Abstract
TLR2 is a pattern recognition receptor that functions in association with TLR1 or TLR6 to mediate innate immune responses to a variety of conserved microbial products. In the present study, the ectodomain of TLR2 was extensively mutated, and the mutants were assessed for their ability to bind and to mediate cellular responses to triacylated lipopeptide Pam3CSK4. This analysis provides evidence that the recently published crystal structure of the TLR2-TLR1-Pam3CSK4 complex represents a functional signal-inducing complex. Furthermore, we report that extended H-bond networks on the surface of TLR2 are critical for signaling in response to Pam3CSK4 and to other di- and tri-acylated TLR2-TLR6 and TLR2-TLR1 ligands. Based on this finding, we suggest a dynamic model for TLR2-mediated recognition of these ligands in which TLR2 fluctuates between a conformation that is more suitable for binding of the fatty acyl moieties of the ligands and a conformation that favors, via a specific orientation of the ligand head group, formation of a signal-inducing ternary complex.
Keywords: Immunology, Immunology/Innate Immunity, Lipoprotein/Lipoprotein-like receptor (LRP), Receptors, Receptors/Structure-Function, Receptors/Toll-like
Introduction
The Toll-like receptor (TLR)2 family endows cells with the ability to induce innate immune responses and activate adaptive immunity to most microbial organisms encountered in life. This remarkable trait for a limited set of 10–13 transmembrane receptors comes at least in part from the versatility of these receptors and from the conserved nature of ligands that are recognized.
TLRs are type I transmembrane proteins that possess an N-terminal ectodomain, a single transmembrane domain, and a C-terminal cytoplasmic Toll/interleukin-1 receptor (TIR) domain. Their TIR domains can, upon ligand-induced dimerization, interact with TIR domain-containing adaptors and induce intracellular signaling. Differential responses mediated by distinct TLRs can be explained in part by the selective use of these adaptor molecules (1–3). The TLRs ectodomains consist of tandem arrays of leucine-rich repeats (LRR) (4). LRRs are 20–29-residue sequence motifs present in a number of proteins with diverse functions (5, 6). The LRR proteins adopt a solenoidal fold (7), in which each LRR corresponds to one coil of the solenoid. The coils consist of a β-strand and mostly α-helical elements connected by loops. The coils are arranged so that all the β-strands and α-helices are parallel to a common axis, resulting in a nonglobular horseshoe-shaped molecule with a curved parallel β-sheet lining the inner circumference of the horseshoe and the helices flanking the outer circumference.
TLRs recognize conserved microbial-associated molecular patterns (MAMPs) that are essential for the survival of the microorganism and are therefore difficult to alter. TLR2, which plays a major role in detecting Gram-positive bacteria, is involved in the recognition of an apparently highly diverse set of MAMPs that includes lipopeptides, lipoteichoic acid, lipoglycans, peptidoglycan, porins, zymosan, and glycosylphosphatidyl myo-inositol anchors (for a recent review see Ref. 1). Several of these MAMPs, which are structurally unrelated, must be acylated to activate immune responses. The ability of TLR2 to function as a heterodimer with either TLR1 or TLR6 allows discrimination between acylation patterns. Tri-acylated lipoproteins, lipoglycans, and glycosylphosphatidyl myo-inositols are preferentially recognized by TLR2-TLR1 complexes, whereas di-acylated lipoproteins, lipoteichoic acids, and glycosylphosphatidyl myo-inositols are recognized by TLR2-TLR6 complexes (8–17). Recently, the crystal structures of a human TLR1-TLR2-triacylated lipopeptide (Pam3CSK4) complex and of mouse TLR2-Pam3CSK4 and TLR2-diacylated lipopeptide (Pam2CSK4) complexes have been determined (18). This shows that the three lipid chains of Pam3CSK4 mediate the heterodimerization of TLR2 and TLR1; the two ester-bound lipid chains are inserted into a hydrophobic pocket of TLR2 formed by an opening between two adjacent repeats at the convex face the horseshoe-like structure, whereas the amide-bound lipid chain is located in a smaller hydrophobic pocket in TLR1. The complex is further stabilized by direct interaction between TLR1 and TLR2. Based on the crystal structure, the authors suggest that lipopeptide-induced dimerization of TLR ectodomains brings the two intracellular TIR domains into close proximity and initiates signaling. Despite this major breakthrough, details of the molecular mechanism of MAMP recognition by TLR2 and most particularly the dynamic aspect of this process remain largely unknown. Whether the crystal structure of the TLR1-TLR2-Pam3CSK4 complex represents the functional signal-inducing complex existing in vivo remains an open question.
