Abstract
Maize eukaryotic translation initiation factor 5A (ZmeIF5A) co-purifies with the catalytic α subunit of protein kinase CK2 and is phosphorylated by this enzyme. Phosphorylated ZmeIF5A was also identified after separation of maize leaf proteins by two-dimensional electrophoresis. Multiple sequence alignment of eIF5A proteins showed that in monocots, in contrast to other eukaryotes, there are two serine/threonine residues that could potentially be phosphorylated by CK2. To identify the phosphorylation site(s) of ZmeIF5A, the serine residues potentially phosphorylated by CK2 were mutated. ZmeIF5A and its mutated variants S2A and S4A were expressed in Escherichia coli and purified. Of these recombinant proteins, only ZmeIF5A-S2A was not phosphorylated by maize CK2. Also, Arabidopsis thaliana and Saccharomyces cerevisiae eIF5A-S2A mutants were not phosphorylated despite effective phosphorylation of wild-type variants. A newly developed method exploiting the specificity of thrombin cleavage was used to confirm that Ser2 in ZmeIF5A is indeed phosphorylated. To find a role of the Ser2 phosphorylation, ZmeIF5A and its variants mutated at Ser2 (S2A and S2D) were transiently expressed in maize protoplasts. The expressed fluorescence labeled proteins were visualized by confocal microscopy. Although wild-type ZmeIF5A and its S2A variant were distributed evenly between the nucleus and cytoplasm, the variant with Ser2 replaced by aspartic acid, which mimics a phosphorylated serine, was sequestered in the nucleus. These results suggests that phosphorylation of Ser2 plays a role in regulation of nucleocytoplasmic shuttling of eIF5A in plant cells.
Keywords: Enzymes/Kinase, Phosphorylation, Protein/Motifs, Protein/Nuclear Translocation, Signal Transduction/Protein Kinases, Translation/Initiation Factors, Protein kinase CK2, Zea mays
Introduction
Protein kinase CK22 (formerly known as casein kinase 2) is a ubiquitously expressed serine/threonine protein kinase present in all eukaryotes that displays extraordinary evolutionary conservation (for review, see Ref. 1). In contrast to most other protein kinases, CK2 is constitutively active, not regulated by a second messenger, and can use both ATP and GTP as phosphate donors. CK2 is most often present as a tetramer composed of two catalytic α and two regulatory β subunits (for review, see Ref. 2). CK2 is a pleiotropic kinase with a growing list of more than 300 substrates (for review, see Ref. 3). A characteristic feature of CK2 phosphorylation sites are multiple acidic residues located mostly downstream of the phosphorylatable amino acid (4). The striking variety of its cellular targets and its absolute requirement for cell viability suggest that CK2 plays essential role(s); it has been implicated in many cellular functions with special reference to proliferation, gene expression, and RNA and protein synthesis (for review, see Ref. 3).
Studies concerning functions of plant CK2s have shown that this enzyme is involved in light-regulated gene expression and growth control (5), cell division, and cell expansion (6–8). CK2 phosphorylates transcription factors (G-box-binding and bZiP factors) involved in photomorphogenesis (9–11). Opaque2, a basic leucine zipper transcriptional activator that binds to the G-box of promoters, is phosphorylated by CK2 during maize seed development. Phosphorylation of Opaque2 is light-dependent and is under diurnal control (12). Also transcription factors CCA1 (circadian clock-associated 1) and LHY (late elongated hypocotyl) are phosphorylated by CK2 and are essential for regulation of the endogenous circadian clock in Arabidopsis (13). Only phosphorylated circadian clock-associated 1 is able to bind promoters and regulate transcription of target genes in the nucleus, whereas the unphosphorylated protein is sequestered in the cytoplasm (14).
The stability of transcription factor HY5 (long hypocotyl 5), which is involved in auxin signaling and photomorphogenesis (15), is regulated both by interaction with a negative regulator of photomorphogenesis (COP1) and phosphorylation by CK2 (9). It has been demonstrated that the maize abscisic acid responsive protein Rab17, a late embryogenesis abundant protein involved in plant responses to stress, is highly phosphorylated by CK2. Rab17 specifically interacts with CK2β (16). In Zea mays, three cDNA clones encoding CK2β and three encoding CK2α (ZmCK2α-1, ZmCK2α-2, and ZmCK2α-3) have been isolated and characterized (17–19), which indicates that in plants several different tetrameric forms of CK2 may be present. Also, enzymatically active monomeric forms of CK2 comprising the catalytic α subunits have been identified in maize (20, 21). A group of low molecular weight proteins were found to co-purify with the monomeric form of CK2. Those proteins migrating at 17–19 kDa were highly phosphorylated by CK2 (22). The present work was undertaken to search for and characterize new substrate(s) of maize CK2 and investigate the role of their phosphorylation.
EXPERIMENTAL PROCEDURES
Materials
Staurosporine, heparin, phosphocellulose, and the HSRT 100 kit were purchased from Sigma. Glutathione-agarose beads, source 15Q, Sephadex G-100, Superdex G-200 10/300 GL column for fast protein liquid chromatography, and pGEX-4T-1 expression vector were obtained from Amersham Biosciences. TOPO® vector was from Invitrogen. Tri® Reagent was obtained from MRC, Cincinnati, OH. Antibodies against ZmeIF5A were purchased from BioGenes. All other chemicals were of the highest grade available.
Plant Growth
Maize seeds (Z. mays cv. Mona), after overnight soaking in water at room temperature, were grown for 72 h at 26 °C on wet paper in the dark. Etiolated apical parts of the seedlings were harvested, immediately frozen in liquid nitrogen, and stored at −80 °C. For leaves, maize plants were cultivated hydroponically for 2 weeks in a growth chamber with a daily cycle of 16 h light at 25 °C and 8 h dark at 20 °C.
Seeds of Arabidopsis thaliana (ecotype Columbia) were sown on soil and maintained at 4 °C for 3 days, and then transferred to a growth chamber with a daily cycle of 16 h light at 23 °C and 8 h dark at 21 °C. Rosette leaves of 2-week-old plants were harvested, frozen in liquid nitrogen, and stored at −80 °C. Saccharomyces cerevisiae strain W303 was grown overnight at 30 °C in YPD medium.
