Abstract
The neuroectodermal tumors neuroblastoma and melanoma represent biologically aggressive and chemoresistant cancers. The chemotherapeutic agents fenretinide and bortezomib induce apoptosis through endoplasmic reticulum (ER) stress in these tumor types. The aim of this study was to test the hypothesis that the early events of ER stress signaling and response pathways induced by fenretinide and bortezomib are mediated by the eukaryotic initiation factor 2α (eIF2α)-ATF4 signaling pathway. Treatment of neuroblastoma and melanoma cell lines with fenretinide, bortezomib, or thapsigargin resulted in induction of eIF2α signaling, characterized by increased expression of phosphorylated eIF2α, ATF4, ATF3, and GADD34. These events correlated with induction of the pro-apoptotic protein Noxa. The cytotoxic response, characterized by up-regulation of Noxa and cell death, was dependent on ATF4, but not the ER-related pro-death signaling pathways involving GADD153 or IRE1. Although PERK-dependent phosphorylation of eIF2α enhanced ATF4 protein levels during ER stress, cell death in response to fenretinide, bortezomib, or thapsigargin was not abrogated by inhibition of eIF2α phosphorylation through PERK knockdown or overexpression of wild-type eIF2α. Furthermore, ATF4 induction in response to ER stress was dependent primarily on transcriptional activation, which occurred in a PERK- and phosphorylated eIF2α-independent manner. These results demonstrate that ATF4 mediates ER stress-induced cell death of neuroectodermal tumor cells in response to fenretinide or bortezomib. Understanding the complex regulation of cell death pathways in response to ER stress-inducing drugs has the potential to reveal novel therapeutic targets, thus allowing the development of improved treatment strategies to overcome chemoresistance.
Keywords: Cancer, Anticancer Drug, Cancer Therapy, ER Stress, Neuroblastoma, Endoplasmic Reticulum Stress, Bortezomib, Fenretinide, Melanoma
Introduction
The development of novel strategies to overcome chemoresistance in cancers, particularly neuroectodermal tumors, which are frequently associated with poor survival despite intensive chemotherapy, would be aided by understanding the complex regulation of cell death programs. The chemotherapeutic agents fenretinide, a synthetic derivative of retinoic acid, and bortezomib, a 26 S proteasome inhibitor, induce endoplasmic reticulum (ER)2 stress, culminating in apoptosis of neuroblastoma and melanoma cells both in vitro (1, 2) and in vivo (3–6). These studies highlight ER stress as an intracellular stress response that can be exploited to promote cancer cell death, providing an opportunity for rational drug design programs and the development of more effective therapeutic strategies.
Normal ER function is required for the regulation of intracellular calcium and correct folding of secretory or cell-surface proteins; ER stress occurs when the protein-folding capacity of the ER is exceeded. Stresses that perturb redox state, energy levels, or calcium homeostasis trigger the accumulation of unfolded proteins within the ER, eliciting a stress response termed the unfolded protein response (UPR). The principles of the UPR are now relatively well defined and are characterized by an inhibition of global protein synthesis in cooperation with the transcriptional activation of UPR target genes to promote protein folding (7). The UPR is primarily an adaptive response to support cell survival, but several lines of evidence suggest that, if homeostasis cannot be re-established, the UPR triggers cell death (8). Three ER transmembrane proteins that mediate distinct arms of the UPR have been identified: IRE1 (inositol-requiring protein-1), ATF6 (activating transcription factor-6), and PERK (protein kinase RNA (PKR)-like ER kinase) (7). All three proteins stimulate expression of pro-apoptotic genes in response to ER stress, although the specific pathways involved are complex and highly context-dependent (9).
Although disturbances in ER homeostasis occur in a variety of cancers (10), whether they arise from cancer-specific mutations or reflect perturbation of ER function within the tumor microenvironment is unclear. From the perspective of cancer therapy, constitutive activation of ER stress pathways may be associated with chemoresistance (11, 12). Moreover, ER-related responses appear to be associated with the activity of a variety of anticancer drugs, including proteasome inhibitors and some DNA-damaging agents. The paradoxical ability of the UPR to engage cell survival and death mechanisms is a critical issue, with recent studies demonstrating that disruption of elements of the survival response can shift the balance toward apoptosis in cells treated with ER stress-inducing agents such as fenretinide and bortezomib (2, 13).
Nevertheless, the relative contribution of pro-apoptotic ER stress signaling to fenretinide- or bortezomib-induced cell death is not clear. Multiple pathways may be involved in ER stress-initiated apoptosis, and in most cases, these converge at the level of the mitochondria (9). The most common upstream pathways identified include calcium signaling, IRE1-JNK (c-Jun N-terminal kinase) activation, and induction of the transcription factor CHOP (C/EBP homologous protein)/GADD153 (growth arrest/DNA damage-inducible 153) (8). However, there is increasing interest in the central role of translation eukaryotic initiation factor 2α (eIF2α) signaling in cell fate determination during ER stress (14, 15). PERK-mediated phosphorylation of eIF2α at serine 51 occurs during ER stress, resulting in inhibition of global translation (15) and enhanced translation of mRNAs such as those encoding ATF4 (16, 17). ATF4 enhances the expression of another member of the same family, the transcriptional repressor ATF3 (18), and the protein phosphatase 1-interacting protein GADD34, which causes protein phosphatase 1 to dephosphorylate eIF2α, thus releasing the translational block and ensuring the transient nature of protein synthesis inhibition (19). ATF4 is a universal stress-responsive gene thought to have a protective role by regulating cellular adaptation to adverse conditions; however, there are studies describing a pro-death role for ATF4 in the context of ER stress (14, 20–22).
Understanding the molecular mechanisms mediating ER stress-induced apoptosis in the context of cancer therapy is essential to promote the pharmacological exploitation of ER stress responses to combat cancer. Both fenretinide and bortezomib induce hallmarks of ER stress (2, 13), and because there is evidence for a central role for eIF2α-ATF4 signaling in ER stress-induced cell death (14, 21, 22), we hypothesize that this pathway is fundamental to the cytotoxicity induced by these agents in neuroectodermal tumor cells.
