Abstract
Cholera toxin travels from the plasma membrane to the endoplasmic reticulum of host cells, where a portion of the toxin, the A1-chain, is unfolded and targeted to a protein-conducting channel for retrotranslocation to the cytosol. Unlike most retrotranslocation substrates, the A1-chain escapes degradation by the proteasome and refolds in the cytosol to induce disease. How this occurs remains poorly understood. Here, we show that an unstructured peptide appended to the N terminus of the A1-chain renders the toxin functionally inactive. Cleavage of the peptide extension prior to cell entry rescues toxin half-life and function. The loss of toxicity is explained by rapid degradation by the proteasome after retrotranslocation to the cytosol. Degradation of the mutant toxin does not follow the N-end rule but depends on the two Lys residues at positions 4 and 17 of the native A1-chain, consistent with polyubiquitination at these sites. Thus, retrotranslocation and refolding of the wild-type A1-chain must proceed in a way that protects these Lys residues from attack by E3 ligases.
Keywords: Proteases/Ubiquitination, Protein/Degradation, Protein/Translocation, Subcellular Organelles/Endoplasmic Reticulum, Toxins, Cholera Toxin, Proteasome
Introduction
Like most toxins, cholera toxin (CT)3 must cross a cell membrane to enter the cytosol of host cells and induce disease. To do this, CT traffics retrograde from the plasma membrane to the endoplasmic reticulum (ER), where it co-opts the machinery for degradation of terminally misfolded proteins in the ER (1). Unlike the misfolded substrates, however, the toxin escapes rapid degradation by the proteasome and refolds to its native conformation so as to act enzymatically in the cytosol (2). This is evidenced most clearly by the lack of sensitivity to chemical inhibition of the proteasome (3). How the A1-chain avoids rapid degradation to induce toxicity, however, is not fully understood. The prevailing view is that the paucity of lysines explains the resistance to ubiquitination and proteasomal degradation (4). Another view is that cytosolic chaperones stabilize the fold of the A1-chain so as to prevent non-ubiquitin-dependent degradation by the 20S proteasome, but degradation of the A1-chain in vivo is slow, occurring after the induction of toxicity (5).
CT typifies the AB5 family of toxins and is the virulence factor responsible for the massive secretory diarrhea seen in Asiatic cholera. The A-subunit is comprised of an enzymatically active A1-chain linked non-covalently to the B-subunit via the A2-chain. The A1- and A2-chains are joined by a flexible loop containing a serine-protease cleavage site bridged by a disulfide bond. Both bonds must be broken before the A1-chain can enter the cytosol of host cells (2). The B subunit consists of five 11.5-kDa peptides assembled non-covalently into a stable homopentamer that binds to the ganglioside GM1 on the plasma membrane. The B-subunit-GM1 complex carries the A-subunit into the ER (6).
In the ER, protein disulfide isomerase (PDI) in its reduced state unfolds and dissociates the A1-chain from the B-subunit. When oxidized, PDI releases the A1-chain to subsequent steps in the retrotranslocation reaction (7). Presumably the A1-chain crosses the ER membrane in an unfolded conformation via transport through a protein-conducting channel, possibly involving the E3 ligase Hrd1, Derlin-1, and Sec61 (8–11). Unlike most other ER-associated degradation (ERAD) substrates, the A1-chain does not require ubiquitination (3) or interaction with the AAA-ATPase P97 for retrotranslocation (12), and the driving force for the retrotranslocation reaction remains unknown. It is possible that rapid and partial refolding of the A1-chain immediately upon transit across the ER membrane might actually drive retrotranslocation by creating the conditions for a molecular ratchet (3). Once in the cytosol, the A1-chain refolds to its native conformation and enzymatically activates adenylyl cyclase. This causes the massive intestinal chloride and water secretion that is the hallmark of cholera.
In an approach to model retrotranslocation of the A1-chain in vitro, we prepared two mutant toxins with essentially the same N- and C-terminal peptide extensions of the A1-chain that would allow for development of a trans-membrane transport assay by protease protection. When tested in vivo, however, we found the toxin with the N-terminally extended A1-chain to be nearly non-functional. Here, we describe how the peptide extension renders the mutant toxin inactive. We find that the addition of an N-terminal peptide allows retrotranslocation but acts as a degron by rendering the two amines present in the native A1-chain (Lys-4 and Lys-17) substrates for ubiquitination. Degradation of the mutant A1-chain is rapid, causing the loss of toxicity, and it is blocked by proteasome inhibitors or by mutation of the Lys residues to Arg. Thus, Lys-4 and Lys-17 of the native A1-chain are capable of being ubiquitinated, implying that retrotranslocation of the wild-type (WT) toxin must proceed in a way that protects these residues from attack by E3 ligases. This allows for escape from degradation by the proteasome and is required for induction of a physiologic response.
EXPERIMENTAL PROCEDURES
Materials
Carboxybenzyl-Leu-Leu-Leu-vinyl sulfone (ZL3VS) was a gift from Dr. H. Ploegh (Whitehead Institute for Biomedical Research). Antibodies were purchased against hemagglutinin (HA) (Roche Applied Science) and BiP (Santa Cruz Biotechnology; Santa Cruz, CA). Antibodies against CT were described previously (13). AcTev protease was from Invitrogen. CT used in the A2-chain experiment was from Calbiochem. Figures were made using BioDraw (Cambridgesoft; Cambridge, MA).
Plasmids
Sequences encoding WT CT, HA-CT, and CT-HA were cloned into pLMP3 and transferred into the arabinose-inducible pARCT plasmid. The non-cleavable mutant, R192G, was previously described (1). The G33D mutant was a gift from Dr. R. Holmes. The E112D, K4R, and K17R mutations were introduced into WT CT and HA-CT within pLMP3 as described previously (3) and transferred into the pARCT.
