Summary
Nedd4-1 is a ‘Neuronal Precursor Cell Expressed and Developmentally Downregulated Protein’ and among the most abundant E3 ubiquitin ligases in mammalian neurons. In analyses of conventional and conditional Nedd4-1 deficient mice, we found that Nedd4-1 plays a critical role in dendrite formation. Nedd4-1, the serine/threonine kinase TNIK, and Rap2A form a complex that controls Nedd4-1-mediated ubiquitination of Rap2A. Ubiquitination by Nedd4-1 inhibits Rap2A function, which reduces the activity of Rap2 effector kinases of the TNIK family and promotes dendrite growth. We conclude that a Nedd4-1/Rap2A/TNIK signaling pathway regulates neurite growth and arborization in mammalian neurons.
Keywords: Dendrite growth, TNIK, ubiquitination, HECT
Introduction
Dendrites are branched extensions of neurons where synapse formation, subsequent functional innervation, and the first steps of input signal integration take place. Dendrite size and complexity of a given neuron are correlated with the numbers of innervating axons and functional synapses (Hume and Purves, 1981; Snider, 1987), and the dendrite structure directly influences synaptogenesis and synapse elimination (Kwon et al., 2006). As dendrites play a critical role in defining the signal input characteristics of neurons, their structure and complexity as well as the processes that control their growth are of profound importance for the establishment of neuronal networks (Parrish et al., 2007; Spruston, 2008).
Multiple neuronal trans-membrane cell surface proteins, including Dscam, Neuropilins, Notch, or Neurotrophin receptors, regulate the morphology of dendrites in a differential manner (Parrish et al., 2007), and numerous neuronal proteins control more subtle aspects of dendrite morphology such as spine formation and stability. The fact that different cell surface proteins control distinct aspects of dendrite morphogenesis implies that distinct intracellular signal transduction cascades exist that translate individual extracellular signals into specific morphogenic changes during dendrite formation. Transcription factors such as Neurogenin 2 or CREB, the tyrosine kinase Fyn, CaMKs, or small GTPases of the Rho and Ras families are likely components of such signaling cascades (Hand et al., 2005; Huang and Reichardt, 2003; Sasaki et al., 2002; Takemoto-Kimura et al., 2007; Yamashita et al., 2007). However, it is largely unknown how these signaling pathways are coordinated to determine specific morphological features of dendrites during different phases of neuronal network formation.
In the present study, we discovered a novel signaling pathway that controls dendrite growth. It involves the E3 ubiquitin ligase Nedd4-1, which acts as an essential positive regulator of dendrite development and operates by mono- or di-ubiquitination of Rap2A and downstream modulation of Rap2A effector kinases of the TNIK family.
Results
Perturbed Embryonic Development of Conventional Nedd4-1-KO Mice
Nedd4-1 mRNA and protein are strongly and ubiquitously expressed in the newborn brain and developmentally downregulated after synapse formation has peaked and dendrite development is throttled (Figure S1), indicating that Nedd4-1 is particularly important in the brain during gestation and early postnatal development. In the adult mouse brain, Nedd4-1 is slightly enriched in ER- and Golgi membranes (Figure S1F).
We generated Nedd4-1-KO mice in which Nedd4-1 protein expression is abolished (Figure S2A-S2E). Although Nedd4-1 protein expression was slightly reduced in heterozygous KOs (Figure S2E), these were viable and showed no obvious phenotypic changes. Offspring of heterozygous interbreedings showed the predicted Mendelian frequency of homozygous KO, heterozygous KO, and wild type (WT) progeny until embryonic day E16.5 (20%:52%:28%), after which the number of homozygous KOs decreased to 3% at E18.5 and 0% at birth, indicating that homozygous KOs die during late gestation. This lethality is likely due to the loss of Nedd4-1 from essential non-neuronal organs. Indeed, starting at E11.5, KO embryos showed a retarded development (Figure S2F), and almost 50% of KOs had subcutaneous bleedings (Figure S2F), indicating an involvement of Nedd4-1 in vasculogenesis or angiogenesis. We did not observe any signs of apoptosis in Nedd4-1-KOs (data not shown). Among the reported conventional KO mice lacking ubiquitously expressed E3 ubiquitin ligases (Bloom et al., 2007; Jiang et al., 1998; Lotz et al., 2004; Montes de Oca Luna et al., 1995; Yamashita et al., 2005; Yao et al., 2007), the Nedd4-1-KO exhibits one of the most severe mutant phenotypes, indicating that Nedd4-1 is a critical E3 ligase in cellular function (Cao et al., 2008; Fouladkou et al., 2008).
Impaired Dendritogenesis in Nedd4-1-KO Neurons
To study the function of Nedd4-1 during nerve cell development, we analyzed the morphology of individual cortical neurons from Nedd4-1-KO and WT brains in autaptic cultures (Bekkers and Stevens, 1991) by plating and culturing identical numbers of KO and WT cells (6,000 cells per 35 mm2) on astrocyte islands. The numbers of surviving neurons were not significantly different between the genotypes at day 10 in vitro (DIV10) (Kolmogorov-Smirnov test; p=0.944) (Table S1), indicating that Nedd4-1 KO does not affect neuron survival. However, KO neurons were smaller than WT cells.
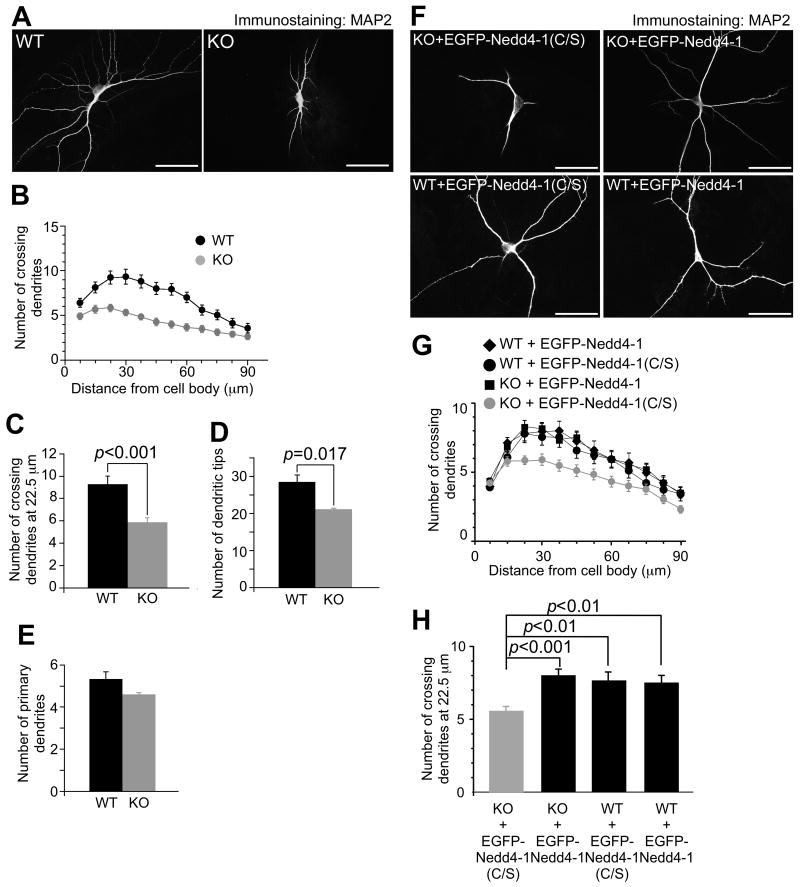
To examine if the smaller size of KO neurons is due to impaired dendrite development, we fixed neurons at DIV10 and immunostained them for MAP2. KO neurons were characterized by more immature dendrites as compared to WT cells (Figure 1A). We quantified this effect by Sholl analysis, where dendrites crossing concentric circles drawn at 7.5 μm intervals around neurons are counted (Sholl, 1953). WT DIV10 neurons showed the highest number of crossings at a 30 μm distance from the cell body (9.36±0.86, n=25) while KO neurons showed a peak at 22.5 μm and overall less crossing dendrites (5.87±0.41, n=38) (Figure 1B). Comparison of numbers of crossing dendrites at a 22.5 μm distance from cell bodies (p<0.001) and numbers of dendritic tips, representing the number of all distal dendrites and dendrite complexity (p=0.017) (Jaworski et al., 2005), yielded significantly lower values for KO neurons (Figures 1C and 1D, and Table S1). However, neither the numbers of primary dendrites emerging from cell bodies at DIV10 (p=0.138) (Figure 1E and Table S1) nor neurite lengths, axon numbers, or total neurite numbers at DIV3 (Figure S3 and Table S1; p>0.05) were different between WT and KO neurons. These results show that Nedd4-1 is dispensable during the acquisition of neuronal cell polarity and initial neuritogenesis but required for subsequent dendrite extension and arborization.
Figure 1. Impaired Dendrite Development in Nedd4-1-KO Neurons.
(A) KO neurons show impaired development of dendrites. Autaptic neurons prepared from cortices of WT and KO embryos were fixed at DIV10 and dendrites were immunostained using an anti-MAP2 antibody. Scale bars, 50 μm.
(B) Sholl analysis of WT and KO neurons. Black circles, WT neurons (n=25); gray circles, KO neurons (n=38).
(C) Numbers of crossing dendrites at 22.5 μm from the cell body in WT and KO neurons.
(D) Numbers of dendritic tips in WT and KO neurons.
(E) Numbers of primary dendrites emerging from WT and KO neurons. Black bars, WT neurons (n=25 for C, n=26 for D and E); gray bars, KO neurons (n=38 for C, n=39 for D and E). Student's t-test revealed significant differences in (C) (p<0.001) and (D) (p=0.017), but not in (E) (p=0.17).
(F) Anti-MAP2 immunostaining of autaptic KO (top two panels) and WT (bottom two panels) neurons overexpressing EGFP fused to the catalytically inactive mutant of Nedd4-1 (C/S) as a negative control (left two panels) or full length EGFP-Nedd4-1 (right two panels). Scale bars, 50 μm.
(G) Sholl analysis of autaptic WT or KO neurons overexpressing EGFP-Nedd4-1 (C/S) or EGFP-Nedd4-1. Black diamonds, WT neurons overexpressing EGFP-Nedd4-1 (n=42); black circles, WT neurons overexpressing EGFP-Nedd4-1 (C/S) (n=40); black rectangles, KO neurons overexpressing EGFP-Nedd4-1 (n=60); gray circles, KO neurons overexpressing EGFP-Nedd4-1(C/S) (n=68).
(H) Numbers of crossing dendrites at 22.5 μm from the cell body in WT and KO neurons expressing EGFP-Nedd4-1(C/S) or EGFP-Nedd4-1. nonparametric ANOVA test revealed a significant effect of interaction between the genotype and expression of EGFP-Nedd4-1s (p=0.0001). Dunn's multiple comparisons test revealed significant differences between KO neurons expressing EGFP-Nedd4-1(C/S) and EGFP-Nedd4-1 (p<0.001) and between EGFP-Nedd4-1(C/S) expressing KO and WT neurons (p<0.01) but not between WT neurons expressing EGFP-Nedd4-1(C/S) and EGFP-Nedd4-1 (p >0.05).
See also Figures S1-S4, and Tables S1 and S2.
To test if the dendrite arborization defect in KO neurons is a specific consequence of Nedd4-1 loss, we tried to rescue this phenotype by reexpressing EGFP-tagged Nedd4-1 in KO cells (Figures 1F-1H). We infected WT and KO neurons 24 h after plating on astrocyte microislands with lentiviruses expressing either EGFP-Nedd4-1 or, as a negative control, EGFP fused to the catalytically inactive point mutant of Nedd4-1 [EGFP-Nedd4-1(C/S)], and fixed cells at DIV10, when EGFP signals were clearly detectable. Upon expression of EGFP-Nedd4-1(C/S) in KO neurons, the dendrite complexity was similar to that of non-infected KO cells (compare Figures 1A and 1F). In contrast, reexpression of EGFP-Nedd4-1 in KO neurons caused a significant increase in the complexity and length of dendrites, which were similar to those in WT cells expressing the negative control construct EGFP-Nedd4-1(C/S) (Figures 1F-1H. Table S2; p<0.001). Sholl analysis revealed that the reintroduction of recombinant Nedd4-1 into KO cells restored the complexity of dendrites (Figure 1G). Overexpression of recombinant Nedd4-1 in WT neurons showed little effect on dendrite complexity (Figures 1F-1H and Table S2; p>0.05), most likely because endogenous Nedd4-1 expression in WT cells is sufficiently high to reach saturation. Corresponding results were obtained when we used EGFP fused to the N-terminus of Nedd4-1 [EGFP-Nedd4-1(1-268)] as a negative control (Table S2), which itself had no catalytic activity or dominant effects (Figure S5A). These results show that the dendrite morphology defect in KO neurons is a specific consequence of Nedd4-1 loss and that the enzymatic activity of Nedd4-1 is crucial for regulation of dendrite development.
