Abstract
The antiapoptotic proteins of the Bcl-2 family are expressed at high levels in many types of cancer. However, the mechanism by which Bcl-2 family proteins regulate apoptosis is not fully understood. Here, we demonstrate the interaction of Bcl-2 with the outer mitochondrial membrane protein, voltage-dependent anion channel 1 (VDAC1). A direct interaction of Bcl-2 with bilayer-reconstituted purified VDAC was demonstrated, with Bcl-2 decreasing channel conductance. Expression of Bcl-2-GFP prevented apoptosis in cells expressing native but not certain VDAC1 mutants. VDAC1 sequences and amino acid residues important for interaction with Bcl-2 were defined through site-directed mutagenesis. Synthetic peptides corresponding to the VDAC1 N-terminal region and selected sequences bound specifically, in a concentration- and time-dependent manner, to immobilized Bcl-2, as revealed by the real-time surface plasmon resonance. Moreover, expression of the VDAC1-based peptides in cells over-expressing Bcl-2 prevented Bcl-2-mediated protection against staurosporine-induced apoptotic cell death. Similarly, a cell-permeable VDAC1-based synthetic peptide was also found to prevent Bcl-2-GFP-mediated protection against apoptosis. These results point to Bcl-2 as promoting tumor cell survival through binding to VDAC1, thereby inhibiting cytochrome c release and apoptotic cell death. Moreover, these findings suggest that interfering with the binding of Bcl-2 to mitochondria by VDAC1-based peptides may serve to potentiate the efficacy of conventional chemotherapeutic agents.
Keywords: Apoptosis, Cancer, Cell/Apoptosis, Membrane/Channels, Cancer Therapy, Bcl-2, Vdac, Mitochondria
Introduction
Mitochondria-mediated apoptosis can occur in response to conditions that result in an increase in outer mitochondrial membrane permeability, allowing for the release of apoptogenic protein to the cytosol including cytochrome c, apoptosis inducing factor, and Smac/Diablo (1, 2), in a manner controlled by pro- and antiapoptotic proteins of the Bcl-2 family. Members of the Bcl-2 family of proteins are central regulators of apoptosis and include proteins essential for cell survival, as well as those required to initiate cell death (3–5). The Bcl-2 family comprises antiapoptotic members, such as Bcl-2, Mcl-1, Bcl-xL, and Bcl2A1; multidomain proapoptotic members, such as Bax and Bak; and proapoptotic BH3-only proteins, including Bad, Bim, Puma, Bid, Bik, Noxa, and Bmf (3, 6–9). Whereas antiapoptotic proteins, such as Bcl-2, are localized to several intracellular membranes, including those of the mitochondria, endoplasmic reticulum, and the nuclear envelope (4, 10), proapoptotic members of the Bcl-2 family, such as Bax, translocate to mitochondria upon sensing a death signal (11). Antiapoptotic members of the Bcl-2-family also contribute to tumor initiation, disease progression, and drug resistance. Indeed, high expression levels of antiapoptotic Bcl-2 family members were shown to be associated with resistance of many tumors to chemotherapy (6, 12, 13).
The mechanisms by which Bcl-2 family proteins regulate apoptosis are still not fully understood, yet it is well established that their activity is mediated via interaction with the mitochondria and by controlling outer mitochondrial membrane permeability (14, 15). Accumulated evidence indicates that both anti- and proapoptotic proteins interact with the mitochondrial protein, the voltage-dependent anion channel (VDAC),2 to regulate mitochondria-mediated apoptosis (5, 16–19). At the outer mitochondrial membrane, VDAC mediates the transport of anions, cations, ATP, Ca2+, and metabolites (20, 21). Thus, VDAC plays an important role in coordinating communication between mitochondrion and cytosol. VDAC1, one of the three isoforms of the protein, is also proposed to serve as a component of the apoptosis machinery participating in the release of cytochrome c (2, 16, 22–27).
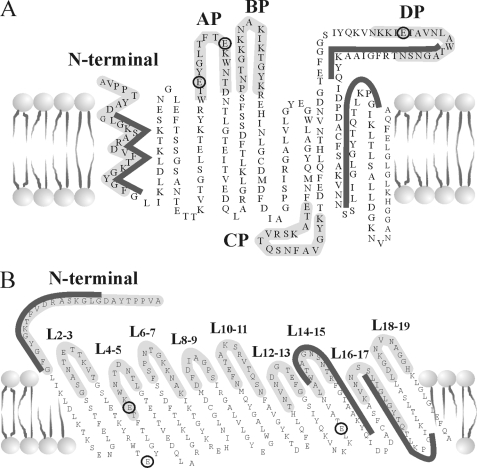
Recently, the transmembrane topology and three-dimensional structures of recombinant human (h)VDAC1 and murine (m)VDAC1 were proposed, based on NMR spectroscopy (28, 29) and x-ray crystallography (30). Still, the proposed structure of the recombinant refolded protein has been questioned (31). Apparently, VDAC1 adopts a β-barrel architecture composed of 19 β-strands with an α-helix positioned horizontally midway within the pore, proposed to regulate the conductance of ions and metabolites passing through the VDAC1 pore (28). However, biochemical and biophysical approaches point to the N-terminal α-helix as alternatively being exposed to the cytoplasm (32), able to interact with anti-VDAC antibodies raised against the N-terminal region of the protein (33–35), as crossing the membrane (20), or lying on the membrane surface (36). These reports and other findings thus suggest that the N-terminal region of VDAC1 constitutes a mobile component of the protein, exhibiting motion during voltage gating (37). Moreover, such mobility of the N-terminal VDAC1 α-helix may modulate the accessibility of apoptosis-regulating proteins of the Bcl-2 family (i.e. Bax and Bcl-xL) to their binding sites on VDAC1 (38). Recently, we have demonstrated that the antiapoptotic proteins, hexokinase (HK) and Bcl-2, interact with the N-terminal region of VDAC1 to inhibit mitochondria-mediated apoptosis (16, 19).
VDAC has been shown to interact with apoptosis-regulating proteins such as Bax/Bak and Bcl-xL (5, 39–41), and with Bax and Bim (41, 42). Cytochrome c release induced by Bax and Bim was found to be inhibited by anti-VDAC antibodies (43), whereas Bid (but not Bax) was shown to modify the conductance of VDAC channels (44). The interaction of Bcl-xL with VDAC1 was also demonstrated using NMR spectroscopy (18, 29). Bcl-xL was further shown to modify the oligomerization state of VDAC1, as revealed by chemical cross-linking of micelle-bound VDAC1, shifting the equilibrium from the trimeric to the dimeric state (18, 29, 67). It has also been suggested that VDAC1 interacts with both Bax and Bcl-xL to form a tertiary complex and that Bcl-xL interacts with VDAC via a putative loop region of VDAC1 (38). In addition, Bcl-2 and Bcl-xL were proposed to interact with VDAC to block As2O3-induced VDAC dimerization (45). These results indicate that Bcl-2 family proteins regulate VDAC-mediated apoptosis and hence, the release of apoptogenic proteins from mitochondria.
In this study, through various experimental approaches, we demonstrate the interaction of Bcl-2 with VDAC and the subsequent modulation of apoptosis. Purified Bcl-2(ΔC23) reduced channel conductance of native but not mutated VDAC1 reconstituted into a planar lipid bilayer (PLB). Using site-directed mutagenesis, we identified VDAC1 domains that are involved in the interaction of the protein with Bcl-2 to confer protection against apoptosis. These VDAC1 domains were used as templates for creating VDAC1-based recombinant and synthetic peptides. The interaction of such peptides with Bcl-2 was demonstrated using surface plasmon resonance or when peptide expression prevented Bcl-2-mediated protection against cell death. These results thus offer new insight into the function of VDAC in mediating Bcl-2 antiapoptotic activity, actions that promote the survival of tumor cells.