Protein Carbamylation and Electrophysiology
performed as described previously (3, 14). Where indicated, intestinal T84 cells were pretreated with 5 μm brefeldin-A (Sigma-Aldrich), 10 μm ZL3VS in dimethyl sulfoxide (DMSO), 10 μm lactacystin (Sigma-Aldrich), or vehicle alone.
Recombinant Toxins and GM1 Beads
Toxins were expressed in Escherichia coli BL21 cells. Cells were grown at 30 °C in 25 μg/ml chloramphenicol to an A600 of 0.6–0.8. Expression was induced with 0.5% arabinose for 3 h. Bacteria were harvested and resuspended in equilibration buffer (50 mm Na2HPO4, 300 mm NaCl, pH 7). Polymyxin B (Sigma-Aldrich) was added (0.5 mg/ml final concentration), incubated for 1 h at room temperature, and the lysate was centrifuged at 10,000 × g (20 min, 4 °C). Imidazole (10 mm) was added to the supernatant and incubated with prewashed TALON® polyhistidine tag purification resin (Clontech) for 1 h at 4 °C. The resin was transferred to a column, washed with equilibration buffer containing 10 mm imidazole, and eluted with equilibration buffer containing 300 mm imidazole. CT was dialyzed into phosphate-buffered saline. Toxin concentration was determined by UV absorbance at λ = 280 using the theoretical molar extinction coefficients for each toxin (15) and comparison on SDS-PAGE with Coomassie Blue. Dynabeads® M-270 carboxylic acid (Invitrogen) were coupled to 1 mg of lyso-monosialoganglioside GM1 (Matreya; Pleasant Gap, PA) according to the manufacturer's instructions.
Retrotranslocation Assay
Retrotranslocation experiments were modified from those previously described (9). Briefly, Vero cells in six-well plates (1.5 × 106 cells/well) were incubated with toxin (10 or 40 nm) in Hanks' balanced salts (modified, without phenol red and sodium bicarbonate (Sigma-Aldrich), 10 mm HEPES pH 7.4) for 90 min at 37 °C. Where indicated, cells were incubated for 4 h in 20 μm ZL3VS, 2 h in 60 μm brefeldin-A, or 1.5 h (or 2.5 h) in 10 μm lactacystin or vehicle control; toxin was present for the last 90 min. Cells were detached with 0.25% trypsin with EDTA, washed, and resuspended in 100 μl of HN buffer (50 mm HEPES pH 7.5, 150 mm NaCl, 0.5 mg/ml EDTA, Complete protease inhibitor cocktail tablet (Roche Applied Science)), incubated on ice for 10 min, and centrifuged at 80,000 rpm (10 min at 4 °C). The supernatant was collected, and the membrane pellet was resuspended in 100 μl of HN buffer. Reducing sample buffer was added, and 25–50% of the cytosol was analyzed by SDS-PAGE and immunoblot.
Release Assay
This assay was modified from that previously described (1). Toxin was biotinylated where indicated, nicked with trypsin, and incubated with 10 μl of GM1 beads (prewashed in binding buffer (50 mm Hepes, 50 mm KOAc, 250 mm sucrose, 2 mm MgCl2)). Beads were washed five times with 100 μl of binding buffer. To dry beads, the following was added to a final volume of 30 μl and incubated for 1 h at 30 °C: 50 eq of cow liver ER lumenal extract (17), 10 mm reduced GSH, 50 mm Hepes, 250 mm sucrose, 50 mm KOAc, 2 mm MgCl2. Mock extract consisted only of the buffer used to prepare the ER lumenal extract (250 mm sucrose, 50 mm triethanolamine, 1 mm dithiothreitol). Beads were collected twice to ensure the purity of the released supernatant fraction. The pellet was resuspended in 30 μl of binding buffer; non-reducing sample buffer was added to both the pellet and the supernatant. 58% of the supernatant and 8.3% of the pellet were loaded on an SDS-PAGE gel and analyzed by immunoblot.
Unfolding Assay
This assay was modified from that previously described (3). It was performed in the same manner as the release assay; however, 3.5 μm PDI (Sigma-Aldrich) was used, and the untreated sample contained phosphate-buffered saline. 30 mm GSSG, or binding buffer, was added to the released A1-chain in the supernatant fraction to allow refolding at 30 °C for the indicated times. 0.1 mg/ml trypsin or phosphate-buffered saline was then added for 1 h on ice, and the reaction was stopped by the addition of 1 mm L1-chlor-3-(4-tosylamido)-7-amino-2-heptanon-hydrochloride (TLCK) for 10 min on ice. Non-reducing sample buffer was added, and 24% of the supernatant was loaded on an SDS-PAGE gel and analyzed by immunoblot.
RESULTS
Structure and Function of the Toxin Mutants
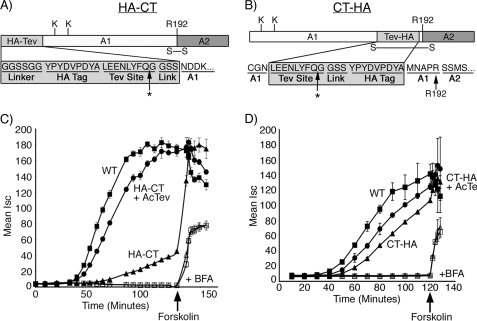
Two mutant toxins were prepared containing the HA epitope linked to the N or C terminus of the A1-chain by a tobacco etch virus protease cleavage site (TEV) and flanked at both ends by Gly and Ser residues (Fig. 1, A and B, named HA-CT and CT-HA, respectively, and see supplemental Fig. 1). Toxin function was examined using intestinal T84 cells grown in monolayer culture, which respond to CT by producing a cAMP-dependent Cl− secretory response. This is measured in real time as a transepithelial short-circuit current (Isc).
