Abstract
Animal cell lines have become very popular substrates for the production of vaccines and biopharmaceuticals. Characterization of candidate production cell lines is central to ensure product safety and maintenance of consistency in the manufacture of biologicals. Nested PCR and isoenzyme analysis have been used widely to prove the identity and purity of various cell lines and primary cells individually and also after deliberate cross-contamination. The nested PCR based on the Cytochrome b (Cyt b) gene of mitochondrial DNA (Mt DNA) was found to be more sensitive than isoenzyme analysis in detecting low levels of contaminants (as low as 1%). Interestingly, competition between different co-cultured cell lines has shown in one case that cross-contamination need not always results in a mixed cell population. The nested PCR technique for the Cyt b gene described in this study appears to be a potential replacement for isoenzyme analysis and here we demonstrate the PCR method used is sensitive and reliable for cell line authentication in a simple, rapid and reliable format to help assure the authenticity of cell substrates for the production of safe vaccines and biopharmaceuticals.
Keywords: Cross-contamination, Cyt b gene, Isoenzyme analysis, Nested PCR
Introduction
In recent years continuous cell lines have become extremely valuable resource and popular substrates for manufacture of vaccines, recombinant proteins and biopharmaceuticals. Authentication of cell substrates is vital to the successful use of a cell line either as a research tool by many different laboratories or as a platform adopted by industries for the commercial production of biopharmaceuticals. A fundamental element of cell line authenticity is correct identity i.e. has not been mixed or switched with other cells (Freshney 2002). Authentication is also important for the evaluation of new cell substrates as candidates for the production of new vaccines and biotherapeutics. It is imperative for the manufacturers to develop and apply the best current technology to assure regulators that the new cell substrates have been thoroughly evaluated from a safety perspective and this includes cell authentication.
Characterization of any cell substrate used in the development and manufacture of vaccines and biopharmaceuticals aims to address some generic issues such as identity, purity, genetic stability, karyotype, tumorigenic phenotype and freedom from infection of microorganisms and potential oncogenic agents that might affect the safety and purity of the biologicals manufactured in them. Cell line cross-contamination is a phenomenon that arises as a result of the continuous cell line culture and handling of many cell lines in the same facility without strictly adhering to good laboratory practices (GLP) (Coecke et al. 2005). Earlier studies on cell line characterization demonstrated that at least one-third of the cell cultures were of a different tissue origin or species to that being claimed (Markovic and Markovic 1998) and many continuous cell lines were cross-contaminated with HeLa cells (Gartler 1967; Nelson-Rees and Flandermeyer 1977). Cross-contaminated or misidentified cultures clearly will invalidate resulting data and economically to a waste of time and money (Stacey et al. 2000). Hence, confirmation of identity of cell lines is a vital part of evaluation of cell substrates and each individual cell bank prepared. Development of new criteria for the acceptability of such cell lines and the improvement of those that already exist has been identified as a priority by several WHO expert committees. Strangely the need for cell line-specific identification or authentication seems to have been a less recognized need and our view has been neglected particularly in research areas using cell culture.
To address the problem of cell line cross-contamination, different methods have been applied for identifying inter-species and intra-species cross-contamination of animal cells. Several methods had been established to monitor cell culture identity using immunological marker (Stulberg et al. 1976; O’Toole et al. 1983), isoenzyme analysis (Halton et al. 1983; Steube et al. 1995) and karyological examination (Miller et al. 1971; Nelson-Rees et al. 1974). Species-specific antibodies are also available commercially and these have been used to confirm species of origin by indirect fluorescence or immunoperoxidase staining (Hay 2002). The advent of the DNA fingerprinting technique offered detection and identification of individual human samples (Jeffreys et al. 1985; Stacey et al. 1992). DNA fingerprinting is also useful to verify the identity and purity of the cell lines (Jeffreys et al. 1985; Stacey et al. 1992) and it is now used as a common method for authentication of human cell lines (Dirks et al. 1999; Masters 2000). PCR based methods for the identification of animal cell lines have also been described (Stacey et al. 1997; Parodi et al. 2002; Liu et al. 2003; Lopez-Andreo et al. 2005; Losi et al. 2008).
Karyological examination remains one of the best techniques for identifying the species of origin of a normal cell line as each animal species has a unique karyotype. However, where transformed cells are used, the incidence of aneuploidy and heteroploidy can make species identification difficult unless specific chromosomal markers are used (Hay 2000; Keith 2000). For this reason, isoenzyme analysis is widely used, which can identify most if not all species currently cultured in vitro. However, each of these techniques has its own disadvantages viz. the need for some specialized reagents, devices and skilled persons to perform the technique. In addition the sensitivity of isoenzyme analysis may be less than other methods for the detection of mixtures of cells derived from other species and generally would not detect mixtures of cell lines derived from the same species (Nims et al. 1998). Hence, the need to develop a simple, robust, sensitive and reliable technique to demonstrate the identity and purity of the cell lines to meet the GMP standards at industries is widely felt. Polymerase chain reaction (PCR) being a simple, robust and sensitive technique is widely used for species identification (Stacey et al. 1997; Parodi et al. 2002; Liu et al. 2003). Recently, Ono et al. (2007) developed a highly sensitive and specific PCR method for amplification of mitochondrial DNA (Mt DNA) sequence that can distinguish cell lines originating from 14 different species commonly used in life science research.
In the present study we attempted amplification of mitochondrial Cytochrome b (Cyt b) gene using a nested PCR technique and speciation using the classical isoenzyme technique to study the identity of various cell lines and primary cells individually and also after deliberate cross-contamination.