Nedd4-1 is a Regulator of Dendrite Arborization In Vivo
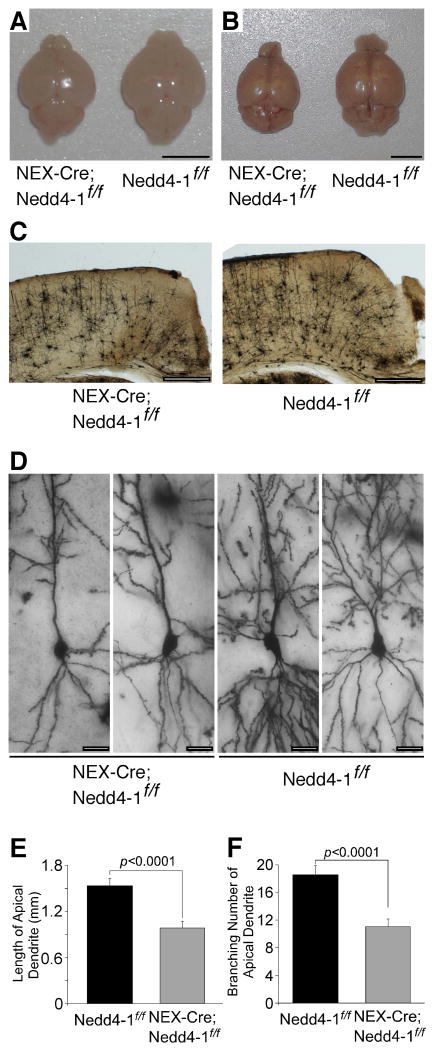
To study the function of Nedd4-1 in vivo, we generated a floxed Nedd4-1 mutant mouse line (Figure S2B). Overexpression of Cre in primary neurons prepared from Nedd4-1flox/flox mice eliminated Nedd4-1 protein expression (Figure S2E). The Nedd4-1flox/flox line was crossed with a NEX-Cre mouse line expressing Cre recombinase in postmitotic cerebral and hippocampal glutamatergic neurons (Goebbels et al., 2006). NEX-Cre;Nedd4-1flox/flox mice were viable and fertile but had a smaller cerebrum than controls, while the size of the cerebellum, where Cre recombinase is expressed only in a minor fraction of neurons, was normal (Figures 2A and 2B). Brains were isolated from NEX-Cre;Nedd4-1flox/flox mice at two to eight months after birth and Golgi stained in order to study dendrite complexity. The thickness of the cerebral cortical temporal lobe was reduced in NEX-Cre;Nedd4-1flox/flox mice (Figure 2C). Upon examination of pyramidal cells in stratum radiatum and stratum lacunosum moleculare of the CA1 region of hippocampus, we found that the extent and branching of apical dendrites were significantly reduced in NEX-Cre;Nedd4-1flox/flox mice (p<0.0001) (Figures 2D-2F, and Table S1). These results show that Nedd4-1 is a critical regulator of dendrite arborization in vivo and determines the neurite arborization of mature neurons.
Figure 2. Effect of Nedd4-1 Loss on Dendrite Development In Vivo.
(A and B) Impaired development of cerebra of NEX-Cre;Nedd4-1flox/flox mice (left) as compared to control Nedd4-1flox/flox mice (right) at immature (P5; A) and mature (8 months; B) stages. Note that the sizes of cerebella are indistinguishable between NEX-Cre;Nedd4-1flox/flox and Nedd4-1flox/flox mice. Scale bars, 5 mm.
(C) Overviews of Golgi stained samples of NEX-Cre;Nedd4-1flox/flox and Nedd4-1flox/flox cerebra. Sections were from two months old littermates. Scale bars, 0.5 mm.
(D) Hippocampal CA1 neurons in NEX-Cre;Nedd4-1flox/flox (left two panels) and Nedd4-1flox/flox (right two panels) mice. Tops of each picture are the apical side. Pictures were taken from three months old littermates. Scale bars, 20 μm.
(E and F) Total lengths (E) and branching numbers (F) of apical dendrites of hippocampal CA1 neurons in NEX-Cre;Nedd4-1flox/flox and Nedd4-1flox/flox mice. Similar numbers of neurons were used for each genotype at the age of two months (NEX-Cre;Nedd4-1flox/flox, n=3; Nedd4-1flox/flox, n=3), three months (NEX-Cre;Nedd4-1flox/flox, n=16; Nedd4-1flox/flox, n=13), and eight months (NEX-Cre;Nedd4-1flox/flox, n=9; Nedd4-1flox/flox, n=9). Student's t-test revealed significant differences in both (E) and (F) (n>24, p<0.0001).
See also Figures S1, S2, and Table S1.
Reduced Synaptic Transmission and Synapse Numbers in Nedd4-1-KO Neurons
To determine the effects of Nedd4-1 KO on synaptic transmission, we studied glutamatergic autaptic cortical neurons from WT, heterozygous KO, and homozygous KO littermate embryos using electrophysiological methods. Because data obtained from WT and heterozygous KO neurons were indistinguishable, we pooled them and used them as the WT control reference.
We first recorded resting membrane potentials and input membrane resistances, which did not differ between KO and WT cells (Figures S3E and S3F, and Table S3; p>0.1). Next, we quantified the delay between stimulus-onset and glutamate-mediated evoked excitatory postsynaptic currents (EPSC) (Stimulus-Peak time), which was almost identical in WT and KO cells (Table S3; Student's t-test, p>0.1). Paired-pulse response characteristics, which we assessed using two action potentials at a 100 ms interval, were not significantly different between neurons of the two groups (WT, 0.96±0.030, n=91; KO, 0.89±0.024, n=68; Student's t-test, p>0.05). These results indicate that neuronal viability and presynaptic release probability are not affected by Nedd4-1 KO.
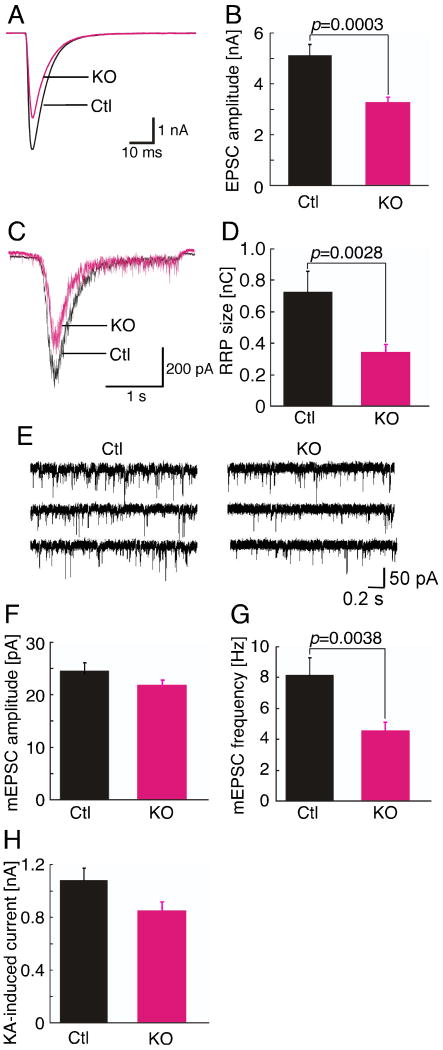
EPSC amplitudes in KO neurons were reduced to about 62% of control values (Figures 3A and 3B, and Table S3; p<0.001). This was paralleled by a similar reduction in the size of the readily releasable vesicle pool in KO neurons (Figures 3C and 3D, and Table S3; p<0.005), which we determined by measuring transmitter release in response to hypertonic sucrose buffer (Rosenmund and Stevens, 1996).
Figure 3. Reduced Synaptic Transmission in Nedd4-1-KO Neurons.
(A) Sample traces of evoked EPSCs from control (Ctl) and KO neurons.
(B) Averaged evoked EPSC amplitudes of control (Ctl) (n=70) and KO (n=82) neurons. Student's t-test, p=0.0003.
(C) Sample traces of postsynaptic currents evoked by the application of hypertonic sucrose solution from control (Ctl) and KO neurons.
(D) Mean readily releasable vesicle pool sizes as estimated by the charge integral measured after release induced by application of 0.5 M sucrose solution in control (Ctl) (n=58) and KO (n=61) neurons. Student's t-test, p=0.0028.
(E) Sample traces of miniature EPSCs from control (Ctl) and KO neurons.
(F) Averaged miniature EPSC amplitudes (Ctl, n=32; KO, n=40). Student's t-test, p>0.1.
(G) Averaged miniature EPSC frequencies (Ctl, n=32; KO, n=40). Student's t-test, p=0.0038.
(H) Averaged kainic acid-induced currents in control (Ctl, n=29) and KO neurons (n=32). Student's t-test, p>0.1.
See also Figures S3 and S4 and Table S3.
Such phenotypic changes in synaptic transmission can be caused by a reduction of the number of synapses formed by a given mutant neuron, which would be expected based on our morphological analysis (Figure 1), or by functional defects at the level of individual synapses. To distinguish these possibilities, we analyzed miniature EPSCs (mEPSC) originating from spontaneous single synaptic vesicle fusion events and postsynaptic detection of released transmitter. mEPSC amplitudes were not different between control and KO neurons, while the frequency of mEPSCs in KO neurons was reduced to 55% of control values (Figures 3E-3G, and Table S3; amplitudes, p>0.05; frequencies, p<0.005). The fact that mEPSC amplitudes were similar in control and KO neurons indicates that the numbers and density of postsynaptic glutamate receptors at existing synapses are normal in KO cells. That the cell surface density of glutamate receptors is indeed unaltered in KO cells was verified by experiments, in which we applied kainic acid to cultured neurons in order to stimulate all cell surface glutamate receptors. Kainic acid-induced currents did not differ significantly between control and KO neurons (Figure 3H and Table S3; p>0.05). The tendency towards lower kainic acid-induced currents in KO neurons is likely due to the reduced cell surface area, which is caused by the impaired development of dendrites.
The fact that AMPA receptor densities at existing synapses are unaltered in KO neurons indicates that AMPA receptor exo- and endocytosis, which can in principle be modulated by ubiquitination (Guo and Wang, 2007), are normal upon Nedd4-1 KO. Consequently, the reduced evoked EPSC amplitudes and mEPSC frequencies in KO neurons are most likely due to a reduction in the numbers of functionally normal synapses, as is expected based on the dendritogenesis defect in KO cells (Figure 1).
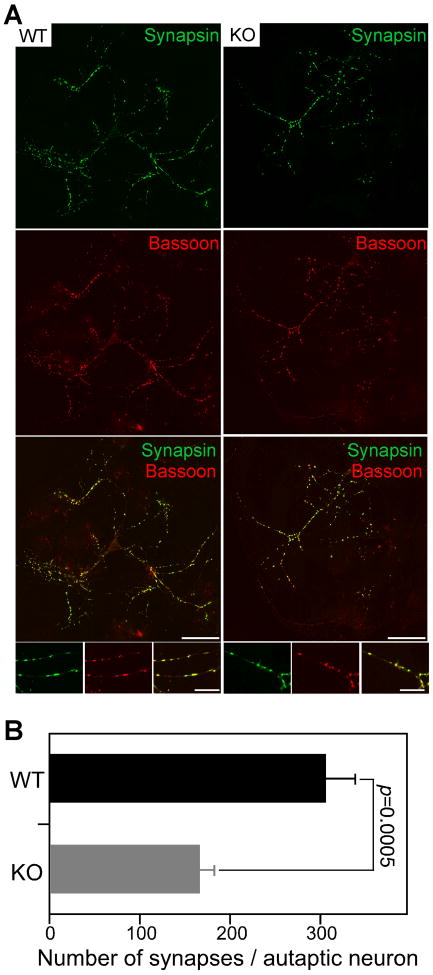
To test this, we measured synapse numbers in autaptic WT and KO neurons at DIV10 after immunostaining for the synapse markers Synapsin and Bassoon (Figure 4). The fluorescence intensities of synapses stained for Synapsin did not differ between KO and WT cells (Figure 4A). However, the total number of Synapsin-positive presynaptic puncta was reduced in KO cells. Quantification of Synapsin and Bassoon double positive synaptic puncta showed that KO neurons had 47% less synapses than WT controls (Figure 4B and Table S3; p<0.001), which is in accord with the reduced evoked EPSC amplitudes and mEPSC frequencies in KO cells (Figures 3A-3G, and Table S3). Reexpression of recombinant EGFP-Nedd4-1 restored synapse numbers and function in autaptic KO neurons (Figure S4 and Table S2). Importantly, the synapse density per denrite length (Figures S3G and S3H and Table S3), presynaptic maturation of synapses (Table S3), and spatial matching of pre- and postsynapses (Figures S3I and S3J) were normal in KO neurons.