FIGURE 1.
Functional analysis of HA-CT and CT-HA. A and B, schematics of the N- and C-terminal peptide extensions are shown. Arg-192 (R192) is located at the cleavage site separating the A1- and A2-chains and bridged by a disulfide bond. Lys-4 and Lys-17 are indicated. * indicates the cleavage site for AcTev protease. C, time course of CT-induced Cl− secretion (Isc) induced by WT CT (filled square), HA-CT (filled triangle), or HA-CT pretreated with AcTev protease to remove the HA tag (filled circle). Some monolayers were pretreated with brefeldin A (indicated by + BFA) followed by application of WT CT (open square) or CT-HA pretreated with AcTev protease to remove the HA tag (open triangle). The viability of monolayers was demonstrated by applying the cAMP agonist, forskolin. D, exactly as for panel C but using the mutant toxin CT-HA. Error bars in C and D indicate S.D.
The secretory response induced by the HA-CT mutant toxin was strongly attenuated when compared with WT CT (Fig. 1C, compare filled triangles with squares). Cleavage of the HA peptide by TEV protease restored toxicity to near wild-type levels (Fig. 1C, circles). The Isc induced by the analogous C-terminal mutant, CT-HA, was only marginally attenuated (Fig. 1D, compare triangles with squares). In all cases, treatment of T84 cells with brefeldin A completely inhibited the Cl− secretory response, indicating that both mutant toxins required retrograde vesicular transport from plasma membrane to ER to induce toxicity. Application of the cAMP agonist forskolin at the end of selected experiments (Fig. 1, C and D, arrow) rapidly induced an increase in Isc, showing that the cells were intact and fully capable of responding to intoxication. Thus, the peptide appended to the N terminus of the A1-chain strongly and reversibly inhibits toxin function, but the analogous peptide appended to the C terminus does not.
Retrotranslocation of the A1-chain in Intact Cells
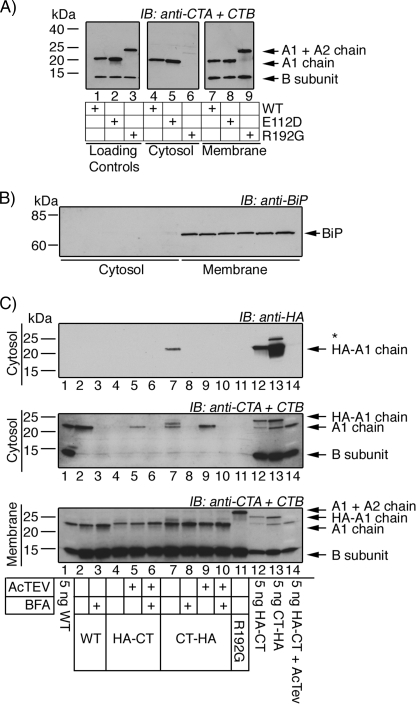
To understand why the N-terminal mutant lacked activity, we modified an assay recently reported to measure retrotranslocation of the A1-chain from ER to cytosol in intact cells (9). The assay isolates cytosol from cell membranes and subcellular organelles by centrifugation. Retrotranslocation is measured by the presence of the A1-chain in the cytosolic fraction by SDS-PAGE and immunoblot using antibodies against the CT A- and B-subunits or the HA tag. After 90-min incubations, the A1-chain of WT CT was found in the cytosol of host cells (African green monkey kidney Vero cells), but the 11.5-kDa monomers of the B-subunit were absent (Fig. 2A, lane 4). The membrane pellet, however, contained both the A-subunits and the B-subunits; this shows specificity for retrotranslocation of only the A1-chain and not the B-subunit or intact A-subunit. We confirmed that the A1-chain is the only retrotranslocation substrate in separate experiments using WT CT by testing for the ability of PDI to dissociate the A1- and A2-chains from the B-subunit in vitro. The results show that only the A1-chain is released by PDI and that the A2-chain remains with the B-subunit (supplemental Fig. 2).
FIGURE 2.
N-terminally tagged A1-chain does not appear to retrotranslocate from ER to cytosol. Retrotranslocation is measured in Vero cells by isolating cytosol from cell membranes and subcellular organelles via centrifugation. The presence of the A1-chain in the cytosolic fraction is analyzed by SDS-PAGE and immunoblot (IB) using antibodies recognizing the A-subunit (CTA) and B-subunit (CTB) or HA tag. A, retrotranslocation results are shown for WT CT (lane 4), the enzymatically inactive E112D mutant (lane 5), and the non-cleavable CT mutant R192G (lane 6). Lanes 1–3 are controls, and lanes 7–9 are the membrane fraction. B, SDS-PAGE and immunoblot of cytosolic and membrane fractions of the retrotranslocation assays using an antibody recognizing ER lumenal marker BiP. C, retrotranslocation assay using WT CT, HA-CT, and CT-HA as indicated. Lanes 1 and 12–14 are controls. * indicates an unidentified band. BFA, brefeldin A.
Molecular controls further verify the retrotranslocation assay. The A1-chain of the mutant toxin containing Gly for Arg at position 192 (R192G), which prevents cleavage of the A-subunit and thus release of the A1-chain from the B-subunit (18, 19), is not seen in the cytosolic fraction (Fig. 2A, lane 6). The A1-chain of another mutant holotoxin G33D, which is deficient in binding GM1 and cannot traffic into the ER of host cells (20), is also absent from the cytosol (data not shown). Finally, a toxin mutated within its enzymatic active site, E112D, but otherwise capable of ER transport is seen in the cytosol as predicted (Fig. 2A, lane 5). In all cases, the B-subunit monomer is found only in the membrane fraction (except for the G33D mutant, which cannot bind GM1), and the ER lumenal chaperone BiP is found only in the membrane fraction, indicating that lumenal ER proteins are not released (Fig. 2B). Thus, the assay is robust and specific for measuring retrotranslocation of the A1-chain (Fig. 2A, compare middle and right panels).
