Morris DL, Groh V, Lanier LL, Strong RK, et al. Interactions of human NKG2D with its ligands MICA, MICB, and homologs of the mouse RAE-1 protein family. Immunogenetics. 2001;53:279–87. doi: 10.1007/s002510100325. [DOI] [PubMed] [Google Scholar]
- 31.Bashirova AA, Martin MP, McVicar DW, Carrington M. The killer immunoglobulin-like receptor gene cluster: tuning the genome for defense. Annu Rev Genomics Hum Genet. 2006;7:277–300. doi: 10.1146/annurev.genom.7.080505.115726. [DOI] [PubMed] [Google Scholar]
- 32.Ahmad A, Menezes J. Antibody-dependent cellular cytotoxicity in HIV infections. FASEB J. 1996;10:258–66. doi: 10.1096/fasebj.10.2.8641559. [DOI] [PubMed] [Google Scholar]
- 33.Carson WE, Parihar R, Lindemann MJ, Personeni N, Dierksheide J, Meropol NJ, et al. Interleukin-2 enhances the natural killer cell response to Herceptin-coated Her2/neu-positive breast cancer cells. Eur J Immunol. 2001;31:3016–25. doi: 10.1002/1521-4141(2001010)31:10<3016::aid-immu3016>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- 34.Weng WK, Levy R. Two immunoglobulin G fragment C receptor polymorphisms independently predict response to rituximab in patients with follicular lymphoma. J Clin Oncol. 2003;21:3940–7. doi: 10.1200/JCO.2003.05.013. [DOI] [PubMed] [Google Scholar]
- 35.Vilches C, Parham P. KIR: diverse, rapidly evolving receptors of innate and adaptive immunity. Annu Rev Immunol. 2002;20:217–51. doi: 10.1146/annurev.immunol.20.092501.134942. [DOI] [PubMed] [Google Scholar]
- 36.Lopez-Botet M, Llano M, Navarro F, Bellon T. NK cell recognition of non-classical HLA class I molecules. Semin Immunol. 2000;12:109–19. doi: 10.1006/smim.2000.0213. [DOI] [PubMed] [Google Scholar]
- 37.Lopez-Botet M, Bellon T. Natural killer cell activation and inhibition by receptors for MHC class I. Curr Opin Immunol. 1999;11:301–7. doi: 10.1016/s0952-7915(99)80048-x. [DOI] [PubMed] [Google Scholar]
- 38.Binstadt BA, Brumbaugh KM, Dick CJ, Scharenberg AM, Williams BL, Colonna M, et al. Sequential involvement of Lck and SHP-1 with MHC-recognizing receptors on NK cells inhibits FcR-initiated tyrosine kinase activation. Immunity. 1996;5:629–38. doi: 10.1016/s1074-7613(00)80276-9. [DOI] [PubMed] [Google Scholar]
- 39.Campbell KS, Dessing M, Lopez-Botet M, Cella M, Colonna M. Tyrosine phosphorylation of a human killer inhibitory receptor recruits protein tyrosine phosphatase 1C. J Exp Med. 1996;184:93–100. doi: 10.1084/jem.184.1.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yusa S, Catina TL, Campbell KS. KIR2DL5 can inhibit human NK cell activation via recruitment of Src homology region 2-containing protein tyrosine phosphatase-2 (SHP-2) J Immunol. 2004;172:7385–92. doi: 10.4049/jimmunol.172.12.7385. [DOI] [PubMed] [Google Scholar]
- 41.Le Drean E, Vely F, Olcese L, Cambiaggi A, Guia S, Krystal G, et al. Inhibition of antigen-induced T cell response and antibody-induced NK cell cytotoxicity by NKG2A: association of NKG2A with SHP-1 and SHP-2 protein-tyrosine phosphatases. Eur J Immunol. 1998;28:264–76. doi: 10.1002/(SICI)1521-4141(199801)28:01<264::AID-IMMU264>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 42.Uhrberg M, Valiante NM, Young NT, Lanier LL, Phillips JH, Parham P. The repertoire of killer cell Ig-like receptor and CD94:NKG2A receptors in T cells: clones sharing identical alpha beta TCR rearrangement express highly diverse killer cell Ig-like receptor patterns. J Immunol. 2001;166:3923–32. doi: 10.4049/jimmunol.166.6.3923. [DOI] [PubMed] [Google Scholar]