Materials and methods
Cell lines and mitochondrial DNA isolation
All the cell lines used in this study (Table 1) were procured either from ATCC, Manassas, VA, USA or ECACC, Salisbury, UK. These cell lines were certified to be free from adventitious agents such as Mycoplasma sp, bacteria, fungi, yeast and species had been identified by isoenzyme analysis at the respective repository. The cell lines were grown at 37 °C in a humidified incubator with 5% CO2 using appropriate culture media (recommended by the supplier and obtained from Invitrogen, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum (Biochrom, Berlin, Germany) and other supplements as applicable. Mitochondrial DNA for PCR was extracted using a commercial mitochondrial DNA extraction kit (Biovision, Mountainview, CA, USA) according to the manufacturer’s instruction.
Table 1.
List of cell lines taken up for this study
| Sl.No | Cell lines | Species |
|---|---|---|
| 1 | A72 | Dog |
| 2 | BHK-21 | Syrian hamster |
| 3 | Hep2 | Human |
| 4 | IBRS-2 | Pig |
| 5 | MDBK | Bovine |
| 6 | MDCK | Canine |
| 7 | MRC-5 | Human |
| 8 | VERO | African green monkey |
| 9 | MNA | Mouse |
Primary cells and mitochondrial DNA isolation
Kidney and peripheral blood samples collected from a feral dog and cross-bred cattle reared in holding farm (IIL, Hyderabad, India) were used for the preparation of primary kidney cells and primary nucleated blood cells, respectively. Kidney collected from Syrian hamster obtained from National center for laboratory animal science (NIN, Hyderabad, India) was the source of primary hamster kidney cells. Primary kidney cell suspension was prepared by a combination of mechanical and enzymatic disaggregation of kidney tissue samples (Freshney 2006). Peripheral blood of Macaca mulatta monkey was obtained from National center for laboratory animal science (NIN, Hyderabad, India). Nucleated blood cells were harvested by pelleting the anticoagulated blood followed by RBC lysis using RBC lysis buffer (Sigma, St. Louis, USA) (Forsell et al. 2005). Mitochondrial DNA of primary kidney and nucleated blood cells was extracted using a commercial kit (Biovision, Mountainview, CA, USA) according to the manufacturer’s instruction.
Nested PCR amplification
The full-length and partial Mt DNA sequences including primer sequence information were taken essentially from Ono et al. (2007). The details of external and internal primer sequences are listed in Table 2. An external primer which was complementary to conserved sequences within Cyt b (forward primer) and 16S ribosomal RNA (rRNA) genes (reverse primer) of 8 different species of interest acted as a universal primer pair. The amplified product encompasses a portion of cytochrome b, d loop, 12S and 16S rRNA genes. Internal primer pairs complementary to the species specific sequences within the region amplified by the universal primer pair acted as nested primer pairs that were designed in such a way to get the products exhibiting length polymorphism for species identification with ease. The reaction mixture (50 μL volume) contained Taq polymerase (3 units, Genei, Bangalore, India), Taq buffer 10× (Mg2+: 2 mM), dNTPs (200 μM each), 20 pmol of each primer and 100–150 ng of template DNA. The thermal cycling was carried out essentially following Ono et al. (2007) with slight modification.
Table 2.
Details of primers used for PCR based speciation of cell lines
| Sl.No | Species | Primer sequence (5′–3′) (internal primers) | |
|---|---|---|---|
| Forward | Reverse | ||
| 1 | Dog | GCCCAACTAACCCCAAACTTA | GGTTAACAATGGGGTGGATAAG |
| 2 | Syrian hamster | GACCTCTTAGGTGTATTCCTAC | GTATGAAGAAGGGGTAGAGCA |
| 3 | Human | TATTGCAGCCCTAGCAGCACTCCA | AGAATGAGGAGGTCTGCGGC |
| 4 | Pig | CCTATATTCAATTACACAACCATGC | GCGTGTGCGAGGAGAAAGGC |
| 5 | Cow | CCTAGATGAGTCTCCCAACTC | GTTGTTTAGTCGAGAGGGTATC |
| 6 | African green monkey | CCAGAAGACCCACGATAACTCTCA | TGTTAGCTCAAGGTAATCGAGTTGTAC |
| 7 | Mouse | GCACTGAAAATGCTTAGATGGATAATTG | CCTCTCATAAACGGATGTCTAG |
| 8 | Cat | TAGAACACCCACGAAGATCC | CATATGGTCTCTTTGGGTCG |
| 9 | Chinese hamster | CCGGCGTAAAACGTGTTATAGACT | GTATTAGGTATAATATCGGCAGTC |
| 10 | Chicken | GTATTCCCGTGCAAAAACGAG | CTTAGTGAAGAGTTGTGGTCTG |
| 11 | External Primer (Universal Primer) |
THGTHSAATGAATCTGAGGVGGVT | CGATGTTGGATCAGGACATC |
Briefly, the PCR cycling involved initial hot start step (94 °C for 1 min) and initial denaturation at 94 °C for 5 min, at 59 °C for 5 min, followed by 35 cycles of elongation at 72 °C for 2.5 min, denaturation at 94 °C for 30 s, annealing at 59 °C for 45 s, with elongation at 72 °C for 10 min in the last cycle. The PCR product (1–3 μL) amplified by universal primer pair acted as a template for the nested PCR. The nested PCR involved initial hot start step and denaturation at 94 °C for 5 min, maintained at 60 °C for 5 min, followed by 35 cycles of elongation at 72 °C for 1.5 min, denaturation at 94 °C for 45 s, annealing at 60 °C for 30 s, with elongation at 72 °C for 10 min in the last cycle. The nested PCR product was analyzed by gel electrophoresis using 2% agarose (Invitrogen, USA) and visualized in a UV transilluminator after staining with ethidium bromide and the image captured using a gel documentation system. A 100 bp and 1 Kb DNA Ladder (MBI Fermantas, Hanover, MD, USA) were used as markers to determine the size of the PCR products.