When tested in this assay, the A1-chain of the C-terminally extended mutant toxin CT-HA was found in the cytosol of host cells, but the A1-chain of the N-terminally extended HA-CT was not (Fig. 2C, middle and top panels, compare lane 7 with lane 4). Cleavage of the peptide extensions by treatment with TEV protease rescued the retrotranslocation reaction for the HA-CT mutant (Fig. 2C, middle panel, compare lanes 4 and 5), consistent with rescue of toxin function by TEV protease (Fig. 1C). Immunoblots using antibodies against the HA epitope are negative in these rescue experiments because the tag has been cleaved. Recombinant WT CT was used as a positive control (Fig. 2C, middle panel, lane 2). In all cases, the B-subunit monomers were absent from the cytosolic fractions, providing an internal control showing specificity for retrotranslocation of the A1-chain. Also, treatment of host cells with brefeldin A completely inhibited the retrotranslocation reaction as expected for blockade of toxin entry into the ER (Fig. 2C, top and middle panels, lanes 3, 6, 8, and 10). Thus, the A1-chain of the N-terminally extended mutant HA-CT was either blocked in the retrotranslocation reaction or rapidly degraded after arrival in the cytosol.
Inhibition of the Proteasome Rescues Function of the HA-CT Mutant
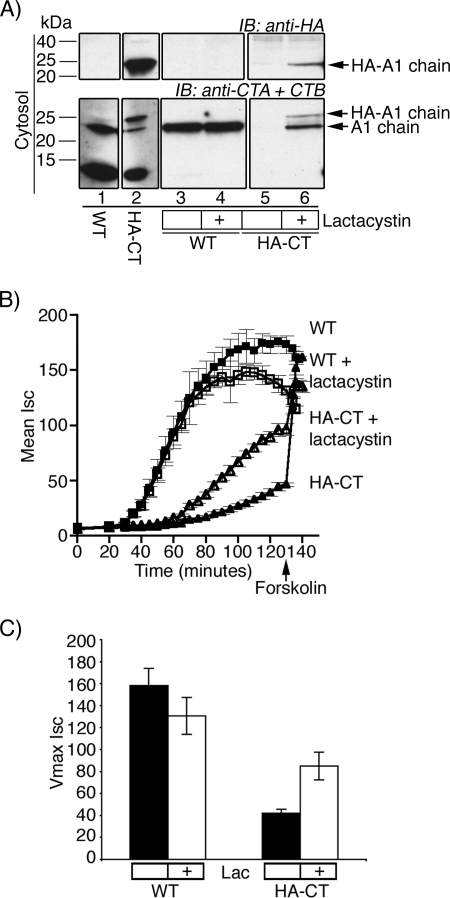
To test for rapid degradation of the A1-chain in the cytosol, we pretreated host cells with the proteasome inhibitor lactacystin or vehicle control alone and assayed for retrotranslocation. Lactacystin blocked proteasome function in Vero cells, as assayed by SDS-PAGE of total cell lysates and immunoblot for polyubiquitinated proteins (data not shown). As predicted by our previous studies (3), lactacystin had no effect on the appearance of the WT A1-chain in the cytosol after retrotranslocation (Fig. 3A, lower panel, compare lanes 3 and 4). For the N-terminally extended HA-CT mutant, however, the inhibitor rescued the appearance of the A1-chain in the cytosol, where no A1-chain was present in the absence of lactacystin (Fig. 3A, compare lanes 5 and 6). The same results were obtained using the proteasome inhibitor ZL3VS (data not shown). Thus, unlike the WT A1-chain, the N-terminal mutant A1-chain is a substrate for proteasomal degradation after arrival in the cytosol.
FIGURE 3.
HA-CT A1-chain is rapidly degraded by the proteasome. A, cells were pretreated with lactacystin or vehicle alone before performing retrotranslocation experiments. Cytosolic fractions were analyzed by SDS-PAGE and immunoblotted (IB) using antibodies recognizing the A-subunit (CTA) and B-subunit (CTB) or HA tag. Lanes 1 and 2 are controls. B, in a time course of CT-induced Cl− secretion (Isc), T84 cells were incubated with WT CT (filled square) or HA-CT (filled triangle) or pretreated with lactacystin followed by WT CT (open square) or HA-CT (open triangle). The viability of monolayers was demonstrated by applying the cAMP agonist, forskolin. C, summarized data from two independent experiments, each done in triplicate, of maximal Isc for untreated or lactacystin (Lac) pretreated T84 cells is shown. Error bars in C and D indicate S.D.
These results were confirmed for the N-terminally extended HA-CT mutant using functional studies for toxin-induced Cl− secretion (Isc). Treatment of T84 cell monolayers with lactacystin (or ZL3VS, data not shown) had minimal or no effect on the time course of Isc induced by WT CT, consistent with our previous studies (3) (Fig. 3B, compare filled and open squares). In contrast, lactacystin partially rescued the Isc induced by HA-CT (Fig. 3B, compare filled and open triangles). The same result was found for ZL3VS (data not shown). Treatment of these monolayers with forskolin at the end of the time course (arrows) induced a further increase in Isc, showing that the cells were physiologically fully intact. The effect of lactacystin on maximal Isc for six independent studies is summarized in Fig. 3C. These results show that the mutant A1-chain of HA-CT undergoes retrotranslocation but is rapidly degraded by the proteasome after arrival in the cytosol. Rescue of toxin function by the proteasome inhibitors, however, was not complete, so the N-terminally extended mutant A1-chains might also have a defect in retrotranslocation or in enzymatic activity after reaching the cytosol, or both.