- 43.Carrington M, Martin MP. The impact of variation at the KIR gene cluster on human disease. Curr Top Microbiol Immunol. 2006;298:225–57. doi: 10.1007/3-540-27743-9_12. [DOI] [PubMed] [Google Scholar]
- 44.Uhrberg M, Valiante NM, Shum BP, Shilling HG, Lienert-Weidenbach K, Corliss B, et al. Human diversity in killer cell inhibitory receptor genes. Immunity. 1997;7:753–63. doi: 10.1016/s1074-7613(00)80394-5. [DOI] [PubMed] [Google Scholar]
- 45.Kikuchi-Maki A, Yusa S, Catina TL, Campbell KS. KIR2DL4 is an IL-2-regulated NK cell receptor that exhibits limited expression in humans but triggers strong IFN-gamma production. J Immunol. 2003;171:3415–25. doi: 10.4049/jimmunol.171.7.3415. [DOI] [PubMed] [Google Scholar]
- 46.Rajagopalan S, Fu J, Long EO. Cutting edge: induction of IFN-gamma production but not cytotoxicity by the killer cell Ig-like receptor KIR2DL4 (CD158d) in resting NK cells. J Immunol. 2001;167:1877–81. doi: 10.4049/jimmunol.167.4.1877. [DOI] [PubMed] [Google Scholar]
- 47.Yusa S, Campbell KS. Src homology region 2-containing protein tyrosine phosphatase-2 (SHP-2) can play a direct role in the inhibitory function of killer cell Ig-like receptors in human NK cells. J Immunol. 2003;170:4539–47. doi: 10.4049/jimmunol.170.9.4539. [DOI] [PubMed] [Google Scholar]
- 48.McVicar DW, Burshtyn DN. Intracellular signaling by the killer immunoglobulin-like receptors and Ly49. Sci STKE 2001. 2001:RE1. doi: 10.1126/stke.2001.75.re1. [DOI] [PubMed] [Google Scholar]
- 49.Kikuchi-Maki A, Catina TL, Campbell KS. Cutting edge: KIR2DL4 transduces signals into human NK cells through association with the Fc receptor gamma protein. J Immunol. 2005;174:3859–63. doi: 10.4049/jimmunol.174.7.3859. [DOI] [PubMed] [Google Scholar]
- 50.Graham DB, Cella M, Giurisato E, Fujikawa K, Miletic AV, Kloeppel T, et al. Vav1 controls DAP10-mediated natural cytotoxicity by regulating actin and microtubule dynamics. J Immunol. 2006;177:2349–55. doi: 10.4049/jimmunol.177.4.2349. [DOI] [PubMed] [Google Scholar]
- 51.Trowsdale J. Genetic and functional relationships between MHC and NK receptor genes. Immunity. 2001;15:363–74. doi: 10.1016/s1074-7613(01)00197-2. [DOI] [PubMed] [Google Scholar]
- 52.Parham P. MHC class I molecules and KIRs in human history, health and survival. Nat Rev Immunol. 2005;5:201–14. doi: 10.1038/nri1570. [DOI] [PubMed] [Google Scholar]
- 53.Boyington JC, Sun PD. A structural perspective on MHC class I recognition by killer cell immunoglobulin-like receptors. Mol Immunol. 2002;38:1007–21. doi: 10.1016/s0161-5890(02)00030-5. [DOI] [PubMed] [Google Scholar]
- 54.Maenaka K, Juji T, Nakayama T, Wyer JR, Gao GF, Maenaka T, et al. Killer cell immunoglobulin receptors and T cell receptors bind peptide-major histocompatibility complex class I with distinct thermodynamic and kinetic properties. J Biol Chem. 1999;274:28329–34. doi: 10.1074/jbc.274.40.28329. [DOI] [PubMed] [Google Scholar]
- 55.Hansasuta P, Dong T, Thananchai H, Weekes M, Willberg C, Aldemir H, et al. Recognition of HLA-A3 and HLA-A11 by KIR3DL2 is peptide-specific. Eur J Immunol. 2004;34:1673–9. doi: 10.1002/eji.200425089. [DOI] [PubMed] [Google Scholar]
