Abstract
AIM: To investigate the expression of microRNA155 (miRNA155) in trinitrobenzene sulphonic acid (TNBS)-induced colitis and the relationship between miRNA155 and tumor necrosis factor (TNF) expressions.
METHODS: In TNBS colitis mice, miRNA155 and TNF mRNA expressions were measured in colons and CD4+ T cells of draining lymph nodes (LNs). CD4+ T cells were cultured in vitro with or without anti-CD3/CD28 antibody, and the expressions of miRNA155 and TNF mRNA in cells and TNF concentration in culture media were examined.
RESULTS: miRNA155 and TNF mRNA expressions in colons and in cells of LNs were significantly increased in TNBS colitis compared with controls. In TNBS colitis, miRNA155 and TNF mRNA expressions in CD4+ T cells of LNs and TNF concentration in CD4+ T cells culture media increased compared with controls. When cultured with anti-CD3/CD28 antibody, miRNA155 and TNF mRNA expressions in CD4+ T cells and TNF concentration in the CD4+ T cells culture media were significantly higher than those cultured without anti-CD3/CD28 antibody. Following analysis using the Pearson’s correlation coefficient, miRNA155 expression had a significant positive correlation with either TNF mRNA expression in CD4+ T cells (r = 0.860, P < 0.05) or TNF concentration in CD4+ T cells culture media (r = 0.892, P < 0.05).
CONCLUSION: miRNA155 is induced in colons and activated CD4+ T cells in TNBS colitis, and the levels of miRNA155 and TNF expressions have a significant positive correlation.
Keywords: Colitis, Crohn’s disease, Lymph nodes, microRNA, Tumor necrosis factor
INTRODUCTION
Crohn’s disease (CD) is a chronic relapsing inflammatory disorder of the gastrointestinal tract. CD is thought to be a multifactorial, polygenic disease, however, the exact pathogenesis of CD is still unclear[1-3]. Some studies have suggested that CD is mediated by T helper type 1 (Th1) cells producing interferon-γ (IFN-γ), tumor necrosis factor (TNF) and interleukin (IL)-12[4-8]. TNF, which is a proinflammatory cytokine secreted by monocytes/macrophages, T cells, B cells, NK cells and mast cells, is necessary for both the initiation and persistence of the Th1 response and contributes to intestinal inflammation in CD patients[9-11]. Today, many animal models of CD are available, and each model has reflected one aspect of human CD. Hapten-induced colitis, in which trinitrobenzene sulfonic acid (TNBS) is delivered intrarectally to rodents, displays Th1 activity of local CD4+ T cells and is considered to closely resemble CD[12-14].
MicroRNAs (miRNAs) are a group of small noncoding RNAs which posttranscriptionally regulate gene expression[15,16]. More than 700 miRNAs have been identified in mammals and are involved in a wide variety of biological processes[15,17]. They are transcribed as primary transcripts by RNA polymerase II, cleaved into a precursor miRNA by the Drosha nuclease, and exported from the nucleus by exportin 5. In the cytoplasm, the precursor miRNAs are further processed by the Dicer nuclease and are incorporated into the RNA-induced silencing complexes. The miRNAs guide the RNA-induced silencing complexes binding to the mRNAs 3'-untranslated regions (UTRs), resulting in either the mRNAs degradation or translational inhibition[18-22].
Although the exact functions of most miRNAs have yet to be elucidated, many studies have suggested that miRNAs have been implicated in many aspects of innate and acquired immunity, such as differentiation, survival and functions of immune cells, and the intracellular signaling pathways[23-25]. miRNA155, which was reported to be involved in the production of TNF and regulation of immunity[26,27], is processed from an exon of the noncoding RNA known as bic[27,28]. In this study, we mainly evaluated the expression of miRNA155 in TNBS colitis, and speculated on the relationship between miRNA155 and TNF expressions.
MATERIALS AND METHODS
Mice
Female 6- to 8-wk-old BALB/C mice weighing 18-22 g were obtained from Shanghai Experimental Animals Centre of Chinese Academy of Sciences. The mice were maintained under specific pathogen free conditions in a room at 23 ± 2°C with a 12 h light-dark cycle, and had free access to food and water during the study. They were allowed to acclimate to these conditions for at least seven days before inclusion in an experiment. All procedures were approved by the Investigation and Ethics Committee of Shanghai Jiaotong University School of Medicine.
