Abstract
Soluble amyloid oligomers are potent neurotoxins that are involved in a wide range of human degenerative diseases, including Alzheimer disease. In Alzheimer disease, amyloid β (Aβ) oligomers bind to neuronal synapses, inhibit long term potentiation, and induce cell death. Recent evidence indicates that several immunologically distinct structural variants exist as follows: prefibrillar oligomers (PFOs), fibrillar oligomers (FOs), and annular protofibrils. Despite widespread interest, amyloid oligomers are poorly characterized in terms of structural differences and pathological significance. FOs are immunologically related to fibrils because they react with OC, a conformation-dependent, fibril-specific antibody and do not react with antibodies specific for other types of oligomers. However, fibrillar oligomers are much smaller than fibrils. FOs are soluble at 100,000 × g, rich in β-sheet structures, but yet bind weakly to thioflavin T. EPR spectroscopy indicates that FOs display significantly more spin-spin interaction at multiple labeled sites than PFOs and are more structurally similar to fibrils. Atomic force microscopy indicates that FOs are approximately one-half to one-third the height of mature fibrils. We found that Aβ FOs do not seed the formation of thioflavin T-positive fibrils from Aβ monomers but instead seed the formation of FOs from Aβ monomers that are positive for the OC anti-fibril antibody. These results indicate that the lattice of FOs is distinct from the fibril lattice even though the polypeptide chains are organized in an immunologically identical conformation. The FOs resulting from seeded reactions have the same dimensions and morphology as the initial seeds, suggesting that the seeds replicate by growing to a limiting size and then splitting, indicating that their lattice is less stable than fibrils. We suggest that FOs may represent small pieces of single fibril protofilament and that the addition of monomers to the ends of FOs is kinetically more favorable than the assembly of the oligomers into fibrils via sheet stacking interaction. These studies provide novel structural insight into the relationship between fibrils and FOs and suggest that the increased toxicity of FOs may be due to their ability to replicate and the exposure of hydrophobic sheet surfaces that are otherwise obscured by sheet-sheet interactions between protofilaments in a fibril.
Keywords: Aging, Diseases/Aging, Diseases/Amyloid, Diseases/Neurodegeneration, Peptides/Conformation, Protein/Assembly, Protein/Folding, Fibril Formation, Oligomerization, Seeding
Introduction
The accumulation of aggregated amyloid proteins is a characteristic hallmark of a wide range of human degenerative diseases, including Alzheimer (AD),2 type II diabetes, Huntington, Parkinson, and spongiform encephalopathy (1, 2). Although familial mutations that result in increased production of Aβ42 support a causal role of Aβ peptide, the mechanism of Aβ pathogenesis remains unknown and controversial. Aβ plaques composed of Aβ fibrils are evident in both patients with AD and a significant number of healthy individuals who are cognitively normal (3). Emerging evidence implicate soluble oligomers formed during protein aggregation are the primary toxic species in amyloid pathogenesis (4). Several studies have shown that Aβ oligomers are toxic in cell culture and transgenic animal models (5–10). Aβ oligomers are found in AD brain, where they have a localization pattern that is distinct from diffuse and thioflavin-positive plaques (5). The increased level of soluble oligomers also correlates better than plaque load with cellular dysfunction and the severity of cognitive impairment in AD patients and transgenic animal models (5, 7, 11). The observation that amyloid oligomers derived from protein sequences that are not related to a human disease are toxic to cell culture models (12) suggests an inherent and generic toxicity of amyloid oligomers.
Although these reports of toxic oligomers are in agreement in terms of solubility, their size, morphology, and immunoreactivity varies indicating that amyloid oligomers are structurally diverse (2). Aβ oligomers have been prepared by several different protocols and variously referred to as Aβ-derived diffusible ligands, globulomers, protofibrils, and amyloid spheroids (6, 13–15). Oligomers that correlate with cognitive dysfunction in transgenic animals have been reported to be 56 kDa (Aβ*56) as determined by SDS-PAGE, whereas oligomers purified from cell culture and from human AD brains have been reported to migrate at the position of dimers (9, 16, 17). Synthetic Aβ oligomers between 50 and 100 kDa in size are also reported as toxic and highly correlative to synaptic loss in cell culture models (11, 18). A similar spectrum of soluble oligomers has been observed for other amyloidogenic proteins, including islet amyloid polypeptide, α-synuclein, prion, insulin, and lysozyme (19–23). It is unknown whether these oligomeric species represent the same or distinct structural variants. Discriminating among the different structures of the aggregated species is the first step to advance our understanding of the aggregation mechanism of amyloid proteins and to clarify the role of oligomers in amyloid pathogenesis.
In the absence of high resolution x-ray crystallographic structural data, conformation-specific antibodies provide a means of distinguishing and classifying the different types of amyloid aggregates by their underlying structures. We have previously reported three antibodies, A11, OC, and α-annular protofibrils, that recognize mutually exclusive generic epitopes specifically associated with prefibrillar oligomers (PFOs), fibrils, and annular protofibrils formed of different amyloid proteins, respectively (5, 24, 25). Interestingly, the OC antibody also recognizes small and soluble oligomers that share the same epitope as fibrils (24). The fact that FOs and fibrils share the same epitope suggests that FOs are structurally similar to fibrils and distinct from PFOs. Because fibril formation is known be nucleation-dependent, the simplest interpretation is that FOs represent fibril nuclei or small pieces of fibril filaments organized in the same structural lattice as in fibrils.
In this study, we examined the properties of Aβ fibrillar oligomers and their relationship to Aβ fibrils in greater detail. Although FOs are immunologically related to fibrils, their morphology, size, and tinctorial properties are distinct from fibrils. Surprisingly, we found that Aβ FOs do not seed the formation of ThT-positive fibrils from Aβ monomers. Instead, FOs nucleate the formation of soluble FOs that are ThT-negative, positive for the anti-fibril antibody, OC, and have the same size and morphology as the FOs seeds. Taken together, these data suggest that FOs may represent small pieces of soluble single fibril protofilaments that are capable of replication by the addition of monomers to the ends of FOs and subsequent splitting. This suggests that the replication of FOs may be a key determinant of the enhanced pathogenic activity of amyloid oligomers.