- 56.Rajagopalan S, Long EO. The direct binding of a p58 killer cell inhibitory receptor to human histocompatibility leukocyte antigen (HLA)-Cw4 exhibits peptide selectivity. J Exp Med. 1997;185:1523–8. doi: 10.1084/jem.185.8.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zappacosta F, Borrego F, Brooks AG, Parker KC, Coligan JE. Peptides isolated from HLA-Cw*0304 confer different degrees of protection from natural killer cell-mediated lysis. Proc Natl Acad Sci U S A. 1997;94:6313–8. doi: 10.1073/pnas.94.12.6313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Biassoni R, Falco M, Cambiaggi A, Costa P, Verdiani S, Pende D, et al. Amino acid substitutions can influence the natural killer (NK)-mediated recognition of HLA-C molecules. Role of serine-77 and lysine-80 in the target cell protection from lysis mediated by “group 2” or “group 1” NK clones. J Exp Med. 1995;182:605–9. doi: 10.1084/jem.182.2.605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Colonna M, Borsellino G, Falco M, Ferrara GB, Strominger JL. HLA-C is the inhibitory ligand that determines dominant resistance to lysis by NK1- and NK2-specific natural killer cells. Proc Natl Acad Sci U S A. 1993;90:12000–4. doi: 10.1073/pnas.90.24.12000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Moretta A, Sivori S, Vitale M, Pende D, Morelli L, Augugliaro R, et al. Existence of both inhibitory (p58) and activatory (p50) receptors for HLA-C molecules in human natural killer cells. J Exp Med. 1995;182:875–84. doi: 10.1084/jem.182.3.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Colonna M, Brooks EG, Falco M, Ferrara GB, Strominger JL. Generation of allospecific natural killer cells by stimulation across a polymorphism of HLA-C. Science. 1993;260:1121–4. doi: 10.1126/science.8493555. [DOI] [PubMed] [Google Scholar]
- 62.Winter CC, Long EO. A single amino acid in the p58 killer cell inhibitory receptor controls the ability of natural killer cells to discriminate between the two groups of HLA-C allotypes. J Immunol. 1997;158:4026–8. [PubMed] [Google Scholar]
- 63.Moesta AK, Norman PJ, Yawata M, Yawata N, Gleimer M, Parham P. Synergistic polymorphism at two positions distal to the ligand-binding site makes KIR2DL2 a stronger receptor for HLA-C than KIR2DL3. J Immunol. 2008;180:3969–79. doi: 10.4049/jimmunol.180.6.3969. [DOI] [PubMed] [Google Scholar]
- 64.Winter CC, Gumperz JE, Parham P, Long EO, Wagtmann N. Direct binding and functional transfer of NK cell inhibitory receptors reveal novel patterns of HLA-C allotype recognition. J Immunol. 1998;161:571–7. [PubMed] [Google Scholar]
- 65.Pende D, Marcenaro S, Falco M, Martini S, Bernardo ME, Montagna D, et al. Anti-leukemia activity of alloreactive NK cells in KIR ligand-mismatched haploidentical HSCT for pediatric patients: evaluation of the functional role of activating KIR and redefinition of inhibitory KIR specificity. Blood. 2009;113:3119–29. doi: 10.1182/blood-2008-06-164103. [DOI] [PubMed] [Google Scholar]
- 66.Gumperz JE, Litwin V, Phillips JH, Lanier LL, Parham P. The Bw4 public epitope of HLA-B molecules confers reactivity with natural killer cell clones that express NKB1, a putative HLA receptor. J Exp Med. 1995;181:1133–44. doi: 10.1084/jem.181.3.1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cella M, Longo A, Ferrara GB, Strominger JL, Colonna M. NK3-specific natural killer cells are selectively inhibited by Bw4-positive HLA alleles with isoleucine 80. J Exp Med. 1994;180:1235–42. doi: 10.1084/jem.180.4.1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gumperz JE, Barber LD, Valiante NM, Percival L, Phillips JH, Lanier LL, et al. Conserved and variable residues within the Bw4 motif of HLA-B make separable contributions to recognition by the NKB1 killer cell-inhibitory receptor. J Immunol. 1997;158:5237–41. [PubMed] [Google Scholar]