We have previously shown that a recombinant purified soluble TLR2 ectodomain could bind PAM3CSK4 directly and with high affinity (19). In addition, we have shown that TLR1 was unable to mediate binding of lipopeptide in the absence of TLR2. TLR2 thus appeared as the primary mediator of lipopeptide binding at the surface of cells expressing a functional TLR2 receptor complex. To better understand the molecular bases of the TLR2-lipopeptide interaction and the effects of this interaction on TLR1- and TLR6-mediated immune responses, we undertook site-directed mutagenesis of TLR2 followed by a series of tests for the functional ability of the mutant proteins.
EXPERIMENTAL PROCEDURES
Materials
Recombinant human soluble CD14 (sCD14) was purified as described (19). Synthetic Pam3CSK4 and MALP-2 lipopeptides come from EMC Microcollections (Tübingen, Germany). Alexa Fluor 488-labeled Pam3CSK4 (A488-Pam3CSK4) was prepared as described (19). Lipomannan from Mycobacterium bovis (BCGLM) was purified as described (26). Complexes between sCD14 and Pam3CSK4, A488-Pam3CSK4, MALP-2, and BCGLM were formed by incubating Pam3CSK4 (8 μg/ml), MALP-2 (8 μg/ml), or BCGLM (22 μg/ml) with sCD14 (100 μg/ml) overnight at 37 °C in Dulbecco's PBS containing 0.05% pyrogen-free human serum albumin (HSA).
Plasmid Preparations
TLR2 tagged with an N-terminal FLAG epitope was cloned into pcDNA3.1 as described (24). FLAG-TLR2 mutants were generated by PCR using the wild type construct. All of the constructs were verified by sequencing. The 3 × NF-κB-driven luciferase construct pNF-κB-luc and the plasmid pEGFP-N1, which constitutively express EGFP, were obtained from Stratagene and Clontech, respectively.
Binding of A488-Pam3CSK4 to the Surface of Cells
293 transfectants were cultured for 24–48 h before the experiment in complete culture medium on glass coverslips precoated with 0.5% gelatin. The cells were washed twice with PBS containing 0.05% HSA and incubated at 37 °C for 15 min with or without A488-Pam3CSK4-sCD14. At the end of incubation, the coverslips were washed several times in PBS containing 0.05% HSA, fixed for 20 min in PBS containing 4% paraformaldehyde, stained for TLR2 by successive incubation for 30 min at 4 °C with a mouse anti-FLAG® M2 monoclonal antibody and a Cy3-labeled anti-mouse antibody from Sigma, and mounted. The images were obtained using a fluorescence microscope equipped with charge-coupled device camera, and cell surface fluorescence intensity ratios of A488-Pam3CSK4 to FLAG-TLR2 variants were measured from CCD images manually using Metamorph software. The data are expressed as arbitrary units.
Flow Cytometry
293 cells cultured on 6-well plates were transfected at semi-confluence with TLR2 constructs or a vector control. At 24 h post-transfection, the cells were detached by incubation in PBS containing 0.5 mm EDTA and held on ice while being labeled sequentially with the M2 anti-FLAG mAb (10 μg/ml in PBS containing 0.05% HSA) and a fluorescein isothiocyanate-conjugated secondary antibody. The cells were then fixed, and fluorescence was measured in a Becton Dickinson FACScalibur analyzer. The data are expressed as percentages of cells with fluorescence above the vector control.
