Abstract
Assembly of TCRα and δ genes from the TCRα/δ locus is tightly controlled for the proper generation of αβ and γδ T cells. Of more than 100 shared variable gene segments in the TCRα/δ locus, only a few are predominantly used for the TCRδ gene assembly while majority are for TCRα. However, importance and mechanisms of the selective variable gene rearrangement for T cell development are not fully understood. We report here that the development of a tissue-specific γδ T cell population is critically affected by recombination signal sequences-associated restriction on the variable gene usage for TCRδ assembly. We found that the development of substitute skin γδ T cells in mice deficient of TCRγ3 gene, which is used in wild-type skin γδ T cells, was drastically affected by the strain background. A Vγ2+ skin γδT cell population developed in mice of the B6 but not 129 strain backgrounds, due to a difference in the rearrangement of endogenous Vδ7+ TCRδ genes, which paired with the Vγ2+ TCRγ gene to generate the Vγ2/Vδ7+ skin γδ T cell precursors in fetal thymi of the B6 background mice. The defective TCRδ rearrangement of the 129-“Vδ7” gene was associated with specific variations in its recombination signal sequence, which renders it poorly compatible for rearrangement to Dδ genes. These findings provide the first direct evidence that recombination signal sequence-associated restriction on the variable gene usage for TCRα/δ gene assembly plays an important role in the T cell development.
Introduction
T cells can be distinguished by their expression of distinct forms of T cell receptors (TCR). αβ T cells express the αβTCR, a heterodimer of TCRα and TCRβ chains and γδ T cells express the γδTCR of TCRγ and TCRδ chains. Although both αβ and γδ T cells are generated in the thymus, they have different peripheral tissue distribution and function. While αβ T cells are predominantly localized in secondary lymphoid organs after exiting the thymus, γδ T cells are more often localized to epithelial tissues, such as the skin and reproductive tracts (1, 2). The epithelial tissue-specific γδ T cells usually display limited diversities in their TCR sequences and function as innate immune cells in the first line of defense (3-5). In mice, nearly all γδ T cells in the skin epidermis, called dendritic epidermal T cells (DETC), express canonical Vγ3/Vδ1+ TCRs while vaginal epithelial γδ T cells express Vγ4/Vδ1+ TCRs. By comparison, γδ T cells in secondary lymphoid organs usually express more diverse TCRs, predominantly of Vγ2 and Vγ1.1 associated with several Vδ chains.
Development of the various tissue-specific γδ T cell subsets is orchestrated at different stages of ontogeny, presumably to optimize the production of different specialized sets of T cells required for host defense. The first subsets to arise in the early E15 fetal thymus assemble Vγ3 and Vγ4 gene segments (1, 6, 7). The resulting T cells include precursors of DETC (Vγ3/Vδ1) and the vaginal intraepithelial T cells (Vγ4/Vδ1), which subsequently migrate to their peripheral destinations. After birth, Vγ3 and Vγ4 rearrangements are suppressed and rearrangements of Vγ2, Vγ1.1 and others predominate (8-10). The resulting T cells preferentially emigrate to secondary lymphoid organs, among other tissues. TCRδ genes are rearranged coordinately with the TCRγ genes. The Vδ1 gene is predominantly rearranged and expressed at the same early fetal thymic stage as Vγ3 and Vγ4 genes (11), enabling the production of Vγ3/Vδ1 and Vγ4/Vδ1 TCRs, the forms expressed in DETCs and vaginal intraepithelial γδ T cells, respectively. In the adult stage, a distinct set of Vδ gene segments are predominantly rearranged (11).
The TCRδ locus is embedded within the TCRα locus, and the two loci share the same set of variable gene segments (Vα/δ). Although all the variable gene segments could potentially rearrange to Dδ/Jδ or Jα segments for assembly of TCRδ or TCRα genes, the actual usage of Vα/δ genes in the TCRδ and TCRα assembly is very restricted. Of more than 100 Vα/δ gene segments in the murine TCRα/δ loci, less than ten of them (so-called Vδ genes) are predominantly used in the TCRδ gene assembly while the majority (Vα genes) are preferentially in rearrangements of TCRα genes (12). The preferential usage of different Vα/δ gene segments for the TCRδ and TCRα gene rearrangement is genetically programmed and independent of cellular selection processes, as it applies even in CD3ε-/- mice, in which cellular selection is defective (13, 14). However, importance and mechanisms of the regulated Vα/δ gene rearrangement for the T cell development are not fully understood.
Multiple mechanisms underlie the regulated TCR gene rearrangements, including gene accessibility, germline transcription, V gene competition and preferential pairing of specific recombination signal sequences (RSS). The accessibility of V, D and J segments in chromatin to the recombinase apparatus is regulated at different ontogenic and developmental stages and mediated by transcription factors through cis-acting regulatory elements located in the TCR loci. Recent evidence suggests that the capacity of the gene segments to undergo germline transcription, which may in turn be regulated by the gene accessibility, is a direct determinant of rearrangement efficiency (15, 16). Accessibility of Vγ gene segments has been found to be an important determinant of the rearrangement pattern in the adult thymus, where the frequently rearranged Vγ2 segment resides in acetylated (active) chromatin, and the rarely rearranged Vγ3 segment resides in inactive chromatin (17). This difference is determined in large part by the promoter regions of the Vγ3 and Vγ2 gene segments (9, 18).
Gene accessibility also plays a role in the selective Vα/δ gene usage for TCRδ and TCRα gene assembly. At the early CD4-CD8- (DN) stage of thymic T cell development, Jα gene segments are inaccessible for rearrangement due to the inactivity of their associated cis-regulatory elements such as the Eα enhancer and TEA promoter while Dδ and Jδ gene segments are rendered accessible by their associated Eδ enhancer. Therefore, only TCRδ genes are assembled at the DN stage (19, 20). Most (but not all) of the “Vδ” genes are located at the 3′ region of the Vα/δ gene cluster, near the Dδ-Jδ region and within the effect range of the Eδ enhancer (21, 22). Consistent with this, many of the proximal “Vδ” genes were present in a hyper-acetylated (active) chromatin status at the DN stage (23). At the later CD4+CD8+ (DP) stage (for αβ T cell development), the more powerful and αβ T lineage specific Eα enhancer becomes active and promotes the accessibility of Jα genes and more distal “Vα” genes and TCRα gene assembly (21). However, several “Vδ” genes are located as far as 1Mb away from the Dδ/Jδ region while some “Vα” genes lies within 100kb from the Dδ-Jδ region, suggesting that the enhancers-regulated accessibility alone could not account completely for the selective usage of Vα/δ genes in the TCRδ and TCRα gene assembly.
Competition among different V gene segments is another mechanism that regulates their usage in the TCR gene assembly. In the TCRγ locus, both Vγ2 and Vγ3 segments are accessible at the early fetal stage, yet Vγ3 is preferentially rearranged to Jγ1 gene. Transgenic and gene targeting studies showed that the preference for Vγ3 is due in large part to its proximal location to the Jγ1 gene (10, 18). A competition mechanism might also play a role in determining Vδ gene usage in fetal thymocytes, especially considering that the fetal-specific Vδ1 gene is most proximal to the Dδ-Jδ segments.
A recombination signal sequence (RSS)-mediated restriction might also play a role in the regulated TCR gene arrangement. An RSS is composed of a semi-conserved heptamer and nonamer, separated by a poorly conserved 23 or 12 bp spacer, and is the target site of Rag1/2 recombinases for cleaving and joining of the V-D-J segments. Rearrangements occur only between an RSS with a 12 bp spacer (12-RSS) and another with a 23 bp spacer (23-RSS), the so-called 12/23 rule. However, certain rearrangements, such as those between the 23-RSS flanked Vβ segments and the 12-RSS flanked Jβ segments, occur rarely if not at all, even though they are allowed by the 12/23 rule. The restriction beyond the 12/23 rule was suggested to be important for the ordered Vβ–Dβ–Jβ rearrangement to generate vast TCRβ repertoires and was reportedly enforced by the in-compatibilities of the Vβ-RSSs with Jβ-RSSs for RAG-mediated pairing and cleavage (24-26). The RSS-mediated restriction might also play a role in the selective V gene usage for the TCRδ/α gene rearrangement. A “motif” found in spacers of many “Vα”- RSSs was suggested for their preferential rearrangement to Jα fragments (27). However, this motif was found only in 80% of known “Vα” genes and has not been directly demonstrated to be involved in restricting the “Vα” genes for TCRα rearrangement. No motif has been discerned in the RSSs of “Vδ” genes and there is currently no direct evidence that rearrangement of Vδ segments is restricted by the RSS beyond the 12/23 rule. However, considering that no motifs have been defined in RSSs of Vβ/Dβ/Jβ fragments, where the restriction was demonstrated, the possibility of RSS-restricted Vδ usage remains open.