- 69.Litwin V, Gumperz J, Parham P, Phillips JH, Lanier LL. NKB1: a natural killer cell receptor involved in the recognition of polymorphic HLA-B molecules. J Exp Med. 1994;180:537–43. doi: 10.1084/jem.180.2.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Carr WH, Pando MJ, Parham P. KIR3DL1 polymorphisms that affect NK cell inhibition by HLA-Bw4 ligand. J Immunol. 2005;175:5222–9. doi: 10.4049/jimmunol.175.8.5222. [DOI] [PubMed] [Google Scholar]
- 71.Dohring C, Scheidegger D, Samaridis J, Cella M, Colonna M. A human killer inhibitory receptor specific for HLA-A1,2. J Immunol. 1996;156:3098–101. [PubMed] [Google Scholar]
- 72.Pende D, Biassoni R, Cantoni C, Verdiani S, Falco M, di Donato C, et al. The natural killer cell receptor specific for HLA-A allotypes: a novel member of the p58/p70 family of inhibitory receptors that is characterized by three immunoglobulin-like domains and is expressed as a 140-kD disulphide-linked dimer. J Exp Med. 1996;184:505–18. doi: 10.1084/jem.184.2.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Saulquin X, Gastinel LN, Vivier E. Crystal structure of the human natural killer cell activating receptor KIR2DS2 (CD158j) J Exp Med. 2003;197:933–8. doi: 10.1084/jem.20021624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Vales-Gomez M, Reyburn HT, Erskine RA, Strominger J. Differential binding to HLA-C of p50-activating and p58-inhibitory natural killer cell receptors. Proc Natl Acad Sci U S A. 1998;95:14326–31. doi: 10.1073/pnas.95.24.14326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Stewart CA, Laugier-Anfossi F, Vely F, Saulquin X, Riedmuller J, Tisserant A, et al. Recognition of peptide-MHC class I complexes by activating killer immunoglobulin-like receptors. Proc Natl Acad Sci U S A. 2005;102:13224–9. doi: 10.1073/pnas.0503594102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Campbell KS, Cella M, Carretero M, Lopez-Botet M, Colonna M. Signaling through human killer cell activating receptors triggers tyrosine phosphorylation of an associated protein complex. Eur J Immunol. 1998;28:599–609. doi: 10.1002/(SICI)1521-4141(199802)28:02<599::AID-IMMU599>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 77.Katz G, Markel G, Mizrahi S, Arnon TI, Mandelboim O. Recognition of HLA-Cw4 but not HLA-Cw6 by the NK cell receptor killer cell Ig-like receptor two-domain short tail number 4. J Immunol. 2001;166:7260–7. doi: 10.4049/jimmunol.166.12.7260. [DOI] [PubMed] [Google Scholar]
- 78.Katz G, Gazit R, Arnon TI, Gonen-Gross T, Tarcic G, Markel G, et al. MHC class I-independent recognition of NK-activating receptor KIR2DS4. J Immunol. 2004;173:1819–25. doi: 10.4049/jimmunol.173.3.1819. [DOI] [PubMed] [Google Scholar]
- 79.Shilling HG, Young N, Guethlein LA, Cheng NW, Gardiner CM, Tyan D, et al. Genetic control of human NK cell repertoire. J Immunol. 2002;169:239–47. doi: 10.4049/jimmunol.169.1.239. [DOI] [PubMed] [Google Scholar]
