Abstract
Equilibrative nucleoside transporters play essential roles in nutrient uptake, cardiovascular and renal function, and purine analog drug chemotherapies. Limited structural information is available for this family of transporters; however, residues in transmembrane domains 1, 2, 4, and 5 appear to be important for ligand and inhibitor binding. In order to identify regions of the transporter that are important for ligand specificity, a genetic selection for mutants of the inosine-guanosine-specific Crithidia fasciculata nucleoside transporter 2 (CfNT2) that had gained the ability to transport adenosine was carried out in the yeast Saccharomyces cerevisiae. Nearly all positive clones from the genetic selection carried mutations at lysine 155 in transmembrane domain 4, highlighting lysine 155 as a pivotal residue governing the ligand specificity of CfNT2. Mutation of lysine 155 to asparagine conferred affinity for adenosine on the mutant transporter at the expense of inosine and guanosine affinity due to weakened contacts to the purine ring of the ligand. Following systematic cysteine-scanning mutagenesis, thiol-specific modification of several positions within transmembrane domain 4 was found to interfere with inosine transport capability, indicating that this helix lines the water-filled ligand translocation channel. Additionally, the pattern of modification of transmembrane domain 4 suggested that it may deviate from helicity in the vicinity of residue 155. Position 155 was also protected from modification in the presence of ligand, suggesting that lysine 155 is in or near the ligand binding site. Transmembrane domain 4 and particularly lysine 155 appear to play key roles in ligand discrimination and translocation by CfNT2.
Keywords: Membrane/Proteins, Metabolism/Nucleotide, Nucleoside/Nucleotide/Analogs, Nucleoside/Nucleotide/Purine, Nucleoside/Nucleotide/Transport, Organisms/Parasite, Organisms/Protozoan, Sulfhydryls/Thiol
Introduction
Both hydrophilic nutrients and nutrient analog drugs gain access to target cells via membrane-spanning protein transporters. One family of such proteins, the equilibrative nucleoside transporters (ENTs2; SLC29), transports purine and pyrimidine nucleobases and nucleosides into eukaryotic cells (1). In parasitic protozoa, such as Leishmania, Plasmodium, and trypanosomes, ENTs serve an essential function, because purines cannot be synthesized de novo by these organisms and must be obtained from the environment (2). Mammalian ENTs are of particular interest in renal and cardiovascular function (adenosine transport (3, 4)) and in the pharmacogenomics of nucleoside analog drug uptake in antiparasitic, antiviral, and anticancer chemotherapies (5). Although many ENT genes and cDNAs from diverse organisms have been cloned and biochemically characterized in recent years, and insight into the structure of the ENT family is emerging from recent threading (6, 7) and ab initio (8) structural models, a detailed understanding of how ENTs recognize their ligands has remained elusive.
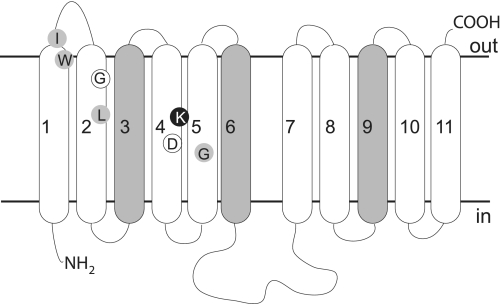
All ENTs studied to date are predicted to be composed of 11 transmembrane-spanning segments (TMs), with a large loop between TM6 and -7 that, like the N terminus, is intracellular, whereas the C terminus projects extracellularly (Fig. 1) (9, 10). All extracellular loops except the first are believed to be quite short, suggesting that ligand discrimination and binding probably depend on amino acid residues within the TMs that project into a water-filled channel. An ab initio structural model of LdNT1.1 predicts that all TMs except TM3, TM6, and TM9 are arranged about this central ligand translocation channel (8). Additionally, mutational analysis by many groups indicates that TMs in the N-terminal half of ENTs (TM1, -2, -4, and -5) (Fig. 1) contribute to ligand affinity and specificity because mutations have been identified in these TMs that confer gain-of-function (increased affinity for particular ligands or inhibitors) or selective loss of affinity for a subset of ligands (Fig. 1). Many other point mutations have been described that lead to partial or complete loss of ENT function (8, 9, 11–16). However, the mutated residues have not been shown to specifically affect ligand binding rather than transporter structure or conformational changes required for ligand translocation. The most intriguing candidate for a ligand binding residue to date is Lys153 in TM4 of LdNT1.1, which when mutated to arginine confers a novel inosine affinity on this adenosine transporter (15). This residue was not shown to be located in the ligand binding site, however.
FIGURE 1.
Predicted topology of residues that influence ENT ligand specificity. The generic topology of an ENT family member is depicted, and N (NH2) and C (COOH) termini as well as intracellular (in) and extracellular (out) sides are indicated. Transmembrane helices predicted to lie on the exterior of the protein structure are shaded. Residues identified in this work are indicated in black (Lys155) and white (Gly86 and Asp159) circles. The predicted locations of residues described in the literature as influencing ligand specificity are indicated by gray circles (TM1, hENT2-I33 (51, 52) and hENT1-W29 (53); TM2, hENT1-L92 (54); TM4, LdNT1.1-K153 (15); TM5, LdNT1.1-G183 (55) and rENT1/rENT2 chimeras (56)).
Here we show that transmembrane domain 4 lines the ligand translocation channel and helps to define the ligand binding site of an equilibrative nucleoside transporter, valuable information in the structural characterization of this ubiquitous and pharmacologically important protein family. We have identified Lys155 in TM4 (see Fig. 1, black circle) of the Crithidia fasciculata inosine-guanosine transporter CfNT2 (17) as a key residue influencing CfNT2 ligand specificity using an unbiased genetic selection for change-of-specificity mutations. Substitutions of asparagine and alanine at this position conferred adenosine transport capability on this transporter and reduced affinity for inosine and guanosine by weakening contacts between the transporter and the purine ring of the ligand, indicating a relaxing of specificity and a phenotype somewhat distinct from that of the orthologous LdNT1.1-K153R mutant (15). Cysteine-scanning mutagenesis and thiol-specific modification (SCAM) of CfNT2 TM4 established that this TM lines the ligand translocation channel, which is consistent with the ab initio ENT model (8). SCAM also demonstrated that Lys155 is water-exposed, lies near the center of TM4 in CfNT2, and is protected from modification by the presence of substrate, intimating that it is located in or near the ligand binding site. Interestingly, the SCAM pattern of TM4 was not strictly helical between residues 152 and 156, opening up the possibility that TM4 is a “broken” helix that contacts ligand at this flexible linker, similar to some sodium-coupled transporters (SLC6), such as LeuT (18).
EXPERIMENTAL PROCEDURES
Cell Culture and Other Reagents
Escherichia coli DH5α and TOP10 strains were used throughout (Invitrogen), and standard methods for recombinant DNA work were employed (19). The purine auxotrophic Saccharomyces cerevisiae strain YPH499 (MATa ura3-52 lys2-801 ade2-101 trp1-Δ63 leu2Δ1) was constructed by Sikorski and Hieter (20). Synthetic defined (SD) media were prepared according to standard methods. Microbiological media and phosphate-buffered saline (PBS) tablets were obtained from Fisher and MP Biomedicals (Irvine, CA). Yeast were transformed by either the rapid or the high efficiency lithium acetate method (21). The Leishmania donovani Δldnt1/Δldnt2 line, which lacks all purine nucleoside uptake capability (22), was used for expression and biochemical characterization of cfnt2 mutant proteins. Parasites were transfected with plasmid DNA according to the method of Robinson and Beverley (23). Parasites were maintained in modified M199 medium as described by Goyard et al. (24) supplemented with 5% fetal bovine serum (Invitrogen), 50 μg/ml hygromycin (Roche Applied Science), 50 μg/ml phleomycin (RPI Research, Mt. Prospect, IL), and 25 μg/ml G418 (BioWhittaker, Walkersville, MD) at 26 °C using adenine as a purine source. Hygromycin and phleomycin were the drugs employed in the selection of L. donovani Δldnt1/Δldnt2 (22). Oligonucleotides were obtained from Invitrogen. 2-Aminoethyl methanethiosulfonate (MTSEA), sodium (2-sulfonatoethyl)methanethiosulfonate (MTSES), and isoguanosine were purchased from Toronto Research Chemicals, Inc. (North York, Ontario, Canada), 7-deazaguanosine was from ChemGenes (Wilmington, MA), and 2′,3′-dideoxyinosine was from ICN. [3H]Adenosine (30 Ci/mmol) and [3H]inosine (15 Ci/mmol) were obtained from American Radiolabeled Chemicals, Inc. (St. Louis, MO). Virtually all other chemicals were purchased from Sigma and were of the highest grade available.
