Abstract
The zinc finger antiviral protein (ZAP) is an interferon-stimulated gene that restricts the replication of retroviruses, alphaviruses, and filoviruses. Relatively little is known, however, regarding the detailed mechanism of ZAP induction during viral infections. We show that, although being inducible by either interferon or virus, expression of ZAP is more efficiently activated by virus than are several other classical interferon-stimulated genes and that viral induction of ZAP occurs under the direct control of interferon regulatory factor 3 (IRF3) independent of interferon paracrine/autocrine signaling. ZAP was up-regulated in cells unresponsive to type I and III interferons upon engagement of TLR3, retinoic inducible gene I/melanoma differentiation-associated gene 5 pathways, or ectopic expression of a constitutively active IRF3 mutant. Conversely, induction of ZAP by virus or dsRNA was severely impaired in cells expressing a dominant-negative mutant IRF3 and completely abrogated in cells lacking IRF3. In contrast to IRF3, ZAP induction was independent of NF-κB activity. Mutational analysis of the human ZAP promoter revealed that multiple interferon-stimulated response elements far distal to the transcription start site serve redundantly to control IRF3-dependent induction of ZAP transcription. Chromatin immunoprecipitation assays demonstrated that IRF3 selectively binds the distal interferon-stimulated response elements in human ZAP promoter following viral infection. Collectively, these data suggest that ZAP is a direct target gene of IRF3 action in cellular antiviral responses.
Keywords: Double-stranded RNA, Innate Immunity, Interferon, Signal Transduction, Virus, IRF3, NF-κB, STAT, ZAP, Interferon
Introduction
Mammalian cells respond to viral infections by the rapid induction of a family of pleiotropic cytokines known as interferons (IFNs).3 Once secreted, IFNs act in an autocrine and/or paracrine manner through the Jak-STAT pathway to stimulate the expression of hundreds of IFN-stimulated genes (ISGs), thereby establishing a cellular antiviral state. IFNs also exert critical immune regulatory functions, shaping subsequent adaptive immunity that helps clear the infection and establishes long term protective immunity (1, 2). The IFN family consists of three types, i.e. types I (mainly IFNβ and IFNα), II (IFNγ), and III (IFNλs). Although IFNγ production is restricted to cell types of lymphocyte origin, type I and III IFNs can be induced in most cell types (1). The importance of type I IFNs in the control of viral infection has been vividly demonstrated in IFNα/β receptor knock-out mice (3).
Upstream, IFNβ and IFNλ genes are transcriptionally activated by a latent cellular transcription factor, IFN regulatory factor 3 (IRF3) (4–6). Predominantly residing in the cytoplasm in quiescent cells, IRF3 undergoes specific carboxyl-terminal serine phosphorylation mediated by the noncanonical IKK kinases, TBK1 or IKKϵ (7, 8), following viral engagement of the toll-like receptor 3 (TLR3) and/or retinoic acid inducible gene I (RIG-I)/melanoma differentiation-associated gene 5 (MDA5) pathways. Phosphorylated IRF3 then dimerizes and translocates into the nucleus, where it associates with p300/cAMP-responsive element-binding protein-binding protein and binds to the IRF element (IRF-E) present in IFNβ and IFNλ gene promoters, activating their transcription (1, 4). Because IRF-Es and the IFN-stimulated responsive elements (ISREs) present in some ISG promoters overlap in sequence, IRF3 is known to directly regulate a subset of ISGs, such as ISG56 and ISG54 (9, 10), independent of the IFN autocrine/paracrine loop. Given the pivotal role of IRF3 in antiviral innate immunity, IRF3 target genes may play important roles in the early host responses to viral infections. For this reason, identification of direct IRF3 target genes is important.
During our investigation of hepatitis C virus (HCV)-mediated disruption of global antiviral gene expression, we observed that viral induction of hundreds of cellular genes (using Sendai virus (SeV) as an inducer) was blocked by HCV in Huh7 cells stably replicating genome length HCV RNAs.4 Because many of the virus-inducible genes were well known ISGs, it was unclear whether the disruption of their induction was a direct result of inhibition of IRF3 activation by HCV or secondary to the blockade of induction of type I/III IFNs, which signal through the JAK-STAT pathway to regulate the expression of these genes. In our efforts to determine the subset of genes whose transcription is directly controlled by IRF3, we identified the zinc finger antiviral protein (ZAP) as a novel IRF3 target gene. Previously known as an ISG, ZAP possesses antiviral activity against viruses of multiple families, including Retroviridae, Togaviridae, and Filoviridae (11–15). The experimental studies reported here reveal that ZAP expression is directly regulated by IRF3 following virus infection or stimulation of cells with dsRNA or dsDNA and is not dependent on NF-κB. Although IFN can also stimulate ZAP expression, our data indicate that ZAP is a primary IRF3 response gene in the innate antiviral signaling pathway.
EXPERIMENTAL PROCEDURES
Cells
HeLa, A549, MRC-5, human hepatoma Huh7, human endometrial carcinoma Hec1B, and HEK293FT cells were maintained by conventional techniques. PH5CH8 is a non-neoplastic human hepatocyte cell line that was transformed with SV40 T antigen (16) and supports a robust response to extracellular poly(I·C) that is dependent on TLR3 signaling (17). HeLa IκBMut is a tetracycline-regulated cell line that conditionally expresses a dominant-negative form of IκBα (bearing alanine mutations at both serine 32 and 36 residues) and was cultured as described (18). HeLa cells stably expressing amino-terminal protease (Npro) of bovine viral diarrhea virus were generated by infecting HeLa cells with replication-incompetent retroviruses carrying dually FLAG- and HA-tagged Npro (pCX4pur-FHNpro), followed by selection with puromycin, as described previously (19).
Plasmids
Conventional PCR and mutagenesis techniques were used to clone various wild type and mutant human ZAP and OASL promoter sequences into the promoter-less and enhancer-less pGL3-basic vector (Promega) using genomic DNA from Huh7 and HEK293 cells as the template and the indicated primer oligonucleotides (supplemental Tables S2–S4). pCX4pur-FHNpro was constructed by inserting dually FLAG- and HA-tagged Npro into pCX4pur (a gift from Tsuyoshi Akagi). The cDNA expression constructs for RIG-I, MDA5 (20, 21), TRIF (22), MAVS (23), IKKϵ (7), RelA (24), IRF1 and IRF3/5D (25, 26), IRF7 (Δ238–408) (27), and the promoter reporter constructs, hIL-8 (−162/+44)-Luc (28) and PRDII-Luc (29) have been described. pOAS2(p69)-GL2 contained the human OAS2 (p69) promoter upstream of the firefly luciferase reporter gene (30). pRL-TK, pRL-CMV, (Promega), or pCMVβgal (Clontech) was used to normalize transfection efficiencies in reporter gene assays. Plasmid DNAs were transfected into cells using TransIT-LT1 (Mirus) (for Huh7, 293FT, and PH5CH8 cells), Optifect (Invitrogen, for Hec1B cells), or Lipofectamine 2000 (Invitrogen, for HeLa and A549 cells), following the manufacturers' instructions.
Northern Blot Analysis
Northern blots were performed using a NorthernMax® kit (Applied Biosystems) as previously described (19). Briefly, total RNA was isolated from cells using the RNeasy mini kit (Qiagen). The RNA samples (15 μg each) were resolved on 0.9% denaturing formaldehyde agarose gel, transferred to BrightStar-Plus nylon membranes (Applied Biosystems), and probed with 32P-labeled antisense riboprobes specific to ZAP, OASL, and ISG56 at 68 °C. After extensive washing, the membranes were scanned with a STORM 860 molecular imager (Molecular Dynamics). Ethidium bromide-stained 28 and 18 S rRNAs were used as loading controls.
Quantitative Reverse Transcription-PCR
Total RNA was isolated from cells grown in 6-well plates following the indicated treatments using TRIzol (Invitrogen) as per the manufacturer's instructions. One microgram of RNA was programmed for synthesis of cDNA, one-fiftieth of which was subsequently used for analysis of abundance of human ZAP, MxA, OAS1, IFN-β, and 28 S (as an internal control for normalization) transcripts by quantitative PCR (Q-PCR) using gene-specific primers (available online in supplemental Table S5) and iQ SYBR Green Supermix (Bio-Rad) on an iCycler IQ5 real time PCR system (Bio-Rad).
