Abstract
The ligand-binding domain of Fbl (the fibrinogen binding protein from Staphylococcus lugdunensis) shares 60% sequence identity with ClfA (clumping factor A) of Staphylococcus aureus. Recombinant Fbl corresponding to the minimum fibrinogen-binding region (subdomains N2N3) was compared with ClfA for binding to fibrinogen. Fbl and ClfA had very similar affinities for fibrinogen by surface plasmon resonance. The binding site for Fbl in fibrinogen was localized to the extreme C terminus of the fibrinogen γ-chain at the same site recognized by ClfA. Isothermal titration calorimetry showed that Fbl and ClfA had very similar affinities for a peptide mimicking the C-terminal segment of the fibrinogen γ-chain. The peptide also inhibited binding of Fbl and ClfA to fibrinogen. A series of substituted γ-chain variant peptides behaved very similarly when used to inhibit ClfA and Fbl binding to immobilized fibrinogen. Both ClfA and Fbl bound to bovine fibrinogen with a lower affinity compared with human fibrinogen and did not bind detectably to ovine fibrinogen. The structure of the N2N3 subdomains of Fbl in complex with the fibrinogen γ-chain peptide was modeled based on the crystal structure of the N2N3 subdomains of the ClfA-γ-chain peptide complex. Residues in the putative binding trench likely to be involved in fibrinogen binding were identified. Fbl variant proteins with alanine substitutions in key residues had reduced affinities for fibrinogen. Thus Fbl and ClfA bind the same site in fibrinogen by similar mechanisms.
Keywords: Bacteria, Crystal Structure, Fibrinogen, Protein Domains, Protein Structure, Adhesin, Pathogen, Staphylococci, Surface Protein, virulence
Introduction
Staphylococcus aureus is an important human pathogen that colonizes the moist squamous epithelium in the anterior nares. It causes both superficial skin infections and more invasive diseases such as endocarditis, osteomyelitis, and septic arthritis. Staphylococcus lugdunensis is a coagulase negative staphylococcus that is a commensal of the human skin. It can occasionally cause serious infections similar to those caused by S. aureus (1).
S. lugdunensis expresses a fibrinogen-binding surface protein (Fbl) with considerable similarity to ClfA (clumping factor A) of S. aureus (2, 3). ClfA is a microbial surface component recognizing adhesive matrix molecules (MSCRAMM)2 that binds to fibrinogen (Fg) and fibrin (4). ClfA promotes bacterial adhesion to immobilized Fg, to blood clots, to ex vivo biomaterial conditioned with plasma proteins, and to sterile thrombi on the heart valves of rabbits and rats in models of endocarditis (5, 6). ClfA is also antiphagocytic and protects bacteria from opsonophagocytosis (7, 8), which might explain its role as a virulence factor in infection models of sepsis and arthritis (9).
The structure and organization of Fbl are very similar to ClfA (2, 3). The N-terminal A domain of ClfA binds to Fg (10). It is composed of three separately folded subdomains: N1, N2, and N3 (11). Subdomains N1 of ClfA and Fbl have 19% amino acid identity, whereas the subdomains N2N3 share almost 60% identity (2). The A domains are linked to the cell wall-anchoring domain by serine-rich repeats (tandem SD repeats in the case of ClfA and SDSDSA repeats for Fbl).
The N2N3 subdomains form the minimum fibrinogen-binding site of ClfA. ClfA binds to a peptide sequence comprising the extreme C terminus of the γ-chain of Fg that protrudes from the D domain (12). The x-ray crystal structure of ClfA N2N3 has been solved both as an apoprotein and in complex with a peptide mimicking the C terminus of the γ-chain (11, 13). The γ-chain peptide binds predominantly in a hydrophobic trench formed between the separately folded N2 and N3 subdomains.
ClfA binds to fibrinogen by a variation of the “dock, lock, and latch” mechanism of ligand binding (13). The dock, lock, and latch mechanism was proposed and validated for the structurally related SdrG from Staphylococcus epidermidis, which binds to the N-terminal β-chain peptide protruding from domain E of fibrinogen (14, 15). The apo-form of the protein adopts an open conformation. Docking of the Fg peptide in the hydrophobic trench between the N2 and N3 subdomains induces a redirection of the C-terminal extension of subdomain N3, resulting in it covering the trench with the bound peptide, thus locking it in place and interacting with the subdomain N2 by making β-strand complementation (14, 15). For SdrG, an open form of the protein is required to bind its ligand. ClfA can bind to the Fg γ-chain even in an artificially constrained closed form where the C-terminal extension of subdomain N3 is held in place in N2 by a disulfide bond (13). Residues at the extreme C terminus of the γ-chain of Fg are crucial for ligand binding because removal of two residues from the C terminus of a 17-mer γ-chain peptide abolished binding to ClfA. The model was supported by substituting several residues in the binding trench of ClfA that were predicted to be involved in ligand binding.
This study describes a detailed comparison of the mechanism of binding to Fg by Fbl and ClfA and concludes that the two staphylococcal proteins are functionally very similar. This striking similarity suggests that new anti-staphylococcal compounds mimicking the C terminus of the γ-chain may be designed to simultaneously target both ClfA and Fbl.
MATERIALS AND METHODS
Bacterial Strains and Growth Conditions
Escherichia coli strain XL-1 Blue (Stratagene) was used as the host for selecting recombinant plasmids following cloning or mutagenesis. E. coli strain TOPP 3 (Stratagene) was used for expression of recombinant proteins and grown in Luria broth supplemented with ampicillin (100 μg/ml) at 37 °C.