Purification of Native CK2 from Maize Seedlings
All purification steps were performed at 4 °C. Frozen maize seedlings (40 g) were ground with liquid nitrogen in a mortar. Proteins were extracted by homogenization for 15 min with 80 ml of Buffer 1 (50 mm Tris-HCl (pH 7.8), 6 mm MgCl2, 10 mm β-mercaptoethanol, 0.1 mm PMSF, 250 mm (NH4)2SO4). After sonication (3 times for 20 s), the suspension was centrifuged at 10,000 × g for 10 min and filtered through glass wool. Proteins were precipitated from the supernatant with (NH4)2SO4 (up to 60% of saturation). After 90 min of gentle stirring, the suspension was centrifuged at 12,000 × g for 45 min. The supernatant was discarded and the collected precipitate was dissolved in Buffer 2 (50 mm Tris-HCl (pH 7.8), 6 mm MgCl2, 10 mm β-mercaptoethanol, 0.1 mm PMSF, 10% (v/v) ethylene glycol) to conductivity of the same buffer containing 40 mm (NH4)2SO4. After equilibration, 120 ml of phosphocellulose resin (pre-equilibrated with 10 volumes of Buffer 2 containing 40 mm (NH4)2SO4) was added. Binding was carried out for 1 h with constant stirring. Then, the phosphocellulose resin with bound proteins was washed with 1,200 ml (3 × 400 ml) of Buffer 2 containing 0.1 m NaCl. The beads were packed into a column and a linear gradient of 0.1–1 m NaCl (2 × 150 ml) in Buffer 2 was applied. Fractions containing CK2 activity, a total volume of 80 ml, were collected, concentrated by Amicon ultrafiltration to 20 ml (Amicon Ultra, Millipore), and dialyzed against Buffer 3 (50 mm Tris-HCl (pH 7.8), 10 mm β-mercaptoethanol, 0.1 mm PMSF, 10% (v/v) ethylene glycol), 3 × 1 liter for 15 h. The obtained sample was loaded on a 5-ml Source 15Q column pre-equilibrated with 10 volumes of Buffer 3 and the column was washed with 50 ml of Buffer 3. A linear gradient of 0–0.6 m NaCl (2 × 30 ml) in Buffer 3 was applied for enzyme elution. Active fractions were collected (30 ml), concentrated by Amicon ultrafiltration to 1 ml, adjusted to a final salt concentration of 0.5 m NaCl, loaded on a Sephadex G-100 column (1.7 × 86.5 cm), and eluted with Buffer 4 (100 mm Tris-HCl (pH 7.5), 0.5 m NaCl, 0.01% Brij 35, 0.05 mm PMSF); 1-ml fractions were collected. Two peaks with CK2 activity (named I and II, 8.0 ml each) were collected separately, concentrated 8-fold by Amicon ultrafiltration, and dialyzed against Buffer 5 (50 mm Tris-HCl (pH 7.5), 0.2 m NaCl, 0.05 mm PMSF) for 4 h (2 × 1 liter). Glycerol was added to the final concentration of 10% to the enzyme preparations and the samples were frozen in liquid nitrogen.
Separation of Maize Leaf Proteins by Two-dimensional Gel Electrophoresis and Identification of Phosphoproteins
Total protein was prepared from 2-week-old maize leaves according to a modified phenol-based procedure as described previously (23). The extracted proteins were precipitated with 100% methanol containing 0.1 m ammonium acetate, and after centrifugation, the obtained pellet was dissolved in isoelectrofocusing extraction buffer (8 m urea, 2 m thiourea, 2% (w/v) CHAPS, 2% (v/v) Triton X-100, 50 mm dithiothreitol). For the first dimension, the sample was loaded on a 13-cm strip (pH 3–10) and focused on a Multiphor II apparatus (Amersham Biosciences). For the second dimension, the strip was equilibrated, placed on an SDS-polyacrylamide gel, and run as described previously (23). For phosphoprotein detection, the gel was stained with Pro-Q® Diamond phosphoprotein gel stain according to Ref. 24 and imaged using a Molecular Imager PharosFX system (Bio-Rad). For detection of all proteins the gel was stained with colloidal Coomassie Brilliant Blue G-250. Phosphoprotein spots were cut out from the gel and analyzed by mass spectrometry (MS).
Mass Spectrometry
Protein bands or spots were excised from the SDS-polyacrylamide gel, reduced, alkylated, digested overnight with trypsin, and then analyzed by tandem mass spectrometry using a Q-TOF 1 mass spectrometer (Micromass, Manchester, UK) combined with a nano-high pressure liquid chromatography system (LC Packings, Amsterdam, The Netherlands). The MaxEnt3 program (Micromass) was used to identify the obtained peptide sequences.
Immunoblot Analysis
Proteins were separated on 12% SDS-polyacrylamide gels and transferred to polyvinylidene difluoride membranes (ImmobilonTM P, Millipore) overnight at 15 V using 25 mm Tris-HCl, 192 mm glycine. The membranes were blocked for 2 h at room temperature in TBST buffer (10 mm Tris-HCl (pH 7.5), 100 mm NaCl, 0.1% Tween 20) containing 5% dry milk, and incubated for 1.5 h in TBST buffer with antibody against eIF5A at a 1:5000 dilution. Anti-eIF5A antibodies were raised against a specific peptide (LTSDGNTKDDLRLPT). After removing the unbound antibodies by washing (5 times for 5 min each) with TBST buffer, the blots were incubated with alkaline phosphatase-conjugated anti-rabbit antibodies at a 1:2000 dilution. After washing (as above) with TBST buffer, immunoreactive proteins were visualized by reaction with 0.3 mg/ml of nitro blue tetrazolium and 0.16 mg/ml of 5-bromo-4-chloro-3-indolyl phosphate in 100 mm Tris-HCl (pH 9.5), containing 100 mm NaCl and 5 mm MgCl2.
Cloning, Expression, and Purification of ZmCK2α-1
The pGEX-4T-1 expression vector was used to express CK2α-1 in E. coli. Plasmid pT7-7 for maize full-length CK2α-1 as a template and primers 1 and 2 (for sequences of primers, see supplemental Table S1) were used for PCR. The PCR product was cloned into the TOPO® vector for amplification and subsequently subcloned into the SmaI/SalI sites of the pGEX-4T-1 expression vector. The GST:ZmCK2α-1 construct was transformed into E. coli BL21(DE3). For induction of GST:ZmCK2α-1 expression, 1 mm isopropyl β-d-1-thiogalactopyranoside was added for 4 h at 37 °C. The expressed protein was purified using glutathione-Sepharose 4B resin according to the manufacturer's instruction. To remove the GST tag the fusion protein bound on the glutathione affinity resin was treated with thrombin. After overnight cleavage, pure ZmCK2α-1 without the GST tag was separated from the resin by centrifugation. The yield was 0.1 mg from 1 liter of bacterial culture.
Cloning of ZmeIF5A (Z. mays), AteIF5A (A. thaliana), and SceIF5A (S. cerevisiae)
Total RNA was isolated from Z. mays seedlings or A. thaliana leaves using TRI Reagent according to the manufacturer's instructions and used as a template for eIF5A cloning. Total RNA was reverse transcribed using the HSRT 100 kit. The reaction was performed for 50 min at 47 °C in a final volume of 20 μl containing 1 unit of enhanced avian reverse transcriptase, 500 μm each dNTP, 1 unit of RNase inhibitor, and 3.5 μm oligo(dT) primer. Two microliters of the reverse transcription reaction was used for PCR in a 20-μl volume containing 0.4 unit of JumpStart AccuTaq LA DNA polymerase, 100 μm each dNTP, 1.5 mm MgCl2, and 300 nm primers 3 and 7 for ZmeIF5A or 15 and 17 for AteIF5A. PCR conditions were: 3 min at 94 °C (first cycle), 30 s at 94 °C, 30 s at 55 °C, 1 min at 68 °C (25–35 cycles), and 10 min at 68 °C (final cycle). The PCR product was cloned into TOPO vector. For amplification of SceIF5A, genomic DNA isolated from S. cerevisiae according to Ref. 25 and primers 12 and 14 were used for PCR at the above conditions. The obtained product was cloned into TOPO vector.
Site-directed Mutagenesis of ZmeIF5A, AteIF5A, and SceIF5A
The TOPO vector containing eIF5A was used as the templates for site-directed mutagenesis. Two constructs of ZmeIF5A were generated with substitutions ZmeIF5A-S2A and ZmeIF5A-S4A. Primers 4 and 7, and 5 and 7 were used in the PCR for the S2A and S4A mutations, respectively. To generate the S2A substitution of AteIF5A and SceIF5A, primers 16 and 17, and 13 and 14 were used, respectively.
To create ZmeIF5A-ΔM, primers 6 and 7 were used (supplemental Fig. S1, panel A, rows 1 and 3). (Deletion of the first Met from the ZmeIF5A sequence moves Ser2 to position +2 in the thrombin cleavage site in the GST-ZmeIF5A-ΔM fusion protein.) The PCR product was cloned into TOPO vector.