HA-CT Contains an N-terminally Located Degron
The first step in degradation by the proteasome involves tethering of the substrate to the proteasome itself, usually mediated by an appended polyubiquitin chain (21). The A1-chains of both WT CT and HA-CT contain three potential sites for polyubiquitination, all present in the native toxin: the extreme N-terminal amine and the two lysines at positions 4 and 17. Thus, to test whether ubiquitination is involved, we substituted the two Lys residues with Arg and blocked the N-terminal α-amine by carbamylation. The Arg substitutions were made in both the WT CT (WT K4RK17R) and the N-terminally extended HA-CT mutant (HA-CT K4RK17R).
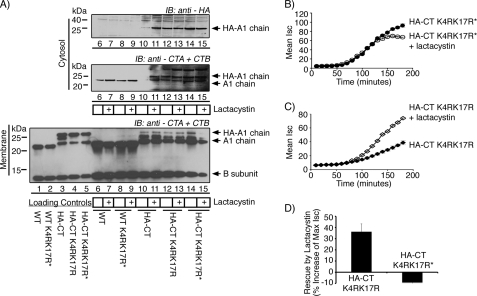
As before, the A1-chain of WT CT appears in the cytosol following incubation with host cells, and lactacystin has no measurable effect (Fig. 4A, middle panel, compare lanes 6 and 7). The A1-chain of the HA-CT mutant does not appear in the cytosol unless the cells are pretreated with lactacystin (Fig. 4A, top and middle panels, compare lanes 10 and 11). Substitution of the two Lys residues in the HA-CT mutant (HA-CT K4RK17R), however, rescues the presence of the A1-chain in the cytosolic fractions, independent of lactacystin treatment (Fig. 4A, middle and upper panels, lanes 12 and 13). The same results were obtained after carbamylation of the HA-CT K4RK17R mutant to block the N-terminal α-amine from ubiquitination (HA-CT K4RK17R*, Fig. 4A, middle and upper panels, lanes 14 and 15). In control studies, Arg substitutions and blockade of the N-terminal α-amine in the WT toxin (WT K4RK17R*) had no effect (Fig. 4A, middle panel, lanes 8 and 9), consistent with our previous experiments (3). Thus, the addition of a small peptide to the N terminus somehow renders Lys-4 and Lys-17 of the native toxin substrates for ubiquitination.
FIGURE 4.
Mutation of lysines in the native A1-chain rescues HA-CT from the proteasome. A, cells were pretreated with lactacystin or vehicle alone before retrotranslocation experiments. WT toxin (lanes 6 and 7) and carbamylated WT CT with Lys-4 and Lys-17 mutations (WT K4RK17R*) (lanes 8 and 9) are shown. HA-CT (lanes 10 and 11), HA-CT K4RK17R (lanes 12 and 13), or the carbamylated HA-CT K4RK17R mutant (HA-CT K4RK17R*) (lanes 14 and 15) are shown. Lanes 1–5 are controls. All samples were analyzed by SDS-PAGE and immunoblotted (IB) using antibodies recognizing the A-subunit (CTA) and B-subunit (CTB) or HA tag. B, in a time course of CT-induced Cl− secretion (Isc), T84 cells were either treated with HA-CT K4RK17R* (filled circle) or pretreated with lactacystin followed by HA-CT K4RK17R* (open circle). C, this was performed as in panel B, but HA-CT K4RK17R (filled diamond) or pretreated HA-CT K4RK17R (open diamond) was tested. D, summarized data from three independent experiments, each done in triplicate, of rescue by lactacystin is shown. The percentage of increase in maximal Isc for lactacystin-pretreated T84 cells versus untreated cells was plotted on the y axis for both HA-CT K4RK17R and HA-CT K4RK17R*. Error bars in indicate S.D.
We did not observe any evidence for ubiquitinated A1-chain in any of our immunoblots, and our attempt to demonstrate ubiquitination of the A1-chain using Vero cells expressing Myc-ubiquitin were not successful. These results typify most studies on ubiquitinated proteins degraded by the proteasome, even in the presence of proteasome inhibitors. Thus, we sought to confirm the role of Lys-4 and Lys-17 using the electrophysiology assay for toxin function in T84 cells. As predicted by our biochemical studies, we found that lactacystin had no further effect on the toxicity of the HA-CT K4RK17R mutant when carbamylated at the N-terminal α-amine (Fig. 4, B and D). The Isc induced by the HA-CT K4RK17R mutant that was not carbamylated at the N terminus, however, was enhanced in cells pretreated with lactacystin (Fig. 4, C and D). Thus, mutation of Lys-4 and Lys-17 does not fully rescue the HA-CT mutant from attack by the proteasome, implying that the α-amine of the N-terminal residue in the HA-CT K4RK17R mutant can also act as a site for ubiquitination.
HA-CT Refolds Poorly after Release from PDI in Vitro
How does the N-terminal peptide act as a degron? Certain E3 ligases recognize substrates that are unfolded or have unfolded domains (21). Because the peptide appended to the A1-chain is likely unstructured (as evidenced by protease degradation in vitro, data not shown), it is possible that the peptide extension itself might target the refolded A1-chain for ubiquitination after retrotranslocation. Alternatively, the peptide extension might block the normal refolding of the A1-chain as it emerges into the cytosol.