Construction of Yeast and Leishmania CfNT2 Expression Vectors and Site-directed Mutants
All expression of CfNT2 derivatives and CfNT1 in yeast was driven from the high copy pRS426-Cu yeast-E. coli shuttle vector (25), which allows copper-inducible expression of the inserted gene and contains a URA3-selectable marker. The construction of the pRS426-Cu-CfNT2 and pRS426-Cu-CfNT1 expression vectors is described by Liu et al. (17). Leishmanial expression of CfNT2 and its cfnt2 mutant variants was from the pALTneo vector (26) with an N-terminal HA tag, pALTneo-HA, as described (27). The construction of the cysteineless cfnt2 gene (cfnt2ΔCys) is described by Arendt et al. (27). All point mutants were constructed by site-directed mutagenesis of pALTneo-HA-CfNT2 or pALTneo-HA-cfnt2ΔCys performed according to the QuikChange mutagenesis protocol (Stratagene, La Jolla, CA) with primer design based on the method of Zheng et al. (28).
Random Mutagenesis of CfNT2 and Gain-of-function Screening in Yeast
Libraries of mutant cfnt2 molecules carried in the pRS426-Cu vector were constructed by mutagenic PCR followed by in vivo recombination and reconstitution of the expression vector within the yeast strain YPH499. The open reading frames of CfNT2 and cfnt2-G86V were subcloned into the E. coli vector pBluescript II KS(+) to provide PCR templates devoid of yeast sequences. Mutagenic PCR was conducted using the Diversify PCR random mutagenesis kit (Clontech) under conditions that were expected to produce 2.0 base changes per open reading frame (ORF), using pBluescript-CfNT2 (first and second rounds) or pBluescript-cfnt2-G86V (second round) as template. Two independent reaction mixtures were pooled, precipitated with ethanol in the presence of Pellet Paint (Invitrogen), and resuspended in a small volume of TE (10 mm Tris-HCl, 1 mm EDTA, pH 8) buffer to a concentration of ∼0.25 mg/ml. Within the pRS426-Cu-CfNT2 yeast expression vector, BglII and SphI restriction sites were engineered into the CfNT2 ORF ∼30 bp from the 5′- and 3′-ends, respectively. Digestion with BglII and SphI followed by gel purification yielded linear vector DNA containing 30 bp of 5′ and 3′ CfNT2 sequence, enough to allow homologous recombination with mutagenized PCR products and formation of a circular expression vector following co-transformation into yeast. YPH499 cells were transformed by the high efficiency lithium acetate method (21) using 0.25–1.0 μg of PCR-derived mutagenesis library material to 1 μg of linear vector fragment. Cells were immediately plated on SD ura− ade−, 200 μm adenosine, 100 μm CuSO4 (first round of selection) or SD ura− ade−, 100 μm adenosine, 100 μm guanosine, 100 μm CuSO4 (second round of selection). An aliquot of cells was plated on SD ura− to allow quantitation of transformation yield. Initial positive clones were restreaked on the same medium originally used for the selection to verify colony formation. Testing of some positive clones on SD ade− medium showed that growth on adenosine-containing medium was not due to reversion of the ade2 locus. Additionally, plating on 5-fluoroorotic acid to select against the URA3-containing plasmid, followed by streaking on SD ade−, 200 μm adenosine, 100 μm CuSO4 revealed that growth on adenosine was dependent on the presence of the CfNT2 plasmid. pRS426-Cu-cfnt2 plasmids from promising clones were rescued by the smash and grab method (29), followed by transformation of electrocompetent E. coli. Clones were sequenced to identify mutations, and plasmids of interest were retransformed into YPH499 for further study. All data presented were obtained with retransformed yeast strains.
Cell Surface Labeling
Log phase parasites (∼1 × 107 cells/ml) were washed twice with ice-cold PBS plus 10 mm glucose, brought to a density of 4 × 108 cells/ml, and incubated on ice with 1 mg/ml EZ-Link Sulfo-NHS-LC-Biotin (sulfosuccinimidyl-6-(biotinamido)hexanoate; Pierce and Thermo Fisher Scientific) in PBS plus 10 mm glucose for 1 h with occasional mixing. Cells were then washed three times with ice-cold 50 mm glycine in PBS and lysed in 1 ml of lysis buffer (20 mm Tris (pH 8.0), 100 mm NaCl, 10% glycerol, 1% Nonidet P-40, and protease inhibitors) for 1 h on a rotator at 4 °C. After centrifugation for 10 min at 16,000 × g at 4 °C, the supernatant was collected and incubated with washed streptavidin-agarose beads (Pierce) for 1 h at 4 °C. Beads were washed three times with 1 ml of lysis buffer and resuspended in a small volume of 2× Laemmli loading buffer. After separation of proteins by 10% SDS-PAGE, transfer to a polyvinylidene fluoride membrane, and blocking in 5% nonfat dry milk in PBS plus 1% Tween (PBST), the membrane was probed with anti-HA monoclonal antibodies (16B12, Covance (Princeton, NJ)) and by goat anti-mouse IgG1-HRP (Roche Applied Science) in 1% milk, PBST, followed by chemiluminescent detection (Western Lightning-ECL, PerkinElmer Life Sciences). For detection of biotin-conjugated myo-inositol transporter (MIT) as a loading control, blots were reprobed in 1% milk, PBST with anti-L. donovani MIT polyclonal antiserum (a gift of Dr. S. Landfear, Oregon Health and Science University) and goat anti-rabbit horseradish peroxidase (Pierce). Densitometry was performed on developed films using AlphaEaseFC (version 4.0.0, Alpha Innotech Corp., San Leandro, CA). Relative intensities of HA-cfnt2 spots compared with the HA-CfNT2 spot on each film were estimated after correcting for differences in intensities of MIT (loading control).
Nucleoside Uptake in Leishmania
Log phase parasites (∼1 × 107 cells/ml) were washed three times in ice-cold PBS plus 10 mm glucose and resuspended in the same buffer at a density of 2–4 × 108/ml. Cells were allowed to warm to room temperature for 15 min, and uptake of [3H]inosine or [3H]adenosine was measured by the oil-stop method (30). Briefly, 100 μl of the cell suspension were mixed with 100 μl of a 2× solution of ligand above a 200-μl cushion composed of a 33:7 mixture of 550 oil (Dow Corning, Midland, MI) to light paraffin oil (catalog number O-119, Fisher). At the desired intervals, cells were centrifuged through the oil cushion at 16,000 × g for 1 min. Following freezing of the sample on dry ice/ethanol, the bottoms of the tubes were clipped into scintillation tubes, and the cells were solubilized in 3 ml of Cytoscint (MP Biomedicals). Scintillation counting was performed after 24–48 h of solubilization and thorough mixing of the contents.
For Km determinations, linear rates of uptake for Δldnt1/Δldnt2 pALTneo and Δldnt1/Δldnt2 pALTneo-HA-cfnt2-K155N cells were determined in parallel at six concentrations of [3H]inosine or [3H]adenosine over 2 min. Rates of uptake by vector-transfected cells were subtracted from cfnt2 rates at each concentration, and the corrected rates were fitted to the Michaelis-Menten equation and plotted in Prism 4.0 (Graphpad Software, Inc., La Jolla, CA).
For competition assays, duplicate tubes containing 5 μm [3H]inosine plus a 500 μm concentration of a potential inhibitor and buffer-matched duplicate control tubes without inhibitor were assayed in parallel at a single time point within the linear range of uptake by the oil-stop method (10 s for Δldnt1/Δldnt2 pALTneo-HA-CfNT2 and 60 s for Δldnt1/Δldnt2 pALTneo-HA-cfnt2-K155N). Samples containing 5 mm unlabeled inosine were used to determine transporter-independent background uptake. Following scintillation counting, corrected uptake levels of parallel samples with and without inhibitor were compared with determined percentage inhibition as follows, (uptake with inhibitor − transporter-independent uptake)/(uptake without inhibitor − transporter-independent uptake).
Relative inosine and adenosine uptake rates by various cfnt2-K155 mutant transporters, as summarized in Table 1, were measured and analyzed as follows. Uptake of 5 μm [3H]inosine into Δldnt1/Δldnt2 pALTneo-HA-cfnt2-K155 cells was determined in duplicate over a 4-min time course for each cell line, and rates were determined (pmol/min/107 cells) by linear regression analysis in Prism 4.0. The rates of 5 μm [3H]inosine uptake in the presence of 1 mm unlabeled inosine by each cell line were measured in parallel and subtracted from the rate of uptake in the absence of cold competitor. This transporter-mediated uptake was compared qualitatively among cell lines. Adenosine uptake by cfnt2-K155 cells was low, and the rate of 5 μm [3H]adenosine uptake was not significantly inhibited by the presence of 1 mm unlabeled adenosine. Therefore, linear rates of 5 μm [3H]adenosine uptake over 5 min were measured in duplicate for each cfnt2-K155 cell line and compared with the rate of uptake by vector-transfected cells (consistently 0.2–0.3 pmol/min/107 cells). Those cell lines that consistently had a linear rate of adenosine uptake more than 2 times higher than that of the vector-transfected cells scored at least one “+” on the qualitative scale. Those with a variable response from experiment to experiment were scored as “+/−”, and those that failed to show significant uptake above the vector background were scored as “−”.
TABLE 1.