MATERIALS AND METHODS
Materials
Soybean asolectin, carboxymethyl-cellulose, n-decane, Hepes, leupeptin, mannitol, phenylmethylsulfonyl fluoride, propidium iodide, staurosporine (STS), sucrose, and Tris were purchased from Sigma. ProteOn GLM sensor chips and hydroxyapatite (Bio-Gel HTP) were purchased from Bio-Rad. Monoclonal anti-VDAC antibodies prepared against the N-terminal region of 31HL human porin came from Calbiochem-Novobiochem (Nottingham, UK). Monoclonal antibodies against actin and Bcl-2 were obtained from Santa Cruz Biotechnology (Santa Cruz, CA). Monoclonal anti-cytochrome c antibodies were obtained from BD Biosciences Pharmingen (San Jose, CA). Horseradish peroxidase (HRP)-conjugated anti-mouse and anti-goat antibodies were obtained from Promega (Madison, WI). Celluspots peptide arrays were obtained from INTAVIS Bioanalytical Instruments (Koeln, Germany). Dulbecco's modified Eagle's medium growth media and the supplements fetal calf serum, l-glutamine, and penicillin-streptomycin, were purchased from Biological Industries (Beit Haemek, Israel). Blasticidin and zeocin were purchased from InvivoGen (San Diego, CA). N,N-Lauryl-(dimethyl)-amineoxide was obtained from Fluka (Buchs, Switzerland). Puromycin was purchased from ICN Biomedicals (Eschwege, Germany).
Plasmids and Site-directed Mutagenesis
Five mVDAC1-based peptide-encoding sequences (AP-W (Trp63–Asn78), BP-W (Pro105–Lys119), CP-W (Glu117–Thr134), DP-W (Lys199–Asn215), and the peptide corresponding to Met1–Gly26 N-terminal residues) were generated by standard PCR and cloned into the BamHI/EcoRI sites of the tetracycline-inducible pcDNA4/TO vector (Invitrogen), as described previously (19). The four VDAC1 loop-shaped peptides, i.e. AP-W to DP-W, were designed to be flanked by a tryptophan zipper motif, namely the SWTWE amino acid sequence, at the N terminus of the peptide and the KWTWK sequence at the C terminus of the peptide. The presence of these sequences induces the formation of a stable β-hairpin. See supplemental data for additional information.
A second set of peptides generated were designed according to the recently published VDAC1 topology model (28–30). These are termed L4-5 (Thr71–Leu80), L14-15, (Leu207–Arg217), L16-17 (Lys235–Gln248), and L18-19 (Leu262–His272) and correspond to loops connecting the indicated β-strands. Synthetic oligonucleotides corresponding to these sequences and containing AgeI and NotI restriction sites were cloned into the GFP-less pEGFP plasmid. Sequence fidelity was confirmed by sequencing.
Cell Lines and Growth
T-REx-293 cells, corresponding to transformed primary human embryonal kidney cells expressing the tetracycline repressor (Invitrogen), were maintained in a humidified atmosphere at 37 °C with 5% CO2. The cells were maintained in Dulbecco's modified Eagle's medium, supplemented with 10% fetal calf serum, 2 mm l-glutamine, 100 units/ml penicillin, 100 μg ml streptomycin, and 5 μg/ml blastocydin. hVDAC1-shRNA-T-REx-293 cells represent T-REx-293 cells silenced for the expression of endogenous human (h)VDAC1 using a shRNA-expressing vector (33).
Cell Transfection
Logarithmically growing T-REx-293 cells were transiently transfected with plasmid pEGFP-Bcl-2 alone or with a pcDNA4/TO plasmid encoding for one of the mVDAC1-peptides, or for native or mutated mVDAC1. The transfections were performed using the metafectene transfection reagent, according to the manufacturer's instructions or using calcium phosphate. Expression was induced for 48–72 h in cells grown in the presence of tetracycline (1–1.5 μg/ml). Cells were exposed to STS (1.25 μm) for the times indicated in Figs. 3, 5, and 7.
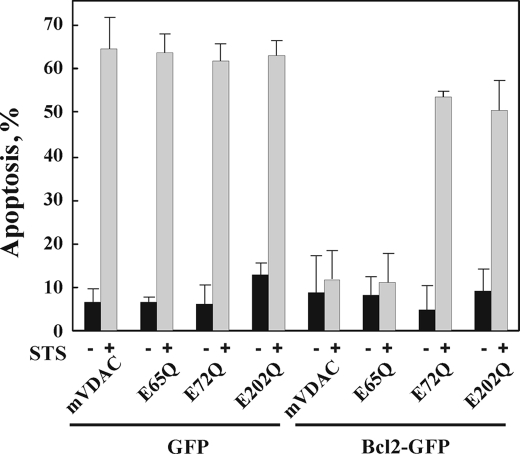
FIGURE 3.
Overexpression of Bcl-2 protects against apoptosis induced by STS in T-REx-293 cells overexpressing native mVDAC1 but not E65Q-, E72Q- or E202Q-mVDAC1 mutants. shRNA-T-Rex-293 cells were transiently cotransfected with plasmids pEGFP-Bcl-2 or pEGFP and pcDNA4/TO, respectively, encoding native, E65Q-, E72Q-, or E202Q-mVDAC1 (or pcDNA4/TO as control) and grown for 48 h in the presence of tetracycline (1 μg/ml, to induce expression) prior to exposure to STS (1.25 μm) for 4 h. Apoptosis was quantitatively analyzed by acridine orange/ethidium bromide staining. Data represent mean values ± S.E. (n = 3).
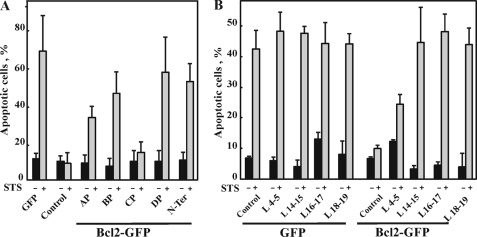
FIGURE 5.
Expression of VDAC1-based peptides in T-Rex-293 cells prevented Bcl-2-mediated protection against cell death induced by STS. T-REx-293 cells were transfected to express Bcl-2 and/or the indicated VDAC1-based peptide designed according to the original (AP, Trp63–Asn78; BP, Pro105–Lys119; CP, Glu117–Thr134; BP, Pro105–Lys119) (A) or newly proposed (L4-5, Thr71–Leu80; L14-15, Leu207–Arg217; L16-17, Lys235–Gln248) and (L18-19, Leu262-His272) (B) VDAC1 transmembrane topology models. VDAC1-based peptide expression was induced by tetracycline (1.5 μg/ml). Apoptosis in the different cell types was induced by incubation with STS (1.25 μm, 4 h) and quantitatively analyzed by acridine orange/ethidium bromide staining. Data represent mean ± S.E. (n = 3). N-ter, N-terminal.
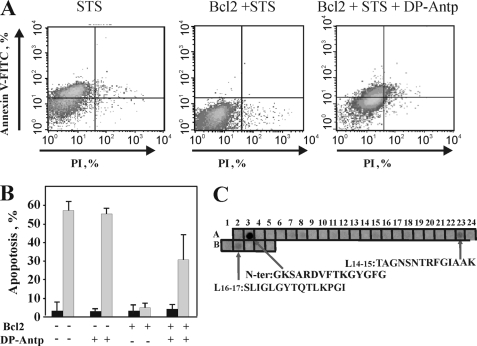
FIGURE 7.