Checking the identity and purity of various cell lines and primary cells
In order to confirm the identity, the Mt DNA extracted from various cell lines and primary cells were subjected to PCR followed by nested PCR using appropriate species specific primer pairs. After confirming the identity, the purity of various cell lines was checked by subjecting the extracted DNA to PCR followed by nested PCR using primer pairs specific for most commonly handled cell lines from ten different species including African green monkey, cattle, cat, dog, Chinese hamster, Syrian hamster, chicken, mouse, human and pig. The nested PCR products were then analyzed by standard agarose gel electrophoresis to determine the species specific amplicon (Sambrook et al. 1989). The PCR products obtained thus were also gel purified and sequenced with an objective of confirming their authenticity. The species specific internal primer pairs were used for nucleotide sequencing.
Deliberate cross-contamination of cell lines
Three different studies were carried out with deliberately cross-contaminated cell lines and primary cells to assess the analytical sensitivity and specificity of nested PCR technique in comparison with isoenzyme analysis. In the first study, MDCK cells were deliberately cross-contaminated with 1% of BHK-21 cells and passaged thrice at 48 h intervals. The cells were observed for their morphology on a daily basis. At every passage, 500 × 105 and 100 × 105 deliberately cross-contaminated cells were taken for Mt DNA analysis by PCR and isoenzyme analyses, respectively. The PCR was done, firstly, with universal primers and, secondly, with the nested PCR using dog and Syrian hamster specific primer pairs. The isoenzyme analysis was performed using AuthentiKit (Innovative chemistry, Marshfield, MA, USA) targeting the isoenzymes including nucleotide phosphorylase (NP), glucose-6-phosphate dehydrogenase (G6PD), malate dehydrogenase (MD) and mannose phosphate isomerase (MPI). In the second study, VERO cells were deliberately cross-contaminated separately with 1% of MDBK and 1% of MDCK and passaged thrice at 48 h interval. The morphology of cells was observed by microscopical examination. At every passage 500 × 105 and 100 × 105 deliberately cross-contaminated cells were taken for PCR and isoenzyme analyses, respectively. In the third study, primary cattle kidney cells were mixed with BHK-21 cell line at 1 and 5% levels and vice versa. Unlike the previous two studies, the deliberately contaminated cells were directly subjected to Mt DNA extraction and nested PCR without passaging. The PCR in the second and third studies was done, firstly, with universal primer pairs and secondly the nested PCR using African green monkey, bovine, dog and Syrian hamster specific primer pairs as appropriate.
Results and discussion
Confirmation of identity and purity of animal cells by nested PCR
The identity of various cell lines such as BHK-21, VERO, MRC-5, MDCK, MDBK, MNA, IBRS-2, Hep-2, A72 and the primary cells of cattle, Syrian hamster, monkey and dog origin could be proven by nested PCR based on the amplicon specific to the given species (Table 3 and Fig. 1a). The results of nested PCR also clearly showed that the primers were specific enough in demonstrating the identity of various cell lines and primary cells of dog, cattle, Syrian hamster and monkey origin. The nucleotide sequences of BHK-21, MDBK, MDCK and VERO cell lines when subjected to BLAST analysis revealed 99–100% homology with the respective published sequences. The results of nested PCR performed on DNA extracted from BHK-21, VERO, MRC-5, MDCK, MNA, IBRS-2, Hep-2, A72 and MDBK cell lines to demonstrate the purity clearly indicated that these cell lines available in our repository were free from other cellular cross-contamination (Table 3).
Table 3.
Results of nested PCR done to demonstrate the identity and purity of various cell lines
| Sl.No | Species | Amplicon size | Identity | Purity | |
|---|---|---|---|---|---|
| Expected (bp) | Actual (bp) | ||||
| 1 | Dog | 755 | 755 | + | + |
| 2 | Syrian hamster | 245 | 245 | + | + |
| 3 | Human | 441 | 441 | + | + |
| 4 | Pig | 819 | 819 | + | + |
| 5 | Cow | 1090 | 1090 | + | + |
| 6 | African green monkey | 301 | 301 | + | + |
| 7 | Mouse | 948 | 948 | + | + |
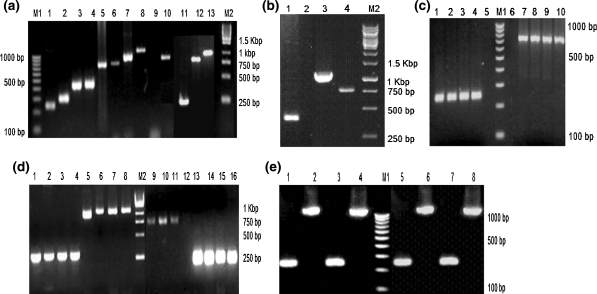
Fig. 1.