Figure 4. Reduced Synapse Numbers in Nedd4-1-KO Neurons.
(A) Immunocytochemical analysis of synapses in autaptic WT and KO neurons using anti-Synapsin and anti-Bassoon antibodies. Cortical neurons were prepared from embryos at E13 and fixed at DIV10. Note the smaller size of the KO neuron. Scale bars, 50 μm (low magnification) and 20 μm (high magnification insets).
(B) Quantification of numbers of Synapsin and Bassoon double positive synaptic puncta in individual autaptic KO (n=37) and WT neurons (n=42). Student's t-test, p=0.0005.
See also Figures S3 and S4 and Table S3.
Taken together, these results show that Nedd4-1 loss causes impaired dendrite development, which in turn leads to a reduction in the number of otherwise normal synapses. However, we cannot exclude that subtle synaptic defects in KO cells went undetected in our analyses.
Rap2 is a Nedd4-1 Substrate
Multiple mammalian proteins were identified as Nedd4-1 substrates, including VEGF-R2, CNrasGEF, ENaC, and PTEN (Murdaca et al., 2004; Pham and Rotin, 2001; Staub et al., 1996; Wang et al., 2007), but the role of their ubiquitination in neurons has not been analyzed. Among these substrates, PTEN could in principle be relevant for the present findings (Jaworski et al., 2005; Kwon et al., 2006), but we found earlier that it is not a Nedd4-1 target in vivo (Fouladkou et al., 2008).
To identify Nedd4-1 targets responsible for the KO phenotype, we performed affinity chromatography using the Nedd4-1 WW domains as affinity matrix and a Triton X-100 rat brain synaptosome extract as the source of Nedd4-1 targets. We identified fifteen proteins from twelve protein bands that bound to the Nedd4-1 affinity column (Figure 5A and Table S4). Among these, our interest was attracted by TNIK. TNIK was originally identified as a Traf2- and Nck-Interacting Kinase (Fu et al., 1999), is part of a protein family that also contains NIK (Nck-Interacting Kinase) and MINK (Misshapen/NIKs-related Kinase)(Figure S5B)(Dan et al., 2000), and was shown to regulate actin dynamics as an effector of the small GTPase Rap2 (Taira et al., 2004). Given that TNIK binds to the WW domains of Nedd4-1 and regulates actin dynamics in conjunction with Rap2, we focused our analysis on the interaction between Nedd4-1, TNIK, and Rap2, whose perturbation might be the cause for the dendritogenesis defects seen in Nedd4-1-KO neurons.
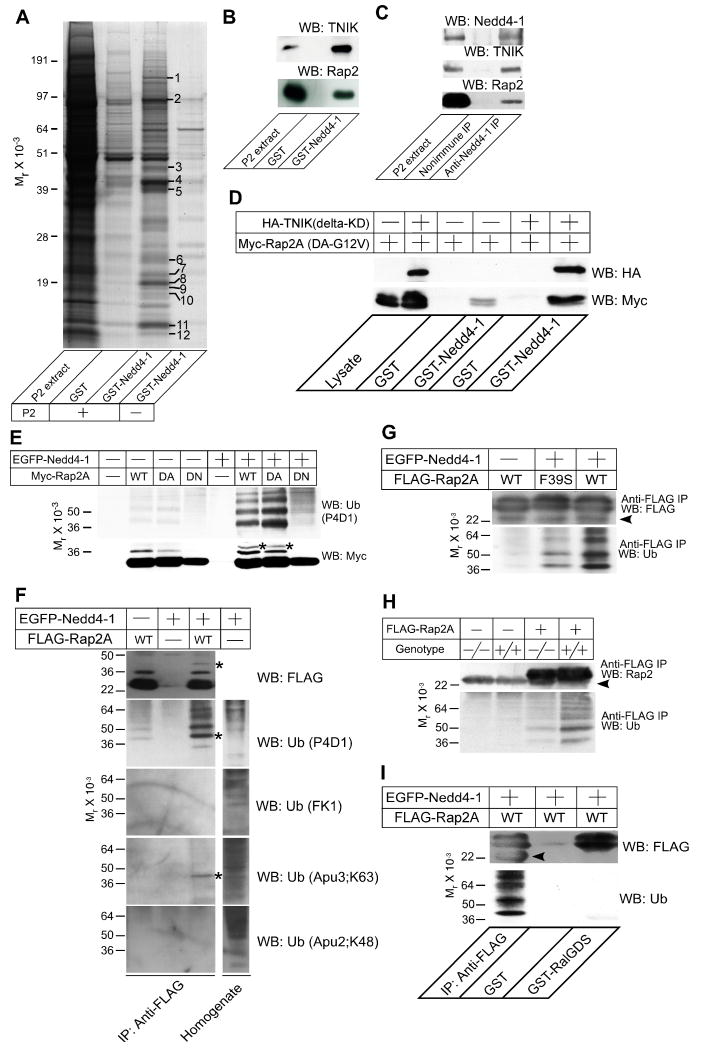
Figure 5. Rap2 is a Target of Nedd4-1.
(A) Affinity purification of TNIK as a binding partner of Nedd4-1. 40 μg of GST or GST Nedd4-1 (residues 217-549) were immobilized on glutathione Sepharose beads and a Triton X-100 extract of rat brain synaptosomes (P2 extract, +) or buffer (-) were applied. After washing the beads, bound proteins were eluted with 1 M NaCl. Protein bands that appeared to be enriched in the eluate from the GST Nedd4-1 column were analyzed by mass spectrometry. Protein identification was successful for the twelve marked bands. The results of the mass spectrometric analysis are given in Table S4.
(B) Complex formation of Rap2 with TNIK and GST-Nedd4-1. Samples eluted from the Nedd4-1 beads (see A) were blotted for TNIK (top) and Rap2 (bottom). Neither antibody cross-reacted with samples purified on GST beads that were used as a negative control.
(C) Complex formation of endogenous Rap2, TNIK, and Nedd4-1. Nedd4-1 was immunoprecipitated using a rabbit polyclonal anti-Nedd4-1 antibody from mouse brain membranes after treatment with a thiol-cleavable chemical crosslinker. Precipitates were boiled in Laemmli buffer with 50 mM DTT, loaded to SDS-PAGE gels, and analyzed by Western blotting using anti-Nedd4-1, anti-TNIK, or anti-Rap2 antibodies. Note that TNIK and Rap2 were coimmunoprecipitated with Nedd4-1 but not with the negative control IgG.
(D) TNIK dependent interaction of Rap2A with Nedd4-1. GST or GST-Nedd4-1 (residues 217-549) were immobilized on glutathione Sepharose beads and recombinant Myc-Rap2A(DA-G12V) was loaded in the presence (+) or absence (-) of HA-TNIK (delta-KD). Interaction was detected by Western blotting using anti-HA or anti-Myc antibodies.
(E) Ubiquitination of Rap2A by Nedd4-1. EGFP-Nedd4-1 (+) or EGFP alone (-) were coexpressed with WT, dominant active (DA), or dominant negative (DN) mutants of Myc-tagged Rap2A. Myc-Rap2A was immunoprecipitated using an anti-Myc antibody. Immunoprecipitates were blotted for Myc (lower panel) and ubiquitin (upper panel).
(F) Mono- and di-ubiquitination of Rap2A by Nedd4-1. FLAG-Rap2A was immunoprecipitated from HEK cells expressing FLAG-Rap2A with or without EGFP-Nedd4-1. FLAG-Rap2A was eluted from anti-FLAG antibody coupled beads with 3×FLAG peptides and immunoblotted using four different monoclonal mouse or rabbit anti-ubiquitin antibodies, P4D1, FK1, Apu3, and Apu2. P4D1 recognizes both poly- and monoubiquitin conjugated proteins while FK1 recognizes only polyubiquitin conjugated ones. Apu3 and Apu2 recognize K63-linked and K48-linked polyubiquitin chains, respectively. The lysate of HEK cells overexpressing EGFP-Nedd4-1 was also blotted using the four anti-ubiquitin antibodies in order to show that the antibody titers are comparable.
(G) Loss of TNIK interaction in the F39S point mutant of Rap2 reduces the ubiqutination of Rap2 by Nedd4-1. WT or F39S point mutant FLAG-Rap2A were overexpressed in HEK cells together with EGFP (-) or EGFP-Nedd4-1 (+). Efficiencies of immunoprecipitation of FLAG-Rap2A were comparable as seen in the Western blot using the anti-FLAG antibody (upper panel). FLAG-Rap2A(F39S) showed clearly weaker intensities of ladder patterns than FLAG-Rap2A(WT) in the blot using the anti-ubiquitin antibody (lower panel).
(H) Ubiquitination of Rap2A by endogenous Nedd4-1 and ubiquitin. WT (+/+) and Nedd4-1-KO (-/-) MEFs were transfected with a mammalian expression vector encoding FLAG-Rap2A. Proteins from lysed cells were immunoprecipiated with an anti-FLAG antibody and blotted for Rap2A with an anti-Rap2 antibody (upper panel) and an anti-ubiquitin antibody (lower panel).
(I) Ubiquitination of Rap2A blocks the Rap2A function. FLAG-Rap2A(WT) expressed in HEK cells together with EGFP-Nedd4-1 was precipitated using anti-FLAG antibodies (first lane) or GST-RalGDS-coupled beads (third lane), and blotted for FLAG (upper panel) or ubiquitin (lower panel). Note that ubiquitinated FLAG-Rap2A was efficiently precipitated only with the anti-FLAG antibody while total amounts of precipitated FLAG-Rap2A were comparable between lanes 1 and 3.
Arrowheads in (G-I) indicate the light chain of the anti-FLAG IgG used for immunoprecipitation. All results shown are representative of two to three independent experiments. Asterisks in (E) and (F) indicate bands with a molecular weight corresponding to Myc-Rap2A or FLAG-Rap2A conjugated with two ubiquitin moieties.
See also Figures S1, S5, and S6, and Table S4.
Western blotting of eluates from GST-Nedd4-1 beads using anti-TNIK and anti-Rap2 antibodies showed that TNIK and Rap2 are present in the affinity purified material, indicating that Nedd4-1, TNIK, and Rap2 may form a complex (Figure 5B). To detect complex formation between Nedd4-1, TNIK, and Rap2, a mouse brain membrane fraction was incubated with a thiol-cleavable membrane-permeable cross-linker before immunoprecipitation with an anti-Nedd4-1 antibody. Under these conditions, but not without crosslinking (not shown), TNIK and Rap2 were coimmunoprecipitated with Nedd4-1 (Figure 5C). In addition, Rap2A and TNIK are enriched and colocalized with Nedd4-1 in the perinuclear region of neurons, and the three proteins partially cosegregate into ER and Golgi enriched fractions upon subcellular fractionation of brain tissue (Figures S1E and S1F). These data show that Nedd4-1, TNIK, and Rap2 partially colocalize in neurons and form a ternary complex.
We mapped the Nedd4-1-binding region of TNIK to the central part (Figures S5C and S5D), which is only moderately conserved in MINK (53% identity) and NIK (57% identity), while the kinase and citron-homology (CNH) domains are very similar in all family members (90-92% identity). The isoform specificity of Nedd4-1 interactions with TNIK, MINK, and NIK was studied by affinity pull-down assays (Figure S5B). We found that the affinity of NIK to GST-Nedd4-1 was as strong as that of TNIK, while MINK binding to GST-Nedd4-1 was barely detectable.