Establishment of TNBS-induced colitis
Colitis was induced in BALB/C mice as described previously with some modification[13,29]. For sensitization, a 2 cm × 2 cm field of the abdominal skin was shaved and 150 μL of 2.5% TNBS (Sigma Chemical Co., St. Louis, MO, USA) in 50% ethanol was applied. Five days after sensitization, mice were anesthetized slightly with an intraperitoneal injection of xylazine (10 mg/kg) and ketamine (50 mg/kg), and then intrarectally administered 100 μL solution of 1% TNBS dissolved in 45% ethanol via a 3.5-French catheter equipped with a 1 mL syringe. The tip of the catheter was inserted 4 cm proximal to the anal verge. Mice were held in a vertical position for 1 min after the intrarectal injection. Control mice were given 100 μL 45% ethanol solution without TNBS using the same technique.
Clinical observations and histologic assessments of colitis
Daily body weight, stool consistency, and occult blood (measured by the guaiac reaction, hemoccult) were assessed. Three days after intrarectal injection, mice were killed by cervical dislocation after being anesthetized with diethyl ether and entire colons were removed from the cecum to the anus, and flushed with saline. Colon specimens located 2 cm above the anal verge were achieved. One section of the specimen was fixed overnight in 4% paraformaldehyde and embedded in paraffin, and then sections stained with hematoxylin and eosin were examined. The other sections of the colon were immediately frozen in liquid nitrogen after dissection and used for quantification of miRNA155, IL-1β, IL-6, TNF and IFN-γ mRNA.
Cell preparation
Three days after intrarectal injection, colon draining lymph nodes (LNs) were aseptically removed. Single-cell suspensions were prepared by pressing LNs through a 40 μm cell strainer using the plunger of a 1 mL syringe. CD4+ T cells were isolated from the cell suspensions with magnetic beads labeled with anti-CD4 (L3T4) monoclonal antibodies (Miltenyi Biotec Inc, Bergisch Gladbach, Germany). Cells were incubated in media (RPMI 1640 supplemented with 100 U/mL penicillin/streptomycin, 2 mmol/L L-glutamine, 50 mol/L 2-mercaptoethanol, and 10% fetal calf serum) at 8 × 104 cells in 150 μL media per well in 96-well plates for 48 h in the absence or presence of dynabeads CD3/CD28 T cells activator (Invitrogen, Carlsbad, CA, USA) at a concentration of 2 μL/well.
Enzyme-linked immunosorbent assay (ELISA)
After incubation for 48 h, the supernatants of the culture media were harvested and assayed for TNF concentration by ELISA using an ELISA kit (R&D Systems, Minneapolis, MN, USA).
Quantitative real-time polymerase chain reaction (qPCR) analysis of mRNA detection
Total RNA from cells and colon samples were extracted using the TRIzol reagent (Invitrogen, Carlsbad, CA, USA). RNA concentrations were determined with a spectrophotometer (Eppendorf, Hamburg, Germany). 0.2-0.5 μg of total RNA was reverse transcribed, and RNA expression levels were quantified by sybergreen-based qPCR using a sequence detection system (Prism 7500; Applied Biosystems Inc., Foster City, USA). β-actin served as the endogenous control. Gene-specific primers for the reported genes are indicated in Table 1. To evaluate the relative expression of each target gene, the comparative threshold (Ct) cycle method was used according to the manufacturer’s manual. The threshold cycle (Ct) for each gene was determined as the cycle number at which the reaction crossed an arbitrarily placed threshold, and the relative amount of each mRNA to β-actin was described using the formula 2-∆Ct where ∆Ct = (CtmRNA - Ctβ-actin).
Table 1.
Primers used for RT or PCR of mRNA or miRNA
| Gene name | Primer sequences (5’-3’) | |
| IL-1β | Sense | GCAACTGTTCCTGAACTCAACT |
| Antisense | ATCTTTTGGGGTCCGTCAACT | |
| IL-6 | Sense | CCACTTCACAAGTCGGAGGCTTA |
| Antisense | GCAAGTGCATCATCGTTGTTCATAC | |
| IFN-γ | Sense | TCAAGTGGCATAGATGTGGAAGAA |
| Antisense | TGGCTCTGCAGGATTTTCATG | |
| TNF | Sense | CCACCACGCTCTTCTGTCTAC |
| Antisense | TGGGCTACAGGCTTGTCACT | |
| β-actin | Sense | CTAGGCACCAGGGTGTGAT |
| Antisense | TGCCAGATCTTCTCCATGTC | |
| miRNA155 | Stem-loop primer | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACACCCCT |
| Sense | GCCCGCTTAATGCTAATTGTGAT | |
| Antisense | GTGCAGGGTCCGAGGT | |
| U6 | Sense | CTCGCTTCGGCAGCACA |
| Antisense | AACGCTTCACGAATTTGCGT |
RT: Reverse transcription; PCR: Polymerase chain reaction.