EXPERIMENTAL PROCEDURES
Aβ Preparations
For preparation of monomers, synthetic Aβ40 (0.3 mg) was solubilized in 150 μl of hexafluoro-2-propanol (HFIP), evaporated to dryness under a stream of N2, diluted in H2O, and used immediately. FOs were prepared by first dissolving Aβ in HFIP at a concentration of 420 μm for 25 min at room temperature. Then the peptide solution was diluted into double distilled H2O (0.02% sodium azide), 70 μm, and the pH was adjusted to 7.4 with NaOH. The sample was stirred at 500 rpm with a Teflon-coated micro stir bar for 2–3 days at room temperature with a cap that has holes. PFOs were prepared as described previously (5). The preparation was similar to that of FOs except the pH was adjusted to 2.5 with HCl. Fibrils were prepared by dissolving Aβ (2 mm) in 100 mm NaOH, followed by bath sonication for 30 s, and then stirring in 10 mm HEPES, 10 mm NaCl, 0.02% sodium azide, pH 7.4, at 500 rpm for 9–10 days. Solubility was assayed by centrifuging Aβ solutions at 100,000 × g for 1 h at room temperature. The pellets were collected, washed three times in double distilled H2O, and resuspended in desired buffers.
Dot Blots
Dot blot assays were performed to detect Aβ PFOs, FOs, and fibrils as described previously (5). In the case of the FO seeding experiment, the fibril-specific antibody LOC was used. The LOC antibody was generated against the islet amyloid polypeptide fibrils, and like OC, it recognizes a variety of fibrils and FOs formed from different amyloidogenic proteins and peptides, including Aβ, but it does not detect Aβ monomers or PFOs (24). In using the LOC antibody, we further ensured that only Aβ fibrils and FOs, and not Aβ monomers, will be detected. Briefly, 1-μl spots containing 0.3 μg of Aβ40 were applied to a nitrocellulose membrane and blocked in 10% nonfat dry milk in Tris-buffered saline (TBS-T) containing 0.01% Tween 20 at room temperature for 1 h. After three 5-min TBS-T washes, the membranes were probed with conformation-specific antibodies (A11, 1:2000; OC, 1:10,000; LOC, 1:2000) overnight at 4 °C. The membranes were then incubated with anti-rabbit IgG conjugated with horseradish peroxidase (1:10,000, Promega) at room temperature for 1 h. The blots were developed with ECL chemiluminescence kit from Amersham Biosciences.
Western Blot
Samples containing 4 μg of Aβ40 were dissolved in SDS treatment buffer, boiled for 5 min, and electrophoresed on 10–20% Tris-HCl (Bio-Rad) gels. Proteins were electrophoretically transferred onto nitrocellulose membranes and developed with conformation-specific antibodies as described above.
ThT Fluorescence Assay
ThT fluorescence was used to monitor Aβ fibril formation, as described previously (26). Samples of Aβ conformers were mixed with ThT solution (3 μm in 10 mm Na2HPO4, pH 7.4), and fluorescence was measured in a Gemini XPS plate reader (Molecular Devices, Sunnyvale, CA). For seeding experiments, reactions were performed in 96-well plates, and the ThT fluorescence signals were read at 10-min intervals. All experiments were performed in triplicate.
Electron Microscopy
One-μl aliquots of Aβ monomers, PFOs, FOs, and fibrils were adsorbed onto 200-mesh Formvar/carbon-coated nickel grids until dry. The grids were then washed with water, stained with 2% uranyl acetate, and washed again. The grids were allowed to dry between all steps and were viewed in a Phillips CM 12 microscope operated at 65 kV.
Atomic Force Microscopy
AFM analysis was carried out using a Nanoscope III multimode instrument (Veeco Instruments, Santa Barbara, CA). 1.5 μl of sample solutions were deposited on freshly cleaved mica coated with poly-l-lysine (0.01% w/v) for 1–2 min. Then the sample was rinsed with double distilled H2O and dried under a stream of N2 gas. Imaging was done in tapping mode with a silicon cantilever at a scan frequency of 1 Hz and an oscillation frequency of 165 kHz. The heights of individual fibrils were measured perpendicular to the fibrillar axis (in case of fibrils) using the profile tool in SPIP software (Image Metrology) and using scan sizes of less than 3 μm2. Height analysis was calculated using Voss volume software (27).
FTIR
FTIR spectra were measured with a Bruker Equinox 55 FTIR instrument (Bruker Optics, Billerica, MA) equipped with a deuterated triglycine sulfate detector. Aqueous solutions of either fibrils or FOs of Aβ40 were deposited on the calcium fluoride glass and dried under nitrogen. 512 scans at 2 cm−1 resolution were collected for each sample under constant purging with nitrogen and corrected for water vapor, and background spectra were subtracted.
Preparation of Spin-labeled Aβ Peptides
Spin-labeled Aβ40 derivatives were generated through labeling of single cysteine mutants. Single cysteine mutants were synthesized, purified, and verified by mass spectrometry as described previously (28). For labeling, lyophilized Aβ40 cysteine mutants were initially dissolved in 100% DMSO and then diluted to 50% with Milli-Q water. 3-Fold molar excess of (1-oxy-2,2,5,5-tetramethypyrroline-3-methyl)-methanethiosulfonate spin label was used to label the peptide for 1 h at room temperature. Hydrochloric acid was then added to a final concentration of 1 mm, and excess spin label was removed using a cation exchange column (Toyopearl resin, Tosoh Corp.). The peptides were washed with a minimum of 10 column volumes of 1% acetic acid or until no more free spin label was detectable by EPR spectroscopy. The spin-labeled peptides were eluted from the column using buffer containing 6 m guanidine HCl in 100 mm Tris, pH 7.4. Subsequently, C-18 Macro SpinColums (Harvard Apparatus) were used for desalting, washing, and eluting of the spin-labeled peptides in 100% HFIP. Finally, the concentration was determined using absorbance at 280 nm and an extinction coefficient of 1280 for the single tyrosine. Samples were aliquoted and lyophilized for later use.