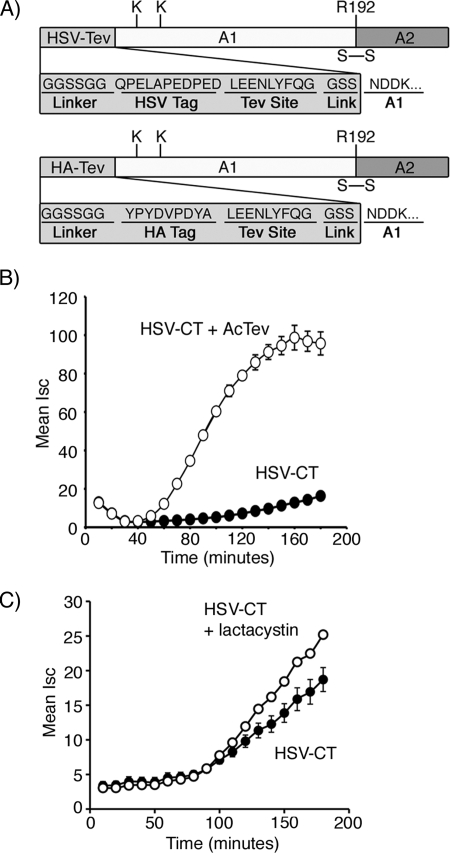
We first tested whether the amino acid sequence of the N-terminal extension is essential for activity by replacing the HA epitope with the epitope tag derived from Herpes simplex virus (Fig. 5A). Like HA-CT, this mutant toxin (HSV-CT) was strongly attenuated in function as determined by assay of toxin-induced Cl− secretion in T84 intestinal cells (Fig. 5B). Proteolytic cleavage of the HSV tag restored toxin activity, exactly as for our results with the HA-CT mutant. Lactacystin also partially rescued toxin function (Fig. 5C). Thus, the specific amino acid sequence of the N-terminal extension is dispensable for activity as a degron. Like the HA peptide, the HSV peptide is also predicted to be unstructured, so it is possible that this feature is essential.
FIGURE 5.
Development and functional analysis of HSV-CT. A, a schematic of the N-terminal peptide extensions, HA-CT and HSV-CT, is shown. Arg-192 (R192) is located at the cleavage site separating the A1- and A2-chains. A disulfide bond, in addition to other non-covalent interactions, links together the two chains. B, in a time course of CT-induced Cl− secretion (Isc), T84 cells were incubated with HSV-CT (filled circle) or HSV-CT pretreated with AcTev protease to remove the tag (HSV-CT, open circle). C, this was performed as in panel B, but cells were treated with HSV-CT (filled circle) or lactacystin-pretreated HSV-CT (open circle). Error bars in B and C indicate S.D.
To test whether refolding of the A1-chain was inhibited in the mutant HA-CT, we modified a protease protection assay previously used to measure refolding of the A1-chain after dissociation from PDI in vitro (3). Here, we incubated the toxins with GM1-coupled beads and PDI or isolated ER lumenal proteins. PDI binds, unfolds, and releases the A1-chain from the B-subunit-GM1 bead complex. This is measured by pelleting the beads and assaying the supernatant for the A1-chain by SDS-PAGE and immunoblot using antibodies against the A- and B-subunits (supplemental Fig. 3A). To assay for the A1-chain in its unfolded conformation, trypsin was added to the supernatant, which digests the A1-chain when bound to PDI and unfolded (supplemental Fig. 3B). The addition of excess oxidized glutathione (GSSG) releases the A1-chain from PDI, allowing it to refold and rendering it resistant to trypsin digestion (supplemental Fig. 3B).
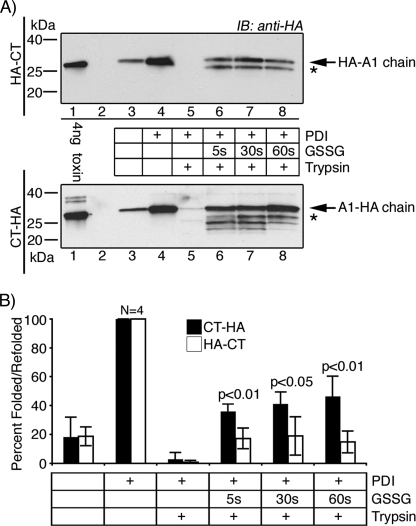
The fraction of A1-chain that refolded to a trypsin-resistant conformation after release from PDI was defined for HA-CT using the CT-HA mutant as a control because of the greater sensitivity in detection by immunoblot using anti-HA antibodies (Fig. 6A, upper versus lower panels, respectively). In the absence of trypsin, PDI released the A1-chain of both mutant toxins into the supernatant (Fig. 6A, compare lane 4 with lane 3). For unexplained reasons, a small amount of A1-chain was also released into the supernatant in the absence of PDI in these experiments. The addition of trypsin completely degraded the A1-chains of both HA-CT and CT-HA, showing that in the presence of reduced PDI, both A1-chains are in unfolded conformations when released from the B-subunit (Fig. 6A, lane 5). After the addition of GSSG, however, a greater fraction of A1-chain from the C-terminal mutant CT-HA was found to be resistant to trypsin when compared with the N-terminal mutant HA-CT. Data from four independent experiments at three time points are summarized in Fig. 6B. Thus, the N-terminal extension inhibits spontaneous refolding of the A1-chain after release from PDI in vitro. These results suggest that the N-terminal extension might act in vivo as a degron by inhibiting the refolding of the A1-chain as it emerges from the ER, thus rendering it a substrate for ubiquitination.
FIGURE 6.