Reporter Gene Assay
Cells plated in 48-well plates were co-transfected with 75 ng of the indicated promoter-luc reporter plasmid, 25 ng of pRL-TK or pRL-CMV (Promega), and, if applicable, 200 ng of the indicated IFN signaling protein encoding vector. Where indicated, at 24 h post-transfection, the cells were mock treated, stimulated with 500 units/ml of recombinant human IFNα-2b (Raybiotech) or by the addition of 50 μg/ml poly(I·C) (Sigma) to the culture medium (M-pIC), infection with 100–200 HAU/ml of SeV (Charles River Laboratories), or transfection with 0.5 μg of poly(I·C) (T-pIC) or poly(dA·dT) (GE Healthcare; T-pdAdT) using Lipofectamine 2000 (Invitrogen). After 6–20 h, the cells were lysed in passive lysis buffer and subjected to dual luciferase assay (Promega). The ratio of firefly luciferase activity to that of Renilla luciferase was used for comparison of promoter activity. In some experiments, pCMVβgal was used in place of pRL-TK for normalization of firefly luciferase activities, as described (31).
IFN Bioassay
IFN bioactivity in cell culture supernatants was determined by a standard microtiter plaque reduction assay using vesicular stomatitis virus on Vero cells, as described previously (32).
Immunoblot Analysis
Cellular extracts were prepared and subjected to SDS-PAGE and immunoblot analysis as previously described (17, 19). The following monoclonal or polyclonal antibodies were used: anti-actin and anti-FLAG monoclonal antibodies (Sigma); anti-HA monoclonal antibody (Invivogen); anti-ISG56 polyclonal antibody (33); anti-ISG15 polyclonal antibody (Rockland); anti-IRF3 polyclonal antibody (a gift from Michael David); anti-Sendai virus polyclonal antibody (a gift from Ilkka Julkunen); anti-phospho specific STAT1 and STAT2 (Cell Signaling Technology); and peroxidase-conjugated secondary anti-mouse and anti-rabbit polyclonal antibodies (Southern Biotech). The protein bands were visualized by enhanced chemiluminescence (Millipore), followed by exposure to x-ray films.
Chromatin Immunoprecipitation (ChIP) Assays
HeLa cells (∼5 × 106) infected with SeV for 8 h or treated with IFN (400 units/ml, 1 h) were cross-linked with 1% formaldehyde (10 min at room temperature). ChIP assays were performed using the ChIP-ITTM Express kit (Active Motif, Carlsbad, CA) according to the manufacturer's instructions. In brief, chromatin was sheared to an average size of ∼300 bp with a Bioruptor UCD-200 (Diagenode Inc., Sparta, NJ). Sheared chromatin was incubated with 2 μg of control IgG (Active Motif) or specific antibody to IRF3, STAT1, or STAT2 (Santa Cruz), and 25 μl of protein G magnetic beads overnight at 4 °C. After reversal of cross-linking and protein digestion with protease K, immunoprecipitated DNA was purified with the MiniElute PCR purification kit (Qiagen), and PCR was performed using GoTaq Q-PCR mixture (Promega) using the following parameters (enzyme activation, 95 °C for 2 min and then 40 cycles of 95 °C for 15 s and 60 °C for 1 min). The sense and antisense primers for the putative ISRE and STAT sites present within the ZAP promoter were designed using Primer 3 plus program and shown in supplemental Table S6.
RESULTS
Induction of ZAP and Other Cellular mRNAs by a Constitutively Active IRF3 in Cells Unresponsive to Type I and III IFNs
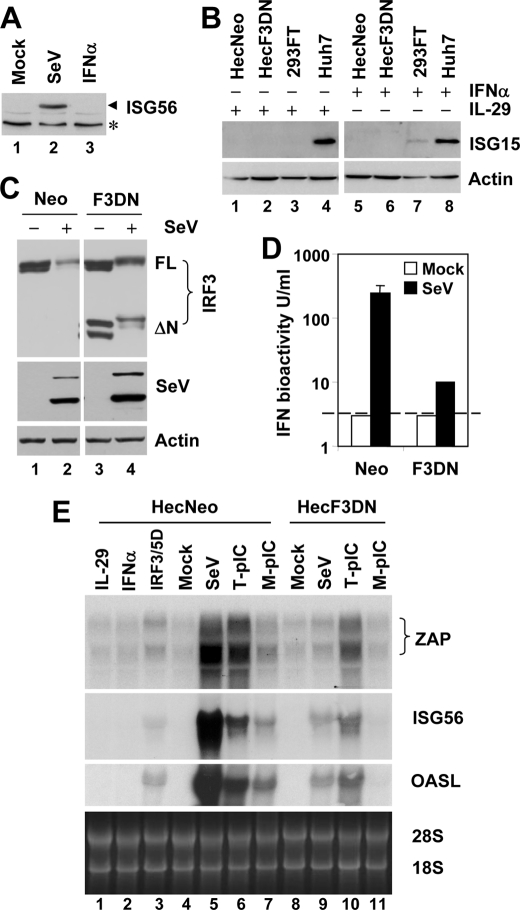
To determine the subset of cellular genes for which transcription is directly controlled by IRF3, we studied cells capable of virus-induced IRF3 activation but lacking the IFN feedback loop. The human endometrial adenocarcinoma cell line, Hec1B, is deficient in autocrine/paracrine type I IFN signaling (34, 35). However, it has not been reported whether these cells respond to type III IFNs by up-regulation of ISGs. We first confirmed the type I IFN-unresponsive phenotype of Hec1B cells, by showing that SeV, but not IFNα, strongly induced ISG56 (Fig. 1A) and ISG15 (data not shown) expression. Furthermore, we found that stimulation with IL-29, a type III IFN (IFNλ1), did not induce ISG15 expression in Hec1B (data not shown) and its stable derivatives, HecNeo (expressing a control vector) and HecF3DN (expressing a dominant-negative mutant IRF3, DN-IRF3, which lacks the amino-terminal 133 amino acids containing the DNA-binding domain) (Fig. 1B). In contrast, ISG15 was strongly induced by IFNα or IL-29 in human hepatoma Huh7 cells, consistent with the ability of Huh7 cells to respond to both types of IFNs (38–40). Unlike type I IFN signaling that utilizes IFNAR1 and IFNAR2, type III IFNs signal through distinct cell surface receptors, IL-28R1 and IL-10R2 (1, 36, 37). The lack of ISG induction following type III IFN stimulation of Hec1B cells most likely reflects a lack of IL-28R1, a receptor that has a highly tissue-restricted distribution and is not expressed in the human uterus (37) from which Hec1B cells were derived. These data indicate that Hec1B cells are a suitable model for exploring virus-regulated cellular genes independent of the IFN feedback loop, because they are deficient in ISG induction in response to both type I and III IFNs.
FIGURE 1.
IRF3-dependent induction of ZAP and OASL mRNAs by virus or dsRNA in cells deficient in both type I and III IFN signaling. A, immunoblot analysis of ISG56 in Hec1B cells mock treated, infected with 100 HAU/ml of SeV, or treated with 1000 units/ml of IFNα for 16 h. The asterisk denotes a nonspecific band. B, immunoblot analysis of ISG15 in Hec1B cells stably expression the control vector (HecNeo) or DN IRF3 (HecF3DN), 293FT, and Huh7 cells treated with 10 ng/ml of recombinant human IL-29 (lanes 1–4) or 500 units/ml of recombinant human IFNα (lanes 5–8) for 18 h. C, HecNeo and HecF3DN cells were mock infected or infected with SeV and subsequently immunoblotted for IRF3, SeV, and actin. In the IRF3 panel, FL denotes full-length IRF3, whereas ΔN denotes DN IRF3. D, IFN production in culture supernatants of HecNeo and HecF3DN cells that were mock infected or infected with SeV for 24 h, determined by VSV plaque reduction assay. The dashed line indicates the detection limit of the assay (∼3 units/ml). E, Northern blot analysis of ZAP, ISG56, and OASL mRNAs in HecNeo (lanes 1–7) and HecF3DN (lanes 8–11) cells growing in 100-mm dishes following the indicated treatments: IL-29 (10 ng/ml); IFNα (1000 units/ml); poly I:C transfection (T-pIC, 6 μg); poly(I·C) added to culture medium (M-pIC, 50 μg/ml); SeV (300 HAU); and transfection of a vector expressing IRF3/5D (6 μg). All of the treatments were done for 8 h except IRF3/5D, which was transfected into cells for 20 h before cell lysis and RNA extraction.