EPR Analysis of Aβ Oligomers
To prepare fibrillar oligomers for EPR analysis, lyophilized spin-labeled peptide were resuspended in HFIP (stock concentration of 1 mm peptide) for 30 min at room temperature. This stock solution was then diluted 1 to 4 into a 10% HFIP solution, giving a final peptide concentration of ∼250 μm and an HFIP concentration of greater than 20%. HFIP was then removed by evaporation with a stream of N2 gas for about 10 min. Small aliquots (1–2 μl) were saved for dot blot analysis with OC and A11 antibodies. Oligomers prepared in this way were positive to the OC antibodies but not to the A11 antibodies. Prefibrillar oligomers were made and assayed by dot blot as described previously (5). EPR spectra were recorded at room temperature with a Bruker EMX X-band EPR spectrometer fitted with an ER4119HS resonator. Some spectra contained up to 2% spectral components arising from monomeric unfolded peptide. These components were subtracted, and the resulting spectra were normalized to same number of spins and base-line corrected. All spectra are shown with a magnetic field scan width of 150 G.
RESULTS
Kinetics of Soluble Oligomer Formation
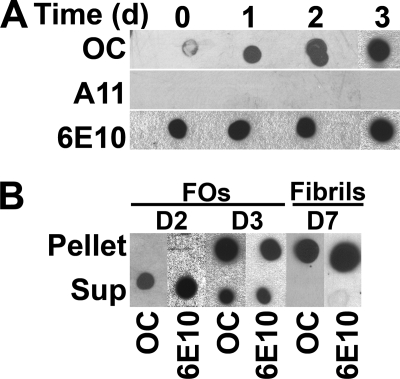
We have previously reported that the fibril-specific, conformation-dependent antibody, OC, also recognizes 100,000 × g soluble oligomers that range in size from dimer to greater than 250 kDa on Western blots (24). To characterize the relationship of these oligomers to fibrils, we investigated the kinetics of FO in comparison with fibril formation. FO formation was monitored by OC immunoreactivity, whereas fibril formation was measured by centrifugation at 100,000 × g (Fig. 1, A and B). OC immunoreactivity of Aβ monomer was minimal at the initial time point and increased after 24 and 48 h (Fig. 1A). No A11 immunoreactivity was detected under these conditions, indicating that PFOs are not formed under these conditions. 6E10 immunoreactivity was uniform over the time course, indicating that equal amounts of peptide were spotted and demonstrating the conformation-specific nature of the OC epitope. OC immunoreactive Aβ42 FOs are completely soluble at 100,000 × g after 48 h of incubation, whereas most of the OC immunoreactive material sediments after 72 h of incubation (Fig. 1B). These data demonstrate that soluble FOs appear in solution prior to the formation of sedimentable fibrils and that FOs eventually form fibrils.
FIGURE 1.
Kinetics and solubility of Aβ42 FO assembly. Aβ42 was dissolved in HFIP and incubated in double distilled H2O, pH 7.4, with stirring (70 μm final concentration). At the time indicated, aliquots were removed and analyzed by dot blot and ultracentrifugation. A, aliquots of Aβ42 were spotted onto nitrocellulose membrane and probed with OC, A11, and 6E10 antibodies. FO-specific immunoreactivity formed within 1 day (d) of aggregation. Under this condition, the FOs formed are homogenous and did not react with the anti-PFO antibody A11. B, aggregation assay and fibrils were also examined by ultracentrifugation at 100,000 × g for 1 h. The supernatant (Sup) and pellet fractions were separated and resuspended in equal volumes of desired buffers. Aβ FOs were soluble in contrast to mature fibrils that sedimented at high speed.
Fibrillar Oligomers Are Rich in β-Sheet Structure but Weakly Bind ThT
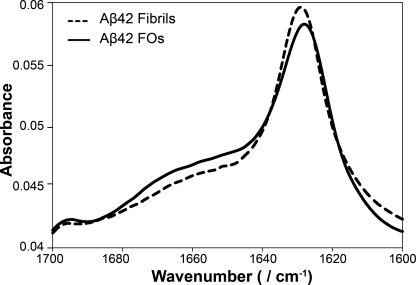
We further analyzed the secondary structure of Aβ FOs using FTIR spectroscopy. The FTIR spectrum of Aβ42 FOs showed the presence of a high content of β-sheet structure with prominent bands at 1629 and 1698 cm−1 (Fig. 2). Additionally, FOs exhibited a broader shoulder at 1640–1670 cm−1 indicative of a higher content of random coil, α-helices, and β-turns. The spectrum of FOs was very similar to the spectrum of Aβ42 fibrils except the intensity of the shoulder at 1640–1670 cm−1 was lower in fibrils indicating that they contain a higher percentage of β-sheets. The spectral similarity between the spectra of FOs and fibrils is consistent with their common immunoreactivity with the fibril-specific conformation-dependent antibody, OC, which indicates that they display the same conformational epitope.
FIGURE 2.
FTIR spectroscopy reveals structural similarity between fibrils and FOs. FTIR spectra of Aβ42 fibrils (dashed line) and FOs (solid line) indicate similar β-sheet-rich secondary structures.
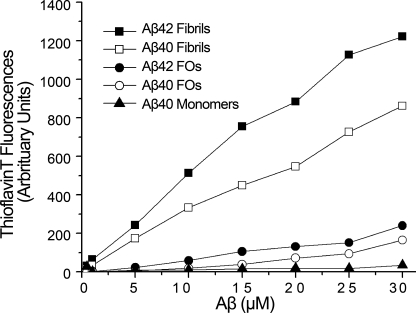
To further compare the properties of FOs and fibrils, we examined their ability to bind to ThT. ThT is a fluorophore that selectively exhibits fluorescence at 480 nm upon binding specifically to amyloid fibrils in vitro and in vivo and not to amyloid precursor protein, monomers, or other β-sheet-rich amyloid aggregates (29). We freshly prepared Aβ42 and Aβ40 monomers, FOs, and fibrils and measured the dependence of ThT fluorescence on the concentration of these Aβ aggregates. Aβ40 monomers exhibited base-line ThT fluorescence as expected (Fig. 3). In comparison, both Aβ40 and Aβ42 FOs weakly enhance the fluorescence of ThT. The extent of fluorescence enhancement by FOs is similar to that of PFO, which has been reported previously (30). However, the fluorescence enhancement of fibrils is 33-fold higher than FOs (Fig. 3). This difference suggests that there is a structural difference between fibrils and FOs that is not apparent by OC immunoreactivity.