qPCR analysis of miRNA detection
Total RNA from cells and colon samples were isolated using the TRIzol reagent. Real time quantitative analyses for miRNAs were performed using stem-loop RT-PCR[30,31]. 0.2-0.5 μg of total RNA was reverse transcribed to cDNA using a target-specific stem-loop primer indicated in Table 1. qPCR was performed on a sequence detection system (Prism 7500; Applied Biosystems Inc., Foster City, USA). In brief, cDNA in water was added to 5 μL of the 2 × SYBR green master mix (Applied Biosystems Inc., Foster City, USA), 400 nmol/L of gene-specific primer and water to 10 μL. The reactions were amplified at 95°C for 10 min followed by 40 cycles at 95°C for 15 s and 60°C for 60 s. U6 small nuclear RNA (U6) served as the endogenous control. At the end of the qPCR, the thermal denaturation protocol was run to determine the number of products that were present in the reaction. The relative amount of miRNA to U6 was calculated using the Ct cycle method. The relative amount of each miRNA to U6 was described using the formula 2-∆Ct where ∆Ct = (CtmiRNA - CtU6)[30,31].
Statistical analysis
Each group contained 5-8 mice, and results were expressed as the mean ± SD. A comparison between the two groups was made using the Student’s t-test. The relationship between the two targets was tested with Pearson’s correlation coefficient. Differences were considered significant at P < 0.05.
RESULTS
Successful establishment of experimental colitis
Administration of TNBS to presensitized mice resulted in a severe illness characterized by bloody diarrhea, rectal prolapse accompanied by sustained weight loss. At day 3-4, the disease reached a peak. Histologic examination of the colons showed severe depletion of mucin-producing goblet and epithelial cells, large areas of ulceration, a marked increase in the thickness of the muscular layer, and transmural inflammation involving all colon wall layers with infiltration of lymphocytes, macrophages and neutrophils extending from the mucosa into the muscular and serosal layers (Figure 1).
Figure 1.
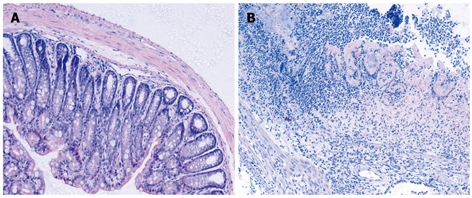
Representative microphotographs of the colon sections stained with hematoxylin and eosin (original magnification, × 100). A: Control colon; B: Colon was obtained on day 3 after intrarectal TNBS.
Increased expressions of miRNA155, IL-1β, IL-6, TNF, and IFN-γ mRNA in the colons of TNBS-induced colitis mice
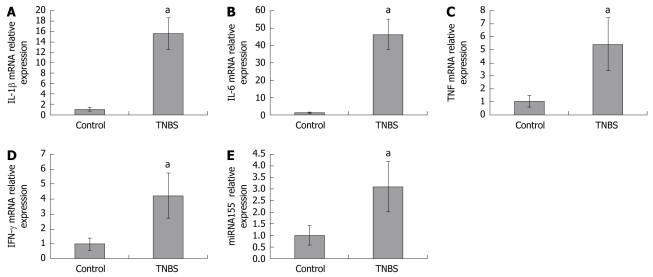
To investigate miRNA155 and cytokine expressions in colons, we assessed miRNA155, IL-1β, IL-6, TNF and IFN-γ mRNA in colon homogenates by qPCR. We found that miRNA155 expression in colon homogenates was significantly increased in TNBS-induced colitis, which was 3.10-fold higher than in control mice. IL-1β, IL-6, TNF, and IFN-γ mRNA expressions in colon homogenates were also significantly increased in TNBS-induced colitis, and were 15.58-, 46.34-, 5.43-, and 4.23-fold higher than in control mice, respectively (Figure 2).
Figure 2.
IL-1β (A), IL-6 (B), TNF (C), IFN-γ (D) mRNA and miRNA155 (E) expressions in colon homogenates of control and TNBS colitis groups. qPCR-derived IL-1β, IL-6, TNF, IFN-γ, and miRNA155 expressions in colon homogenates were significantly increased in TNBS-induced colitis. aP < 0.05 vs control.