We next determined the effect of ectopic expression of IRF3/5D (26), a constitutively active phospho-mimetic IRF3 mutant on the cellular transcriptome in Hec1B cells. Overexpression of IRF3/5D mimics virus-induced IRF3 activation and has proven useful in exploring IRF3 target genes (9). We tested total cellular RNAs from Hec1B cells transiently expressing IRF3/5D or the control vector for 20 h by hybridization to Affymetrix human GeneChip oligonucleotide microarrays (HG®-U133A) that contain probe sets representing 22,283 known human genes. We found that ectopic expression of IRF3/5D significantly up-regulated only nine genes ≥2-fold and did not down-regulate any cellular mRNAs (supplemental Table S1) when compared with control vector-expressing Hec1B cells. The up-regulated genes included ISG56 (14.9-fold), ISG15 (9.2-fold), ISG54 (5.3-fold), Noxa (also known as phorbol 12-myristate 13-acetate-induced protein 1, 3.2–3.5-fold), and ISG60 (3-fold), all of which have been shown previously to be regulated by IRF3 (9, 10, 41). Other genes induced by IRF3/5D were: lymphocyte cytosolic protein 2 (also known as SLP-76, 3.5-fold), ZAP (also known as ZC3HAV1, 3.5-fold), OASL (splice variants, p30 and p59, were up-regulated by 2.8- and 2.5-fold, respectively), and zinc finger protein 287 (ZNF287, 2.3-fold), whose function is unknown. Among these latter genes, lymphocyte cytosolic protein 2 is a cytosolic adaptor protein essential for thymocyte development and T cell activation (42), which was shown to be induced by retinoic acid during myeloid differentiation of human myeloblastic leukemia cells (43). However, the expression of lymphocyte cytosolic protein 2 has not been linked to IRF3 activation or IFN signaling previously. OASL is an atypical member of the OAS family, because it lacks the characteristic 2′-5′ OAS activity shared by OAS1, OAS2, and OAS3 (44). Long known as an ISG, the antiviral role of OASL p59 had not been recognized until the recent observation that overexpression of OASL inhibited encephalomyocarditis virus replication (45). Very recently, it was shown that viral induction of OASL depends on IRF3 (46). ZAP was originally isolated as a host factor that prevents cells from infection by Moloney murine leukemia virus (11). Subsequently, it was demonstrated that overexpression of ZAP restricts the replication of RNA viruses of multiple families, including Retroviridae, Togaviridae, and Filoviridae (12–15). ZAP specifically binds to cytoplasmic viral RNAs and promotes their decay by recruiting cellular RNA degradation machinery (47). Although ZAP is known as an ISG (38, 48), transcriptional regulation of ZAP remains poorly characterized. Because our gene profiling experiment identified ZAP as an IRF3/5D-responsive gene in the IFN-unresponsive Hec1B cells, we focused our investigation on characterizing the mechanism by which IRF3 regulates ZAP expression.
Induction of ZAP mRNA by Virus or dsRNA Is Inhibited by a Dominant-negative (DN) Form of IRF3
To confirm the role of IRF3 in regulation of ZAP expression in a physiologic setting, we compared the induction of endogenous ZAP mRNAs following virus infection or dsRNA stimulation, in HecNeo and HecF3DN cells. Confirming its function, DN-IRF3 suppressed SeV-induced IFN production by 96% in stably transfected cells (Fig. 1D, compare F3DN versus Neo) without negatively regulating SeV replication, the stimulus (Fig. 1C). Northern blot analysis revealed that human ZAP (hZAP) mRNA was basally expressed as two different transcripts, a pattern similar to that previously described for rat ZAP (11), and that both hZAP mRNA species were strongly up-regulated by either SeV or intracellular dsRNA (transfected poly(I·C)), which trigger RIG-I and MDA5 pathways, respectively (21, 49) (Fig. 1E, lanes 5 and 6 versus lane 4). Although its effect was less robust, induction of TLR3 signaling by extracellular dsRNA stimulation also up-regulated hZAP mRNA expression (lane 7 versus 4). Importantly, similar to what was observed with OASL and ISG56, up-regulation of hZAP mRNAs by each of these stimuli was substantially reduced in cells stably expressing DN-IRF3 (Fig. 1E, compare lane 9 versus lane 5, lane 10 versus lane 6, and lane 11 versus 7, respectively). As expected, neither IL-29 nor IFNα had a demonstrable effect on expression of ZAP, OASL, and ISG56 mRNAs in HecNeo cells (Fig. 1E, lanes 1 and 2 versus lane 4). In contrast, all three ISG mRNAs were up-regulated upon ectopic expression of IRF3/5D (lane 3 versus lane 4). These results confirm the microarray data and indicate that ZAP and OASL are IRF3 target genes and that their induction is independent of the autocrine/paracrine action of IFN.
IRF3 Deficiency Abrogates Virus-induced ZAP Expression
To further corroborate the role of IRF3 in regulating ZAP expression, we determined how ZAP is induced in cells deficient in IRF3 expression. We took advantage of the ability of pestivirus Npro to induce proteasome-dependent IRF3 degradation (19, 50–52) to establish HeLa cells devoid of IRF3. As shown in Fig. 2A, whereas IRF3 protein was expressed abundantly in parental HeLa cells, it was undetectable in HeLaNpro cells that stably express bovine viral diarrhea virus Npro (Fig. 2A, left panel). As expected, treatment with epoxomicin, a potent proteasome inhibitor, stabilized IRF3 protein expression in HeLaNpro cells (Fig. 2A, right panel). IRF3 deficiency in HeLaNpro cells abrogated ZAP induction by SeV infection or transfection of HCV RNA (Fig. 2B, left panel), as well as the induction of two well studied IRF3-dependent genes, ISG56 and IFN-β (9, 10), and a classical ISG, OAS1 (53) (Fig. 2). In contrast, ZAP and OAS1 induction by IFN was not affected in HeLaNpro cells (Fig. 2B), because neither IRF3 deficiency nor Npro expression impairs Jak-STAT signaling (52, 53). Taken together, these data corroborate the results obtained in DN-IRF3-expressing cells (Fig. 1) that IRF3 is essential for viral induction of ZAP.
FIGURE 2.
IRF3 deficiency abrogates viral induction of ZAP. A, left panel, HeLa and HeLa cells stably expressing BVDV Npro (HeLaNpro) were mock infected or infected with 100 HAU/ml of SeV for 16 h. The cell lysates were immunoblotted for Npro (using anti-HA monoclonal antibody), IRF3, actin, and ISG56. Right panel, HeLa and HeLaNpro cells were mock treated or treated with 100 nm of epoxomicin for 12 h prior to immunoblot analysis of Npro, IRF3, and actin. B, Q-PCR analysis of ZAP, IFN-β, and OAS1 mRNA levels in HeLa and HeLaNpro cells mock treated or stimulated with 500 units/ml IFNα, 100 HAU/ml of SeV or transfected with 2 μg of in vitro transcribed HCV RNA for 8 h. mRNA abundance was normalized to cellular 28 S ribosomal RNA. Fold changes were calculated by dividing normalized mRNA abundance following various treatments by that of the mock treated HeLa cells.