Considering that Rap2 binds the C-terminus of TNIK and Nedd4-1 the central linker domain, TNIK might mediate an indirect interaction between Nedd4-1 and Rap2 (Figure S5E). To test this, lysates of HEK293FT (HEK) cells expressing Myc-tagged dominant active mutant Rap2A (DA-G12V) alone or Myc-tagged Rap2A (DA-G12V) together with the HA-tagged kinase domain deletion mutant (delta KD) of TNIK, which contains the Rap2- and Nedd4-1-binding regions, were loaded onto GST-Nedd4-1-coupled beads (Figure 5D). We found that the interaction of Myc-Rap2A (DA-G12V) with GST-Nedd4-1 was dependent on HA-TNIK (delta KD) (lanes 4 and 6 in Figure 5D) while HA-TNIK (delta KD) interacted with GST-Nedd4-1 in a Myc-Rap2A (G12V)-independent manner (Figure S5D). The slight interaction between Myc-Rap2 (DA-G12V) and GST-Nedd4-1 in the absence of HA-TNIK (delta KD) is likely mediated by endogenous TNIK in HEK cells (lane 4 in Figure 5D).
Next, we tested if the interaction with Nedd4-1 leads to ubiquitination of TNIK or Rap2. Expression vectors for EGFP-tagged full-length Nedd4-1, His6-tagged ubiquitin, and Myc-tagged WT Rap2A, DA-G12V Rap2A, or the dominant negative mutant (DN-S17N) Rap2A were cotransfected in HEK cells, and Myc-tagged proteins were immunoprecipiated from cell extracts using anti-Myc antibodies. Ubiquitination of immunoprecipitated protein was detected by immunoblotting with an anti-ubiquitin antibody. Immunoprecipitates from Rap2A (WT) and Rap2A (DA-G12V), but not Rap2A (DN-S17N) expressing HEK cells showed a ladder of protein bands that were detected by an anti-ubiquitin antibody only in the presence of the overexpressed EGFP-tagged WT Nedd4-1 (Figure 5E). These ubiquitin-reactive bands were preserved when Myc-Rap2A was extracted under denaturing conditions (100°C, 1% SDS), indicating that Rap2A itself, and not associated proteins, is ubiquitinated by Nedd4-1 (Figure S5F). The ubiquitination of Rap2A was abolished when catalytically inactive instead of WT Nedd4-1 was used (Figure S5G). Unexpectedly, we did not detect any ubiquitination of TNIK (data not shown).
Recently, di-ubiquitination of HRas and polyubiquitination of Rap1B were reported (Jura et al., 2006; Schwamborn et al., 2007). Mutation of eight lysines of HRas abolishes di-ubiquitination, indicating that at least some of these lysines are di-ubiquitination targets. Three of these eight lysines are conserved in Rap2A (Figure S5H), which indicates that Rap2A might be di-ubiquitinated by Nedd4-1. However, a single lysine (K5) of Rap1B, which is also conserved in Rap2A (Figure S5H), is polyubiquitinated by the HECT type E3 ligase Smurf2. To investigate if Rap2A is mono-, di-, or polyubiquitinated, we immunoblotted ubiquitinated Rap2A (WT) using four anti-ubiquitin monoclonal antibodies, which allow to discriminate between mono- and polyubiquitinated proteins: P4D1 detects poly- and monoubiquitin conjugates, FK1 detects only polyubiquitin conjugates (Arevalo et al., 2006; Fujimuro et al., 1994), and Apu2 and Apu3 detect K48- and K63-linked polyubiquitin chains, respectively (Newton et al., 2008). The protein ladder representing ubiquitinated Rap2A was detected only by P4D1 but not by FK1, indicating monoubiquitination of Rap2A by EGFP-Nedd4-1 (Figure 5F). Apu3 detected a single faint band with a molecular weight equivalent to Rap2A conjugated with two ubiquitins, indicating that a minor fraction of Rap2A is modified by Nedd4-1 via K63-linked di-ubiquitination. Apu2 did not detect any specific bands in these experiments, indicating that Rap2A ubiquitination by Nedd4-1 is not K48-linked, which would cause proteasome dependent degradation. Indeed, the appearance of ubiquitinated Rap2A as a ladder of protein bands indicates that multiple lysine residues are mono- or di-ubiquitinated. To confirm the fidelity of the monoclonal anti-ubiquitin antibodies used, a total lysate of HEK cells expressing EGFP-tagged Nedd4-1 was immunoblotted using these antibodies, which all detected a complex pattern of blurred bands that likely represent multiple polyubiquitinated proteins (Figure 5F). Western blotting with FK1, Apu3, and Apu2 yielded more complex patterns than P4D1, indicating that the titers of these antibodies are higher than that of P4D1. With FK1, Apu3, or Apu2, we did not detect any specific Nedd4-1 dependent bands, not even after long exposures, except for di-ubiquitinated Rap2 crossreacting with the Apu3 antibody (data not shown). To test if Nedd4-1 dependent ubiquitination is specific for Rap2A, we examined FLAG-tagged Rap1A, Rap2A, HRas, and KRas in the HEK cell ubiquitination assay. We found that only Rap2A was prominently ubiquitinated (Figure S5I), indicating that Rap2A is a specific Nedd4-1substrate.
We next analyzed if the ternary complex formation of Nedd4-1, TNIK, and Rap2 is critical for Rap2 ubiquitination. To block the interaction of endogenous TNIK with Rap2 in our ubiqutination assays, we used an F39S point mutant of Rap2A whose affinity to TNIK family proteins is specifically reduced while the interaction with another target, Raf, is unaffected (Taira et al., 2004). FLAG-tagged Rap2A or Rap2A (F39S) were transfected into HEK cells together with His6-ubiquitin and EGFP-Nedd4-1. FLAG-Rap2A was immunoprecipitated using an anti-FLAG antibody and ubiquitination was detected by an anti-ubiquitin polyclonal antibody. Ubiquitination of FLAG-Rap2A (F39S) was reduced compared to FLAG-Rap2A (WT) (Figure 5G), indicating that complex formation of Rap2, TNIK, and Nedd4-1 is critical for proper Rap2 ubiquitination. This result and the specific interaction of Rap2 with TNIK family proteins also explain the specific ubiquitination of Rap2 by Nedd4-1 as compared to other Ras superfamily GTPases (Figure S5I).
To test if endogenous Nedd4-1 can ubiquitinate Rap2A with endogenous ubiquitin, we used WT and Nedd4-1-KO embryonic fibroblasts (MEFs). Because no anti-Rap2 antibodies suitable for immunoprecipitation are available, we expressed FLAG-tagged Rap2A in WT and KO MEFs, and immunoprecipitated FLAG-Rap2A using anti-FLAG antibodies (Figure 5H). Western blotting with an anti-ubiquitin antibody revealed a 70% reduction of ubiquitination of FLAG-Rap2A in KO MEFs as compared to WT cells, indicating that Rap2A is a physiological Nedd4-1 substrate.
To study the effect of ubiquitination on Rap2 function, we purified and immobilized a GST fusion of the Rap2-binding region of RalGDS and used this to pull down active GTP-bound Rap2 (Herrmann et al., 1996). FLAG-tagged Rap2A and EGFP-Nedd4-1 were then cotransfected to HEK cells and the cell lysate was loaded onto GST-RalGDS or onto anti-FLAG antibody coupled beads. The anti-FLAG antibody efficiently immunoprecipitated ubiquitinated and non-ubiquitinated FLAG-Rap2A, while GST-RalGDS coupled beads pulled down only non-ubiquitinated FLAG-Rap2A (Figure 5I), indicating that ubiquitination blocks Rap2 function by perturbing Rap2 interaction with effector proteins.
We next tested if Rap2 function is upregulated by Nedd4-1 KO using WT and KO MEFs. For this purpose, we performed FRET analyses using Raichu-Rap2A (DA-G12V) as a probe (i.e. a fusion protein composed of Venus as a FRET acceptor, Rap2A (DA-G12V), the Rap2A-binding region of RalGDS, SECFP as a FRET donor, and the consensus motif for lipid modification for Ras family proteins) (Mochizuki et al., 2001). DA-G12V Rap2A instead of WT Rap2A was used to discriminate between effects of Nedd4-1 loss on the GTPase activity of Rap2A from effects on target protein recognition. We examined the activities of Rap2A at the plasma membrane of MEFs because it is known that Ras family small GTPases are activated at the plasma membrane. Figures S6A and S6B show an enhanced FRET signal from the KO cells compared to WT cells (p<0.001). Interestingly, the expression level of endogenous Rap2 was downregulated in KO MEFs, which indicates that overactivation of Rap2 in KO cells due to reduced Nedd4-1 mediated ubiquitination causes a compensatory reduction of Rap2 expression (Figure S5J). In summary, the RalGDS pull-down and FRET data further support the conclusion that Rap2A is a physiological target of Nedd4-1 and that Nedd4-1 mediated ubiquitination perturbs Rap2A function.
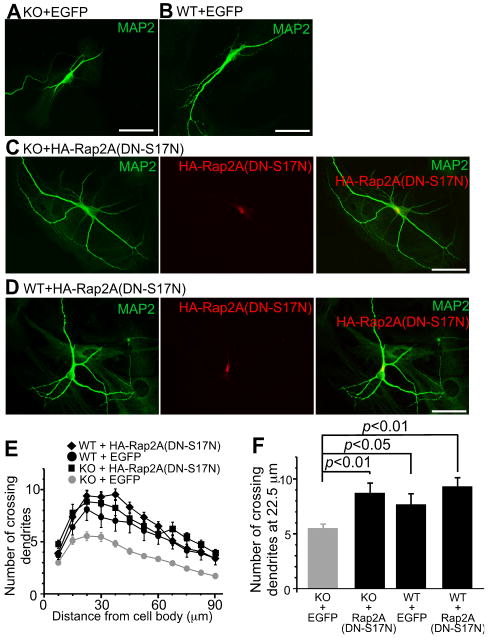
Dominant Negative Rap2A or TNIK Mutants Rescue the Dendritogenesis Defect in Nedd4-1-KO Neurons
Rap1 and Rap2 share 60% identity at the amino acid level and form a Ras-like GTPase subfamily implicated in axon and dendrite extension. During initial development in culture (DIV2-DIV3), polarized neurons extend a single axon. The tip of the axonal growth cone contains endogenous Rap1, which in turn recruits Cdc42 (Schwamborn and Puschel, 2004). This polarized localization of Rap1 is regulated by Smurf2, which polyubiquitinates GDP-bound Rap1 and accelerates its degradation at non-polarized neurites. The role of Rap GTPase subfamily members in dendrites appears to be different from that in axons. When DN mutant Rap1 or Rap2 are overexpressed in DIV14-DIV15 neurons, they promote neurite extension while DA mutant Rap2 reduces it, indicating that Rap1 and Rap2 interfere negatively with neurite growth and arborization (Fu et al., 2007).
So far, three Rap2 proteins (Rap2A/B/C) have been cloned. They are encoded by independent genes, which makes it difficult to analyze their shared functions by gene targeting or RNAi. To investigate if ubiquitination and regulation of Rap2 by Nedd4-1 modulate dendritogenesis and if the perturbation of these processes contribute to the neuronal Nedd4-1-KO phenotype, HA-tagged DN-S17N Rap2A or EGFP, as a negative control, were overexpressed in autaptic Nedd4-1-KO neurons (Figure 6) using neuron-specific recombinant adenovirus vectors. We pursued this approach because we assumed that DN Rap2A would ultimately interfere with the cellular function of all known Rap2 variants. The adenovirus was applied at DIV1 and expression of proteins was first detectable at DIV3-4. Cells were fixed and analyzed at DIV10. Overexpression of HA-Rap2A(DN-S17N) resulted in increased length, number, and complexity of dendrites in infected Nedd4-1-KO neurons (Figure 6C). Sholl analysis revealed an almost perfect cross-rescue of the dendritogenesis defect of KO cells after overexpression of HA-Rap2A(DN-S17N) (Figure 6E). Statistical analysis of the dendrite complexity (Figures 6E and 6F, Table S2) showed that the effect of HA-Rap2A(DN-S17N) was stronger in KO than in WT cells (p<0.01).