EXPERIMENTAL PROCEDURES
Cell Culture, Drug Treatment, and Analysis of Apoptosis
Melanoma cell lines A375 and SK-MEL-28, obtained from the American Type Culture Collection, and the neuroblastoma cell line SH-SY5Y were cultured as described previously (1). Fenretinide (Janssen-Cilag Ltd., Bassersdorf, Switzerland) was added in ethanol; bortezomib (Janssen-Cilag Ltd., High Wycombe, UK), thapsigargin (Sigma, Poole, UK), actinomycin D (Sigma), and salubrinal (Alexis Biochemicals and Enzo Life Sciences Ltd.) were added in Me2SO with an equal volume of vehicle used to treat control cells. Flow cytometry of propidium iodide-stained cells was used to estimate cell death (apoptosis) from the percentage of cells in the sub-G1 fraction (13). Caspase-3/7 activation was measured by the Caspase-Glo 3/7 assay (Promega, Southampton, UK) according to the manufacturer's instructions.
Western Blotting
Total protein was extracted from cell pellets, separated by electrophoresis through 4–20% SDS-polyacrylamide gels (20 μg/lane), and blotted onto polyvinylidene difluoride membranes (1). Blots were probed with antibodies to ATF4 (Calbiochem); ATF3 and GADD153 (B-3, Santa Cruz Biotechnology); Noxa (Alexis Biochemicals); c-Myc, eIF2α, phospho-eIF2α, phospho-c-Jun, and IRE1α (Cell Signaling Technology); and spliced XBP-1 (Cambridge BioScience). An anti-β-actin antibody (Sigma) was used as a loading control. The binding of primary antibodies was detected with secondary peroxidase-conjugated antibodies (Upstate Biotechnology and Vector Laboratories) and visualized using the ECL system (Amersham Biosciences).
Gene Expression Analysis
Total RNA was isolated from cell pellets using the RNeasy mini kit (Qiagen, Crawley, UK) and reverse-transcribed using a high capacity reverse transcription kit (Applied Biosystems, Warrington, UK) according to the manufacturers' instructions. Real-time PCR was performed on 20 ng of cDNA using validated TaqMan gene expression assays for human Noxa, ATF4, ATF3, GADD34, GADD153, PERK, and p8 in combination with the TaqMan Universal PCR Master Mix (Applied Biosystems). Appropriate controls for nonspecific amplification and contamination were included. A GeneAmp 7500 sequence detection system was used for real-time PCR amplification. As an internal standard, β-actin was measured simultaneously using the endogenous control assay provided by Applied Biosystems. PCR amplification procedures followed the manufacturer's instructions. The data were analyzed using the GeneAmp sequence detection system software, and the comparative Ct method (2−ΔΔCt) was used for relative quantification of gene expression.
Transfection Experiments
RNA interference-mediated gene knockdown was achieved using prevalidated Qiagen HP small interfering RNAs (siRNAs) for ATF4 (SI03019345), PPP1R15A (GADD34; SI02659132), EIF2AK3 (PERK; SI02223725), c-myc (SI00300902, SI02662611), and ERN1 (IRE1α; SI00605255) or Invitrogen siRNAs for ATF3 (HSS100778) and GADD153 (DDIT3 validated Stealth siRNA, duplex 2; (GADD153-1). Additional siRNAs targeting GADD153 (GADD153-2) (23) and p8 (20) (Eurogentec, Southampton, UK) were also used. All siRNA experiments incorporated a validated negative control siRNA (Qiagen AllStars negative control siRNA). Expression vectors for ATF4 (pATF4, NM_001675) and the empty vector pXL4 were from OriGene (Cambridge Bioscience). Wild-type eIF2α in pcDNA3.CD2 was provided by David Ron (Skirball Institute for Biomolecular Medicine, New York University School of Medicine, New York) and subcloned into pcDNA4 (Invitrogen). siRNA knockdown experiments were carried out by plating 0.25–0.4 × 106 cells in 6-well plates overnight before transfection with 40 nm siRNA using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. Cells were then cultured in normal growth medium for 24 h prior to drug treatment. For overexpression of ATF4 or eIF2α, transient transfection of 2 μg of expression vector or empty control vector was performed using Lipofectamine 2000 as described above.
DNA-dependent Promoter Pulldown Assays
Genomic DNA from A375 cells was used to amplify a DNA fragment corresponding to bases −110 to +40 of the Noxa promoter (24). Promoter templates biotinylated at the 5′-end of the upper strand were generated by PCR and purified using QIAquick spin columns (Qiagen). Whole cell extracts (400 μg) were precleared with streptavidin-agarose beads prior to incubation for 2 h at 4 °C with the DNA template in Buffer A (20 mm HEPES, pH 7.9, 100 mm KCl, 6 mm MgCl2, 1 mm dithiothreitol, 20% glycerol, 0.01% Nonidet P-40, 0.15 mg/ml poly(dI/dC), 0.15 mg/ml single-stranded salmon testis DNA, and EDTA-free protease inhibitor mixture (Roche Diagnostics Ltd., Burgess Hill, UK)). The DNA template was precipitated using streptavidin-agarose beads for 2 h at 4 °C, and beads were washed three times with Buffer A containing 1 mg/ml bovine serum albumin. Proteins were eluted with SDS-PAGE sample buffer prior to gel electrophoresis and Western blotting.
Statistical Analysis
Statistical analysis of apoptosis was performed using SPSS Version 15.0 (SPSS Inc., Chicago, IL). For experiments on siRNA-mediated knockdown, the effects on apoptosis induced by fenretinide, bortezomib, or thapsigargin were tested by two-way analysis of variance (ANOVA; GLM) with siRNA type (control or gene-specific) and treatment (fenretinide, bortezomib, or thapsigargin) main effects. The specific effects of siRNA type (when more than one gene-specific siRNA was used) or treatments were tested, when appropriate, using simple contrasts.
RESULTS
Kinetics of eIF2α Pathway Activation during Fenretinide- and Bortezomib-induced Apoptosis
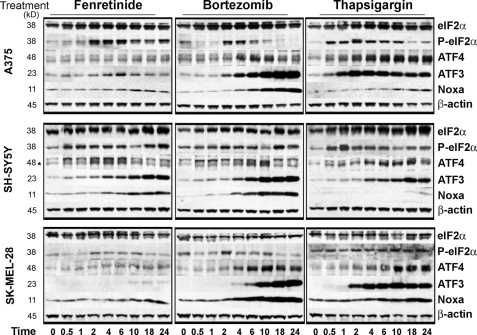
Fenretinide and bortezomib induce caspase-dependent cell death in neuroblastoma and melanoma cells (25, 26), and induction of caspase-3/7 activity by clinically achievable concentrations of fenretinide and bortezomib (2) was confirmed in SH-SY5Y neuroblastoma and A375 and SK-MEL-28 melanoma cells (supplemental Fig. 1). Caspase-3 activation reflected the enhanced sensitivity to bortezomib compared with fenretinide at these concentrations. The ER stress-inducing agent thapsigargin, included as a positive control for ER stress, also induced caspase-3/7 activity, although the level of induction was lower in the melanoma cell lines. To investigate the kinetics of activation of eIF2α-ATF4 signaling during ER stress in neuroectodermal tumor cells, time course experiments were performed over 0–24 h in SH-SY5Y, A375, and SK-MEL-28 cells after treatment with fenretinide, bortezomib, or thapsigargin. Western blot analysis revealed that phosphorylation of eIF2α was transient, and the kinetics varied between drug treatment and cell line (Fig. 1). The response to bortezomib was the most short-lived, with the greatest increase in eIF2α phosphorylation seen between 2 and 6 h of drug treatment. A more rapid response to thapsigargin treatment was observed, with eIF2α phosphorylation first detected at 0.5 h and phosphorylation maintained for up to 10 h. Phosphorylation of eIF2α was observed 0.5 or 2 h after fenretinide treatment in SH-SY5Y or melanoma cells, respectively, and this was maintained for an additional 8 h.