Expression and Purification of ZmeIF5A, AteIF5A, SceIF5A, and Their Mutated Variants
The pGEX-4T-1 expression vector was used to express eIF5As and its mutated variants in E. coli. ZmeIF5A-wt, ZmeIF5A-S2A, ZmeIF5A-S4A, AteIF5A-wt, AteIF5A-S2A, SceIF5A-wt, and SceIF5A-S2A were subcloned from TOPO vector into the EcoRI/SalI or EcoRI/XhoI sites, whereas ZmeIF5A-ΔM was subcloned into the BamHI/SalI sites of pGEX-4T-1 (see supplemental Table S1). The constructs were transformed into E. coli BL21(DE3), expressed, and purified using glutathione-Sepharose 4B according to the manufacturer's instructions. For induction of expression, 0.5 mm isopropyl β-d-1-thiogalactopyranoside was added for 4 h at 37 °C. Cleavage of GST from the recombinant proteins using thrombin was carried out overnight at 4 °C in thrombin cleavage buffer (100 units/ml of thrombin, 50 mm Tris-HCl (pH 8.0), 100 mm NaCl, 2.5 mm CaCl2). GST bound to the resin was removed by centrifugation. Glycerol was added to a final concentration of 10% and the obtained proteins were frozen in liquid nitrogen. The yield was about 1 mg from 1 liter of bacterial culture. When cleavage by thrombin was used to check for Ser2 phosphorylation, glutathione-Sepharose (500 μg) with bound GST-ZmeIF5A-ΔM was divided into halves that were separately incubated for 1 h at 37 °C: in 20 mm Tris-HCl (pH 7.5), containing 20 mm MgCl2 (nonphosphorylated); and the second half in the same buffer with ATP (50 μm) and recombinant ZmCK2α-1 (15 μg) (phosphorylated). After incubation, both portions of the resin, containing, respectively, nonphosphorylated and phosphorylated GST-ZmeIF5A-ΔM, were separately washed 5 times with 1 ml of 50 mm Tris-HCl (pH 8), 100 mm NaCl, and 2.5 mm CaCl2. Cleavage by thrombin (100 units/ml) was carried out overnight at 4 °C in the above buffer.
Constructs of Wild Type and Mutated ZmeIF5A Tagged with Enhanced Yellow Fluorescent Protein (EYFP)
Reverse primer 9 and forward primers 3, 4, or 8 were used for amplification of full-length cDNA of ZmeIF5A-wt, ZmeIF5A-S2A, and ZmeIF5A-S2D, respectively. For amplification of full-length EYFP cDNA, primers 10 and 11 were used. The PCR products were cloned into TOPO vector. ZmeIF5A-wt, ZmeIF5A-S2A, or ZmeIF5A-S2D were separately connected with EYFP by the restriction site for HindIII and inserted into the pLNU expression vector using EcoRI and XhoI/SalI sites. pLNU is a pUC-based vector carrying a constitutively active maize ubiquitin Ubi-1 promoter with the first Ubi-1 intron (supplemental Fig. S2, panel A). The Ubi-1 intron was used to enhance transgene expression in monocots (26). The Ubi-1 intron sequence ends with a PstI restriction site containing the 3′ splice junction. The nucleotide sequence of the 3′-region of the Ubi-1 intron and the 5′-polylinker sequence of pLNU vector is presented in supplementary data (supplemental Fig. S2, panel B). The ZmeIF5A-EYFP sequence begins with ATG, therefore expression of the ZmeIF5A-EYFP protein starts from the native methionine codon. All constructs were sequenced to confirm the correctness and the introduced changes. Sequences of primers 1–17 used for cloning are given in supplemental Table S1.
The numbering of amino acid residues in recombinant eIF5A is in accordance with numbering of the encoded amino acid sequence of native ZmeIF5A. Detailed information concerning the additional N-terminal residues coming from the vector is provided in supplemental Fig. S1, panel A.
Protoplast Transient Expression Assay
The preparation and electroporation of maize mesophyll protoplasts was performed according to the protocol described previously (27). Maize protoplasts (about 2 × 105) were transformed separately with 50 μg of plasmid DNA (ZmeIF5A-wt-EYFP, ZmeIF5A-S2A-EYFP, and ZmeIF5A-S2D-EYFP) and about 50 μg of carrier DNA. The transformed protoplasts were incubated at 22 °C for 16 h, and the fluorescent labeled proteins visualized by confocal microscopy (Eclipse TE200-E, Nikon).
In Vitro Phosphorylation Assay
Recombinant proteins, ZmeIF5A-wt, ZmeIF5A-S2A, ZmeIF5A-S4A, AteIF5A-wt, AteIF5A-S2A, SceIF5A-wt, SceIF5A-S2A, or casein as a positive control (3 μg of each) were separately incubated for 20 min at 30 °C with recombinant ZmCK2α-1 at the concentration of the enzyme giving Vmax (0.1 μg of protein) and [γ-32P]ATP (50 μm, 100–200 cpm pmol−1) in 50 μl of 20 mm Tris-HCl (pH 7.5), 20 mm MgCl2. After incubation, 40 μl of the assay mixture was spotted onto a square (2 × 2 cm) of 3MM Whatman paper, which was then immediately immersed in cold 5% (w/v) trichloroacetic acid containing 0.3% o-phosphoric acid, and washed five times for 10 min each. Then the squares were washed in 96% ethanol and allowed to dry. The radioactivity was quantified using a liquid scintillation counter. Efficiency of phosphorylation was calculated from the specific radioactivity of 32P incorporated into the given amounts of proteins, and was expressed as mole of phosphate/mole of ZmeIF5As and mole of phosphate/mole of casein. Alternatively, the reaction products were subjected to SDS-PAGE, visualized by Coomassie Brilliant Blue, and analyzed by autoradiography.
Gel Filtration of ZmeIF5A
Filtration was performed at 4 °C on a Superdex G-200 10/300 GL FPLC column calibrated with molecular mass markers. Recombinant ZmeIF5A (100 μg) was applied on the column previously equilibrated with 100 mm Tris-HCl (pH 7.5), 0.01% Tween, 1 mm EDTA containing 0.3 m NaCl. Filtration was performed with the same buffer.
RESULTS
Identification of Maize eIF5A (ZmeIF5A)
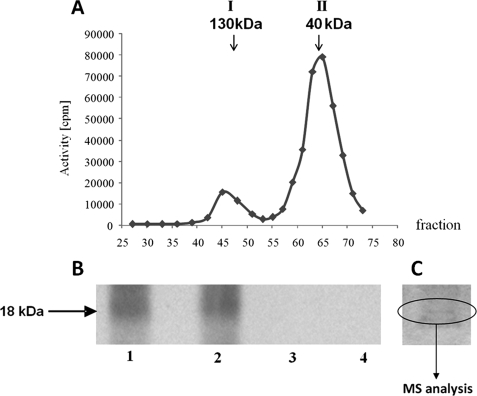
After gel filtration, the last step of purification of native CK2 from maize seedlings (described under “Experimental Procedures”), two peaks exhibiting CK2 activity were eluted: the first peak of about 130 kDa (designated I130), and the second one of about 40 kDa (designated II40) (Fig. 1A). After electrophoresis in 12% SDS-PAGE, gel fragments with proteins from both peaks (stained with Coomassie Brilliant Blue) were cut out and the proteins were identified by MS (Table 1). The analysis demonstrated that peak I130 contains three CK2β (β1, β2, β3) and one isoform of CK2α. The calculated mass of the eluted proteins and MS results indicate that the holoenzyme of CK2 (α2β2) is present in this peak. In peak II40, containing proteins of lower molecular masses, three isoforms of the CK2α (α1, α2, and α3) were detected only, which confirmed the previous supposition (20) that in plants CK2 may be present also as an active free CK2α.