FIGURE 3.
FOs are distinct from fibrils by thioflavin T binding. Aliquots of Aβ42 and Aβ40 were added to 3 μm ThT in 10 mm sodium phosphate, pH 7.4. ThT fluorescence was measured as described under “Experimental Procedures.” Significant dosage-dependent increase in ThT binding is only observed in fibrils but not in FOs or monomers.
Fibrillar Oligomers Are Structurally Related to Fibrils and Distinct from PFOs
To compare the structural features of the different oligomer preparations, we generated spin-labeled Aβ oligomers that were either positive to the OC (FOs) or A11 antibody (PFOs) and recorded their EPR spectra (Fig. 4). We have previously shown that spin-labeled Aβ peptides retain the ability to form amyloid fibrils and co-assemble normally with unlabeled Aβ (28). As illustrated with the example of Aβ labeled at position 14 (14R1), FOs and PFOs give rise to distinctly different spectra. The most striking difference is the reduced signal amplitude of OC-positive FOs, which is caused by significant line broadening that extends beyond the typical ∼70 G width of a nitroxide spectrum (Fig. 4B). This broadening is a clear indicator of strong spin-spin interaction that is present in the OC-positive oligomers. Although some spin-spin interaction is also visible in the A11-positive PFOs, it is clearly much smaller and only has minor spectral contributions. These results indicate that the spin labels in PFOs are farther apart than in fibrils and FOs. To further demonstrate the spin-spin interaction in the OC-positive FOs, we performed dilution experiments containing only 10% R1-labeled proteins. Under these conditions, the effects of spin-spin interaction should be strongly alleviated, and in fact, a pronounced increase in signal intensity and loss of the broadening can be observed (Fig. 4C).
FIGURE 4.
Comparison of EPR spectra for Aβ labeled at position 14 in different oligomeric or fibrillar forms. A shows the EPR spectrum of the fibrils, which was reported previously and is reproduced here for comparison. The EPR spectra for FOs (B, solid line) are of much lower amplitude than those of PFOs (B, dashed line). The reduction in amplitude coincides with significant spectral broadening beyond 70 G indicating the presence of strong spin-spin interaction. The presence of spin-spin interaction is illustrated in C, which overlays the spectrum of FOs labeled at 100% (solid line) with the spectrum of FOs generated from 10% of spin-labeled peptide and 90% of wild-type peptide (dashed line). In contrast to the FOs, the spectra of the fibrils contain a clear component of spin exchange, which is characterized by single line EPR spectra (D) (32). The spectral component was obtained by subtraction of the hyperfine structure from the spectrum in A. Similar spectral subtractions did not yield any evidence for single line, exchange narrowed EPR spectra for FOs. All spectra were obtained at the X-band, normalized to the same number of spins, and are shown at a scan width of 150 G.
Overall, the strong spin-spin interaction makes the EPR spectra of the OC-positive oligomers more similar to those of the fibrils, whose parallel, in-register structure brings neighboring labeled sites into close contact (Fig. 4A). Despite these similarities, however, analysis of the EPR spectra for fibrils and FOs reveals some differences also. The EPR spectrum for the fibril contains a significant spectral component of spin exchange narrowing (shown after spectral subtraction in Fig. 4D). This component indicates the simultaneous contact (orbital overlap) of multiple spin labels in agreement with a parallel in-register structure formed by multiple strands (31, 32). No such component can readily be detected in the case of the OC-positive FOs. Moreover, comparison of the EPR spectra in Fig. 4C reveals that there is a significant distribution of distances between labeled side chains from close to contacts to distances that are in the 10–20 Å range. Thus, although the FOs are likely structurally related to the fibrils, there appears to be significantly more heterogeneity in spin-spin interaction. This notion is further supported by spectral analysis of several other labeled sites that display the same spectral features (supplemental Fig. 1). In general, the EPR spectra from all sites indicate that the OC-positive FOs (and fibrils) have significantly more spin-spin interaction than the A11-positive PFOs. The only site that gives rise to relatively little spin-spin interaction for all of the forms is the carboxyl-terminal residue at position 40.
Fibrillar Oligomers Are One-half to One-third the Height of Fibrils
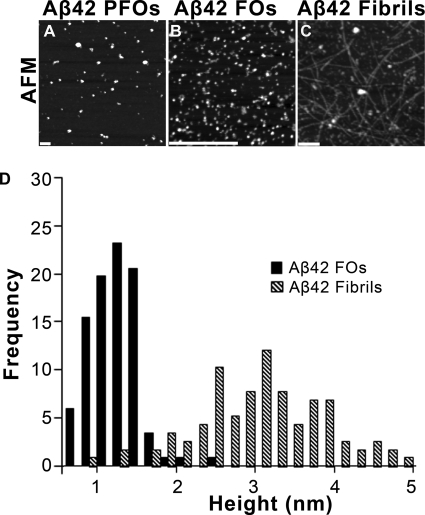
To gain insight into the morphological relationship between PFOs, FOs, and fibrils, we examined them by atomic force microscopy. PFOs appeared as large globular aggregates that are 20–40 nm in diameter, whereas FOs are smaller aggregates that are 5–15 nm in diameter (Fig. 5, A and B). Fibrils, on the other hand, were straight, 10–20 nm in width, 500–900 nm long, and consisted of multiple filaments (Fig. 5C). Fibrils also exhibit morphological diversity with respect to their width, length, and degree of twisting, consistent with the well known structural polymorphism of fibrils (33). Height analysis reveals that the height of FOs is distributed within a narrow range (1.13 ± 0.08 nm) that is consistent with the presence of a single sandwich of two β-sheets (Fig. 5D). Based on their dimensions in AFM, FOs are calculated to contain 3–10 Aβ monomers in agreement with their size distribution on Western blots (supplemental Fig. 2). For the calculation, the volume of the oligomer was obtained from the AFM and EM data, and the volume of each monomer was estimated assuming peptide partial specific volume of 0.73 cm3/g (27). AFM analysis revealed the heights of Aβ42 fibrils are in a broad range of 2–5 nm (Fig. 5D). The fact that FOs are one-half to one-third the height of fibrils suggests that they may represent small segments of the protofilaments that make up mature fibrils.