Figure 6. Overexpression of a Dominant Negative Mutant of Rap2A Rescues the Dendritogenesis Defect in Nedd4-1-KO Neurons.
(A-D) Anti-MAP2 immunostaining of KO or WT neurons overexpressing EGFP (A and B), and anti-MAP2 and anti-HA immunostaining of KO or WT neurons overexpressing dominant negative HA-Rap2A(DN-S17N) (C and D). HA-Rap2A(DN-S17N) overexpressing neurons showed longer and more complex dendrites. This effect was more evident in KO neurons. Scale bars, 50 μm.
(E) Sholl analysis of KO neurons overexpressing EGFP (gray circles, n=68) or HA-Rap2ADN (black rectangles, n=22), and of WT neurons overexpressing EGFP (black circles, n=20) or HA-Rap2ADN (black diamonds, n=12).
(F) Numbers of crossing dendrites at 22.5 μm from the cell body in WT and KO neurons overexpressing EGFP or HA-Rap2ADN. Nonparametric ANOVA test revealed a significant effect of interaction between the genotype and expression of HA-Rap2A(DN-S17N) (p<0.0001). Dunn's multiple comparisons test revealed significant differences between KO neurons expressing EGFP and HA-Rap2A(DN-S17N) (p<0.01), and between KO and WT neurons expressing EGFP (p<0.05), but not between WT neurons expressing EGFP and HA-Rap2A(DN-S17N) (p >0.05).
See also Figures S4 and S6-S8, and Table S2.
We next tested if the number of functional synapses is restored by expression of HA-Rap2A(DN-S17N) in Nedd4-1-KO neurons. Evoked EPSC amplitudes and miniature EPSC frequencies were significantly smaller in KO neurons expressing EGFP (p<0.05), while HA-Rap2(DN-S17N) expressing KO neurons were restored to levels of WT neurons expressing either EGFP or HA-Rap2(DN-S17N) (Figures S4I-S4K, Table S2). When the dominant active mutant (DA-G12V) of HA-tagged Rap2A was overexpressed in KO neurons, we could barely detect an additive effect of Rap2A overexpression (Figures S6C-S6H and Table S2; p>0.05). These results indicate that Rap2 is highly activated in the absence of Nedd4-1 and that the function of Rap2 is regulated by Nedd4-1.
Above findings further support our notion that Rap2 is functioning as a downstream target of Nedd4-1 in neurite development. Several different target proteins of Rap2 have been identified, including TNIK family proteins (Nonaka et al., 2008; Taira et al., 2004), RalGDS, and Raf (Ohba et al., 2000). Among these, TNIK is most likely to play a role in the Nedd4-1 and Rap2 effects we have studied, since a pathway involving TNIK-family members and Rap2 was shown to play a key role in actin cytoskeleton rearrangements and in cell morphology regulation (Taira et al., 2004). To specifically promote or block downstream effects of TNIK-family proteins, we expressed WT or kinase-dead mutant (KM-K54R) TNIK in the presence or absence of (DA-G12V) Rap2A in mass cultures of WT rat neurons (Figure S7 and Table S2). The (KM-K54R) TNIK mutant was chosen because it is predicted to have a dominant negative effect on all pathways involving TNIK protein family members (Kolch et al., 1991). In these experiments, a CaMKII promoter-driven EGFP expression vector (Dittgen et al., 2004) was cotransfected with TNIK or Rap2A expression vectors to visualize neurite arborization. Overexpression of Myc-Rap2A (DA-G12V) or HA-TNIK (WT) resulted in indistinguishable phenotypes with short and simple neurites resembling the Nedd4-1-KO phenotype (Figures S7A-S7G). On the other hand, overexpression of HA-TNIK (KM-K54R) showed a slight but non-significant enhancement of dendrite branching and extension (Figures S7H-S7J). Coexpression of Myc-Rap2A (DA-G12V) with HA-TNIK (WT) led to a slight but non-significant shortening of dendrites as compared to single expression of Myc-Rap2A (DA-G12V) or HA-TNIK (WT) (Figures S7K-S7N, S7S), while coexpression of Myc-Rap2A (DA-G12V) and HA-TNIK (KM-K54R) showed the opposite phenotype, similar to overexpression of HA-TNIK (KM-K54R) alone (Figures S7H-S7J, S7O-S7R, S7S). That Rap2A(DA-G12V) overexpression did not compensate the effect of overexpression of HA-TNIK (KM-K54R) indicates that Rap2A is an upstream regulator of TNIK [Figures S7S and S7T; compare EGFP+HA-TNIK(KM-K54R) and EGFP+HA-TNIK(KM-K54R)+Myc-Rap2A(DA-G12V)]. These data and the published information (Taira et al., 2004) indicate that TNIK is regulated by Rap2A and that it is unlikely that Rap2 target proteins other than TNIK-family proteins are relevant for the neurite development processes studied here.
We next studied if interference with the kinase activities of TNIK family proteins can rescue the loss-of-function phenotype of Nedd4-1-KO neurons. We prepared mass cultures of WT and KO mouse neurons and transfected the HA-TNIK (KM-K54R) expression vector together with the CaMKII promoter-driven EGFP expression vector (Figure S8 and Table S2). Sholl analysis of WT neurons expressing EGFP revealed a peak dendrite density at 52.5 μm from the cell center (Figure S8E). We thus decided to quantify numbers of crossing dendrites at 52.5 μm from the somata (Figure S8F). KO neurons showed simpler dendrites than WT neurons (p<0.01) (Figures S8A and S8B), reproducing the data obtained in autaptic neurons (Figure 1). Overexpression of HA-TNIK (KM-K54R) in KO cells efficiently restored this defect (p<0.001), and the increase in dendrite complexity caused by HA-TNIK (KM-K54R) was more profound in KO neurons than in WT cells (Figures S8C-S8F). These results show that not only Rap2 but also TNIK-family proteins are involved in Nedd4-1-dependent regulation of neurite formation, supporting the view that TNIK-family proteins are overactivated in Nedd4-1-KO neurons, which results in an impairment of dendrite development.
Impairment of Rap2A Ubiquitination Results in a Gain of Rap2A Function in Dendrite Development
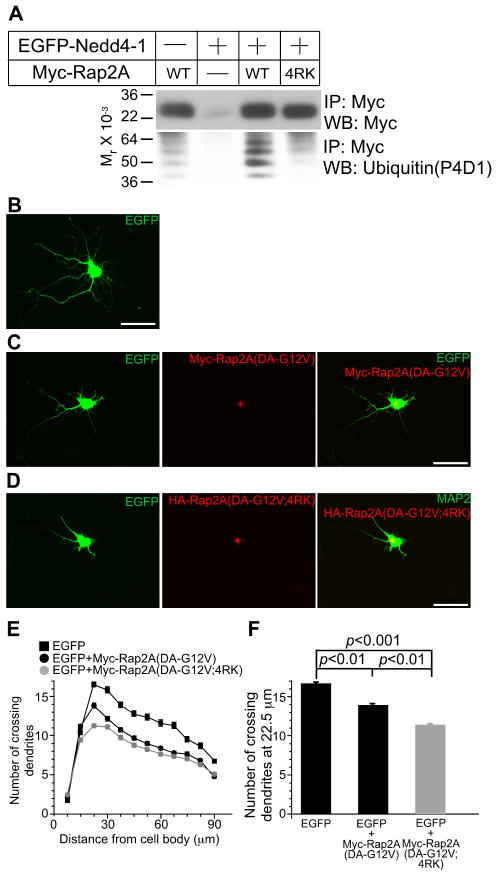
So far, our experiments indicated that the Rap2/TNIK signaling pathway in neuritogenesis is negatively regulated by Nedd4-1. We next tested if inhibition of Rap2 ubiquitination results in a gain of Rap2 function. Based on the crystal structure of Rap2A (Cherfils et al., 1997), we identified nine lysines that are likely to be surface exposed and thus might be conjugated to ubiquitin (Figure S5H). Assuming that the combined mutation of all of these nine lysine residues would cause a loss of enzymatic activity, we screened multiple compound point mutants of Rap2A to minimize the number of lysine mutations. Mutation of lysine residues 5, 94, 148, and 150 to arginine (4RK) efficiently reduced Rap2 ubiquitination (Figure 7A) but did not affect RalGDS binding (data not shown), indicating that the 4KR mutant Rap2 retains basic functionality and can be used for the analysis of the role of Rap2A ubiquitination in dendrite development. To maximize the dynamic range of our assay for effects of Rap2 ubiquitination on neurite development, we used Rap2A(DA-G12V) or Rap2A(DA-G12V;4RK) instead of the corresponding WT variants for transfection of rat hippocampal neurons as Nedd4-1 ubiquitinates DA mutant Rap2A more robustly than WT Rap2A (Figure 5E). We transfected neurons at DIV5 and fixed them at DIV8, two days earlier than in the experiments shown in Figures 8 and S7-S8 because the activity of overexpressed Rap2A was already saturated at DIV10 (data not shown). Upon overexpression of Rap2A (DA-G12V), WT rat hippocampal neurons showed impaired neurite development (Figures 7B, 7C, 7E, and 7F). This effect was enhanced upon Rap2A (DA-G12V;4RK) overexpression (Figures 7B-7F, Table S2), indicating that the function of Rap2 is blocked by ubiquitination of K5, K94, K148 and/or K150.
Figure 7. Perturbation of Ubiquitination of Rap2A Enhances Rap2A Function in Cultured Neurons.
(A) HEK cells were transfected with Myc-tagged WT or 4RK mutant variants of Rap2A, with or without EGFP-Nedd4-1. Myc-Rap2A was immunoprecipitated with anti-Myc antibodies, and ubiquitination was detected with the anti-ubiquitin P4D1 monoclonal antibody (Jura et al., 2006).
(B-D) Overexpression of Myc-Rap2A(DA-G12V;4RK) reveals a gain-of-function phenotype. Myc-Rap2A (DA-G12V) and EGFP coexpressing neurons shows impaired development of neurites as compared to EGFP expressing neurons (B, C). This effect of Rap2A was more pronounced when Myc-Rap2A(DA-G12V;4RK) was coexpressed with EGFP (D). Scale bar, 50 μm.
(E) Sholl analysis of Rap2A overexpressing neurons. Black rectangles, EGFP- expressing neurons (n=80); black circles, EGFP- and Myc-Rap2A(DA-G12V) coexpressing neurons (n=109); gray circles, EGFP- and Myc-Rap2A(DA-G12V;4RK) coexpressing neurons (n=109).
(F) Numbers of crossing dendrites at 22.5 μm from the cell body in EGFP-, EGFP- and Myc-Rap2A(G12V)-, or EGFP- and Myc-Rap2A(DA-G12V;4RK)-over expressing neurons. Nonparametric ANOVA test revealed a significant effect of expression of Myc-Rap2 mutants (p<0.0001). Dunn's multiple comparisons test revealed significant differences between groups linked with black lines.
See also Figures S6-S8 and Table S2.
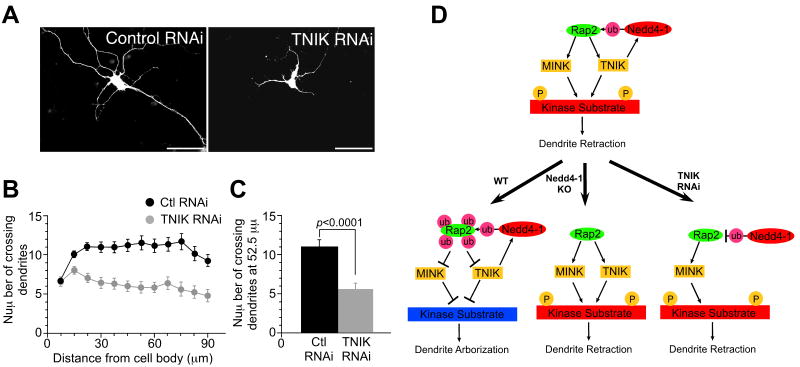
Figure 8. RNAi Silencing of Endogenous TNIK Phenocopies the Effect of Nedd4-1 Loss in Neurons.