Adenosine and inosine uptake and analog toxicity by a panel of cfnt2-K155 mutants in L. donovani Δldnt1/Δldnt2
| Cell line | Adenosine, uptakea | Inosine uptakeb | Tubercidin EC50c | Formycin B EC50c |
|---|---|---|---|---|
| Vector | − | − | 89.1 ± 13 | 18.9 ± 4.9 |
| CfNT1 | ++++ | − | 0.0139 ± 0.0046 | 1.18 ± 0.14 |
| CfNT2 | − | ++++ | 3.23 ± 1.2 | 0.0163 ± 0.0064 |
| cfnt2-K155N | + | ++ | 0.733 ± 0.23 | 0.363 ± 0.11 |
| cfnt2-K155A | ++ | + | 0.610 ± 0.20 | 0.0485 ± 0.018 |
| cfnt2-K155Q | +/− | + | 0.773 ± 0.30 | 0.0828 ± 0.032 |
| cfnt2-K155R | + | +++ | 5.74 ± 1.3 | 0.185 ± 0.085 |
| cfnt2-K155T | − | + | 1.62 ± 0.61 | 0.102 ± 0.037 |
| cfnt2-K155E | − | − | 1.90 ± 0.41 | 0.322 ± 0.11 |
| cfnt2-K155M | − | − | 3.01 ± 1.2 | 0.102 ± 0.043 |
| cfnt2-K155Y | − | − | 6.67 ± 0.91 | 2.41 ± 1.0 |
| cfnt2-K155L | − | − | 110. ± 16 | 33.6 ± 5.4 |
a Uptake of 5 μm [3H]adenosine relative to vector cells (set to “−”), n = 2.
b Uptake of 5 μm [3H]inosine relative to uptake in the presence of 1 mm unlabeled inosine, n = 2.
c EC50 values are expressed in μm (n = 3–5).
Modification with Thiol-specific Reagents
For SCAM assays, aliquots of cells were treated at room temperature with 2.5 mm MTSEA or 10 mm MTSES in PBS plus 10 mm glucose for 10 min and used immediately for uptake experiments as described above. Triplicate measurements of uptake of 5 μm [3H]inosine were taken at a single time point within the linear range of uptake (1.5–5 min, depending on the particular mutant). Background uptake of 5 μm [3H]inosine by untreated cells in the presence of 5 mm unlabeled inosine was subtracted from all other values. In experiments in which protection by ligand was examined, experimental samples were supplemented with 1 mm inosine (from a 50 mm stock in PBS) 2 min prior to MTSES or MTSEA treatment. After 10 min of incubation, unreacted reagent and unlabeled ligand were removed by three washes with 1.5 ml of ice-cold PBS plus 10 mm glucose. Cell density was adjusted to 3–4 × 108/ml, and uptake of 1 μm [3H]inosine and 1 μm [3H]inosine plus 1 m unlabeled inosine was measured in triplicate at a single time point within the linear range of uptake as described above.
Growth Inhibition Assays with Alamar Blue
In a 96-well plate, cells at 1 × 105 cells/ml were mixed with tubercidin or formycin B that had been diluted 2-fold over 11–15 wells. After ∼6 days of growth at 26 °C, Alamar Blue (Invitrogen) was added, and absorbance at 570 and 600 nm was measured at several time points. The percentage reduction of Alamar Blue was calculated as recommended by the manufacturer, and the time point that gave ∼100% reduction at low concentrations of drug (between 1.5 and 4.5 h) was used for calculations. The effective concentration of drug inhibiting growth by 50% (EC50) was determined by fitting the log-plotted data to a sigmoidal dose-response curve in Prism 4.0.
Computational Modeling of Bader Atomic Charges
Chemical modeling was performed with ADF® version 2008.01 from Scientific Computing and Modeling (Amsterdam) (31, 32) on a Mac Pro 8 core computer. Molecular geometries were optimized using density functional theory with a BLYP exchange correlation functional (33, 34), triple-zeta single polarization atomic basis, and COSMO (35) simulation of a water solvent. Bader atomic charge analysis (36, 37) was then performed using B3LYP hybrid exchange correlation (38), triple-zeta double polarization atomic basis, and COSMO simulation of water.
RESULTS
Genetic Selection of Gain-of-function Mutants of Inosine/Guanosine Transporter CfNT2 Identified Residues in TM2 and TM4
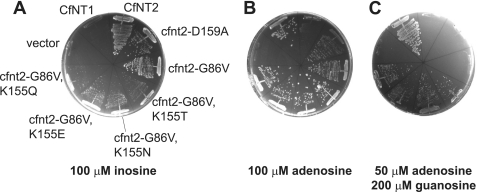
Mutation of ligand-binding residues can result in loss of ligand affinity and therefore loss of transporter function; however, many other types of mutations can also give a loss-of-function phenotype. Therefore, a selection for rare mutants that led to a gain of function, namely a change in ligand specificity, was used to identify regions of the C. fasciculata inosine/guanosine transporter CfNT2 that were specifically required for ligand discrimination rather than ligand translocation and/or proper protein folding. The genetic selection was performed using the yeast S. cerevisiae, an organism that lacks endogenous purine nucleoside transport activity (6, 39), has an available mutant strain that relies on exogenous purines for growth (ade2), and has genetic tools that facilitate rapid and efficient screening of plasmid libraries. Expression of CfNT2 conferred upon yeast the ability to grow on medium containing inosine as the sole purine source (Fig. 2A) (17, 27) but not on adenosine (Fig. 2B), which is not a ligand of the wild-type transporter (compare with robust growth of cells expressing CfNT1, a C. fasciculata adenosine transporter (17) (Fig. 2B)). Following PCR mutagenesis of the CfNT2 ORF, transformants were selected directly on medium containing 100 μm adenosine as the sole purine source to identify cfnt2 mutants that had gained the ability to transport adenosine.
FIGURE 2.
Substitutions at Lys155 of CfNT2 confer adenosine transport activity. Purine auxotrophic yeast cells (Δade2) expressing the indicated ORFs under the control of the yeast CUP1 promoter were grown on SD ura− ade− medium supplemented with 100 μm CuSO4 and inosine (A), adenosine (B), or adenosine and guanosine (C) as purine source(s). Plates were photographed after 3 days of growth at 30 °C.
Thirty positive clones were subjected to plasmid sequencing, and the majority carried a mutation of Gly86 (to Val or Ser; cfnt2-G86V and cfnt2-G86S, respectively), which maps to predicted TM2, or of Asp159 (to Ala, Gly, or Asn), which is in the predicted TM4 (Fig. 1). Although the mutant transporters facilitated growth of ade2 yeast on adenosine (Fig. 2B), little uptake of [3H]adenosine by these cells could be measured biochemically by the oil-stop method (data not shown). The cfnt2-G86V- and cfnt2-D159A-expressing cells exhibited robust growth on inosine (Fig. 2A), suggesting that the encoded transporters retained considerable inosine/guanosine uptake capability. Following random mutagenesis of cfnt2-G86V and cfnt2-D159A ORFs, a second round of selection was performed on plates containing both 100 μm adenosine and 100 μm guanosine, conditions under which cells expressing CfNT2 and the parental mutants could not grow (data not shown). Because yeast cannot utilize guanosine as their sole source of purines (they lack GMP reductase, which converts GMP to IMP (40)), this selection required that mutants gain adenosine affinity and/or lose guanosine affinity relative to the parental background in order to better proliferate on the selective medium. Eighteen positive clones obtained from mutagenesis of the cfnt2-G86V ORF that exhibited increased adenosine transport based on the ability to grow on medium containing 50 μm adenosine and 200 μm guanosine and uptake of 40 μm [3H]adenosine were sequenced. Seventeen of these eighteen positive clones (94%) encoded four different substitutions at Lys155 (Asn, Thr, Glu, and Gln) (Fig. 2C). These double mutant transporters allowed enhanced growth of yeast on 100 μm adenosine compared with the parental cfnt2-G86V-expressing strain (Fig. 2B) and weaker growth on 100 μm inosine (Fig. 2A). The cfnt2-D159A parental line did not yield positive clones that were able to grow on plates containing 50 μm adenosine and 200 μm guanosine as purine sources, and these clones were therefore not studied further.
Mutation of CfNT2 Lys155 Can Broaden Ligand Specificity of the Transporter
The biochemical characterization of cfnt2 mutant transporters was undertaken in an organism more closely related to C. fasciculata, L. donovani, using a mutant cell line (Δldnt1/Δldnt2) that lacked endogenous purine nucleoside transport (22). Mutation of Gly86 alone or in combination with Lys155 did not have a measurable effect on adenosine uptake by CfNT2 in the L. donovani expression system; therefore, further studies focused exclusively on Lys155 single mutants. Nine cfnt2 mutant genes were constructed that substituted a variety of amino acid residues at position 155: positively charged (Arg), negatively charged (Glu), neutral hydrophilic (Asn, Gln, and Thr), small neutral (Ala), and large hydrophobic (Met, Leu, and Tyr). The linear rate of uptake of 5 μm [3H]inosine or [3H]adenosine of cells expressing each mutant transporter was measured, and results are compared qualitatively in Table 1. Substitution of Arg for Lys resulted in an essentially wild-type phenotype, with the cfnt2-K155R transporter driving inosine uptake with high affinity (Km ∼2 μm) (data not shown) but little adenosine uptake. Only substitution of Lys155 with Asn or Ala resulted in consistent uptake of adenosine above background in the parasite expression system (Table 1). Mutant transporters carrying neutral amino acids, such as Gln and Thr, at position 155 also displayed adenosine uptake in some experiments. These same substitutions (Asn, Ala, Gln, and Thr) also allowed for the most significant inosine uptake of any of the cfnt2-K155 mutant proteins (Table 1). By contrast, little uptake of either inosine or adenosine was measured in cfnt2-K155E-, cfnt2-K155M-, cfnt2-K155Y-, or cfnt2-K155L-expressing cells.