- 80.Hsu KC, Liu XR, Selvakumar A, Mickelson E, O’Reilly RJ, Dupont B. Killer Ig-like receptor haplotype analysis by gene content: evidence for genomic diversity with a minimum of six basic framework haplotypes, each with multiple subsets. J Immunol. 2002;169:5118–29. doi: 10.4049/jimmunol.169.9.5118. [DOI] [PubMed] [Google Scholar]
- 81.Goodridge JP, Lathbury LJ, Steiner NK, Shulse CN, Pullikotil P, Seidah NG, et al. Three common alleles of KIR2DL4 (CD158d) encode constitutively expressed, inducible and secreted receptors in NK cells. Eur J Immunol. 2007;37:199–211. doi: 10.1002/eji.200636316. [DOI] [PubMed] [Google Scholar]
- 82.Toneva M, Lepage V, Lafay G, Dulphy N, Busson M, Lester S, et al. Genomic diversity of natural killer cell receptor genes in three populations. Tissue Antigens. 2001;57:358–62. doi: 10.1034/j.1399-0039.2001.057004358.x. [DOI] [PubMed] [Google Scholar]
- 83.Whang DH, Park H, Yoon JA, Park MH. Haplotype analysis of killer cell immunoglobulin-like receptor genes in 77 Korean families. Hum Immunol. 2005;66:146–54. doi: 10.1016/j.humimm.2004.10.013. [DOI] [PubMed] [Google Scholar]
- 84.Yawata M, Yawata N, McQueen KL, Cheng NW, Guethlein LA, Rajalingam R, et al. Predominance of group A KIR haplotypes in Japanese associated with diverse NK cell repertoires of KIR expression. Immunogenetics. 2002;54:543–50. doi: 10.1007/s00251-002-0497-x. [DOI] [PubMed] [Google Scholar]
- 85.Yawata M, Yawata N, Draghi M, Little AM, Partheniou F, Parham P. Roles for HLA and KIR polymorphisms in natural killer cell repertoire selection and modulation of effector function. J Exp Med. 2006;203:633–45. doi: 10.1084/jem.20051884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Gardiner CM, Guethlein LA, Shilling HG, Pando M, Carr WH, Rajalingam R, et al. Different NK cell surface phenotypes defined by the DX9 antibody are due to KIR3DL1 gene polymorphism. J Immunol. 2001;166:2992–3001. doi: 10.4049/jimmunol.166.5.2992. [DOI] [PubMed] [Google Scholar]
- 87.Martin MP, Pascal V, Yeager M, Phair J, Kirk GD, Hoots K, et al. A mutation in KIR3DS1 that results in truncation and lack of cell surface expression. Immunogenetics. 2007;59:823–9. doi: 10.1007/s00251-007-0240-8. [DOI] [PubMed] [Google Scholar]
- 88.Pando MJ, Gardiner CM, Gleimer M, McQueen KL, Parham P. The protein made from a common allele of KIR3DL1 (3DL1*004) is poorly expressed at cell surfaces due to substitution at positions 86 in Ig domain 0 and 182 in Ig domain 1. J Immunol. 2003;171:6640–9. doi: 10.4049/jimmunol.171.12.6640. [DOI] [PubMed] [Google Scholar]
- 89.Khakoo SI, Geller R, Shin S, Jenkins JA, Parham P. The D0 domain of KIR3D acts as a major histocompatibility complex class I binding enhancer. J Exp Med. 2002;196:911–21. doi: 10.1084/jem.20020304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sanjanwala B, Draghi M, Norman PJ, Guethlein LA, Parham P. Polymorphic sites away from the Bw4 epitope that affect interaction of Bw4+ HLA-B with KIR3DL1. J Immunol. 2008;181:6293–300. doi: 10.4049/jimmunol.181.9.6293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Valiante NM, Uhrberg M, Shilling HG, Lienert-Weidenbach K, Arnett KL, D’Andrea A, et al. Functionally and structurally distinct NK cell receptor repertoires in the peripheral blood of two human donors. Immunity. 1997;7:739–51. doi: 10.1016/s1074-7613(00)80393-3. [DOI] [PubMed] [Google Scholar]