(A) WT mouse neurons transfected with the vector encoding an shRNA expression cassette for TNIK knock-down showed impaired development of neurites (right panel) while control shRNA expressing neurons showed normal morphology (left panel). Scale bar; 50 μm.
(B) Sholl analysis of TNIK knock-down neurons. Black circles, control knockdown neurons (n=37); gray circles, TNIK-knock-down neurons (n=26).
(C) Numbers of crossing dendrites at 52.5 mm from the cell body in control and TNIK-knock-down neurons. Student's t-test; p<0.0001.
(D) Model of Nedd4-1 function in the regulation of neurite development. Given that only a fraction of total Rap2 is ubiquitinated, the depicted signaling pathway must be restricted (i.e. subcellularly compartmentalized) to growing neurite compartments by an as yet unknown mechanism. See Discussion for details.
See also Figures S6-S8 and Table S2.
RNAi Knock-Down of TNIK Phenocopies the Loss of Nedd4-1 Function
Both TNIK and MINK, but not NIK, are preferentially expressed in the brain and thus likely to be redundant downstream effectors of Rap2. However, due to its different linker domain and low Nedd4-1 affinity MINK should not form trimeric complexes with Rap2 and Nedd4-1 (Figure 8). We generated vectors encoding an shRNA expression cassette together with the EGFP-expression cassette, which was used to visualize the morphology of neurites, and isolated an shRNA vector that efficiently knocked down TNIK expression (Figure S5K). Upon knock down of TNIK expression using this vector, WT mouse neurons showed impaired neurite development (p<0.0001), resembling the phenotype of Nedd4-1-KO neurons, while expression of the control shRNA did not affect neurite morphology (Figures 8A-C and Table S2). This finding and the data described above (Figures 6 and S8) indicate that Nedd4-1 regulates the growth and arborization of neurites by modulating the activity of the Rap2-TNIK/MINK signal transduction cascade through complex formation with Rap2 and TNIK and monoubiquitination of Rap2.
Discussion
Ubiquitination is a key regulatory process in mammalian brain function. Aberrant activity of the ubiquitin-proteasome system causes multiple neurodegenerative diseases (DiFiglia et al., 1997; Kuzuhara et al., 1988) and specific ubiquitination defects are involved in several neurodevelopmental disorders (Kishino et al., 1997; Matsuura et al., 1997). More importantly in the context of the present study, ubiquitination processes can directly regulate mammalian nerve cell development. RNAi knock-down of the Ring Finger type E3 ligase APC or its coactivator Cdc20, for example, causes an impairment of dendrite growth due to aberrant ubiquitination of the centrosomal protein Id1 (Kim et al., 2009). Our present study demonstrates that Nedd4-1 is a novel positive regulator of dendrite extension and arborization (Figure 1) that operates by monoubiquitinating Rap2 (Figures 5, 6, 7, S6, S7). Thus, by regulating neurite development through the control of Rap2A function, Nedd4-1 complements the HECT type E3 ubiquitin ligases Smurf1 and Smurf2, which regulate axon extension by polyubiquitinating GDP-bound RhoA and Rap1B (Schwamborn et al., 2007; Wang et al., 2003).
Nedd4-1 KO leads to the development of shorter and less complex dendrites (Figure 1), which in turn causes a decrease in the number of functional synapses and a concomitant reduction in synaptic transmission (Figures 3 and 4). The cellular KO phenotype is rescued by reexpression of recombinant Nedd4-1 (Figure 1), indicating that it is due to the loss of a cell-autonomous Nedd4-1 function and not to secondary effects. This is supported by the fact that glutamatergic neuron specific conditional Nedd4-1-KO mice show related morphological changes (Figure 2). In addition, the survival of cultured Nedd4-1-KO neurons (Table S1), their passive membrane properties (Figure S3), and their neurite development at DIV3 (Figure S3) were all comparable to WT data, indicating that the viability and basic cell biological functions of KO neurons are normal. The fact that mEPSC amplitudes and responses to exogenously applied glutamate receptor agonists are normal in KO neurons (Figures 3E-3H) indicates that the formation and function of the remaining synapses and the cell surface trafficking of glutamate receptors are not perturbed in KO cells. We conclude that the role of Nedd4-1 in mouse neurons is neurite specific and that the functional changes in KO neurons can be explained by a specific developmental defect of neuritogenesis.
The establishment of neuronal dendritic fields is controlled by intrinsic (e.g. transcription factors, secretory pathway components, Rho and Ras family GTPases, or Ca2+ mediated signal transduction cascades) and extrinsic factors (e.g. neurotrophins and Trk receptors, Sema3A, Slit/Roundabout, or Notch/Delta). The Nedd4-1 homolog Nedd4-2 ubiquitinates TrkA in response to NGF stimulation, and TrkA expression is upregulated after RNAi knock-down of Nedd4-2 (Arevalo et al., 2006). This indicates that endogenous Nedd4-1 cannot compensate for the loss of Nedd4-2 function and is therefore unlikely to be redundant with Nedd4-2 with respect to TrkA regulation or to be otherwise involved in TrkA function. Likewise, Nedd4-1 function differs from that of Drosophila DNedd4, which downregulates Notch (Sakata et al., 2004; Wilkin et al., 2004), because the DNedd4-binding PPXY motif of Drosophila Notch is not conserved in mouse Notch1, and we did not identify any Notch related proteins among the affinity purified Nedd4-1 binding partners (Figure 5A and Table S4).
Instead of TrkA, Notch1, or any other previously published Nedd4 substrate, except Connexin43 (Leykauf et al., 2006), we affinity purified TNIK as a binding partner of Nedd4-1 WW domains. TNIK had previously been identified as a target of Rap2, and both TNIK and Rap2 are modulators of the cytoskeleton and cell morphology (Fu et al., 2007; Taira et al., 2004). Our subsequent biochemical analysis showed that Nedd4-1 forms a complex with TNIK and Rap2 and that Nedd4-1 ubiquitinates Rap2, but not TNIK (Figure 5), indicating that TNIK acts as an adaptor protein that mediates an indirect interaction between Nedd4-1 and its substrate Rap2. This notion is in accord with the domain structure of TNIK and the binding specificities of its linker domain for Nedd4-1 and of its Citron homology domain for Rap2. Moreover, the interaction between Nedd4-1 and Rap2 is dependent upon TNIK expression (Figure 5D) and the ternary complex formation is important for ubiquitination of Rap2 by Nedd4-1 (Figure 5G).
Recently, di-ubiquitination of eight lysine residues of HRas was reported (Jura et al., 2006). Rap2A and HRas share two of these lysine residues in their N-termini, which are critical for the recognition of downstream target proteins. We found that GTP-bound DA mutant Rap2A and WT Rap2A, but not a GDP-bound DN mutant Rap2A, are robustly ubiquitinated by Nedd4-1 (Figure 5E), indicating that ubiquitination of Rap2 is selective for GTP-bound Rap2. Given that the affinity of ubiquitinated Rap2A to the target protein RalGDS is reduced to undetectable levels (Figure 5I), Nedd4-1 dependent ubiquitination likely blocks the function of Rap2A by perturbing interactions between GTP-bound Rap2A and downstream targets. We cannot exclude that ubiquitination of Rap2A also affects GTP hydrolysis or GTP binding to Rap2, which is hard to test directly in our biochemical experimental systems. However, we found Rap2A function to be upregulated in Nedd4-1-KO MEFs using a FRET assay (Figure S6). As we used DA mutant but not WT Rap2A for this experiment, this result indicates that Rap2 ubiquitination affects its interaction with other proteins, including RalGDS (Figure S6) and TNIK/MINK, unlike di-ubiquitination of HRas, which regulates the subcellular compartmentalization of HRas. A likely explanation for this difference is that di- and monoubiquitination or different sites of ubiquitination exert differential effects (Jura et al., 2006).
Our overexpression experiments employing DN-S17N Rap2A and KM-K54R TNIK mutants show that inactivation of Rap2A or TNIK signaling can rescue the Nedd4-1-KO phenotype. In addition, the effects of overexpression of these mutants were more drastic in KO neurons than in WT cells, indicating that DN-S17N Rap2A and KM-K54R TNIK are epistatic to the Nedd4-1 loss-of-function mutation in cortical neurons (Figures 6 and S8). In contrast, overexpression of DA-G12V Rap2A showed almost no effect on neurite complexity in KO neurons while it efficiently reduced neurite complexity in WT cells (Figure S6), indicating that endogenous Rap2 is already overactivated in KO neurons. Accordingly, overexpression of dominant active Myc-Rap2A(G12V) in WT rat or mouse neurons phenocopied the effect of Nedd4-1 KO (Figures S6C-S6H and S7). The effect of KM-K54R TNIK overexpression on dendrite complexity was enhanced in WT rat neurons when DA-G12V Rap2A was coexpressed (Figures S7S and S7T). This corroborates the effects of KM-K54R TNIK on dendrite complexity in mouse neurons, which were weaker in WT neurons than in Nedd4-1-KO neurons that already have increased Rap2 function (Figure S8).
We tested for a direct involvement of ubiqutination in the regulation of Rap2 function by studying the effects of overexpression of a ubiquitination-compromised Rap2 point mutant in cultured neurons. The corresponding data showed that Rap2 function is increased upon mutation of putative ubiquitination sites (Figure 7). Together, these findings provide strong support for a model, according to which Nedd4-1-mediated di- and multi-monoubiquitination of Rap2 inhibits the Rap2-TNIK/MINK signaling cascade (Figure 8D).
In WT neurons, in which the two Rap2 targets TNIK and MINK are strongly expressed, downregulation of TNIK expression by RNAi phenocopied the denritogenesis defect of Nedd4-1-KO cells (Figure 8). At first glance, this finding is unexpected as overexpression of the dominant negative KM-K54R kinase mutant of TNIK in Nedd4-1-KO neurons rescues the KO phenotype (Figure S8). However, the two datasets can be reconciled by taking into account that the kinase activities of both TNIK and MINK are involved in the signaling pathway that controls neurite growth, whereas only TNIK is able to bind Nedd4-1 via its linker domain and to mediate a negative feedback loop by allowing Rap2 ubiquitination and inactivation (Figures S5B and 8D). Thus, the dominant negative KM-K54R TNIK mutant is predicted to block phosphorylation of all joint substrates of TNIK and MINK, and to thereby shut down the negative effect that TNIK and MINK exert on dendrite extension, leading to increases in dendrite growth. In contrast, the selective knock-down of only TNIK is expected to inhibit Rap2 ubiquitylation by Nedd4-1, leading to an increase in the kinase activity of MINK, and a reduction in neurite growth.
Experimental Procedures
Expression Vectors and Antibodies
Expression vectors and antibodies used in the present study are described in detail in the Supplemental Data.
In Situ Hybridization
In situ hybridization was performed as described previously (Varoqueaux et al., 2006). Details are given in the Supplemental Data.
Mutant Mice
In conventional Nedd4-1-KOs, exons 7 and 8 were replaced by a Neomycin resistance cassette (Figure S2A). The Nedd4-1 conditional allele contained a Neomycin resistance cassette flanked by frt sites in intron 8, two loxP sites flanking exon 9, and two herpes simplex virus thymidine kinase expression cassettes for negative selection (Figure S2B). The conditional Nedd4-1flox/+ line was first crossed with a FLP1-expressing deleter line [B6;SJL-Tg(ACTFLPe)9205Dym/J; Jackson Laboratory] to remove the Neomycin resistance cassette and subsequently with a NEX-Cre line (Goebbels et al., 2006) to obtain the NEX-Cre;Nedd4-1flox/+ line. Details are given in the Supplemental Data.
Microisland and Continental Neuron Culture, Staining, and FRET Experiments
Microisland and continental neuron cultures were prepared, transfected, and immunostained as described previously (Dresbach et al., 2003). Methodological details, including descriptions of the Golgi staining and the FRET experiments using Rauchu-Rap2A(DA-G12V) are provided in the Supplemental Data.
Electrophysiology
Autaptic neurons were analyzed electrophysiologically as described previously (Bekkers and Stevens, 1991; Jockusch et al., 2007; Pyott and Rosenmund, 2002). Details are provided in the Supplemental Data.