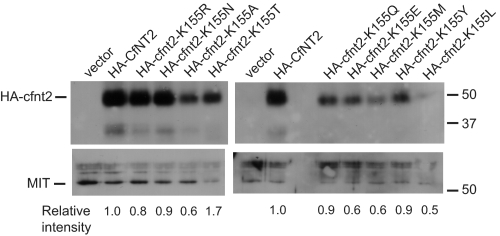
To confirm the nucleoside uptake phenotypes, EC50 values for Δldnt1/Δldnt2 cells expressing each of the mutant transporters were measured for the toxic adenosine analog tubercidin and the toxic inosine analog formycin B over a 5–6-day growth period (Table 1). Cells carrying the empty vector were very resistant to both drugs, whereas CfNT1-expressing cells were exquisitely sensitive to tubercidin and resistant to formycin B, and CfNT2-expressing cells displayed the opposite phenotype. Cells expressing cfnt2-K155R, cfnt2-K155M, or cfnt2-K155Y were as resistant to tubercidin as parasites expressing wild-type CfNT2, consistent with their lack of measurable [3H]adenosine uptake. By contrast, cfnt2-K155N-, cfnt2-K155A-, cfnt2-K155Q-, cfnt2-K155T-, and cfnt2-K155E-expressing cells were somewhat more sensitive to tubercidin than wild-type CfNT2-expressing cells, suggesting slow uptake of the drug by cells harboring the mutant transporters, which mostly mirrored the [3H]adenosine results. Cfnt2-K155R, cfnt2-K155A, cfnt2-K155Q, cfnt2-K155T, and cfnt2-K155M mutant cells retained the most sensitivity to formycin B, implying that these mutant transporters are functional and have some affinity for this inosine isomer. All of these cell lines except cfnt2-K155M displayed measurable [3H]inosine transport (Table 1, column 2). cfnt2-K155N and cfnt2-K155E cells were somewhat less sensitive to formycin B, whereas cfnt2-K155Y cells were as refractory as CfNT1-expressing parasites. Notably, cells harboring the cfnt2-K155L mutant transporter were nearly as resistant to both tubercidin and formycin B as vector-transfected cells, intimating that this transporter is completely non-functional and/or is not expressed at the plasma membrane. Cell surface labeling experiments showed that all tested substitutions at Lys155, including K155L, retained significant cell surface expression compared with wild type CfNT2 (Fig. 3).
FIGURE 3.
Cell surface labeling of cfnt2-K155 mutants expressed in L. donovani. 4 × 108 Δldnt1/Δldnt2 cells transfected with pALTneo carrying the indicated allele or with empty plasmid were treated with biotin as described under “Experimental Procedures.” Biotinylated proteins were bound to streptavidin beads and separated on a 10% SDS-polyacrylamide gel. Western blots were performed with anti-HA and anti-MIT antibodies, and proteins were visualized with chemiluminescence. Locations of size markers in kDa are indicated on the right, and the positions of HA-cfnt2 and the MIT protein (loading control) are indicated on the left. Relative intensities of the HA-cfnt2 bands were estimated from densitometry of scanned films.
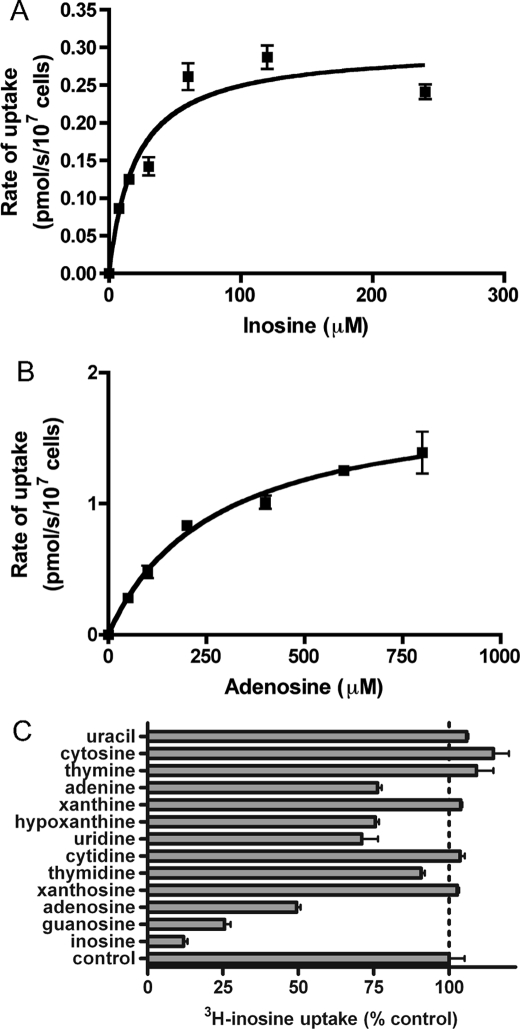
Of all of the cfnt2-K155-expressing lines that were permissive for adenosine uptake, the cfnt2-K155N cells showed the most significant inosine uptake. Therefore, these cells were chosen for more detailed biochemical analysis. Uptake of [3H]inosine, [3H]guanosine, and [3H]adenosine was measured over a range of concentrations, allowing ligand affinity to be estimated. Although CfNT2-expressing cells displayed an inosine Km of ∼1 μm and guanosine Km of 5 μm (17), the affinity of cfnt2-K155N-expressing parasites for both ligands was decreased significantly, as demonstrated by their ∼20-fold higher apparent Km values of 20.7 ± 10.1 and 84.9 ± 24 μm for inosine and guanosine, respectively (Fig. 4A) (data not shown). The cfnt2-K155N transporter also displayed the desired gain of function that motivated the genetic selection, facilitating saturable adenosine uptake with a high capacity, low affinity phenotype (Km = 374 ± 101 μm; Fig. 4B). Measurement of uptake of [3H]inosine in the presence of a 100-fold excess of several unlabeled purines and pyrimidines showed significant inhibition by all three cfnt2-K155N ligands (inosine, guanosine, and adenosine) and a small but reproducible level of inhibition by the pyrimidine nucleoside uridine and the purine nucleobases adenine and hypoxanthine (Fig. 4C), which may reflect low level binding and/or translocation of these compounds.
FIGURE 4.
Biochemical and kinetic characterization of cfnt2-K155N. Δldnt1/Δldnt2 pALTneo-HA-cfnt2-K155N cells were incubated with varying concentrations of [3H]inosine (A) or [3H]adenosine (B), and transport was measured over linear 2-min time courses by the oil-stop method. Michaelis-Menten kinetics of representative experiments (n = 3) are depicted. C, the same cell line was incubated with 5 μm [3H]inosine in the presence of 500 μm unlabeled purine or pyrimidine, and triplicate transport measurements were made at 30 s. A representative experiment is shown (n = 2).
Transmembrane Helix 4 of CfNT2 Lines the Ligand Translocation Channel, and Lys155 Is in or Near the Ligand Binding Site
The effect on ligand specificity of substitutions at Lys155 intimated that this residue may be located in or near the ligand binding site. For this hypothesis to be true, Lys155 would have to be exposed to the water-filled translocation channel, accessible to ligand from the extracellular side of the membrane, and should be protected from chemical modification in the presence of ligand. Lys155 is predicted to lie within TM4 based on all published sequence alignments and predictions of ENT topology (Fig. 5A). However, topology predictions based on protozoan ENT sequences place Lys155 very near the intracellular end of the helix (15, 17), whereas others indicate that it should be near the center of the helix (10, 11), where ligand-binding residues reside in other solute transporters, such as lactose permease (41). To experimentally determine the environmental context of residues within TM4, and especially Lys155, SCAM (42) was utilized. Using the cysteineless variant of CfNT2, cfnt2ΔCys (27), as a starting point, a cysteine codon was introduced at each position predicted to be part of TM4 in any of the published ENT topology predictions. Genes expressing cfnt2 mutants containing a single introduced cysteine residue (cfnt2ΔCys-H138C through cfnt2ΔCys-K172C) were expressed in the Δldnt1/Δldnt2 cell line, and uptake of [3H]inosine by these lines in the presence and absence of 1000× unlabeled inosine was assayed. Measurable uptake above the background level of the inhibited control cells was observed for all single-cysteine mutants, although four (G152C, S154C, K155C, and A156C) exhibited low rates of uptake, suggesting that the mutated residues affected protein function and/or expression levels (Table 2, column 2, open circles). All other positions, especially at the extracellular end of the predicted helix, were tolerant of substitution with cysteine.