FIGURE 5.
Morphological analysis of Aβ42 PFOs, Fos, and fibrils. A–C, AFM shows that Aβ42 PFOs, FOs, and fibrils have different distinct morphologies. FOs are small spherical aggregates that are 5–10 nm in diameter. Mature fibrils are filaments that are 6–10 nm in diameter and 1–3 μm long (scale bars, 200 nm). D, height distribution of Aβ42 FOs (solid) and fibrils (dashed) as analyzed by AFM. The data were collected by an AFM operating in air using tapping mode by measuring the heights in cross-section of >100 oligomers or fibrils.
Fibrillar Oligomers Nucleate the Oligomerization of Monomeric Aβ and Do Not Seed Fibrils
Because FOs are small and soluble oligomers that share secondary structure and immunoreactivity with fibrils, it is possible that they may represent fibril nuclei or seeds. Alternatively, FOs may represent a unique type of Aβ oligomers that share conformational similarity to fibrils. To address this question, we analyzed the seeding properties of FOs. The assembly of amyloid fibrils is a nucleation-dependent process, and assembly reactions display a characteristic lag phase prior to initiation of fibril formation that reflects the time required for the spontaneous formation of fibrils (34). This lag phase is reduced or eliminated by seeding the assembly reaction with preformed fibrils. Mature Aβ40 fibrils that have been sheared by sonication were used to seed monomeric Aβ40 as a positive control, and Aβ40 monomers without any seeds served as a control. In the absence of preformed seeds, monomeric Aβ40 assembled into ThT-positive fibrils with a lag phase of 2.79 ± 0.33 h (Fig. 6A). The presence of 5 and 10% of fibrils reduced this lag phase to 2.64 ± 0.64 and 1.14 ± 0.13 h, respectively (supplemental Table 1). In contrast, the addition of 5 and 10% of FOs slightly prolonged the lag phase of fibril assembly to 3.03 ± 0.29 and 3.01 ± 0.34 h (Fig. 6B). This indicates that FOs do not seed fibril formation and are not fibril nuclei.
FIGURE 6.
Aβ40 FOs do not serve as nuclei for fibril formation. Aβ40 fibrils (A) and FOs (B) were sonicated for 5 min in a water bath and incubated at 25 °C with 45 μm Aβ40 monomers (Mon). The addition of fibril seeds to monomers accelerated fibril formation (A). In contrast, when Aβ40 monomers were seeded with FOs, the lag phase of fibril formation increased slightly in a seed percentage-dependent manner (B). Experiments were performed in triplicate. ThT data were normalized by subtracting ThT fluorescence values of fibril seeds and FO seeds incubated alone and buffer, respectively. AU, arbitrary units.
Because FOs do not seed fibrils, we then examined whether FOs seed the formation of FOs using the fibril-specific and conformation-specific antibody LOC as a read-out of FO formation (24). Assembly reactions containing 2% of Aβ40 FOs and fibrils were assayed at different time points for fibril-specific immunoreactivity (Fig. 7A). Aβ40 monomers with no seeds were used as controls. For Aβ40 monomers seeded with 2% of FOs, significant LOC immunoreactivity was observed as early as 0.5 h and increased up to 2 h after the initiation of aggregation (Fig. 7A). In contrast, unseeded assembly reactions and solutions seeded with Aβ fibrils did not develop LOC immunoreactivity until 1.5–2 h (Fig. 7A). The increase in fibril immunoreactivity is due to the conversion of monomer into FOs, because the added seeds are below the limit of detection at this dilution (Fig. 7B). In contrast, Aβ40 monomers incubated without seeding exhibited slower aggregation kinetics. LOC immunoreactivity was minimal at 1.5-h time point and became strong only at 7 h after initiation of aggregation (Fig. 7, A and B).
FIGURE 7.
Aβ40 FOs seed the formation of FOs from monomers. Aβ40 fibrils and FOs were sonicated and incubated at 25 °C with 45 μm Aβ40 monomers. At the time indicated, aliquots were spotted onto nitrocellulose membrane and probed with the fibril- and FO-specific OC-like antibody LOC. A, Aβ40 monomers seeded with 2 and 5% of FOs formed LOC-immunoreactive aggregates within 1.5 h. In contrast, Aβ40 monomers in the absence of seeds developed LOC immunoreactivity after 1.5 h. Monomers seeded with fibrils developed LOC reactivity at 1.5 and 2 h. B, Aβ40 FO and fibril seeds control. Immunoreactivity of seeds diluted to 2% is below the limit of LOC detection. C, quantification of LOC immunoreactivity of Aβ40 monomers alone and monomers seeded with 2% FOs. The amount of LOC reactivity represents the amount of FOs or fibrils in each reaction. AU, arbitrary units.