ZAP Is More Efficiently Induced by Virus than the Classical ISGs, MxA, and OAS1
MxA and OAS1 represent classical ISGs whose induction by virus requires the paracrine/autocrine action through Jak-STAT signaling downstream of the IFN receptor (53, 54). The transcription of these genes is not stimulated directly by activated IRF3. Consistent with this, we found that IFNα treatment activated OAS1 expression more strongly than did SeV or HCV RNA in HeLa cells (Fig. 2B, right panel). In contrast, induction of ZAP by SeV or HCV RNA was similar to that by IFN (Fig. 2B, left panel). This pattern is consistent with direct IRF3-dependent induction of ZAP. To characterize ZAP induction by virus versus IFN in nonmalignant cell types, we performed Q-PCR analysis of ZAP mRNA levels following treatment with IFNα (50 and 500 units/ml) and SeV infection in human normal lung fibroblast MRC-5 and non-neoplastic hepatocyte PH5CH8 cells (Fig. 3, left panel). We found that ZAP induction by SeV was consistently higher than that by 500 units/ml of IFNα in both cell types (12.1- versus 9.3-fold and 6.7- versus 4.6-fold, in MRC-5 and PH5CH8 cells, respectively). In contrast, MxA expression was induced to a greater extent by 50 units/ml of IFNα than by SeV (109.4- versus 61.1-fold and 48.8- versus 21.4-fold, in MRC-5 and PH5CH8, respectively) (Fig. 3, right panel). Of note, PH5CH8 cells expressed a 6.8-fold higher basal level of ZAP mRNA than did MRC-5 cells. These results suggest that, although ZAP is induced by both virus and IFN, ZAP is preferentially induced by virus when compared with classical ISGs whose expression is not activated directly by IRF3.
FIGURE 3.
Induction of ZAP and MxA by IFNα and virus in nonmalignant human cells. MRC-5 and PH5CH8 cells were mock treated or treated with the indicated stimuli for 8 h. Total cellular RNA were subjected to Q-PCR analysis of ZAP (left panel) and MxA (right panel) transcripts. mRNA abundance was normalized to cellular 28 S ribosomal RNA. Fold changes were calculated by dividing normalized mRNA abundance following various treatments by that of the mock treated MRC-5 cells. Note that PH5CH8 cells expressed 6.8- and 6.4-fold higher basal level of ZAP and MxA mRNAs, respectively, than did MRC-5 cells.
IRF3 Controls ZAP Promoter Activity Following Engagement of RIG-I/MDA5 and TLR3 Pathways
To determine whether IRF3 regulates hZAP expression at the transcriptional level, we generated luciferase reporter constructs that contain different hZAP promoter fragments inserted upstream of firefly luciferase gene in the promoter-less and enhancer-less plasmid, pGL3-basic (pGL3b, Promega) (Fig. 4A). We also cloned the human OASL (hOASL) promoter sequence (−795 to −1 fragment) into pGL3b. hZAP(−2881)-GL3, which contains an ∼2.9-kb sequence upstream of the hZAP transcription start site, demonstrated basal promoter activity in transfected HeLa, Hec1B, and PH5CH8 cells, as compared with pGL3b, which had negligible activity. Infection with SeV strongly stimulated the hZAP(−2881) promoter in both HeLa and PH5CH8 cells, as did IFNα (Fig. 4, A, left panel, and B). However, although the hZAP(−2881) promoter was activated by SeV, it was not activated by IFNα in Hec1B cells, consistent with an IFN-independent mechanism of activation in these cells (Fig. 4A, right panel). The hZAP(−2881) promoter was also up-regulated by stimulation of PH5CH8 cells with extracellular poly(I·C) (Fig. 4B), consistent with the presence of a functional TLR3 pathway in these hepatocyte-derived cells (17). Similar results were obtained with the hOASL promoter (supplemental Fig. S2, B and C, and data not shown). As a control for specificity, the hOAS2 p69 promoter, which requires IFN signaling through the Jak-STAT pathway for optimal induction (55, 56), was not activated by SeV in Hec1B cells (supplemental Fig. S2B).
FIGURE 4.
Activation of hZAP and hOASL promoters by induction of the TLR3 and RIG-I/MDA5 signaling pathways. A and B, promoter activities of hZAP(−2881) and pGL3basic in HeLa and Hec1B (A) and PH5CH8 cells (B) following indicated treatments. M-pIC, poly(I·C) added to culture medium. C, activities of hZAP(−2881) and hOASL promoters in Hec1B cells transiently expressing an control vector or individual signaling components in the RIG-I/MDA5 or TLR3 pathways. N-RIG and N-MDA5 denote the constitutively active CARD domain of RIG-I and MDA5, respectively. D, activation of hOASL promoter in Hec1B cells by ectopic expression of various forms of IRFs or the constitutively active RelA. IRF7Δ is a constitutive IRF7 mutant which has a internal deletion (deletion of amino acids 238–408). E, activation of wild type (WT) or the indicated mutant hOASL promoters or human OAS2 p69 promoter in Hec1B cells by IRF3/5D. mISREp and mISREd denote mutation in the proximal (−291 to −271) and distal (−335 to −315) ISRE, respectively (see supplemental Fig. S2A for further details). F, activation of various hZAP promoters by IRF3/5D in Hec1B cells. The wild type hZAP(−2881) promoter was also tested for its activation by other indicated IRFs or RelA.
RIG-I/MDA5 and TLR3 represent two major parallel viral dsRNA sensing pathways in mammalian cells. They are linked to the downstream IRF3 kinases, TBK1/IKKϵ (7, 8) via the adaptor proteins MAVS (also known as IPS-1, Cardif, VISA) and TRIF (also known as TICAM1), respectively (57–62). We found ectopic expression of MAVS or the CARD domain of RIG-I (N-RIG) or MDA5 (N-MDA5), manipulations that mimic activation of the RIG-I/MDA5 pathway, all effectively activated the hZAP(−2881) and hOASL promoters in Hec1B cells, although the extent of the induction varied (Fig. 4C). Overexpression of TRIF (mimicking activation of the TLR3 pathway) or IKKϵ (that phosphorylates and directly activates IRF3) (Fig. 4C) was also able to activate expression of both promoters. These data, in agreement with those shown in Fig. 1E, suggest that activating IRF3 by engagement of all the known viral RNA sensing pathways up-regulates hZAP and hOASL expression at the promoter level.
In addition to IRF3, IRF7 and IRF1 also regulate the virus-induced expression of IFN genes and ISGs (4, 63–65). When ectopically expressed in Hec1B cells, the constitutively active IRF3/5D strongly activated both hOASL (Fig. 4D) and hZAP(−2881) (Fig. 4F) promoters. A constitutively active form of IRF7, in which amino acids 238–408 were deleted (IRF7Δ) (27), was also able to activate both promoters, albeit less effectively. In contrast, neither promoter was activated by ectopic expression of IRF1 or DN-IRF3 in Hec1B cells (Fig. 4F). We conclude from these experiments that IRF3 and, to a lesser extent, IRF7 control virus-induced activation of hZAP and hOASL promoters. Importantly, IRF7 is only constitutively expressed in plasmacytoid dendritic cells, and its expression requires IFN induction in most parenchymal cell types.
The hOASL promoter contains two putative ISREs, located in positions (−335 to −315) and (−291 to −271), respectively (supplemental Fig. S2A). A recent study suggested that the distal ISRE (ISREd, −335 to −315) was responsible for IRF3 induction of the hOASL promoter (46). However, mutation of this distal ISRE had no demonstrable effect on induction of hOASL promoter by virus, IFN, or IRF3/5D, whereas mutation of the proximal ISRE (ISREp, −291 to −271) drastically reduced promoter activation by all three stimuli (supplemental Fig. S2 and Fig. 4E). The importance of ISREp in regulating hOASL induction was confirmed in multiple cell lines, Hec1B, Huh7, and 293FT cells (supplemental Fig. S2 and data not shown). Further studies will be needed to address the discrepancy between these and previous reported data (46).