Expression and Purification of Recombinant Proteins
Plasmid pKS80::fbl (2) containing the entire coding region of the fbl gene of S. lugdunensis strain N920143 was used as a template for amplification of the region encoding amino acids 206–533 and 40–533, respectively, using primers incorporating BglII and HindIII restriction sites. Plasmid pQE30 (Qiagen) was manipulated to replace the BamHI site of the multiple cloning site with a BglII site. PCR products were cloned into this modified vector allowing N-terminal hexahistidine-tagged proteins to be expressed. Plasmids pCF41 and pCF40 are derivatives of pQE30 containing codons for ClfA amino acids 221–559 and 40–559, respectively (16). Recombinant His-tagged proteins were expressed and purified by Ni2+ chelate chromatography as described previously (16).
Generation of a Three-dimensional Model of rFbl206–533
Sequence alignment using the LALIGN server (17) showed no gaps in the sequences between ClfA and Fbl. A homology model of the Fbl-Fg γ-chain peptide complex in closed conformation was generated using the crystal structure of the ClfA-Fg γ-chain D410A peptide complex (13) as template. The structure of the Fbl N2N3 subdomains (Fbl206–533) was modeled with the HOMOLOGY module of InsightII software (Accrelys Inc.) using the sequence alignment from LALIGN. The wild-type sequence of γ-chain rather than the D410A variant was used for modeling. The resulting model did not show any steric violation between the γ-chain peptide and Fbl. The stereochemical parameters of the model were checked using PROCHECK (18). The figures with ribbon models were generated using RIBBONS (19).
Site-directed Mutagenesis
Site-directed mutagenesis was performed using the QuikChange method (Stratagene). Overlapping complementary primers containing the appropriate base changes (Table 1) were used to amplify the pQE30 plasmid containing the DNA encoding amino acids 206–533 of Fbl. Cycling conditions used were as outlined by the QuikChange protocol. The products were digested with DpnI to eliminate parental DNA and transformed into E. coli XL-1 Blue. The mutations were verified by sequencing (GATC Biotech).
TABLE 1.
Oligonucleotides used in this study
| Name | Sequencea |
|---|---|
| Fbl40–533 forward | GGGAGATCTGAAGAAGTGGAGCGTAATTTG |
| Fbl reverse | GGGAAGCTTATTTGTTGCTTCAACTTTTG |
| Fbl206–533 forward | GAGAGATCTACAGATAATAACGTTACTC |
| N270A forward | CATTAAACTTGCTGGTGTAACAG |
| N270A reverse | CTGTTACACCAGCAAGTTTAATG |
| Y322A forward | CAATTCCTGGGGCTATTGATCCTAAAATG |
| Y322A reverse | CATTTTAGGATCAATAGCCCCAGGATTG |
| W507A forward | GTTTCTATGGCAGCGGATAATGAAG |
| W507A reverse | CTTCATTATCCGCTGCCATAGAAAC |
| E510A forward | CATGGGATAATGCAGTAGAATATC |
| E510A reverse | GATATTCTACTGCATTATCCCATG |
a The restriction sites are in bold type.
Synthesis of Peptides
A synthetic peptide comprising the 17 C-terminal residues (residues 395–411) of the γ-chain of human Fg was synthesized by Genscript (Piscataway, NJ). The human 15-mer and the putative ovine 15-mer were synthesized by Biomatik (Wilmington, DE). Variant human Fg γ-chain peptides with alanine (or serine) substitutions at each position (Table 2) were synthesized as previously described (12) and purified using high pressure liquid chromatography.
TABLE 2.
Synthetic peptides
| Peptide name | Substitution | Sequencea |
|---|---|---|
| Human 17-mer | GEGQQHHLGGAKQAGDV | |
| P1 | G395A | AEGQQHHLGGAKQAGDV |
| P2 | E396A | GAGQQHHLGGAKQAGDV |
| P3 | G397A | GEAQQHHLGGAKQAGDV |
| P4 | Q398A | GEGAQHHLGGAKQAGDV |
| P5 | Q399A | GEGQAHHLGGAKQAGDV |
| P6 | H400A | GEGQQAHLGGAKQAGDV |
| P7 | H401A | GEGQQHALGGAKQAGDV |
| P8 | L402A | GEGQQHHAGGAKQAGDV |
| P9 | G403A | GEGQQHHLAGAKQAGDV |
| P10 | G404A | GEGQQHHLGAAKQAGDV |
| P11 | A405S | GEGQQHHLGGSKQAGDV |
| P12 | K406A | GEGQQHHLGGAAQAGDV |
| P13 | Q407A | GEGQQHHLGGAKAAGDV |
| P14 | A408S | GEGQQHHLGGAKQSGDV |
| P15 | G409A | GEGQQHHLGGAKQAADV |
| P16 | D410A | GEGQQHHLGGAKQAGAV |
| P17 | V411A | GEGQQHHLGGAKQAGDA |
| Ovine 15-mer | GQQHHLGGAKKAGDV | |
| Human 15-mer | GQQHHLGGAKQAGDV |
a Deviations from the human sequence are in underlined.
Inhibition Assays
Recombinant Fbl or ClfA were preincubated with a range of concentrations of a synthetic peptide comprising the 17 C-terminal residues of the γ-chain of Fg or variant peptides with alanine (or serine) substitutions at each position (Table 2) for 1 h at room temperature. A solution of Fg in PBS (10 μg/ml) was used to coat microtiter wells (Sarstedt) for 18 h at 4 °C. The wells were washed three times with PBS and blocked with 5% (w/v) skimmed milk in PBS for 2 h at 37 °C. The wells were washed again, and recombinant proteins were added. The plates were incubated for 1.5 h at 37 °C. Unbound protein was removed by washing with PBS. Bound protein was detected by incubation with anti-His6-peroxidase (Roche Applied Science) at 37 °C for 1 h and detected as described previously (20). The percentage of inhibition was calculated relative to the level of bound protein detected in the absence of inhibitor peptide.