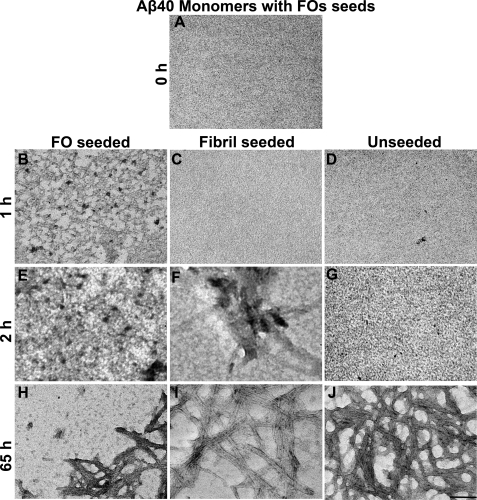
Fibril immunoreactivity could be indicative of the formation of either small FOs or fibrils; however, the lag time for fibril formation as judged by ThT fluorescence under the same conditions is 2.6 h, suggesting that fibrillar oligomers had formed rather than mature fibrils. These results suggest that FOs seed the formation of FOs rather than fibrils, and the kinetics of FO formation are faster than fibril formation in seeded reactions. To confirm this, TEM at the 1-h time points revealed the appearance of small FOs (5–15 nm in diameter) that are not observed at time 0 in Aβ40 monomers seeded with FOs (Fig. 8, A and B). After 2 h of aggregation, the number of oligomers increased. We have observed that the morphology and size (6–15 nm) are faithfully propagated from the parental FO seeds onto the resulting daughter FOs (Fig. 8 and Fig. 9). No fibrils were observed in FO-seeded samples over the time frame of 0–2 h (Fig. 8, A, B, and E). Consistent with the dot blot and ThT assays, TEM analysis revealed that the unseeded Aβ40 sample did not contain oligomers over the same times of incubation (Fig. 8, A–D and G). The appearance of fibrils in fibril-seeded samples was evident at 2 h after initiation of aggregation and is correlated by TEM with the appearance of fibrils (Fig. 8, A, C, and F). At longer incubation times, all reactions displayed the presence of mature fibrils in TEM images (Fig. 8, H–J). The presence of FOs in FO-seeded samples and the absence of observable fibrils provide further confirmation for the interpretation that FOs seed FO formation but not fibrils.
FIGURE 8.
TEM analysis of seeded assembly reactions. Aβ40 monomers alone, monomers seeded with FOs, and monomers seeded with fibrils were analyzed by TEM at times indicated. FOs seeded formation of more FOs from monomers after 1 h of incubation. At the same time point, monomers alone did not form oligomers. The fibril-seeded reaction formed fibrils after 2 h. Scale bar = 100 nm.
FIGURE 9.
Size distribution of FO seeds (gray) and the resulting seeded FOs (black). Data represent measurements from two EM images and >150 oligomers.
DISCUSSION
These results demonstrate that FOs represent a novel class of Aβ oligomers that are distinct from both PFOs and fibrils. The results show that Aβ FOs are soluble oligomeric species that are structurally and immunologically related to fibrils and are conformationally distinct from PFOs. FOs and PFOs are both soluble at 100,000 × g, exhibit a high content of β-structure, are weakly thioflavin T-positive, and appear as spherical oligomers in EM and AFM images (5, 24, 30). However, FOs are smaller than PFOs by AFM analysis (Fig. 4) and are differentially recognized by A11 and OC antibodies, suggesting that they have a fundamental structural difference. Because both A11 and OC recognize generic epitopes that are displayed by many different amyloid sequences, these structural differences appear to represent fundamental classes of amyloid oligomer structures. These structural differences are readily apparent by EPR spectroscopy, where FOs display significantly stronger spin-spin interaction between adjacent spin labels, indicating that the labels are more closely spaced than PFOs (Fig. 4 and supplemental Fig. 1). The lack of strong spin coupling from adjacent spin labels in PFOs suggests that the β-strands may be antiparallel or staggered or both. FOs share the same conformation-specific, generic epitope that is displayed in Aβ fibrils, which are known to be organized as parallel in-register β-sheets (28, 35–37). The EPR spectra of FOs and fibrils are very similar, but FOs lack evidence of spin exchange that arises from the molecular contact of multiple spin labels in linear arrays. Although FOs show some strong spin-spin interactions, the spin coupling is more heterogeneous than that observed in fibrils. Whether this heterogeneity is due to reduced coupling at the probes located at the ends of the small sheets or whether the FOs are polymorphic or structurally heterogeneous is not yet clear. Aβ fibrils are known to be polymorphic, and this variation arises from differences in the location of the bend in the β-hairpin (33, 37), although it is not obvious that this type of polymorphism would account for the observed heterogeneity in spin coupling.
In contrast to fibrils, FOs are soluble at 100,000 × g, are spherical in morphology, and bind ThT significantly less than fibrils. AFM analysis indicates that FOs are approximately one-third the heights of fibrils (1.13 nm for FOs and 3 nm for fibrils; Fig. 5). Although the absolute height estimates of proteins by AFM may be too low due to deformation of the protein by the AFM tip, the ratio of heights of FOs and fibrils should be an accurate reflection of their relative heights. In Aβ40 fibrils, each peptide molecule is folded into a hairpin with β-strands (13–24 and 28–40) separated by a loop, and these molecules are intermolecularly hydrogen-bonded to form a protofilament, with two or three protofilaments in a fibril, depending on their particular morphology (33). FOs are approximately one-half to one-third the height of fibrils by AFM image analysis. Taken together, these results suggest that FOs represent small segments of a protofilament (Fig. 10). Based on these results, we hypothesized that FOs may represent fibril nuclei, but surprisingly, we found that FOs are unable to nucleate fibril formation. Instead, FOs seed the formation of more FOs. The fact that FOs seed the replication of FOs but not fibrils is consistent with the suggestion that they have distinct growing ends or lattices. Because FO formation precedes fibril formation, this suggests that the formation of a protofilament lattice consisting of intermolecularly hydrogen-bonded Aβ hairpins is kinetically more favorable than the formation of the fibril lattice, which contains two or more protofilaments that form sheet stacking interactions via their carboxyl-terminal sheet surfaces (Fig. 10). After 72 h of incubation, the majority of Aβ has assembled into insoluble fibrils, which indicates that fibrils are thermodynamically more favorable than fibrillar oligomers. Presumably, this increased stability results from the additional sheet stacking interactions of the hydrophobic surfaces of the carboxyl-terminal sheets of two or more adjacent protofilaments. The finding that FOs are weakly thioflavin-positive suggests that thioflavin binding may depend on the sheet stacking interface between the protofilaments.