FIGURE 1.
Fenretinide and bortezomib regulate eIF2α signaling in neuroectodermal tumor cells. Shown are Western blots for eIF2α, phospho-eIF2α, ATF4 (indicated by the asterisk), ATF3, Noxa, and β-actin in SH-SY5Y, A375, and SK-MEL-28 cells treated with fenretinide (SH-SY5Y, 5 μm; and A375/SK-MEL-28, 10 μm), bortezomib (SH-SY5Y, 5 nm; and A375/SK-MEL-28, 30 nm), or thapsigargin (SH-SY5Y, 1.5 μm; and A375/SK-MEL-28, 7.5 μm) for 0–24 h.
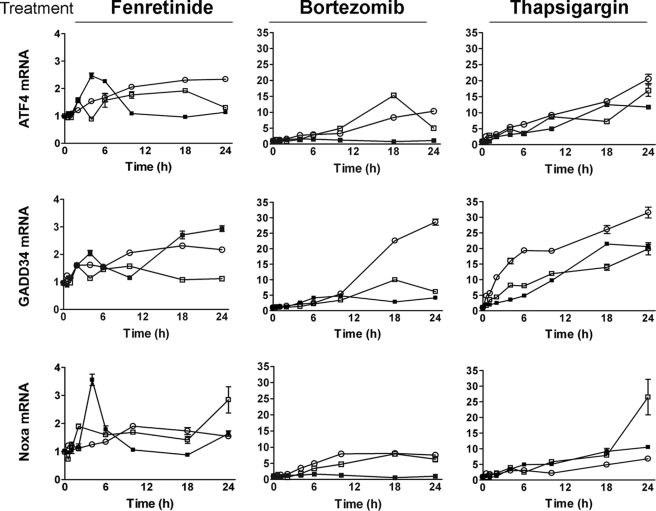
Increased phosphorylation of eIF2α was associated with increased expression of ATF4 protein, consistent with the reported enhancement of ATF4 translation under conditions of ER stress (Fig. 1). Treatment of A375 or SK-MEL-28 melanoma cells with bortezomib or thapsigargin resulted in the greatest induction of ATF4 protein expression, which began to diminish between 10 and 18 h after bortezomib treatment compared with between 18 and 24 h after thapsigargin treatment. These findings are consistent with the more transient nature of bortezomib-induced eIF2α phosphorylation. In SH-SY5Y cells, ATF4 expression was also up-regulated to the greatest extent in response to thapsigargin and to a lesser extent in response to fenretinide or bortezomib. However, ATF4 expression was only weakly up-regulated in response to fenretinide in A375 and SK-MEL-28 cells despite strong phosphorylation of eIF2α; this is consistent with weak ATF3 induction under these conditions in these melanoma cell lines. ATF4 mRNA was also increased during drug treatment, with the magnitude of induction greatest in response to bortezomib in the melanoma cell lines or thapsigargin treatment in all three cell lines, consistent with the observed protein levels (Fig. 2).
FIGURE 2.
Fenretinide and bortezomib regulate eIF2α signaling in neuroectodermal tumor cells. ATF4, GADD34, or Noxa mRNA was measured by real-time PCR relative to β-actin as an internal control in SH-SY5Y (■), A375 (□), and SK-MEL-28 (○) cells treated with fenretinide (SH-SY5Y, 5 μm; and A375/SK-MEL-28, 10 μm), bortezomib (SH-SY5Y, 5 nm; and A375/SK-MEL-28, 30 nm), or thapsigargin (SH-SY5Y, 1.5 μm; and A375/SK-MEL-28, 7.5 μm) for 0–24 h. Gene expression is expressed relative to control untreated cells, and the y axis scale is 10-fold greater for the melanoma cell lines A375 and SK-MEL-28. Data are expressed as the mean ± S.E. (n = 3) and are a complete time course, consistent with n ≥ three independent experiments at 0, 6, 18, and 24 h.
ATF3 was induced in response to fenretinide, bortezomib, and thapsigargin, but induction was more marked compared with ATF4, except in response to fenretinide in melanoma cells, in agreement with the observed weak ATF4 induction (Fig. 1). We and others (1, 26) have shown that apoptosis in response to fenretinide, bortezomib, or thapsigargin is dependent on the transcriptional and translational up-regulation of the pro-apoptotic BH3-only protein Noxa. In agreement with previous data (1, 26), Noxa was up-regulated 6–10 h after drug treatment (Fig. 1), with the greatest induction observed in response to bortezomib. However, although thapsigargin was a weaker inducer of Noxa protein expression, it considerably enhanced Noxa mRNA levels over 24 h (Fig. 2). Increased levels of Noxa mRNA were also observed after fenretinide or bortezomib treatment, with the magnitude of induction greatest in response to bortezomib in the melanoma cells (Fig. 2). In addition, up-regulation of GADD34 mRNA was seen across all cell lines and drug treatments (Fig. 2), consistent with this gene being a target of ATF4 (27).