The synthetic VDAC1-based peptide, DP-Antp, prevented Bcl-2-mediated protection against apoptosis and binding of Bcl-2(ΔC23) to a cellulose-bound VDAC1-based peptide array. T-REx-293 cells were transfected with pcDNA3.1 vector either empty or encoding Bcl-2. 48 h post-transfection, the cells were incubated with the synthetic peptide, DP-Antp (20 μm) for 90 min and/or STS (1.25 μm) for 3 h. Apoptotic cell death was analyzed using Annexin V-fluorescein isothiocyanate (V-FITC) and propidium iodide (PI) staining and FACS analysis, as described under “Materials and Methods.” A, representative FACS analysis data for cells treated with and without STS, and Bcl-2-expressing cells treated with STS or with both DP-Antp and STS. B, quantitative analysis of FACS experiments. Data represent mean values ± S.E. (n = 3). C, cellulose-bound peptide arrays consisting of overlapping peptides derived from VDAC1 sequence were incubated overnight with recombinant Bcl-2(ΔC23) (30 μg/ml) and blotted with anti-Bcl-2 antibodies, followed by incubation with HRP-conjugated anti-mouse IgG and detected using a chemiluminescence kit. Dark spots represent binding of Bcl-2(ΔC23) to the specific peptide corresponding to the N-terminal domain (N-Ter), and to peptide sequences overlapping with the L14-15 and L16-17 sequences (see Fig. 2). A representative blot of three similar experiments is shown.
VDAC Purification
VDAC was purified from sheep liver mitochondria after solubilization with N,N-lauryl-(dimethyl)-amineoxide and purified using hydroxyapatite and methylcellulose columns chromatography as described previously (21). For recombinant VDAC1 purification, mVDAC1 and E72Q-mVDAC1 were expressed in the Saccharomyces cerevisiae M22-2 por1 mutant strain (46) and purified from mitochondria as was rat liver mitochondria VDAC.
Bcl-2(ΔC23) Expression and Purification
DNA encoding Bcl-2(ΔC23) cloned in the pHisparallel1 vector and expressed in Escherichia coli BL21 cells. Bacteria were grown at 37 °C to an A600 of 0.4, exposed to a heat shock of 40 °C for 2 h, followed by induction of expression with isopropyl β-d-1-thiogalactopyranoside for 30–60 min at 20 °C. Following sonication and centrifugation, the soluble fraction (∼5–10% of the total expressed Bcl-2(ΔC23)) was purified by chromatography using nickel-nitrilotriacetic acid resin (5 ml). Bcl-2(ΔC23) was eluted from the column by a linear gradient of imadazol (10–500 mm). Purified Bcl-2(ΔC23) in 300 mm NaCl was stored in aliquots at −80 °C.
Reconstitution of Purified VDAC into a PLB
Single and multiple channel current recording and data analysis were carried out as described previously (47). Briefly, a PLB was prepared from soybean asolectin dissolved in n-decane (50 mg/ml). Purified VDAC (∼1–10 ng) was added to the chamber defined as the cis side containing 1 or 0.5 m NaCl. After one or a few channels were inserted into the PLB, excess protein was removed by prefusing the cis chamber with ∼20 volumes of solution to prevent further incorporation. Following several recordings of channel activity at different voltages, purified Bcl-2(ΔC23) (∼1–2 μg) was added, and currents through the channel were again recorded. Recordings were made under voltage clamp using a Bilayer Clamp BC-535B amplifier (Warner Instruments, Hamden, CT). Currents were measured with respect to the trans side of the membrane (ground). The currents were low pass-filtered at 1 kHz and digitized on-line using a Digidata 1440-interface board and Clampex 10.2 software (Axon Instruments, Union City, CA).
Bcl-2(Δ23) Binding to Cellulose-bound Peptides
The interaction of purified Bcl-2(ΔC23) with membrane-bound peptide arrays was assayed following membrane washing (3 times, 10 min each) with Tris-buffered saline (150 mm NaCl, 50 mm Tris-HCl, pH 7.4), followed by a 3-h incubation with blocking buffer (Tris-buffered saline containing low-fat dry milk, 3.5%, w/v). The membranes were then incubated overnight with purified Bcl-2(ΔC23) (2 μm in blocking buffer) at 4 °C. Following extensive membrane washing with Tris-buffered saline containing 0.05% Tween 20, Bcl-2(ΔC23) binding was detected using monoclonal anti-Bcl-2 antibodies and HRP-conjugated anti-mouse IgG as a secondary antibody. The blots were developed using EZ-ECL (Biological Industries), according to the manufacturer's instructions.
Peptide Synthesis
Synthetic N-terminal AP (Trp63-Asn78), BP (Pro105-Lys119), CP (Glu117-Thr134) and DP (Lys199-Asn215) VDAC1-based peptides, as well as the DP-Antp-looped peptide comprising peptide DP fused to the cell-penetrating peptide (Antp) (48, 49), were synthesized by GL Biochem (Shanghai, China). The N-terminal and DP peptides were dissolved in water, whereas the other peptides were dissolved in dimethyl sulfoxide.
Real-time Surface Plasmon Resonance
Surface plasmon resonance (SPR), using the ProteOn-XPR36 system (Bio-Rad) was employed to study the interactions of VDAC1-based synthetic peptides with purified Bcl-2(CΔ23). Purified Bcl-2(CΔ23) and rabbit IgG were immobilized onto a GLM sensor surface, according to the manufacturer's instructions. The peptides were diluted in running buffer (150 mm NaCl, 0.005% Tween 20, 4% (v/v) dimethyl sulfoxide, phosphate-buffered saline, pH 7.4) and injected onto the sensor chip at varying concentrations, at a flow rate of 40 μl/min. All experiments were carried out at 25 °C. Responses (resonance units) were monitored using the ProteOn imaging system and related software tools. Signals were normalized using appropriate controls.
Cell Death Analysis
To determine the extent of apoptosis, cells were subjected to staining with acridine orange and ethidium bromide, as described previously (46). Cells were visualized by fluorescence microscopy (Olympus IX51), and images were recorded with an Olympus DP70 camera, using a superwide band filter. In each independent experiment, ∼300 live, early, and late apoptotic cells were counted. Apoptosis was also analyzed by flow cytometric analysis using propidium iodide and Annexin V-FITC. Cells (2–4 × 106) were exposed to an apoptosis inducer, collected (1,500 × g for 5 min), washed twice with phosphate-buffered saline and resuspended in 400 μl of binding buffer (10 mm Hepes/NaOH, pH 7.4, 140 mm NaCl, and 2.5 mm CaCl2). Annexin V-FITC was added to a final concentration of 4.5 μg/ml, and the cells were incubated in the dark for 15 min. The cells were then washed twice and resuspended in 400 μl of binding buffer, to which propidium iodide was added immediately before analysis using ModFIT-lt2.0 software (FACScan, Beckton-Dickinson, San Jose, CA).
DP-Antp Treatment of Bcl-2-overexpressing Cells
T-REx-293 cells were transiently transfected with plasmid pcDNA3.1 Bcl-2 or the control plasmid, pcDNA3.1. Control or 48 h post-transfection cells were incubated for 90 min with the DP-Antp (Antennapedia homeodomain protein) peptide at a final concentration of 10 μm in serum-free Dulbecco's modified Eagle's medium and then exposed to STS (1.25 μm, 3 h). Cells were analyzed for cell death by FACS, using Annexin V-FITC and propidium iodide.