Results of nested PCR. M1—100 bp DNA ladder. M2—1 kb DNA ladder. a Results of nested PCR done to prove the identity of various cell lines and primary kidney cells. Species specific primers were used to amplify the Cyt b gene of mitochondrial DNA. Lane 1 to 10 shows species specific amplicon of BHK-21 (245 bp), VERO (301 bp), MRC-5 (441 bp), Hep-2 (441 bp), MDCK (755 bp), A72 (755 bp), MNA (948 bp), MDBK (1090 bp), Negative control, IBRS-2 (819 bp) cell lines, respectively. Lanes 11–13 shows species specific amplicons of primary Syrian hamster kidney (245 bp), Primary dog kidney (755 bp) and Primary cattle kidney (1090 bp) cells, respectively. b Results of nested PCR done on the primary blood cells. Lanes 1, 3 and 4 show species specific amplicons of monkey (301 bp), cattle (1090 bp) and dog (755 bp) blood cells, respectively. Lane 2 is negative control. c Nested PCR result of the MDCK cell line deliberately contaminated with 1% of BHK-21. Lane 1–4 shows Syrian hamster specific amplicon (245 bp) and Lanes 7–10 shows canine specific amplicon (755 bp) of passages P0, P1, P2 and P3, respectively. Lanes 5 and 6 are negative controls. d Nested PCR result of the Vero cell line deliberately contaminated separately with 1% of MDBK and 1% of MDCK. Lane 1–4 shows monkey specific amplicon (301 bp) and Lane 5–8 shows bovine specific amplicon (1090 bp) of passages P0, P1, P2 and P3, respectively. Lanes 9 to 11 show canine specific amplicon (755 bp) at P0, P1 and P2, respectively. Canine specific amplicon at P3 is missing in lane 12. Lanes 13 to 16 show African green monkey specific (301 bp) amplicon at P0, P1, P2 and P3, respectively. eLanes 1 and 3 show Syrian hamster specific amplicons (245 bp) and Lanes 2 and 4 show bovine specific amplicons (1090 bp) of the primary cattle kidney cells deliberately contaminated with 1 and 5% of BHK-21 cell line. Lanes 5 and 7 show Syrian hamster specific amplicons (245 bp) and lanes 6 and 8 shows bovine specific amplicons (1090 bp) of the BHK-21 cell line deliberately contaminated with 1 and 5% of primary cattle kidney cells
Deliberate cross-contamination of animal cells
Morphological examination
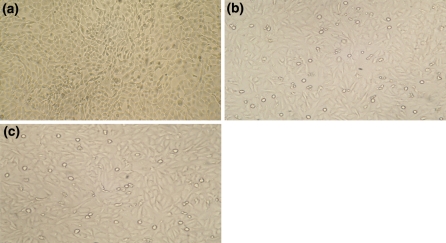
Daily microscopic examination of MDCK cells deliberately cross-contaminated with 1% of BHK-21 cells, showed morphology typical of MDCK cells. The BHK-21 morphology could not be discerned in all the three passages (Fig. 2a). Morphological examination of VERO cells deliberately cross-contaminated separately with 1% of MDCK and 1% of MDBK revealed the predominating morphology typical of VERO cells (Fig. 2b, c). The MDBK and MDCK cell morphologies could not be discerned in all the three passages. The results of morphological examination of deliberately cross-contaminated cell lines clearly shows that it is probably not possible to discern the presence of low level contaminating MDCK and MDBK cells in VERO cell culture and BHK-21 cells in MDCK cell cultures. In these deliberately contaminated cultures, the contaminant cell line could not be seen by visual microscopic observation in all the three cases (Fig. 2a–c). It is therefore necessary to verify the identity and purity of production cell lines by a standard technique. Such cross-contaminations may go unnoticed or overlooked even by experienced laboratory personnel because of the morphological resemblance of the cell lines under study. Difference in the morphology and patchy growth may at times give suspicion regarding cross-contamination. Also morphology can be ambivalent as similarities can exist between cells of very different origins (Freshney 2006). Hence, morphological examination alone cannot be considered as a reliable method for identification of cross-contamination of cell lines. It might rather mislead as far as the identification of cross-contamination at an early stage is concerned. Sensitivity of detection of cell mixtures is important to exclude the possibility of cell line cross-contamination. The problem of cross-contamination occurs most often in cultures which undergo innumerable passages or where spontaneous transformation of slow growing cultures is observed (Stacey et al. 1992).
Fig. 2.
Morphology of deliberated contaminated animal cells. a Monolayer of MDCK cell line contaminated with 1% of BHK-21 cells not showing the contaminant cell line morphology—Unstained preparation (100× magnification). b Monolayer of VERO cell line contaminated with 1% of MDCK cells not showing the contaminant cell line morphology—Unstained preparation (100× magnification). c Monolayer of VERO cell line contaminated with 1% MDBK cells not showing the contaminant cell line morphology—Unstained preparation (100× magnification)
Species identification by the nested PCR
The nested PCR performed on DNA extracted from MDCK cells deliberately cross-contaminated with 1% of BHK-21 cells and passaged thrice clearly proved not only the identity of MDCK cell line but also the presence of contaminant BHK-21 cells (Fig. 1c; Table 4). Similarly, the nested PCR performed on DNA extracted from VERO cells deliberately cross-contaminated separately with 1% of MDCK cells and 1% of MDBK cells and passaged thrice clearly proved the identity of VERO cell line and also the presence of MDBK contaminant in all the three passages (Fig. 1d; Table 4). However, it revealed the existence of MDCK contaminant only in initial two passages (Fig. 1d; Table 4). The present study indicated the usefulness of the nested PCR based amplification of mitochondrial Cyt b gene, in identifying various individual and deliberately contaminated cell lines and also the primary cells of the same species of cell lines used for cross contamination viz. MDBK, MDCK, BHK-21 and VERO of bovine, dog, Syrian hamster and monkey origin, respectively. The nested PCR technique was sensitive enough in demonstrating the presence of low level of contamination (as low as 1% level) as strong product bands, both in continuous cell line and primary cells except in the case of MDCK contaminant in VERO cell line at third passage (Table 4; Fig. 1c–e). The nested PCR performed on DNA extracted from the primary cattle kidney cells deliberately contaminated with BHK-21 cell line and vice versa clearly revealed the existence of contaminant at 1 and 5% levels and proved the species of origin of the continuous cell line of hamster origin (Fig. 1e). This clearly shows that this nested PCR based authentication could be suitable for both continuous cell line and primary cells. It was not difficult establishing the species identity of cell lines and primary cells since the PCR products exhibited the appropriate length polymorphism. This is in agreement with Ono et al. (2007) who clearly demonstrated the sensitive and specific nature of nested PCR in species identification of cell lines. In addition, the authenticity of the nested PCR amplicon was also proven by BLAST analysis of the nucleotide sequences which revealed 99–100% homology with the respective published sequences. The nested PCR technique could be used for in-process quality control where long culture periods are involved such as continuous multi-harvest bioreactor systems.
Table 4.