miRNA155 and TNF mRNA expressions in the CD4+ T cells of colon draining LNs and TNF concentration in CD4+ T cells culture media
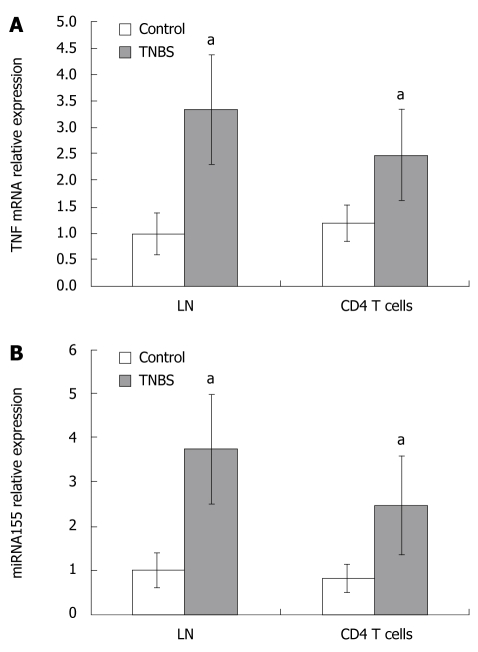
As CD4+ T cells play a central role in Th1 response, we evaluated the levels of miRNA155 and TNF mRNA expressions in LNs and CD4+ T cells from LNs. In TNBS colitis, miRNA155 in draining LNs or CD4+ T cells from LNs increased and were 3.74- and 3.07-fold higher than in controls, respectively (Figure 3). TNF mRNA in draining LNs or CD4+ T cells from LNs of TNBS colitis were 3.34- and 2.06-fold higher than in controls (Figure 3). In the CD4+ T cells culture media, the concentration of TNF protein increased in TNBS colitis and was 4.19-fold higher than in controls (128.04 ± 38.71 pg/mL vs 30.55 ± 8.37 pg/mL, P < 0.05, Figure 4).
Figure 3.
TNF mRNA (A) and miRNA155 (B) expressions in LNs and CD4+ T cells in control and TNBS colitis groups. TNF mRNA and miRNA155 expressions in LNs and CD4+ T cells were significantly increased in TNBS-induced colitis. aP < 0.05 vs control.
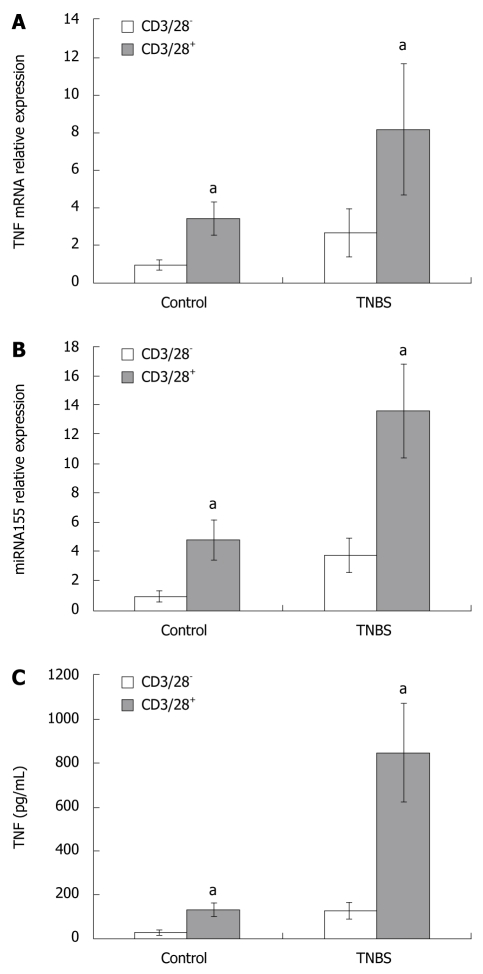
Figure 4.
TNF mRNA (A) and miRNA155 (B) expressions in CD4+ T cells cultured with or without anti-CD3/CD28 antibody, and the TNF protein expression (C) in the supernatants of CD4+ T cells culture media with or without anti-CD3/CD28 antibody. TNF mRNA and miRNA155 expressions in CD4+ T cells cultured with anti-CD3/CD28 antibody, and the TNF concentration in the supernatants of culture media containing anti-CD3/CD28 antibody increased in comparison to those in cells or media cultured without anti-CD3/CD28 antibody. aP < 0.05 vs control.
Anti-CD3/CD28 antibody promoted the miRNA155 and TNF mRNA expressions in the CD4+ T cells and the TNF concentration in the supernatants of CD4+ T cells culture media
To study whether the T cell receptor (TCR) and the costimulatory receptor were involved in miRNA155 expression, and to study the relationship between miRNA155 and TNF production, we used anti-CD3/CD28 antibody to stimulate CD4+ T cells. When cultured with anti-CD3/CD28 antibody, the CD4+ T cells became larger and displayed an activated appearance. In control and TNBS colitis, the miRNA155 expressions in CD4+ T cells cultured with anti-CD3/CD28 antibody were 4.72- and 3.61-fold higher than cells cultured without anti-CD3/CD28 antibody, and the TNF mRNA expression in CD4+ T cells cultured with anti-CD3/CD28 antibody were 3.42- and 3.03-fold higher than cells cultured without anti-CD3/CD28 antibody, respectively (Figure 4). The TNF concentration in the supernatants of culture media which contained anti-CD3/CD28 antibody increased both in control and TNBS colitis mice, and were 4.28- and 6.87-fold higher than in media cultured without anti-CD3/CD28 antibody (135.66 ± 32.11 pg/mL vs 30.55 ± 8.37 pg/mL, P < 0.05; 850.94 ± 219.49 pg/mL vs 128.04 ± 38.71 pg/mL, P < 0.05, Figure 4).