- 92.Fauriat C, Andersson S, Bjorklund AT, Carlsten M, Schaffer M, Bjorkstrom NK, et al. Estimation of the size of the alloreactive NK cell repertoire: studies in individuals homozygous for the group A KIR haplotype. J Immunol. 2008;181:6010–9. doi: 10.4049/jimmunol.181.9.6010. [DOI] [PubMed] [Google Scholar]
- 93.Freud AG, Caligiuri MA. Human natural killer cell development. Immunol Rev. 2006;214:56–72. doi: 10.1111/j.1600-065X.2006.00451.x. [DOI] [PubMed] [Google Scholar]
- 94.Parham P. Taking license with natural killer cell maturation and repertoire development. Immunol Rev. 2006;214:155–60. doi: 10.1111/j.1600-065X.2006.00462.x. [DOI] [PubMed] [Google Scholar]
- 95.Fischer JC, Ottinger H, Ferencik S, Sribar M, Punzel M, Beelen DW, et al. Relevance of C1 and C2 epitopes for hemopoietic stem cell transplantation: role for sequential acquisition of HLA-C-specific inhibitory killer Ig-like receptor. J Immunol. 2007;178:3918–23. doi: 10.4049/jimmunol.178.6.3918. [DOI] [PubMed] [Google Scholar]
- 96.Khakoo SI, Carrington M. KIR and disease: a model system or system of models? Immunol Rev. 2006;214:186–201. doi: 10.1111/j.1600-065X.2006.00459.x. [DOI] [PubMed] [Google Scholar]
- 97.Korbel DS, Norman PJ, Newman KC, Horowitz A, Gendzekhadze K, Parham P, et al. Killer Ig-like receptor (KIR) genotype predicts the capacity of human KIR-positive CD56dim NK cells to respond to pathogen-associated signals. J Immunol. 2009;182:6426–34. doi: 10.4049/jimmunol.0804224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ahlenstiel G, Martin MP, Gao X, Carrington M, Rehermann B. Distinct KIR/HLA compound genotypes affect the kinetics of human antiviral natural killer cell responses. J Clin Invest. 2008;118:1017–26. doi: 10.1172/JCI32400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Arnheim L, Dillner J, Sanjeevi CB. A population-based cohort study of KIR genes and genotypes in relation to cervical intraepithelial neoplasia. Tissue Antigens. 2005;65:252–9. doi: 10.1111/j.1399-0039.2005.00359.x. [DOI] [PubMed] [Google Scholar]
- 100.Carrington M, Wang S, Martin MP, Gao X, Schiffman M, Cheng J, et al. Hierarchy of resistance to cervical neoplasia mediated by combinations of killer immunoglobulin-like receptor and human leukocyte antigen loci. J Exp Med. 2005;201:1069–75. doi: 10.1084/jem.20042158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Naumova E, Mihaylova A, Stoitchkov K, Ivanova M, Quin L, Toneva M. Genetic polymorphism of NK receptors and their ligands in melanoma patients: prevalence of inhibitory over activating signals. Cancer Immunol Immunother. 2005;54:172–8. doi: 10.1007/s00262-004-0575-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Verheyden S, Bernier M, Demanet C. Identification of natural killer cell receptor phenotypes associated with leukemia. Leukemia. 2004;18:2002–7. doi: 10.1038/sj.leu.2403525. [DOI] [PubMed] [Google Scholar]
- 103.Besson C, Roetynck S, Williams F, Orsi L, Amiel C, Lependeven C, et al. Association of killer cell immunoglobulin-like receptor genes with Hodgkin’s lymphoma in a familial study. PLoS One. 2007;2:e406. doi: 10.1371/journal.pone.0000406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Martin MP, Qi Y, Gao X, Yamada E, Martin JN, Pereyra F, et al. Innate partnership of HLA-B and KIR3DL1 subtypes against HIV-1. Nat Genet. 2007;39:733–40. doi: 10.1038/ng2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Ruggeri L, Aversa F, Martelli MF, Velardi A. Allogeneic hematopoietic transplantation and natural killer cell recognition of missing self. Immunol Rev. 2006;214:202–18. doi: 10.1111/j.1600-065X.2006.00455.x. [DOI] [PubMed] [Google Scholar]