Gel Electrophoresis and Immunoblot Analyses
SDS-PAGE was performed according to Laemmli (50). Gels were stained with Coomassie Brilliant Blue. For immunostaining, membranes containing electrotransferred proteins were blocked with 5% nonfat dry milk and 0.1% Tween 20 in Tris-buffered saline, incubated with monoclonal anti-VDAC antibodies (1:10,000), and then with HRP-conjugated anti-mouse IgG (1:10,000). Enhanced chemiluminescent substrate (Pierce, Rockford, IL) was used for detection of HRP.
RESULTS
Members of the Bcl-2 family of proteins, such as Bcl-2, Bcl-xL, and Bax, have been proposed to interact with VDAC (5, 16–19). In this study, we addressed the interaction of Bcl-2 with VDAC using several approaches, including site-directed mutagenesis, modulation of VDAC channel activity, through the use of synthetic and recombinant VDAC1-based peptides, and by assessing the modulation of Bcl-2 antiapoptotic activity by such peptides.
Bcl-2 Interacts with VDAC to Induce Channel Closure
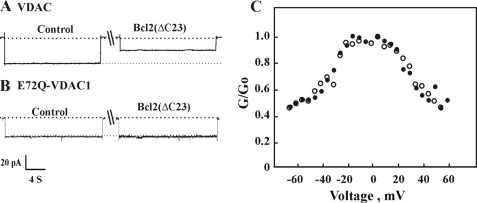
First, we demonstrated that purified recombinant Bcl-2 interacts directly with purified planar lipid bilayer-reconstituted VDAC to reduce channel conductance (Fig. 1). Recombinant Bcl-2(ΔC23) was expressed and purified from E. coli BL21 cells using a Hi-trap nickel column (Fig. 1D). Purified mitochondrial VDAC (Fig. 1D) was reconstituted into a PLB, and channel activity was studied under voltage-clamp conditions (47, 51). We found that the interaction of Bcl-2(ΔC23) with VDAC requires exposure of VDAC to high voltages, as demonstrated in the following experiment (Fig. 1A). When reconstituted VDAC was exposed to Bcl-2(ΔC23) at a holding potential of −10 mV, no effect on VDAC channel activity was observed (even after a 27-min incubation). However, when the voltage was stepped to −60 mV for 2 min, and channel conductance was recorded 3 min later at −10 mV, a decrease in channel conductance was observed, with the channel being stabilized in a low conducting state (Fig. 1A). Such voltage dependence suggests that Bcl-2 interacts with a high voltage-stabilized VDAC conformation. Thus, in all experiments involving the Bcl-2 interaction with bilayer-reconstituted VDAC, a 2-min exposure to high voltage was applied before current recording. Upon addition of purified Bcl-2(ΔC23) to bilayer-reconstituted VDAC, the current produced in response to voltage steps from a holding potential of 0 mV to −10 mV was recorded before and 10–15 min after the addition of Bcl-2(ΔC23). Channel conductance was reduced by Bcl-2(ΔC23), and the channel was stabilized in a low conducting state (Fig. 1B). Moreover, removing Bcl-2(ΔC23) upon replacement of bath solution with Bcl-2(ΔC23)-free solution restored channel conductance, as demonstrated in single channel (Fig. 1B) or multichannel (Fig. 1C) recordings. Bcl-2(ΔC23)-based modification of VDAC conductance was only observed when Bcl-2(ΔC23) was added to the cis side of the bilayer in which VDAC was reconstituted, indicating that Bcl-2(ΔC23) interacts with the cytosolic face of VDAC.
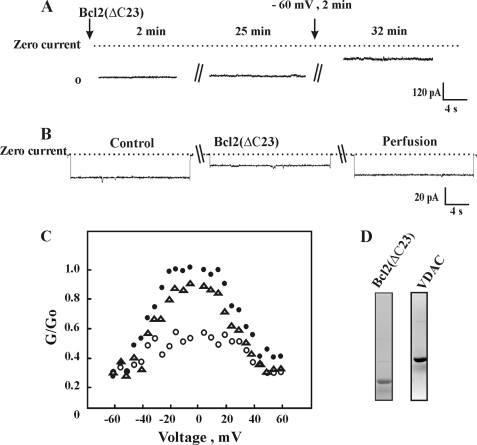
FIGURE 1.
Bcl-2(ΔC23) reduces bilayer-reconstituted VDAC conductance. A, VDAC was reconstituted into a PLB and current in response to a voltage step from 0 to −10 mV was recorded 2 and 25 min after the addition of purified Bcl-2(ΔC23) (1 nm). Thereafter, the voltage was switched for 2 to −60 mV, and current was recorded at −10 mV (total incubation time with Bcl-2(ΔC23) was 32 min). B, VDAC was reconstituted into a PLB, and currents through VDAC in response to a voltage step from 0 to −10 mV were recorded before and 15 min after the addition of purified Bcl-2(ΔC23) (1 nm) and after exposing the protein to high negative voltage for 2–3 min. Following current recording, the cis compartment was perfused to highly decrease the concentration of Bcl-2(ΔC23) by severalfold. The dashed lines indicate the zero current levels. C, shown are multichannel recordings as a function of the voltage (4 s at each voltage), and the average steady-state conductance of mVDAC1 before (●) and 30 min after (○) the addition of Bcl-2(ΔC23) (1 nm) and after exchanging the solution with Bcl-2(ΔC23)-free medium (▵) are presented. Relative conductance was determined as the ratio of the conductance at a given voltage (G) to the maximal conductance (Go). D, a Coomassie-stained purified VDAC and Bcl-2 proteins are shown.
Bcl-2(ΔC23) decreased VDAC conductance in multichannel experiments at all voltages tested and stabilized VDAC at a low conductance state, regardless of the voltage gradient applied. Channel conductance was restored close to the level noted before Bcl-2(ΔC23) addition upon removal of the added protein (Fig. 1C). These results suggest that Bcl-2 interacts reversibly and in a voltage-dependent manner with VDAC to modify VDAC conductance.
Mapping the Bcl-2-binding Site of VDAC1
To identify VDAC1 amino acid residues and domains involved in the interaction with the Bcl-2 protein, site-directed mutagenesis of VDAC1 and analysis of the antiapoptotic effect of Bcl-2 on cells expressing the mutations was carried out. The VDAC1 mutations E65Q, E72Q, and E202Q were selected based on their roles of these residues in preventing hexokinase isoforms, HK-I- and HK-II-mediated protection against apoptosis (16, 19, 67). The location of the selected mutated amino acids in both the previous (20) and newly proposed VDAC1 topology models (28–30) is presented in Fig. 2. The effects of these mVDAC1 mutations on Bcl-2-mediated protection against cell death were then analyzed by expression of native or mutated mVDAC1 in hVDAC1-shRNA T-REx-293 cells, where the endogenous VDAC1 level was suppressed (by ∼ 85%) or in the same cells overexpressing Bcl-2 (Fig. 3).
FIGURE 2.
VDAC1 transmembrane topology models showing the sites of mutated amino acids, the sequences used for synthetic peptide synthesis, and the proposed domains interacting with Bcl-2. The previously proposed transmembrane topology of VDAC1 (modified from Ref. 20) (A) and a revised model based on the newly proposed structure (28–30) are shown (B). The sequences of peptides (labeled in light gray) expressed or synthesized as synthetic peptides and tested in this work are illustrated. The black circles around Glu65, Glu72, and Glu202 and E65Q indicate mutations in mVDAC1 used in this study and were found to eliminate Bcl-2-mediated antiapoptotic activity. The dark gray lines indicate VDAC1 sequences interacting with Bcl-2 in the peptide array experiment (Fig. 7C).
hVDAC1-shRNA-T-REx-293 cells were transfected to express native, E65Q-, E72Q-, or E202Q-mVDAC1 under the control of tetracycline, alone or together with Bcl-2-GFP (Fig. 3). STS induced apoptosis in ∼65% of the control cells. This was almost completely prevented upon expression of Bcl-2-GFP. Protection against STS-induced apoptosis by Bcl-2-GFP was observed only in cells expressing native or E65Q-mVDAC1 but not in cells expressing E72Q- or E202Q-mVDAC1 (Fig. 3A). These results indicate that Bcl-2 interacts with native mVDAC1 but not with certain mVDAC1 mutations.