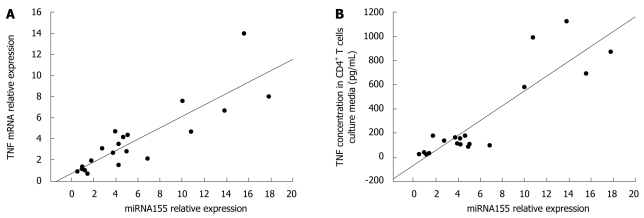
Relationship between miRNA155 and TNF gene expressions
Since TNF plays an important role in the pathogenesis of CD, we evaluated the potential correlation between miRNA155 and TNF gene expressions. Our data indicated that there was a significant positive correlation between miRNA155 and TNF mRNA expressions in CD4+ T cells (r = 0.860, P < 0.05, Figure 5), and miRNA155 expression in CD4+ T cells and TNF protein concentration in CD4+ T cells culture media (r = 0.892, P < 0.05, Figure 5).
Figure 5.
Relationships between miRNA155 expression in CD4+ T cells and TNF mRNA expression in CD4+ T cells (A) or TNF concentration in CD4+ T cells culture media (B). Data analyzed by Pearson’s correlation coefficient.
DISCUSSION
As the exact etiology of CD is still unclear, TNBS-induced colitis was used to study many important aspects of the pathogenesis in CD. TNBS colitis is thought to resemble CD because of the mucosal inflammation mediated by excessive IFN-γ, TNF and other proinflammatory cytokine production[12-14]. In agreement with previous results, our data also showed that TNBS colitis is a Th1 model with elevated IFN-γ and TNF expressions in colon. In this study, we found that miRNA155 was increased in colons and in CD4+ T cells of LNs in TNBS colitis and the levels of miRNA155 and TNF expressions had a significant positive correlation.
MiRNAs are a group of small noncoding RNAs which are thought to posttranscriptionally regulate gene expressions. Dysregulation of miRNAs has been associated with several autoimmune diseases[32,33]. miRNA155 is processed from an exon of the noncoding RNA. Some studies have shown that miRNA155 is required for normal innate and acquired immunity[26,27,34,35].
In macrophages, miRNA155 was reported to enhance the production of TNF, but may target transcripts encoding for several proteins, such as IκBepsilon kinase and Fas-associated death domain protein whose ultimate function results in the activation of the lipopolysaccharide (LPS)/TNF pathway[26]. Therefore, miRNA155 may exert both positive and negative effects on the activation of innate immunity. In acquired immunity, miRNA155 was reported to affect lymphoid cell development[27,34,36]. In miRNA155-/- mice, CD4+ T cells are intrinsically biased toward Th2 differentiation. miRNA155 modulates the level of a transcription factor c-Maf in T cells and is likely to induce the attenuation of Th2 cell responses in vivo[34]. With regard to B cells, several studies have proved that B cells require miRNA155 for normal production of isotype-switched, high-affinity antibodies and for a memory response[27,36]. However, the expression of miRNA155 and its relationship with CD remains unclear. In this paper, we reported that miRNA155 expression is increased in the colon and it’s draining LNs in TNBS colitis. This result hints that miRNA155 may have a role in the pathogenesis of TNBS colitis. As CD4+ T cells play a central role in Th1 response, we determined the level of miRNA155 expression in the CD4+ T cells from LNs in TNBS colitis and found that miRNA155 was increased in CD4+ T cells in TNBS colitis.
Considering TNF plays an important role in the pathogenesis of CD, we evaluated the relationship between miRNA155 and TNF expression in CD4+ T cells. Our data indicated that the gene expression of miRNA155 in CD4+ T cells of draining LNs in TNBS colitis has a significant positive correlation with either TNF mRNA in CD4+ T cells or the concentration of TNF protein in the culture media. Some reports have shown that miRNA155 may be involved in TNF production[26,27]. Eu-miR-155 transgenic mice which specifically overexpress miRNA155 in B cells produced more TNF when challenged with LPS, and were hypersensitive to LPS/D-galactosamine-induced septic shock[26]. In addition, miRNA155−/− B cells produce less TNF when activated in vitro by BCR cross-linking[27]. Scientists have found that the miRNA155 effects on TNF production are at both the transcriptional and posttranscriptional level[26,27]. More TNF transcripts were observed in wild-type mice compared with miRNA155-/- mice[27]. At the posttranscriptional level, miRNA155 may target the 3’-UTR of TNF transcripts to increase the stability of transcripts and enhance translation[26]. Our results showed that the TNF protein concentration in the CD4+ T cells culture media increased more than the TNF mRNA in CD4+ T cells, which is in agreement with the proposition that miRNA155 regulated TNF production at both the transcriptional and posttranscriptional levels. These findings suggested that miRNA155 may prompt the production of TNF in some types of immune cells. From our results, miRNA155 increased and had a positive relationship with TNF expression in CD4+ T cells, therefore we suppose that miRNA155 may influence TNF expression in CD4+ T cells and is possibly involved in the pathogenesis of TNBS colitis.
