Abstract
The unfolded protein response (UPR) or endoplasmic reticulum (ER) stress response is a physiological process enabling cells to cope with altered protein synthesis demands. However, under conditions of obesity, prolonged activation of the UPR has been shown to have deteriorating effects on different metabolic pathways. Here we identify Bax inhibitor-1 (BI-1), an evolutionary conserved ER-membrane protein, as a novel modulator of the obesity-associated alteration of the UPR. BI-1 partially inhibits the UPR by interacting with IRE1α and inhibiting IRE1α endonuclease activity as seen on the splicing of the transcription factor Xbp-1. Because we observed a down-regulation of BI-1 expression in liver and muscle of genetically obese ob/ob and db/db mice as well as in mice with diet-induced obesity in vivo, we investigated the effect of restoring BI-1 expression on metabolic processes in these mice. Importantly, BI-1 overexpression by adenoviral gene transfer dramatically improved glucose metabolism in both standard diet-fed mice as well as in mice with diet-induced obesity and, critically, reversed hyperglycemia in db/db mice. This improvement in whole body glucose metabolism and insulin sensitivity was due to dramatically reduced gluconeogenesis as shown by reduction of glucose-6-phosphatase and phosphoenolpyruvate carboxykinase expression. Taken together, these results identify BI-1 as a critical regulator of ER stress responses in the development of obesity-associated insulin resistance and provide proof of concept evidence that gene transfer-mediated elevations in hepatic BI-1 may represent a promising approach for the treatment of type 2 diabetes.
Keywords: Carbohydrate, Diabetes, Gluconeogenesis, Glucose, Insulin, Metabolic Diseases, Obesity
Introduction
The prevalence of obesity steadily increases worldwide (1). Obesity causes insulin resistance, thus predisposing to the development of type 2 diabetes (2). Therefore, research on the molecular mechanisms responsible for obesity-associated inhibition of insulin signal transduction has rapidly expanded in recent years. One mechanism of insulin resistance that appears to be of central importance is the activation of inflammatory and stress pathways by endoplasmic reticulum (ER)4 stress (3–5). ER stress stimulates three distinct unfolded protein response (UPR) signaling pathways through sensors that include inositol-requiring enzyme 1 (IRE1α), PKR-like ER kinase, and activating transcription factor 6 (ATF6) (6, 7). These sensors couple the identification of misfolded proteins in the ER to the activation of signal transduction processes promoting expression of genes required for folding of newly synthesized proteins and degradation of the unfolded proteins in an effort to reestablish homeostasis and normal ER function (for review, see Ref. 7). Indeed, a variety of conditions linked to obesity including lipid accumulation, high demand for protein synthesis, and glucose deprivation have been described to trigger ER stress in vitro and in vivo (6, 8). Moreover, signs of ER stress were found in liver and adipose tissue of genetically obese mice and mice exposed to a high fat diet, indicating that the metabolic abnormalities associated with obesity cause ER stress in vivo (3). Importantly, improving the folding capacity of the ER by administration of chemical chaperones has been shown to have beneficial effects on the glucose metabolism of genetically obese mice (4). Altogether, these studies show that the effect of chronic ER stress on glucose homeostasis in obesity may represent a central and integrating mechanism underlying both peripheral insulin resistance and impaired insulin secretion, leading to the development of type 2 diabetes as a consequence of obesity.
The anti-apoptotic protein Bax Inhibitor-1 (BI-1) (9) was originally identified as an inhibitor of ER stress-induced apoptosis. BI-1 contains six transmembrane regions and localizes to ER membranes, and its cytoprotective function is well conserved in plants and mammals (10–12). We have recently described that BI-1 deficiency promotes ER stress in the context of ischemia-reperfusion (13). Besides its cytoprotective function upon ER stress, BI-1 also inhibits the splicing of Xbp-1 by IRE1α and was shown to manipulate calcium release upon ER stress (14–16). IRE1α is a serine-threonine protein kinase and endoribonuclease that, upon activation, initiates the unconventional splicing of the mRNA encoding X-box-binding protein 1 (XBP-1) (17–19). Spliced XBP-1 is a potent transcriptional activator that increases expression of a subset of UPR-related genes involved in efficient protein folding, maturation, and degradation in the ER (20). Besides exhibiting an endonuclease activity, the cytosolic domain of activated IRE1α binds tumor necrosis factor-associated factor 2 and triggers the activation of the c-Jun N-terminal kinase (JNK) signaling pathway (21). IRE1α-deficient cells remain insulin sensitive when challenged with chemical agent-induced-ER stress, underscoring the importance of this UPR branch in insulin resistance (3). Liver tissue of BI-1-deficient mice show evidence of increased stress kinase activity (JNK, p38 mitogen-activated protein kinase) implying BI-1 is required to surpass signal transduction downstream of IRE1α kinase activity. Although BI-1 seems to show specificity for inhibiting the IRE1α branch of the UPR, its exact mechanism of action remains unclear (14).
Because BI-1 was shown to be an inhibitor of the IRE1α branch of the UPR and because this branch is known to be involved in development of high fat diet-induced insulin resistance, we investigated the role of BI-1 in vivo in the regulation of metabolic processes. Here we show that both in diet and genetic models of obesity and glucose intolerance, hepatic (and skeletal muscle) Bi-1 mRNA expression is reduced by more than 40%. Restoration of hepatic BI-1 expression by adenoviral gene transfer dramatically ameliorates glucose intolerance in leptin receptor-deficient (db/db) and high fat diet (HFD)-fed mice and improves fasting as well as fed blood glucose levels. We demonstrate that this improvement in whole body glucose metabolism is caused by inhibition of hepatic gluconeogenesis. Additionally, we show that restoration of BI-1 expression severely blunts the IRE1α-dependent splicing of XBP-1 mRNA by forming a complex with IRE1α. Taken together, we identify BI-1 as a critical regulator of IRE1α-dependent splicing and major hepatic regulator of glucose homeostasis.
EXPERIMENTAL PROCEDURES
Animals
All animal procedures were conducted in compliance with protocols approved by local government authorities (Bezirksregierung Köln) and were in accordance with National Institutes of Health guidelines. Mice were housed in groups of 3–5 at 22–24 °C in a 12-h light/12-h dark cycle with lights on at 6 a.m. Animals were fed either normal chow food (Global Rodent T.2018.R12 from Harlan Teklad containing 12% of calories from fat) or a high fat diet (C1057 from Altromin, containing 55.2% of calories from fat, with linoleic acid as the most abundant fatty acid). Water was available ad libitum, and food was only withdrawn if required for an experiment. At the end of the study period, the animals were killed under CO2 anesthesia.