Multiple ISRE/IRF-Es within a Distal Region Upstream of the ZAP Promoter Transcription Start Site Control Virus-induced, IRF3-dependent Promoter Activation
We next characterized the role of the cis-acting regulatory elements in hZAP promoter in controlling the viral induction of ZAP expression. Examination of putative transcription binding sites by TFSEARCH revealed five STAT binding sites (hereafter referred as STAT I–V) and five ISRE/IRF-Es (hereafter referred as ISRE1–5) in the hZAP(−2881) promoter. Interestingly, the five ISREs are clustered in a region far upstream of the transcription start site (−2486 to −2214). The five STAT sites, however, are more scattered throughout the hZAP promoter (supplemental Fig. S1 and Fig. 5A). Deletion of the STAT I site (−2486) or STAT II-III sites (−2881/ΔXma) did not affect viral induction of hZAP promoter activity, nor did a series of 5′ deletions that cumulatively removed ISRE1 (−2469), ISRE1–2 (−2339), ISRE1–3 (−2302), or ISRE1–4 (−2268) (Fig. 5B). However, deletion of all five ISREs (−2214) severely impaired SeV induction of the hZAP promoter (Fig. 5B) and completely ablated its activation by IRF3/5D (Fig. 4F) in Hec1B cells. The essential role of the distal ISREs in viral activation of hZAP promoter was also confirmed in A549 and HeLa cells, in which deletion of all five ISREs (−2214) ablated SeV activation of the hZAP promoter (Fig. 6B). Nonetheless, analysis of additional mutants revealed that deletions of ISRE5, ISRE4–5, and ISRE3–5, respectively, had no demonstrable effect on SeV induction of hZAP promoter when compared with the wild type or the promoter containing only ISRE5 (−2268) (Fig. 5, D and E). Therefore, the five distal ISREs appear to function redundantly in regulating IRF3-dependent viral activation of hZAP promoter. Thus, only one of the distal ISREs is required for maximal responses of the hZAP promoter to viral infection, and there is no apparent preferential usage of a specific ISRE.
FIGURE 5.
The five ISREs distal to transcription start site are essential and function in redundancy for viral induction of hZAP promoter, whereas they are dispensable for activation the promoter by IFN. A, schematic representation of the hZAP promoter reporter plasmids with various length of 5′-flanking sequence. The putative ISRE/IRF-E and STAT binding sites were indicated as filled circles and hatched bars, respectively. B and C, activation of various hZAP promoters by SeV in Hec1B cells (B) and by IFNα in Huh7 cells (C). D and E, activities of various hZAP promoter deletion mutants in Hec1B cells (D) and Huh7 cells (E) following mock treatment, SeV infection, or IFNα stimulation for 17 h. Note in B and C that the hZAP(−1438) always had a lower basal activity than other mutants, indicating the presence of a negatively regulatory element between positions −1438 and −800.
FIGURE 6.
The proximal STAT site (STAT V) is important for both virus- and IFN-induced activation of hZAP promoter. A, schematic representation of the hZAP promoter reporter plasmids bearing mutations at the STAT IV (mST4) or STAT V (mST5) sites or both (mST4 + 5). B, activities of various hZAP promoters in A549 (upper panel) and HeLa (lower panel) cells following IFNα stimulation (400 units/ml) or SeV infection (200 HAU/ml) for 8 h.
All Five Distal ISRE/IRF-Es in the Human ZAP Promoter Are Dispensable for IFN-induced ZAP Transcription
Next, we investigated the role of the ISREs in induction of the hZAP promoter by IFNα. We first utilized Huh7 cells for these experiments, which respond robustly to IFN by ISG induction (Fig. 1B). Huh7 cells also elicit a weak response to SeV that depends on RIG-I (66). Consistent with the data obtained in Hec1B cells (Fig. 5, B and D), only one copy of the distal ISRE of the hZAP promoter was needed for viral induction of the promoter, which was no longer responsive to SeV infection when all five ISREs were deleted (Fig. 5E, compare (−2214) versus (−2268) and other ISRE deletion mutants in the hZAP(−2486) backbone). In contrast, deletion of all five ISREs had a negligible effect on IFNα induction of the hZAP promoter (Fig. 5C and 5E, compare hZAP(−2268) versus (−2214)). Furthermore, a 5′ deletion up to (−800) in the hZAP promoter (retaining only the two proximal STAT sites) still allowed efficient induction of the promoter by IFNα (Fig. 5C). Comparable induction of hZAP(−2486), hZAP(−2214), and hZAP(−800) promoters by IFNα was also observed in A549 and HeLa cells (Fig. 6B). These data suggest that the distal ISREs, although essential for IRF3-dependent viral activation of the hZAP promoter, are not absolutely required for IFN up-regulation of hZAP transcription through the Jak-STAT pathway.
Among the hZAP promoter deletion mutants, the hZAP(−1438) promoter consistently demonstrated lower basal activity than hZAP(−2214) and hZAP(−800) promoters (Fig. 5, B and C). However, the magnitude of induction (fold increase) of the three promoters by IFN or SeV was similar. Most likely, one or more negative regulatory motifs are present between −1438 and −800 positions of the hZAP promoter and can be overwhelmed by inclusion of upstream promoter sequences. This hypothesis will need to be investigated in future studies.
The Most Proximal STAT Site (STAT V) Is Important for Both Virus- and IFN-induced Activation of the Human ZAP Promoter
Because the hZAP(−800) promoter retained the same potency as the hZAP(−2486) promoter in its inducibility by IFNα (Figs. 5E and 6B), we postulated that the two proximal STAT motifs (i.e. STAT IV and V sites), either alone or in combination, mainly determine ZAP induction by IFN. To test this hypothesis, we generated additional mutant promoter constructs with mutation of the STAT IV (mST4) and/or STAT V (mST5) motifs, in the hZAP(−2486), hZAP(−2214), and hZAP(−800) backbones, respectively (Fig. 6A). In transfected A549 and HeLa cells (Fig. 6B), mutation of the STAT IV site had a negligible effect on IFNα induction of hZAP(−2486) promoter, whereas mutation of the STAT V site reduced the ability of IFNα to activate the promoter by ∼60%. Mutations at both STAT IV and V sites did not further impair IFN activation of hZAP(−2486) promoter. Thus, the most proximal STAT motif (STAT V, −111 to −91) is essential for up-regulation of human ZAP transcription by IFN. The distal ISREs, and possibly the STAT II and III motifs as well, may also contribute to ZAP induction by IFN, especially when the proximal STAT V site is disrupted. Further evidence confirming the importance of the STAT V site in IFN activation of human ZAP transcription was obtained from reporter assays examining the STAT V site-disrupted hZAP(−2214) and hZAP(−800) reporter constructs, which lost most of the IFN responsiveness as compared with their wild type counterparts (Fig. 6B).
Interestingly, mutation of the STAT V site, but not STAT IV, reduced SeV up-regulation of hZAP(−2486) promoter by ∼60% in both A549 and HeLa cells (Fig. 6B), a phenomenon that closely mimicked the effect of this mutation on IFN induction of the promoter. Most likely, the STAT V motif regulates viral activation of hZAP promoter by interacting with STAT proteins that mediate the autocrine/paracrine action of IFN produced during early viral infection.
IRF3 Binds to the Distal ISRE/IRF-Es of the Human ZAP Promoter Following Virus Infection
To characterize IRF3 binding to the hZAP promoter, we conducted ChIP analysis of IRF3 binding to various regions of the hZAP promoter following SeV infection or IFN stimulation of HeLa cells. Q-PCR was conducted to quantify ChIP-enriched DNA using primers flanking ISRE1–2, ISRE3–5, and STAT V motifs, respectively (Fig. 7B). We found that SeV infection potently induced IRF3 binding to both the distal ISRE1–2 and ISRE3–5 clusters but not to the proximal STAT V site (upper panel). As expected, IFNα stimulation did not induce IRF3 binding to any of the three sites, consistent with the fact that IFN does not activate IRF3.
FIGURE 7.
Differential binding of IRF3 and STAT transcription factors to the hZAP promoter following virus infection or IFN stimulation. A, immunoblot analysis of ISG56 expression and phosphorylation status of STAT1 and STAT2 in HeLa cells following stimulation of IFNα (500 units/ml) for 1 and 8 h or infection with SeV (100 HAU/ml) for 16 h. Actin blot was shown as a loading control. B, ChIP analyses of IRF3 and STAT binding to the ISRE1–2, ISRE3–5, and STAT V sites within hZAP promoter in HeLa cells mock treated, stimulated with IFNα (400 units/ml) for 1 h, or infected with SeV (200 HAU/ml) for 8 h. The ChIP-enriched DNA levels analyzed by Q-PCR were normalized to input DNA, followed by subtraction of nonspecific binding determined with control IgG.