Next, the interaction of Bcl-2 with mutated E72Q-mVDAC1 was addressed using bilayer-reconstituted recombinant native or mutated mVDAC1. Native or E72Q-mVDAC1 were expressed in porin-less yeast, purified from their mitochondria, and reconstituted into a PLB (Fig. 4). The currents produced in response to voltages stepped from a holding potential of 0 mV to −10 were recorded before and 5 min after the addition of Bcl-2(ΔC23) (Fig. 4A). Upon addition of Bcl-2(ΔC23) to native mVDAC1, channel conductance was reduced, and the channel was stabilized in a low conducting state. On the other hand, Bcl-2(ΔC23) addition had no effect on the conductance of E72Q-mVDAC1 assayed at the single channel (Fig. 4B) or multichannel levels at all voltages tested (−60 to +60 mV) (Fig. 4C). These results suggest that Glu72 is essential for the interaction of VDAC1 with Bcl-2(ΔC23).
FIGURE 4.
Bcl-2(ΔC23) reduces channel conductance of bilayer-reconstituted native but not E72Q- mVDAC1. Purified recombinant, native (A) or E72Q-mVDAC1 (B) was reconstituted into a PLB in symmetrical solutions of 0.5 m NaCl, and currents through the VDAC1 channel in response to a voltage step from 0 to −10 were recorded before and 5 min after the addition Bcl-2(ΔC23) (1 nm), after applying the high voltage step (−60 mV, 2 min). The dashed lines indicate the zero and maximal current levels. C, multichannel recordings of the average steady-state conductance of mVDAC1 (●) or E72Q-mVDAC1 (○) 5 min after the addition of Bcl-2(ΔC23) (1 nm).
VDAC1-based Peptides Prevent Bcl-2-mediated Protection against Apoptosis
Next, based on the results obtained with mutated VDAC1 (Figs. 3 and 4), we constructed and expressed several VDAC1-based peptides and tested their effects on the antiapoptotic effects of Bcl-2 (Fig. 5). VDAC1-based peptides were expressed alone or together with Bcl-2, and apoptosis, as induced by STS, was analyzed in the various transfected cells (Fig. 5). Bcl-2 overexpression prevented apoptotic cell death as induced by STS. When cells expressing Bcl-2 were also transfected to express VDAC1-based peptides, designed according to the previously proposed VDAC1 topology model (19) (see Fig. 2), it was observed that the N-terminal, PA-W, PB-W, or PD-W but not the PC-W peptide prevented the protection (25–70%) afforded by Bcl-2 against STS-induced cell death (Fig. 5A).
VDAC1-based peptides (9–15 amino acids) designed according to recent x-ray crystallography-based and NMR-based topology models for recombinant VDAC1 were also tested (28–30) (see Fig. 2). These peptides correspond to loops connecting β-strands 4 and 5 (L4-5), β-strands 14 and 15 (L14-15), and β-strands 18 and 19 (L18-19), loops all facing the cytosol. Expression of peptides L14-15 and L18-19 completely (95%) prevented Bcl-2-mediated protection against STS-induced cell death, whereas peptide L4–5 conferred protection to a lesser extent (40%) (Fig. 5B). These results suggest that the VDAC1-based recombinant peptides interact with Bcl-2 and prevent its interaction with VDAC1, thereby preventing Bcl-2-mediated protection against cell death, as induced by STS.
Bcl-2 Interaction with VDAC1-based Peptides
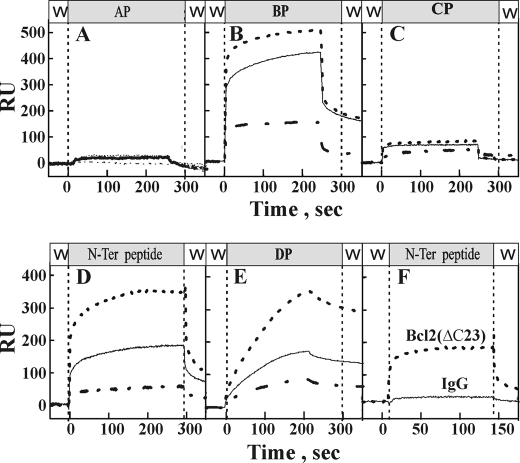
Based on the results obtained with mutated VDAC1 (Figs. 3 and 4) and the expressed peptides (Fig. 5), we generated the N-terminal, PA, PB, PD, and PC peptide sequences as synthetic peptides and analyzed their direct interactions with purified Bcl-2(ΔC23) using SPR technology (Fig. 6). Purified Bcl-2(ΔC23) was coupled to a SPR biosensor, the ProteOn GLM chip, and increasing concentrations (50–400 μm) of the VDAC1-based peptides were injected onto the sensor chips. Their binding to the immobilized Bcl-2(ΔC23) was then monitored. The N-terminal peptide and peptides PB and PD strongly bound to immobilized Bcl-2(ΔC23) in a concentration- and time-dependent manner (Fig. 6, B, D, and E). In contrast, the two other VDAC1-based peptides, i.e. peptides PA and PC, did not interact with Bcl-2(ΔC23) (Fig. 6, A and C). The binding of the PB, PD, and N-terminal peptides to Bcl-2(ΔC23) was specific, as no signal was obtained with a IgG-immobilized control chip (Fig. 6F). All Bcl-2-interacting VDAC1-based peptides associated relatively strongly with the immobilized Bcl-2. However, whereas the PB and N-terminal peptides showed rapid dissociation, the PD peptide showed slow dissociation from the immobilized Bcl-2 (ΔC23). The results thus demonstrate the direct and specific interaction of selected VDAC1-based peptides with Bcl-2(ΔC23).
FIGURE 6.
Selected VDAC1-based peptides interact with Bcl-2(ΔC23) in a specific and dose-dependent manner. Interaction of purified Bcl-2(ΔC23) with VDAC1-based peptides was revealed using real-time SPR. Bcl-2 immobilized onto a GLM sensor surface was exposed to VDAC1-based peptides: A, AP (Trp63–Asn78; 50, 100, and 200 μm); B, BP (Pro105–Lys119; 50, 100, and 200 μm); C, CP (Glu117–Thr134; 50, 100, and 200 μm); D, DP (Lys199–Asn215; 50, 100, and 200 μm); and E, N-terminal domain (Met1–Phe25; 50, 100, and 200 μm). To control for nonspecific binding, the interaction of the N-terminal peptide (N-ter; F, 200 μm) with surface strips of IgG was analyzed. The peptides were run in parallel over surface strips of Bcl-2(ΔC23) and responses (resonance units, RU) as a function of peptide concentration were monitored using the ProteOn imaging system and related software tools. All experiments were carried out at 25 °C.
For further confirmation of those VDAC1 sequences that interact with Bcl-2, a cellulose-bound peptide array consisting of overlapping peptides derived from human VDAC1 was used. The immobilized peptide array was exposed to purified Bcl-2(ΔC23), and interactions were revealed by immunostaining using anti-Bcl-2 antibodies. Interactions of Bcl-2(ΔC23) with three peptides corresponding to the N-terminal region and to sequences overlapping with L14-15 and L16-17 were noted (see Fig. 2), with the strongest interaction being with the N-terminal region peptide (Fig. 7A). These results are in correlation with the findings attained using SPR technology (Fig. 6).