- 106.Moretta A, Pende D, Locatelli F, Moretta L. Activating and inhibitory killer immunoglobulin-like receptors (KIR) in haploidentical haemopoietic stem cell transplantation to cure high-risk leukaemias. Clin Exp Immunol. 2009;157:325–31. doi: 10.1111/j.1365-2249.2009.03983.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Ruggeri L, Mancusi A, Capanni M, Urbani E, Carotti A, Aloisi T, et al. Donor natural killer cell allorecognition of missing self in haploidentical hematopoietic transplantation for acute myeloid leukemia: challenging its predictive value. Blood. 2007;110:433–40. doi: 10.1182/blood-2006-07-038687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ruggeri L, Capanni M, Urbani E, Perruccio K, Shlomchik WD, Tosti A, et al. Effectiveness of donor natural killer cell alloreactivity in mismatched hematopoietic transplants. Science. 2002;295:2097–100. doi: 10.1126/science.1068440. [DOI] [PubMed] [Google Scholar]
- 109.Shlomchik WD, Couzens MS, Tang CB, McNiff J, Robert ME, Liu J, et al. Prevention of graft versus host disease by inactivation of host antigen-presenting cells. Science. 1999;285:412–5. doi: 10.1126/science.285.5426.412. [DOI] [PubMed] [Google Scholar]
- 110.Farag SS, Bacigalupo A, Eapen M, Hurley C, Dupont B, Caligiuri MA, et al. The effect of KIR ligand incompatibility on the outcome of unrelated donor transplantation: a report from the center for international blood and marrow transplant research, the European blood and marrow transplant registry, and the Dutch registry. Biol Blood Marrow Transplant. 2006;12:876–84. doi: 10.1016/j.bbmt.2006.05.007. [DOI] [PubMed] [Google Scholar]
- 111.Ottinger HD, Ferencik S, Beelen DW, Lindemann M, Peceny R, Elmaagacli AH, et al. Impact of HLA-A,B,C allele mismatches on outcome after unrelated blood stem cell transplantation in whites. Transplantation. 2004;78:1077–80. doi: 10.1097/01.tp.0000137791.28140.93. [DOI] [PubMed] [Google Scholar]
- 112.Bornhauser M, Schwerdtfeger R, Martin H, Frank KH, Theuser C, Ehninger G. Role of KIR ligand incompatibility in hematopoietic stem cell transplantation using unrelated donors. Blood. 2004;103:2860–1. doi: 10.1182/blood-2003-11-3893. author reply 2. [DOI] [PubMed] [Google Scholar]
- 113.Giebel S, Locatelli F, Lamparelli T, Velardi A, Davies S, Frumento G, et al. Survival advantage with KIR ligand incompatibility in hematopoietic stem cell transplantation from unrelated donors. Blood. 2003;102:814–9. doi: 10.1182/blood-2003-01-0091. [DOI] [PubMed] [Google Scholar]
- 114.Miller JS, Cooley S, Parham P, Farag SS, Verneris MR, McQueen KL, et al. Missing KIR ligands are associated with less relapse and increased graft-versus-host disease (GVHD) following unrelated donor allogeneic HCT. Blood. 2007;109:5058–61. doi: 10.1182/blood-2007-01-065383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Leung W, Iyengar R, Turner V, Lang P, Bader P, Conn P, et al. Determinants of antileukemia effects of allogeneic NK cells. J Immunol. 2004;172:644–50. doi: 10.4049/jimmunol.172.1.644. [DOI] [PubMed] [Google Scholar]
- 116.Shilling HG, McQueen KL, Cheng NW, Shizuru JA, Negrin RS, Parham P. Reconstitution of NK cell receptor repertoire following HLA-matched hematopoietic cell transplantation. Blood. 2003;101:3730–40. doi: 10.1182/blood-2002-08-2568. [DOI] [PubMed] [Google Scholar]