ATF4 Mediates Fenretinide- and Bortezomib-induced Cell Death
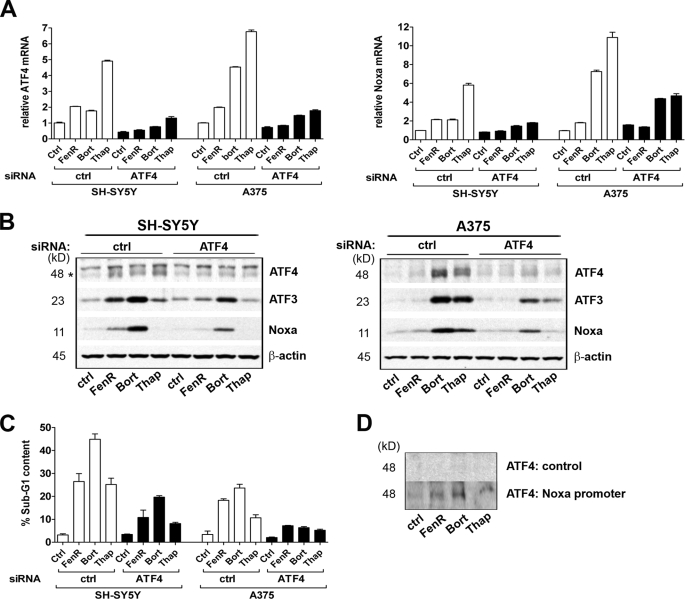
The early up-regulation of phosphorylated eIF2α and ATF4 expression in response to different ER stress-inducing drugs is consistent with this pathway being a potential mediator of ER-related apoptotic signaling. To test the hypothesis that ATF4 promotes cell death in this context, ATF4 expression was down-regulated by siRNA in a neuroblastoma (SH-SY5Y) and a melanoma (A375) cell line prior to treatment with fenretinide, bortezomib, or thapsigargin. ATF4 knockdown was confirmed at both the mRNA and protein levels in response to drug treatment (Fig. 3, A and B). siRNA-mediated knockdown of ATF4 resulted in a reduced induction of ATF3 (Fig. 3B), supporting existing evidence that ATF3 is a target of ATF4. A pro-apoptotic function of ATF4 was demonstrated by a significant reduction in fenretinide-, bortezomib-, or thapsigargin-induced cell death in both SH-SY5Y and A375 cells with reduced ATF4 expression (siRNA main effect, F1,31 > 11, p < 0.001) (Fig. 3C), and there was evidence for increased death of A375 cells overexpressing ATF4 in response to treatment with these agents (effect of ATF4 overexpression, F1,22 = 3.68, p = 0.068) (supplemental Fig. 2).
FIGURE 3.
ATF4 mediates fenretinide- and bortezomib-induced cell death. A and B, SH-SY5Y and A375 cells were transfected with siRNAs for ATF4 or with a non-silencing control siRNA (ctrl) prior to treatment with fenretinide (FenR) (SH-SY5Y, 5 μm; and A375, 10 μm), bortezomib (Bort) (SH-SY5Y, 5 nm; and A375, 30 nm), or thapsigargin (Thap) (SH-SY5Y, 1.5 μm; and A375, 7.5 μm) for 6 h. ATF4 or Noxa mRNA was measured by real-time PCR relative to β-actin as an internal control (A). ATF4 (lower band indicated by the asterisk in SH-SY5Y cells), ATF3, Noxa, and β-actin expression was determined by Western blotting (B). C, SH-SY5Y and A375 cells were transfected with siRNAs for ATF4 or with a non-silencing control siRNA prior to treatment with fenretinide (SH-SY5Y, 10 μm; and A375, 15 μm), bortezomib (SH-SY5Y, 5 nm; and A375, 50 nm), or thapsigargin (SH-SY5Y, 3 μm; and A375, 10 μm) for 24 h. Apoptosis was measured by flow cytometry of propidium iodide-stained cells to determine the sub-G1 fraction. Data are expressed as the percentage total population or relative to control untreated cells; each point is the mean ± S.E. (n ≥ 3). D, A375 cells were treated with fenretinide (15 μm), bortezomib (50 nm), or thapsigargin (10 μm) for 6 h. Recruitment to the Noxa promoter was determined by promoter pulldown assays in the absence (control) or presence of the Noxa promoter DNA fragment, followed by Western blotting for ATF4. Data are shown for whole cell extracts; similar results were obtained with nuclear extracts (not shown).
In support of a pro-death role for ATF4 during ER stress, ATF4 knockdown resulted in inhibition of both Noxa mRNA and protein induction in response to drug treatment (Fig. 3, A and B). Together, these data indicate that ATF4 mediates ER stress-induced cell death in neuroectodermal tumor cells. To test the association of ATF4 with the Noxa promoter, pulldown assays were performed using either whole cell or nuclear extracts and a biotinylated DNA probe corresponding to bases −110 to +40 relative to the Noxa transcription start site, the fragment previously reported to recruit ATF4 (24). Endogenous ATF4 was demonstrated to bind to the Noxa promoter following treatment of A375 cells with fenretinide, bortezomib, or thapsigargin (Fig. 3D), suggesting that ATF4 is a direct transcriptional activator of Noxa.
These data are in contrast to a previous report in which c-Myc was determined to be responsible for bortezomib-induced Noxa expression (28). Although c-Myc expression was up-regulated in response to bortezomib treatment in SK-MEL-28 cells, a cell line included in the study by Nikiforov et al. (28), siRNA-mediated knockdown of c-Myc expression did not abrogate fenretinide- or bortezomib-induced Noxa (supplemental Fig. 3A). Conversely, c-Myc expression was down-regulated in response to fenretinide, bortezomib, or thapsigargin treatment in SH-SY5Y and A375 cells (supplemental Fig. 3A), suggesting differential regulation of c-Myc in response to ER stress between neuroectodermal tumor cell lines.
The role of ATF3 induction is less clear, with no significant effect of ATF3 knockdown on fenretinide-, bortezomib-, or thapsigargin-induced cell death overall (F1,26 = 1.71, p = 0.203) (supplemental Fig. 3B). However, in the analysis of the apoptosis data by ANOVA, the interaction term (siRNA*treatment) was significant (F2,26 = 15.99, p < 0.001), with fenretinide-induced apoptosis significantly increased (F1,26 = 26.2, p < 0.001) and bortezomib-induced apoptosis significantly decreased (F1,26 = 7.03, p = 0.013), albeit to a small extent, and with no effect on thapsigargin-induced apoptosis (F1,26 = 0.102, p = 0.75). In addition, ATF3 knockdown enhanced ER stress-induced Noxa transcription, although an increase in Noxa protein induction was evident only after fenretinide treatment (supplemental Fig. 3C). The effect on Noxa mRNA is consistent with a role for ATF3 as a transcriptional repressor under these conditions, whereas the effects on cell death suggest more complex controls of cell death regulation in response to different ER stress inducers.