Enzyme-linked Immunosorbent Assay
Recombinant ClfA or Fbl was coated onto microtiter plates (Nunc) in sodium carbonate buffer (pH 9.6) for 18 h at 4 °C. The wells were washed three times with PBS and blocked with 5% (w/v) skimmed milk proteins in PBS for 2 h at 37 °C. The wells were washed again, and varying dilutions of polyclonal rabbit anti-ClfA or anti-Fbl antibodies were added to the wells and incubated for 1 h at room temperature. The wells were washed three times with PBS, and goat anti-rabbit IgG horseradish peroxidase-conjugated antibodies (Dako) were added to the wells and incubated for 1 h. After washing three times with PBS, bound horseradish peroxidase-conjugated antibodies were detected as described previously (20).
Isothermal Titration Calorimetry
ITC was performed with a VP-ITC or ITC200 microcalorimeter (MicroCal Inc., Northampton, MA) stirring at 1000 rpm at 30 °C. In a typical experiment, the cell contained 30 μm rFbl 206–533 or rClfA 221–559, and the syringe contained 0.5–1 mm peptide. Both solutions were in HBS buffer (10 mm HEPES, 150 mm NaCl, pH 7.4). To take into account the heat of dilution, two blank titrations were performed, one by injecting peptide into buffer and the other by injecting buffer into the protein solution. The averaged heats of dilution were subtracted. The data were analyzed using MicroCal Origin software (version 5.0), fitting them to a single site model.
Surface Plasmon Resonance
SPR was performed using the BIAcore X100 system (GE Healthcare). Human Fg purified from contaminating fibronectin, von Willebrand factor, and plasminogen (Enzyme Research Laboratories, South Bend, IN) was covalently immobilized on CM5 sensor chips using amine coupling. This was performed using 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide hydrochloride, followed by N-hydroxysuccinimide and ethanolamine hydrochloride, as described by the manufacturer. Fg (10 μg/ml) was dissolved in 10 mm sodium citrate at pH 5.5 and then immobilized on the flow cell at a flow rate of 30 μl/min in PBS. On another flow cell, the dextran matrix was treated as described above but without Fg present to provide an uncoated reference flow cell. Increasing concentrations of recombinant proteins were flowed over immobilized Fg at a rate of 5 μl/min.
For murine monoclonal antibody 12-9, a capture approach was used. Rabbit anti-mouse Fc IgG (Pierce) was diluted in 10 mm sodium acetate buffer at pH 4.5 and immobilized on two flow cells by amine coupling as described above. The 12-9 murine monoclonal antibody (100 μg/ml) in PBS was passed over the anti-mouse Fc IgG surface of one flow cell, and the other flow cell served as a reference. Recombinant ClfA221–559 (100 nm) and rFbl206–533 (100 nm) were flowed over the surface at a rate of 5 μl/min.
All of the sensorgram data presented were subtracted from the corresponding data from the reference flow cell. The response generated from injection of buffer over the chip was also subtracted from all of the sensorgrams. The data were analyzed using the BIAevaluation software version 3.0. A plot of the level of binding (response units) at equilibrium against concentration of analyte was used to determine the KD.
RESULTS
Measurement of the Dissociation Constant for Fibrinogen by Surface Plasmon Resonance
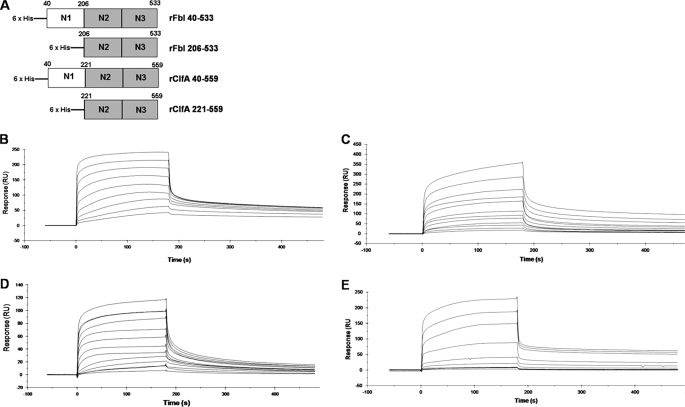
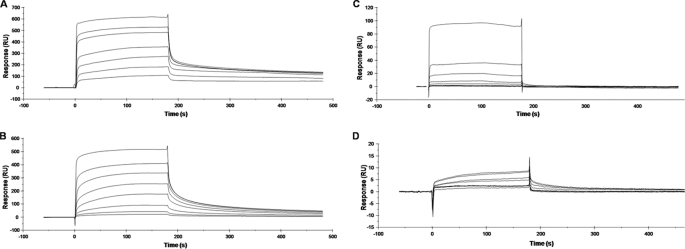
The full-length recombinant Fbl A domain (N1N2N3; rFbl40–533) and the N2N3 subdomains (rFbl206–533) were expressed with N-terminal His6 tags (Fig. 1A). The affinity of these proteins for fibrinogen was measured and compared with recombinant His6-tagged ClfA N1N2N3 domains (rClfA40–559) and ClfA N2N3 domains (rClfA221–559) using SPR (Fig. 1, B and C). Increasing concentrations of recombinant proteins were flowed over a chip that had been coated with Fg. From analysis of the equilibrium binding data, the dissociation constant (KD) of the interaction between rFbl40–533 and Fg was determined to be 0.79 ± 0.09 μm, and for rFbl206–533 and Fg the KD was 0.59 ± 0.1 μm, indicating that these proteins bind to fibrinogen with similar affinities. Similarly, the recombinant His6-tagged ClfA N1N2N3 domains (rClfA40–559) bound to Fg with a KD of 0.8 ± 0.11 μm and ClfA N2N3 domains (rClfA221–559) with a KD of 0.77 ± 0.06 μm. This demonstrates that the Fg binding activities of both Fbl and ClfA lie solely in the N2N3 subdomains and that the N1 subdomains have no discernable role. Furthermore, ClfA and Fbl have very similar affinities for Fg. Similar results were obtained in solid phase fibrinogen binding assays (data not shown).