Adenovirus-mediated Gene Transfer in Vivo
The adenoviral vector expressing BI-1 (Ad BI-1) was generated using the Gateway® system (Invitrogen). Sequence-specific recombination between BI-1 expression plasmid cassette and adenovirus backbone plasmid pAd/PL-DESTTM (Invitrogen) was carried out using the Gateway® LR ClonaseTM enzyme mix (Invitrogen). Linearized recombinant adenoviral genomes were transfected into 293 cells allowing growth of recombinant adenoviruses. Adenoviral stocks were purified twice by CsCl gradient centrifugation, dialyzed extensively against virus storage buffer (137 mm NaCl, 5 mm KCl, 10 mm Tris, pH 7.4, 1 mm MgCl2), and stored in small aliquots at −80 °C until further use. Titers of purified adenoviral vectors were determined as infection units by immunofluorescence staining using fluorescein isothiocyanate-conjugated anti-Ad5-hexon antibody K6100 (DakoCytomation). For adenovirus-mediated gene transfer in vivo, C57Bl/6 mice on a HFD or normal diet (ND) and db/db mice were injected in the tail vein with 10e9 infectious units of the respective adenovirus as previously described (22).
Glucose and Insulin Tolerance Tests
Glucose tolerance tests were performed on animals that had been fasted overnight for 16 h. After determination of fasted blood glucose levels, each animal received an intraperitoneal injection of 2 mg/g of body weight of glucose (20% glucose; Delta Select). Blood glucose levels were detected after 15, 30, 60, and 120 min.
Insulin tolerance tests were performed on random-fed animals. After determination of random fed blood glucose levels, each animal received an intraperitoneal injection of 0.75 milliunits/g of body weight of insulin (Actrapid®; Novo Nordisk).
Analytical Procedures
Blood glucose values were determined from whole venous blood using an automatic glucose monitor (GlucoMen® GlycÓ; A. Menarini Diagnostics). Serum levels of cholesterol, triglycerides, aspartate aminotransferase and alanine aminotransferase were determined in a diagnostic laboratory by standard procedures.
In Vivo Insulin Signaling
In vivo insulin signaling was performed on animals that had been fasted overnight for 16 h. Mice were anesthetized by intraperitoneal injection of avertin (tribromoethyl alcohol and tert-amyl alcohol; Sigma), and adequacy of the anesthesia was ensured by loss of pedal reflexes. The abdominal cavity of the mice was opened, and 100-μl samples containing 13 milliunits of regular human insulin (Actrapid; Novo Nordisk) diluted in 0.9% saline were injected into the vena cava inferior. Sham injections were performed with 100 μl of 0.9% saline. Samples of liver were harvested 2 min after injection and snap-frozen in liquid nitrogen.
Histology
Liver tissue was excised and snap-frozen in Jung Tissue Freezing Medium® (Leica Microsystems), transferred to a cryostat (CM3050S; Leica Microsystems), and cut into 7-μm-thin sections. Specimens were collected on clean poly-l-lysine-coated glass slides (PolysineTM; Menzel), dried at room temperature overnight, and then stained using hematoxylin and eosin or Oil red O.
Immunoblot Analysis
Liver samples were homogenized in lysis buffer (50 mm HEPES, pH 7.4, 1% Triton X-100, 0.1 m sodium fluoride, 10 mm EDTA, 50 mm sodium chloride, 10 mm sodium orthovanadate, 0.1% SDS, 10 μg/ml aprotinin, 2 mm benzamidine, and 2 mm phenylmethylsulfonyl fluoride) using a Polytron homogenizer (IKA Werke). Particulate matter was removed by centrifugation at 13,000 rpm for 1 h at 4 °C. Western blot analyses were performed according to standard protocols using the following primary antibodies: anti-CHOP (Santa Cruz, #7351); anti-ATF6(N) (Santa Cruz, #22799); anti-XBP-1 (Santa Cruz, #7160); anti-GRP78 (Santa Cruz, #13968); anti-actin (Sigma, #5441); anti-HA (Santa Cruz, #805); anti-GFP (Mobitec, #a6455); anti-phospho-AKT (Ser-473), anti-phospho-AKT (Thr-308), and anti-AKT (Cell Signaling, #9271, #4056, #4685); anti-glycogen synthase kinase (GSK) 3β and pGSK3α/β (Cell Signaling, #9315 and #9331); anti-glucose-6-phosphatase (G6P) (Santa Cruz, #25840); anti-C/EBPα (Santa Cruz, #61); anti-IRE1α (Abcam, #37073); anti-SREBP1 (Santa Cruz, #8984); anti-fatty acid synthase (Santa Cruz, #20140); anti-PPARγ (Upstate, #07-466). Equal loading was assured by Ponceau S staining. Antibody detection was accomplished using horseradish peroxidase-conjugated secondary antibodies (anti-mouse immunoglobulin G or anti-rabbit immunoglobulin G, Sigma) and an enhanced chemiluminescence method (Amersham Biosciences). Immunoblots were scanned, and signals were quantified using ImageQuant software.
Immunoprecipitation
For co-immunoprecipitation, HeLa cells were lysed (0.1% Triton X-100, 0.5% sodium deoxycholate, 100 mm KCl, 50 mm Tris, pH 7.6) and precipitated with anti-HA beads (Roche Applied Science, #11815016001) for 4 h at 4 °C. The beads were washed five times with lysis buffer, and the bound proteins were analyzed by immunoblotting using anti-IRE1α and anti-HA antibodies.
Gene Expression Analysis
Gene expression was analyzed using quantitative reverse transcription-PCR. RNA was extracted from liver using Trizol (Invitrogen) according to the manufacturer's instructions. RNA was reverse-transcribed with SuperScript reverse transcriptase (Invitrogen) and amplified using TaqMan Universal PCR Master Mix. Quantitative PCR was performed on an ABI-PRISM 7700 sequence detector (Applied Biosystems). Calculations were performed by a comparative cycle threshold (Ct) method; the starting copy number of test samples was determined in comparison with the known copy number of the calibrator sample (ddCt). The relative gene copy number was calculated as 2 to the power of −ddCt. Assays were linear over 4 orders of magnitude. Probes for target genes were from TaqMan Assay-on-Demand kits (Applied Biosystems). Expression analysis of Xbp1 was performed using the following oligonucleotides (Eurogentec): 5′-CGC AGC AGG TGC AGG CCC A-3′ (spliced Xbp1 probe); 5′-GCA GCA CTC AGA CTA TGT GCA CCT CT-3′ (unspliced Xbp1 probe); 5′-GAA TGG ACA CGC TGG ATC CT-3′ (sense primer); 5′-TCA GAA TCT GAA GAG GCA ACA G-3′ (antisense primer). Samples were adjusted for total RNA content by hypoxanthine-guanine phosphoribosyltransferase (Hprt).
Statistical Analysis
Statistical significance was determined by two-tailed unpaired Student's t test. All p values below 0.05 were considered significant, denoted by asterisks (*, p ≤ 0.05; **, p ≤ 0.01; ***, p ≤ 0.001).
RESULTS
BI-1 Gene Expression Is Down-regulated in Murine Models of Obesity and Diabetes
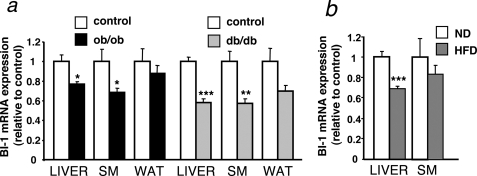
To determine whether BI-1 is potentially involved in the ER stress response dysregulation associated with obesity, we first analyzed Bi-1 mRNA expression in different tissues of genetic models of murine obesity (db/db and ob/ob mice) compared with lean control mice. This analysis revealed significantly reduced Bi-1 mRNA expression in liver and skeletal muscle (but not white adipose tissue) of db/db- and ob/ob-mice as compared with controls (Fig. 1a). Furthermore, also diet-induced obese mice showed a reduction in Bi-1 mRNA expression in liver (Fig. 1b). Although Bi-1 mRNA expression was reduced in all models, this effect appeared even more pronounced in the obese and diabetic db/db mice compared with obese ob/ob mice, possibly indicating that the reduction in BI-1 expression parallels the severity of glucose intolerance or insulin resistance.