To examine the potential contribution of STAT proteins in IFN- and virus-induced signaling pathways, we determined STAT1 and STAT2 activation in HeLa cells following viral infection or IFN stimulation. Upon IFN-induced tyrosine phosphorylation, STAT1 and STAT2 form a heterotrimer complex with IRF9 known as ISGF3, a major transcriptional activator complex in the classical Jak-STAT signaling pathway. As shown in Fig. 7A, we found that IFNα strongly induced both STAT1 and STAT2 phosphorylation, whereas SeV infection only induced STAT1 phosphorylation. This suggests that virus-induced STAT signaling mainly involves activated STAT1 in HeLa cells. Consistent with the data that the proximal STAT site plays an important role in activation of hZAP promoter by both virus and IFN (Fig. 6B), ChIP assay indicated that STAT1 was recruited to the proximal promoter region comprising only the STAT V site following either IFNα treatment or SeV infection (Fig. 7B, lower panel). Most importantly, IFNα treatment, but not SeV infection, induced the selective recruitment of STAT2 as well as STAT1 to the proximal STAT V site, which is consistent with the data shown in Fig. 6.
Activation of NF-κB Is Neither Necessary nor Sufficient for ZAP Induction
NF-κB contributes to the optimal expression of many ISGs (67). Although we did not find candidate NF-κB binding sites in the hZAP promoter, further experiments were carried out to ascertain whether NF-κB plays a role in regulating hZAP expression. Although TNFα activated the NF-κB-dependent PRDII promoter in Hec1B cells, it had no effect on either of the hZAP or hOASL promoters, although the latter two were strongly activated by SeV (Fig. 8A). These data indicate that activation of NF-κB is alone not sufficient to activate the hZAP and hOASL promoters. This interpretation is consistent with our earlier findings that ectopic expression of constitutively active RelA failed to activate either promoter (Fig. 4, D and F). To investigate whether activation of NF-κB was required for induction of hZAP and hOASL, we took advantage of a HeLa cell line with tetracycline-regulated, conditional expression of a dominant-negative, nondegradable IκBα mutant (IκBαMut). In these cells, withdrawal of tetracycline from the culture medium results in expression of IκBαMut, which in turn prevents NF-κB activation by sequestering NF-κB in the cytoplasm (18). Induction of IκBαMut completely ablated activation of the NF-κB-dependent hIL8 and PRDII promoters by TNFα, SeV, or transfection of poly(dA·dT), a dsDNA analog (Fig. 8C). However, it did not affect activation of the hZAP promoter by SeV or poly(dA·dT) (Fig. 8B). Collectively, these data suggest that activation of NF-κB is neither necessary nor sufficient for activation of the hZAP promoter by viral stimuli. Furthermore, these results demonstrate that the hZAP promoter can also be activated by cytosolic dsDNA, a pathogen-associated molecular pattern generated during replication of DNA viruses and some intracellular bacteria, by a NF-κB-independent pathway.
FIGURE 8.
NF-κB activation is neither sufficient nor required for induction of hZAP promoter by virus or dsDNA. A, promoter activities of hZAP(−2881), hOASL(−795), and PRDII in Hec1B cells mock treated or treated with TNFα (10 ng/ml), SeV (50 HAU), or IFNα (1000 units/ml) for 17 h. B and C, promoter activities of hZAP(−2486) (B) and hIL-8 (−162/+44) and PRDII (C) in HeLa Tet-Off cells conditionally expressing the NF-κB super-suppressor, IκBαMut (bearing S32A and S36A double mutations) that were manipulated to induce or repress IκBαMut expression. Where indicated, the cells were treated with TNFα, infected with SeV, or transfected with poly(dA·dT) for 17 h. Note in B that although the basal activity of hZAP promoter was lower in cells expressing IκBαMut, induction of the promoter by SeV or poly(dA·dT) was actually more than cells without IκBαMut expression (16.9- versus 9.1-fold and 9.9- versus 6.0-fold by SeV and poly(dA·dT), respectively).
DISCUSSION
ZAP is an antiviral host factor that prevents cytoplasmic accumulation of viral RNAs by promoting their decay by the exosome (47) and thereby restricts the replication of many RNA viruses, especially those within the families Retroviridae, Togaviridae, and Filoviridae (11–15). Although ZAP is an ISG induced early after viral infection (38, 48), how ZAP is transcriptionally regulated has not been well studied. The present study identifies ZAP as a gene that is directly regulated by IRF3. IRF3 activation, which occurs via several distinct signaling pathways when cells sense virus infection, is both necessary and sufficient for up-regulation of ZAP transcription in the context of virus infection (Figs. 1 and 4). Conversely, disruption of IRF3 function by DN-IRF3 significantly attenuated up-regulation of ZAP by virus or dsRNA (Fig. 1). In addition, IRF3 deficiency completely abrogated viral induction of ZAP (Fig. 2). The IRF3-dependent viral induction of ZAP is independent of IFN paracrine/autocrine action, because it occurs in cells either capable of (Fig. 2) or deficient in IFN signaling (Figs. 1 and 4). Furthermore, disruption of NF-κB activation, which is known to inhibit IFN-β induction, does not impair activation of ZAP promoter by virus (Fig. 8). The latter data provide additional support for the concept that viral induction of ZAP does not rely on IFNs released by infected cells early after infection. Taken together, these data place ZAP in a subset of ISGs that are induced as a result of direct IRF3 action or IFN-induced Jak-STAT signaling, such as ISG56 and ISG15.
OAS1 and MxA represent classical ISGs that require autocrine/paracrine action of IFN for induction during viral infections (53, 54). As a result they are more efficiently induced by IFN than by virus (Figs. 2B and 3). In contrast, ZAP induction by virus is at least as efficient, if not better, as induction by high IFN concentrations (Figs. 2B and 3). This is consistent with direct regulation of ZAP expression by IRF3 without a requirement for IFN signaling. Importantly, we have demonstrated this to be the case not only in cancer cell lines of various tissue origins (HeLa and A549) but also in non-neoplastic hepatocytes (PH5CH8) and in normal lung fibroblasts (MRC-5). An exception was the Huh7 hepatoma cell line, in which IFN seemed to be a more efficient inducer than virus for ZAP expression. This is not surprising, because Huh7 cells have an impaired RIG-I pathway and only mount a relatively weak IRF3 activation response following SeV infection (17, 68).
Mutational analyses of the human ZAP promoter led us to identify five ISRE/IRF-Es far upstream of the transcription start site (−2.5 to −2.2 kb) that serve in a redundant role to regulate IRF3-dependent viral induction of ZAP (Fig. 5). In agreement with this, IRF3 was found to bind strongly and specifically to these distal ISREs following virus infection as determined by ChIP assays (Fig. 7B). This is in contrast to the transcriptional control of most other well known IRF3 direct target genes, such as IFN-β and ISG56, in which the ISRE/IRF-Es are proximal to the transcription start site (4, 9). Whether this unique arrangement in transcriptional regulation of ZAP reflects an evolutionary advantage remains to be determined.
Our study also identifies a proximal STAT site (STAT V, −111 to −91) that is critical for both IFN- and virus-induced hZAP promoter activity (Fig. 6). STAT1/STAT2 preferentially bound to this site following IFN stimulation in ChIP assays (Fig. 7). The distal ISREs are apparently dispensable for IFN stimulation of hZAP promoter (Figs. 5, C and E, and 6), because deletion of these ISREs did not affect the promoter activation. However, our data do not rule out the possibility that other distal STAT sites as well as other STAT proteins may also contribute to IFN regulation of hZAP promoter.