Cellular selection further shapes the development of T cell populations resulting from the regulated TCR rearrangement. Although γδ T cell selection processes are not as well understood as those of αβ T cells, recent studies found that a positive selection is critical, at least, for development of the skin-specific DETCs (28-30). We reported that positive selection of the DETC precursors Vγ3/Vδ1+ γδ T cells in the fetal thymus resulted in a coordinate switch in expression of several chemokine receptors and cytokine receptors important for their localization and expansion in the skin (28). In absence of the positive selection, such as in a sub-strain of FVB mice (FVB/Taconic), few of the fetal thymic Vγ3/Vδ1+ γδ T cells developed into DETCs even though they are generated in normal numbers in the fetal thymus (29).
Although the Vγ3/Vδ1+ γδ T cells are the dominant population of DETCs in wild type mice, other (but not all) subsets of γδ T cells could substitute in the skin, especially in mice deficient of the native Vγ3/Vδ1+ DETCs (31, 32). We reported that a rearranged TCRγ2 transgene (TCRγ2Tg) could restore the development of DETCs in 234JCγ1-kockout (Cγ1-/-) mice that are deleted of the Cγ1 cluster of TCRγ locus (including the DETC-specific Vγ3, as well as Vγ2, 4 and 5 genes) and, therefore, lack the native Vγ3+ DETCs (28). The transgenic Vγ2+ DETCs predominantly expressed endogenous TCRδ genes of Vδ7-Dδ2-Jδ1 rearrangements (TCRδ7) (28), suggesting that they were selected for the DETC development. Supporting this, of multiple subsets of fetal thymic transgenic Vγ2+ γδ T cells that paired with different endogenous TCRδ chains, only the Vγ2/Vδ7+ subset preferentially underwent the positive selection-associated cytokine and chemokine receptor expression, which was important for their homing and expansion in the skin (28). However, factors that affect the DETC development are not fully understood. For example, although the Vγ3-/- mice reportedly had abundant substitute DETCs, of which many expressed Vγ1.1+ γδ TCRs (31), nearly no DETCs developed in the Cγ1-/- mice that generated the Vγ1.1+ γδ T cells normally (28). These observations suggested the existence of additional factors that affect the development of DETCs.
We report here that differential TCRδ gene rearrangement of different strain backgrounds affect the DETC development significantly. Particularly, we found that variations in recombination signal sequences of the Vδ7 genes of different strains were associated with their selective usage for the TCRδ gene rearrangement, which in turn had a dramatic effect on development of a skin-specific γδ T cell population. These findings provide the first compelling evidence that the RSS-associated restriction on TCRδ gene assembly plays an important role in the T cell development.
Materials and Methods
Mice
Cγ1-/-, Vγ3-/- and TCRγ2Tg mice have been described (28, 31). C57BL/6 (B6) and 129 mice were purchased from Jackson laboratory (Maine). The Cγ1-/- mice were originally generated on a pure 129 background (Cγ1-/-129) (28, 31). To move Cγ1-/- on B6 background (Cγ1-/-B6), the Cγ1-/-129 mice were backcrossed to B6 mice for 7-8 generations. Vγ3-/-, TCRγ2Tg and TCRγ2Tg+Cγ1-/- mice, originally on the mixed 129×B6 background (28, 31), were backcrossed to B6 or 129 mice for 7-8 generations to place them on the dominant B6 or 129 backgrounds.
Cell Preparation
Epidermal cells were prepared from the skin as described (33). Thymocytes were prepared by a standard method (34). The specific subsets of skin γδ T cells of TCRγ2Tg mice used for RT-PCR were sorted from the epidermal cells on an EPICS Elite ESP sorter (Beckman-Coulter, Miami, FL) following staining with appropriate antibodies.
Antibodies and flow cytometry
FITC-, biotin-conjugated anti-γδ TCR (GL3), PE-Cy5-conjugated anti-CD3, FITC-conjugated anti-Vγ3 TCR (F536) and anti-Vγ2 TCR (UC3) antibodies were purchased from BD Biosciences (San Diego, CA). PE- or biotin conjugated CD122 antibody was from Ebioscience (San Diego, CA). Biotin-conjugated anti-Vγ3 TCR (F536) and anti-Vγ2 TCR (UC3) antibodies were prepared in house. PE-Cy5- and PE-conjugated streptavidin were purchased from Invitrogen (Eugene, OR). Expression of TCR and other markers on cells were analyzed by multiple color flow cytometry on an EPICS XL instrument (Coulter, Hialeah, FL) following staining with appropriate antibodies. The flow cytometry data were analyzed by FlowJo software (Tree Star Inc, San Carlos, CA).
DNA and RNA preparation
Thymic genomic DNA was prepared as described (9). Total RNA was prepared from sorted γδ T cells by Trizol according to the manufacture's instruction (Invitrogen). To prevent genomic DNA contamination, the RNA samples were treated with RNase-free DNase1 before being used for RT-PCR analysis.
Semi-quantitative PCR and RT-PCR
The semi-quantitative PCR and RT-PCR assays were performed essentially as described (18, 28). To assess TCRδ rearrangements of Vδ7 and its TRAV13 family members, thymic genomic DNA was serially diluted and subjected to PCR with primers TRAV13-FP: CTTGGTTCTGCAGGAGGGGGAGAACGCAGAGC and Jδ1-RP: AATGACTTACTTGGTTCCACAGTCACTTGGGT. The sequence of TRAV13-FP primer complements all TRAV13 (Vδ7) family members while the sequence of Jδ1-RP complements the Jδ1 gene.
For the RT-PCR assessment of Vδ gene usage in sorted skin γδ T cells, RNA samples were reverse transcribed (RT) with Superscript II RNase H- reverse transcriptase using oligo-dT primers. The RT products were serially diluted (indicated in Figures) and subjected to PCR with proper sets of primers. PCR primers for detection of transcripts of various rearranged TCRδ genes and tubulin were described (7, 28).
Sequencing analysis of TCRδ gene rearrangements of the Vδ7 genes
PCR products with the TRAV13-FP/Jδ1-RP primer set were inserted into TOPO TA cloning vectors (Invitrogen), which were used to transform competent E. coli cells. Plasmids containing inserts of the PCR products were extracted from subclones of the transformed E. coli and sequenced for the inserts. Based on the sequence alignment, frequencies of Vδ7 and the other family members in the subclones were calculated. In addition, frequencies of in-frame and out-of-frame rearrangements in the rearranged TCRδ7 genes were also obtained based on the sequencing. Sequence analysis was carried out using NCBI blast, Expasy and Clustal W.
Computation analysis of TRAV13 (Vδ7) family members of TCRα/δ Loci
Genomic DNA sequences of TCRα/δ loci of both B6 and 129 strains are available in public database. The call number for the TCRα/δ sequences of B6 strain is NT_039606 and for 129 is NT_039614. In order to identify all the members of the Vδ7 (TRAV13) family on TCRα/δ loci of B6 and 129 strains, a sequence probe corresponding to the coding sequence of the B6-Vδ7 gene segment was used to blast the genomic sequences of both strains. The hits were aligned to each other for comparison of coding and recombination signal sequences with Clustal W, Expasy and the NCBI sequence alignment programs.
Construction of recombinant substrates and cell-based in vitro recombination assay
Recombinant substrates that contain nucleotide sequences corresponding to B6-Vδ7 RSS and 129-Vδ7 RSS, along with one of 5′Dδ2, 5′Dδ1, Jα48 and Jα56 RSSs, were constructed from a pCMV-based competitive recombinant substrate (35). The basic structure of the competitive recombinant substrate is same as the original construct except that the original RSSs were replaced with RSSs of the two competing Vδ7 and Dδ2 (or Dδ1, Jα48, Jα56) genes, each with 6 nucleotides of their respective endogenous coding flank. There are several nucleotide variations in Vδ7-RSSs of B6 and 129 strains while RSSs of Dδ2, Dδ1, Jα48 and Jα56 genes of the two strains are identical (refer to the Result Section for details). All the substrates were sequence confirmed before used in a cell-based in vitro recombination assay.
The cell-based recombination assay was also performed as previously reported (35). Briefly, HEK293T cells were transiently transfected with the recombinant substrate and Rag1 and Rag2-expressing constructs. Two-three days after the transfection, plasmid DNAs were recovered from the transfected cells and used in a PCR reaction to detect rearranged Vδ7 genes. The two primers used in the PCR are complementary to common sequences of the pCMV vector that flank the recombinant substrates (35). The PCR products were run on 2% agarose gel and intensities of the bands corresponding to rearrangements of the two competing Vδ7 RSSs were compared.