Nested PCR results of the deliberately contaminated cell lines
| Cross contaminated cell Line | Passages | Nested PCR result | |
|---|---|---|---|
| MDCK (Dog) | BHK-21 (Syrian hamster) | ||
| MDCK + BHK-21 (99% MDCK + 1% BHK-21) | P0 | * | * |
| P1 | * | * | |
| P2 | * | * | |
| P3 | * | * | |
| VERO (African green monkey) | MDCK (Dog) | ||
|---|---|---|---|
| VERO + MDCK (99% VERO + 1% MDCK) | P0 | * | * |
| P1 | * | * | |
| P2 | * | * | |
| P3 | * | – |
| VERO (African green monkey) | MDBK (Bovine) | ||
|---|---|---|---|
| VERO + MDBK (99% VERO + 1% MDBK) | P0 | * | * |
| P1 | * | * | |
| P2 | * | * | |
| P3 | * | * |
| Primary Cattle Kidney (PCK) + BHK-21 cell line | PCK (Bovine) | BHK-21 (Syrian hamster) | |
|---|---|---|---|
| 99% PCK + 1% BHK-21 | P0 | * | * |
| 95% PCK + 5% BHK-21 | P0 | * | * |
| BHK-21 cell line + Primary Cattle Kidney (PCK) | BHK-21 (Syrian hamster) | PCK (Bovine) | |
|---|---|---|---|
| 99% BHK-21 + 1% PCK | P0 | * | * |
| 95% BHK-21 + 5% PCK | P0 | * | * |
* Species specific amplicons seen
Species identification by isoenzyme analysis
Isoenzyme analysis was done targeting the isoenzymes including NP, G6PD, MD and MPI to detect cross-contamination of MDCK cells with 1% of BHK-21 cells, VERO cells with 1% of MDBK and 1% of MDCK cells and passaged thrice. Species identity of pure, uncontaminated MDBK, VERO, BHK-21 and MDCK cell lines were confirmed by Isoenzyme analysis (Fig. 3a, b). A faint band of NP and MD isoenzymes suggestive of bovine species revealed the existence of MDBK contaminant in VERO cell culture in all the three passages. However, bovine species specific G6PD and MPI isoenzyme bands could not be seen in VERO cell culture contaminated with MDBK cells (Fig. 4a; Table 5). But, even faint bands of NP, G6PD, MD, and MPI isoenzymes suggestive of canine species could not be seen in VERO cell line contaminated with MDCK cells in all the three passages (Fig. 4b; Table 5). In contrast, Syrian hamster specific bands of all the four isoenzymes targeted could not be seen in MDCK cell line contaminated with BHK-21 in all the three passages. Examination of MDCK cells contaminated with BHK-21 cells indicated the presence of standard canine specific bands of all the four isoenzymes in all the three passages and also proved the canine identity of the MDCK line (Fig. 4c; Table 5). However, isoenzyme analysis failed to reveal the presence of BHK-21 contamination at 1% level in all the three passages. The techniques which have been most commonly applied to authentication of cell culture are isoenzyme analysis and cytogenetics and have been historically used to highlight the significant problem of cross-contaminated and mis-identified cell cultures (O’Brien et al. 1980; Nelson-Rees et al. 1989). Isoenzyme analysis is one of the standard techniques recognized by the regulatory authorities and was therefore used as the reference technique for analysis of species. The failure to detect isoenzymes in cultures with 1% cross-contamination is in agreement with Nims et al. (1998) who reported that isoenzyme technique is sensitive only at 10% level of cross-contamination. In isoenzyme analysis, the sensitivity of detection appeared to be influenced by the species of cells involved and the enzyme analyzed (Stacey et al. 1997) but may also relate to biochemical activity of the cells analyzed. In isoenzyme analysis, close migration of bands from different cells may leave the two populations unresolved and the cell mixture could remain unidentified (Stacey et al. 1997). Isoenzyme analysis also requires optimization of the testing strategy with regard to colour development time, correction of migration distance and selection of the appropriate enzymes.
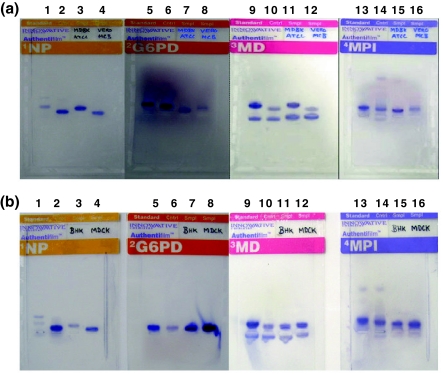
Fig. 3.
Results of Isoenzyme analyses done for species identification of animal cells. The species specific migration distances were calculated using enzyme migration data and decision tree analysis forms supplied by the manufacturer. a Isoenzyme analysis of MDBK and VERO cell lines showing species specific bands of NP, G6PD, MD and MPI enzymes. Lanes 1 to 4 show NP enzyme specific bands of standard, control, MDBK (Bovine) and VERO (African green monkey), respectively. Lanes 5 to 8 show G6PD enzyme specific bands of standard, control, MDBK (Bovine) and VERO (African green monkey), respectively. Lanes 9 to 12 show MD enzyme specific bands of standard, control, MDBK (Bovine) and VERO (African green monkey), respectively. Lanes 13 to 16 show MPI enzyme specific bands of standard, control, MDBK (Bovine) and VERO (African green monkey), respectively. b Isoenzyme analysis of BHK-21 and MDCK cell lines showing species specific bands of NP, G6PD, MD and MPI enzymes. Lanes 1 to 4 show NP enzyme specific bands of standard, control, BHK-21 (Syrian hamster) and MDCK (canine), respectively. Lanes 5 to 8 show G6PD enzyme specific bands of standard, control, BHK-21 (Syrian hamster) and MDCK (canine), respectively. Lanes 9 to 12 show MD enzyme specific bands of standard, control, BHK-21 (Syrian hamster) and MDCK (canine), respectively. Lanes 13 to 16 show MPI enzyme specific bands of standard, control, BHK-21 (Syrian hamster) and MDCK (canine), respectively
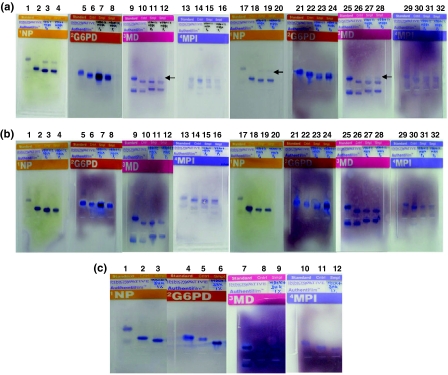
Fig. 4.