Affinity Purification of GST-Nedd4-1 Binding Proteins
Affinity purification of GST-Nedd4-1 binding proteins was performed based on a published protocol (Ingham et al., 2005) using recombinant GST-Nedd4-1 (residues 217-549). Proteins were separated by SDS-PAGE on pre-cast NuPAGE 10% Bis-Tris gels (Invitrogen) and proteins were processed and analyzed by MALDI-TOF mass spectrometry as described previously (Jahn et al., 2006). Further details are given in the Supplemental Data.
Protein Chemical Procedures and In Vitro Ubiquitination Assays
Affinity pull-down assays with GST-Nedd4-1 and GST-RalGDS were performed according to standard protocols. For chemical crosslinking experiments we used 1 mM dithiobis succinimidyl propionate (DSP) (Pierce) for 30 min on ice. Details on crosslinking and subcellular fractionation protocols are given in the Supplemental Data. Ubiquitination assays were performed on transfected HEK cells employing published procedures. MEFs prepared from WT and Nedd4-1-KO embryos were immortalized by stably overexpressing large T-antigen using a retrovirus system. Details are given in the Supplemental Data.
shRNA Experiments
The targeting sequence of shRNA for TNIK corresponds to bp 191-211 of human TNIK (AF172264) and bp 191-211 of mouse TNIK (BC137800). The sequence of luciferase was chosen for the negative control shRNA based on a published report (Barnes et al., 2007). Hairpin sequences and vector design for the TNIK knock-down are given in the Supplemental Data.
Statistics
Analyses were performed using Kaleidagraph or InStat software. Data are expressed as mean±SEM. The normality of the distribution was tested with the Kolmogorov-Smirnov test or the Dallal and Wilkinson approximation to Lilliefor's method. For comparison of two groups with normal distributions, Student's t-test was applied. For comparison of two groups that failed normality tests, the Kolmogorov-Smirnov test was used. Analyses of more than three groups with normal distributions were done by one-way ANOVA with post hoc Tukey-HSD test. For comparison of more than three groups that fail normality tests, the Kruskal-Wallis (nonparametric ANOVA) test with Dunn's multiple comparisons test was used.
Supplementary Material
Acknowledgments
We thank B. Hesse-Niessen, S. Wenger, S. Handt, F. Scheve, F. Benseler, I. Thanhäuser, D. Schwerdtfeger, A. Zeuch, M. Dörre, A. Galinski, I. Herfort, and T. Wiles for excellent technical assistance, and the staff of the animal facility at the Max Planck Institute of Experimental Medicine for maintenance of the mouse colony. We are grateful to D.A. Gray, R. Nehring, A. Betz, T. Nagase, A. Kusumi, P. Osten, R.L. Huganir, M. Matsuda, R. Brandt, A. Wittinghofer, and N. Ziv for reagents and software, and to G. Meyer, S. Bungers, M. Kishimoto-Suga, V. Tarabykin, K. Chowdhury, D. Sargin, M. Scheffner, S. Yamagishi, F. Nakamura, I. Vetter, A. Püschel, D. Rotin, S. Okabe, D. Yokomaku, and A.M. Craig for discussions and advice. This study was supported by the Deutsche Forschungsgemeinschaft (SFB271/B11; N.B.), the Japan Society for the Promotion of Science Postdoctoral Fellowship for Research Abroad (H.K.), the Yamanouchi Foundation for Research on Metabolic Disorders (H.K.), the Mochida Memorial Foundation for Medical and Pharmaceutical Research (H.K.), the Alexander von Humboldt Foundation (S.M.Y.), National Institutes of Health grants NIH R01GM66955 (D.E.Z.), the Max Planck Society (N.B.) and the European Commission (EU Synapse grant LSHM-CT-2005-019055; S.M.Y. and N.B.).
Footnotes
E-TOC Alert Paragraph: This study identifies a novel signaling pathway that regulates neuronal dendrite growth. The pathway involves the E3 ubiquitin ligase Nedd4-1, the small GTPase Rap2, and the serine/threonine kinase TNIK. Nedd4-1 promotes neuronal dendrite growth by mono/di-ubiquitinating Rap2, thus blocking the Rap2 interaction with its downstream effector TNIK and reducing the activity of TNIK, which is a negative regulator of cell spreading and dendrite growth.
Highlights
- The E3 ubiquitin ligase Nedd4-1 mono/di-ubiquitinates the small GTPase Rap2.
- Ubiquitinated Rap2 cannot activate the kinase TNIK, a dendrite growth regulator.
- Nedd4-1/Rap2/TNIK signaling in neurons was perturbed by genetic methods.
- Perturbation of Nedd4-1/Rap2/TNIK signaling reduces neuronal dendrite growth.
Supplemental Data: The Supplemental Data include Supplemental Experimental Procedures, 8 figures, and 4 tables, and can be found at http://www.neuron.org.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Arevalo JC, Waite J, Rajagopal R, Beyna M, Chen ZY, Lee FS, Chao MV. Cell survival through Trk neurotrophin receptors is differentially regulated by ubiquitination. Neuron. 2006;50:549–559. doi: 10.1016/j.neuron.2006.03.044. [DOI] [PubMed] [Google Scholar]
- Barnes AP, Lilley BN, Pan YA, Plummer LJ, Powell AW, Raines AN, Sanes JR, Polleux F. LKB1 and SAD kinases define a pathway required for the polarization of cortical neurons. Cell. 2007;129:549–563. doi: 10.1016/j.cell.2007.03.025. [DOI] [PubMed] [Google Scholar]
- Bekkers JM, Stevens CF. Excitatory and inhibitory autaptic currents in isolated hippocampal neurons maintained in cell culture. Proc Natl Acad Sci U S A. 1991;88:7834–7838. doi: 10.1073/pnas.88.17.7834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom AJ, Miller BR, Sanes JR, DiAntonio A. The requirement for Phr1 in CNS axon tract formation reveals the corticostriatal boundary as a choice point for cortical axons. Genes Dev. 2007;21:2593–2606. doi: 10.1101/gad.1592107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao XR, Lill NL, Boase N, Shi PP, Croucher DR, Shan H, Qu J, Sweezer EM, Place T, Kirby PA, et al. Nedd4 controls animal growth by regulating IGF-1 signaling. Sci Signal. 2008;1:ra5. doi: 10.1126/scisignal.1160940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherfils J, Menetrey J, Le Bras G, Janoueix-Lerosey I, de Gunzburg J, Garel JR, Auzat I. Crystal structures of the small G protein Rap2A in complex with its substrate GTP, with GDP and with GTPgammaS. Embo J. 1997;16:5582–5591. doi: 10.1093/emboj/16.18.5582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dan I, Watanabe NM, Kobayashi T, Yamashita-Suzuki K, Fukagaya Y, Kajikawa E, Kimura WK, Nakashima TM, Matsumoto K, Ninomiya-Tsuji J, et al. Molecular cloning of MINK, a novel member of mammalian GCK family kinases, which is up-regulated during postnatal mouse cerebral development. FEBS Lett. 2000;469:19–23. doi: 10.1016/s0014-5793(00)01247-3. [DOI] [PubMed] [Google Scholar]
- DiFiglia M, Sapp E, Chase KO, Davies SW, Bates GP, Vonsattel JP, Aronin N. Aggregation of huntingtin in neuronal intranuclear inclusions and dystrophic neurites in brain. Science. 1997;277:1990–1993. doi: 10.1126/science.277.5334.1990. [DOI] [PubMed] [Google Scholar]
- Dittgen T, Nimmerjahn A, Komai S, Licznerski P, Waters J, Margrie TW, Helmchen F, Denk W, Brecht M, Osten P. Lentivirus-based genetic manipulations of cortical neurons and their optical and electrophysiological monitoring in vivo. Proc Natl Acad Sci U S A. 2004;101:18206–18211. doi: 10.1073/pnas.0407976101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dresbach T, Hempelmann A, Spilker C, tom Dieck S, Altrock WD, Zuschratter W, Garner CC, Gundelfinger ED. Functional regions of the presynaptic cytomatrix protein bassoon: significance for synaptic targeting and cytomatrix anchoring. Mol Cell Neurosci. 2003;23:279–291. doi: 10.1016/s1044-7431(03)00015-0. [DOI] [PubMed] [Google Scholar]
- Fouladkou F, Landry T, Kawabe H, Neeb A, Lu C, Brose N, Stambolic V, Rotin D. The ubiquitin ligase Nedd4-1 is dispensable for the regulation of PTEN stability and localization. Proc Natl Acad Sci U S A. 2008;105:8585–8590. doi: 10.1073/pnas.0803233105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu CA, Shen M, Huang BC, Lasaga J, Payan DG, Luo Y. TNIK, a novel member of the germinal center kinase family that activates the c-Jun N-terminal kinase pathway and regulates the cytoskeleton. J Biol Chem. 1999;274:30729–30737. doi: 10.1074/jbc.274.43.30729. [DOI] [PubMed] [Google Scholar]
- Fu Z, Lee SH, Simonetta A, Hansen J, Sheng M, Pak DT. Differential roles of Rap1 and Rap2 small GTPases in neurite retraction and synapse elimination in hippocampal spiny neurons. J Neurochem. 2007;100:118–131. doi: 10.1111/j.1471-4159.2006.04195.x. [DOI] [PubMed] [Google Scholar]
- Fujimuro M, Sawada H, Yokosawa H. Production and characterization of monoclonal antibodies specific to multi-ubiquitin chains of polyubiquitinated proteins. FEBS Lett. 1994;349:173–180. doi: 10.1016/0014-5793(94)00647-4. [DOI] [PubMed] [Google Scholar]
- Goebbels S, Bormuth I, Bode U, Hermanson O, Schwab MH, Nave KA. Genetic targeting of principal neurons in neocortex and hippocampus of NEX-Cre mice. Genesis. 2006;44:611–621. doi: 10.1002/dvg.20256. [DOI] [PubMed] [Google Scholar]
- Guo L, Wang Y. Glutamate stimulates glutamate receptor interacting protein 1 degradation by ubiquitin-proteasome system to regulate surface expression of GluR2. Neuroscience. 2007;145:100–109. doi: 10.1016/j.neuroscience.2006.11.042. [DOI] [PubMed] [Google Scholar]
- Hand R, Bortone D, Mattar P, Nguyen L, Heng JI, Guerrier S, Boutt E, Peters E, Barnes AP, Parras C, et al. Phosphorylation of Neurogenin2 specifies the migration properties and the dendritic morphology of pyramidal neurons in the neocortex. Neuron. 2005;48:45–62. doi: 10.1016/j.neuron.2005.08.032. [DOI] [PubMed] [Google Scholar]
- Herrmann C, Horn G, Spaargaren M, Wittinghofer A. Differential interaction of the ras family GTP-binding proteins H-Ras, Rap1A, and R-Ras with the putative effector molecules Raf kinase and Ral-guanine nucleotide exchange factor. J Biol Chem. 1996;271:6794–6800. doi: 10.1074/jbc.271.12.6794. [DOI] [PubMed] [Google Scholar]
- Huang EJ, Reichardt LF. Trk receptors: roles in neuronal signal transduction. Annu Rev Biochem. 2003;72:609–642. doi: 10.1146/annurev.biochem.72.121801.161629. [DOI] [PubMed] [Google Scholar]
- Hume RI, Purves D. Geometry of neonatal neurones and the regulation of synapse elimination. Nature. 1981;293:469–471. doi: 10.1038/293469a0. [DOI] [PubMed] [Google Scholar]