FIGURE 1.
Analysis of CK2 activity in maize seedlings. A, separation on Sephadex G-100 of two peaks (I130 and II40) of the kinase activity. CK2 activity was determined as described under “Experimental Procedures” using casein as a substrate (see “In Vitro Phosphorylation Assay”). B, phosphorylation of proteins in peak II40 was performed in 50 μl of reaction mixture containing 50 μm ATP, 20 mm Tris-HCl (pH 7.5), 20 mm MgCl2; alone (lane 1), in the presence of 10 μm staurosporine (lane 2), 10 μm heparin (lane 3), or 50 μm 4,5,6,7-tetrabromobenzotriazole (lane 4). [γ-32P]ATP was used for detection of phosphorylation, whereas nonradioactive ATP was used for MS analysis. After 20 min of incubation at 30 °C the reaction was stopped by addition of Laemmli buffer, heated at 95 °C for 5 min, and subjected to SDS-PAGE. In the case of proteins phosphorylated with [γ-32P]ATP, the gel was dried and exposed to autoradiography. C, SDS-PAGE of proteins from peak II40. For MS analysis, the appropriate protein band (18 kDa) stained with Coomassie Brilliant Blue was cut from the wet gel.
TABLE 1.
Identification of CK2 subunits in peaks I130 and II40 by MS
| Accession No. | Name | Mascot score | Queries matched |
|---|---|---|---|
| Peak I130 | |||
| gi|11526999 | ZmCK2β-1 | 152 | 3 |
| gi|11527001 | ZmCK2β-2 | 130 | 4 |
| gi|11527003 | ZmCK2β-3 | 160 | 5 |
| gi|22116 | ZmCK2α-1 | 66 | 2 |
| Peak II40 | |||
| gi|22116 | ZmCK2α-1 | 1035 | 44 |
| gi|3821792 | ZmCK2α-2 | 882 | 33 |
| gi|162463502 | ZmCK2α-3 | 797 | 31 |
For identification of endogenous CK2α substrates, the proteins that co-purified with CK2α in peak II40 were allowed to undergo phosphorylation by endogenous protein kinase(s) using [γ-32P]ATP as the phosphate donor. After separation of the phosphorylated proteins by SDS-PAGE, the gel was dried and exposed to autoradiography. A protein of about 18 kDa was phosphorylated. To demonstrate specificity of the phosphorylation, the following inhibitors of protein kinases were added to the phosphorylation assay (see legend to Fig. 1): heparin (28) and 4,5,6,7-tetrabromobenzotriazole (for review, see Ref. 29), which selectively inhibits CK2 activity; and staurosporine, an inhibitor of many protein kinases but not CK2 (30). Phosphorylation of the protein(s) was not decreased by staurosporine, whereas heparin and 4,5,6,7-tetrabromobenzotriazole abolished phosphorylation completely (Fig. 1B). The results clearly indicate that phosphorylation is catalyzed by CK2. The fragment of the gel that contained the 18-kDa protein(s) was cut out from the wet gel as indicated in Fig. 1C and subjected to MS analysis. Two high mobility group (HMG) proteins characterized before as CK2 substrates were identified: HMGB1 and HMGc1 (accession numbers CAA46876 and CAA69605, respectively). Phosphorylation of cytosolic HMG homologues of the animal HMG-1 subgroup by CK2 was observed previously in broccoli (31). Phosphorylation of HMGBI by CK2 modulates its stability and DNA binding properties (32). Among the 18-kDa proteins, we also identified eIF5A by the presence of two peptides (VVEVSTSK and LPTDETLVAQIK). Maize eIF5A (accession number YO7920) consists of 480 nucleotides encoding 160 amino acids.
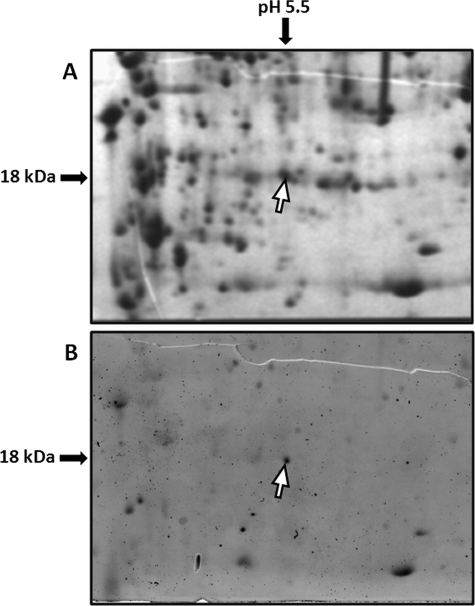
To check whether ZmeIF5A is phosphorylated in vivo, total proteins from maize leaves were separated by two-dimensional electrophoresis (Fig. 2). Among the phosphorylated proteins detected by Pro-Q Diamond phosphoprotein staining (Fig. 2B) we identified one spot containing a protein of about 18 kDa with a pI of about 5.5. This spot was analyzed by MS and five peptides of the ZmeIF5A sequence were identified (Table 2). The result indicates that ZmeIF5A is phosphorylated in vivo.
FIGURE 2.
Two-dimensional gel electrophoresis of proteins from maize leaves. A, gel stained with Coomassie Brilliant Blue. B, phosphoproteins visualized with Pro-Q Diamond phosphoprotein. ZmeIF5A is indicated by a white arrow.
TABLE 2.
Identification of maize eIF5A peptides by MS
| Peptide sequence | Localization of peptide in ZmeIF5A sequence | Mascot score | Queries matched |
|---|---|---|---|
| TYPQQAGTVR | 20–29 | 64 | 94 |
| PTDETLVAQIK | 119–131 | 99 | 121 |
| AMGEEQICALK | 145–155 | 67 | 128 |
| LPTDETLVAQIK | 118–131 | 53 | 141 |
| DLVVTVQSAMGEEQICALK | 137–155 | 101 | 228 |
A comparison of eIF5A sequences from different organisms showed that eIF5A proteins are highly conserved but have lower amino acid similarity at the terminal positions (Fig. 3). It should be stressed that in nine analyzed plants, worm (Caenorhabditis elegans) and yeast (S. cerevisiae), but not in another yeast (Schizosaccharomyces pombe) or animals (Drosophila melanogaster and Homo sapiens), eIF5As contain a serine residue at position 2. Moreover, in all five analyzed monocots, additional phosphorylatable residues (serine or threonine), surrounded by acidic amino acids, are present at position 4 (Fig. 3).
FIGURE 3.
Alignment of eIF5A sequences from different kingdoms. The sequences are derived from: Z. mays (Y07920), O. sativa (NP0011051336), T. aestivum (AAZ95173), S. sylvestre (ABB29987), D. sinicus (ABW78939), A. thaliana (AAF87023), M. trunculata (ACJ85877), N. tabacum (CAA45105), S. tuberosum (BAA20876), S. cerevisiae (NP010880), C. elegans (NP495807), S. pombe (NP596130), H. sapiens (NP001961), and D. melanogaster (NP611878). Identical residues are indicated by black background, the dash indicates the gap introduced to maximize alignment.