The exact mechanism of miRNA155 expression regulation in CD4+ T cells remains unclear. To study the role of TCR in miRNA155 expression and the relationship between miRNA155 expression and the activation of CD4+ T cells, we used anti-CD3/CD28 antibody to stimulate CD4+ T cells, and found that the miRNA155 expression level was elevated in CD4+ T cells when cultured with the antibody. Stimulation of the TCR/CD3 complex and costimulatory receptor CD28 in CD4+ T cells by anti-CD3/CD28 antibody can lead to activation of multiple transcription factors, including NF-AT and NF-κB, which ultimately control transcription of cytokines and T-cell proliferation[37]. miRNA155 level was reported to have a link with the NF-κB pathway. NF-κB activity is required for the change in miRNA155 levels following TNF stimulation in macrophages, however, this relationship remains elusive[26]. The precise mechanism of the regulation of miRNA155 expression may be a new issue for future research.
In conclusion, miRNA155 expression was found to increase in colons and activated CD4+ T cells in TNBS colitis. A significant positive correlation was observed between miRNA155 expression in CD4+ T cells and the expression of TNF mRNA in CD4+ T cells and the TNF protein concentration in CD4+ T cells culture media. miRNA155 may be involved in the activation and TNF production of CD4+ T cells in TNBS colitis.
COMMENTS
Background
MicroRNAs (miRNAs) are a group of small noncoding RNAs which posttranscriptionally regulate gene expression. miRNA155 is reported to be involved in the production of TNF and the regulation of immunity. Tumor necrosis factor (TNF) plays an important role in the pathogenesis of Crohn’s disease (CD). However, the expression of miRNA155 and its relationship with TNF production in CD remains unclear.
Research frontiers
The exact etiology of CD is still unclear. MiRNA research may represent a new way to explore the pathogenesis of CD. The expressions and functions of most miRNAs in CD remain a mystery.
Innovations and breakthroughs
The study found that miRNA155 is increased in colons and draining lymph nodes. In CD4+ T cells which play a central role in Th1 response, miRNA155 expression is higher in TNBS colitis than in controls. As CD4+ T cells were activated by anti-CD3/CD28 antibody, the miRNA155 expression was higher than that when cultured without anti-CD3/CD28 antibody. In addition, the levels of miRNA155 and TNF expressions had a significant positive correlation.
Applications
The results of this study may enhance our understanding of the pathogenesis of CD. By investigating the exact function of miRNA155 in the pathogenesis of CD, the findings may contribute to improvements in future drug therapies.
Terminology
MiRNA: MicroRNAs (miRNAs) are a group of small noncoding RNAs which post-transcriptionally regulate gene expressions. More than 700 miRNAs have been identified in mammals and are involved in a wide variety of biological processes. Stem-loop RT-PCR: RNA is reverse transcribed to cDNA using a gene-specific stem-loop RT primer, and then the RT products are quantified using conventional real time PCR.
Peer review
This paper reports a set of relatively new data about the pathogenic mechanism of IBD that may be referred by those investigators working on IBD.