FIGURE 5.
Predicted topology of CfNT2 TM4. A, alignment of protozoan and human ENT proteins around predicted TM4. The filled arrowheads indicate residues in CfNT2 shown by SCAM to be water-accessible; the open arrowhead indicates Lys155, which influences ligand specificity. The line below the alignment indicates a published prediction for the residues included in TM4 in human transporters hENT1 and hENT2 (11), which is consistent with these results. The line above the alignment indicates a previous prediction for the topology of CfNT2 TM4 (17). GenBankTM accession numbers are as follows: CfNT2 (AAG22611), LdNT2 (AAF74264), LdNT1.1 (AAC32597), TbNT2 (AAF04490), TbAT1 (AAB93848), hENT1 (AAC51103), hENT2 (NP_001523). The GeneDB accession number for LmajNT3 is LmjF13.1210. B, helical wheel diagram of residues 151–170 from CfNT2. Positions that were observably modified with MTSEA and/or MTSES in the SCAM analysis are highlighted in gray. Those whose modification could be partially blocked by the presence of ligand are highlighted in black.
TABLE 2.
Inosine uptake by single cysteine mutants of TM4 and change in inosine uptake following MTSEA or MTSES treatment of Δldnt1/Δldnt2 cells
| Residuea | Inosine uptakeb | Uptake inhibition by |
|
|---|---|---|---|
| MTSESc | MTSEAc | ||
| ΔCys | ● | N | N |
| H138C (out) | ●● | N | N |
| G139C | ● | N | N |
| A140C | ●● | N | N |
| I141C | ● | N | N |
| A142C | ●● | N | N |
| V143C | ●
|
N | N |
| I144C | N | N | |
| M145C | ● | N | N |
| V146C | ● | N | N |
| V147C | N | N | |
| A148C | N | N | |
| C149C | N | N | |
| V150C | N | N | |
| G151C | N | N | |
| G152C (n = 2) | ● | −35% | −93% |
| F153C | ● | N | N |
| S154C (n = 2) | ○ | N | −77% |
| K155C (n = 3) | ○ | N | +360% |
| A156C (n = 2) | ○ | −30% | −95% |
| L157C | N | N | |
| C158C | N | N | |
| D159C (n = 2) | −65% | −80% | |
| S160C | ●●● | N | N |
| C161C | ●●● | N | N |
| T162C | ● | N | N |
| N163C (n = 3) | N | N | |
| A164C | ●●● | N | N |
| L165C | ● | N | N |
| V166C | N | N | |
| G167C (n = 2) | N | −65% | |
| P168C | ● | N | N |
| F169C | N | N | |
| P170C (n = 2) | N | −80% | |
| T171C | ● | N | N |
| K172C (in) | ● | N | N |
a Position of single cysteine residue within the cfnt2-ΔCys background. n indicates the number of times a ligand protection experiment was performed for the indicated construct. “Out” indicates the extracellular end of the helix, and “in” indicates the intracellular end.
b Each  and ● indicates 5 and 10 pmol of [3H]inosine/min/107 cells transported, respectively (e.g. ●
and ● indicates 5 and 10 pmol of [3H]inosine/min/107 cells transported, respectively (e.g. ● indicates 15 pmol of [3H]inosine/min/107 cells). ○ indicates transport of approximately 1 pmol of [3H]inosine/min/107 cells.
indicates 15 pmol of [3H]inosine/min/107 cells). ○ indicates transport of approximately 1 pmol of [3H]inosine/min/107 cells.
c N indicates inhibition of [3H]inosine uptake similar to that measured for cfnt2-ΔCys cells. Italic type indicates that there was no protection offered by the presence of 1 mm inosine during treatment, and boldface type indicates that there was ∼50% protection.
The ability of each mutant transporter to take up [3H]inosine was then measured following pretreatment of cells with MTSEA or MTSES. An effect on inosine uptake following treatment was expected only if 1) the cysteine residue was water-exposed within the protein structure and accessible to the modifying reagent and 2) modification affected uptake of inosine in some way (steric hindrance, etc.). Modification that did not result in a change in ligand uptake could not be observed by this method, so any positive result was limited to those residues that lined a narrow portion of the substrate channel and/or were in the ligand binding site. MTSES is negatively charged and membrane-impermeant, whereas MTSEA can be positively charged but is membrane-permeable in its neutral state and can modify residues from either side of the membrane (43). A limited number of mutant transporters was detectably modified by both MTSEA and MTSES, in each case leading to a decrease in [3H]inosine uptake (G152C, A156C, and D159C; Table 2). Cysteine at position 159 could be protected by 1 mm inosine from modification by MTSES but not by MTSEA. Additional residues were observably reactive with only MTSEA, including S154C, K155C, G167C, and P170C. Notably, modification of K155C with MTSEA was partially blocked by the presence of ligand (Table 2). MTSEA modification of N163C resulted in a variable amount of inhibition of inosine uptake in several experiments, although the amount of inhibition did not meet the level of statistical significance.
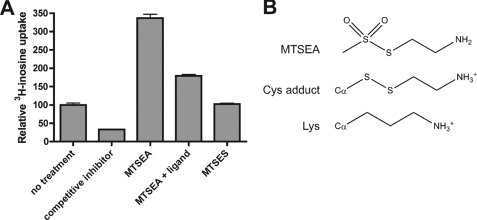
Strikingly, although thiol-specific modification of most TM4 single-cysteine mutants resulted in a loss of inosine transport capability, modification of cfnt2ΔCys-K155C with MTSEA resulted in a large augmentation of activity (Table 2 and Fig. 6A). Treatment with MTSEA resulted in a 3.5-fold increase in [3H]inosine uptake, which could be partially prevented by treating with MTSEA in the presence of 1 mm unlabeled inosine (Fig. 6A). The addition of the MTSEA thioethylamino group to a protein cysteinyl residue produces a side chain with a terminal primary amine that is only slightly longer than that of lysine (Fig. 6B). Although treatment with MTSES triggered no change in activity, the possibility that K155C could be modified with MTSES and that modification of K155C with thioethylsulfonate did not detectably interfere with uptake of [3H]inosine cannot be ruled out.
FIGURE 6.
Effect of MTSEA treatment of cfnt2ΔCys-K155C on inosine uptake capability. A, uptake of over 3 min was measured in triplicate by Δldnt1/Δldnt2 pALTneo-HA-cfnt2ΔCys-K155C cells under a variety of conditions. All cells were first treated or mock-treated with 2.5 mm MTSEA or 10 mm MTSES for 10 min in the absence or presence (+ ligand) of 1 mm inosine. Following three washes of the cells to remove unreacted reagent and excess unlabeled ligand, uptake of 5 μm [3H]inosine was measured. No treatment, cells not treated with thiol-modifying reagents (relative uptake set to 100%); competitive inhibitor, mock-treated cells whose uptake of 5 μm [3H]inosine was challenged with 5 mm unlabeled inosine. B, structures of MTSEA as well as MTSEA-modified cysteine (Cys adduct) and lysine side chains.
CfNT2 Contacts Purine Nucleosides on Each of the Three Rings
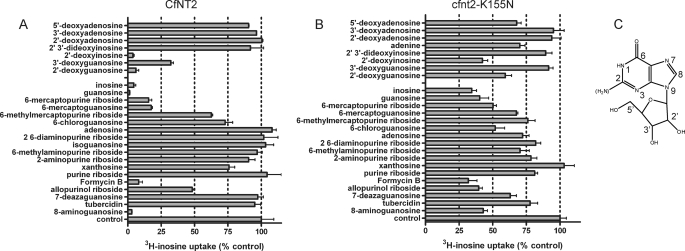
Published data (17) show that CfNT2 is specific for 6-oxopurine nucleosides because [3H]inosine uptake is only inhibited by inosine and guanosine and to a lesser extent xanthosine but not by any other naturally occurring purine or pyrimidine nucleobase or nucleoside. In order to determine which moieties of the ligand are important for transporter affinity and specificity, a structure-activity analysis of CfNT2 was performed by evaluating the ability of purine nucleoside analogs to inhibit [3H]inosine transport of CfNT2-expressing Δldnt1/Δldnt2 cells (Fig. 7A).
FIGURE 7.
Structure-activity analysis. Δldnt1/Δldnt2 cells expressing CfNT2 (A) or cfnt2-K155N (B) were incubated with 5 μm [3H]inosine in the presence of a 500 μm concentration of the indicated purine analog in parallel with uninhibited cells (control) in duplicate at a time point within the linear range of uptake (7 s for CfNT2 and 60 s for cfnt2-K155N). Relative uptake compared with uninhibited cells is shown. Data represent at least two independent determinations with each inhibitor. C, the structure of inosine with purine nucleoside numbering scheme indicated. The 2-amino group of guanosine is indicated in parentheses.