Results of Isoenzyme analysis done for detecting deliberate contamination of animal cells. The species specific migration distances were calculated using enzyme migration data and decision tree analysis forms supplied by the manufacturer. aLanes 1 to 16 show isoenzyme analysis of VERO cell line contaminated with 1% of MDBK showing species specific bands of NP, G6PD, MD and MPI enzymes of cells at P0 and P1, respectively. Standards are shown in lanes 1, 5, 9, and 13, controls are shown in lanes 2, 6, 10, and 14, samples of VERO + 1% MDBK (P0) are shown in lanes 3, 7, 11, and 15, and samples of VERO + 1% MDBK (P1) are shown in lanes 4, 8, 12, and 16. Lanes 17 to 32 show isoenzyme analysis of VERO cell line contaminated with 1% of MDBK showing species specific bands of NP, G6PD, MD and MPI enzymes of cells at P2 and P3, respectively. A faint band of NP and MD isoenzymes specific to bovine species (contaminant MDBK cell line) is seen but G6PD and MPI specific bands are not seen in all three passages. The identity of VERO cell line is confirmed by the four isoenzymes. Standards are shown in lanes 17, 21, 25, and 29, controls are shown in lanes 18, 22, 26, and 30, samples of VERO + 1% MDBK (P2) are shown in lanes 19, 23, 27, and 31, and samples of VERO + 1% MDBK (P3) are shown in lanes 20, 24, 28, and 32. Lanes 1 to 4, 17 to 20 show specific bands of NP, lanes 5 to 8, 21 to 24 show specific bands of G6PD, lanes 9 to 12, 25 to 28 show specific bands of MD, and lanes 13 to 16, 29 to 32 show specific bands of MPI. bLanes 1 to 16 show isoenzyme analysis of VERO cell line contaminated with 1% of MDCK showing species specific bands of NP, G6PD, MD and MPI enzymes of cells at P0 and P1, respectively. Standards are shown in lanes 1, 5, 9, and 13, controls are shown in lanes 2, 6, 10, and 14, samples of VERO + 1% MDCK (P0) are shown in lanes 3, 7, 11, and 15, and samples of VERO + 1% MDCK (P1) are shown in lanes 4, 8, 12, and 16. Lanes 17 to 32 show isoenzyme analysis of VERO cell line contaminated with 1% of MDCK showing species specific bands of NP, G6PD, MD and MPI enzymes of cells at P2 and P3, respectively. Canine species specific isoenzyme bands are not seen in all the three passages but the identity of VERO cell line is confirmed by the four isoenzymes. Standards are shown in lanes 17, 21, 25, and 29, controls are shown in lanes 18, 22, 26, and 30, samples of VERO + 1% MDCK (P2) are shown in lanes 19, 23, 27, and 31, and samples of VERO + 1% MDCK (P3) are shown in lanes 20, 24, 28, and 32. Lanes 1 to 4, 17 to 20 show specific bands of NP, lanes 5 to 8, 21 to 24 show specific bands of G6PD, lanes 9 to 12, 25 to 28 show specific bands of MD, and lanes 13 to 16, 29 to 32 show specific bands of MPI. c Isoenzyme analysis of MDCK cell line contaminated with 1% of BHK-21 cells. The contaminant BHK-21 cell line specific isoenzyme bands are not seen in all the three passages but the identity of MDCK cell line is confirmed by all the four isoenzymes. Lanes 1 to 3 show NP enzyme specific standard, control and MDCK (canine) bands, respectively. Lanes 4 to 6 show G6PD enzyme specific standard, control and MDCK (canine) bands, respectively. Lanes 7 to 9 show MD enzyme specific standard, control and MDCK (canine) bands, respectively. Lanes 10 to 12 show MPI enzyme specific bands of standard, control and MDCK (canine) bands, respectively
Table 5.
Results of isoenzyme analysis for species identification of the deliberately contaminated cell lines
| Cell line | Passage | Isoenzyme | |||||||
|---|---|---|---|---|---|---|---|---|---|
| NP | G6PD | MD | MPI | ||||||
| MDCK + BHK-21 | MDCK | BHK-21 | MDCK | BHK-21 | MDCK | BHK-21 | MDCK | BHK-21 | |
| 99% MDCK + 1% BHK-21 | P0 | * | – | * | – | * | – | * | – |
| P1 | * | – | * | – | * | – | * | – | |
| P2 | * | – | * | – | * | – | * | – | |
| P3 | * | – | * | – | * | – | * | – | |
| VERO + MDCK | VERO | MDCK | VERO | MDCK | VERO | MDCK | VERO | MDCK | |
|---|---|---|---|---|---|---|---|---|---|
| 99% VERO + 1% MDCK | P0 | * | – | * | – | * | – | * | – |
| P1 | * | – | * | – | * | – | * | – | |
| P2 | * | – | * | – | * | – | * | – | |
| P3 | * | – | * | – | * | – | * | – |
| VERO + MDBK | VERO | MDBK | VERO | MDBK | VERO | MDBK | VERO | MDBK | |
|---|---|---|---|---|---|---|---|---|---|
| 99% VERO + 1% MDBK | P0 | * | * (very faint) | * | – | * | * (very faint) | * | – |
| P1 | * | D | * | – | * | D | * | – | |
| P2 | * | * | * | – | * | * | * | – | |
| P3 | * | * | * | – | * | * | * | – |
* Species specific mobility pattern observed. D-Doubtful
The nested PCR technique was found to be more sensitive than isoenzyme analysis for species identification in that it was capable of demonstrating the presence of BHK-21 cell contamination in the presence of predominating MDCK cells and MDBK cell contamination in the presence of predominating VERO cells in all the three passages. Surprisingly, the nested PCR technique could demonstrate the presence of 1% MDCK cell in the presence of predominating VERO cells contamination only in first and second passages. However, in third passage MDCK specific PCR amplicon could not be seen. Failure of detection of MDCK cells by the nested PCR at third passage could possibly be due to its inability to get established resulting in its extinction. Different cell lines may proliferate with varying growth rates such that a single cell from a rapidly growing line, introduced into a culture of slower growing cells can overtake the original culture in the course of a few passages (Nims and Herbstritt 2005). This is an interesting observation which is in contrast to the general perception that cross-contamination in continuous cell lines will always result in purity problems. Cell line cross contamination is a concern when two cell types of similar growth characteristics such as doubling time and degree of contact inhibition are contaminated.