FIGURE 1.
Bi-1 expression is decreased in mouse models of obesity and diabetes. a, shown is relative expression of Bi-1 mRNA in the liver, skeletal muscle (SM), and white adipose tissue (WAT) from diabetic (db/db) (filled gray bars), obese (ob/ob) (filled black bars), and age-and sex-matched lean control mice (open bars) (mean ± S.E. of 15 animals in each group). Statistical significance was determined by t test and is denoted by asterisks (* p ≤ 0.05; ** p ≤ 0.01; *** p ≤ 0.001). b, shown is relative expression of Bi-1 mRNA in the liver and skeletal muscle from mice HFD (n = 12) (gray bars)- and ND (n = 8) (white bars)-fed C57Bl/6 mice (mean ± S.E. is shown of each group). Statistical significance was determined by t test and is denoted by asterisks (***, p ≤ 0.001).
Increased Hepatic BI-1 Expression Improves Insulin Sensitivity and Glucose Homeostasis in Vivo
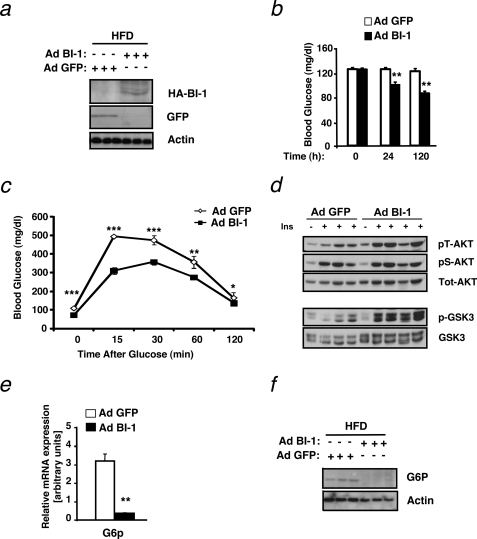
Given that obesity-induced insulin resistance is associated with a condition of increased ER stress (3), that BI-1 is an inhibitor of chemical-induced ER stress (15), and that hepatic expression of BI-1 is down-regulated in mouse models of obesity (Fig. 1), we designed gene transfer experiments to directly address whether overexpression of BI-1 provides a novel approach to ameliorate obesity-induced insulin resistance in vivo. To this end, we employed a recombinant adenovirus-expressing HA-tagged human BI-1 (Ad BI-1) and confirmed its ability to drive expression of HA-tagged BI-1-protein in HeLa cells in culture (supplemental Fig. S1a). To investigate the effect of BI-1 overexpression on ER stress regulation, insulin sensitivity, and systemic glucose metabolism in vivo, we injected mice exposed to HFD (feeding for 12 weeks) with either a recombinant adenovirus expressing green fluorescent protein (Ad GFP) or recombinant Ad BI-1. Before injection, we confirmed that HFD feeding induced insulin resistance and glucose intolerance in these mice (data not shown). At 6 days after adenovirus delivery, expression of the HA-tagged human BI-1 or GFP was detectable in the liver of these mice by immunoblotting (Fig. 2a). Concomitantly, hepatic human BI-1 mRNA levels were detectable in the livers of mice injected with Ad BI-1 but not in those injected with Ad GFP (supplemental Fig. S1b), further confirming adenovirus-mediated expression of BI-1 in the liver of Ad BI-1 injected mice in vivo.
FIGURE 2.
Hepatic BI-1 overexpression ameliorates HFD-induced insulin resistance and glucose metabolism. a, immunoblot analysis was performed for BI-1 or GFP and actin in liver lysates from mice provided HFD and infected with Ad BI-1 or Ad GFP. Livers were collected from mice after an overnight fast, and proteins were extracted and processed as described under “Experimental Procedures.” Each lane represents liver lysates from a different mouse. b, shown are blood glucose concentrations (mg/dl) in HFD-fed mice immediately before and 24 and 120 h after adenovirus treatment. The results are the mean ± S.E. of n = 9 mice per group (**, p ≤ 0.01). c, glucose tolerance tests (0.5 g/kg, intraperitoneal) were performed on HFD-fed mice 60 h after adenovirus injection. Results are given as the mean ± S.E., n = 9 per group (*, p ≤ 0.05; **, p ≤ 0.01; ***, p ≤ 0.001). d, shown is insulin-stimulated phosphorylation of AKT (Ser-473 and Thr-308) and GSK3 was measured in liver tissues of Ad BI-1- and Ad GFP-treated HFD-fed mice upon intravenous insulin (Ins, 13 milliunits) injection. e, shown is relative expression of G6p mRNA in the liver of Ad BI-1- or Ad GFP-infected HFD-fed mice (n = 5 per group; **, p ≤ 0.01). f, G6P and actin protein levels in the liver of Ad BI-1- or Ad GFP-infected HFD-fed mice are assessed by immunoblotting (n = 3 of each group).
Next, we measured blood glucose concentrations in random fed mice injected with either Ad GFP or Ad BI-1. BI-1 gene therapy significantly reduced blood glucose concentrations in the fed state as early as 24 h after adenoviral gene delivery, and this reduction was even more prominent 5 days (120 h) after injection (Fig. 2b). To further investigate the effect of BI-1 gene transfer on glucose metabolism in these HFD fed mice, we performed intraperitoneal glucose tolerance tests. Strikingly, adenovirus-mediated BI-1 expression significantly reduced fasting glycemia levels in these mice compared with mice injected with Ad GFP (Fig. 2c).
To test whether improved insulin signaling was responsible for the enhanced glucose metabolism of HFD-fed mice treated with Ad BI-1, we investigated insulin signaling in the liver of these mice. One of the main effects of insulin in the liver is the inhibition of gluconeogenesis via a pathway that depends on the kinases AKT and GSK3. Intravenous administration of insulin in Ad GFP-treated HFD mice modestly activated insulin signaling in the liver, as seen by AKT phosphorylation on both serine as well as threonine residues (Fig. 2d). Strikingly, mice overexpressing BI-1 showed enhanced threonine phosphorylation of AKT (Fig. 2d). Because phosphorylation and, thus, activation of AKT is responsible for the phosphorylation of GSK3 (23, 24), we analyzed whether GSK3 phosphorylation levels are altered in Ad BI-1-treated mice compared with control mice. In line with increased AKT phosphorylation levels, BI-1-overexpressing mice exhibited higher GSK3 phosphorylation levels than Ad GFP-treated mice fed HFD (Fig. 2d). Insulin-mediated GSK3 phosphorylation and, hence, inactivation leads to inhibition of gluconeogenesis by inhibiting the expression of G6P and phosphoenolpyruvate carboxykinase (PEPCK) (25). In line with the enhanced phosphorylation of GSK3, we also observed a marked decrease in the expression of G6P both at the mRNA and at the protein level (Fig. 2, e and f). In summary, these results reveal that BI-1 improves glucose metabolism and ameliorates insulin sensitivity in hepatocytes of mice exposed to HFD.