Footnotes
Peer reviewers: Pingchang Yang, MD, PhD, Department of Pathology & Molecular Medicine, McMaster University, BBI-T3330, 50 Charlton Ave East, Hamilton, L8N 4A6, Canada; Mitsunori Yamakawa, Professor, Department of Pathological Diagnostics, Yamagata University, Faculty of Medicine, 2-2-2 Iida-Nishi, Yamagata 990-9585, Japan
S- Editor Tian L L- Editor Webster JR E- Editor Ma WH
References
- 1.Podolsky DK. Inflammatory bowel disease. N Engl J Med. 2002;347:417–429. doi: 10.1056/NEJMra020831. [DOI] [PubMed] [Google Scholar]
- 2.Kucharzik T, Maaser C, Lügering A, Kagnoff M, Mayer L, Targan S, Domschke W. Recent understanding of IBD pathogenesis: implications for future therapies. Inflamm Bowel Dis. 2006;12:1068–1083. doi: 10.1097/01.mib.0000235827.21778.d5. [DOI] [PubMed] [Google Scholar]
- 3.Xavier RJ, Podolsky DK. Unravelling the pathogenesis of inflammatory bowel disease. Nature. 2007;448:427–434. doi: 10.1038/nature06005. [DOI] [PubMed] [Google Scholar]
- 4.Strober W, Fuss IJ, Blumberg RS. The immunology of mucosal models of inflammation. Annu Rev Immunol. 2002;20:495–549. doi: 10.1146/annurev.immunol.20.100301.064816. [DOI] [PubMed] [Google Scholar]
- 5.Strober W, Fuss I, Mannon P. The fundamental basis of inflammatory bowel disease. J Clin Invest. 2007;117:514–521. doi: 10.1172/JCI30587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mizoguchi A, Mizoguchi E, Bhan AK. Immune networks in animal models of inflammatory bowel disease. Inflamm Bowel Dis. 2003;9:246–259. doi: 10.1097/00054725-200307000-00005. [DOI] [PubMed] [Google Scholar]
- 7.Elson CO, Cong Y, McCracken VJ, Dimmitt RA, Lorenz RG, Weaver CT. Experimental models of inflammatory bowel disease reveal innate, adaptive, and regulatory mechanisms of host dialogue with the microbiota. Immunol Rev. 2005;206:260–276. doi: 10.1111/j.0105-2896.2005.00291.x. [DOI] [PubMed] [Google Scholar]
- 8.Neurath MF, Finotto S, Glimcher LH. The role of Th1/Th2 polarization in mucosal immunity. Nat Med. 2002;8:567–573. doi: 10.1038/nm0602-567. [DOI] [PubMed] [Google Scholar]
- 9.Braegger CP, MacDonald TT. Immune mechanisms in chronic inflammatory bowel disease. Ann Allergy. 1994;72:135–141. [PubMed] [Google Scholar]
- 10.Neurath MF, Fuss I, Pasparakis M, Alexopoulou L, Haralambous S, Meyer zum Büschenfelde KH, Strober W, Kollias G. Predominant pathogenic role of tumor necrosis factor in experimental colitis in mice. Eur J Immunol. 1997;27:1743–1750. doi: 10.1002/eji.1830270722. [DOI] [PubMed] [Google Scholar]
- 11.Strober W, Fuss IJ, Blumberg RS. The immunology of mucosal models of inflammation. Annu Rev Immunol. 2002;20:495–549. doi: 10.1146/annurev.immunol.20.100301.064816. [DOI] [PubMed] [Google Scholar]
- 12.Morris GP, Beck PL, Herridge MS, Depew WT, Szewczuk MR, Wallace JL. Hapten-induced model of chronic inflammation and ulceration in the rat colon. Gastroenterology. 1989;96:795–803. [PubMed] [Google Scholar]
- 13.Neurath MF, Fuss I, Kelsall BL, Stüber E, Strober W. Antibodies to interleukin 12 abrogate established experimental colitis in mice. J Exp Med. 1995;182:1281–1290. doi: 10.1084/jem.182.5.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Neurath MF, Pettersson S, Meyer zum Büschenfelde KH, Strober W. Local administration of antisense phosphorothioate oligonucleotides to the p65 subunit of NF-kappa B abrogates established experimental colitis in mice. Nat Med. 1996;2:998–1004. doi: 10.1038/nm0996-998. [DOI] [PubMed] [Google Scholar]
- 15.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 16.Engels BM, Hutvagner G. Principles and effects of microRNA-mediated post-transcriptional gene regulation. Oncogene. 2006;25:6163–6169. doi: 10.1038/sj.onc.1209909. [DOI] [PubMed] [Google Scholar]
- 17.Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34:D140–D144. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cai X, Hagedorn CH, Cullen BR. Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. RNA. 2004;10:1957–1966. doi: 10.1261/rna.7135204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Han J, Lee Y, Yeom KH, Nam JW, Heo I, Rhee JK, Sohn SY, Cho Y, Zhang BT, Kim VN. Molecular basis for the recognition of primary microRNAs by the Drosha-DGCR8 complex. Cell. 2006;125:887–901. doi: 10.1016/j.cell.2006.03.043. [DOI] [PubMed] [Google Scholar]