- 117.Miller JS, Soignier Y, Panoskaltsis-Mortari A, McNearney SA, Yun GH, Fautsch SK, et al. Successful adoptive transfer and in vivo expansion of human haploidentical NK cells in patients with cancer. Blood. 2005;105:3051–7. doi: 10.1182/blood-2004-07-2974. [DOI] [PubMed] [Google Scholar]
- 118.Hsu KC, Keever-Taylor CA, Wilton A, Pinto C, Heller G, Arkun K, et al. Improved outcome in HLA-identical sibling hematopoietic stem-cell transplantation for acute myelogenous leukemia predicted by KIR and HLA genotypes. Blood. 2005;105:4878–84. doi: 10.1182/blood-2004-12-4825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Leung W, Handgretinger R, Iyengar R, Turner V, Holladay MS, Hale GA. Inhibitory KIR-HLA receptor-ligand mismatch in autologous haematopoietic stem cell transplantation for solid tumour and lymphoma. Br J Cancer. 2007;97:539–42. doi: 10.1038/sj.bjc.6603913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Cooley S, Trachtenberg E, Bergemann TL, Saeteurn K, Klein J, Le CT, et al. Donors with group B KIR haplotypes improve relapse-free survival after unrelated hematopoietic cell transplantation for acute myelogenous leukemia. Blood. 2009;113:726–32. doi: 10.1182/blood-2008-07-171926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Ljunggren HG, Malmberg KJ. Prospects for the use of NK cells in immunotherapy of human cancer. Nat Rev Immunol. 2007;7:329–39. doi: 10.1038/nri2073. [DOI] [PubMed] [Google Scholar]
- 122.Binyamin L, Alpaugh RK, Hughes TL, Lutz CT, Campbell KS, Weiner LM. Blocking NK cell inhibitory self-recognition promotes antibody-dependent cellular cytotoxicity in a model of anti-lymphoma therapy. J Immunol. 2008;180:6392–401. doi: 10.4049/jimmunol.180.9.6392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Sola C, Andre P, Lemmers C, Fuseri N, Bonnafous C, Blery M, et al. Genetic and antibody-mediated reprogramming of natural killer cell missing-self recognition in vivo. Proc Natl Acad Sci U S A. 2009;106:12879–84. doi: 10.1073/pnas.0901653106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Hatjiharissi E, Xu L, Santos DD, Hunter ZR, Ciccarelli BT, Verselis S, et al. Increased natural killer cell expression of CD16, augmented binding and ADCC activity to rituximab among individuals expressing the Fc{gamma}RIIIa-158 V/V and V/F polymorphism. Blood. 2007;110:2561–4. doi: 10.1182/blood-2007-01-070656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Weng WK, Czerwinski D, Levy R. Humoral immune response and immunoglobulin G Fc receptor genotype are associated with better clinical outcome following idiotype vaccination in follicular lymphoma patients regardless of their response to induction chemotherapy. Blood. 2007;109:951–3. doi: 10.1182/blood-2006-03-013136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Romagne F, Andre P, Spee P, Zahn S, Anfossi N, Gauthier L, et al. Preclinical characterization of 1-7F9, a novel human anti-KIR receptor therapeutic antibody that augments natural killer-mediated killing of tumor cells. Blood. 2009;114:2667–77. doi: 10.1182/blood-2009-02-206532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Gomez-Lozano N, Gardiner CM, Parham P, Vilches C. Some human KIR haplotypes contain two KIR2DL5 genes: KIR2DL5A and KIR2DL5B. Immunogenetics. 2002;54:314–9. doi: 10.1007/s00251-002-0476-2. [DOI] [PubMed] [Google Scholar]