Adenoviral BI-1 Expression Improves Glucose Homeostasis of Genetically Obese Mice
Having observed a significant effect of BI-1 overexpression in the liver on HFD-induced alterations of glucose metabolism, we next addressed whether the same approach could also improve the glucose metabolism in genetically obese mice. To this end, db/db mice, which are massively obese and suffer from diabetes due to genetic ablation of the leptin receptor, were injected with Ad BI-1 or Ad GFP as described above. Similar to HFD-fed mice overexpressing BI-1, db/db mice injected with Ad BI-1 had markedly lower random fed blood glucose levels compared with db/db mice injected with Ad GFP 5 days after injection (Fig. 3a). Importantly, fasted blood glucose levels of Ad BI-1-treated db/db mice were significantly decreased as well and reached levels usually seen in healthy control mice (Fig. 3b).
FIGURE 3.
Hepatic BI-1 expression improves insulin sensitivity and glucose metabolism in db/db mice. a, shown are blood glucose concentrations (mg/dl) in db/db mice immediately before and 24 and 120 h after Ad BI-1 or Ad GFP adenovirus treatment. The results are the mean ± S.E. of n = 10 mice per group (**, p ≤ 0.01). b, shown are overnight fasted blood glucose concentrations (mg/dl) in db/db mice 144 h after adenovirus treatment. The results are the mean ± S.E. of n = 10 mice per group (***, p ≤ 0.001). c, insulin tolerance tests were performed db/db mice 120 h after adenovirus injection. Results are given as the mean ± S.E.; n = 10 per group (*, p ≤ 0.05; **, p ≤ 0.01). d, shown is relative expression of G6p and Pepck mRNA in the liver of Ad BI-1 or Ad GFP infected db/db mice (n = 5 per group; *** p ≤ 0.001). e, pyruvate tolerance tests were performed on db/db mice 120 h after adenovirus injection. Results are given as the mean ± S.E., n = 7–8 per group (*, p ≤ 0.05; ***, p ≤ 0.001). f, daily food intake was measured 24, 48, and 72 h after adenovirus injection. Results are given as the mean ± S.E. (n = 7–8 per group). g, body weight was measured 72 h after adenovirus injection. Results are given as the mean ± S.E., n = 7–8 per group. h, relative fat mass was determined by NMR analysis 128 h after adenovirus injection. Results are given as the mean ± S.E., n = 7–8 per group.
Next, we addressed whether these BI-1-overexpressing mice exhibit a better responsiveness to insulin in an insulin tolerance test. As seen in Fig. 3c, Ad BI-1-injected mice clearly demonstrated improved insulin sensitivity compared with Ad GFP-injected mice, which were completely insulin-resistant. This improved insulin signaling was further confirmed by markedly lower expression levels of the gluconeogenic enzymes G6p and Pepck upon overexpression of BI-1 (Fig. 3d). To directly address the role of hepatic glucose production in Ad BI-1-treated mice, we performed pyruvate tolerance tests in Ad BI-1 and Ad GFP injected db/db mice. Injection of pyruvate increased blood glucose concentrations in Ad GFP-treated db/db mice, which was drastically ameliorated in Ad BI-1-treated db/db animals (Fig. 3e).
Furthermore, improved glucose tolerance was not due to general sickness or reduced feeding, as body weight, fat content, and food intake was unchanged between Ad BI-1- and Ad GFP- treated animals (Fig. 3, f–h). Taken together, these data indicate that BI-1 overexpression not only improves the glucose metabolism in diet-induced obese mice but also restores insulin sensitivity of db/db mice, a mouse model with genetically induced severe diabetes and obesity.
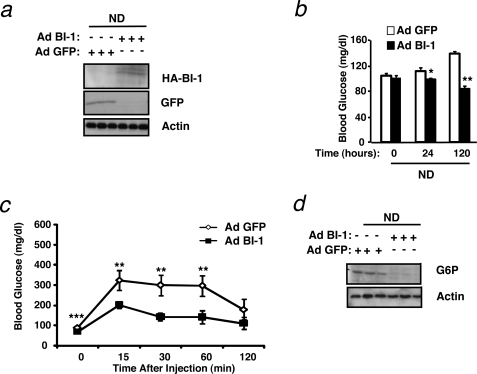
To investigate whether the BI-1-mediated effect on glucose metabolism was a general mechanism, we also analyzed normal chow diet-fed mice. Strikingly, Ad BI-1-injected mice on a regular diet showed decreased blood glucose levels 5 days after injection and increased glucose clearance compared with Ad GFP-injected mice in a glucose tolerance test (Fig. 4, a–c). Again, this was reflected by a drastic decrease in expression of G6P on mRNA and protein levels (data not shown and Fig. 4d). Altogether, these data clearly indicate that BI-1 exhibits an important role in glucose homeostasis in normal as well as diet-induced and genetically obese mice.
FIGURE 4.
Hepatic BI-1 overexpression increases glucose tolerance in mice fed a normal chow diet. a, immunoblot analysis was performed for BI-1 or GFP and actin on liver lysates from mice provided a normal chow diet and infected with Ad BI-1 or Ad GFP. Livers were collected from mice after an overnight fast, and proteins were extracted and processed as described under “Experimental Procedures.” Each lane represents liver lysates from a different mouse. b, blood glucose concentrations (mg/dl) in ND-fed mice immediately before and 24 and 120 h after adenovirus treatment are shown. The results are the mean ± S.E. of n = 9 mice per group (*, p ≤ 0.05; **, p ≤ 0.01). c, glucose tolerance tests (0.5 g/kg, intraperitoneal) were performed on ND-fed mice 60 h after adenovirus injection. Results are given as the mean ± S.E., n = 9 per group (**, p ≤ 0.01; ***, p ≤ 0.001). d, shown are G6P and actin protein levels in the liver of Ad BI-1- or Ad GFP-infected ND-fed mice as assessed by immunoblotting (n = 3 of each group).