HA-CT refolds poorly after release from PDI in vitro. Briefly, we incubated all toxins with GM1-coupled beads and PDI in reducing conditions. PDI binds, unfolds, and releases the A1-chain into the supernatant from the B-subunit-GM1 magnetic beads. The supernatants were left untreated or were treated with GSSG to induce release of the A1-chain from PDI, and refolding was measured as resistance to trypsin degradation. All samples were analyzed by SDS-PAGE under non-reducing conditions, except the 4-ng control, which was reduced to separate the A1-chain. A, the unfolding and refolding of HA-CT (upper panel) and CT-HA (lower panel) were analyzed by immunoblot (IB) using an antibody against the HA tag. * indicates partially refolded toxin fragments. B, summarized data from four independent experiments. Compare HA-CT (white bars) with CT-HA (black bars). Band intensities were acquired using GeneSnap (GeneGnome HR, Syngene) and normalized to the total fraction of A1-chain released by PDI in the absence of trypsin (lanes 4). Error bars indicate S.D.
DISCUSSION
Most ERAD substrates in mammalian cells and yeast are retrotranslocated to the cytosol and targeted to the proteasome by polyubiquitination (22). Ubiquitination, in fact, may contribute fundamentally to the retrotranslocation reaction by allowing for stable interaction between the ERAD substrate and the cytosolic AAA-ATPase P97 (22, 23). P97 provides the driving force for these reactions by pulling the ERAD substrate into the cytosol from the ER membrane (23). The toxins, however, are an exception. They do not require P97 and are presumed to escape ubiquitination and proteasomal degradation long enough to act enzymatically in the cytosol and induce disease. This is partially explained by the paucity of lysine residues in the enzymatic chains of the toxins (4). Here, we find that other factors must also be involved.
CT contains two Lys residues in the N-terminal domain of the A1-chain that are conserved in the closely related E. coli heat labile toxin type I (24). Both are dispensable for toxin function (3). In this study, we show that a short peptide extension to the N terminus of the A1-chain of CT inhibits toxin action by rendering these Lys substrates for ubiquitination, thus causing rapid degradation by the proteasome. In nature, however, retrotranslocation of the WT A1-chain does not result in rapid degradation. Thus, these same residues must somehow be protected against attack by E3 ligases, either during or very quickly after the A1-chain enters the cytosol. Our results show that this is required for induction of a physiologic response. Such escape from ubiquitination for WT CT is now of particular interest because recent studies implicate the E3 ligases Hrd1 and GP78 and an E2 ubiquitin-conjugating enzyme in the retrotranslocation reaction itself (10).
How Does the N Terminus of the Mutant A1-chain Act as a Degron?
The N-terminal residue of the HA mutant A1-chain is glycine, which is predicted to stabilize the peptide against degradation according to the N-end rule (25, 26). Also, cleavage of the peptide by TEV protease leaves the activated A1-chain with the same residue (Gly) at the N terminus, and this renders the mutant toxin fully active. So the N-end rule cannot explain our results. Rather, it is the presence of an additional peptide chain, located at the N terminus, which induces ubiquitination. The primary structure of the peptide extension is dispensable as another peptide of different amino acid sequence elicits the same effect. Both peptide extensions tested are predicted to be loosely structured (27, 28), so this feature might be an essential factor. Placement of the degron at the N terminus in close proximity to the two Lys residues (29) also appears to be required as the analogous peptide appended to the C terminus has little effect on toxicity. Furthermore, recent studies show that ubiquitination may not be sufficient to ensure degradation by the proteasome and that an unstructured domain is needed to initiate unfolding and physical entry into the proteasome channel (16, 29). The unstructured peptide extension of HA-CT might also act in this way.
What Protects Lys-4 and Lys-17 of the Native Toxin from Ubiquitination after Retrotranslocation to the Cytosol?
Because retrotranslocation of the A1-chain does not require P97 in intact cells (12), we previously speculated that rapid and partial refolding of the A1-chain immediately upon transit across the ER membrane might drive the retrotranslocation reaction by creating the conditions for a molecular ratchet (3). These ideas led us to test whether the N-terminal peptide might act as a degron by interfering with refolding of the A1-chain as it emerges from the ER into the cytosol, thus rendering the amines of the A1-chain substrates for attack by E3 ligases. Our in vitro results are consistent with this hypothesis. The peptide extension of the HA-CT mutant inhibits spontaneous refolding of the A1-chain after release from PDI.
Circular dichroism and fluorescence spectroscopy studies by Pande et al. (5), however, show that the A1-chain on its own is thermally unstable in vitro and may not refold spontaneously after retrotranslocation in vivo. This study also shows that binding of ADP-ribosylating factor 6 (ARF6), an enzymatic cofactor of CT, stabilizes the fold of the A1-chain, and the authors raise the important idea that cytosolic chaperones might be required for refolding of the A1-chain in the cytosol. Chaperone binding might then, directly or indirectly, protect the A1-chain from ubiquitination and rapid degradation immediately upon exit from the ER. Extension of the A1-chain at the N terminus could block this reaction, especially if the N terminus leads the peptide through the retrotranslocation channel.
We also note that inhibition of the proteasome does not fully rescue retrotranslocation of the A1-chain nor its toxicity. These results might be explained by the inability of lactacystin or ZL3VS to fully inhibit proteasome function, but they are also consistent with an inhibitory effect of the N-terminal peptide on the retrotranslocation reaction itself. Unfolding and dissociation of the mutant A1-chains by PDI appear to be intact, at least as assessed in vitro. Perhaps the N terminus of the A1-chain helps target the protein to the retrotranslocation channel or to another ER chaperone essential for the reaction, and this is blocked in our N-terminal peptide mutant. However, the primary sequence of the extreme N- or C-terminal domains of the A1-chain cannot be required because both mutant A1-chains (HA-CT and CT-HA) clearly enter the cytosol from the ER when degradation by the proteasome is inhibited.