Both the IRE1-XBP-1/JNK axis and GADD153 can influence UPR-mediated cell survival and death in response to ER stress (29, 30). Therefore, the role of these alternative ER-related apoptotic pathways was investigated in A375 cells. Knockdown of IRE1α by siRNA and subsequent inhibition of XBP-1 splicing (demonstrated by a decrease in XBP-1s protein produced from the spliced mRNA) did not abrogate fenretinide-, bortezomib-, or thapsigargin-induced Noxa expression or cell death in response to these agents (two-way ANOVA, effect of control, IRE1α, or GADD153 siRNA, F2,36 = 5.165, p = 0.011; contrast on main effect, control siRNA versus IRE1α siRNA, p = 0.55, non-significant) (supplemental Fig. 4, A and B). Similarly, abrogation of JNK activity using a small molecule inhibitor (SP600125) did not affect the cell death response (data not shown). These data indicate that IRE1α signaling is not the predominant UPR branch regulating cell death in neuroectodermal tumor cells. Moreover, knockdown of GADD153 revealed that this transcription factor was not essential for fenretinide-, bortezomib-, or thapsigargin-induced Noxa induction (supplemental Fig. 4C); unexpectedly, GADD153 knockdown slightly increased cell death in response to these agents (contrast on main effect, control siRNA versus GADD153 siRNA, p = 0.008) (supplemental Fig. 4A). Similar results were obtained in SH-SY5Y cells (data not shown).
Regulation of ATF4 in Response to Fenretinide or Bortezomib in Neuroectodermal Tumor Cells
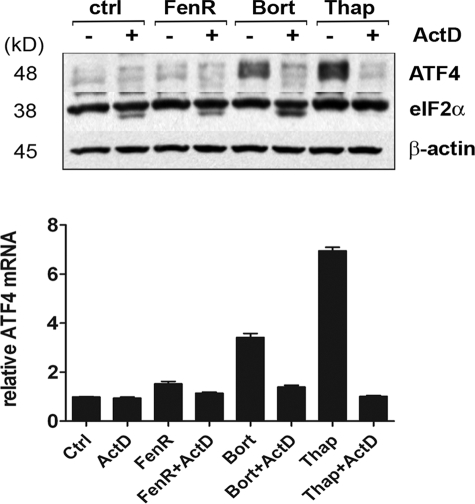
To determine whether ATF4 induction is dependent on transcriptional control during ER stress, A375 cells were treated with ER stress inducers in the absence or presence of actinomycin D. Both ATF4 mRNA and protein induction in response to drug treatment were severely abrogated in the presence of actinomycin D, whereas eIF2α and β-actin protein levels remained relatively constant (Fig. 4). These results suggest that ATF4 is regulated transcriptionally under these conditions and that increased levels of mRNA contribute to the observed protein levels. Although the transcriptional control of ATF4 is not well understood, the stress-regulated transcription factor p8 (candidate of metastasis-1) has been implicated in the up-regulation of ATF4 mRNA during cannabinoid-induced apoptosis in tumor cells, a process involving ER stress (20, 31). Although the expression of p8 mRNA was induced in A375 cells in response to bortezomib or thapsigargin treatment and to a much a lesser extent in response to fenretinide, the siRNA-mediated knockdown of p8 resulted in enhanced ATF4 mRNA levels, indicating that p8 acts as a negative regulator of ATF4 in this context (supplemental Fig. 5).
FIGURE 4.
Transcriptional regulation of ATF4. A375 cells were treated with fenretinide (FenR; 10 μm), bortezomib (Bort; 30 nm), or thapsigargin (Thap; 7.5 μm) in the absence or presence of actinomycin D (ActD; 0.5 μm) for 10 h. ATF4, eIF2α, and β-actin expression was determined by Western blotting, and ATF4 mRNA was measured by real-time PCR and is expressed relative to β-actin. ctrl, control.
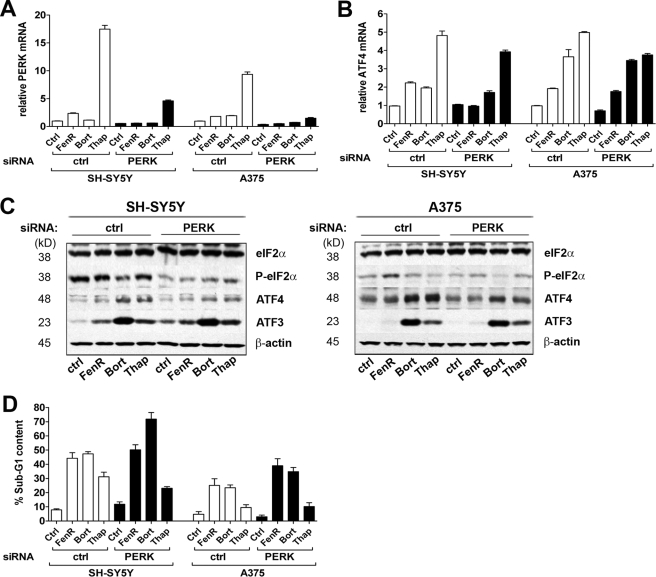
Evidence for the transcriptional regulation of ATF4 during ER stress is in contrast to the idea that enhanced ATF4 expression chiefly results from eIF2α phosphorylation and delayed translation re-initiation at upstream open reading frames within the 5′-end of ATF4 mRNA (16, 17). Central to this response are eIF2α kinases, which are activated by a variety of stimuli and form part of an integrated stress response. The ER-resident protein PERK is the principle Ser/Thr protein kinase that phosphorylates eIF2α during ER stress. To determine the role of PERK in the response of neuroectodermal tumor cells to fenretinide, bortezomib, or thapsigargin, PERK expression was abrogated using siRNA (Fig. 5A). Down-regulation of PERK resulted in a reduction in eIF2α phosphorylation in SH-SY5Y and A375 cells after drug treatment (Fig. 5C), demonstrating that PERK mediates eIF2α phosphorylation during ER stress. However, although PERK knockdown resulted in a small reduction in ATF4 protein induction, the levels of ATF4 mRNA after drug treatment were not affected (with the exception of fenretinide-induced ATF4, which was inhibited) (Fig. 5, B and C), suggesting that there are PERK-independent mechanisms of ATF4 regulation. Conversely, there was no inhibitory effect of PERK knockdown on ER stress-induced ATF3 expression (Fig. 5C), implying that the levels of ATF4 sustained during this time were sufficient to promote transcriptional activation of ATF3. However, PERK knockdown increased cell death in response to ER stress induction (two-way ANOVA, effect of siRNA, F1,25 = 7.72 (p = 0.01) for A375 cells and F1,26 = 6.22 (p = 0.019) for SH-SY5Y cells) (Fig. 5D).
FIGURE 5.