Results
Differential development of skin γδ T cells in Vγ3 or Cγ1 knockout mice of the B6 versus 129 genetic background
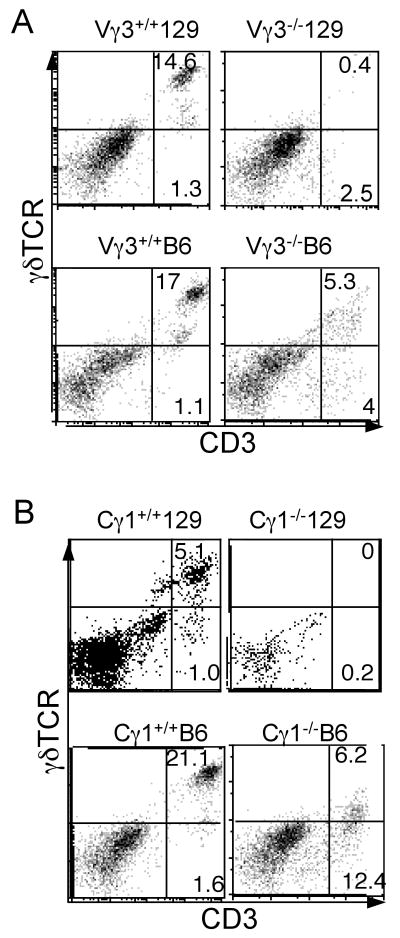
To determine whether genetic background affects the development of substitute DETC in mice deficient of the native skin-specific Vγ3+ γδ T cells, we backcrossed Vγ3-/- mice onto the C57BL/6 (B6) and 129 strain background and compared the development of their skin γδ T cells. Similarly, we compared the development of skin γδ T cells in Cγ1-/- mice of the B6 vs. 129 background. Wild-type littermates of both the B6 and 129 backgrounds contained abundant γδTCR+ DETCs, as expected (Fig. 1). However, the development of the skin γδ T cells was strikingly different in the Vγ3-/- (or Cγ1-/-) mice of B6 and 129 backgrounds. Compared to those of their respective wild type littermates, skin γδ T cells were nearly absent (>30 fold reduction) in the Vγ3-/- (or Cγ1-/-) mice of the 129 background (Fig. 1A&B, top panels). In contrast, skin γδ T cells were readily detectable in Vγ3-/- (or Cγ1-/-) mice of the B6 background, whose numbers were only reduced 3-4 fold compared to their wild-type littermate controls (Fig. 1A&B, bottom panels). Hence, substitute DETCs developed efficiently in Vγ3-/- (or Cγ1-/-) mice of the B6 but not 129 strain background, suggesting that the strain genetic background is an important factor in determining development of the skin-specific γδ T cells.
Figure 1.
Different developmental potentials of skin γδ T cells in Vγ3-/- or Cγ1-/- mice of 129 vs. B6 background. Epidermal cells were prepared from the skin of Vγ3 knockout (panel A) or 234JCγ1 (Cγ1) knockout mice (panel B) of 129 and B6 background and analyzed by flow cytometry for CD3 and γδTCR expression. The data shown in this figure, as well as in the other figures, are representatives of at least two independent experiments with similar results.
Differential development of transgenic Vγ2+ skin γδ T cells in mice of the B6 versus 129 background
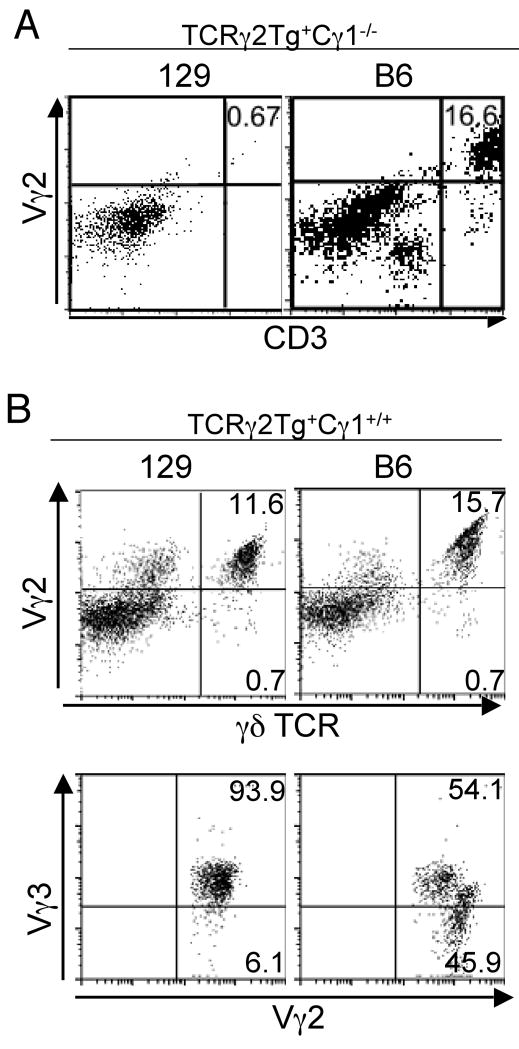
Published results found that the substitute DETCs in Vγ3 knockout mice were somewhat heterogeneous populations (31). We reasoned that efforts to dissect the role of background genes in the development of substitute DETCs would be facilitated by simplifying the repertoire of these cells. Since the transgenic Vγ2+ γδ T cells could develop into DETCs which predominantly co-expressed the endogenous TCRδ7 gene in a mixed B6×129 background (28), we compared development of the transgenic Vγ2+ DETCs in TCRγ2Tg+Cγ1-/- mice of the B6 and 129 background (TCRγ2Tg+Cγ1-/-B6 or TCRγ2Tg+Cγ1-/-129). The transgenic Vγ2+ DETCs developed efficiently in the TCRγ2Tg+Cγ1-/- mice of the B6 background but not in those of the 129 background (Fig. 2A). Thus the strain background also affected development of the transgenic Vγ2+ DETCs.
Figure 2.
Differential development of transgenic Vγ2+ skin γδ T cells in TCRγ2 transgenic mice of the 129 vs. B6 background. A. Development of transgenic Vγ2+ skin γδ T cells is efficient in TCRγ2Tg+Cγ1-/- mice of B6 background but defective in those of 129 background. Epidermal cells were prepared from TCRγ2Tg+Cγ1-/- mice of the indicated backgrounds and assessed for percentages of the transgenic Vγ2+ γδ T cells by flow cytometric analysis of anti-TCRδ and Vγ2 antibody staining. B. The Vγ2+Vγ3- skin γδ cells developed only in TCRγ2Tg mice of B6 background while the Vγ2+Vγ3+ skin γδ T cells developed in TCRγ2Tg mice of both B6 and 129 backgrounds. Epidermal cell preparations were stained and analyzed by flow cytometry for Vγ2+ and total γδ T cells (top panels). The Vγ2+ γδ T cells were further gated for analysis of Vγ3 expression (bottom panels).
Analysis of TCRγ2Tg+Cγ1+/+ mice of the B6 or 129 background that retain the endogenous TCRγ locus provided additional information. Unlike in the TCRγ2Tg+Cγ1-/-129 mice, abundant Vγ2+ DETCs developed in the TCRγ2Tg+Cγ1+/+ mice of the 129 background, which co-expressed the endogenous Vγ3 (Fig. 2B, left panels), suggesting that their development was dependent on co-expression of the endogenous Vγ3, reminiscent of a previous report that the development of transgenic TCRδ6.3+ DETCs depended on co-expression of endogenous TCRδ genes (36). In contrast, many Vγ2+ DETCs in TCRγ2Tg+Cγ1+/+ mice of the B6 background did not co-express the Vγ3, which could be also distinguished from the Vγ2+Vγ3+ DETC subset by their higher levels of Vγ2 staining (Fig. 2B, right panels). These data demonstrate that the Vγ3 expression is critical for the DETC development in the 129 strain background, but the transgenic Vγ2+ γδ T cells can substitute Vγ3 cells for the DETC development in the B6 background.
Defective development of the transgenic Vγ2+ skin γδ T cells on the 129 background results from absence of their positively selected precursors in the fetal thymus
Since DETCs originate from their precursor cells in the fetal thymus, which are positively selected to express the chemokine receptors, such as CCR10, and cytokine receptors, such as CD122, that promote their epidermal localization and proliferation (28), we analyzed selection status of the transgenic Vγ2+ fetal thymic cells of the B6 and 129 backgrounds, using the CD122 upregulation as a positive selection marker (28).