FIGURE 1.
Surface plasmon resonance analysis of recombinant proteins binding to fibrinogen. Recombinant truncates of ClfA or Fbl (A) were compared for binding to fibrinogen immobilized on the surface of a CM5 sensor chip. Sensorgrams of the binding to fibrinogen were obtained by passing increasing concentrations of rFbl40–533 (15.625–4000 nm) (B), rFbl206–533 (25–6400 nm) (C), rClfA40–559 (10–5120 nm) (D), or rClfA221–559 (5–5120 nm) (E) over the surface. Injections began at 0 s and ended at 180 s. The results shown are representative of three independent experiments performed on at least two different fibrinogen-coated sensor chips.
Isothermal Titration Calorimetry
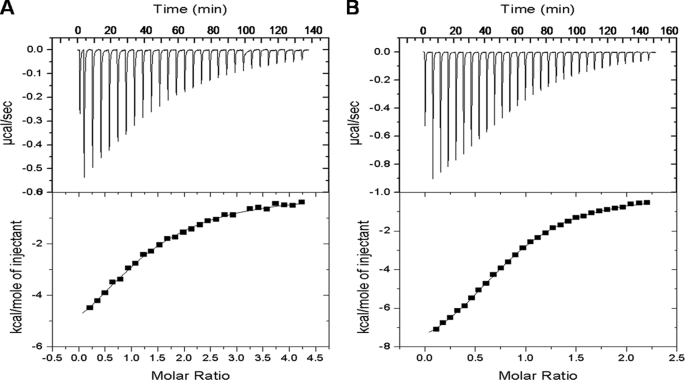
ClfA binds to the extreme C terminus of the γ-chain of Fg (12). Because recombinant ClfA could block binding of Fbl to Fg (2) and given the high level of similarity between ClfA and Fbl binding domains, it seemed likely that the two proteins would bind to the same region of Fg. Thus the affinities of rClfA221–559 and rFbl206–533 for a synthetic peptide corresponding to the C-terminal 17 amino acids of the γ-chain were measured by ITC. Recombinant Fbl206–533 bound with a KD of 9 ± 0.6 μm (Fig. 2A), whereas rClfA221–559 had a KD of 15 ± 2 μm (Fig. 2B). These experiments showed that Fbl bound to the C terminus of the Fg γ-chain and that the affinity of Fbl for the peptide is very similar to that of ClfA.
FIGURE 2.
Isothermal titration calorimetry analysis of binding of rFbl206–533 and rClfA221–559 to the fibrinogen γ-chain peptide. The upper panels show enthalpic heat released/second at 30 °C during titration of peptide into the cell containing rFbl206–533 (A) or rClfA221–559 (B). The lower panels show integrated binding isotherms of the titration and the best fit to a single-site model. The best fit yielded parameters as follows: for Fbl, the dissociation constant KD = 15.0 × 10−6 m, enthalpy ΔH = −7.7 kcal/mol, and molar binding stoichiometry n = 1.26; for ClfA, the dissociation constant KD = 9.1 × 10−6 m, enthalpy ΔH = −3.7 kcal/mol, and molar binding stoichiometry n = 1.2.
Inhibition of Binding
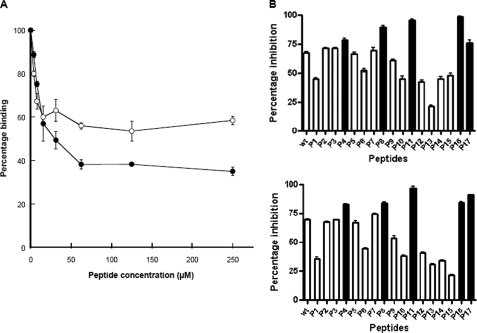
To further investigate the interaction between Fbl and the C-terminal residues of Fg, the 17-mer γ-chain peptide was tested for its ability to inhibit the binding of rFbl206–533 to immobilized Fg. This peptide was previously shown to inhibit recombinant ClfA and FnBPA A domain binding to Fg (21). Preincubation of rFbl206–533 or rClfA221–559 with increasing concentrations of peptide resulted in dose-dependent inhibition of binding (Fig. 3A). This confirms that the binding site for Fbl in Fg is the C-terminal residues of the γ-chain. The maximum inhibition achieved by the γ-chain peptide was ∼50% for Fbl and ∼60% for ClfA.
FIGURE 3.
Inhibition of rFbl206–533 and rClfA221–559 binding to immobilized fibrinogen by γ-chain peptide. Recombinant Fbl206–533 (100 nm) (○) and rClfA221–559 (100 nm) (●) were incubated with increasing concentrations of the Fg γ-chain peptide for 1 h before being added to the wells of microtiter plates coated with Fg (A). Alternatively, rFbl206–533 (60 nm) (B, upper panel) or rClfA221–559 (100 nm) (B, lower panel) was incubated with γ-chain peptide variants P1–P17 (Table 2) for 1 h before being added to Fg-coated plates. Bound proteins were detected using anti-His6-peroxidase antibody. The values are expressed as percentages of control wells lacking inhibitor peptide. The filled boxes (B) represent peptides showing increased inhibition compared with wild-type (wt) peptide. The results shown are the mean values of triplicate samples. These experiments were performed three times with similar results.
Inhibition of rFbl206–533 and rClfA221–559 binding to Fg was measured using γ-chain peptides with single alanine (or serine) substitutions in each of the terminal 17 residues (13). The variant peptides had the same effect on rFbl206–533 and rClfA221–559 binding to Fg. Peptides with G395A, H400A, G403A, G404A, K406A, Q407A, A408S, and G409A substitutions had reduced ability to inhibit both ClfA and Fbl compared with the native peptide (Fig. 3B). Variants Q398A, L402A, A405S, D410A, and V411A had an increased inhibitory effect on both ClfA and Fbl binding to Fg. Amino acid substitutions that increase or reduce inhibition by the peptide presumably alter the affinity of the peptide for ClfA and Fbl, indicating that these residues are important in the interaction with the MSCRAMM. Substitutions E396A, G397A, Q399A, and H401A had little effect on the inhibition of either protein binding to Fg. This indicates that the interaction of ClfA and Fbl with the C-terminal residues of the γ-chain of Fg is very similar.