BI-1 Inhibits Xbp-1 Splicing in Vivo and Interacts with IRE1α
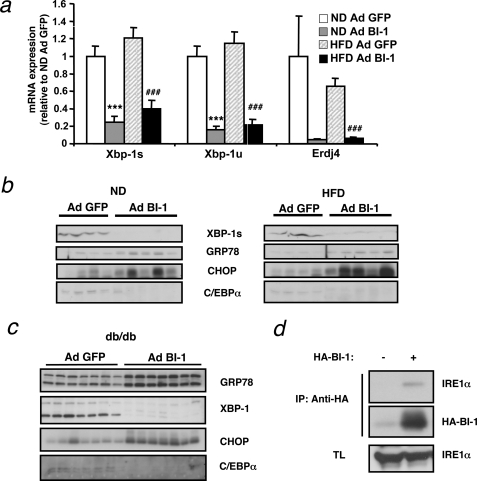
Overexpression of BI-1 in vitro inhibits the UPR in response to chemical ER stress-inducing agents such as thapsigargin or tunicamycin (Ref. 15 and supplemental Fig. S2, a and b). Because diet-induced obesity is accompanied by increased ER stress in the liver, we asked whether BI-1 overexpression affected the ER stress response in vivo. Compared with Ad GFP-injected mice, adenoviral transfer of BI-1 clearly reduced levels of both unspliced and spliced Xbp-1, a target of the IRE1α branch of the UPR, both under normal as well as HFD conditions (Fig. 5a). In line with this result, expression of the specific XBP-1 target gene Erdj4 was also strongly inhibited by overexpression of BI-1. Accordingly, we could detect a marked decrease in protein levels of spliced XBP-1 upon BI-1 gene transfer in both HFD and db/db mice (Fig. 5. b and c). Interestingly, regular diet-fed mice also showed basal levels of XBP-1 splicing that were blunted by overexpression of BI-1, indicating that BI-1 overexpression not only lowers the obesity-induced ER stress response but also the physiological UPR response. On the other hand, BI-1 overexpression does not lower the overall UPR, as both GRP78 and CHOP were significantly elevated in Ad BI-1-injected mice compared with control mice both in HFD and ND fed mice as well as in db/db mice (Fig. 5, b and c). The latter indicates that, at least in vivo, BI-1 does not block all branches of the UPR but exhibits specificity for the IRE1α branch that mediates Xbp-1 splicing.
FIGURE 5.
Adenovirus-mediated expression of BI-1 in the liver inhibits IRE-1α-mediated XBP-1 processing. a, shown is relative expression of spliced Xbp-1 (Xbp-1s), unspliced Xbp-1u, and the XBP-1 target Erdj4 in the liver of Ad BI-1-or Ad GFP-infected ND and HFD mice as measured by real-time PCR (n = 5 per group). b, shown are XBP-1s, GRP78, CHOP, and C/EBPα protein levels in the liver of Ad BI-1- or Ad GFP-infected ND- and HFD-fed mice as assessed by immunoblotting (n = 4–5 of each group). c, shown are XBP-1, GRP78, CHOP, and C/EBPα protein levels in the liver of Ad BI-1- or Ad GFP-infected db/db mice as assessed by immunoblotting (n = 7 of each group). d, shown is co-immunoprecipitation (IP) of BI-1 and IRE1α. HeLa cells with tetracycline (TL)-inducible HA-tagged BI-1 expression were either stimulated with (+) or without (−) doxycycline. BI-1 was immunoprecipitated using α-HA antibody, and its interaction with IRE1α was assessed by immunoblotting.
BI-1 and IRE1α are transmembrane ER-residing proteins that have recently been reported to complex in cultured cells (14). We independently confirmed this result. To this end we made use of a doxycycline-inducible BI-1 expression system in HeLa cells. After administration of doxycycline to both control cells and BI-1-overexpressing cells, we immunoprecipitated the HA-tagged BI-1 with anti-HA beads and immunoblotted for IRE1α. Only HA-BI-1-expressing cells allowed pull down of IRE1α protein, confirming that these proteins interact (Fig. 5d). Because spliced Xbp-1 binds to and activates the XBP-1 promoter and, thus, increases its own expression, reduced splicing of Xbp-1 may be responsible for the parallel reduction in unspliced Xbp-1 (26). Together these data show that whereas the IRE1α branch of the UPR is suppressed by overexpression of BI-1, these mice exhibited an increased level of ER stress shown by increased levels of CHOP and GRP78 expression.
Hepatic BI-1 Overexpression Predisposes for the Development of Steatosis in Mice
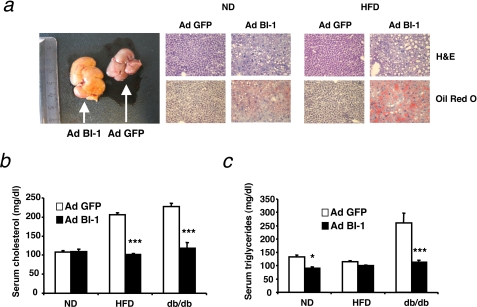
It has been shown that interfering with individual branches of the UPR predisposes mice to the development of hepatic steatosis when these mice are triggered with chemical ER stress-inducing agents such as tunicamycin (27). In line with this, livers of BI-1-overexpressing mice showed an altered lipid metabolism both at a macroscopic and at microscopic level (Fig. 6a). Ad BI-1-expressing livers were much lighter in color than their Ad GFP-injected control counterparts, possibly indicating lipid accumulation. Both ND and HFD livers of Ad BI-1-injected mice showed structural differences compared with control mice upon hematoxylin and eosin staining, although the effect was clearly more pronounced in the HFD-fed mice. In addition, Oil red O staining clearly confirmed the accumulation of fat deposits in BI-1-overexpressing livers compared with control livers. Nonetheless, in db/db mice, BI-1 overexpressing livers did not exhibit a clear enhanced accumulation of fat in the liver compared with control livers, likely due to the fact that db/db mice showed massive steatosis even before adenoviral treatment (supplemental Fig. S3a).
FIGURE 6.
Adenovirus-mediated expression of BI-1 in the liver induces steatosis and decreases circulating cholesterol and triglycerides. a, representative livers of Ad BI-1- or Ad GFP-infected HFD-fed mice are shown on the left. Hematoxylin and eosin (H&E) and Oil Red O stainings of liver sections of Ad BI-1- or Ad GFP-infected ND- and HFD-fed mice are shown on the right. b, shown are fasting serum cholesterol concentrations from Ad BI-1- or Ad GFP-infected ND, HFD, and db/db mice (n = 9 per group; ***, p ≤ 0.001). c, shown is fasting serum triglyceride concentrations from Ad BI-1- or Ad GFP-infected ND, HFD, and db/db mice (n = 9 per group; *, p ≤ 0.05; ***, p ≤ 0.001).
Because the absence of components of the UPR response has been shown to affect the serum lipid content of mice (27, 28), we analyzed fasted serum levels of cholesterol and triglycerides in all treatment groups. In line with the observations for tunicamycin-challenged ATF6-deficient mice, serum levels of cholesterol were reduced by overexpression of BI-1 in diet-induced or genetically obese animals (Fig. 6b). Whereas plasma triglyceride levels in HFD-fed mice remained unchanged, we additionally observed a clear reduction of triglyceride levels in the serum of both normal chow diet-fed mice as well as in db/db mice upon adenoviral gene transfer of BI-1 (Fig. 6c). Altogether these data indicate that adenoviral gene transfer of BI-1 reduces circulating levels of lipids but induces hepatic steatosis.