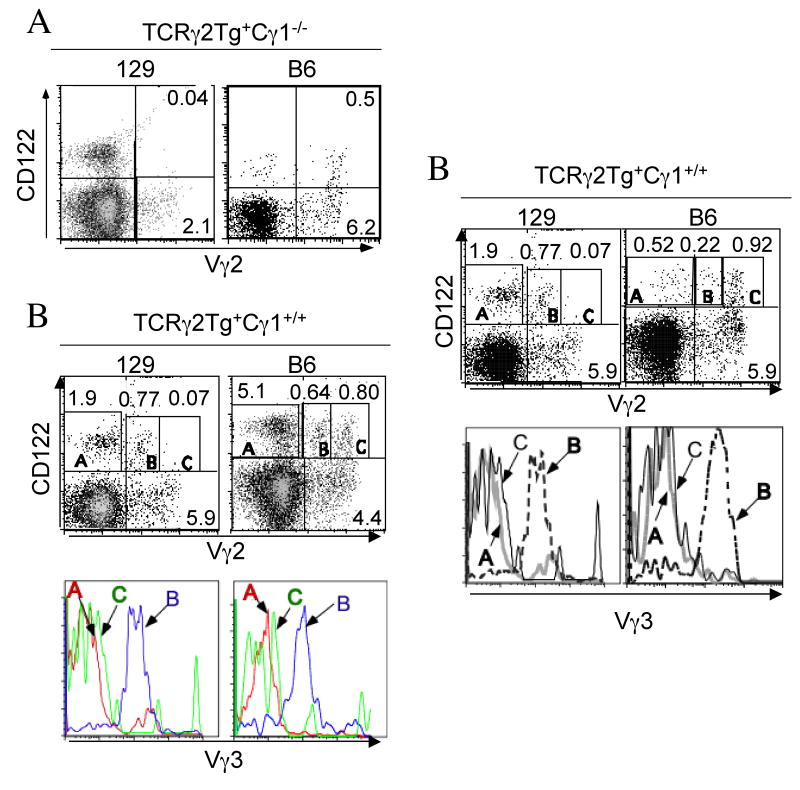
In the TCRγ2Tg+Cγ1-/-129 mice, there was essentially no CD122 upregulation on the transgenic Vγ2+ fetal thymic T cells (Fig. 3A, left panel), consistent with their absence in the skin (Fig. 2A). In contrast, CD122 was upregulated in a significant fraction of the transgenic Vγ2+ fetal thymocytes of the TCRγ2Tg+Cγ1-/-B6 mice (Fig. 3A, right panel), in which the transgenic Vγ2+ DETCs developed (Fig. 2A).
Figure 3.
Different positive selection processes of the transgenic Vγ2+ γδ T cells in fetal thymi of 129 vs. B6 background. A. Presence of positively selected transgenic Vγ2+ γδ T cells in the TCRγ2Tg+Cγ1-/- mice of the B6 but not 129 background. E16 fetal thymocytes of the TCRγ2Tg+Cγ1-/- mice of the 129 or B6 background were analyzed by flow cytometry for expression of CD122 on the transgenic Vγ2+ γδ T cells. B. The positively selected CD122+ Vγ2high+Vγ3- fetal thymic T cells were detected only in the TCRγ2Tg mice of the B6 background while the Vγ2medium+Vγ3+ fetal thymic T cells were positively selected in the TCRγ2Tg mice of both B6 and 129 backgrounds. E16 fetal thymocytes of the TCRγ2Tg mice of both backgrounds were analyzed by flow cytometry for expression of Vγ2, Vγ3 and CD122. The histographs in the top panels were gated on total thymocytes while the histographs in the bottom panels were gated on the CD122+ Vγ2− (A), CD122+Vγ2medium+ (B) or CD122+Vγ2high+ (C) population of the top panels (as indicated) and analyzed for Vγ3 TCR expression. Note that only the CD122+Vγ2medium+ population co-expressed Vγ3+ TCR. The CD122+ Vγ2- population (A) was mostly NK cells.
Analysis of the TCRγ2Tg+Cγ1+/+ mice further confirmed the correlation between positive selection of the specific subsets of transgenic Vγ2+ fetal thymic T cells and their development into DETCs. Although CD122 was upregulated on significant percentages of Vγ2+ fetal thymic γδ T cells in the TCRγ2Tg+Cγ1+/+ mice of both B6 and 129 backgrounds, nearly all the CD122+ fetal thymic Vγ2+ cells of the 129 background had intermediate levels of Vγ2 expression and co-expressed Vγ3 (Fig. 3B, left panels, Population B), consistent with development of the Vγ2+Vγ3+ DETCs in these mice (Fig. 2B). On the other hand, in the TCRγ2Tg+Cγ1+/+B6 mice, besides the CD122+ Vγ2medium+Vγ3+ fetal thymic T cell population, there was an additional CD122+ Vγ2high+Vγ3- population (Fig. 3B, right panels, Population C), consistent with the development of such a subset of DETCs in these mice (Fig. 2B). The fact that the Vγ2+Vγ3+ but not Vγ2+Vγ3- fetal thymic population underwent the positive selection and developed into DETCs in the TCRγ2Tg+129 mice indicates that the defective DETC development of the latter is not likely due to any defective signaling process downstream of the TCR-mediated positive selection of the fetal thymic DETC precursors. Instead, these data suggest that the defect is in generation of the Vγ2+Vγ3− DETC precursors or/and molecules that mediate their selection.
Mice of the 129 background have an intrinsic defect in the rearrangement of TCRδ7 genes that were expressed in the transgenic Vγ2+ skin γδ T cells of the B6 background
Since the transgenic Vγ2+(Vγ3−) skin γδ T cells in the TCRγ2Tg+Cγ1-/- mice on a mixed B6×129 background predominantly expressed an endogenous TCRδ7 gene and originated from their positively selected fetal thymic precursors (28), we decided to test the possibility that the differential development of these cells in the B6 and 129 background mice is due to differential generation or selection of the fetal thymic Vγ2/Vδ7+ γδT cells between the two backgrounds. First, we confirmed association of TCRδ7 expression with the Vγ2+Vγ3− DETCs in the B6 background mice by analyzing TCRδ gene usage in sorted skin γδ T cell subsets from TCRγ2Tg+Cγ1+/+ mice of the B6 or 129 background (Fig. 4A). The Vγ2+Vγ3+ skin γδ T cells of both backgrounds expressed same levels of TCRδ1 transcripts as the Vγ3+ DETCs of wild-type B6 mice but rarely any other Vδ transcripts (Fig. 4A), suggesting that the development of this DETC subset is mediated by the endogenous Vγ3/Vδ1+ TCR even though the transgenic TCRγ2 was co-expressed. In contrast, the Vγ2+Vγ3− DETC subset of the B6 background had at least 20-fold reduction in TCRδ1 transcripts but predominantly expressed TCRδ7 (Fig. 4A). These data confirm that the Vγ2/Vδ7+ γδ T cell population is selected for development into skin γδ T cells in the B6 background mice and are consistent with the idea that absence of such a population in the 129 background mice is due to defective TCRδ7 rearrangement/expression or failed positive selection of the Vγ2/Vδ7+ population if they are ever generated.
Figure 4.
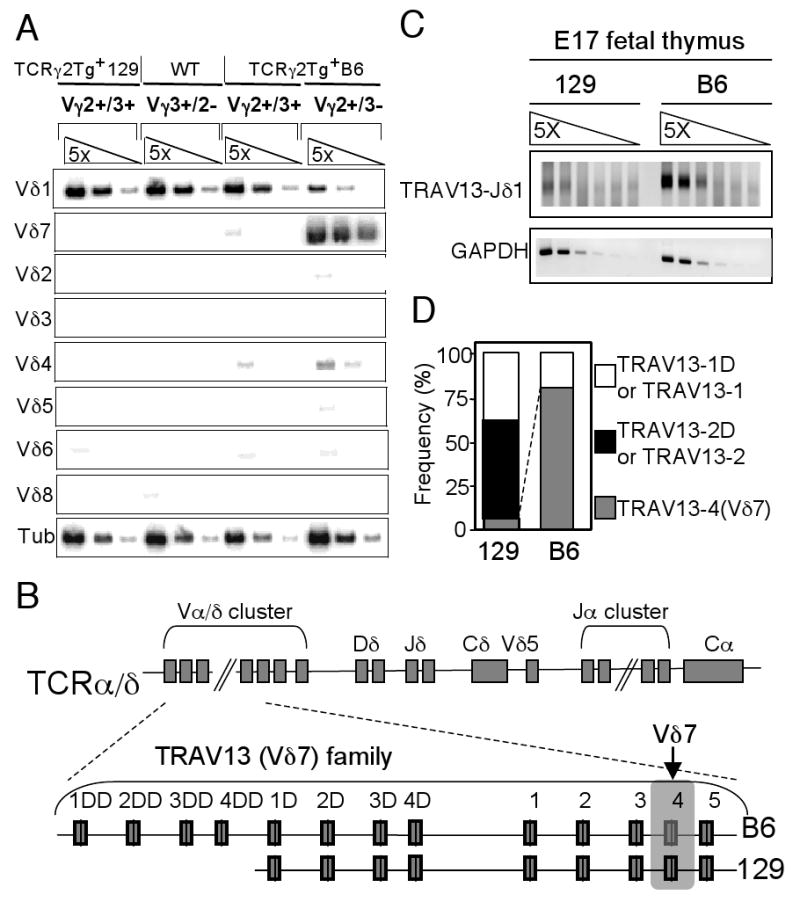
Intrinsically defective rearrangement of TCRδ7 gene in mice of the 129 background. A. Vδ7+ TCRδ gene is predominantly expressed only in the transgenic Vγ2+Vγ3 skin γδ T cells of the TCRγ2Tg+B6 mice while Vδ1+ TCRδ gene is predominantly expressed in the Vγ2+Vγ3+ skin γδ T cells of both B6 and 129 backgrounds. RNA was prepared from sorted populations of skin γδ T cells with different Vγ2 and Vγ3 expression of the indicated mouse strains and analyzed for Vδ gene usage by semi-quantitative RT-PCR. RT-PCR for β-tubulin (tub) was used as loading controls. B. Genomic organization of TRAV13 (Vδ7) family members in the TCRα/δ loci of the B6 vs. 129 strain. The Vδ7 gene is the second from the right in the family, as indicated. C. Comparison of TCRδ gene rearrangements of the TRAV13 (Vδ7) family members in the TCRγ2Tg mice of the 129 and B6 background. The genomic DNAs of E17 fetal thymocytes were serially diluted and analyzed by semi-quantitative PCR. GAPDH was used as loading controls. D. 129-“Vδ7” was rarely used in TCRδ rearrangements of the TRAV13 family members while B6-Vδ7 gene was predominantly used. Subcloned PCR products of 129 or B6 genomic DNAs in the panel C were sequenced and frequencies of the TCRδ rearrangements of Vδ7 genes and the other family members were plotted.