Next, to allow passive penetration of PD (80% overlap with the L14-15 sequence) into the cell, we synthesized the PD-Antp peptide in which PD was flanked by a tryptophan zipper motif (see “Materials and Methods”) and attached at the N terminus to a 16-amino acid sequence representing the Antennapedia peptide Antp. Antp is a well known, nontoxic cell-penetrating peptide, which is able to facilitate the translocation of fused peptides across cell membrane (48, 49, 52, 53). STS treatment induced cell death in control but not in cells overexpressing Bcl-2-GFP. However, Bcl-2-mediated protection against STS-induced apoptosis was significantly reduced upon incubation with the PD-Antp peptide (Fig. 7, B and C). Although Bcl-2-GFP protected against STS-induced apoptosis by about 95% in cells exposed to the PD-Antp peptide, protection was reduced by 50% in the presence of the PD-Antp peptide.
DISCUSSION
A number of studies have elucidated the roles of pro- and antiapoptotic Bcl-2 family members in tumor pathogenesis, as well as expanding our understanding of how Bcl-2-like proteins maintain or perturb mitochondrial integrity (54). Moreover, high levels of antiapoptotic Bcl-2 family members are associated with resistance of many tumors to chemotherapy (6, 12, 13). Based on these and other studies, antiapoptotic Bcl-2 family members offer an attractive but challenging target for the development of anti-cancer agents. Indeed, numerous attempts have been made toward developing rational design-based anti-cancer therapies that directly target Bcl-2-regulated events at the level of mitochondria (55–58). However, while interfering with the prosurvival functions of Bcl-2 has long been considered an attractive manner to kill tumor cells, Bcl-2 proteins were simply deemed nondeliverable by pharmacological means. On the other hand, antisense-based strategies to reduce expression of Bcl-2 or Bcl-xL, such as Genasense (G3139/Oblimersen), an 18-mer phosphorothioate oligonucleotide targeted to Bcl-2 mRNA, were found to effectively inhibit the proliferation of human lymphoma cells (55, 57, 58). However, these antisense compounds were also reported to trigger inflammatory responses (57) and can only target one, or at best, two highly homologues Bcl-2 proteins at a time. Certain green tea catechins and black tea theaflavins were reported to be potent inhibitors of Bcl-xL and Bcl-2 (59). Other Bcl-2 inhibitors, known as “BH3 mimetics,” are nonpeptidic compounds rationally designed on the basis of protein-protein interactions between anti- and proapoptotic proteins of the Bcl-2 family. Such a putative BH3 mimetic, TW-37, was designed based on the structure of gossypol. A Bim-derived BH3 peptide (TW-37) bound to Bcl-2 was found to displace Bid from its binding to Bcl-2, Bcl-xL, and Mcl-1 and is proposed to kill in a Bak/Bax-dependent manner (56). The peptide was, however, also found to induce activation of the proapoptotic functions of p53 in melanoma cell lines (60). TW-37 was found to disrupt heterodimer formation between Bax or truncated-Bid and antiapoptotic proteins in the order Mcl-1 > Bcl-2 ≫ Bcl-xL) and proposed to be administrated together with standard chemotherapy as an effective strategy in the treatment of B-cell lymphoma (61).
These various approaches target the Bcl-2 family of proteins and either affect their expression levels or interactions between them. In this study and others (5, 16–19), we have reported another strategy for modifying the antiapoptotic activity of Bcl-2, namely interfering with the interaction of Bcl-2 with VDAC1. Initially, the interaction of Bcl-2 with VDAC1 was confirmed using several approaches. Purified Bcl-2 directly interacted with purified VDAC, as reflected in the decrease in conductance of bilayer-reconstituted native but not mutated VDAC1 in the presence of purified Bcl-2(ΔC23) (Figs. 1 and 4). Bcl-2 also interacted with immobilized VDAC1-derived peptides (Fig. 7A) (and VDAC1-based peptides interacted with immobilized Bcl-2), as revealed by surface plasmon resonance technology (Fig. 6). In addition, a VDAC1-based peptide linked to a cell-penetrating peptide derived from Antp, added to facilitate translocation across the cell membrane, was found to attenuate Bcl-mediated protection against STS-induced cell death (Fig. 7, B and C).
Based on the results obtained with site-directed mutations of VDAC1 eliminating protection against apoptosis induced by Bcl-2 (Fig. 3) and VDAC1-based peptide expression preventing Bcl-2 antiapoptotic activity (Figs. 5 and 7), possible interaction sites of VDAC1 with Bcl-2 were localized to sequences represented by peptides. These corresponding to residues Trp209–Gln224, located in β-strand 15, and residues Leu241–Gly253, located in β-strand 17, as well as the VDAC1 N-terminal domain (see Fig. 7 and Ref. 16). Interestingly, the peptide corresponding to Leu241–Gly253 represents VDAC1 sequences within β-strand 17, which, according to both the previous (20) and the revised (28–30) VDAC1 topology models, span the membrane (see Fig. 2). Because Bcl-2 is associated with the membrane and is suggested to be inserted into the membrane via its hydrophobic C-terminal domain (4), it is possible that Bcl-2 interacts with membrane-embedded VDAC1 sequences within β-strands 15–17. When studying the interaction of Bcl-2(ΔC23), which does not include the transmembrane domain, with VDAC1 or VDAC1-based peptides, the only possible interactions with VDAC1 would involve those channel amino acid residues facing the cytosolic surface. Thus, Bcl-2 interaction involves membranal and cytosolic VDAC1 sequences.
This study also explored the N-terminal region of VDAC1 as the interaction site of Bcl-2 (Figs. 5, 6, and 7C). This is of considerable of interest, given recent VDAC1 structural studies proposing that the N-terminal region of VDAC1 (residues 1–23) is nestled within the pore (28–30). These and other studies (62) further indicate that only part of the N-terminal is in the form of an α-helix. By contract, other approaches point to the N-terminal α-helix as being exposed to the cytoplasm (32), crossing the membrane (20), or lying on the membrane surface (36). Further evidence suggests that the N-terminal region is involved in channel voltage gating (63, 64) and exhibits motion during voltage changes (37). Thus, the N-terminal of VDAC1 might adopt different conformations and locations with respect to the VDAC1 β-strands and pore, depending on the apoptosis signaling conditions. This proposal is supported by the finding that exposure of bilayer-reconstituted VDAC to high voltages is required for Bcl-2-mediated modification of channel conductance. This suggest that the mobility of the VDAC1 N-terminal α-helix may modulate the accessibility of other VDAC1 domains for interaction with Bcl-2 (Fig. 1) and/or other apoptosis-regulating proteins of the Bcl-2 family (i.e. Bax and Bcl-xL) (16, 38). In addition, we have shown that the N-terminal region of VDAC1 is required for apoptosis induction, suggesting the it adopts different conformations in response to apoptotic signals (16). Our results demonstrating Bcl-2 interaction with the N-terminal region of VDAC1 (Figs. 5, 6, and 7C) suggest that such an interaction would prevent the N-terminal domain of VDAC1 from adapting an apoptosis-induced position, thus inhibiting induction of apoptosis.