Binding to Fibrinogen from Different Species
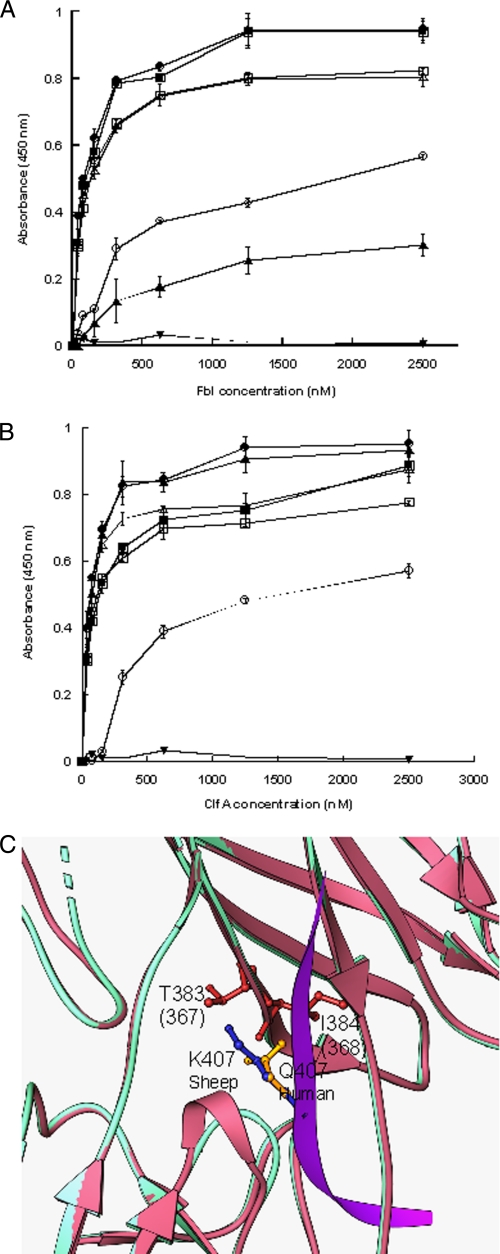
The ability of rClfA and rFbl to bind Fg from different species was compared. Differences in the C-terminal γ-chain residues might influence the affinity of ClfA and Fbl and could be informative. Microtiter plates were coated with human, bovine, ovine, murine, feline, canine, and porcine Fg. Both ClfA and Fbl bound to canine, murine, and porcine Fg at similar levels to human Fg (Fig. 4). In contrast, both ClfA and Fbl bound weakly to bovine Fg and did not bind to ovine Fg. The one difference between the two MSCRAMMs was the weak binding of Fbl to feline Fg compared with ClfA, which bound with the same avidity as it did to human Fg (Fig. 4). Thus Fbl and ClfA behave almost identically with the exception of the binding to feline Fg.
FIGURE 4.
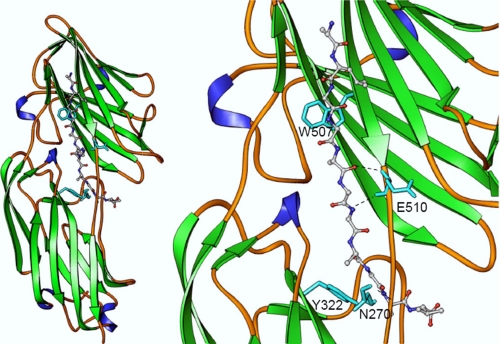
Binding of rClfA221–559 and rFbl206–533 to fibrinogen from different species. Microtiter plates were coated with human (●), bovine (○), murine (■), canine (○), feline (▴), porcine (▵), or ovine (▾) fibrinogen. Increasing concentrations of rFbl206–533 (A) or rClfA221–559 (B) were added. Bound protein was detected using anti-His6-Peroxidase antibody. The results shown are the mean values of triplicate samples. The error bars show the standard deviation. The results are representative of three independent experiments. C, ribbon representation of a homology model of Fbl N2N3 (pink) overlaid on to the crystal structure of the ClfA (cyan)-γ-chain peptide (magenta) complex. Side chain atoms of glutamine (yellow) from the human sequence (crystal structure) and lysine (blue) from the ovine sequence (model) at position 407 are shown as ball and stick objects. Thr383 and backbone atoms of Ile384 that could make severe steric clashes with lysine are shown in red. Residue numbers corresponding to Thr383 and Ile384 of Fbl are shown in parentheses.
The ovine Fg γ-chain is not annotated in the public databases. A bioinformatic search was performed for ovine expressed sequence tag clones using BLAST (tblastn module) (22). The sequence of the C-terminal residues of the human γ-chain (residues 351–411) was used as template. This generated a liver cDNA sequence (GenBankTM accession number gi114476568) with a matching stop codon and 90% identity to the human C-terminal sequence. When the sequences were aligned, the putative ovine sequence had an apparent gap at human position 395 and substitutions E396D and Q407K. The crystal structure of the ClfA-Fg γ-chain complex (13) showed that Gln407 of Fg makes key interactions with ClfA and is completely buried in the complex. Furthermore, the alanine scan of γ-chain peptide (Fig. 3) showed that substitution of a bulky glutamine residue with alanine (Q407A) reduced binding to ClfA or Fbl. To determine whether the Q407K variation is responsible for the inability of ClfA and Fbl to bind ovine Fg, a peptide corresponding to the C-terminal 15 residues of the predicted ovine Fg γ-chain was compared with the equivalent human Fg 15-mer (Table 2). The human peptide bound to ClfA with a KD of 44 ± 8.6 μm and to Fbl with a KD of 40 ± 6.9 μm (data not shown), but the 15-mer ovine peptide did not bind detectably to either MSCRAMM (data not shown). This suggests that the failure of Fbl and ClfA to bind to the ovine γ-chain may be due to the Q407K substitution in the C terminus of the ovine Fg γ-chain.