To distinguish these two possibilities, we compared levels of TCRδ7 rearrangements in fetal thymi of the TCRγ2Tg+ mice of 129 and B6 backgrounds by semi-quantitative PCR. The Vδ7 gene belongs to a closely related family of Vα/δ genes (TRAV13 family according to the IMGT nomenclature), of which only Vδ7 (also designated TRAV13-4) is known to rearrange predominantly to Dδ-Jδ gene segments for the TCRδ assembly in adult wild-type B6 mice while all the other family members are preferentially used for the TCRα rearrangements (14, 22). The TRAV13 family consists of 5 founding members, located at the 3′ region of the Vα/δ gene cluster, four of which were duplicated once in the 129 strain and twice in the B6 strain (Fig. 4B, assembled based on mouse genomic database, refer to Materials and Methods) (Nucleotides sequences of these family members are also compiled in an online database of http://imgt.cines.fr). The Vδ7 gene is the second to last member of the founding group and is therefore the second most proximal of the entire family in position to the Dδ/Jδ (Fig. 4B). The coding sequence of the B6-Vδ7 gene is 98% identical to that of the 129-“Vδ7” gene and all the family members are at least 81% identical to each other in both strains (Supplementary Figure 1). We used a forward primer complementary to all members of the TRAV13 family (TRAV13 primer) and a common reverse Jδ1 primer for the PCR, which would amplify rearrangements of all the TRAV13 family members, including Vδ7. The rearrangements to Jδ1 were assessed because the Vγ2/Vδ7+ DETCs expressed TCRδ of Vδ7-Dδ2-Jδ1 rearrangements (28). Remarkably, the TRAV13-Jδ1 rearrangements were approximately 25-fold less abundant in the 129 background mice than in the B6 (Fig. 4C), suggesting a severe defect in the TCRδ gene rearrangement of TRAV13 family members, most likely of the Vδ7, in the fetal thymi of 129 background mice.
There was a possibility that the difference in TRAV13-Jδ1 rearrangements between the B6 and 129 strains was due not to their differential rearrangement efficiencies, but rather due to differences in the positive selection of the transgenic Vγ2+ fetal thymic cells expressing the rearranged TCRδ7. If this is the case, the higher level of rearrangements in the B6 background mice should result from the positive selection-mediated enrichment of the Vγ2/Vδ7+ population, and, consequently, most of the rearranged TCRδ7 genes should be in-frame and productive. On the other hand, if the positive selection does not result in the enrichment of the Vγ2/Vδ7+ fetal thymic population, it is predicted that only 1/3 of the rearrangements should be in-frame. Therefore, we sequenced subcloned PCR products of the TRAV13-Jδ1 rearrangements to determine frequencies of in-frame versus out-of-frame rearrangements. The sequencing also allowed us to determine percentages of the rearrangements of Vδ7 gene vs. the other members of the TRAV13 family. Based on the sequencing, a vast majority of the rearrangements, ∼83% (10/12 subclones), of the B6 genomic DNA were of Vδ7 (Fig. 4D, right column), demonstrating that Vδ7 is the only member of the TRAV13 family that predominantly rearranges for TCRδ genes in the fetal thymus. Importantly, 70% (7/10) of the Vδ7-Jδ1 rearrangements were out of frame, arguing strongly against the possibility that the higher level of Vδ7 rearrangements in B6 fetal thymocytes is due to the positive selection-mediated enrichment of cells bearing this receptor chain. Instead, the data suggest that the Vδ7 rearrangement efficiency for TCRδ gene assembly in the 129 fetal thymocytes is intrinsically lower than in the B6. Confirming this, we sequenced subcloned PCR products of TRAV13-Jδ1 rearrangements of the 129 background mice and found that only 4% (1/24 subclones) of the rearrangements was of the 129-“Vδ7” gene (Fig. 4D, left panel). Together, these data demonstrate that the absence of Vγ2/Vδ7+ DETCs in the 129 background mice is due to an intrinsic defect in the rearrangement of TCRδ7 genes and, consequently, the failed generation of such a γδ T cell population in the fetal thymus.
Association of variations in recombination signal sequences of Vδ7 genes of the B6 and 129 strains with their different preferences for TCRδ rearrangement
The impaired TCRδ rearrangement of the “Vδ7”gene in the 129 background mice is likely due to local Vδ7 gene variations but not global TCRδ gene rearrangement deficiency, since percentages of total transgenic Vγ2+ fetal thymocytes were not significantly different in the TCRγ2Tg+ mice of 129 and B6 backgrounds (Fig. 3A and B). Therefore, we compared genomic sequences of the Vδ7 genes of 129 and B6 strains for variations. Interestingly, there were significant differences in their recombination signal sequences (RSSs), which became very striking when aligned with RSSs of other members of the TRAV13 family of both strains (Table 1). Compared to those of the 129-“Vδ7” and all the other TRAV13 family members, RSS of the B6-Vδ7 is remarkably unique. Specifically, the first nucleotide of the 23bp spacer in the B6-Vδ7 RSS is an adenine (A), whereas the corresponding site of the 129-“Vδ7” and other family members is a cytosine (C). In addition, the fourth nucleotide in the nonamer of the B6-Vδ7 RSS is a thymine (T) while the corresponding site is a cytosine (C) in all the others. The RSS of the 129-“Vδ7” gene is more similar to those of the other TRAV13 family members than that of the B6-Vδ7. In contrast to the significant variations in their Vδ7 RSSs, there were no differences at all in RSSs of Dδ and Jδ gene segments between the 129 and B6 strains (Supplementary Figure 2). Since the recombination signal nonamer and spacer have been reported to be able to mediate restriction on the TCRβ gene rearrangement (37), the variations in the 129-“Vδ7” RSS might be responsible for its impaired TCRδ rearrangement.
Table 1.
Sequence alignment of recombination signal sequences of Vδ7 and the other TRAV13 family members of B6 and 129 strains
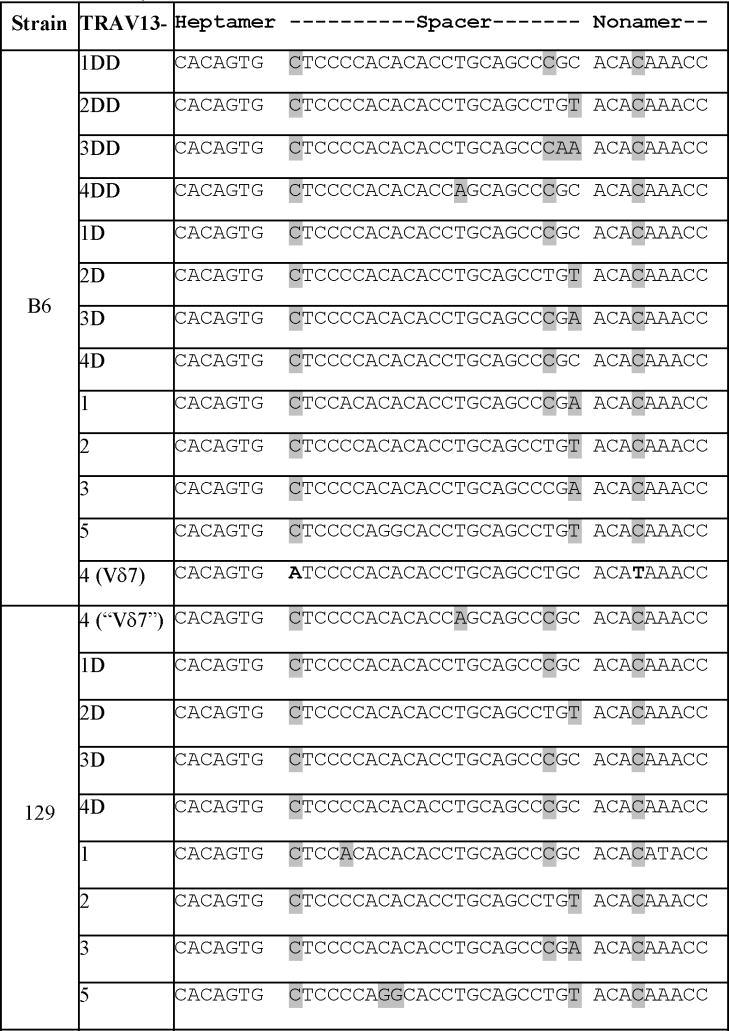 |
Note: The shadowed letters represent nucleotides that differ from those of the B6-Vδ7 RSS.