FIGURE 10.
Schematic diagram of amyloid aggregation pathways. Misfolded or random coil monomers can assemble along two divergent pathways into two distinct types of oligomeric intermediates as follows: prefibrillar oligomers and fibrillar oligomers. Although the structure of PFOs is unknown, we propose that FOs are pieces of single fibril protofilament. Fibrils, on the other hand, contain two or three protofilaments composed of Aβ organized in parallel in-register β-sheets. Only the end of the aggregates are shown, but they contain multiple intermolecularly hydrogen-bonded strands. Once FOs or fibrils have formed, monomer can add on to the ends, resulting in elongation of fibrils and replication of FOs that appear to reach a size at which they become unstable and break into smaller units. It is not yet clear whether FOs coalesce via stacking of the two carboxyl-terminal sheets or whether they dissociate into monomer prior to fibril formation.
FOs also nucleate the conformational conversion of monomer in a fashion distinct from that of fibrils. In fibril-seeded reactions, monomer conversion results in fibril elongation, which is monitored by an increase in thioflavin T fluorescence. If fibril nuclei contain intermolecular sheet stacking between 2 or more β-sheets, this suggests that the sheet stacking interaction between FOs that would be necessary to create a fibril nucleus is the rate-limiting step for fibril nucleation. However, FOs seed the conversion of Aβ monomer into FOs that are thioflavin-negative and are detected as an increase in OC immunoreactivity. Unlike fibrils, which appear to grow by addition of monomers, the FOs resulting from seeded reactions are not detectably larger than the FO seeds. This suggests that FOs grow to a size where their lattice becomes unstable and then split to produce additional growing ends. This implies that unlike Aβ fibrils, FOs are capable of replicating in a binary fashion that is similar to yeast prions.
The identification of Aβ FOs as small pieces of an individual protofilament suggests that FOs may be a generic oligomer class that is readily adapted by other amyloidogenic proteins, including islet amyloid polypeptide and expanded polyglutamine repeat protein peptides (24). Therefore, the ability of amyloid oligomers to propagate by seeding could also be a general property, intrinsic to other amyloidogenic proteins and other types of oligomers. Because amyloid oligomers have been reported to be more toxic than amyloid fibrils and they are widely believed to represent the primary pathological species in neurodegenerative disease (5, 6, 38), seeding could be one of the mechanisms by which amyloid formation spreads in various neurodegenerative diseases. For yeast prions, the propensity of the aggregates to fragment and generate new seeds is a key determinant of the ability of the prions to infect and propagate efficiently (39). By extension, the ability of FOs to fragment and generate new seeds may also be important for the enhanced pathogenic significance of oligomers as compared with fibrils in AD. The small size of FOs may also be important for toxicity as their ability to diffuse may be important for the spread of disease through the brain. The structure of FOs as pieces of amyloid protofilaments may also contribute significantly to their pathogenic activities. In fibrils, the hydrophobic carboxyl-terminal sheet surface from residues 28–40 is shielded by sheet stacking interaction with the same surface from adjacent protofilaments, but in FOs, this hydrophobic surface is exposed. This exposed hydrophobic surface may be able to interact with other hydrophobic surfaces, like cell membranes, and this interaction may explain the ability of amyloid oligomers to interact adventitiously with hydrophobic interfaces and permeabilize cell membranes (40–42).
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grants NS 38298 and AG00538 and Grant AG027936 (to R. L.). This work was also supported by the Cure Alzheimer Fund (to C. G.) and a grant from the Larry L. Hillblom Foundation (to C. G. and R. L.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1 and 2 and Table 1.
- AD
- Alzheimer disease
- FO
- fibrillar oligomer
- PFO
- prefibrillar oligomer
- TEM
- transmission electron microscopy
- AFM
- atomic force microscopy
- ThT
- thioflavin T
- A.U.
- arbitrary units
- FTIR
- Fourier transform infrared spectroscopy
- Aβ
- amyloid β
- HFIP
- hexafluoro-2-propanol.
REFERENCES
- 1.Baglioni S., Casamenti F., Bucciantini M., Luheshi L. M., Taddei N., Chiti F., Dobson C. M., Stefani M. (2006) J. Neurosci. 26, 8160–8167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Glabe C. G. (2008) J. Biol. Chem. 283, 29639–29643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Terry R. D. (1996) J. Neuropathol. Exp. Neurol. 55, 1023–1025 [PubMed] [Google Scholar]
- 4.Walsh D. M., Selkoe D. J. (2007) J. Neurochem. 101, 1172–1184 [DOI] [PubMed] [Google Scholar]
- 5.Kayed R., Head E., Thompson J. L., McIntire T. M., Milton S. C., Cotman C. W., Glabe C. G. (2003) Science 300, 486–489 [DOI] [PubMed] [Google Scholar]
- 6.Lambert M. P., Barlow A. K., Chromy B. A., Edwards C., Freed R., Liosatos M., Morgan T. E., Rozovsky I., Trommer B., Viola K. L., Wals P., Zhang C., Finch C. E., Krafft G. A., Klein W. L. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 6448–6453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Oddo S., Caccamo A., Shepherd J. D., Murphy M. P., Golde T. E., Kayed R., Metherate R., Mattson M. P., Akbari Y., LaFerla F. M. (2003) Neuron 39, 409–421 [DOI] [PubMed] [Google Scholar]