To examine this in more detail, structural models of both Fbl and ClfA in complex with the Q407K variant peptide were generated. The crystal structure of the ClfA-Fg γ-chain peptide complex (13) was used as template. Visual examination of the model suggests that the bulky Lys407 residue (human Fg numbering) of the ovine Fg sequence clashes with Thr383 and the backbone atoms of Ile384 of ClfA (Fig. 4C), preventing the ovine sequence from fitting into the binding trench and abolishing the interaction with ClfA. In addition, changing a polar uncharged glutamine to a positively charged lysine could impose charge-charge repulsion with any of the surrounding positively charged residues such as His252 or Lys381. Thus the loss of binding of ClfA and Fbl to ovine Fg may be due to the Q407K substitution in the C terminus of the ovine Fg γ-chain.
Amino Acid Substitution Mutants of Fbl
The homology model of the Fbl-Fg γ-chain peptide complex (based on the structure of the ClfA-Fg γ-chain D410A peptide complex) (13) was used to identify residues in the putative ligand-binding trench of Fbl that might be involved in Fg binding (Fig. 5). Residues lining the trench region located between N2 and N3 are completely conserved between ClfA and Fbl in contrast to residues located elsewhere where 40% divergence occurs. Amino acid substitutions were made in four residues in the putative ligand-binding trench of Fbl that could possibly interact with the γ-chain of Fg (Asn270, Tyr322, Trp507, and Glu510; Fig. 5, right panel). The same residues are located at equivalent positions (Asn286, Try338, Trp523, and Glu526) in ClfA. The crystal structure of the ClfA-Fg γ-chain complex demonstrates a direct interaction between the side chains of residues Tyr338 and Trp523 and the Fg peptide (13). The backbone atoms of the residues Glu526 and Asn286 of ClfA play a role in anchoring the peptide through hydrogen bonding interactions (13). Previous studies have demonstrated the importance of residues Tyr338 and Glu526 in fibrinogen binding by ClfA (11, 23, 24).
FIGURE 5.
Structural model of Fbl N2N3 subdomains in complex with γ-chain peptide. The fibrinogen-derived peptide is shown as a ball and stick object colored by atom type (carbon, gray; nitrogen, blue; oxygen, red). Residues Asn270, Tyr322, Trp507, and Glu510 are shown as stick objects in cyan. As in the case of ClfA, the conserved residues Tyr322 and Trp507 could help Fbl anchor the peptide through stacking interaction. Side chain atoms of Lys406 and Gln407 are not shown in the close-up view of the figure (right panel) for clarity. The backbone atoms of Asn270 and Glu510 of Fbl interact through hydrogen bonding interactions, which are shown as dotted lines.
The affinity of each of the rFbl mutants for Fg was determined by SPR (Fig. 6). Analysis of the equilibrium binding data showed that rFbl N270A and rFbl E510A had slightly reduced affinity for Fg compared with rFbl206–533 (KD of 1.17 ± 0.29 and 1.74 ± 0.17 μm, respectively compared with 0.59 ± 0.11 μm for wild type). Amino acids in the loop regions around the ligand-binding trench are critical for optimum binding even when their side chains are not directly involved. The P336A substitution near the binding site in ClfA reduced Fg binding (25) but did not participate directly in the interaction (13). Residue Asn270 in Fbl is located in a loop in the N2 domain (Fig. 5), and the N270A substitution at the binding site could alter the conformation around the residue and also affect the interaction through the main chain atoms.
FIGURE 6.
Surface plasmon resonance of rFbl206–533 mutants binding to fibrinogen. Sensorgrams of binding to fibrinogen were obtained by passing increasing concentrations of rFbl N270A (0.1–25.6 μm) (A), rFbl E510A 206–533 (0.1–25.6 μm) (B), rFbl W507A (0.03125–16 μm) (C), or rFbl Y322A (0.25–16 μm) (D) over the surface. The injections began at 0 s and ended at 180 s. The results shown are representative of two independent experiments on two different fibrinogen-coated sensor chips.
The E526A substitution is at the linker region before the latch and could affect optimal redirection of the latch segment and backbone hydrogen bonding interaction (13). The rFbl W507A and rFbl Y322A mutants had much weaker affinity for Fg. Recombinant Fbl W507A was estimated to have a KD of ∼28.65 μm for fibrinogen (Fig. 6C), whereas the response for rFbl Y322A was so low that the KD could not be determined with any reliability (Fig. 6D). Thus Fbl residues Tyr322 and Trp507 appear to play an important role in Fg binding by Fbl.
Cross-reactivity of Polyclonal Anti-ClfA and Anti-Fbl Antibodies
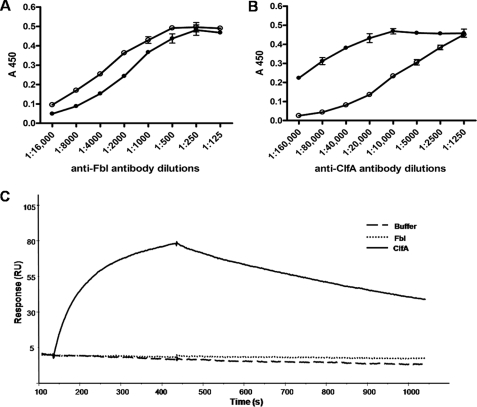
Comparison of the x-ray crystal structure of ClfA with the modeled structure of Fbl indicated that the majority of variant residues are located on the surfaces of the proteins (data not shown). The cross-reactivity of rabbit polyclonal anti-ClfA and anti-Fbl antibodies was measured. Anti-ClfA A domain antibodies had a 32-fold lower titer for rFbl206–533, whereas polyclonal anti-Fbl A domain antibodies had a 2-fold lower titer for rClfA221–559 (Fig. 7). Both recombinant proteins reacted equally with monoclonal anti-His6-peroxidase antibody (data not shown), indicating that they coated the dish similarly. In addition, an important function-blocking anti-ClfA monoclonal antibody 12-9 (26) that binds with high affinity to ClfA did not bind detectably to Fbl when analyzed by SPR (Fig. 7C) or enzyme-linked immunosorbent assay (data not shown).