BI-1 Overexpression Affects Hepatic Lipid Metabolism at Different Levels
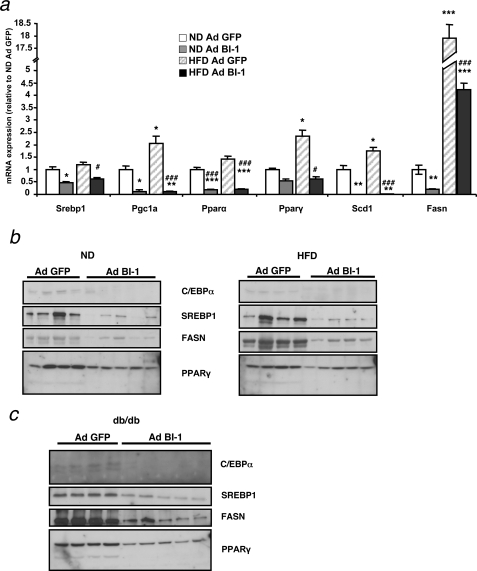
Interference with one of the branches of the UPR predisposes mice to develop hepatosteatosis upon chemical induction of ER stress by injection of tunicamycin (27). These mice show an altered lipid metabolism due to a down-regulation of genes involved in lipogenesis, lipid oxidation, and lipid mobilization. Because adenoviral transfer of BI-1 diminishes the activation of the IRE1α branch of the UPR while inducing and/or predisposing for steatosis, we investigated if Ad BI-1-injected mice showed similar defects in lipid metabolism as reported for IRE1α knock-out mice. We analyzed the expression levels of key enzymes in fatty acid metabolism at the mRNA level. HFD induces the expression of Pgc1α and PPAR family members, which are involved in fatty acid oxidation, and enhances expression of genes involved in fatty acid synthesis, such as fatty acid synthase (Fasn) and stearoyl-CoA desaturase 1 (Scd1) (Fig. 7a). Similar to IRE1α knock-out mice challenged with tunicamycin (27), repression of the IRE1α branch of the UPR by overexpressing BI-1 resulted in a down-regulation of mRNAs corresponding to pathways involved in lipid metabolism both in normal-fed as well as in HFD mice (Fig. 7a). A similar decrease of expression of SREBP1, FASN, and PPARγ was observed at the protein level (Fig. 7, b and c).
FIGURE 7.
Effect of adenovirus-mediated expression of BI-1 on hepatic liver metabolism. a, shown is relative mRNA expression of sterol response element-binding protein 1 (Srebp1), PPARγ coactivator 1α (Pgc1α), peroxisome proliferator-activated receptor α and γ (Pparα, Pparγ), stearoyl-coenzyme A desaturase 1 (Scd1), and fatty acid synthase (Fasn) in the liver of Ad BI-1- or Ad GFP-infected ND and HFD mice as measured by real-time PCR (n = 5 per group). b, shown are C/EBPα, SREBP1, FASN, and PPARγ protein levels in the liver of Ad BI-1- or Ad GFP-infected ND- and HFD-fed mice as assessed by immunoblotting (n = 4–5 of each group). c, shown are C/EBPα, SREBP1, FASN, and PPARγ protein levels in the liver of Ad BI-1- or Ad GFP-infected db/db mice as assessed by immunoblotting (n = 4–5 of each group).
UPR is known to induce increased CHOP levels, which in turn interfere with the function of C/EBPα, a transcription factor involved in hepatic gluconeogenesis and lipid homeostasis. Because Ad BI-1-injected mice show decreased IRE1α-mediated Xbp-1 splicing in combination with an increase in CHOP protein levels, we investigated a possible effect of BI-1 overexpression on C/EBPα expression. Indeed, C/EBPα levels were decreased in ND, HFD, and db/db mice treated with Ad BI-1 compared with Ad GFP-injected mice (Fig. 7, b and c). Altogether, these data indicate that overexpression of BI-1 alters the normal lipid homeostasis in mice both under normal conditions as well as under the altered metabolic conditions of HFD or db/db mice.
DISCUSSION
ER stress has recently emerged as a central pathophysiological mechanism in the development of obesity-associated impairment of glucose homeostasis (3). Obese mice and humans have been shown to have increased activation of the different branches of the UPR (3, 29–31). Moreover, administration of chemical chaperones dampens the ER stress response and improves glucose metabolism in genetically obese mice by restoring insulin action in liver (4). Interestingly, analysis of ER stress-activated indicator transgenic mice has revealed early activation of ER stress in liver but not white adipose tissue when animals are exposed to high fat/high sucrose diet (32). Thus, exaggerated ER stress occurs early in liver tissue after exposure to HFD; this is the same tissue where we find BI-1 most prominently down-regulated in genetically obese or HFD-fed animals.
Because BI-1 was shown to be an inhibitor of ER stress in vitro (11, 15), we questioned whether down-regulation of BI-1 expression levels is linked to an increased ER stress status in obesity. Specifically we asked whether restoration of BI-1 expression levels in models of obesity and glucose intolerance has beneficial metabolic effects. We report that overexpression of BI-1 in the liver results in dramatically improved glucose metabolism in HFD or db/db mice as measured by fasting glucose levels and glucose uptake in glucose tolerance tests. Interestingly, we also observed an improved glucose metabolism even in normal diet-fed mice (see below).
Recent evidence shows that BI-1 is an inhibitor of the IRE1α branch of the UPR interfering with IRE1α endonuclease activity (14). Consistent with this notion, we observed markedly reduced splicing of the transcription factor Xbp-1, a known target of IRE1α, in mice overexpressing BI-1 in liver. IRE1α-deficient cells have been shown to exhibit improved insulin signaling when challenged with chemical agent-induced ER stress such as tunicamycin, thus implicating IRE1α signaling in the mechanism of insulin resistance (3).
Supporting a role for BI-1 as a key inhibitor of the IRE1α branch of the UPR, we observed enhanced phosphorylation of AKT and GSK3 in the livers of BI-1-overexpressing mice exposed to HFD, thus phenocopying the effects observed with IRE1α deficiency. In line with this increased phosphorylation of these two key downstream signaling molecules in the insulin pathway, we also observed significantly increased insulin sensitivity in db/db mice with hepatic BI-1 overexpression.
Besides this increased insulin signaling potential, it is likely that the dramatic improvement in fasted and fed blood glucose as well as overall glucose tolerance is also due to decreased C/EBPα expression in BI-1-overexpressing livers. C/EBPα is indispensable for the induction of two key gluconeogenic enzymes, PEPCK and G6P, which explains why mice deficient in C/EBPα die at birth due to fasting hypoglycemia (33). Our findings with BI-1, thus, support the recent report that mice deficient for IRE1α in the liver exhibit a decrease in hepatic C/EBPα levels and down-regulation of PEPCK expression (27). Interestingly, impairment of the PKR-like ER kinase or the ATF6 branch of the UPR also results in down-regulation of C/EBPα and increased glucose metabolism (5). Possibly, blockage of one of the UPR pathways leads to unresolved ER stress, which in turn overactivates the other branches of the UPR as seen by increased induction of CHOP and GRP78 in the liver of Ad BI-1-injected mice. As such, normal gene expression of key metabolic enzymes is impaired, leading to fasting hypoglycemia and impaired gluconeogenesis as shown in pyruvate tolerance tests. At this point, we cannot exclude the possibility that hepatic BI-1 regulates glucose uptake in other tissues by unknown means or modulates primarily hepatic glucose release compared with glucose production. Nonetheless, several lines of experimental evidence including RNA and protein expression of gluconeogenic enzymes as well as results from pyruvate tolerance tests argue for reduced gluconeogenesis, largely accounting for the phenotype of Ad BI-1-injected mice.