Conclusions
Cultured cell lines have become an extremely valuable resource, the successful use of which both in academic research and in industrial biotechnology depends on the authentication of their identity to ensure the validity of the derived experimental data. For a cell line to be authentic it must display characteristics that confirm its identity beyond question (Freshney 2002). The sensitivity for detection of cross-contaminated cells is important as species confirmation is usually only done on cell banks and a rapid growing contaminant could expand during production and would not be detected until end of production run cells were analyzed. Therefore characterization of the cell banks to assure the identity and purity of a cell line is of paramount importance to ensure safety and purity of the biological products derived from those cell lines. Primary cells obtained directly from chicken and duck embryos and kidney of monkeys, hamsters, cattle etc. have played a pivotal role for the major successes in the control of viral diseases by acting as cell substrates for production of vaccines. Among several methods available for identifying the species of origin, isoenzyme and PCR based analyses have been widely used. In the present study we attempted amplification of mitochondrial cytochrome b (Cyt b) gene using a nested PCR technique and speciation using the classical isoenzyme technique to study the identity of various cell lines and by deliberately contaminating with other cell lines and primary cells derived from various animal species.
The nested PCR and isoenzyme analyses described in this study are useful in proving the identity of various cell lines and primary cells. The nested PCR technique was found to be more sensitive than isoenzyme analysis in demonstrating the presence of low levels of contamination (as low as 1% level). Morphological examination of cell lines alone can not be considered as a reliable method for identification of cross-contamination of cell lines although at times it gives a suspicion of cross-contamination. A cellular contaminant failed to get established and became extinct in the face of stiff competition for solid phase to adhere and grow thereby proved a point that cross-contamination need not always result in purity problems. This study demonstrates usefulness of the nested PCR technique based on the Cyt b gene of Mt DNA as a potential alternative for sensitive and reliable cell line authentication. This method needs no subsequent sequencing step or DNA restriction and no computer-based analysis system. The nested PCR technique can be extensively used for identification of inter-species contamination of cell lines to ensure the production of safe vaccines and biopharmaceuticals. Isoenzyme analysis continues to remain the most commonly and widely used technique for cell line authentication worldwide by all cell culture repositories, industries and academia in spite of its poor sensitivity in comparison with nested PCR technique.
Acknowledgments
Our thanks are due to Dr. Glyn N. Stacey for his valuable suggestions, constant support and critical reading of the manuscript. We also thank Dr. Rajan Sriraman and Dr. Mohanasubramanian of Plant Biotechnology and Dr. S. B. Nagendrakumar and Dr. M. Madhanmohan of Foot- and- mouth disease virus laboratory at R and D center, Indian Immunologicals Limited, Hyderabad for their excellent technical support and assistance.
References
- Coecke S, Balls M, Bowe G, Davis J, Gstraunthaler G, Hartung T, Hay R, Merten OW, Price A, Schechtman L, Stacey G, Stokes W. Guidance on good cell culture practice. A report of the second ECVAM task force on good cell culture practice. Altern Lab Anim. 2005;33:261–287. doi: 10.1177/026119290503300313. [DOI] [PubMed] [Google Scholar]
- Dirks WG, MacLeod RAF, Jaeger K, Milch H, Drexler HG. First searchable data base for DNA profile of human cell lines; sequential use of fingerprint techniques for authentication. Cell Mol Biol. 1999;45:841–853. [PubMed] [Google Scholar]
- Forsell MNE, Li Y, Sundback M, Svehla K, Liljestorm P, Masera JR, Wyatt R, Hedestam GBK. Biochemical and immunogenic characterization of soluble human immunodeficiency virus type 1 envelope glycoprotein trimers expressed by Semliki Forest virus. J Virol. 2005;79:10902–10914. doi: 10.1128/JVI.79.17.10902-10914.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freshney RI. Cell line provenance. Cytotechnology. 2002;39:55–67. doi: 10.1023/A:1022949730029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freshney RI. Culture of cells for tissue engineering. Hoboken: Wiley; 2006. [Google Scholar]
- Gartler SM. Genetic markers as tracers in cell culture. Natl Cancer Inst Monogr. 1967;26:167–195. [PubMed] [Google Scholar]
- Halton DM, Peterson WD, Jr, Hukku B. Cell culture quality control by rapid isoenzymatic characterization. In Vitro. 1983;19:16–24. doi: 10.1007/BF02617989. [DOI] [PubMed] [Google Scholar]