FIGURE 7.
Cross-reactivity of anti-Fbl region A and anti-ClfA region A antibodies. Microtiter wells were coated with rClfA221–559 (250 nm) (●) or rFbl206–533 (250 nm) (○). Increasing dilutions of polyclonal rabbit anti-Fbl region A antibodies (A) or polyclonal rabbit anti-ClfA region A (B) antibodies were added to the wells. Bound antibody was detected using peroxidase-conjugated goat anti-rabbit IgG antibody. The results shown are the mean values of triplicate samples. The error bars show the standard deviation. For surface plasmon resonance analysis of rClfA221–559 and rFbl206–533 binding to murine monoclonal antibody 12-9, the antibody was captured on the surface of a CM5 sensor chip by a rabbit anti-mouse Fc antibody. The same concentration (100 nm) of rClfA221–559 and rFbl206–533 were passed over the surface (C). Injections started at 130 s and ended at 430 s.
DISCUSSION
S. lugdunensis expresses a fibrinogen-binding surface protein called Fbl that is similar in structure and organization to ClfA of S. aureus. In particular the minimum ligand-binding region of ClfA, subdomains N2 and N3, is 60% identical to the same region of Fbl. This study set out to compare Fbl and ClfA to gain further insight into their mechanism of binding to Fg.
We have shown using SPR that the affinities of ClfA and Fbl for human fibrinogen are very similar. This contradicts an earlier report suggesting that Fbl had a lower affinity for Fg than ClfA (2). This can be attributed to the use of polyclonal antibodies to measure Fbl and ClfA binding to immobilized Fg in a solid phase binding assay in the earlier study. The polyclonal antibodies have different affinities for the two antigens and are likely to contain some antibodies with the ability to displace the bound MSCRAMM.
The data reported in this paper directly demonstrate that Fbl binds to the C terminus of the γ-chain of Fg in a very similar fashion to ClfA. The proteins bound to the synthetic 17-mer γ-chain peptide. The KD determined here for rClfA221–559 binding to γ-chain by ITC (15 μm) is similar to the KD previously determined for rClfA229–550 binding to γ-chain peptide by ITC (5.8 μm) (13) and rClfA221–559 binding to fluorescein-labeled γ-chain peptide by fluorescence polarization (15 μm (16) and 8 μm (11)).
A synthetic γ-chain peptide inhibited the binding of Fbl to Fg by ∼50%. Previous reports have noted incomplete inhibition of ClfA by this peptide (60–70%) (12, 21). In contrast binding to Fg by the recombinant A domain of fibronectin-binding protein A was completely inhibited by the peptide (21). Wann et al. (21) suggested that ClfA recognizes an additional site in Fg, whereas FnBPA binds only to the γ-chain.
Two different approaches were taken to probing the interaction of Fbl and ClfA with Fg. First, a set of synthetic 17-mer peptides with alanine (or serine) substitutions in each of the 17 positions at the C terminus of the Fg γ-chain were used to compare inhibitory effects on rClfA221–559 and rFbl206–533 binding to immobilized Fg. Each of the peptides had the same effect on ligand binding by the two MSCRAMMs, suggesting that the mechanism of binding of ClfA and Fbl to Fg is very similar. Some peptides inhibited binding of both proteins more weakly than the parental peptide, which could indicate that the residue replaced is directly involved in binding. Several peptides inhibited binding more strongly than the parental peptide, indicating that they have a higher affinity for the MSCRAMM.
Information about the mechanism of ligand binding was also obtained by measuring the ability of ClfA and Fbl to bind to Fg purified from different species. This was particularly informative where the sequences of the C termini of the γ-chains are known.
ClfA and Fbl did not bind detectably to ovine Fg. The ovine Fg γ-chain sequence is not annotated, but because the putative ovine γ-chain 15-mer with a single Q407K substitution compared with the human sequence lost binding to both MSCRAMMs, this result may explain the lack of binding to full-length ovine Fg. It will be of particular interest to see whether ClfA from ovine strains bind to ovine Fg more strongly than ClfA from human strains. Fbl bound weakly to feline Fg compared with ClfA, which bound with the same affinity as to human Fg. This indicates a subtle difference between the two MSCRAMMs, but unfortunately a molecular explanation is lacking because the amino acid sequence of the γ-chain of feline Fg is not known. The reduced binding of ClfA and Fbl to Fg from certain animals could represent adaptation to the human host.
The three-dimensional structure of the apo-protein and ligand bound form of ClfA has been solved. It is possible to model the structure of Fbl based on its amino acid sequence similarity and structural similarity to ClfA. Amino acid residues in the ligand-binding trench are completely conserved in contrast to residues located elsewhere on the protein. Four residues in the putative ligand-binding trench of Fbl were substituted, and the variant proteins showed a reduction in ligand binding by SPR. This confirms that Fbl and ClfA bind to Fg similarly and that the Fbl ligand binding site is the trench located between subdomains N2 and N3. Solving the crystal structures of Fbl in complex with γ-chain peptide will provide further insight into the mechanism of Fg binding by Fbl. Overall it appears that the ClfA and Fbl interaction with Fg is very similar. This similarity may be used to design new anti-staphylococcal compounds mimicking the Fg γ-chain C terminus that would have an effect against both S. aureus and S. lugdunensis.