Strikingly, we found improved glucose tolerance to occur not only when mice were under metabolically induced ER stress upon HFD feeding but also that animals fed a normal diet showed lower glucose levels when injected with Ad BI-1. Importantly, livers of Ad BI-1-injected mice did not show increased apoptosis as determined by terminal dUTP nick-end labeling staining (data not shown) nor were circulating concentrations of liver enzymes such as alanine aminotransferase and aspartate aminotransferase increased (supplemental Fig. S3b), indicating that general liver function is not impaired by adenoviral BI-1 expression.
Although exhibiting improved glucose metabolism, BI-1-overexpressing mice showed a marked accumulation of lipids in the liver, leading to hepatosteatosis. Recently Rutkowski et al. (27) showed that acute induction of ER stress by injection of tunicamycin in mice leads to steatosis when any of the three branches of the UPR is blocked. Here we show that the same holds true under more physiological conditions of ER stress. Upon high fat feeding, mice overexpressing BI-1 in the liver showed increased accumulation of lipids compared with Ad GFP-injected mice. Interestingly, however, functional down-regulation of the IRE1α pathway by overexpression of BI-1 results in down-regulation of key enzymes of lipid homeostasis pathways not only under conditions of increased ER stress as seen in HFD mice but also in normal fed mice. The latter possibly indicates that the IRE1α branch of the UPR is also active (at low levels) under normal physiological conditions and as such contributes to a normal metabolic homeostasis. In this respect it is interesting to note that hepatic lack of XBP-1, a downstream target of IRE1α, also affects lipid metabolism in the liver under normal-fed conditions (28). The latter mice show decreased plasma levels of triglycerides and cholesterol, which was also observed in Ad BI-1-injected mice, although to a lesser extent. Similar to the phenotype of mice lacking XBP-1 in liver (28), BI-1 overexpression in the liver leads to a down-regulation of Scd1, a key lipogenic enzyme. Hepatic lack of XBP-1, however, does not lead to hepatosteatosis nor does it lead to a down-regulation of enzymes involved in lipid oxidation or lipid synthesis such as PPARα and PPARγ. This difference in the phenotypes of XBP-1 deficiency versus BI-1 overexpression in liver suggests that 1) BI-1 blocks the IRE1α pathway upstream of Xbp-1 splicing and 2) impacts other substrates of IRE1α endoribonuclease activity or that 3) the BI-1 effects on stress kinase activation downstream of IRE1α may contribute to hepatosteatosis. So far, hepatic ablation of XBP-1 has only been achieved by using an inducible promoter (MX) (28), which might partially induce deletion in other cell types. Thus, it will be highly informative to examine the effect of BI-1 expression on hepatic metabolism in mouse models lacking XBP-1 and/or IRE1α specifically in liver. Last, the molecular signaling mediators by which UPR affects lipid oxidation, generation, and release remain to be clarified in greater detail.
Our data support the involvement of BI-1 in down-regulating IRE1α signaling in vivo and link this activity to metabolic homeostasis. Moreover, our findings together with others suggests that a normal balance of activation of IRE1α signaling is of major importance for metabolic homeostasis; over-activation of IRE1α signaling can lead to insulin resistance and impaired glucose homeostasis (3), whereas lower than “normal” IRE1α signaling, as seen in IRE1α knock-out mice or Ad BI-1-overexpressing mice, leads to hepatosteatosis but improves glucose metabolism. Additionally, it is likely that the hepatosteatosis observed in Ad BI-1-treated mice occurs partially due to increased hepatic insulin signaling and, thus, inhibition of gluconeogenesis. Insulin is a key negative regulator of Pepck and G6p expression in a FOXO1-dependent manner (34). Importantly, mice with liver-specific ablation of the key gluconeogenic enzyme PEPCK suffer from hepatic steatosis, which is phenocopied in mice with hepatic overexpression of BI-1 (35). This indicates that inhibition of gluconeogenesis alone is sufficient to increase hepatic lipid content. Application of current pharmacological insulin sensitizers (such as thiazolidinediones and metformin) likely does not cause hepatic steatosis but instead reduces steatosis, because these agents increase insulin sensitivity in all other major insulin target tissues such as muscle and white adipose tissue, thus increasing peripheral glucose (and lipid) uptake and subsequently increasing hepatic glucose and lipid output to maintain glucose and lipid homeostasis (36). Indeed, it has been proposed that increasing insulin sensitivity in the liver alone will negatively affect systemic glucose and lipid metabolism, due to severely increased lipogenesis (37). In contrast, reducing systemic ER stress, for example by application of chemical chaperones, does not induce hepatic steatosis (4). Because we also detected decreases in BI-1 expression in skeletal muscle of diabetic mice, future studies should examine the effects of BI-1 re-expression in multiple organs with regard to their impact on hyperglycemia and hepatic dyslipidemia. In parallel, the search for potentially novel BI-1 interaction partners might increase our understanding of its molecular functions.
We hypothesize that the reduction in BI-1 expression described here in diabetic mice represents a pathophysiological event, as acute BI-1 re-expression dramatically and within 24 h ameliorates hyperglycemia in both diet-induced and genetic models of obesity and glucose intolerance. Thus, we define BI-1 as a critical regulator of IRE1α activity, insulin sensitivity, and glucose and lipid metabolism. Future work will be necessary to address the mechanisms regulating BI-1 transcription and to explore pharmacological re-activation of BI-1 expression in amelioration of metabolic disorders.
Supplementary Material
Acknowledgments
We thank G. Schmall and T. Rayle for excellent secretarial assistance and S. Irlenbusch for outstanding technical assistance. B. Bailly-Maitre thanks the Fondation pour le Recherche Medicale.
This work was supported, in whole or in part, by National Institutes of Health Grant AG15393 (to J. C. R.). This work was also supported by funding from the Cologne Cluster of Excellence on Cellular Stress Responses in Aging-associated Diseases and grants from the Center of Molecular Medicine Cologne (TV A1; to J. C. B.), European Community 7th Framework Programme (FP7/2007-2013) Grant 201608, acronym “TOBI” (to J. C. B.), German Research Foundation (Deutsche Forschungsgemeinschaft) Grants 1492/7-1, SFB 612 (TP B16), and SFB 670 (TP 17), and the Californian Breast Cancer Research Program.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–3.
- ER
- endoplasmic reticulum
- UPR
- unfolded protein response
- IRE1α
- inositol-requiring enzyme 1
- ATF6
- activating transcription factor 6
- BI-1
- Bax inhibitor-1
- XBP-1
- X-box-binding protein 1
- JNK
- c-Jun N-terminal kinase
- Ad
- adenoviral
- ND
- normal diet
- HFD
- high fat diet
- GSK
- glycogen synthase kinase
- G6P
- glucose-6-phosphatase
- SREBP1
- sterol response element-binding protein 1
- PEPCK
- phosphoenolpyruvate carboxykinase
- HA
- hemagglutinin
- PPAR
- peroxisome proliferator-activated receptor
- FASN
- fatty acid synthase
- CHOP
- C/EBP homologous protein.