NF-κB Reporter Assay
293 cells were plated on a 96-well plate at 0.3 × 105 cells/well the day before transfection. Semiconfluent cells were transfected with a DNA-Lipofectamine mixture containing 25 ng of luciferase reporter construct, 50 ng of pEGFP-N1 plasmid, and 25 ng of pTLR2 (mutant or wild type) or control pcDNA3/well. At 24 h post-transfection, the cells were incubated for 5 h at 37 °C with various agonists and then lysed using 100 μl/well cell culture lysis reagent (Promega). EGFP fluorescence was measured using a microplate reader to normalize for transfection efficiencies. A luciferase assay reagent (50 μl; catalog number E1483; Promega) was then added to each well, and the luminescence was measured immediately in a microtiter plate luminometer. The relative luminescence units are expressed as luminescence/(10 × EGFP fluorescence).
RESULTS AND DISCUSSION
Design of TLR2 Mutants
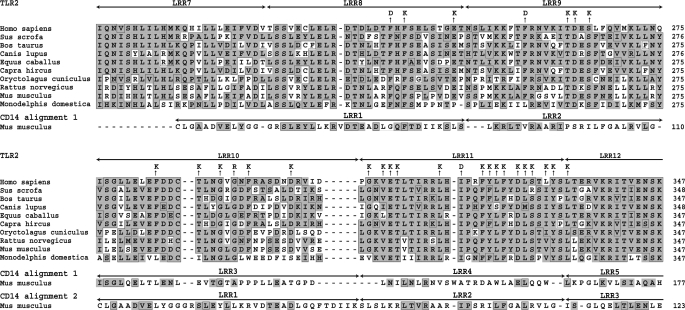
When the present study was started, the crystal structure of TLR2 was not known. Residues of TLR2 were thus selected for site-directed mutagenesis based on the following considerations. (i) The TLR2 ectodomain consists essentially of LRRs. The concave face of the LRR horseshoe-like structures is the typical region where protein-protein interactions occur (6). However, it seemed unlikely that the smooth concave β-structural surface of TLR2 may form a deep complementary pocket for small bacterial ligands such as lipopeptides. (ii) Examination of the crystal structure of CD14 (20), a LRR-containing protein that binds bacterial lipids and promotes TLR2- and TLR4-mediated responses to these lipids, suggested an alternative hypothesis. The crystal structure of CD14 shows a hydrophobic pocket that is formed through an opening between two adjacent repeats (LRR1 and LRR2) at the convex face of the horseshoe structure and is large enough to accommodate a variety of lipids. This suggested that binding of lipidic ligands by TLR2 could also occur via an opening between two repeats on the convex face. To test this hypothesis, we aligned the amino acid sequence of CD14 with those of TLR2 proteins from different organisms and found significant similarity (BLAST E-value ∼ e−11) between certain regions of these proteins including the repeats (LRR1–3) of CD14 involved in the formation of the lipid-binding pocket and three central repeats (LRR8–10) of TLR2 (Fig. 1). Starting from this alignment and shifting sequences of TLR2 and CD14, we manually found an alternative alignment (Fig. 1). Plausibility of the alignment was verified by molecular modeling and multiple sequence alignments. Based on these alignments, we designed a series of 28 TLR2 mutants (Fig. 1) by selecting residues within LRR8–11 that were mostly predicted to be located on the convex face and were conserved throughout evolution. Most of these residues were either hydrophobic or negatively charged. Therefore, to ensure a drastic change in their properties, we replaced them in most cases by a positively charged lysine residue.
FIGURE 1.
Alignment of TLR2 and CD14. Alignment of TLR2 from different organisms was obtained by using the CLUSTALW program (27). The first alignment of CD14 (amino acids 46–288) with the TLR2 sequences (amino acids 223–469 for human protein) was made by using BLAST (28) and has E values in the range of e−11 (only the N-terminal part of the alignment is shown). The alternative alignment of CD14 with TLR2 was made manually after examination of the first alignment. The boxed residues are identical to the consensus for the multiple alignment. Similar residues are shaded.