Fenretinide- and bortezomib-induced cell death is independent of PERK. A and B, SH-SY5Y and A375 cells were transfected with siRNAs for PERK or with a non-silencing control siRNA (ctrl) prior to treatment with fenretinide (FenR) (SH-SY5Y, 5 μm; and A375, 10 μm), bortezomib (Bort) (SH-SY5Y, 5 nm; and A375, 30 nm), or thapsigargin (Thap) (SH-SY5Y, 1.5 μm; and A375, 7.5 μm) for 6 h. PERK or ATF4 mRNA was measured by real-time PCR relative to β-actin as an internal control (A). eIF2α, phospho-eIF2α, ATF4, ATF3, and β-actin expression was determined by Western blotting (B). C, SH-SY5Y and A375 cells were transfected with siRNAs for PERK or with a non-silencing control siRNA prior to treatment with fenretinide (SH-SY5Y, 10 μm; and A375, 15 μm), bortezomib (SH-SY5Y, 5 nm; and A375, 50 nm), or thapsigargin (SH-SY5Y, 3 μm; and A375, 10 μm) for 24 h. D, apoptosis was measured by flow cytometry of propidium iodide-stained cells to determine the sub-G1 fraction. Data are expressed as the percentage total population; each point is the mean ± S.E. (n ≥ 3).
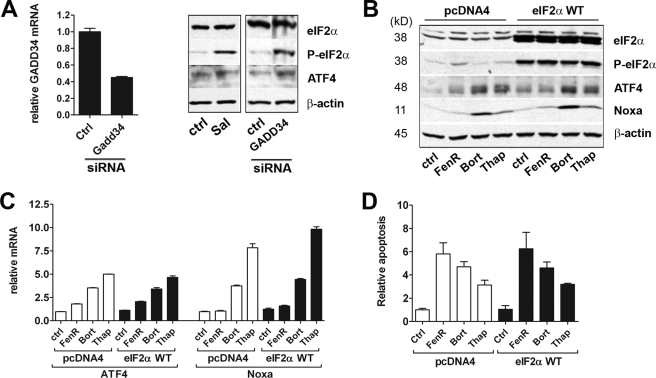
Enhancement of eIF2α phosphorylation through use of salubrinal (an inhibitor of GADD34-protein phosphatase 1 complex assembly), siRNA-mediated knockdown of GADD34, or overexpression of wild-type eIF2α in A375 cells resulted in increased basal levels of phosphorylated eIF2α and ATF4 protein (Fig. 6, A and B). Nevertheless, overexpression of wild-type eIF2α did not significantly affect fenretinide-, bortezomib-, or thapsigargin-induced ATF4 or Noxa induction at either the protein (Fig. 6B) or mRNA (Fig. 6C) level or in terms of the extent of observed cell death (two-way ANOVA on data relative to the control, effect of siRNA, F1,15 = 0.05, p > 0.8; siRNA by treatment interaction, F2,15 = 0.07, p > 0.9) (Fig. 6D). These data suggest that, although phosphorylation of eIF2α is able to enhance ATF4 protein expression, it is not the principle driving force behind ATF4 induction during ER stress in neuroectodermal tumor cells.
FIGURE 6.
Role of eIF2α in fenretinide- and bortezomib-induced cell death. A, A375 cells were treated with salubrinal (Sal; 20 μm) for 18 h or transfected with siRNAs for GADD34 or with a non-silencing control siRNA (ctrl). Proteins were blotted and probed as described below; GADD34 mRNA knockdown was confirmed by real-time PCR. B and C, A375 cells were transfected with a wild-type eIF2α expression vector (eIF2α WT) or control vector pcDNA4 prior to treatment with fenretinide (FenR; 10 μm), bortezomib (Bort; 30 nm), or thapsigargin (Thap; 7.5 μm) for 6 h. eIF2α, phospho-eIF2α, ATF4, Noxa, and β-actin expression was determined by Western blotting (A and B). GADD34, ATF4, or Noxa mRNA was measured by real-time PCR relative to β-actin as an internal control (A and C). D, A375 cells were transfected with a wild-type eIF2α expression vector or control vector pcDNA4 prior to fenretinide (15 μm), bortezomib (50 nm), or thapsigargin (10 μm) treatment for 24 h. Apoptosis was measured by flow cytometry of propidium iodide-stained cells to determine the sub-G1 fraction. Data are expressed relative to control untreated cells; each point is the mean ± S.E. (n ≥ 3).
DISCUSSION
This study shows that the ER stress-induced transcription factor ATF4 is a key mediator of the cytotoxic response of neuroectodermal tumor cells to fenretinide or bortezomib. Characterization of eIF2α signaling revealed a similar stress response in terms of eIF2α phosphorylation and up-regulation of ATF3, ATF4, and GADD34 expression; however, the kinetics and magnitude of the response varied between drug treatment and cell line. The mechanism of ER stress induction in response to fenretinide or bortezomib is likely to be very different. Fenretinide induces oxidative stress via ceramide metabolism prior to ER stress (13), whereas bortezomib mediates its effects primarily through inhibition of the 26 S proteasome (32). At clinically achievable concentrations of fenretinide and bortezomib, used in this study, a superior response to bortezomib was observed in terms of the degree of cell death and stress signaling compared with fenretinide. This may be due, in part, to the ability of bortezomib to inhibit protein degradation and increase protein stability, thereby having a positive feedback effect. Furthermore, the UPR is also able to engage a cell survival mechanism, and we have demonstrated recently that inhibition of elements of the survival response can enhance both ER stress signaling and apoptosis in cells treated with fenretinide or bortezomib (2, 13). Taken together, these data suggest that ER stress is a major mechanism of fenretinide and bortezomib action. In contrast, thapsigargin was less effective at inducing cell death, consistent with previous data (1, 33). In general, thapsigargin induced a more rapid and pronounced phosphorylation of eIF2α, which may contribute to cell survival through inhibition of translation of pro-apoptotic factors such as Noxa and highlights the concept that the kinetics of ER stress signaling are important in the determination of cell fate.
The role of ATF4 as a protective factor during cellular stress is well documented (reviewed in Ref. 34); nevertheless, there are reports describing a pro-death role for ATF4 during certain conditions, e.g. during oxidative stress in neurons (35), proteasome inhibition of squamous cell carcinoma cells (14, 21), or ER stress in tumor cells including neuroblastoma and melanoma (20, 22). Our present study provides evidence that ATF4 functions in a pro-death manner in response to fenretinide, bortezomib, or thapsigargin treatment of neuroblastoma and melanoma cell lines. Originally described as a transcriptional repressor, ATF4 also functions as a transcriptional activator. Transcriptional activity and selectivity of ATF4 are tightly controlled by its ability to form heterodimers with multiple C/EBP bZIP or AP-1 family members, through interaction with general transcriptional machinery, and through post-translational modulation of protein stability (34). Although, in some cases, the pro-death function of ATF4 has been attributed to its regulation of GADD153, the data presented in this study suggest that GADD153 is dispensable for cell death in response to ER stress in neuroectodermal tumor cells.