In summary, ZAP is a primary response ISG in cellular antiviral responses. Its transcription can be directly controlled by IRF3 or as a result of IFN autocrine/paracrine signaling in viral infections. We propose the following model of ZAP induction. Early after infection, IRF3 is activated and translocated into the nucleus, whereupon it binds to the distal ISRE/IRF-Es in hZAP promoter to up-regulate ZAP transcription. This step is independent of IFN signaling. In the later phase, IFN-β is secreted by infected cells and activates, in turn, Jak-STAT signaling in infected cells and/or uninfected neighboring cells, resulting in the activation of STATs that translocate into the nucleus. These STATs interact with the proximal STAT V site and possibly other STAT sites in the hZAP promoter and sustain ZAP induction. It remains to be determined whether IRF7, induced by autocrine/paracrine IFN, may also contribute to the late phase of ZAP induction in parenchymal cells via the distal ISREs.
Supplementary Material
Acknowledgments
We thank R. Lin for providing the IRF3/5D and IRF-1 constructs, S. Akira and T. Kawai for the IRF7Δ238–408 construct, A. Kumar and I. Riz for the OAS2p69-GL2 construct, K. Narayanan for Hec1B cells, and R. Davey for 293FT cells.
This work was supported in part by National Institutes of Health Grants R21-DA018054 and R01-AI069285 (to K. L.), U19-AI066316 (to L. M. P.), and U19-AI40035 and R21-AI081058 (to S. M. L.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1–S6 and Figs. S1 and S2.
K. Li and S. M. Lemon, unpublished data.
- IFN
- interferon
- ZAP
- zinc finger antiviral protein
- IRF
- interferon regulatory factor
- IRF-E
- IRF element
- NF-κB
- nuclear factor κB
- STAT
- signal transducers and activators of transcription
- ISG
- interferon stimulated gene
- ISRE
- interferon-stimulated response element
- TLR3
- Toll-like receptor 3
- RIG-I
- retinoic inducible gene I
- MDA5
- melanoma differentiation-associated gene 5
- MAVS
- mitochondrial antiviral signaling protein, also known as IPS-1/Cardif/VISA
- TRIF
- Toll-IL1 receptor homology domain containing adaptor inducing interferon-β, also known as TICAM1
- OASL
- 2′,5′-oligoadenylate synthetase like gene
- ChIP
- chromatin immunoprecipitation
- HCV
- hepatitis C virus
- SeV
- Sendai virus
- ds
- double-stranded
- Q-PCR
- quantitative PCR
- DN
- dominant-negative
- h
- human
- IKK
- inhibitor of κB kinase
- HAU
- hemagglutinin units.
REFERENCES
- 1.Borden E. C., Sen G. C., Uze G., Silverman R. H., Ransohoff R. M., Foster G. R., Stark G. R. (2007) Nat. Rev. Drug. Discov. 6, 975–990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sen G. C. (2001) Annu. Rev. Microbiol. 55, 255–281 [DOI] [PubMed] [Google Scholar]
- 3.Müller U., Steinhoff U., Reis L. F., Hemmi S., Pavlovic J., Zinkernagel R. M., Aguet M. (1994) Science 264, 1918–1921 [DOI] [PubMed] [Google Scholar]
- 4.Hiscott J. (2007) J. Biol. Chem. 282, 15325–15329 [DOI] [PubMed] [Google Scholar]
- 5.Osterlund P. I., Pietilä T. E., Veckman V., Kotenko S. V., Julkunen I. (2007) J. Immunol. 179, 3434–3442 [DOI] [PubMed] [Google Scholar]
- 6.Onoguchi K., Yoneyama M., Takemura A., Akira S., Taniguchi T., Namiki H., Fujita T. (2007) J. Biol. Chem. 282, 7576–7581 [DOI] [PubMed] [Google Scholar]
- 7.Fitzgerald K. A., McWhirter S. M., Faia K. L., Rowe D. C., Latz E., Golenbock D. T., Coyle A. J., Liao S. M., Maniatis T. (2003) Nat. Immunol. 4, 491–496 [DOI] [PubMed] [Google Scholar]
- 8.Sharma S., tenOever B. R., Grandvaux N., Zhou G. P., Lin R., Hiscott J. (2003) Science 300, 1148–1151 [DOI] [PubMed] [Google Scholar]
- 9.Grandvaux N., Servant M. J., tenOever B., Sen G. C., Balachandran S., Barber G. N., Lin R., Hiscott J. (2002) J. Virol. 76, 5532–5539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Peters K. L., Smith H. L., Stark G. R., Sen G. C. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 6322–6327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gao G., Guo X., Goff S. P. (2002) Science 297, 1703–1706 [DOI] [PubMed] [Google Scholar]
- 12.Müller S., Möller P., Bick M. J., Wurr S., Becker S., Günther S., Kümmerer B. M. (2007) J. Virol. 81, 2391–2400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bick M. J., Carroll J. W., Gao G., Goff S. P., Rice C. M., MacDonald M. R. (2003) J. Virol. 77, 11555–11562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang Y., Burke C. W., Ryman K. D., Klimstra W. B. (2007) J. Virol. 81, 11246–11255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.MacDonald M. R., Machlin E. S., Albin O. R., Levy D. E. (2007) J. Virol. 81, 13509–13518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ikeda M., Sugiyama K., Mizutani T., Tanaka T., Tanaka K., Sekihara H., Shimotohno K., Kato N. (1998) Virus Res. 56, 157–167 [DOI] [PubMed] [Google Scholar]
- 17.Li K., Chen Z., Kato N., Gale M., Jr., Lemon S. M. (2005) J. Biol. Chem. 280, 16739–16747 [DOI] [PubMed] [Google Scholar]
- 18.Tian B., Zhang Y., Luxon B. A., Garofalo R. P., Casola A., Sinha M., Brasier A. R. (2002) J. Virol. 76, 6800–6814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen Z., Rijnbrand R., Jangra R. K., Devaraj S. G., Qu L., Ma Y., Lemon S. M., Li K. (2007) Virology 366, 277–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yoneyama M., Kikuchi M., Matsumoto K., Imaizumi T., Miyagishi M., Taira K., Foy E., Loo Y. M., Gale M., Jr., Akira S., Yonehara S., Kato A., Fujita T. (2005) J. Immunol. 175, 2851–2858 [DOI] [PubMed] [Google Scholar]
- 21.Yoneyama M., Kikuchi M., Natsukawa T., Shinobu N., Imaizumi T., Miyagishi M., Taira K., Akira S., Fujita T. (2004) Nat. Immunol. 5, 730–737 [DOI] [PubMed] [Google Scholar]
- 22.Li K., Foy E., Ferreon J. C., Nakamura M., Ferreon A. C., Ikeda M., Ray S. C., Gale M., Jr., Lemon S. M. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 2992–2997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen Z., Benureau Y., Rijnbrand R., Yi J., Wang T., Warter L., Lanford R. E., Weinman S. A., Lemon S. M., Martin A., Li K. (2007) J. Virol. 81, 964–976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lu Y., Jamieson L., Brasier A. R., Fields A. P. (2001) Oncogene 20, 4777–4792 [DOI] [PubMed] [Google Scholar]
- 25.Lin R., Mustafa A., Nguyen H., Gewert D., Hiscott J. (1994) J. Biol. Chem. 269, 17542–17549 [PubMed] [Google Scholar]
- 26.Lin R., Heylbroeck C., Pitha P. M., Hiscott J. (1998) Mol. Cell. Biol. 18, 2986–2996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kawai T., Sato S., Ishii K. J., Coban C., Hemmi H., Yamamoto M., Terai K., Matsuda M., Inoue J., Uematsu S., Takeuchi O., Akira S. (2004) Nat. Immunol. 5, 1061–1068 [DOI] [PubMed] [Google Scholar]