Overall, our results show that the primary effect of the appended peptide is to target the A1-chain for ubiquitination and degradation by the proteasome. Both the N-terminal α-amine and the two Lys residues near the N terminus provide reactive sites. Ubiquitination, however, does not occur for the WT toxin (3). Thus, we propose that the structure of the N-terminal domain in the native toxin allows for protection of the A1-chain against attack by E3 ligases. This could occur by allowing for rapid refolding of the peptide as it emerges from the ER, perhaps by association with cytosolic factors. Whatever the mechanism, such protection against ubiquitination explains how the A1-chain escapes rapid degradation by the proteasome after retrotranslocation, and this is essential for toxin function in the pathogenesis of disease.
Supplementary Material
Acknowledgments
We give our thanks to members of the Lencer laboratory and Fred Goldberg and Henrike Besche for helpful discussions and to David Saslowsky and Edda Fiebiger for reading the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grants RO1 DK48106 (to W. I. L.) and F32 DK078426 (N. L. B. W.). This work was also supported by Harvard Digestive Diseases Center Grant P30 DK034854.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–3.
- CT
- cholera toxin
- ER
- endoplasmic reticulum
- ERAD
- ER-associated degradation
- PDI
- protein disulfide isomerase
- WT
- wild-type
- HA
- hemagglutinin
- TEV
- tobacco etch virus
- HSV
- Herpes simplex virus.
REFERENCES
- 1.Tsai B., Rodighiero C., Lencer W. I., Rapoport T. A. (2001) Cell 104, 937–948 [DOI] [PubMed] [Google Scholar]
- 2.Lencer W. I., Tsai B. (2003) Trends Biochem. Sci. 28, 639–645 [DOI] [PubMed] [Google Scholar]
- 3.Rodighiero C., Tsai B., Rapoport T. A., Lencer W. I. (2002) EMBO Rep. 3, 1222–1227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hazes B., Read R. J. (1997) Biochemistry 36, 11051–11054 [DOI] [PubMed] [Google Scholar]
- 5.Pande A. H., Scaglione P., Taylor M., Nemec K. N., Tuthill S., Moe D., Holmes R. K., Tatulian S. A., Teter K. (2007) J. Mol. Biol. 374, 1114–1128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fujinaga Y., Wolf A. A., Rodighiero C., Wheeler H., Tsai B., Allen L., Jobling M. G., Rapoport T., Holmes R. K., Lencer W. I. (2003) Mol. Biol. Cell 14, 4783–4793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tsai B., Rapoport T. A. (2002) J. Cell Biol. 159, 207–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dixit G., Mikoryak C., Hayslett T., Bhat A., Draper R. K. (2008) Exp. Biol. Med. (Maywood) 233, 163–175 [DOI] [PubMed] [Google Scholar]
- 9.Bernardi K. M., Forster M. L., Lencer W. I., Tsai B. (2008) Mol. Biol. Cell 19, 877–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bernardi K. M., Williams J. M., Kikkert M., van Voorden S., Wiertz E. J., Ye Y., Tsai B. (2010) Mol. Biol. Cell 21, 140–151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schmitz A., Herrgen H., Winkeler A., Herzog V. (2000) J. Cell Biol. 148, 1203–1212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kothe M., Ye Y., Wagner J. S., De Luca H. E., Kern E., Rapoport T. A., Lencer W. I. (2005) J. Biol. Chem. 280, 28127–28132 [DOI] [PubMed] [Google Scholar]
- 13.Lencer W. I., Moe S., Rufo P. A., Madara J. L. (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 10094–10098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lencer W. I., Delp C., Neutra M. R., Madara J. L. (1992) J. Cell Biol. 117, 1197–1209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gill S. C., von Hippel P. H. (1989) Anal. Biochem. 182, 319–326; Correction: (1990) (1990) Anal. Biochem.189, 283 [DOI] [PubMed] [Google Scholar]
- 16.Takeuchi J., Chen H., Coffino P. (2007) EMBO J. 26, 123–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Walter P., Blobel G. (1983) Methods Enzymol. 96, 84–93 [DOI] [PubMed] [Google Scholar]
- 18.Mekalanos J. J., Collier R. J., Romig W. R. (1979) J. Biol. Chem. 254, 5855–5861 [PubMed] [Google Scholar]
- 19.Lencer W. I., Constable C., Moe S., Rufo P. A., Wolf A., Jobling M. G., Ruston S. P., Madara J. L., Holmes R. K., Hirst T. R. (1997) J. Biol. Chem. 272, 15562–15568 [DOI] [PubMed] [Google Scholar]
- 20.Jobling M. G., Holmes R. K. (1991) Mol. Microbiol. 5, 1755–1767 [DOI] [PubMed] [Google Scholar]
- 21.Glickman M. H., Ciechanover A. (2002) Physiol. Rev. 82, 373–428 [DOI] [PubMed] [Google Scholar]
- 22.Meusser B., Hirsch C., Jarosch E., Sommer T. (2005) Nat. Cell Biol. 7, 766–772 [DOI] [PubMed] [Google Scholar]
- 23.Ye Y., Meyer H. H., Rapoport T. A. (2001) Nature 414, 652–656 [DOI] [PubMed] [Google Scholar]
- 24.Spangler B. D. (1992) Microbiol. Rev. 56, 622–647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mogk A., Schmidt R., Bukau B. (2007) Trends Cell Biol. 17, 165–172 [DOI] [PubMed] [Google Scholar]
- 26.Varshavsky A. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 12142–12149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Linding R., Jensen L. J., Diella F., Bork P., Gibson T. J., Russell R. B. (2003) Structure 11, 1453–1459 [DOI] [PubMed] [Google Scholar]
- 28.Linding R., Russell R. B., Neduva V., Gibson T. J. (2003) Nucleic Acids Res. 31, 3701–3708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Prakash S., Tian L., Ratliff K. S., Lehotzky R. E., Matouschek A. (2004) Nat. Struct. Mol. Biol. 11, 830–837 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.