Phosphorylation of eIF5A by CK2 and Identification of Phosphorylated Residue by Site-directed Mutagenesis
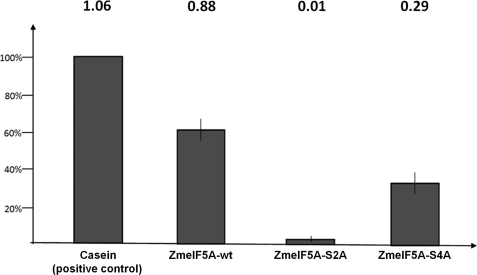
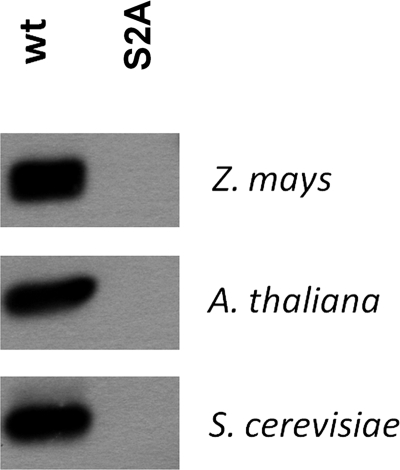
Because eIF5A had not been identified before as a CK2 substrate, to investigate its phosphorylation by CK2α in an in vitro system, recombinant ZmeIF5Awt, AteIF5Awt, SceIF5Awt, and ZmCK2α-1 proteins were produced in E. coli (see “Experimental Procedures”). To identify which serine residue(s) (Ser2 and/or Ser4) is phosphorylated in eIF5As, these serine residues were individually mutated to alanine, which cannot undergo phosphorylation, and the resulting mutated proteins ZmeIF5A-S2A, ZmeIF5A-S4A, AteIF5A-S2A, and SceIF5A-S2A were expressed in bacteria. Wild and mutated variants of eIF5A were phosphorylated by recombinant ZmCK2α-1, using casein as a positive control. ZmeIF5Awt and casein were phosphorylated efficiently. The efficiency of ZmeIF5Awt phosphorylation was 0.88 mol of phosphate/mol of protein indicating that wild-type ZmeIF5A is a good substrate for CK2α. The protein with Ser2 converted to Ala was not phosphorylated by CK2 (Fig. 4). The lack of phosphorylation of the mutated S2A variant indicates that only Ser2, and no other Ser/Thr residue present in ZmeIF5A, undergoes phosphorylation by CK2. The protein with Ser4 converted to Ala was phosphorylated less efficiently (0.29 mol of phosphate/mol of ZmeIF5A-S4A) than the wild-type of ZmeIF5A. The decreased incorporation of phosphate into the S4A mutated variant may indicate that the presence of the hydrophilic, hydroxyl group of Ser4 facilitates accessibility of Ser2 for CK2. Similarly to ZmeIF5Awt, eIF5As from the other organisms (A. thaliana and S. cerevisiae) were efficiently phosphorylated by CK2, whereas their mutated variants (S2A) were unable to accept phosphate groups (Fig. 5). The above results demonstrate that Ser2 present in plants and yeast of eIF5A is phosphorylated by CK2. To verify the site of phosphorylation, these phosphoproteins were analyzed by MS. However, we were unable to detect any phosphorylation (data not shown), most probably because the phosphorylated residue is located at the terminal position of the protein. The failure to identify by MS an N-terminal peptide containing modified residue(s) has prompted us to elaborate a new method to confirm the phosphorylation of Ser2 in ZmeIF5A.
FIGURE 4.
Phosphorylation of ZmeIF5Awt and its mutated variants. Efficiency of phosphorylation is shown relative to casein phosphorylation taken as 100%. Values (in mole of phosphate/mole of protein) are given above the bars. In vitro phosphorylation was performed in three independent experiments as described under “Experimental Procedures.”
FIGURE 5.
Phosphorylation of recombinant eIF5A from Z. mays, A. thaliana, and S. cerevisiae, and their S2A mutants by CK2.
Identification of Phosphorylated Serine by Exploiting the Specificity of Thrombin
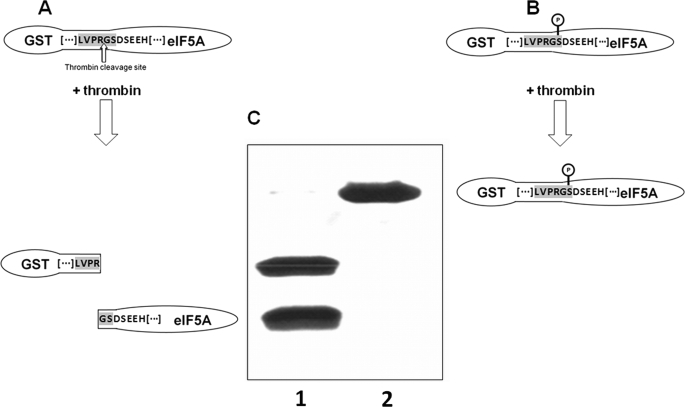
The method elaborated for this purpose exploits the specific proteolytic cleavage by thrombin. The minimal consensus recognized by thrombin is BBPR↓UU (−4−3−2−1↓+1+2) (B, hydrophobic amino acid; P, proline; R, arginine; U, non-acidic amino acid), where the cleavage occurs after the arginine. In the GST-ZmeIF5A fusion protein the C-terminal part of the GST tag (LVPRG) and the N-terminal amino acid of ZmeIF5A (Met) create a sequence recognized and cleaved by thrombin. After thrombin cleavage the recombinant protein contains an additional glycine residue derived from the GST tag at the N terminus (Gly-Met-Ser). For thrombin to be used as a probe of Ser2 phosphorylation, the assumption was as follows. Removal of methionine from GST-ZmeIF5A-wt (GST-ZmeIF5A-ΔM) moves serine 2 to the critical thrombin cleavage position at +2, which has to be occupied by a non-acidic amino acid. When this serine is not modified, thrombin will recognize and cleave the sequence. However, if there is the negatively charged phosphoserine at position +2, the site no longer fulfils the consensus criteria for thrombin and the protease should not be able to cleave the sequence (Fig. 6A). Therefore, construction of the modified fusion protein containing at position +2 a residue susceptible to phosphorylation should allow distinguishing between a nonphosphorylated (cleaved by thrombin) and a phosphorylated (not cleaved by thrombin) protein.
FIGURE 6.
Effect of GST-ZmeIF5A-ΔM phosphorylation on thrombin cleavage. Consensus sequence recognized by thrombin is in gray. The construct encoding the GST-ZmeIF5A-ΔM fusion protein was obtained and expressed as described under “Experimental Procedures.” A, predicted effect of thrombin action on nonphosphorylated GST-ZmeIF5A-ΔM. B, predicted effect of thrombin action on phosphorylated GST-ZmeIF5A-ΔM. C, SDS-PAGE visualization of thrombin treatment of nonphosphorylated (lane 1) and phosphorylated (lane 2) GST-ZmeIF5A-ΔM.
After thrombin treatment, phosphorylated and nonphosphorylated GST-ZmeIF5A-ΔM were analyzed by SDS-PAGE. In the experiment where GST-ZmeIF5A-ΔM was not phosphorylated, thrombin action produced two proteins, as expected: GST and ZmeIF5A-ΔM (Fig. 6, A and C). When eIF5A was phosphorylated, only one protein band of higher molecular mass (45 kDa) was detected (Fig. 6, B and C) indicating that the phosphorylated fusion protein was resistant to thrombin. This approach employing the specificity of protease cleavage can be applied for confirmation of postulated phosphorylation sites also in other proteins.
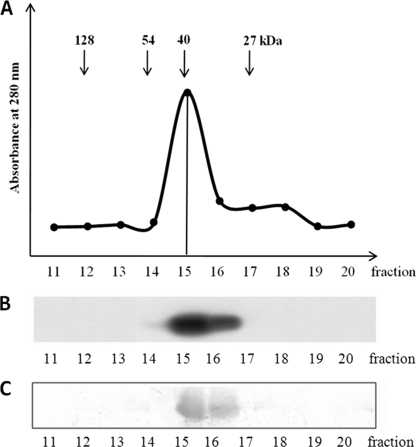
Dimerization of ZmeIF5A
To check whether ZmeIF5A assumes a monomeric or oligomeric form, gel filtration of the recombinant protein was carried out on Superdex G-200 in a buffer containing 0.3 m NaCl to preclude nonspecific ionic interactions (Fig. 7A). ZmeIF5A was detected in the eluted fractions by its phosphorylation by recombinant ZmCK2α-1 (Fig. 7B) and Western blotting using anti-eIF5A antibodies (Fig. 7C). ZmeIF5A was present in fraction 15 containing proteins of about 40 kDa (Fig. 7). This indicates that ZmeIF5A occurs in a dimeric form.