- 28.Brasier A. R., Jamaluddin M., Casola A., Duan W., Shen Q., Garofalo R. P. (1998) J. Biol. Chem. 273, 3551–3561 [DOI] [PubMed] [Google Scholar]
- 29.Fredericksen B., Akkaraju G. R., Foy E., Wang C., Pflugheber J., Chen Z. J., Gale M., Jr. (2002) Viral. Immunol. 15, 29–40 [DOI] [PubMed] [Google Scholar]
- 30.Krasnoselskaya-Riz I., Spruill A., Chen Y. W., Schuster D., Teslovich T., Baker C., Kumar A., Stephan D. A. (2002) AIDS Res. Hum. Retroviruses 18, 591–604 [DOI] [PubMed] [Google Scholar]
- 31.Li K. (2009) Methods Mol. Biol. 510, 211–226 [DOI] [PubMed] [Google Scholar]
- 32.Langford M. P., Weigent D. A., Stanton G. J., Baron S. (1981) Methods Enzymol. 78, 339–346 [DOI] [PubMed] [Google Scholar]
- 33.Wang N., Liang Y., Devaraj S., Wang J., Lemon S. M., Li K. (2009) J. Virol. 83, 9824–9834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Weaver B. K., Kumar K. P., Reich N. C. (1998) Mol. Cell. Biol. 18, 1359–1368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Verhaegen M., Divizia M., Vandenbussche P., Kuwata T., Content J. (1980) Proc. Natl. Acad. Sci. U.S.A. 77, 4479–4483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kotenko S. V., Gallagher G., Baurin V. V., Lewis-Antes A., Shen M., Shah N. K., Langer J. A., Sheikh F., Dickensheets H., Donnelly R. P. (2003) Nat. Immunol. 4, 69–77 [DOI] [PubMed] [Google Scholar]
- 37.Sheppard P., Kindsvogel W., Xu W., Henderson K., Schlutsmeyer S., Whitmore T. E., Kuestner R., Garrigues U., Birks C., Roraback J., Ostrander C., Dong D., Shin J., Presnell S., Fox B., Haldeman B., Cooper E., Taft D., Gilbert T., Grant F. J., Tackett M., Krivan W., McKnight G., Clegg C., Foster D., Klucher K. M. (2003) Nat. Immunol. 4, 63–68 [DOI] [PubMed] [Google Scholar]
- 38.Marcello T., Grakoui A., Barba-Spaeth G., Machlin E. S., Kotenko S. V., MacDonald M. R., Rice C. M. (2006) Gastroenterology 131, 1887–1898 [DOI] [PubMed] [Google Scholar]
- 39.Zhu H., Butera M., Nelson D. R., Liu C. (2005) Virol. J. 2, 80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Robek M. D., Boyd B. S., Chisari F. V. (2005) J. Virol. 79, 3851–3854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Elco C. P., Guenther J. M., Williams B. R., Sen G. C. (2005) J. Virol. 79, 3920–3929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Clements J. L. (2003) Immunol. Rev. 191, 211–219 [DOI] [PubMed] [Google Scholar]
- 43.Yen A., Varvayanis S., Smith J. L., Lamkin T. J. (2006) Eur. J. Cell Biol. 85, 117–132 [DOI] [PubMed] [Google Scholar]
- 44.Rebouillat D., Marié I., Hovanessian A. G. (1998) Eur. J. Biochem. 257, 319–330 [DOI] [PubMed] [Google Scholar]
- 45.Marques J., Anwar J., Eskildsen-Larsen S., Rebouillat D., Paludan S. R., Sen G., Williams B. R., Hartmann R. (2008) J. Gen. Virol. 89, 2767–2772 [DOI] [PubMed] [Google Scholar]
- 46.Melchjorsen J., Kristiansen H., Christiansen R., Rintahaka J., Matikainen S., Paludan S. R., Hartmann R. (2009) J. Interferon Cytokine Res. 29, 199–208 [DOI] [PubMed] [Google Scholar]
- 47.Guo X., Ma J., Sun J., Gao G. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 151–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ryman K. D., Meier K. C., Nangle E. M., Ragsdale S. L., Korneeva N. L., Rhoads R. E., MacDonald M. R., Klimstra W. B. (2005) J. Virol. 79, 1487–1499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kato H., Takeuchi O., Sato S., Yoneyama M., Yamamoto M., Matsui K., Uematsu S., Jung A., Kawai T., Ishii K. J., Yamaguchi O., Otsu K., Tsujimura T., Koh C. S., Reis e Sousa C., Matsuura Y., Fujita T., Akira S. (2006) Nature 441, 101–105 [DOI] [PubMed] [Google Scholar]
- 50.Bauhofer O., Summerfield A., Sakoda Y., Tratschin J. D., Hofmann M. A., Ruggli N. (2007) J. Virol. 81, 3087–3096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hilton L., Moganeradj K., Zhang G., Chen Y. H., Randall R. E., McCauley J. W., Goodbourn S. (2006) J. Virol. 80, 11723–11732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Seago J., Hilton L., Reid E., Doceul V., Jeyatheesan J., Moganeradj K., McCauley J., Charleston B., Goodbourn S. (2007) J. Gen. Virol. 88, 3002–3006 [DOI] [PubMed] [Google Scholar]
- 53.Nakaya T., Sato M., Hata N., Asagiri M., Suemori H., Noguchi S., Tanaka N., Taniguchi T. (2001) Biochem. Biophys. Res. Commun. 283, 1150–1156 [DOI] [PubMed] [Google Scholar]
- 54.Ronni T., Matikainen S., Lehtonen A., Palvimo J., Dellis J., Van Eylen F., Goetschy J. F., Horisberger M., Content J., Julkunen I. (1998) J. Interferon Cytokine Res. 18, 773–781 [DOI] [PubMed] [Google Scholar]
- 55.Wang Q., Floyd-Smith G. (1998) Gene 222, 83–90 [DOI] [PubMed] [Google Scholar]
- 56.Floyd-Smith G., Wang Q., Sen G. C. (1999) Exp. Cell Res. 246, 138–147 [DOI] [PubMed] [Google Scholar]
- 57.Kawai T., Takahashi K., Sato S., Coban C., Kumar H., Kato H., Ishii K. J., Takeuchi O., Akira S. (2005) Nat. Immunol. 6, 981–988 [DOI] [PubMed] [Google Scholar]
- 58.Meylan E., Curran J., Hofmann K., Moradpour D., Binder M., Bartenschlager R., Tschopp J. (2005) Nature 437, 1167–1172 [DOI] [PubMed] [Google Scholar]
- 59.Seth R. B., Sun L., Ea C. K., Chen Z. J. (2005) Cell 122, 669–682 [DOI] [PubMed] [Google Scholar]
- 60.Xu L. G., Wang Y. Y., Han K. J., Li L. Y., Zhai Z., Shu H. B. (2005) Mol. Cell 19, 727–740 [DOI] [PubMed] [Google Scholar]
- 61.Oshiumi H., Matsumoto M., Funami K., Akazawa T., Seya T. (2003) Nat. Immunol. 4, 161–167 [DOI] [PubMed] [Google Scholar]
- 62.Yamamoto M., Sato S., Mori K., Hoshino K., Takeuchi O., Takeda K., Akira S. (2002) J. Immunol. 169, 6668–6672 [DOI] [PubMed] [Google Scholar]
- 63.Pitha P. M., Au W. C., Lowther W., Juang Y. T., Schafer S. L., Burysek L., Hiscott J., Moore P. A. (1998) Biochimie 80, 651–658 [DOI] [PubMed] [Google Scholar]
- 64.Taniguchi T., Ogasawara K., Takaoka A., Tanaka N. (2001) Annu. Rev. Immunol. 19, 623–655 [DOI] [PubMed] [Google Scholar]
- 65.Honda K., Takaoka A., Taniguchi T. (2006) Immunity 25, 349–360 [DOI] [PubMed] [Google Scholar]
- 66.Sumpter R., Jr., Loo Y. M., Foy E., Li K., Yoneyama M., Fujita T., Lemon S. M., Gale M., Jr. (2005) J. Virol. 79, 2689–2699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Du Z., Wei L., Murti A., Pfeffer S. R., Fan M., Yang C. H., Pfeffer L. M. (2007) J. Cell. Biochem. 102, 1087–1094 [DOI] [PubMed] [Google Scholar]
- 68.Foy E., Li K., Wang C., Sumpter R., Jr., Ikeda M., Lemon S. M., Gale M., Jr. (2003) Science 300, 1145–1148 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.