- 8.Roselli F., Tirard M., Lu J., Hutzler P., Lamberti P., Livrea P., Morabito M., Almeida O. F. (2005) J. Neurosci. 25, 11061–11070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Walsh D. M., Klyubin I., Fadeeva J. V., Cullen W. K., Anwyl R., Wolfe M. S., Rowan M. J., Selkoe D. J. (2002) Nature 416, 535–539 [DOI] [PubMed] [Google Scholar]
- 10.Walsh D. M., Selkoe D. J. (2004) Protein Pept. Lett. 11, 213–228 [DOI] [PubMed] [Google Scholar]
- 11.Gong Y., Chang L., Viola K. L., Lacor P. N., Lambert M. P., Finch C. E., Krafft G. A., Klein W. L. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 10417–10422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bucciantini M., Giannoni E., Chiti F., Baroni F., Formigli L., Zurdo J., Taddei N., Ramponi G., Dobson C. M., Stefani M. (2002) Nature 416, 507–511 [DOI] [PubMed] [Google Scholar]
- 13.Hoshi M., Sato M., Matsumoto S., Noguchi A., Yasutake K., Yoshida N., Sato K. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 6370–6375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gellermann G. P., Byrnes H., Striebinger A., Ullrich K., Mueller R., Hillen H., Barghorn S. (2008) Neurobiol. Dis. 30, 212–220 [DOI] [PubMed] [Google Scholar]
- 15.Harper J. D., Wong S. S., Lieber C. M., Lansbury P. T. (1997) Chem. Biol. 4, 119–125 [DOI] [PubMed] [Google Scholar]
- 16.Shankar G. M., Li S., Mehta T. H., Garcia-Munoz A., Shepardson N. E., Smith I., Brett F. M., Farrell M. A., Rowan M. J., Lemere C. A., Regan C. M., Walsh D. M., Sabatini B. L., Selkoe D. J. (2008) Nat. Med. 14, 837–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lesné S., Koh M. T., Kotilinek L., Kayed R., Glabe C. G., Yang A., Gallagher M., Ashe K. H. (2006) Nature 440, 352–357 [DOI] [PubMed] [Google Scholar]
- 18.Deshpande A., Mina E., Glabe C., Busciglio J. (2006) J. Neurosci. 26, 6011–6018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nimmrich V., Grimm C., Draguhn A., Barghorn S., Lehmann A., Schoemaker H., Hillen H., Gross G., Ebert U., Bruehl C. (2008) J. Neurosci. 28, 788–797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Danzer K. M., Haasen D., Karow A. R., Moussaud S., Habeck M., Giese A., Kretzschmar H., Hengerer B., Kostka M. (2007) J. Neurosci. 27, 9220–9232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Janson J., Ashley R. H., Harrison D., McIntyre S., Butler P. C. (1999) Diabetes 48, 491–498 [DOI] [PubMed] [Google Scholar]
- 22.Janson J., Laedtke T., Parisi J. E., O'Brien P., Petersen R. C., Butler P. C. (2004) Diabetes 53, 474–481 [DOI] [PubMed] [Google Scholar]
- 23.Gharibyan A. L., Zamotin V., Yanamandra K., Moskaleva O. S., Margulis B. A., Kostanyan I. A., Morozova-Roche L. A. (2007) J. Mol. Biol. 365, 1337–1349 [DOI] [PubMed] [Google Scholar]
- 24.Kayed R., Head E., Sarsoza F., Saing T., Cotman C. W., Necula M., Margol L., Wu J., Breydo L., Thompson J. L., Rasool S., Gurlo T., Butler P., Glabe C. G. (2007) Mol. Neurodegener. 2, 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kayed R., Pensalfini A., Margol L., Sokolov Y., Sarsoza F., Head E., Hall J., Glabe C. (2009) J. Biol. Chem. 284, 4230–4237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.LeVine H., 3rd (1993) Protein Sci. 2, 404–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vincent V. A., DeVoss J. J., Ryan H. S., Murphy G. M., Jr. (2002) J. Neurosci. Res. 69, 578–586 [DOI] [PubMed] [Google Scholar]
- 28.Török M., Milton S., Kayed R., Wu P., McIntire T., Glabe C. G., Langen R. (2002) J. Biol. Chem. 277, 40810–40815 [DOI] [PubMed] [Google Scholar]
- 29.LeVine H., 3rd, Scholten J. D. (1999) Methods Enzymol. 309, 467–476 [DOI] [PubMed] [Google Scholar]
- 30.Kayed R., Glabe C. G. (2006) Methods Enzymol. 413, 326–344 [DOI] [PubMed] [Google Scholar]
- 31.Margittai M., Langen R. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 10278–10283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Margittai M., Langen R. (2008) Q. Rev. Biophys. 41, 265–297 [DOI] [PubMed] [Google Scholar]
- 33.Petkova A. T., Leapman R. D., Guo Z., Yau W. M., Mattson M. P., Tycko R. (2005) Science 307, 262–265 [DOI] [PubMed] [Google Scholar]
- 34.Harper J. D., Lansbury P. T., Jr. (1997) Annu. Rev. Biochem. 66, 385–407 [DOI] [PubMed] [Google Scholar]
- 35.Benzinger T. L., Gregory D. M., Burkoth T. S., Miller-Auer H., Lynn D. G., Botto R. E., Meredith S. C. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 13407–13412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Antzutkin O. N., Leapman R. D., Balbach J. J., Tycko R. (2002) Biochemistry 41, 15436–15450 [DOI] [PubMed] [Google Scholar]
- 37.Lührs T., Ritter C., Adrian M., Riek-Loher D., Bohrmann B., Döbeli H., Schubert D., Riek R. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 17342–17347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Haass C., Selkoe D. J. (2007) Nat. Rev. Mol. Cell Biol. 8, 101–112 [DOI] [PubMed] [Google Scholar]
- 39.Tanaka M., Collins S. R., Toyama B. H., Weissman J. S. (2006) Nature 442, 585–589 [DOI] [PubMed] [Google Scholar]
- 40.Kayed R., Sokolov Y., Edmonds B., McIntire T. M., Milton S. C., Hall J. E., Glabe C. G. (2004) J. Biol. Chem. 279, 46363–46366 [DOI] [PubMed] [Google Scholar]
- 41.Demuro A., Mina E., Kayed R., Milton S. C., Parker I., Glabe C. G. (2005) J. Biol. Chem. 280, 17294–17300 [DOI] [PubMed] [Google Scholar]
- 42.Chimon S., Shaibat M. A., Jones C. R., Calero D. C., Aizezi B., Ishii Y. (2007) Nat. Struct. Mol. Biol. 14, 1157–1164 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.