The mechanisms of ATF4-mediated cell death are likely to be complex and context-dependent; however, we and others (21, 24) have shown that ATF4 can regulate expression of the BH3-only protein Noxa in tumor cells. BH3-only proteins are able to initiate apoptosis through inhibition of pro-survival proteins and activation of pro-apoptotic Bax or Bak (36). Recent studies have highlighted an essential role for Noxa in fenretinide-, bortezomib-, and thapsigargin-induced cytotoxicity in neuroectodermal tumors (1, 37). Furthermore, ATF4 can regulate Noxa transcription directly during treatment of cancer cells with bortezomib or an inhibitor of ER-associated protein degradation (24), consistent with our observations that endogenous ATF4 can associate with the Noxa promoter. The involvement of ATF4 in the regulation of Noxa in response to bortezomib is in contrast to a previous report in which the oncogene c-myc was identified as a direct modulator of Noxa mRNA and essential for the regulation of Noxa by bortezomib (28). However, in our hands, c-Myc did not appear to mediate Noxa induction in response to ER stress-inducing agents; therefore, a relationship between c-Myc and ATF4 as a generic component of the ER stress responses in neuroectodermal tumors seems unlikely.
Multiple intracellular stress pathways converge on a single event: phosphorylation of eIF2α and subsequent translational activation of ATF4. This study demonstrates that, although phosphorylation of eIF2α enhances ATF4 protein levels, it is the (PERK-independent) transcriptional up-regulation of ATF4 mRNA that is required for efficient ATF4 protein expression in response to fenretinide, bortezomib, or thapsigargin treatment in neuroectodermal tumor cells. In support of this, PERK knockdown was not sufficient to appreciably inhibit ATF4 induction or to prevent ATF4-mediated ATF3 expression and cell death during ER stress. In contrast, increased cell death occurred during PERK knockdown in response to ER stress, consistent with recent observations that PERK-dependent translation of members of the IAP (inhibitor of apoptosis) family promotes cell survival during ER stress (38).
Although the data presented provide strong evidence for transcriptional regulation of ATF4 during ER stress, the mechanisms of transcriptional regulation are not well understood. However, the stress-regulated protein p8 was identified previously as a regulator of ATF4 transcription during ER stress (20, 31). p8 belongs to a family of high mobility group I/Y transcription factors (39) and is thought to play a role in tumor development (40). Conversely, p8 acts as a negative regulator of ATF4 mRNA in response to fenretinide, bortezomib, or thapsigargin treatment in A375 cells, suggesting that its function is context-dependent. Additional studies are therefore required to elucidate the mechanism of ATF4 activation in response to ER stress-inducing drugs. Moreover, we have shown recently that combined treatment with fenretinide and bortezomib results in a synergistic response in terms of both induction of GADD153 and cell death (3), suggesting that a combination of these agents may be a more effective strategy. Given the complexity of events triggered during ER stress, it would seem unlikely that ATF4 acts as the sole mediator of cytotoxic effects, and alternative pathways of ER stress signaling specific to fenretinide or bortezomib may be responsible for the synergism between these drugs. Studies on cells selected for acquired resistance to fenretinide, bortezomib, or thapsigargin are one approach that may address this question.
In summary, the clinically viable agents fenretinide and bortezomib induce an ER stress response in neuroectodermal tumor cells, culminating in the activation of ATF4 and cell death. Although precise characterization of the regulation of ATF4 is required to elucidate fully its functional role in a particular context, agents that activate ATF4 transcription may regulate a novel ER-related stress pathway that can be exploited for cancer therapy. The concept of promoting ER stress as a therapeutic strategy is attractive as cancer cells appear more susceptible to increased levels of this stress than their normal counterparts (41). In this respect, understanding the complex regulation of cellular programs leading to cancer cell death will allow the identification of novel molecular targets and facilitate the design of therapeutic strategies to specifically target tumor cells.
Supplementary Material
Acknowledgment
We thank David Ron for the wild-type eIF2α plasmid.
This work was supported by Cancer Research UK, the Newcastle Healthcare Charity, and the British Skin Foundation.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–5.
- ER
- endoplasmic reticulum
- UPR
- unfolded protein response
- eIF2α
- eukaryotic initiation factor 2α
- siRNA
- small interfering RNA
- ANOVA
- analysis of variance.