- Ingham RJ, Colwill K, Howard C, Dettwiler S, Lim CS, Yu J, Hersi K, Raaijmakers J, Gish G, Mbamalu G, et al. WW domains provide a platform for the assembly of multiprotein networks. Mol Cell Biol. 2005;25:7092–7106. doi: 10.1128/MCB.25.16.7092-7106.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahn O, Hesse D, Reinelt M, Kratzin HD. Technical innovations for the automated identification of gel-separated proteins by MALDI-TOF mass spectrometry. Anal Bioanal Chem. 2006;386:92–103. doi: 10.1007/s00216-006-0592-1. [DOI] [PubMed] [Google Scholar]
- Jaworski J, Spangler S, Seeburg DP, Hoogenraad CC, Sheng M. Control of dendritic arborization by the phosphoinositide-3′-kinase-Akt-mammalian target of rapamycin pathway. J Neurosci. 2005;25:11300–11312. doi: 10.1523/JNEUROSCI.2270-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang YH, Armstrong D, Albrecht U, Atkins CM, Noebels JL, Eichele G, Sweatt JD, Beaudet AL. Mutation of the Angelman ubiquitin ligase in mice causes increased cytoplasmic p53 and deficits of contextual learning and long-term potentiation. Neuron. 1998;21:799–811. doi: 10.1016/s0896-6273(00)80596-6. [DOI] [PubMed] [Google Scholar]
- Jockusch WJ, Speidel D, Sigler A, Sorensen JB, Varoqueaux F, Rhee JS, Brose N. CAPS-1 and CAPS-2 Are Essential Synaptic Vesicle Priming Proteins. Cell. 2007;131:796–808. doi: 10.1016/j.cell.2007.11.002. [DOI] [PubMed] [Google Scholar]
- Jura N, Scotto-Lavino E, Sobczyk A, Bar-Sagi D. Differential modification of Ras proteins by ubiquitination. Mol Cell. 2006;21:679–687. doi: 10.1016/j.molcel.2006.02.011. [DOI] [PubMed] [Google Scholar]
- Kim AH, Puram SV, Bilimoria PM, Ikeuchi Y, Keough S, Wong M, Rowitch D, Bonni A. A centrosomal Cdc20-APC pathway controls dendrite morphogenesis in postmitotic neurons. Cell. 2009;136:322–336. doi: 10.1016/j.cell.2008.11.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishino T, Lalande M, Wagstaff J. UBE3A/E6-AP mutations cause Angelman syndrome. Nat Genet. 1997;15:70–73. doi: 10.1038/ng0197-70. [DOI] [PubMed] [Google Scholar]
- Kolch W, Heidecker G, Lloyd P, Rapp UR. Raf-1 protein kinase is required for growth of induced NIH/3T3 cells. Nature. 1991;349:426–428. doi: 10.1038/349426a0. [DOI] [PubMed] [Google Scholar]
- Kuzuhara S, Mori H, Izumiyama N, Yoshimura M, Ihara Y. Lewy bodies are ubiquitinated. A light and electron microscopic immunocytochemical study. Acta Neuropathol. 1988;75:345–353. doi: 10.1007/BF00687787. [DOI] [PubMed] [Google Scholar]
- Kwon CH, Luikart BW, Powell CM, Zhou J, Matheny SA, Zhang W, Li Y, Baker SJ, Parada LF. Pten regulates neuronal arborization and social interaction in mice. Neuron. 2006;50:377–388. doi: 10.1016/j.neuron.2006.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leykauf K, Salek M, Bomke J, Frech M, Lehmann WD, Durst M, Alonso A. Ubiquitin protein ligase Nedd4 binds to connexin43 by a phosphorylation-modulated process. J Cell Sci. 2006;119:3634–3642. doi: 10.1242/jcs.03149. [DOI] [PubMed] [Google Scholar]
- Lotz K, Pyrowolakis G, Jentsch S. BRUCE, a giant E2/E3 ubiquitin ligase and inhibitor of apoptosis protein of the trans-Golgi network, is required for normal placenta development and mouse survival. Mol Cell Biol. 2004;24:9339–9350. doi: 10.1128/MCB.24.21.9339-9350.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuura T, Sutcliffe JS, Fang P, Galjaard RJ, Jiang YH, Benton CS, Rommens JM, Beaudet AL. De novo truncating mutations in E6-AP ubiquitin-protein ligase gene (UBE3A) in Angelman syndrome. Nat Genet. 1997;15:74–77. doi: 10.1038/ng0197-74. [DOI] [PubMed] [Google Scholar]
- Mochizuki N, Yamashita S, Kurokawa K, Ohba Y, Nagai T, Miyawaki A, Matsuda M. Spatio-temporal images of growth-factor-induced activation of Ras and Rap1. Nature. 2001;411:1065–1068. doi: 10.1038/35082594. [DOI] [PubMed] [Google Scholar]
- Montes de Oca Luna R, Wagner DS, Lozano G. Rescue of early embryonic lethality in mdm2-deficient mice by deletion of p53. Nature. 1995;378:203–206. doi: 10.1038/378203a0. [DOI] [PubMed] [Google Scholar]
- Murdaca J, Treins C, Monthouel-Kartmann MN, Pontier-Bres R, Kumar S, Van Obberghen E, Giorgetti-Peraldi S. Grb10 prevents Nedd4-mediated vascular endothelial growth factor receptor-2 degradation. J Biol Chem. 2004;279:26754–26761. doi: 10.1074/jbc.M311802200. [DOI] [PubMed] [Google Scholar]
- Newton K, Matsumoto ML, Wertz IE, Kirkpatrick DS, Lill JR, Tan J, Dugger D, Gordon N, Sidhu SS, Fellouse FA, et al. Ubiquitin chain editing revealed by polyubiquitin linkage-specific antibodies. Cell. 2008;134:668–678. doi: 10.1016/j.cell.2008.07.039. [DOI] [PubMed] [Google Scholar]
- Nonaka H, Takei K, Umikawa M, Oshiro M, Kuninaka K, Bayarjargal M, Asato T, Yamashiro Y, Uechi Y, Endo S, et al. MINK is a Rap2 effector for phosphorylation of the postsynaptic scaffold protein TANC1. Biochem Biophys Res Commun. 2008;377:573–578. doi: 10.1016/j.bbrc.2008.10.038. [DOI] [PubMed] [Google Scholar]
- Ohba Y, Mochizuki N, Matsuo K, Yamashita S, Nakaya M, Hashimoto Y, Hamaguchi M, Kurata T, Nagashima K, Matsuda M. Rap2 as a slowly responding molecular switch in the Rap1 signaling cascade. Mol Cell Biol. 2000;20:6074–6083. doi: 10.1128/mcb.20.16.6074-6083.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrish JZ, Emoto K, Kim MD, Jan YN. Mechanisms that regulate establishment, maintenance, and remodeling of dendritic fields. Annu Rev Neurosci. 2007;30:399–423. doi: 10.1146/annurev.neuro.29.051605.112907. [DOI] [PubMed] [Google Scholar]
- Pham N, Rotin D. Nedd4 regulates ubiquitination and stability of the guanine-nucleotide exchange factor CNrasGEF. J Biol Chem. 2001;276:46995–47003. doi: 10.1074/jbc.M108373200. [DOI] [PubMed] [Google Scholar]
- Pyott SJ, Rosenmund C. The effects of temperature on vesicular supply and release in autaptic cultures of rat and mouse hippocampal neurons. J Physiol. 2002;539:523–535. doi: 10.1113/jphysiol.2001.013277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenmund C, Stevens CF. Definition of the readily releasable pool of vesicles at hippocampal synapses. Neuron. 1996;16:1197–1207. doi: 10.1016/s0896-6273(00)80146-4. [DOI] [PubMed] [Google Scholar]
- Sakata T, Sakaguchi H, Tsuda L, Higashitani A, Aigaki T, Matsuno K, Hayashi S. Drosophila Nedd4 regulates endocytosis of notch and suppresses its ligand-independent activation. Curr Biol. 2004;14:2228–2236. doi: 10.1016/j.cub.2004.12.028. [DOI] [PubMed] [Google Scholar]
- Sasaki Y, Cheng C, Uchida Y, Nakajima O, Ohshima T, Yagi T, Taniguchi M, Nakayama T, Kishida R, Kudo Y, et al. Fyn and Cdk5 mediate semaphorin-3A signaling, which is involved in regulation of dendrite orientation in cerebral cortex. Neuron. 2002;35:907–920. doi: 10.1016/s0896-6273(02)00857-7. [DOI] [PubMed] [Google Scholar]
- Schwamborn JC, Muller M, Becker AH, Puschel AW. Ubiquitination of the GTPase Rap1B by the ubiquitin ligase Smurf2 is required for the establishment of neuronal polarity. Embo J. 2007;26:1410–1422. doi: 10.1038/sj.emboj.7601580. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Schwamborn JC, Puschel AW. The sequential activity of the GTPases Rap1B and Cdc42 determines neuronal polarity. Nat Neurosci. 2004;7:923–929. doi: 10.1038/nn1295. [DOI] [PubMed] [Google Scholar]
- Sholl DA. Dendritic organization in the neurons of the visual and motor cortices of the cat. J Anat. 1953;87:387–406. [PMC free article] [PubMed] [Google Scholar]
- Snider WD. The dendritic complexity and innervation of submandibular neurons in five species of mammals. J Neurosci. 1987;7:1760–1768. doi: 10.1523/JNEUROSCI.07-06-01760.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spruston N. Pyramidal neurons: dendritic structure and synaptic integration. Nat Rev Neurosci. 2008;9:206–221. doi: 10.1038/nrn2286. [DOI] [PubMed] [Google Scholar]
- Staub O, Dho S, Henry P, Correa J, Ishikawa T, McGlade J, Rotin D. WW domains of Nedd4 bind to the proline-rich PY motifs in the epithelial Na+ channel deleted in Liddle's syndrome. Embo J. 1996;15:2371–2380. [PMC free article] [PubMed] [Google Scholar]
- Taira K, Umikawa M, Takei K, Myagmar BE, Shinzato M, Machida N, Uezato H, Nonaka S, Kariya K. The Traf2- and Nck-interacting kinase as a putative effector of Rap2 to regulate actin cytoskeleton. J Biol Chem. 2004;279:49488–49496. doi: 10.1074/jbc.M406370200. [DOI] [PubMed] [Google Scholar]
- Takemoto-Kimura S, Ageta-Ishihara N, Nonaka M, Adachi-Morishima A, Mano T, Okamura M, Fujii H, Fuse T, Hoshino M, Suzuki S, et al. Regulation of dendritogenesis via a lipid-raft-associated Ca2+/calmodulin-dependent protein kinase CLICK-III/CaMKIgamma. Neuron. 2007;54:755–770. doi: 10.1016/j.neuron.2007.05.021. [DOI] [PubMed] [Google Scholar]
- Varoqueaux F, Aramuni G, Rawson RL, Mohrmann R, Missler M, Gottmann K, Zhang W, Sudhof TC, Brose N. Neuroligins determine synapse maturation and function. Neuron. 2006;51:741–754. doi: 10.1016/j.neuron.2006.09.003. [DOI] [PubMed] [Google Scholar]
- Wang HR, Zhang Y, Ozdamar B, Ogunjimi AA, Alexandrova E, Thomsen GH, Wrana JL. Regulation of cell polarity and protrusion formation by targeting RhoA for degradation. Science. 2003;302:1775–1779. doi: 10.1126/science.1090772. [DOI] [PubMed] [Google Scholar]
- Wang X, Trotman LC, Koppie T, Alimonti A, Chen Z, Gao Z, Wang J, Erdjument-Bromage H, Tempst P, Cordon-Cardo C, et al. NEDD4-1 is a proto-oncogenic ubiquitin ligase for PTEN. Cell. 2007;128:129–139. doi: 10.1016/j.cell.2006.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkin MB, Carbery AM, Fostier M, Aslam H, Mazaleyrat SL, Higgs J, Myat A, Evans DA, Cornell M, Baron M. Regulation of notch endosomal sorting and signaling by Drosophila Nedd4 family proteins. Curr Biol. 2004;14:2237–2244. doi: 10.1016/j.cub.2004.11.030. [DOI] [PubMed] [Google Scholar]
- Yamashita M, Ying SX, Zhang GM, Li C, Cheng SY, Deng CX, Zhang YE. Ubiquitin ligase Smurf1 controls osteoblast activity and bone homeostasis by targeting MEKK2 for degradation. Cell. 2005;121:101–113. doi: 10.1016/j.cell.2005.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita N, Morita A, Uchida Y, Nakamura F, Usui H, Ohshima T, Taniguchi M, Honnorat J, Thomasset N, Takei K, et al. Regulation of spine development by semaphorin3A through cyclin-dependent kinase 5 phosphorylation of collapsin response mediator protein 1. J Neurosci. 2007;27:12546–12554. doi: 10.1523/JNEUROSCI.3463-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao I, Takagi H, Ageta H, Kahyo T, Sato S, Hatanaka K, Fukuda Y, Chiba T, Morone N, Yuasa S, et al. SCRAPPER-dependent ubiquitination of active zone protein RIM1 regulates synaptic vesicle release. Cell. 2007;130:943–957. doi: 10.1016/j.cell.2007.06.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.