The ribose ring was a likely site of CfNT2-ligand interactions because purine nucleobases, which lack a ribose, do not inhibit [3H]inosine uptake by CfNT2 (17). Inhibition of [3H]inosine transport by guanosine compared with 2′- and 3′-deoxyguanosine and inosine compared with 2′- and 2′,3′-deoxyinosine molecules indicates that the 3′-hydroxyl is very likely a contact site (Fig. 7A) because compounds lacking this moiety were weaker inhibitors than their 2′-deoxy counterparts. However, the large decrease in inhibitory capability of 2′,3′-deoxyinosine compared with 3′-deoxyguanosine suggests that the 2′- and 3′-hydroxyls may be partially redundant with respect to transporter contacts. 5′-Deoxyguanosine and 5′-deoxyinosine compounds were not available, precluding evaluation of the 5′-hydroxyl as a contact site in the wild-type transporter.
Purine riboside, which lacks both the 6-oxo moiety and therefore also the protonated N1 of inosine, was not an inhibitor of [3H]inosine uptake, suggesting that one or both of these functional groups was necessary for CfNT2 binding affinity (Fig. 7A; see Fig. 7C for the numbering scheme). Substitution of sulfur for oxygen at C6 (6-mercaptopurine riboside, 6-mercaptoguanosine), resulting in either a weakly electronegative thioketone and protonated N1 or weakly electropositive thiol and unprotonated N1 (Table 3), led to slightly lower levels of inhibition of [3H]inosine uptake (Fig. 7A). 6-Methylmercaptopurine riboside, which has a sulfur atom that is slightly less electropositive than a thiol (Table 3) but lacks the N1 proton, was a much less efficient inhibitor than the other two mercapto-substituted compounds. 6-Chloroguanosine had a weakly inhibitory effect on [3H]inosine transport by CfNT2 (Fig. 7A). This purine analog lacks an N1 proton, but the chlorine substituent is predicted to be slightly electronegative according to Bader charge analysis (Table 3). In all cases, the presence of a hydrogen bond donor or electropositive substitution at C6 (amino or methylamino) and absence of the N1 proton resulted in an inability to inhibit [3H]inosine uptake by CfNT2, highlighting the importance of the C6 and N1 substituents to ligand/CfNT2 contacts. An amino substituent at the C2-position, as found in guanosine, was tolerated by CfNT2 but probably did not make a meaningful contact with the transporter, since 2-aminopurine riboside was nearly as poor an inhibitor as purine riboside (Fig. 7A). The relatively poor ability of xanthosine (2,6-purinedione riboside) to inhibit [3H]inosine uptake suggests that a ketone at the 2-position had a negative effect on binding. Direct contributions of the N1 proton and N3 to ligand binding were not probed because 1- and 3-deazapurine ribosides were not available as 6-oxo derivatives.
TABLE 3.
Bader charge analysis and N1 protonation status for selected purine analogs
| Molecule | Xa | X charge | Ya | Y charge | N1(H) |
|---|---|---|---|---|---|
| Inosine | =O | −1.173 | N1H | ||
| 6-Mercaptopurine riboside | -S- | +0.164 | H | 0.020 | N1 |
| =S | −0.231 | N1H | |||
| 6-Methylmercaptopurine riboside | -S- | +0.122 | CH3 | 0.005 | N1 |
| 6-Chloroguanosine | -Cl | −0.197 | N1 | ||
| Adenosine | -N- | −1.090 | H | 0.450 | N1 |
a X indicates the non-ring atom connected to C6. Y indicates the atom(s) connected to X, if any.
Purine analogs with altered nitrogen content in the pyrazolo ring were also evaluated for their ability to inhibit [3H]inosine uptake by CfNT2 (Fig. 7A). Formycin B (8-aza-9-deazainosine; see Fig. 7C for numbering scheme) inhibited uptake nearly as well as inosine, suggesting that N9 was not a contact point and that the presence of a nitrogen at the 8-position of the purine ring did not interfere with ligand binding. Substitution of N7 with carbon as in 7-deazaguanosine resulted in a complete lack of inhibitory activity (Fig. 7A). Because N7 to C7 substitution should not significantly affect the planarity of the pyrazolo ring (sp2 N or sp2 C), the data indicate that the ability to form a hydrogen bond with N7 was important for CfNT2 ligand binding. Allopurinol riboside (8-aza-7-deazainosine) was an inefficient inhibitor of CfNT2, with ∼50% of inosine uptake still present, suggesting that lack of N7 could be partially rescued by the presence of N8. Interestingly, 8-aminoguanosine was a very efficient inhibitor of inosine uptake (Fig. 7A), intimating that the C8-position was not particularly hindered sterically and/or that this amino group could participate in a hydrogen bond contact with the transporter. Tubercidin (7-deazaadenosine), like adenosine itself, was not an inhibitor of [3H]inosine uptake by CfNT2.
Mutation of Lys155 Weakens Transporter Contacts to the C6- and N7-positions of the Purine Ring
In order to determine if the loss of specificity of cfnt2-K155N for 6-oxopurine nucleosides could be attributed to loss of a hydrogen bond between one of the key contact points identified above (3′-hydroxyl, N1 hydrogen, 6-oxo, and N7 moieties), a structure-activity relationship analysis was also performed with the mutant transporter (Fig. 7B). In order to maintain a 100-fold excess of inhibitor to [3H]inosine while remaining below the solubility limit of the inhibitor compounds, a concentration of ligand significantly below the Km for the mutant transporter was used for the transport assays. Therefore, inhibition by unlabeled inosine and guanosine did not appear to be as strong as for CfNT2 under the same conditions, and weak inhibitors produced measurable inhibition that would not have been observed in the experiment depicted in Fig. 7A. Overall, the data suggest that contacts made between transporter and ligand are similar but not identical for CfNT2 and cfnt2-K155N (Fig. 7, A and B). The failure of 3′-deoxyguanosine to inhibit [3H]inosine uptake by cfnt2-K155N compared with relatively robust uptake in the presence of 2′-deoxyinosine or 2′-deoxyguanosine (Fig. 7B) indicated that the 3′-hydroxyl of the ribose was more important than the 2′-hydroxyl for cfnt2-K155N binding, just as for CfNT2 (Fig. 7A). 5′-Deoxyadenosine was as efficient an inhibitor of cfnt2-K155N [3H]inosine uptake as adenosine itself, suggesting that the 5′-hydroxyl does not make a contact with the transporter (Fig. 7B). Interestingly, hypoxanthine and adenine were both weak inhibitors (Fig. 4C) and weak ligands (data not shown) of cfnt2-K155N but not of CfNT2 (17).
Overall, the cfnt2-K155N transporter retained a preference for compounds with a protonated N1 and/or an electronegative substituent at C6, because inosine, guanosine, 6-mercaptopurine riboside, 6-mercaptoguanosine, and 6-chloroguanosine were significantly stronger inhibitors than purine riboside (Fig. 7B). Purine riboside, which lacks a substituent at C6, was able to inhibit cfnt2-K155N but not CfNT2 inosine uptake activity (Fig. 7), implying that the contact between cfnt2-K155N and the 6-oxo moiety of inosine was not a major contributor to affinity. Likewise, the mutant transporter showed relaxed specificity toward compounds with bulky and/or less electronegative C6 substitutions and a deprotonated N1 compared with its wild-type counterpart. Nearly all such compounds (e.g. adenosine, 2,6-diaminopurine riboside, and 6-methylaminopurine riboside) produced similar inhibition to purine riboside, suggesting that electropositive moieties at C6 were not actively discriminated against by cfnt2-K155N.
Interestingly, the N7 moiety did not appear to have as significant a role in binding affinity to cfnt2-K155N as to wild-type CfNT2: 1) 7-deazaguanosine was able to inhibit [3H]inosine uptake by the mutant transporter, and 2) allopurinol riboside (7-deaza-8-azainosine) was nearly as effective an inhibitor of cfnt2-K155N as inosine. Likewise, adenosine and tubercidin (7-deazaadenosine) inhibited [3H]inosine uptake to similar extents.
DISCUSSION
A key component of ENT function with respect to uptake of endogenous ligands for nutritional purposes or of drugs for chemotherapeutic uses is the specificity and affinity afforded by noncovalent protein-ligand contacts. Little is known, however, about how ENTs contact their ligands or even which residues are in the proper location to potentially influence ligand binding, and no atomic level structure is yet available for any member of this protein family. Our work provides structural information concerning this protein family that is relevant to ligand binding, specifically that TM4 is located within the water-filled translocation channel, that Lys155 is located in or near the ligand binding site, and that this residue, either directly or indirectly, strongly influences binding of CfNT2 to the purine ring moiety of its ligands. These data indicate the location and orientation of TM4 within the structure of the transporter and predict the location and character of the CfNT2 ligand binding site, which will guide the refinement of future structural models for ENTs.