REFERENCES
- 1.Armstrong J. L., Veal G. J., Redfern C. P., Lovat P. E. (2007) Apoptosis 12, 613–622 [DOI] [PubMed] [Google Scholar]
- 2.Lovat P. E., Corazzari M., Armstrong J. L., Martin S., Pagliarini V., Hill D., Brown A. M., Piacentini M., Birch-Machin M. A., Redfern C. P. (2008) Cancer Res. 68, 5363–5369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hill D. S., Martin S., Armstrong J. L., Flockhart R., Tonison J. J., Simpson D. G., Birch-Machin M. A., Redfern C. P., Lovat P. E. (2009) Clin. Cancer Res. 15, 1192–1198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brignole C., Marimpietri D., Pastorino F., Nico B., Di Paolo D., Cioni M., Piccardi F., Cilli M., Pezzolo A., Corrias M. V., Pistoia V., Ribatti D., Pagnan G., Ponzoni M. (2006) J. Natl. Cancer Inst. 98, 1142–1157 [DOI] [PubMed] [Google Scholar]
- 5.Qin J. Z., Xin H., Sitailo L. A., Denning M. F., Nickoloff B. J. (2006) Cancer Res. 66, 9636–9645 [DOI] [PubMed] [Google Scholar]
- 6.Raffaghello L., Pagnan G., Pastorino F., Cosimo E., Brignole C., Marimpietri D., Montaldo P. G., Gambini C., Allen T. M., Bogenmann E., Ponzoni M. (2003) Int. J. Cancer 104, 559–567 [DOI] [PubMed] [Google Scholar]
- 7.Ron D., Walter P. (2007) Nat. Rev. Mol. Cell Biol. 8, 519–529 [DOI] [PubMed] [Google Scholar]
- 8.Xu C., Bailly-Maitre B., Reed J. C. (2005) J. Clin. Invest. 115, 2656–2664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heath-Engel H. M., Chang N. C., Shore G. C. (2008) Oncogene 27, 6419–6433 [DOI] [PubMed] [Google Scholar]
- 10.Moenner M., Pluquet O., Bouchecareilh M., Chevet E. (2007) Cancer Res. 67, 10631–10634 [DOI] [PubMed] [Google Scholar]
- 11.Virrey J. J., Dong D., Stiles C., Patterson J. B., Pen L., Ni M., Schönthal A. H., Chen T. C., Hofman F. M., Lee A. S. (2008) Mol. Cancer Res. 6, 1268–1275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Linder S., Shoshan M. C. (2005) Drug Resist. Updates 8, 199–204 [DOI] [PubMed] [Google Scholar]
- 13.Corazzari M., Lovat P. E., Armstrong J. L., Fimia G. M., Hill D. S., Birch-Machin M., Redfern C. P., Piacentini M. (2007) Br. J. Cancer 96, 1062–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jiang H. Y., Wek R. C. (2005) J. Biol. Chem. 280, 14189–14202 [DOI] [PubMed] [Google Scholar]
- 15.Wek R. C., Jiang H. Y., Anthony T. G. (2006) Biochem. Soc. Trans. 34, 7–11 [DOI] [PubMed] [Google Scholar]
- 16.Lu P. D., Harding H. P., Ron D. (2004) J. Cell Biol. 167, 27–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vattem K. M., Wek R. C. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 11269–11274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jiang H. Y., Wek S. A., McGrath B. C., Lu D., Hai T., Harding H. P., Wang X., Ron D., Cavener D. R., Wek R. C. (2004) Mol. Cell. Biol. 24, 1365–1377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brush M. H., Weiser D. C., Shenolikar S. (2003) Mol. Cell. Biol. 23, 1292–1303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Carracedo A., Lorente M., Egia A., Blázquez C., García S., Giroux V., Malicet C., Villuendas R., Gironella M., González-Feria L., Piris M. A., Iovanna J. L., Guzmán M., Velasco G. (2006) Cancer Cell 9, 301–312 [DOI] [PubMed] [Google Scholar]
- 21.Fribley A. M., Evenchik B., Zeng Q., Park B. K., Guan J. Y., Zhang H., Hale T. J., Soengas M. S., Kaufman R. J., Wang C. Y. (2006) J. Biol. Chem. 281, 31440–31447 [DOI] [PubMed] [Google Scholar]
- 22.Ohoka N., Yoshii S., Hattori T., Onozaki K., Hayashi H. (2005) EMBO J. 24, 1243–1255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zu K., Bihani T., Lin A., Park Y. M., Mori K., Ip C. (2006) Oncogene 25, 546–554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang Q., Mora-Jensen H., Weniger M. A., Perez-Galan P., Wolford C., Hai T., Ron D., Chen W., Trenkle W., Wiestner A., Ye Y. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 2200–2205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lovat P. E., Ranalli M., Annichiarrico-Petruzzelli M., Bernassola F., Piacentini M., Malcolm A. J., Pearson A. D., Melino G., Redfern C. P. (2000) Exp. Cell Res. 260, 50–60 [DOI] [PubMed] [Google Scholar]
- 26.Qin J. Z., Ziffra J., Stennett L., Bodner B., Bonish B. K., Chaturvedi V., Bennett F., Pollock P. M., Trent J. M., Hendrix M. J., Rizzo P., Miele L., Nickoloff B. J. (2005) Cancer Res. 65, 6282–6293 [DOI] [PubMed] [Google Scholar]
- 27.Ma Y., Hendershot L. M. (2003) J. Biol. Chem. 278, 34864–34873 [DOI] [PubMed] [Google Scholar]
- 28.Nikiforov M. A., Riblett M., Tang W. H., Gratchouck V., Zhuang D., Fernandez Y., Verhaegen M., Varambally S., Chinnaiyan A. M., Jakubowiak A. J., Soengas M. S. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 19488–19493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim R., Emi M., Tanabe K., Murakami S. (2006) Apoptosis 11, 5–13 [DOI] [PubMed] [Google Scholar]
- 30.Lin J. H., Li H., Yasumura D., Cohen H. R., Zhang C., Panning B., Shokat K. M., Lavail M. M., Walter P. (2007) Science 318, 944–949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Carracedo A., Gironella M., Lorente M., Garcia S., Guzmán M., Velasco G., Iovanna J. L. (2006) Cancer Res. 66, 6748–6755 [DOI] [PubMed] [Google Scholar]
- 32.Adams J., Palombella V. J., Sausville E. A., Johnson J., Destree A., Lazarus D. D., Maas J., Pien C. S., Prakash S., Elliott P. J. (1999) Cancer Res. 59, 2615–2622 [PubMed] [Google Scholar]
- 33.Jiang C. C., Chen L. H., Gillespie S., Wang Y. F., Kiejda K. A., Zhang X. D., Hersey P. (2007) Cancer Res. 67, 9750–9761 [DOI] [PubMed] [Google Scholar]
- 34.Ameri K., Harris A. L. (2008) Int. J. Biochem. Cell Biol. 40, 14–21 [DOI] [PubMed] [Google Scholar]
- 35.Lange P. S., Chavez J. C., Pinto J. T., Coppola G., Sun C. W., Townes T. M., Geschwind D. H., Ratan R. R. (2008) J. Exp. Med. 205, 1227–1242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Youle R. J., Strasser A. (2008) Nat. Rev. Mol. Cell Biol. 9, 47–59 [DOI] [PubMed] [Google Scholar]
- 37.Fernández Y., Verhaegen M., Miller T. P., Rush J. L., Steiner P., Opipari A. W., Jr., Lowe S. W., Soengas M. S. (2005) Cancer Res. 65, 6294–6304 [DOI] [PubMed] [Google Scholar]
- 38.Hamanaka R. B., Bobrovnikova-Marjon E., Ji X., Liebhaber S. A., Diehl J. A. (2009) Oncogene 28, 910–920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Encinar J. A., Mallo G. V., Mizyrycki C., Giono L., Gonzalez-Ros J. M., Rico M., Cánepa E., Moreno S., Neira J. L., Iovanna J. L. (2001) J. Biol. Chem. 276, 2742–2751 [DOI] [PubMed] [Google Scholar]
- 40.Iovanna J. L. (2002) Int. J. Gastrointest. Cancer 31, 89–98 [DOI] [PubMed] [Google Scholar]
- 41.Schönthal A. H. (2009) Cancer Lett. 275, 163–169 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.