- Hay RJ. Animal cell culture, a practical approach. Oxford: IRL Press; 2000. [Google Scholar]
- Hay RJ. Methods of tissue engineering. New York: Academic Press; 2002. [Google Scholar]
- Jeffreys AJ, Wilson V, Thein SL. Individual-specific ‘fingerprints’ of human DNA. Nature. 1985;316:76–79. doi: 10.1038/316076a0. [DOI] [PubMed] [Google Scholar]
- Keith WN. Animal cell culture, a practical approach. Oxford: IRL Press; 2000. [Google Scholar]
- Liu MY, Lin SC, Liu H, Candal F, Vafai A. Identification and authentication of animal cell culture by polymerase chain reaction amplification and DNA sequencing. In Vitro Cell Dev Biol Anim. 2003;39:424–427. doi: 10.1290/1543-706X(2003)039<0424:IAAOAC>2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Lopez-Andreo M, Lugo L, Garrido–Pertierra A, Prieto MI, Puyet A. Identification and quantitation of species in complex DNA mixtures by real time polymerase chain reaction. Anal Biochem. 2005;339:73–82. doi: 10.1016/j.ab.2004.11.045. [DOI] [PubMed] [Google Scholar]
- Losi GC, Ferrari S, Sossi E, Villa R, Ferrari M. An alternative method to isoenzyme profile for cell line identification and interspecies cross-contaminations: cytochrome b PCR-RFLP analysis. In Vitro Cell Dev Biol Animal. 2008;44:321–329. doi: 10.1007/s11626-008-9125-x. [DOI] [PubMed] [Google Scholar]
- Markovic O, Markovic N. Cell cross-contamination in cell cultures: the silent and neglected danger. In Vitro Cell Dev Biol Anim. 1998;34:1–8. doi: 10.1007/s11626-998-0040-y. [DOI] [PubMed] [Google Scholar]
- Masters JR. Human cancer cell lines: facts and fantasy. Nat Rev Mol Cell Biol. 2000;1:233–236. doi: 10.1038/35043102. [DOI] [PubMed] [Google Scholar]
- Miller OJ, Miller DA, Allderdice PW, Dev VG, Grewal MS. Quinacrine fluorescent karyotypes of human diploid and heteroploid cell lines. Cytogenetics. 1971;10:338–346. doi: 10.1159/000130152. [DOI] [PubMed] [Google Scholar]
- Nelson-Rees W, Flandermeyer RR. Inter-and intraspecies contamination of human breast tumor cell lines HBC and BrCa5 and other cell cultures. Science. 1977;195:1343–1344. doi: 10.1126/science.557237. [DOI] [PubMed] [Google Scholar]
- Nelson-Rees WA, Flandermeyer RR, Hawthorne PK. Banded marker chromosome as indicators of intraspecies cellular contamination. Science. 1974;184:1093–1096. doi: 10.1126/science.184.4141.1093. [DOI] [PubMed] [Google Scholar]
- Nelson-Rees WA, Daniels DW, Flandermeyer RR. Contamination of cells in culture. Science. 1989;212:446–452. doi: 10.1126/science.6451928. [DOI] [PubMed] [Google Scholar]
- Nims RW, Herbstritt CJ. Cell line authentication using isoenzyme analysis. BioPharm Int. 2005;1:1–5. [Google Scholar]
- Nims RW, Shoemaker AP, Bauernschub MA, Rec LJ, Harbell JW. Sensitivity of isoenzyme analysis for the detection of interspecies cell line cross-contamination. In Vitro Cell Dev Biol Anim. 1998;34:35–39. doi: 10.1007/s11626-998-0050-9. [DOI] [PubMed] [Google Scholar]
- O’Brien SJ, Shannon JE, Gail MH. A molecular approach to the identification and individualization of human and animal cells in culture: isozyme and allozyme genetic signatures. In Vitro. 1980;16:119–135. doi: 10.1007/BF02831503. [DOI] [PubMed] [Google Scholar]
- O’Toole CM, Pavey S, Hepburn P, Frank LM. Identity of some human bladder cancer lines. Nature. 1983;201:429–430. doi: 10.1038/301429a0. [DOI] [PubMed] [Google Scholar]
- Ono K, Satoh M, Yoshida T, Ozawa Y, Kohara A, Takeuchi M. Species identification of animal cells by nested PCR targeted to mitochondrial DNA. In Vitro Cell Dev Biol Anim. 2007 doi: 10.1007/s11626-007-9033-5. [DOI] [PubMed] [Google Scholar]
- Parodi B, Aresu O, Bini D, Lorenzini R, Schena F, Visconti P, et al. Species identification and confirmation of human and animal cell lines: a PCR-based method. Biotechniques. 2002;32:432–440. doi: 10.2144/02322rr05. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: a laboratory manual. New York: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Stacey GN, Bolton BJ, Doyle A. DNA fingerprinting transforms the art of cell authentication. Nature. 1992;357:261–262. doi: 10.1038/357261a0. [DOI] [PubMed] [Google Scholar]
- Stacey GN, Hoelzl H, Stephenson JR, Doyle A. Authentication of animal cell cultures by direct visualization of repetitive DNA, aldolase gene PCR and isoenzyme analysis. Biologicals. 1997;25:75–85. doi: 10.1006/biol.1996.0062. [DOI] [PubMed] [Google Scholar]
- Stacey GN, Masters JR, Hay RJ, Drexler HG, MacLeod RAF, Freshney RI. Cell contamination leads to inaccurate data: we must take action now. Nature. 2000;403:356. doi: 10.1038/35000394. [DOI] [PubMed] [Google Scholar]
- Steube KG, Grunicke D, Drexler HG. Isoenzyme analysis as a rapid method for the examination of the species identity of cell cultures. In Vitro Cell Dev Biol Anim. 1995;31:115–119. doi: 10.1007/BF02633971. [DOI] [PubMed] [Google Scholar]
- Stulberg CS, Peterson WR, Jr, Simpson WF. Identification of cells in culture. Am J Hematol. 1976;1:237–242. doi: 10.1002/ajh.2830010208. [DOI] [PubMed] [Google Scholar]