FIGURE 7.
Gel filtration of recombinant ZmeIF5A. A, elution profile of ZmeIF5A on a Superdex G-200 FPLC column. Fractions of 0.5 ml were collected. B, phosphorylation of ZmeIF5A. Each fraction was incubated with recombinant ZmCK2α-1 and [γ-32P]ATP. After SDS-PAGE, phosphorylated eIF5A was detected by autoradiography. C, immunodetection. Western blotting using anti-eIF5A antibodies. For details see “Experimental Procedures.”
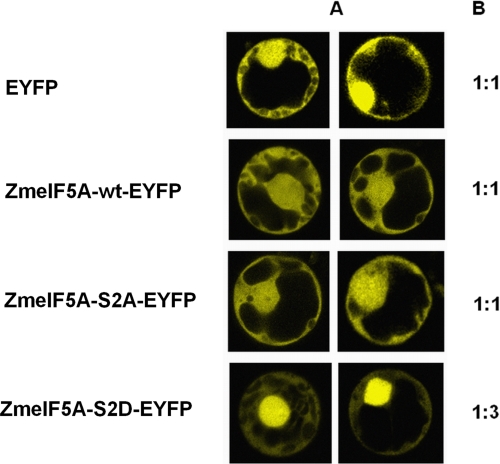
Role of ZmeIF5A Phosphorylation by CK2
To shed light on the physiological significance of phosphorylation of Ser2 in ZmeIF5A, transient expression of wild-type eIF5A and its variants mutated at position 2 of the encoded amino acid sequence was performed in maize protoplasts. In one mutated form the serine was substituted by the neutral alanine, and in the second, by an aspartic acid, which mimics phosphoserine. As a reporter of expression EYFP was used either alone (as a control) or as a C-terminal tag of ZmeIF5A. Four constructs in the pLNU vector were prepared individually (supplemental Table S2). After expression of the proteins in maize protoplasts (see “Experimental Procedures”), the intensity of fluorescence in the nucleus and cytoplasm was measured by confocal microscopy in optical sections of the protoplasts (Fig. 8A). EYFP was located both in the nucleus and cytoplasm (for which the ratio of fluorescence intensity was 1:1). Also ZmeIF5Awt-EYFP (ratio 1:1.21, S.D. 0.23) and ZmeIF5A-S2A-EYFP (ratio 1:1.09, S.D. 0.29) were evenly distributed between the nucleus and cytoplasm. In contrast, the ZmeIF5A-S2D-EYFP protein exhibited about 2–3-fold higher fluorescence in the nucleus (ratio 1:2.91, S.D. 0.61) than in the cytoplasm (Fig. 8B). The acidic group of aspartic acid that mimics phosphorylated serine seems to inhibit nuclear cytoplasmic shuttling, leading to preferential ZmeIF5A accumulation in the nucleus. The obtained results suggest that phosphorylation of Ser2 plays a role in determining the intracellular distribution of ZmeIF5A.
FIGURE 8.
Localization of ZmeIF5A-EYFP and its mutated variants in maize protoplasts. A, optical sections by confocal microscopy. B, relative intensities of fluorescence in the nucleus (taken as 1) and cytoplasm. Constructs are described in supplemental Table S2. The results are representative of eight independent experiments.
DISCUSSION
eIF5A is a highly conserved 18-kDa protein present in all eukaryotic cells (33), involved in a broad spectrum of cellular functions, including proliferation and cell cycle control (for review, see Refs. 34 and 35). eIF5A is the only known cellular protein that contains the unusual positively charged amino acid hypusine, a derivative of lysine. Hypusination is essential for eukaryotic cell proliferation and apoptosis (for review, see Ref. 34).
Currently available results indicate that in plants, like in other eukaryotic organisms, eIF5A is involved in many cellular functions. Three isoforms of eIF5A have been identified in Arabidopsis (36). Isoform 1 (AteIF-5A1) plays a role in xylem formation (37), whereas isoform 2 (AteIF-5A2) regulates programmed cell death caused by pathogen infection (38). Reducing the level of eIF5A in Arabidopsis has dramatic effects on growth and development of the plant. The developmental defects are associated with abnormal cell division and cell growth (39). In tomato (Lycopersicon esculentum), a correlation has been found between expression of eIF5A and programmed cell death (40). In rice (Oryza sativa), two cDNA clones encoding eIF5A have been characterized. Analysis of the mRNA level showed that OseIF5A-1 and OseIF5A-2 were expressed in leaves and panicles, and relatively high amounts of both transcripts were detected in old leaves. In addition, both OseIF5A-1 and OseIF5A-2 levels were regulated during leaf development, sugar starvation, and environmental stresses (41). In maize, eIF5A is expressed in most tissues investigated. Unfertilized egg cells of maize in the G0 phase contain a high level of eIF5A transcript. The transcript is strongly induced during the G1 phase, but continuously decreases during the S, G2, and M phases (42). The authors suggest that unfertilized egg cells of maize are prepared for selective mRNA translation that is quickly triggered after fertilization. The triggering could be regulated by reversible phosphorylation known as the key modification of proteins involved in regulation of cellular activities including gene expression, proliferation, and differentiation (43).
The N-terminal sequence of the plant eIF5A contains a consensus motif for CK2 ((S/T)XXZ ((0)+1+2+3), were S/T is serine or threonine; X, non-basic amino acid; and Z, an acidic or phosphoamino acid), suggesting that this enzyme might be involved in eIF5A regulation by phosphorylation. Indeed, we have shown here that phosphorylation of ZmeIF5A occurs in vivo in maize leaves. Moreover, our results indicate that Ser2 is the target of CK2 not only in the case of ZmeIF5A, but also in eIF5A from other organisms (A. thaliana and S. cerevisiae). It is noteworthy to add that these serine residues are absent in human, insect, and another yeast (S. pombe) eIF5As. Interestingly, in monocots an additional potentially phosphorylatable residue (Ser/Thr) is present in position 4, although the CK2 consensus for this residue is predicted to be weak (3). In the case of ZmeIF5A, in vitro phosphorylation of Ser4 by CK2 was not observed. The role of Ser/Thr4 in monocots might be in enhancing Ser2 phosphorylation by CK2. Ser/Thr4 is located at position +2 in the consensus of Ser2 phosphorylation by CK2, were acidic amino acids (Asp, Glu, Tyr(P), Ser(P), and Thr(P)) predominate (3). One can assume that phosphorylation of Ser4 by other protein kinases could also enhance the phosphorylation of Ser2 by CK2. Other eukaryotic proteins containing CK2-phosphorylated Ser2 are known, e.g. translation initiation factor 2β (eIF2β, MSGDEMI…) or CK2β (MSSSEEV…) (44, 45).