The data presented here establish that Lys155 of CfNT2 is pivotal for selectivity and affinity of the transporter protein for 6-oxopurine nucleosides because its mutation to asparagine either directly or indirectly led to relaxed specificity as well as reduced affinity for its natural ligands (Figs. 4, A and B, and 7B). Lys155 may be the only residue with this strong “ligand selector” function in CfNT2, given the near exclusivity with which mutations at this position were obtained in the genetic selection. The SCAM results demonstrated that K155C was water-accessible and able to be protected from thiol modification by the presence of ligand, leaving open the possibility that Lys155 may be located in the ligand binding site, although a direct role for this residue in ligand binding could not be assessed. Substitution of Lys155 with a negatively charged or large hydrophobic residue interfered with nucleoside uptake, whereas small and neutral residues were tolerated at this position, and a positive charge at position 155 retained the most wild type-like activity (Table 1). Interestingly, MTSEA modification of a cysteinyl residue at position 155 restored both a positively charged side chain (Fig. 6B) and also significant levels of inosine uptake activity (Fig. 6A) to the cfnt2-K155C mutant transporter. A similar effect was noted for MTSEA modification of K157C in the sodium-glucose cotransporter SGLT1 (44). Although mutation of Lys155 of CfNT2 to a variety of non-cationic residues resulted in reduced inosine uptake and/or broadened ligand selectivity (Table 1) but correct cell surface localization (Fig. 3), mutation of the Lys155 ortholog Lys153 in LdNT1.1 to arginine conferred significant affinity for inosine on the mutant transporter with a minor loss of affinity for its native ligands, and its mutation to alanine led to a significant decrease in overall protein level and cell surface localization (15), suggesting that the roles of these two residues in the structure and function of their respective transporters are distinct. The precise mechanism by which Lys155 affects ligand selectivity in CfNT2 awaits experimental structural information on this transporter family.
Structure-activity relationship analysis suggested that there is at least one key contact made by CfNT2 with each of the three rings of the ligand: at the 3′-hydroxyl, at the N1 proton and/or the 6-oxo moiety, and at the N7 moiety (Fig. 7A). These data account for the exquisite specificity of CfNT2 for nucleosides versus nucleobases, purines versus pyrimidines, and 6-oxopurines versus adenosine. Because those compounds with much less electronegative substituents at C6 but some hydrogen bond donor character at N1 were strong inhibitors (e.g. 6-mercaptopurine riboside), whereas those compounds without a protonated N1 (e.g. 6-chloroguanosine) were weaker inhibitors of CfNT2, it is likely that the primary contact by the transporter is to the N1 hydrogen, rather than the 6-oxo substituent, similar to another 6-oxo-specific protozoan ENT (7). Structure-activity relationship analysis of the cfnt2-K155N mutant transporter suggested that contacts between the transporter and the “top” of the purine portion of the ligand were weakened compared with CfNT2; it lacked a contact to the N7 moiety that was made by the wild-type transporter and was less selective for electronegative substituents at the C6-position (Fig. 7), consistent with the increased adenosine and decreased inosine uptake and affinity of cfnt2-K155N- compared with CfNT2-expressing cells (Table 1 and Figs. 2 and 4). The consequences of a lack of the 3′-hydroxyl for binding between ligand and the mutant transporter were quite severe (Fig. 7B). These data suggest that binding of cfnt2-K155N to the ribose may contribute greatly to the affinity between mutant transporter and nucleoside ligands and may in part explain the ability of the pyrimidine nucleosides uridine and thymidine to inhibit inosine uptake by cfnt2-K155N but not by CfNT2 (Fig. 4C) (17).
Three models for the ENT structure have been published in which very similar arrangements of the 11 helices in space are proposed, with all helices except 3, 6, and 9 lining the central ligand translocation channel (6–8). These models are consistent with published mutagenesis studies, which have not indicated a role for TM3, -6, and -9 in ligand binding or specificity, and with previous SCAM data showing that TM5 lines the water-filled channel (45). Interestingly, all three models propose that TM2 and -4 are close neighbors, which is consistent with the synergistic effect of (genetic interaction between) mutation of Gly86 in TM2 and Lys155 in TM4 in the yeast genetic selection. A similar synergistic effect on ligand binding by simultaneous mutation of residues in TM2 and -4 has been observed in studies of the human hENT1 transporter (11).
In addition to the predictions of the structural models, successful modification of Cys140 in rat rENT2 with the membrane-impermeant p-chloromercuriphenyl sulfonate (46) indicated that at least a portion of TM4 was also likely to line the channel. Data presented here using SCAM clearly demonstrate that a helical face stretching the entire likely length of TM4 is indeed water-accessible (Table 2 and Fig. 5) and also critical for ligand translocation through the water-filled channel because modification of residues by MTSEA and/or MTSES generally interfered with [3H]inosine uptake (Table 2). Residues 152, 156, and 159 were accessible to both reagents and therefore define the extracellular end of TM4. Because MTSEA can diffuse through the membrane but MTSES cannot (43), the fact that G167C and P170C could be modified with MTSEA but not MTSES suggests that Gly167 and Pro170 are near the intracellular end of the TM4 helix. The lack of observable reactivity of S154C and K155C with MTSES, in contrast to successful modification of D159C with this reagent, may indicate either that these residues are in a sterically cramped section of the channel that restricts access by the slightly larger MTSES reagent but not by MTSEA or that these residues are on the water-accessible face of TM4 only in the inward facing conformation of the transporter.
The SCAM results indicate that TM4 of CfNT2 extends from approximately Gly152 to Pro170, which places Lys155 in the extracellular half of the TM, where ligand might be expected to bind. This topology for TM4 is consistent with previous computational predictions for mammalian ENTs (Fig. 5A) (10, 11) as well as a recent ab initio model for a protozoan ENT (8) but stands in contrast to previous predictions of parasite ENT topology that placed Asp159 at the intracellular end of the helix (17, 47). In addition, the SCAM data provide strong evidence for the location of Lys155 as well as Asp159 in or near the ligand binding site because their modification with MTSEA or MTSES, respectively, could be partially blocked by the presence of ligand (Table 2 and Fig. 6A).
The SCAM data also present the intriguing possibility that an unwound region in TM4 that contains Lys155 is located within the ligand binding site of CfNT2. Although the results indicate that TM4 presents a generally helical face to the water-filled ligand channel (Fig. 5B), residues 154, 155, and 156 were all modified by MTSEA (Table 2). Similar runs of three adjacent modifiable residues have been observed in the M2 segment of the α subunit of the acetylcholine receptor (48) and TM4 of the Na,K-ATPase α subunit (49), and the recent x-ray crystal structure of the Na,K-ATPase (50) shows that this TM has a non-helical loop structure at the position indicated by the SCAM data. Additionally, Lys155 is located within a GXGXG-like motif, a motif that forms non-helical ligand-binding loops in LeuT, a bacterial homolog of Na+/Cl−-dependent neurotransmitter transporters (18). CfNT2 contains a G152FSKA156 pentapeptide in this key region of TM4 (Lys155 indicated in boldface type; Fig. 5A), and this probably does represent a functionally important motif, because small residues (Gly, Ala, or Ser) are conserved at the underlined positions in other protozoan transporters (Fig. 5A), and these same residues were exquisitely sensitive to mutation to the larger cysteinyl group in CfNT2 (Table 2). The ab initio structural model for LdNT1.1 does not predict unwinding of TM4 near this region (8), and it remains possible that the unexpected accessibility of S154C, which does not lie on the helical face (Fig. 5B), to MTSEA could be due to conformational changes between inward and outward conformations of the transporter. However, we postulate that at least in the protozoan ENTs, the center portion of TM4 containing the ligand specificity determinant Lys155 has a non- helical structure.
Acknowledgments
We extend special thanks to Kevin Johnson of Pacific University for the Bader charge analysis calculations; to Jan Boitz, Philip Yates, and Hoa Lesselroth for assistance with cell culture; to John Harrelson for HPLC analysis of purine analog purity; and to Scott Landfear for the gift of the anti-MIT antibodies. We also thank Nicola Carter, Philip Yates, Scott Landfear, and Ujwal Shinde for helpful discussions.
This work was supported, in whole or in part, by National Institutes of Health Grants AI023682 and AI044138. This work was also supported by American Cancer Society Grant PF-02-097-01-CSM (to C. S. A.).
- ENT
- equilibrative nucleoside transporter
- TM
- transmembrane domain
- SCAM
- substituted cysteine accessibility method
- SD
- synthetic defined medium
- MTSEA
- 2-aminoethyl methanethiosulfonate
- MTSES
- sodium (2-sulfonatoethyl)methanethiosulfonate
- ORF
- open reading frame
- MIT
- myo-inositol transporter
- EC50
- effective concentration of drug inhibiting growth by 50%
- PBS
- phosphate-buffered saline
- HA
- hemagglutinin.