To test this, we used a cell-based in vitro recombination assay to directly compare rearrangement efficiencies of the 129-“Vδ7” RSS and B6-Vδ7 RSS to Dδ2 gene segment, which was used in all TCRδ7 rearrangements of the transgenic Vγ2/Vδ7+ skin γδ T cells (28). To this purpose, we assembled a competitive recombinant substrate (p129/B6) that contains the 129-“Vδ7” RSS, B6-Vδ7 RSS and the 5′RSS of Dδ2 gene, subcloned into a pCMV-based vector (Fig. 5A, top panel) (35). To correct any potential effect of the position on rearrangement efficiencies of the two competing Vδ7 genes, a recombinant substrate containing two identical B6-Vδ7 RSSs (pB6/B6) were used as a control (Fig. 5A, bottom panel). The substrates, along with Rag1 and Rag2-expressing vectors, were transiently transfected into 293 cells, and rearrangements of the two competing Vδ7 RSSs to Dδ2 were assessed by PCR with a primer set located outside the recombinant substrates, which distinguish rearrangements of the two competing Vδ7 RSSs based on their different sizes (Fig. 5A and B).
Figure 5.
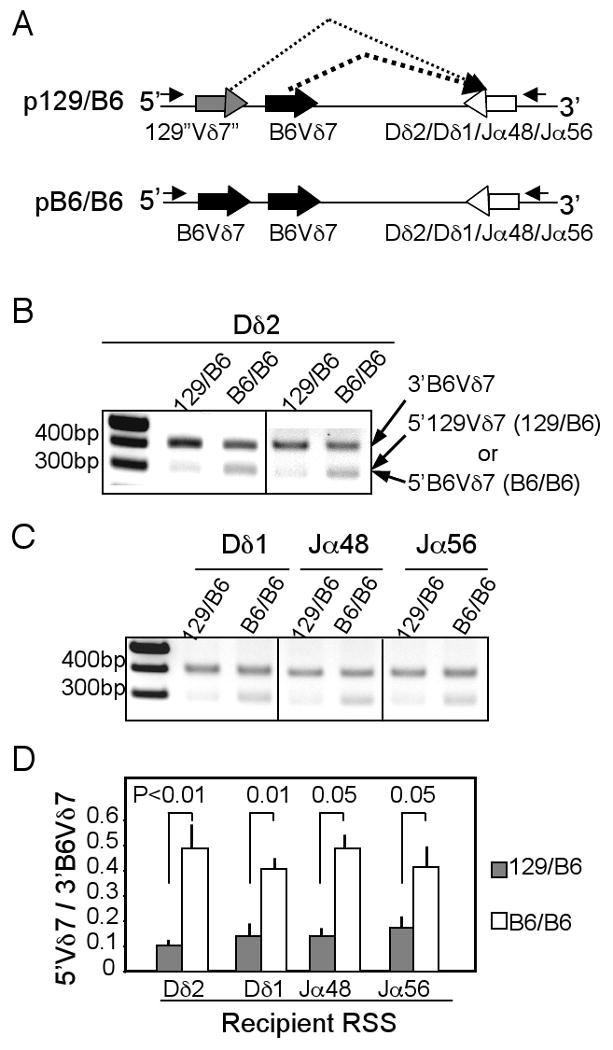
Comparison of rearrangement efficiencies of B6-Vδ7 RSS vs. 129-“Vδ7”RSS in an in vitro recombination assay. A. Schematic depiction of competitive recombination substrate constructs. Two “Vδ7” RSSs were assembled in the same construct with a recipient RSS of Dδ2, Dδ1, Jα48 or Jα56 gene and would compete for rearrangement to the recipient RSS when the construct was transfected into cells expressing Rag1/2 genes. Rearrangements of the two competing Vδ7 RSSs were assessed by PCR with primers located in 5′ and 3′ flanking regions of the substrate (indicated by the two small arrows), which gave different sizes of products. B. Analysis of rearrangements of the two competing “Vδ7” RSSs to the Dδ2 RSS. Plasmid DNAs recovered from recombinant substrates-transfected cells were subject to the PCR as depicted in the panel A and gel analyzed. Bands of 400bp correspond to rearrangements of the 3′ B6-Vδ7 while those of 300bp correspond to rearrangements of the 5′ 129-Vδ7 (129/B6) or 5′ B6-Vδ7 (B6/B6). C. Analysis of rearrangements of two competing “Vδ7” RSSs to the recipient RSS of Dδ1, Jα48 or Jα56 gene, similar as in the penal B. D. Quantification of relative rearrangement efficiencies of the 129-“Vδ7” RSS vs. B6-Vδ7 RSS of the same position to the Dδ2, Dδ1, Jα48 or Jα56 RSS based on analyses of the panels B and C. The efficiency was expressed as a ratio of intensities of the 5′Vδ7 (129 or B6) vs. the common 3′B6Vδ7 rearrangement bands.
In cells transfected with substrates containing the competing 129 and B6-Vδ7 RSSs, the distal 5′129-Vδ7 RSS rearranged much less efficiently (about ten-fold) to the Dδ2 RSS than the proximal 3′B6-Vδ7 RSS did (Fig. 5B and D). In contrast, in cells transfected with substrates containing the two identical B6-Vδ7 RSSs, the distal B6-Vδ7 RSS rearranged to the Dδ2 RSS only slightly less efficiently (about two-fold) than the proximal B6-Vδ7 RSS (Fig. 5B and D), likely due to a minor position effect that was observed previously (37) and reportedly affected by distances between different RSSs (38), among other factors. Normalized on rearrangements of the common proximal B6-Vδ7-RSS to factor in the position effect, the 129-“Vδ7” RSS still rearranged significantly less efficiently than the B6-Vδ7 RSS of same distal position (4.7±1.3 fold reduction, n=3), demonstrating that the variations in 129-“Vδ7” RSS impair its rearrangement efficiency for TCRδ gene assembly.
To test further whether the reduced rearrangement efficiency of 129-Vδ7 RSS is restricted to the Dδ2 RSS, we also constructed competitive recombinant substrates containing the two competing Vδ7-RSSs with an RSS of Dδ1, Jα48 or Jα56 gene fragment (Fig. 5A). The Dδ1, Jα48 or Jα56 RSSs are identical between 129 and B6 strains (Supplementary Fig. 2). Same as for the rearrangement to Dδ2 RSS, the 129-Vδ7 RSS also rearranged less efficiently than the B6-Vδ7 RSS of same position to the Dδ1 RSS (3.0±0.7, n=2) (Fig. 5C and D). In addition, the variations also rendered the 129-Vδ7 RSS less compatible for rearrangement to the tested Jα RSSs (3.5±0.7 and 2.4±0.2 fold reduction to Jα48 and Jα56 respectively, n=2 each), suggesting that the reduced TCRδ gene rearrangement of 129-Vδ7 RSS is probably not due to restriction beyond the 12/23 rule. Together, these results demonstrate that a dysfunctional 129-Vδ7 RSS is at least in part responsible for the defective TCRδ7 rearrangement and transgenic Vγ2/Vδ7+ DETC development in 129 background mice.
Discussion
The proper rearrangement of TCR genes is critical for the T cell development. Since both TCRα and TCRδ loci share the same Vα/δ gene segments, unique questions with these loci are how the shared Vα/δ segments are selectively used for the TCRα/δ gene assembly and its effect on the development of αβ/γδ T cells. Here, we reported that differential TCRδ7 gene rearrangement in fetal thymi of the B6 and 129 backgrounds profoundly affected development of a skin-specific γδ T cell population. Furthermore, we found that the variations in RSSs of the Vδ7 genes of the two strains were associated with their preferential usage for the TCRδ gene assembly, demonstrating for the first time a biologically important role of the RSS-associated restriction of Vα/δ rearrangement in the T cell development. In addition, our findings provided further support for a critical role of positive selection in development of the skin-specific γδ T cells.
Although it has been suggested for a long time that the selective Vα/δ usage plays an important role in T cell development, the evidence is lacking. Our findings provide the first compelling evidence that the selective Vδ gene usage for TCRδ rearrangement affects development of the skin-specific T cell population. However, effects of selective TCRδ gene rearrangement on the development of different tissue-specific T cell populations might vary. Unlike the skin γδ T cells, development of γδ T cells in secondary lymphoid organs such as spleens was not significantly different in the Vγ2 transgenic mice of the 129 and B6 backgrounds (data not shown). Possibly, innate T cell populations, such as the skin-specific γδ T cells that use restricted TCR composition, are more easily affected by the selective Vδ gene usage than T cell populations with diverse TCR composition, such as the splenic γδ T cells. However, even in the latter, the differential usage of Vδ genes might still affect the T cell repertoire formation and possibly immune functions.