Transfection of HeLa cells with human eIF2β (carrying the hemagglutinin tag) or its phosphorylation site mutants showed Ser2 as the main site for constitutive eIF2β phosphorylation by CK2 (44). In CK2β, Ser2, and to a lesser extent Ser3, are phosphorylated; both fulfill the CK2 consensus sequence requirements (3). The N-terminal region of CK2β also contains several docking sites for proteins that are involved in many essential functions, like cellular localization, control of translation, and protein degradation (46). These results suggest that the N terminus of CK2β reflects the supramolecular organization of the native protein complex, in which phosphorylation plays an important regulatory role (45). This suggestion was partially confirmed by Bodenbach et al. (47), which showed that in the recombinant holoenzyme of CK2 (expressed in a tandem arrangement without any tag), Ser2 and Ser3 are phosphorylated. We have demonstrated that CK2 can phosphorylate Ser2 of recombinant ZmeIF5A irrespective of additional residues located upstream (supplemental Fig. S1, panel B).
All three CK2β (β1, β2, β3) identified in maize are able to interact with CK2α subunits as the dimer, which results in formation of a stable tetrameric form of the holoenzyme (19). Similarly to the CK2β, the dimeric form of ZmeIF5A could play a role of a scaffold protein assembling specific partners. Interestingly, such binding ability has been shown for eIF2β (48). Llorens and co-workers (48) have demonstrated that eIF2β, besides serving as a substrate for CK2 holoenzyme, binds free CK2α and affects its enzymatic properties. This suggests that eIF2β might act as a CK2 anchoring protein, locating the kinase in close vicinity of its targets and helping to maintain some components of the translation machinery in a functional conformation.
The role of eIF5A phosphorylation in plants and yeast is not known. Previous results (49) indicate that the phosphorylated and non-phosphorylated forms of yeast eIF5A exhibit virtually identical activities in the mammalian assay for methionyl-puromycin synthesis. However, it is possible that the in vitro assay system used for translation was not sufficiently sensitive to detect any differences, or that phosphorylation of eIF5A regulates other cellular processes. The N-terminal region of eIF5As has a high content of acidic residues, whereas the classical nuclear localization signals are rich in basic residues (50). Using confocal microscopy and different constructs encoding eIF5A-related sequences fused to green fluorescent protein, those authors have shown that the N-terminal region of mammalian eIF5A proteins is essential for localization of the protein to the nucleus. Because the N-terminal region of most eukaryotic eIF5As studied is rich in acidic residues and does not contain phosphorylatable residue(s), those authors (50) did not consider the role of phosphorylation in eIF5A localization. It is known that exportin 4 (Exp4) functions as a nuclear export receptor for eIF5A (51). eIF5A can specifically assemble into a trimeric eIF5A-Exp4-RanGTP export complex, which results in efficient nuclear export. The complex is disassembled when cytoplasmic RanBP1 and RanGAP are both present. The positively charged amino acid hypusine improves eIF5A binding to Exp4, thereby enhancing its nuclear export. It is possible that phosphorylation may cause an opposite effect, the sequestration of eIF5A in the nucleus by inhibiting its interaction with the exporting protein complex. Intramolecular masking of nuclear localization signals within cargo proteins to prevent IMP/EXP recognition is a common mechanism of regulation of the efficiency of nuclear transport. It can be effected by phosphorylation close to the target sequence to prevent IMP/EXP binding (for review, see Ref. 52). There is evidence that phosphorylation by CK2 is mechanistically important in nuclear localization signal recognition and enhances nuclear import by facilitating recognition by the transport receptor (53). Maize CK2 catalytic subunit α contains a single functional nuclear localization signal, consisting of a basic stretch of 20 amino acids (16) sufficient to target the protein to the nucleus of plant cells (19). CK2 trafficking to the nucleus is a signal-mediated process (54, 55), whereas eIF5A can enter the nucleus via passive diffusion, as revealed for a human isoform (56). We suggest that eIF5A can easily enter into the nucleus, but its transport from the nucleus is arrested by the acidic residue of phosphoserine, which in our experiment was mimicked by Asp. The S2D mutation increased the rate of accumulation of eIF5A in the nucleus because it could not be dephosphorylated.
In the case of ZmeIF5A, phosphorylation of Ser2 blocks its export from the nucleus by preventing interaction with the nuclear exporting complex; whereas phosho-Ser2 dephosphorylation would reactivate nuclear export of ZmeIF5A. It is possible that modulation of nucleocytoplasmic shuttling of eIF5A by phosphorylation may be a physiological mechanism unique to plants and yeast regulating specific nuclear export of eIF5A-bound mRNAs.
Supplementary Material
Acknowledgments
We thank Professors S. Sarno (University of Padua) for providing plasmid pT7-7 for maize full-length CK2α-1, D. Becker (University of Wuerzburg) for pLNU vector, and D. Shugar and M. Bretner (IBB PAS, Warsaw) for 4,5,6,7-tetrabromobenzotriazole. We are grateful to Professors G. Dobrowolska (IBB PAS, Warsaw) and Jan Fronk (University of Warsaw) for fruitful discussion, and J. Sheen (Harvard Medical School) and E. Parys and E. Romanowska (University of Warsaw) for valuable suggestions regarding the preparation of maize protoplasts; Dr. A. Anielska-Mazur for assistance with confocal microscopy, and Katarzyna Król for assistance in preparation of the manuscript.
This work was supported by Ministry of Science and Higher Education Grant PBZ-MIN-014/P05/2004.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2 and Tables S1 and S2.
- CK2
- casein kinase 2/protein kinase CK2
- CK2α
- catalytic α subunit of CK2
- CK2β
- regulatory β subunit of CK2
- eIF5A
- eukaryotic translation initiation factor 5A
- Exp4
- exportin 4
- EYFP
- enhanced yellow fluorescent protein
- GST
- glutathione S-transferase
- MS
- mass spectrometry
- WT
- wild-type (without mutations)
- ΔMet
- lack of the first methionine in a protein
- FPLC
- fast protein liquid chromatography
- HMG
- high mobility group
- PMSF
- phenylmethylsulfonyl fluoride
- CHAPS
- 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonic acid.