- 20.Yi R, Qin Y, Macara IG, Cullen BR. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003;17:3011–3016. doi: 10.1101/gad.1158803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chendrimada TP, Finn KJ, Ji X, Baillat D, Gregory RI, Liebhaber SA, Pasquinelli AE, Shiekhattar R. MicroRNA silencing through RISC recruitment of eIF6. Nature. 2007;447:823–828. doi: 10.1038/nature05841. [DOI] [PubMed] [Google Scholar]
- 22.Chendrimada TP, Gregory RI, Kumaraswamy E, Norman J, Cooch N, Nishikura K, Shiekhattar R. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature. 2005;436:740–744. doi: 10.1038/nature03868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Taganov KD, Boldin MP, Baltimore D. MicroRNAs and immunity: tiny players in a big field. Immunity. 2007;26:133–137. doi: 10.1016/j.immuni.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 24.Baltimore D, Boldin MP, O’Connell RM, Rao DS, Taganov KD. MicroRNAs: new regulators of immune cell development and function. Nat Immunol. 2008;9:839–845. doi: 10.1038/ni.f.209. [DOI] [PubMed] [Google Scholar]
- 25.Bi Y, Liu G, Yang R. MicroRNAs: novel regulators during the immune response. J Cell Physiol. 2009;218:467–472. doi: 10.1002/jcp.21639. [DOI] [PubMed] [Google Scholar]
- 26.Tili E, Michaille JJ, Cimino A, Costinean S, Dumitru CD, Adair B, Fabbri M, Alder H, Liu CG, Calin GA, et al. Modulation of miR-155 and miR-125b levels following lipopolysaccharide/TNF-alpha stimulation and their possible roles in regulating the response to endotoxin shock. J Immunol. 2007;179:5082–5089. doi: 10.4049/jimmunol.179.8.5082. [DOI] [PubMed] [Google Scholar]
- 27.Thai TH, Calado DP, Casola S, Ansel KM, Xiao C, Xue Y, Murphy A, Frendewey D, Valenzuela D, Kutok JL, et al. Regulation of the germinal center response by microRNA-155. Science. 2007;316:604–608. doi: 10.1126/science.1141229. [DOI] [PubMed] [Google Scholar]
- 28.Tam W. Identification and characterization of human BIC, a gene on chromosome 21 that encodes a noncoding RNA. Gene. 2001;274:157–167. doi: 10.1016/s0378-1119(01)00612-6. [DOI] [PubMed] [Google Scholar]
- 29.te Velde AA, Verstege MI, Hommes DW. Critical appraisal of the current practice in murine TNBS-induced colitis. Inflamm Bowel Dis. 2006;12:995–999. doi: 10.1097/01.mib.0000227817.54969.5e. [DOI] [PubMed] [Google Scholar]
- 30.Chen C, Ridzon DA, Broomer AJ, Zhou Z, Lee DH, Nguyen JT, Barbisin M, Xu NL, Mahuvakar VR, Andersen MR, et al. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 2005;33:e179. doi: 10.1093/nar/gni178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schmittgen TD, Lee EJ, Jiang J, Sarkar A, Yang L, Elton TS, Chen C. Real-time PCR quantification of precursor and mature microRNA. Methods. 2008;44:31–38. doi: 10.1016/j.ymeth.2007.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tang Y, Luo X, Cui H, Ni X, Yuan M, Guo Y, Huang X, Zhou H, de Vries N, Tak PP, et al. MicroRNA-146A contributes to abnormal activation of the type I interferon pathway in human lupus by targeting the key signaling proteins. Arthritis Rheum. 2009;60:1065–1075. doi: 10.1002/art.24436. [DOI] [PubMed] [Google Scholar]
- 33.Wu F, Zikusoka M, Trindade A, Dassopoulos T, Harris ML, Bayless TM, Brant SR, Chakravarti S, Kwon JH. MicroRNAs are differentially expressed in ulcerative colitis and alter expression of macrophage inflammatory peptide-2 alpha. Gastroenterology. 2008;135:1624–1635.e24. doi: 10.1053/j.gastro.2008.07.068. [DOI] [PubMed] [Google Scholar]
- 34.Rodriguez A, Vigorito E, Clare S, Warren MV, Couttet P, Soond DR, van Dongen S, Grocock RJ, Das PP, Miska EA, et al. Requirement of bic/microRNA-155 for normal immune function. Science. 2007;316:608–611. doi: 10.1126/science.1139253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.O’Connell RM, Taganov KD, Boldin MP, Cheng G, Baltimore D. MicroRNA-155 is induced during the macrophage inflammatory response. Proc Natl Acad Sci USA. 2007;104:1604–1609. doi: 10.1073/pnas.0610731104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vigorito E, Perks KL, Abreu-Goodger C, Bunting S, Xiang Z, Kohlhaas S, Das PP, Miska EA, Rodriguez A, Bradley A, et al. microRNA-155 regulates the generation of immunoglobulin class-switched plasma cells. Immunity. 2007;27:847–859. doi: 10.1016/j.immuni.2007.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang D, You Y, Case SM, McAllister-Lucas LM, Wang L, DiStefano PS, Nuñez G, Bertin J, Lin X. A requirement for CARMA1 in TCR-induced NF-kappa B activation. Nat Immunol. 2002;3:830–835. doi: 10.1038/ni824. [DOI] [PubMed] [Google Scholar]