The finding that the differential rearrangement of the Vδ7 genes significantly affected the skin-specific γδ T cell development also provides further support for a critical role of positive selection in the development of this tissue-specific γδ T cell population. In spite of the defective TCRδ7 rearrangement, abundant transgenic Vγ2+ γδ T cells were still generated in the TCRγ2Tg+Cγ1-/-129 mice. But they were not positively selected and did not develop into the skin γδ T cells (Fig. 2 and 3), demonstrating that only specific γδ T cell populations could be positively selected in the fetal thymus and develop into the skin γδ T cells. Events that impair the generation of these specific γδ T cells in the fetal thymus, as is the case for the Vγ2/Vδ7+ γδ T cells in the TCRγ2Tg+Cγ1-/-129 mice, will therefore impair the skin-specific γδ T cell development.
Cellular selection has been also suggested to explain relatively abundant Vγ2/Vδ7+ γδ T cells in spleens of the adult B6 than in Balb/c mice (39). However, the difference in thymic TCRδ7 gene expression between B6 and Balb/c mice (<2 fold) is much smaller than that of the TCRδ7 rearrangements between B6 and 129 mice, suggesting that different underlying mechanisms might operate in these two settings. Likely, the difference in the splenic Vγ2/Vδ7+ γδ T cells between the B6 and Balb/c mice was resulting from the post-thymic selection events while the impaired TCRδ7 rearrangement and generation of the corresponding Vδ7+ γδ T cells in the fetal thymus is predominantly responsible for the failed development of skin Vγ2/Vδ7+ γδ T cells in the 129 background mice.
Interestingly, the impaired rearrangement of TCRδ7 genes in the 129 background mice is associated with the variations in recombination signal sequences of the Vδ7 gene segments, suggesting a role of the RSS in their restricted usage for TCRδ gene rearrangement. Supporting this, an in vitro rearrangement assay demonstrated directly that the 129-“Vδ7”RSS rearranges to the RSS of Dδ2 segment much less efficiently than the B6-Vδ7 RSS does. This also suggests a possibility that the variations in the B6-Vδ7 RSS may represent a nucleotide “motif” in restricting the variable gene usage for TCRδ assembly. However, such variations were not found in many other known Vδ genes (data not shown). In fact, the B6-Vδ7 RSS, but not the 129-“Vδ7” RSS, contains a palindrome sequence (CTGCAG) in its 23-bp spacer that were previously found in 80% of “Vα” gene segments (27), suggesting that no common Vδ or Vα “motif” exists in determining their preferential TCRα/δ rearrangements. Consistent with this, there was no apparent correlation between reported TCRα/δ rearrangement preferences and RSSs among members of several other Vα/δ gene families (data not shown).
In contrast to the stringent restriction imposed by RSSs on Vβ and Jβ genes to prevent the direct Vβ-Jβ rearrangement, the selective usage of Vα/δ genes for the TCRα and TCRδ gene rearrangement is relatively loose. Although there are apparent preferences in the usage of share Vα/δ segments for TCRα and TCRδ rearrangement, many of the Vα/δ genes could be used for both (22), suggesting compatibility of their RSSs with those of both Dδ/Jδ and Jα segments. However, such promiscuity might make them less fully compatible to a specific type of RSSs and more easily affected by individual variations, which could alter rearrangement preferences of specific Vα/δ genes and affect the development of T cells, especially those that use limited TCR composition, as we report here. Therefore, the variations in RSSs of the Vα/δ gene segments might represent a major factor in determining γδ and αβ T repertoire formation among individuals. Paralleling this, RSS variations were also reported to affect rearrangement efficiencies in immunoglobulin (Ig) gene loci. In one report, it was found that a single base-pair change in the spacers or nonamers could affect recombination frequency of a VH gene with significant biological consequences (40).
Our data demonstrate that the variations in the 129-Vδ7 RSS affect its rearrangement efficiencies for TCRδ gene assembly, which likely contributes to the defective TCRδ7 rearrangements and skin γδ T cell development of the 129 background mice. However, the exact contribution of the RSS variations in these processes remains to be assessed in vivo, by using techniques such as the knocking-in of the B6-Vδ7 RSS into the 129 allele or vice versa. In addition, its effect is superimposed on other regulation mechanisms, such as the gene accessibility, which was known to be a prerequisite for selective usage of Vα/δ genes in the TCRα and TCRδ rearrangement at different developmental stages (23). It is possible that multiple factors operate to regulate the differential Vδ7 gene rearrangement in the different genetic backgrounds.
Supplementary Material
Acknowledgments
We thank Adrian Hayday for the Vγ3 knockout mice, Ferenc Livak for plasmids and advice on the in vitro recombination assay, Hector Nolla for assistance in cell sorting and Christina Saylor for technical support.
This work was supported by grants from the National Institutes of Health (to N.X and D. H. R) and, in part, under a grant with the Pennsylvania Department of Health using Tobacco Settlement Funds (to N.X.). The Department specifically disclaims responsibility for any analyses, interpretations or conclusions.
Footnotes
Disclosures: The authors declare that they have no conflict of interest.
References
- 1.Raulet D, Spencer D, Hsiang YH, Goldman J, Bix M, Liao NS, Zijlstra M, Jaenisch R, Correa I. Control of γδ T cell development. Immunol Rev. 1991;120:185–204. doi: 10.1111/j.1600-065x.1991.tb00592.x. [DOI] [PubMed] [Google Scholar]
- 2.Xiong N, Raulet DH. Development and selection of gammadelta T cells. Immunol Rev. 2007;215:15–31. doi: 10.1111/j.1600-065X.2006.00478.x. [DOI] [PubMed] [Google Scholar]
- 3.Girardi M, Oppenheim DE, Steele CR, Lewis JM, Glusac E, Filler R, Hobby P, Sutton B, Tigelaar RE, Hayday AC. Regulation of cutaneous malignancy by gammadelta T cells. Science. 2001;294:605–609. doi: 10.1126/science.1063916. [DOI] [PubMed] [Google Scholar]
- 4.Jameson J, Ugarte K, Chen N, Yachi P, Fuchs E, Boismenu R, Havran WL. A role for skin gammadelta T cells in wound repair. Science. 2002;296:747–749. doi: 10.1126/science.1069639. [DOI] [PubMed] [Google Scholar]
- 5.Hayday A, Tigelaar R. Immunoregulation in the tissues by gammadelta T cells. Nat Rev Immunol. 2003;3:233–242. doi: 10.1038/nri1030. [DOI] [PubMed] [Google Scholar]
- 6.Havran WL, Allison JP. Origin of Thy-1+ dendritic epidermal cells of adult mice from fetal thymic precursors. Nature. 1990;344:68–70. doi: 10.1038/344068a0. [DOI] [PubMed] [Google Scholar]
- 7.Goldman J, Spencer D, Raulet D. Ordered rearrangement of variable region genes of the T cell receptor gamma locus correlates with transcription of the unrearranged genes. Journal of Experimental Medicine. 1993;177:729–739. doi: 10.1084/jem.177.3.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Itohara S, Mombaerts P, Lafaille J, Iacomini J, Nelson A, Clarke AR, Hooper ML, Farr A, Tonegawa S. T cell receptor δ gene mutant mice: independent generation of αβ T cells and programmed rearrangements of γδ TCR genes. Cell. 1993;72:337–348. doi: 10.1016/0092-8674(93)90112-4. [DOI] [PubMed] [Google Scholar]
- 9.Baker JE, Cado D, Raulet DH. Developmentally programmed rearrangement of T cell receptor Vγ genes is controlled by sequences immediately upstream of the Vγ genes. Immunity. 1998;9:159–168. doi: 10.1016/s1074-7613(00)80598-1. [DOI] [PubMed] [Google Scholar]
- 10.Xiong N, Zhang L, Kang C, Raulet DH. Gene placement and competition control T cell receptor gamma variable region gene rearrangement. J Exp Med. 2008;205:929–938. doi: 10.1084/jem.20071275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weber-Arden J, Wilbert OM, Kabelitz D, Arden B. V delta repertoire during thymic ontogeny suggests three novel waves of gamma delta TCR expression. J Immunol. 2000;164:1002–1012. doi: 10.4049/jimmunol.164.2.1002. [DOI] [PubMed] [Google Scholar]