REFERENCES
- 1.Pinna L. A. (2002) J. Cell Sci. 115, 3873–3878 [DOI] [PubMed] [Google Scholar]
- 2.Litchfield D. W. (2003) Biochem. J. 369, 1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Meggio F., Pinna L. A. (2003) FASEB J. 17, 349–368 [DOI] [PubMed] [Google Scholar]
- 4.Marin O., Meggio F., Marchiori F., Borin G., Pinna L. A. (1986) Eur. J. Biochem. 160, 239–244 [DOI] [PubMed] [Google Scholar]
- 5.Lee Y., Lloyd A. M., Roux S. J. (1999) Plant Physiol. 119, 989–1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Espunya M. C., Combettes B., Dot J., Chaubet-Gigot N., Martínez M. C. (1999) Plant J. 19, 655–666 [DOI] [PubMed] [Google Scholar]
- 7.Espunya M. C., López-Giráldez T., Hernan I., Carballo M., Martínez M. C. (2005) J. Exp. Bot. 56, 3183–3192 [DOI] [PubMed] [Google Scholar]
- 8.Moreno-Romero J., Espunya M. C., Platara M., Ariño J., Martínez M. C. (2008) Plant J. 55, 118–130 [DOI] [PubMed] [Google Scholar]
- 9.Hardtke C. S., Gohda K., Osterlund M. T., Oyama T., Okada K., Deng X. W. (2000) EMBO J. 19, 4997– 5006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Klimczak L. J., Collinge M. A., Farini D., Giuliano G., Walker J. C., Cashmore A. R. (1995) Plant Cell 7, 105–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Menkens A. E., Schindler U., Cashmore A. R. (1995) Trends Biochem. Sci. 20, 506–510 [DOI] [PubMed] [Google Scholar]
- 12.Ciceri P., Gianazza E., Lazzari B., Lippoli G., Genga A., Hoscheck G., Schmidt R. J., Viotti A. (1997) Plant Cell 9, 97–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sugano S., Andronis C., Ong M. S., Green R. M., Tobin E. M. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 12362–12366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Daniel X., Sugano S., Tobin E. M. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 3292–3297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee J., He K., Stolc V., Lee H., Figueroa P., Gao Y., Tongprasit W., Zhao H., Lee I., Deng X. W. (2007) Plant Cell 19, 731–749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Riera M., Figueras M., Lopez C., Goday A., Pages M. (2004) Proc. Natl. Acad. Sci. U.S.A. 29, 9879–9884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dobrowolska G., Boldyreff B., Issinger O. G. (1991) Biochim. Biophys. Acta 1129, 139–140 [DOI] [PubMed] [Google Scholar]
- 18.Peracchia G., Jensen A. B., Culiáñez-Macià F. A., Grosset J., Goday A., Issinger O. G., Pagès M. (1999) Plant Mol. Biol. 40, 199–211 [DOI] [PubMed] [Google Scholar]
- 19.Riera M., Peracchia G., de Nadal E., Ariño J., Pagès M. (2001) Plant J. 25, 365–374 [DOI] [PubMed] [Google Scholar]
- 20.Dobrowolska G., Meggio F., Szczegielniak J., Muszynska G., Pinna L. A. (1992) Eur. J. Biochem. 204, 299–303 [DOI] [PubMed] [Google Scholar]
- 21.Lebska M., Szczegielniak J., Dobrowolska G., Cozza G., Moro S., Muszyńska G. (2009) Physiol. Plant. 136, 251–263 [DOI] [PubMed] [Google Scholar]
- 22.Dobrowolska G., Meggio F., Pinna L. A. (1987) Biochim. Biophys. Acta 931, 188–195 [DOI] [PubMed] [Google Scholar]
- 23.Hajduch M., Ganapathy A., Stein J. W., Thelen J. J. (2005) Plant Physiol. 137, 1397–1419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Agrawal G. K., Thelen J. J. (2006) Mol. Cell Proteomics 5, 2044–2059 [DOI] [PubMed] [Google Scholar]
- 25.Hoffman C. S., Winston F. (1987) Gene 57, 267–272 [DOI] [PubMed] [Google Scholar]
- 26.Christensen A. H., Quail P. H. (1996) Transgenic Res. 5, 213–218 [DOI] [PubMed] [Google Scholar]
- 27.Sheen J. (1990) Plant Cell 2, 1027–1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hathaway G. M., Lubben T. H., Traugh J. A. (1980) J. Biol. Chem. 255, 8038–8041 [PubMed] [Google Scholar]
- 29.Sarno S., Salvi M., Battistutta R., Zanotti G., Pinna L. A. (2005) Biochim. Biophys. Acta 1754, 263–270 [DOI] [PubMed] [Google Scholar]
- 30.Meggio F., Donella Deana A., Ruzzene M., Brunati A. M., Cesaro L., Guerra B., Meyer T., Mett H., Fabbro D., Furet P., Dobrowolska G., Pinna L. A. (1995) Eur. J. Biochem. 234, 317–322 [DOI] [PubMed] [Google Scholar]
- 31.Klimczak L. J., Cashmore A. R. (1994) Plant Physiol. 105, 911–919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stemmer C., Schwander A., Bauw G., Fojan P., Grasser K. D. (2002) J. Biol. Chem. 277, 1092–1098 [DOI] [PubMed] [Google Scholar]
- 33.Jenkins Z. A., Hååg P. G., Johansson H. E. (2001) Genomics 71, 101–109 [DOI] [PubMed] [Google Scholar]
- 34.Park M. H., Lee Y. B., Joe Y. A. (1997) Biol. Signals 6, 115–123 [DOI] [PubMed] [Google Scholar]
- 35.Caraglia M., Marra M., Giuberti G., D'Alessandro A. M., Budillon A., del Prete S., Lentini A., Beninati S., Abbruzzese A. (2001) Amino Acids 20, 91–104 [DOI] [PubMed] [Google Scholar]
- 36.Thompson J. E., Hopkins M. T., Taylor C., Wang T. W. (2004) Trends Plant Sci. 9, 174–179 [DOI] [PubMed] [Google Scholar]
- 37.Liu Z., Duguay J., Ma F., Wang T. W., Tshin R., Hopkins M. T., McNamara L., Thompson J. E. (2008) J. Exp. Bot. 59, 939–950 [DOI] [PubMed] [Google Scholar]
- 38.Hopkins M. T., Lampi Y., Wang T. W., Liu Z., Thompson J. E. (2008) Plant Physiol. 148, 479–489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Feng H., Chen Q., Feng J., Zhang J., Yang X., Zuo J. (2007) Plant Physiol. 144, 1531–1545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang T. W., Lu L., Wang D., Thompson J. E. (2001) J. Biol. Chem. 276, 17541–17549 [DOI] [PubMed] [Google Scholar]
- 41.Chou W. C., Huang Y. W., Tsay W. S., Chiang T. Y., Huang D. D., Huang H. J. (2004) Physiol. Plant. 121, 50–57 [DOI] [PubMed] [Google Scholar]
- 42.Dresselhaus T., Cordts S., Lörz H. (1999) Plant Mol. Biol. 39, 1063–1071 [DOI] [PubMed] [Google Scholar]
- 43.Shugar D. (2005) Biochim. Biophys. Acta 1754, 1–2 [DOI] [PubMed] [Google Scholar]
- 44.Llorens F., Duarri A., Sarró E., Roher N., Plana M., Itarte E. (2006) Biochem. J. 394, 227–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pagano M. A., Sarno S., Poletto G., Cozza G., Pinna L. A., Meggio F. (2005) Mol. Cell. Biochem. 274, 23–29 [DOI] [PubMed] [Google Scholar]
- 46.French A. C., Luscher B., Litchfield D. W. (2007) J. Biol. Chem. 282, 29667–29677 [DOI] [PubMed] [Google Scholar]
- 47.Bodenbach L., Fauss J., Robitzki A., Krehan A., Lorenz P., Lozeman F. J., Pyerin W. (1994) Eur. J. Biochem. 220, 263–273 [DOI] [PubMed] [Google Scholar]
- 48.Llorens F., Sarno S., Sarró E., Duarri A., Roher N., Meggio F., Plana M., Pinna L. A., Itarte E. (2005) Mol. Cell. Biochem. 274, 53–61 [DOI] [PubMed] [Google Scholar]
- 49.Kang H. A., Schwelberger H. G., Hershey J. W. (1993) J. Biol. Chem. 268, 14750–14756 [PubMed] [Google Scholar]
- 50.Parreiras-E-Silva L. T., Gomes M. D., Oliveira E. B., Costa-Neto C. M. (2007) Biochem. Biophys. Res. Commun. 362, 393–398 [DOI] [PubMed] [Google Scholar]
- 51.Lipowsky G., Bischoff F. R., Schwarzmaier P., Kraft R., Kostka S., Hartmann E., Hutay U., Görlich D. (2000) EMBO J. 19, 4362–4371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Poon I. K., Jans D. A. (2005) Traffic 3, 173–186 [DOI] [PubMed] [Google Scholar]
- 53.Hübner S., Xiao C. Y., Jans D. A. (1997) J. Biol. Chem. 272, 17191–17195 [DOI] [PubMed] [Google Scholar]
- 54.Martel V., Filhol O., Nueda A., Cochet C. (2002) Ann. N. Y. Acad. Sci. 973, 272–277 [DOI] [PubMed] [Google Scholar]
- 55.Wang H., Yu S., Davis A. T., Ahmed K. (2003) J. Cell. Biochem. 88, 812–822 [DOI] [PubMed] [Google Scholar]
- 56.Jao D. L., Yu Chen K. (2002) J. Cell. Biochem. 86, 590–600 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.