We have also demonstrated that certain mutations in VDAC1, some which are localized to the membranal domain of a β-strand, such as E72Q (according to the new model, see Fig. 2), diminished Bcl-2 binding to VDAC1 (Fig. 4), whereas other mutations prevented the antiapoptotic effects of Bcl-2 (Fig. 3). This may be explained by the location of these amino acid residues in the Bcl-2-binding site, involving the membrane-inserted sequence of the protein. However, these mutations may modify the Bcl-2-recognized VDAC1 conformation. Indeed, NMR studies showed that the N-terminal domain of VDAC1 and the first four strands of the β-barrel are not stable in detergent and that such structural instability might be influenced by Glu73 (i.e. Glu72, when not counting the initial methionine) (28). Using hydrogen/deuterium exchange measurements coupled to NMR spectroscopy, it was demonstrated that the β-barrel becomes more stable by substitution of Glu73 with valine (28). Substitution of Glu73 with glutamine had a marked effect on VDAC1, abolishing ruthenium red- and HK I-mediated inhibition of VDAC1 channel activity and protection against apoptosis (19, 46, 65). Because the E72Q mutation affects ruthenium red, HK-I, HK-II, as well as Bcl-2-mediated protection against apoptosis, it is most likely that E72Q-VDAC1 adapts a conformation not recognized by these agents.
In summary, because high levels of antiapoptotic Bcl-2 family members are associated with resistance of many tumors to chemotherapy (6, 12, 13), targeting these proteins presents a possible strategy for cancer therapy. Solution of the structure of antiapoptotic Bcl-2 family member proteins has led to the design of novel small molecule inhibitors. Although many such molecules have been synthesized, rigorous verification of their specificity is often lacking (66).
Further studies have revealed that many putative Bcl-2 inhibitors are not specific and have other cellular targets, resulting in nonmechanism-based toxicity. Thus, based on the results obtained with site-directed mutations affecting the antiapoptotic effects of Bcl-2, using VDAC1-based peptides interacting with Bcl-2 and preventing the antiapoptotic activity of Bcl-2 and by considering the interaction of VDAC1-based peptides with purified Bcl-2, we suggest the use of such VDAC1-based peptides as a novel strategy for overcoming apoptosis resistance by interfering with the ability of Bcl-2 to prevent apoptosis and promote survival of tumor cells. Targeting of VDAC1-based peptide to tumor cells overexpressing antiapoptotic proteins, such as Bcl-2, would minimize the self-defense mechanisms of the cancer cells, thereby promoting apoptosis and increasing sensitivity to chemotherapy. Thus, VDAC1-based peptides offer an attractive target for further development as anti-cancer agents.
Supplementary Material
Acknowledgments
We thank Bio-Rad, Israel, for providing ProteOn instrument, Tsafrir Bravman and Dr. Monica Dines for help with the SPR experiments, and Dr. Asaf Friedler for helping with the peptide array experiments.
This work was supported in part by grants from the Israel Science Foundation and the Chief Scientist Office of the Ministry of Health, Israel.

The on-line version of this article (available at http://www.jbc.org) contains supplemental data.
- VDAC
- voltage-dependent anion channel
- hVDAC1
- human VDAC1
- mVDAC1
- murine VDAC1
- HRP
- horseradish peroxidase
- PLB
- planar lipid bilayer
- SPR
- surface plasmon resonance
- STS
- staurosporine
- shRNA
- short hairpin RNA
- FACS
- fluorescence-activated cell sorter
- GFP
- green fluorescent protein.
REFERENCES
- 1.Green D. R., Reed J. C. (1998) Science 281, 1309–1312 [DOI] [PubMed] [Google Scholar]
- 2.Kroemer G., Galluzzi L., Brenner C. (2007) Physiol. Rev. 87, 99–163 [DOI] [PubMed] [Google Scholar]
- 3.Cory S., Adams J. M. (2002) Nat Rev Cancer 2, 647–656 [DOI] [PubMed] [Google Scholar]
- 4.Gross A., McDonnell J. M., Korsmeyer S. J. (1999) Genes Dev. 13, 1899–1911 [DOI] [PubMed] [Google Scholar]
- 5.Shimizu S., Konishi A., Kodama T., Tsujimoto Y. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 3100–3105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Adams J. M., Cory S. (2007) Oncogene 26, 1324–1337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Danial N. N., Korsmeyer S. J. (2004) Cell 116, 205–219 [DOI] [PubMed] [Google Scholar]
- 8.Huang D. C., Strasser A. (2000) Cell 103, 839–842 [DOI] [PubMed] [Google Scholar]
- 9.Youle R. J., Strasser A. (2008) Nat. Rev. Mol. Cell Biol. 9, 47–59 [DOI] [PubMed] [Google Scholar]
- 10.Krajewski S., Tanaka S., Takayama S., Schibler M. J., Fenton W., Reed J. C. (1993) Cancer Res. 53, 4701–4714 [PubMed] [Google Scholar]
- 11.Gross A., Jockel J., Wei M. C., Korsmeyer S. J. (1998) EMBO J. 17, 3878–3885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sentman C. L., Shutter J. R., Hockenbery D., Kanagawa O., Korsmeyer S. J. (1991) Cell 67, 879–888 [DOI] [PubMed] [Google Scholar]
- 13.Miyashita T., Reed J. C. (1993) Blood 81, 151–157 [PubMed] [Google Scholar]
- 14.Scorrano L., Oakes S. A., Opferman J. T., Cheng E. H., Sorcinelli M. D., Pozzan T., Korsmeyer S. J. (2003) Science 300, 135–139 [DOI] [PubMed] [Google Scholar]
- 15.Cheng E. H., Wei M. C., Weiler S., Flavell R. A., Mak T. W., Lindsten T., Korsmeyer S. J. (2001) Mol Cell 8, 705–711 [DOI] [PubMed] [Google Scholar]
- 16.Abu-Hamad S., Arbel N., Calo D., Arzoine L., Israelson A., Keinan N., Ben-Romano R., Friedman O., Shoshan-Barmatz V. (2009) J. Cell Sci. 122, 1906–1916 [DOI] [PubMed] [Google Scholar]
- 17.Tajeddine N., Galluzzi L., Kepp O., Hangen E., Morselli E., Senovilla L., Araujo N., Pinna G., Larochette N., Zamzami N., Modjtahedi N., Harel-Bellan A., Kroemer G. (2008) Oncogene 27, 4221–4232 [DOI] [PubMed] [Google Scholar]
- 18.Malia T. J., Wagner G. (2007) Biochemistry 46, 514–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Arzoine L., Zilberberg N., Ben-Romano R., Shoshan-Barmatz V. (2009) J. Biol. Chem. 284, 3946–3955 [DOI] [PubMed] [Google Scholar]
- 20.Colombini M. (2004) Mol Cell Biochem. 256–257, 107–115 [DOI] [PubMed] [Google Scholar]
- 21.Shoshan-Barmatz V., Gincel D. (2003) Cell Biochem. Biophys 39, 279–292 [DOI] [PubMed] [Google Scholar]
- 22.Crompton M. (1999) Biochem. J. 341, 233–249 [PMC free article] [PubMed] [Google Scholar]
- 23.Halestrap A. P., Doran E., Gillespie J. P., O'Toole A. (2000) Biochem. Soc. Trans. 28, 170–177 [DOI] [PubMed] [Google Scholar]