Acknowledgment
We thank Dr. A. Khan (School of Biochemistry and Immunology, Trinity College, Dublin, Ireland) for use of the ITC200 calorimeter.
This work was supported, in whole or in part, by National Institutes of Health Grant AI20624 (to M. H.). This work was also supported by a Health Research Board of Ireland project grant and a Science Foundation Ireland Programme Investigator grant (to T. J. F.).
- MSCRAMM
- microbial surface component recognizing adhesive matrix molecules
- Fg
- fibrinogen
- ITC
- isothermal titration calorimetry
- PBS
- phosphate-buffered saline.
REFERENCES
- 1.Frank K. L., Del Pozo J. L., Patel R. (2008) Clin. Microbiol. Rev. 21, 111–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mitchell J., Tristan A., Foster T. J. (2004) Microbiology 150, 3831–3841 [DOI] [PubMed] [Google Scholar]
- 3.Nilsson M., Bjerketorp J., Guss B., Frykberg L. (2004) FEMS Microbiol. Lett. 241, 87–93 [DOI] [PubMed] [Google Scholar]
- 4.McDevitt D., Francois P., Vaudaux P., Foster T. J. (1994) Mol. Microbiol. 11, 237–248 [DOI] [PubMed] [Google Scholar]
- 5.Moreillon P., Entenza J. M., Francioli P., McDevitt D., Foster T. J., François P., Vaudaux P. (1995) Infect. Immun. 63, 4738–4743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Que Y. A., Haefliger J. A., Piroth L., François P., Widmer E., Entenza J. M., Sinha B., Herrmann M., Francioli P., Vaudaux P., Moreillon P. (2005) J. Exp. Med. 201, 1627–1635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hair P. S., Ward M. D., Semmes O. J., Foster T. J., Cunnion K. M. (2008) J. Infect Dis. 198, 125–133 [DOI] [PubMed] [Google Scholar]
- 8.Higgins J., Loughman A., van Kessel K. P., van Strijp J. A., Foster T. J. (2006) FEMS Microbiol. Lett. 258, 290–296 [DOI] [PubMed] [Google Scholar]
- 9.Josefsson E., Hartford O., O'Brien L., Patti J. M., Foster T. (2001) J. Infect. Dis. 184, 1572–1580 [DOI] [PubMed] [Google Scholar]
- 10.McDevitt D., Francois P., Vaudaux P., Foster T. J. (1995) Mol. Microbiol. 16, 895–907 [DOI] [PubMed] [Google Scholar]
- 11.Deivanayagam C. C., Wann E. R., Chen W., Carson M., Rajashankar K. R., Höök M., Narayana S. V. (2002) EMBO J. 21, 6660–6672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McDevitt D., Nanavaty T., House-Pompeo K., Bell E., Turner N., McIntire L., Foster T., Höök M. (1997) Eur. J. Biochem. 247, 416–424 [DOI] [PubMed] [Google Scholar]
- 13.Ganesh V. K., Rivera J. J., Smeds E., Ko Y. P., Bowden M. G., Wann E. R., Gurusiddappa S., Fitzgerald J. R., Höök M. (2008) PLoS Pathog. 4, e1000226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ponnuraj K., Bowden M. G., Davis S., Gurusiddappa S., Moore D., Choe D., Xu Y., Hook M., Narayana S. V. (2003) Cell 115, 217–228 [DOI] [PubMed] [Google Scholar]
- 15.Bowden M. G., Heuck A. P., Ponnuraj K., Kolosova E., Choe D., Gurusiddappa S., Narayana S. V., Johnson A. E., Höök M. (2008) J. Biol. Chem. 283, 638–647 [DOI] [PubMed] [Google Scholar]
- 16.O'Connell D. P., Nanavaty T., McDevitt D., Gurusiddappa S., Höök M., Foster T. J. (1998) J. Biol. Chem. 273, 6821–6829 [DOI] [PubMed] [Google Scholar]
- 17.Huang X. M., Miller W. (1991) Adv. Appl. Math 12, 337–357 [Google Scholar]
- 18.Laskowski R. A., Moss D. S., Thornton J. M. (1993) J. Mol. Biol. 231, 1049–1067 [DOI] [PubMed] [Google Scholar]
- 19.Carson M. (1997) J. Mol. Graph. 5, 103–106 [Google Scholar]
- 20.Keane F. M., Loughman A., Valtulina V., Brennan M., Speziale P., Foster T. J. (2007) Mol. Microbiol. 63, 711–723 [DOI] [PubMed] [Google Scholar]
- 21.Wann E. R., Gurusiddappa S., Hook M. (2000) J. Biol. Chem. 275, 13863–13871 [DOI] [PubMed] [Google Scholar]
- 22.Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. (1990) J. Mol. Biol. 215, 403–410 [DOI] [PubMed] [Google Scholar]
- 23.Deivanayagam C. C., Perkins S., Danthuluri S., Owens R. T., Bice T., Nanavathy T., Foster T. J., Höök M., Narayana S. V. (1999) Acta Crystallogr. D Biol. Crystallogr. 55, 554–556 [DOI] [PubMed] [Google Scholar]
- 24.Hartford O. M., Wann E. R., Höök M., Foster T. J. (2001) J. Biol. Chem. 276, 2466–2473 [DOI] [PubMed] [Google Scholar]
- 25.Loughman A., Fitzgerald J. R., Brennan M. P., Higgins J., Downer R., Cox D., Foster T. J. (2005) Mol. Microbiol. 57, 804–818 [DOI] [PubMed] [Google Scholar]
- 26.Hall A. E., Domanski P. J., Patel P. R., Vernachio J. H., Syribeys P. J., Gorovits E. L., Johnson M. A., Ross J. M., Hutchins J. T., Patti J. M. (2003) Infect. Immun. 71, 6864–6870 [DOI] [PMC free article] [PubMed] [Google Scholar]