- 12.Krangel MS, Carabana J, Abbarategui I, Schlimgen R, Hawwari A. Enforcing order within a complex locus: current perspectives on the control of V(D)J recombination at the murine T-cell receptor alpha/delta locus. Immunol Rev. 2004;200:224–232. doi: 10.1111/j.0105-2896.2004.00155.x. [DOI] [PubMed] [Google Scholar]
- 13.Gallagher M, Candeias S, Martinon C, Borel E, Malissen M, Marche PN, Jouvin-Marche E. Use of TCR ADV gene segments by the delta chain is independent of their position and of CD3 expression. Eur J Immunol. 1998;28:3878–3885. doi: 10.1002/(SICI)1521-4141(199811)28:11<3878::AID-IMMU3878>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 14.Pereira P, Hermitte V, Lembezat MP, Boucontet L, Azuara V, Grigoriadou K. Developmentally regulated and lineage-specific rearrangement of T cell receptor Valpha/delta gene segments. European Journal of Immunology. 2000;30:1988–1997. doi: 10.1002/1521-4141(200007)30:7<1988::AID-IMMU1988>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 15.Abarrategui I, Krangel MS. Regulation of T cell receptor-alpha gene recombination by transcription. Nat Immunol. 2006;7:1109–1115. doi: 10.1038/ni1379. [DOI] [PubMed] [Google Scholar]
- 16.Abarrategui I, Krangel MS. Noncoding transcription controls downstream promoters to regulate T-cell receptor alpha recombination. Embo J. 2007;26:4380–4390. doi: 10.1038/sj.emboj.7601866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Agata Y, Katakai T, Ye SK, Sugai M, Gonda H, Honjo T, Ikuta K, Shimizu A. Histone acetylation determines the developmentally regulated accessibility for T cell receptor gamma gene recombination. Journal of Experimental Medicine. 2001;193:873–879. doi: 10.1084/jem.193.7.873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xiong N, Baker JE, Kang C, Raulet DH. The genomic arrangement of T cell receptor variable genes is a determinant of the developmental rearrangement pattern. Proc Natl Acad Sci U S A. 2004;101:260–265. doi: 10.1073/pnas.0303738101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krangel MS, Hernandez-Munain C, Lauzurica P, McMurry M, Roberts JL, Zhong XP. Developmental regulation of V(D)J recombination at the TCR alpha/delta locus. Immunological Reviews. 1998;165:131–147. doi: 10.1111/j.1600-065x.1998.tb01236.x. [DOI] [PubMed] [Google Scholar]
- 20.Monroe RJ, Sleckman BP, Monroe BC, Khor B, Claypool S, Ferrini R, Davidson L, Alt FW. Developmental regulation of TCR delta locus accessibility and expression by the TCR delta enhancer. Immunity. 1999;10:503–513. doi: 10.1016/s1074-7613(00)80050-3. [DOI] [PubMed] [Google Scholar]
- 21.Bassing CH, Tillman RE, Woodman BB, Canty D, Monroe RJ, Sleckman BP, Alt FW. T cell receptor (TCR) alpha/delta locus enhancer identity and position are critical for the assembly of TCR delta and alpha variable region genes. Proc Natl Acad Sci U S A. 2003;100:2598–2603. doi: 10.1073/pnas.0437943100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bosc N, Lefranc MP. The mouse (Mus musculus) T cell receptor alpha (TRA) and delta (TRD) variable genes. Dev Comp Immunol. 2003;27:465–497. doi: 10.1016/s0145-305x(03)00027-2. [DOI] [PubMed] [Google Scholar]
- 23.Hawwari A, Krangel MS. Regulation of TCR delta and alpha repertoires by local and long-distance control of variable gene segment chromatin structure. J Exp Med. 2005;202:467–472. doi: 10.1084/jem.20050680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bassing CH, Alt FW, Hughes MM, D'Auteuil M, Wehrly TD, Woodman BB, Gärtner F, White JM, Davidson L, Sleckman BP. Recombination signal sequences restrict chromosomal V(D)J recombination beyond the 12/23 rule. Nature. 2000;405:583–586. doi: 10.1038/35014635. [DOI] [PubMed] [Google Scholar]
- 25.Tillman RE, Wooley AL, Hughes MM, Khor B, Sleckman BP. Regulation of T-cell receptor beta-chain gene assembly by recombination signals: the beyond 12/23 restriction. Immunol Rev. 2004;200:36–43. doi: 10.1111/j.0105-2896.2004.00156.x. [DOI] [PubMed] [Google Scholar]
- 26.Drejer-Teel AH, Fugmann SD, Schatz DG. The beyond 12/23 restriction is imposed at the nicking and pairing steps of DNA cleavage during V(D)J recombination. Mol Cell Biol. 2007;27:6288–6299. doi: 10.1128/MCB.00835-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Probst J, Blumenthal SG, Tenzer S, Weinschenk T, Dittmer J, Schoor O, Six A, Rammensee HG, Pascolo S. A conserved sequence in the mouse variable T cell receptor alpha recombination signal sequence 23-bp spacer can affect recombination. Eur J Immunol. 2004;34:2179–2190. doi: 10.1002/eji.200424910. [DOI] [PubMed] [Google Scholar]
- 28.Xiong N, Kang C, Raulet DH. Positive selection of dendritic epidermal γδ T cell precursors in the fetal thymus determines expression of skin-homing receptors. Immunity. 2004;21:121–131. doi: 10.1016/j.immuni.2004.06.008. [DOI] [PubMed] [Google Scholar]
- 29.Lewis JM, Girardi M, Roberts SJ, Barbee SD, Hayday AC, Tigelaar RE. Selection of the cutaneous intraepithelial gammadelta+ T cell repertoire by a thymic stromal determinant. Nat Immunol. 2006;7:843–850. doi: 10.1038/ni1363. [DOI] [PubMed] [Google Scholar]
- 30.Boyden LM, Lewis JM, Barbee SD, Bas A, Girardi M, Hayday AC, Tigelaar RE, Lifton RP. Skint1, the prototype of a newly identified immunoglobulin superfamily gene cluster, positively selects epidermal gammadelta T cells. Nat Genet. 2008;40:656–662. doi: 10.1038/ng.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mallick-Wood C, Lewis JM, Richie LI, Owen MJ, Tigelaar RE, Hayday AC. Conservation of T cell receptor conformation in epidermal γδ cells with disrupted primary Vγ gene usage. Science. 1998;279:1729–1733. doi: 10.1126/science.279.5357.1729. [DOI] [PubMed] [Google Scholar]
- 32.Hara H, Kishihara K, Matsuzaki G, Takimoto H, Tsukiyama T, Tigelaar RE, Nomoto K. Development of dendritic epidermal T cells with a skewed diversity of gamma delta TCRs in V delta 1-deficient mice. Journal of Immunology. 2000;165:3695–3705. doi: 10.4049/jimmunol.165.7.3695. [DOI] [PubMed] [Google Scholar]
- 33.Sullivan S, Bergstresser P, Tigelaar R, Streilein JW. FACS purification of bone marrow-derived epidermal populations in mice: Langerhans cells and Thy-I+ dendritic cells. J Invest Derm. 1985;84:491–495. doi: 10.1111/1523-1747.ep12273454. [DOI] [PubMed] [Google Scholar]
- 34.Coligan J, Kruisbeek A, Margulies D, Shevach E, Strober W. Current Protocols in Immunology. Greene Publishing Associates and Wiley & Sons; New York: 2008. [Google Scholar]
- 35.Olaru A, Patterson DN, Villey I, Livak F. DNA-Rag protein interactions in the control of selective D gene utilization in the TCR beta locus. J Immunol. 2003;171:3605–3611. doi: 10.4049/jimmunol.171.7.3605. [DOI] [PubMed] [Google Scholar]
- 36.Ferrero I, Wilson A, Beermann F, Held W, MacDonald HR. T cell receptor specificity is critical for the development of epidermal gammadelta T cells. J Exp Med. 2001;194:1473–1483. doi: 10.1084/jem.194.10.1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hughes MM, Tillman RE, Wehrly TD, White JM, Sleckman BP. The B12/23 restriction is critically dependent on recombination signal nonamer and spacer sequences. J Immunol. 2003;171:6604–6610. doi: 10.4049/jimmunol.171.12.6604. [DOI] [PubMed] [Google Scholar]
- 38.Jung D, Bassing CH, Fugmann SD, Cheng HL, Schatz DG, Alt FW. Extrachromosomal recombination substrates recapitulate beyond 12/23 restricted VDJ recombination in nonlymphoid cells. Immunity. 2003;18:65–74. doi: 10.1016/s1074-7613(02)00507-1. [DOI] [PubMed] [Google Scholar]
- 39.Sperling AI, Decker DC, DiPaolo RJ, Stern DA, Shum A, Bluestone JA. Selective expansion of Vgamma2-Vdelta7 TCR gammadelta cells in C57BL/6 mice is postnatal and extrathymic. J Immunol. 1997;159:86–91. [PubMed] [Google Scholar]
- 40.Feeney AJ, Goebel P, Espinoza CR. Many levels of control of V gene rearrangement frequency. Immunol Rev. 2004;200:44–56. doi: 10.1111/j.0105-2896.2004.00163.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.