REFERENCES
- 1.Haslam D. W., James W. P. (2005) Lancet 366, 1197–1209 [DOI] [PubMed] [Google Scholar]
- 2.Permutt M. A., Wasson J., Cox N. (2005) J. Clin. Invest. 115, 1431–1439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ozcan U., Cao Q., Yilmaz E., Lee A. H., Iwakoshi N. N., Ozdelen E., Tuncman G., Görgün C., Glimcher L. H., Hotamisligil G. S. (2004) Science 306, 457–461 [DOI] [PubMed] [Google Scholar]
- 4.Ozcan U., Yilmaz E., Ozcan L., Furuhashi M., Vaillancourt E., Smith R. O., Görgün C. Z., Hotamisligil G. S. (2006) Science 313, 1137–1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Oyadomari S., Harding H. P., Zhang Y., Oyadomari M., Ron D. (2008) Cell Metab. 7, 520–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ron D., Walter P. (2007) Nat. Rev. Mol. Cell Biol 8, 519–529 [DOI] [PubMed] [Google Scholar]
- 7.Xu C., Bailly-Maitre B., Reed J. C. (2005) J. Clin. Invest. 115, 2656–2664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ma Y., Hendershot L. M. (2001) Cell 107, 827–830 [DOI] [PubMed] [Google Scholar]
- 9.Xu Q., Reed J. C. (1998) Mol. Cell 1, 337–346 [DOI] [PubMed] [Google Scholar]
- 10.Bolduc N., Ouellet M., Pitre F., Brisson L. F. (2003) Planta 216, 377–386 [DOI] [PubMed] [Google Scholar]
- 11.Chae H. J., Kim H. R., Xu C., Bailly-Maitre B., Krajewska M., Krajewski S., Banares S., Cui J., Digicaylioglu M., Ke N., Kitada S., Monosov E., Thomas M., Kress C. L., Babendure J. R., Tsien R. Y., Lipton S. A., Reed J. C. (2004) Mol. Cell 15, 355–366 [DOI] [PubMed] [Google Scholar]
- 12.Chae H. J., Ke N., Kim H. R., Chen S., Godzik A., Dickman M., Reed J. C. (2003) Gene 323, 101–113 [DOI] [PubMed] [Google Scholar]
- 13.Bailly-Maitre B., Fondevila C., Kaldas F., Droin N., Luciano F., Ricci J. E., Croxton R., Krajewska M., Zapata J. M., Kupiec-Weglinski J. W., Farmer D., Reed J. C. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 2809–2814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lisbona F., Rojas-Rivera D., Thielen P., Zamorano S., Todd D., Martinon F., Glavic A., Kress C., Lin J. H., Walter P., Reed J. C., Glimcher L. H., Hetz C. (2009) Mol. Cell 33, 679–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee G. H., Kim H. K., Chae S. W., Kim D. S., Ha K. C., Cuddy M., Kress C., Reed J. C., Kim H. R., Chae H. J. (2007) J. Biol. Chem. 282, 21618–21628 [DOI] [PubMed] [Google Scholar]
- 16.Kim H. R., Lee G. H., Cho E. Y., Chae S. W., Ahn T., Chae H. J. (2009) J. Cell Sci. 122, 1126–1133 [DOI] [PubMed] [Google Scholar]
- 17.Calfon M., Zeng H., Urano F., Till J. H., Hubbard S. R., Harding H. P., Clark S. G., Ron D. (2002) Nature 415, 92–96 [DOI] [PubMed] [Google Scholar]
- 18.Lee K., Tirasophon W., Shen X., Michalak M., Prywes R., Okada T., Yoshida H., Mori K., Kaufman R. J. (2002) Genes Dev. 16, 452–466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yoshida H., Matsui T., Yamamoto A., Okada T., Mori K. (2001) Cell 107, 881–891 [DOI] [PubMed] [Google Scholar]
- 20.Lee A. H., Iwakoshi N. N., Glimcher L. H. (2003) Mol. Cell. Biol. 23, 7448–7459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Urano F., Wang X., Bertolotti A., Zhang Y., Chung P., Harding H. P., Ron D. (2000) Science 287, 664–666 [DOI] [PubMed] [Google Scholar]
- 22.Sprinzl M. F., Oberwinkler H., Schaller H., Protzer U. (2001) J. Virol. 75, 5108–5118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Alessi D. R., Caudwell F. B., Andjelkovic M., Hemmings B. A., Cohen P. (1996) FEBS Lett. 399, 333–338 [DOI] [PubMed] [Google Scholar]
- 24.Cross D. A., Alessi D. R., Cohen P., Andjelkovich M., Hemmings B. A. (1995) Nature 378, 785–789 [DOI] [PubMed] [Google Scholar]
- 25.Lochhead P. A., Coghlan M., Rice S. Q., Sutherland C. (2001) Diabetes 50, 937–946 [DOI] [PubMed] [Google Scholar]
- 26.Reimold A. M., Ponath P. D., Li Y. S., Hardy R. R., David C. S., Strominger J. L., Glimcher L. H. (1996) J. Exp. Med. 183, 393–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rutkowski D. T., Wu J., Back S. H., Callaghan M. U., Ferris S. P., Iqbal J., Clark R., Miao H., Hassler J. R., Fornek J., Katze M. G., Hussain M. M., Song B., Swathirajan J., Wang J., Yau G. D., Kaufman R. J. (2008) Dev. Cell 15, 829–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee A. H., Scapa E. F., Cohen D. E., Glimcher L. H. (2008) Science 320, 1492–1496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Boden G., Duan X., Homko C., Molina E. J., Song W., Perez O., Cheung P., Merali S. (2008) Diabetes 57, 2438–2444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Laybutt D. R., Preston A. M., Akerfeldt M. C., Kench J. G., Busch A. K., Biankin A. V., Biden T. J. (2007) Diabetologia 50, 752–763 [DOI] [PubMed] [Google Scholar]
- 31.Gregor M. F., Yang L., Fabbrini E., Mohammed B. S., Eagon J. C., Hotamisligil G. S., Klein S. (2009) Diabetes 58, 693–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yoshiuchi K., Kaneto H., Matsuoka T. A., Kohno K., Iwawaki T., Nakatani Y., Yamasaki Y., Hori M., Matsuhisa M. (2008) Biochem. Biophys. Res. Commun. 366, 545–550 [DOI] [PubMed] [Google Scholar]
- 33.Wang N. D., Finegold M. J., Bradley A., Ou C. N., Abdelsayed S. V., Wilde M. D., Taylor L. R., Wilson D. R., Darlington G. J. (1995) Science 269, 1108–1112 [DOI] [PubMed] [Google Scholar]
- 34.Nakae J., Kitamura T., Silver D. L., Accili D. (2001) J. Clin. Invest. 108, 1359–1367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.She P., Shiota M., Shelton K. D., Chalkley R., Postic C., Magnuson M. A. (2000) Mol. Cell. Biol. 20, 6508–6517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Anderson N., Borlak J. (2008) Pharmacol. Rev. 60, 311–357 [DOI] [PubMed] [Google Scholar]
- 37.Brown M. S., Goldstein J. L. (2008) Cell Metab. 7, 95–96 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.