REFERENCES
- 1.Hyde R. J., Cass C. E., Young J. D., Baldwin S. A. (2001) Mol. Membr. Biol. 18, 53–63 [PubMed] [Google Scholar]
- 2.Downie M. J., Kirk K., Mamoun C. B. (2008) Eukaryot. Cell. 7, 1231–1237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Elwi A. N., Damaraju V. L., Baldwin S. A., Young J. D., Sawyer M. B., Cass C. E. (2006) Biochem. Cell Biol. 84, 844–858 [DOI] [PubMed] [Google Scholar]
- 4.Young J. D., Yao S. Y., Sun L., Cass C. E., Baldwin S. A. (2008) Xenobiotica 38, 995–1021 [DOI] [PubMed] [Google Scholar]
- 5.Molina-Arcas M., Trigueros-Motos L., Casado F. J., Pastor-Anglada M. (2008) Nucleosides Nucleotides Nucleic Acids 27, 769–778 [DOI] [PubMed] [Google Scholar]
- 6.Arastu-Kapur S., Arendt C. S., Purnat T., Carter N. S., Ullman B. (2005) J. Biol. Chem. 280, 2213–2219 [DOI] [PubMed] [Google Scholar]
- 7.Papageorgiou I., De Koning H. P., Soteriadou K., Diallinas G. (2008) Int. J. Parasitol. 38, 641–653 [DOI] [PubMed] [Google Scholar]
- 8.Valdés R., Arastu-Kapur S., Landfear S. M., Shinde U. (2009) J. Biol. Chem. 284, 19067–19076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Arastu-Kapur S., Ford E., Ullman B., Carter N. S. (2003) J. Biol. Chem. 278, 33327–33333 [DOI] [PubMed] [Google Scholar]
- 10.Sundaram M., Yao S. Y., Ingram J. C., Berry Z. A., Abidi F., Cass C. E., Baldwin S. A., Young J. D. (2001) J. Biol. Chem. 276, 45270–45275 [DOI] [PubMed] [Google Scholar]
- 11.Endres C. J., Unadkat J. D. (2005) Mol. Pharmacol. 67, 837–844 [DOI] [PubMed] [Google Scholar]
- 12.Galazka J., Carter N. S., Bekhouche S., Arastu-Kapur S., Ullman B. (2006) Int. J. Biochem. Cell Biol. 38, 1221–1229 [DOI] [PubMed] [Google Scholar]
- 13.SenGupta D. J., Lum P. Y., Lai Y., Shubochkina E., Bakken A. H., Schneider G., Unadkat J. D. (2002) Biochemistry 41, 1512–1519 [DOI] [PubMed] [Google Scholar]
- 14.Visser F., Sun L., Damaraju V., Tackaberry T., Peng Y., Robins M. J., Baldwin S. A., Young J. D., Cass C. E. (2007) J. Biol. Chem. 282, 14148–14157 [DOI] [PubMed] [Google Scholar]
- 15.Valdés R., Liu W., Ullman B., Landfear S. M. (2006) J. Biol. Chem. 281, 22647–22655 [DOI] [PubMed] [Google Scholar]
- 16.SenGupta D. J., Unadkat J. D. (2004) Biochem. Pharmacol. 67, 453–458 [DOI] [PubMed] [Google Scholar]
- 17.Liu W., Arendt C. S., Gessford S. K., Ntaba D., Carter N. S., Ullman B. (2005) Mol. Biochem. Parasitol. 140, 1–12 [DOI] [PubMed] [Google Scholar]
- 18.Yamashita A., Singh S. K., Kawate T., Jin Y., Gouaux E. (2005) Nature 437, 215–223 [DOI] [PubMed] [Google Scholar]
- 19.Ausubel F. M., Brent R., Kingston R. E., Moore D. D., Seidman J. G., Smith J. A., Struhl K. (2002) Current Protocols in Molecular Biology, Wiley Interscience, New York [Google Scholar]
- 20.Sikorski R. S., Hieter P. (1989) Genetics 122, 19–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gietz R. D., Woods R. A. (2002) Methods Enzymol. 350, 87–96 [DOI] [PubMed] [Google Scholar]
- 22.Liu W., Boitz J. M., Galazka J., Arendt C. S., Carter N. S., Ullman B. (2006) Mol. Biochem. Parasitol. 150, 300–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Robinson K. A., Beverley S. M. (2003) Mol. Biochem. Parasitol. 128, 217–228 [DOI] [PubMed] [Google Scholar]
- 24.Goyard S., Segawa H., Gordon J., Showalter M., Duncan R., Turco S. J., Beverley S. M. (2003) Mol. Biochem. Parasitol. 130, 31–42 [DOI] [PubMed] [Google Scholar]
- 25.Labbé S., Thiele D. J. (1999) Methods Enzymol. 306, 145–153 [DOI] [PubMed] [Google Scholar]
- 26.Laban A., Tobin J. F., Curotto de Lafaille M. A., Wirth D. F. (1990) Nature 343, 572–574 [DOI] [PubMed] [Google Scholar]
- 27.Arendt C. S., Ri K., Yates P. A., Ullman B. (2007) Anal. Biochem. 365, 185–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zheng L., Baumann U., Reymond J. L. (2004) Nucleic Acids Res. 32, e115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hoffman C. S., Winston F. (1987) Gene 57, 267–272 [DOI] [PubMed] [Google Scholar]
- 30.Aronow B., Kaur K., McCartan K., Ullman B. (1987) Mol. Biochem. Parasitol. 22, 29–37 [DOI] [PubMed] [Google Scholar]
- 31.te Velde G., Bickelhaupt F. M., Baerends E. J., Fonseca Guerra C., van Gisbergen S. J., Snijders J. G., Ziegler T. (2001) J. Comput. Chem. 22, 931–967 [Google Scholar]
- 32.Fonseca Guerra C., Snijders J. G., te Velde G., Baerends E. J. (1998) Theor. Chem. Acc. 99, 391–403 [Google Scholar]
- 33.Becke A. D. (1988) Phys. Rev. A 38, 3098–3100 [DOI] [PubMed] [Google Scholar]
- 34.Lee C., Yang W., Parr R. G. (1988) Phys. Rev. B Condens. Matter 37, 785–789 [DOI] [PubMed] [Google Scholar]
- 35.Klamt A. (1995) J. Phys. Chem. 99, 2224–2235 [Google Scholar]
- 36.Rodriguez J. I., Bader R. F., Ayers P. W., Michel C., Gotz A. W., Bo C. (2009) Chem. Phys. Lett. 472, 149–152 [Google Scholar]
- 37.Rodríguez J. I., Köster A. M., Ayers P. W., Santos-Valle A., Vela A., Merino G. (2009) J. Comput. Chem. 30, 1082–1092 [DOI] [PubMed] [Google Scholar]
- 38.Becke A. D. (1993) J. Chem. Phys. 98, 5648 [Google Scholar]
- 39.Mäser P., Sütterlin C., Kralli A., Kaminsky R. (1999) Science 285, 242–244 [DOI] [PubMed] [Google Scholar]
- 40.Breton A., Pinson B., Coulpier F., Giraud M. F., Dautant A., Daignan-Fornier B. (2008) Genetics 178, 815–824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Guan L., Kaback H. R. (2006) Annu. Rev. Biophys. Biomol. Struct. 35, 67–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bogdanov M., Zhang W., Xie J., Dowhan W. (2005) Methods 36, 148–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Holmgren M., Liu Y., Xu Y., Yellen G. (1996) Neuropharmacology 35, 797–804 [DOI] [PubMed] [Google Scholar]
- 44.Liu T., Lo B., Speight P., Silverman M. (2008) Am. J. Physiol. Cell. Physiol. 295, C64–C72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Valdés R., Vasudevan G., Conklin D., Landfear S. M. (2004) Biochemistry 43, 6793–6802 [DOI] [PubMed] [Google Scholar]
- 46.Yao S. Y., Sundaram M., Chomey E. G., Cass C. E., Baldwin S. A., Young J. D. (2001) Biochem. J. 353, 387–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vasudevan G., Carter N. S., Drew M. E., Beverley S. M., Sanchez M. A., Seyfang A., Ullman B., Landfear S. M. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 9873–9878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Akabas M. H., Kaufmann C., Archdeacon P., Karlin A. (1994) Neuron 13, 919–927 [DOI] [PubMed] [Google Scholar]
- 49.Horisberger J. D., Kharoubi-Hess S., Guennoun S., Michielin O. (2004) J. Biol. Chem. 279, 29542–29550 [DOI] [PubMed] [Google Scholar]
- 50.Shinoda T., Ogawa H., Cornelius F., Toyoshima C. (2009) Nature 459, 446–450 [DOI] [PubMed] [Google Scholar]
- 51.Visser F., Baldwin S. A., Isaac R. E., Young J. D., Cass C. E. (2005) J. Biol. Chem. 280, 11025–11034 [DOI] [PubMed] [Google Scholar]
- 52.Visser F., Vickers M. F., Ng A. M., Baldwin S. A., Young J. D., Cass C. E. (2002) J. Biol. Chem. 277, 395–401 [DOI] [PubMed] [Google Scholar]
- 53.Paproski R. J., Visser F., Zhang J., Tackaberry T., Damaraju V., Baldwin S. A., Young J. D., Cass C. E. (2008) Biochem. J. 414, 291–300 [DOI] [PubMed] [Google Scholar]
- 54.Endres C. J., Sengupta D. J., Unadkat J. D. (2004) Biochem. J. 380, 131–137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vasudevan G., Ullman B., Landfear S. M. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 6092–6097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yao S. Y., Ng A. M., Vickers M. F., Sundaram M., Cass C. E., Baldwin S. A., Young J. D. (2002) J. Biol. Chem. 277, 24938–24948 [DOI] [PubMed] [Google Scholar]