- 24.Lemasters J. J. (1999) Cardiovasc. Res. 44, 470–473 [DOI] [PubMed] [Google Scholar]
- 25.Shoshan-Barmatz V., Israelson A., Brdiczka D., Sheu S. S. (2006) Curr. Pharm. Des. 12, 2249–2270 [DOI] [PubMed] [Google Scholar]
- 26.Shoshan-Barmatz V., Keinan N., Zaid H. (2008) J. Bioenerg. Biomembr. 40, 183–191 [DOI] [PubMed] [Google Scholar]
- 27.Tsujimoto Y., Shimizu S. (2002) Biochimie 84, 187–193 [DOI] [PubMed] [Google Scholar]
- 28.Bayrhuber M., Meins T., Habeck M., Becker S., Giller K., Villinger S., Vonrhein C., Griesinger C., Zweckstetter M., Zeth K. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 15370–15375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hiller S., Garces R. G., Malia T. J., Orekhov V. Y., Colombini M., Wagner G. (2008) Science 321, 1206–1210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ujwal R., Cascio D., Colletier J. P., Faham S., Zhang J., Toro L., Ping P., Abramson J. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 17742–17747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Colombini M. (2009) Trends Biochem. Sci 34, 382–389 [DOI] [PubMed] [Google Scholar]
- 32.De Pinto V., Messina A., Accardi R., Aiello R., Guarino F., Tomasello M. F., Tommasino M., Tasco G., Casadio R., Benz R., De Giorgi F., Ichas F., Baker M., Lawen A. (2003) Ital. J. Biochem. 52, 17–24 [PubMed] [Google Scholar]
- 33.Abu-Hamad S., Sivan S., Shoshan-Barmatz V. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 5787–5792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Junankar P. R., Dulhunty A. F., Curtis S. M., Pace S. M., Thinnes F. P. (1995) J. Muscle Res. Cell Motil. 16, 595–610 [DOI] [PubMed] [Google Scholar]
- 35.Shoshan-Barmatz V., Zalk R., Gincel D., Vardi N. (2004) Biochim. Biophys. Acta. 1657, 105–114 [DOI] [PubMed] [Google Scholar]
- 36.Reymann S., Flörke H., Heiden M., Jakob C., Stadtmüller U., Steinacker P., Lalk V. E., Pardowitz I., Thinnes F. P. (1995) Biochem. Mol. Med. 54, 75–87 [DOI] [PubMed] [Google Scholar]
- 37.Mannella C. A. (1997) J. Bioenerg. Biomembr. 29, 525–531 [DOI] [PubMed] [Google Scholar]
- 38.Shi Y., Chen J., Weng C., Chen R., Zheng Y., Chen Q., Tang H. (2003) Biochem. Biophys. Res. Commun. 305, 989–996 [DOI] [PubMed] [Google Scholar]
- 39.De Marchi U., Campello S., Szabò I., Tombola F., Martinou J. C., Zoratti M. (2004) J. Biol. Chem. 279, 37415–37422 [DOI] [PubMed] [Google Scholar]
- 40.Shimizu S., Narita M., Tsujimoto Y. (1999) Nature 399, 483–487 [DOI] [PubMed] [Google Scholar]
- 41.Sugiyama T., Shimizu S., Matsuoka Y., Yoneda Y., Tsujimoto Y. (2002) Oncogene 21, 4944–4956 [DOI] [PubMed] [Google Scholar]
- 42.Shimizu S., Tsujimoto Y. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 577–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shimizu S., Matsuoka Y., Shinohara Y., Yoneda Y., Tsujimoto Y. (2001) J. Cell Biol. 152, 237–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rostovtseva T. K., Antonsson B., Suzuki M., Youle R. J., Colombini M., Bezrukov S. M. (2004) J. Biol. Chem. 279, 13575–13583 [DOI] [PubMed] [Google Scholar]
- 45.Zheng Y., Shi Y., Tian C., Jiang C., Jin H., Chen J., Almasan A., Tang H., Chen Q. (2004) Oncogene 23, 1239–1247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zaid H., Abu-Hamad S., Israelson A., Nathan I., Shoshan-Barmatz V. (2005) Cell Death Differ. 12, 751–760 [DOI] [PubMed] [Google Scholar]
- 47.Gincel D., Zaid H., Shoshan-Barmatz V. (2001) Biochem. J. 358, 147–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Järver P., Langel U. (2004) Drug Discov Today 9, 395–402 [DOI] [PubMed] [Google Scholar]
- 49.Schimmer A. D., Hedley D. W., Chow S., Pham N. A., Chakrabartty A., Bouchard D., Mak T. W., Trus M. R., Minden M. D. (2001) Cell Death Differ. 8, 725–733 [DOI] [PubMed] [Google Scholar]
- 50.Laemmli U. K. (1970) Nature 227, 680–685 [DOI] [PubMed] [Google Scholar]
- 51.Azoulay-Zohar H., Israelson A., Abu-Hamad S., Shoshan-Barmatz V. (2004) Biochem. J. 377, 347–355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dinnen R. D., Drew L., Petrylak D. P., Mao Y., Cassai N., Szmulewicz J., Brandt-Rauf P., Fine R. L. (2007) J. Biol. Chem. 282, 26675–26686 [DOI] [PubMed] [Google Scholar]
- 53.Holinger E. P., Chittenden T., Lutz R. J. (1999) J. Biol. Chem. 274, 13298–13304 [DOI] [PubMed] [Google Scholar]
- 54.Chipuk J. E., Green D. R. (2008) Trends Cell Biol. 18, 157–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dias N., Stein C. A. (2002) Mol. Cancer Ther. 1, 347–355 [PubMed] [Google Scholar]
- 56.Wang G., Nikolovska-Coleska Z., Yang C. Y., Wang R., Tang G., Guo J., Shangary S., Qiu S., Gao W., Yang D., Meagher J., Stuckey J., Krajewski K., Jiang S., Roller P. P., Abaan H. O., Tomita Y., Wang S. (2006) J. Med. Chem. 49, 6139–6142 [DOI] [PubMed] [Google Scholar]
- 57.Kim R., Emi M., Matsuura K., Tanabe K. (2007) Cancer Gene Ther. 14, 1–11 [DOI] [PubMed] [Google Scholar]
- 58.Vidal L., Blagden S., Attard G., de Bono J. (2005) Eur. J. Cancer 41, 2812–2818 [DOI] [PubMed] [Google Scholar]
- 59.Leone M., Zhai D., Sareth S., Kitada S., Reed J. C., Pellecchia M. (2003) Cancer Res. 63, 8118–8121 [PubMed] [Google Scholar]
- 60.Verhaegen M., Bauer J. A., Martín de la Vega C., Wang G., Wolter K. G., Brenner J. C., Nikolovska-Coleska Z., Bengtson A., Nair R., Elder J. T., Van Brocklin M., Carey T. E., Bradford C. R., Wang S., Soengas M. S. (2006) Cancer Res. 66, 11348–11359 [DOI] [PubMed] [Google Scholar]
- 61.Mohammad R. M., Goustin A. S., Aboukameel A., Chen B., Banerjee S., Wang G., Nikolovska-Coleska Z., Wang S., Al-Katib A. (2007) Clin. Cancer Res. 13, 2226–2235 [DOI] [PubMed] [Google Scholar]
- 62.De Pinto V., Reina S., Guarino F., Messina A. (2008) J. Bioenerg. Biomembr. 40, 139–147 [DOI] [PubMed] [Google Scholar]
- 63.Koppel D. A., Kinnally K. W., Masters P., Forte M., Blachly-Dyson E., Mannella C. A. (1998) J. Biol. Chem. 273, 13794–13800 [DOI] [PubMed] [Google Scholar]
- 64.Popp B., Court D. A., Benz R., Neupert W., Lill R. (1996) J. Biol. Chem. 271, 13593–13599 [DOI] [PubMed] [Google Scholar]
- 65.Israelson A., Zaid H., Abu-Hamad S., Nahon E., Shoshan-Barmatz V. (2008) Cell Calcium 43, 196–204 [DOI] [PubMed] [Google Scholar]
- 66.Vogler M., Dinsdale D., Dyer M. J., Cohen G. M. (2009) Cell Death Differ. 16, 360–367 [DOI] [PubMed] [Google Scholar]
- 67.Abu-Hamad S., Zaid H., Israelson A., Nahon E., Shoshan-Barmatz V. J. (2008) J. Biol. Chem. 283, 13482–13490 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.