Ability of TLR2 Mutants to Mediate Responses to Pam3CSK4
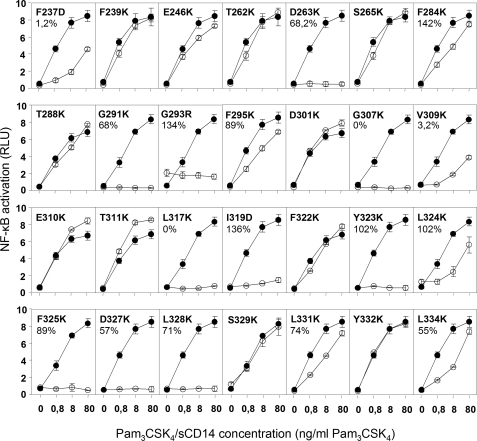
To test mutants, we used human epithelial kidney 293 cells. These cells express low levels of TLR1 and TLR6 and no TLR2. Upon transfection with TLR2, they are able to activate cell signaling in response to TLR2-TLR1 or TLR2-TLR6 agonists (10, 21). In addition, the sensitivity of their response to TLR2-inducing agonist is greatly increased when they also express membrane-bound CD14 (22, 23) or when agonists are incubated in the presence of sCD14 (24). Mutants were thus first tested for their ability to activate signal in response to TLR2-TLR1 agonist Pam3CSK4-sCD14 complexes. Mutants with defective responses were then further investigated for their expression at the cell surface. Of 28 mutants tested, nine lost completely or partially the ability to induce NF-κB in response to Pam3CSK4-sCD14 complexes while being expressed at the cell surface at levels similar to wild type TLR2 (Fig. 2 and Table 1). Moreover, they were unable to mediate CD14-independent responses to high concentrations of Pam3CSK4 (supplemental Fig. S1), indicating that the TLR2 defect was CD14-independent.
FIGURE 2.
Activation of NF-κB by TLR2 mutants in response to a synthetic tri-acylated lipopeptide presented under a complexed form to sCD14. 293 cells were incubated with increasing concentrations of Pam3CSK4-sCD14 complexes for 5 h at 37 °C. At the end of the incubation, the cells were lysed and assayed for expression of a NF-κB reporter construct as described under “Experimental Procedures.” The results are expressed as the means ± S.D. and are representative of two independent experiments. On each panel, responses of 293 cells expressing the indicated mutant (○) is compared with that of cells expressing wild type TLR2 (●).
TABLE 1.
Phenotypic characterization of TLR2 mutants
| Group | Mutants | Response to PAM3CSK4a | Response to MALP-2a | Expressionb | Binding of PAM3CSK4c |
|---|---|---|---|---|---|
| % of wild type | % of wild type ± S.E. | ||||
| Apolar residues with side chains inside the lipid-binding pocket | F284K | ++ | ++ | NDd | 142 ± 12 |
| F295K | ++ | ++ | ND | 89 ± 7,2 | |
| I319D | − | + | 136 | 3,4 ± 4 | |
| F325K | − | − | 89,4 | 65 ± 4,8 | |
| L328K | − | − | 70,6 | 46 ± 6 | |
| L331K | + | + | 74 | 82 ± 8,7 | |
| Residues on the surface near the entrance into the binding pocket | F322K | ++ | ++ | ND | 76,8 ± 6,4 |
| Y323K | − | − | 102 | 42 ± 4,8 | |
| L324K | + | + | 102 | 75 ± 8,4 | |
| D327K | − | + | 56,5 | 106 ± 8,5 | |
| Residues not in contact with either TLR1 or lipopeptide Pam3CSK4 | F237D | + | + | 1,2 | ND |
| F239K | ++ | ++ | ND | ND | |
| E246K | ++ | ++ | ND | ND | |
| T262K | ++ | ++ | ND | ND | |
| D263K | − | − | 68 | 119 ± 9,7 | |
| S265K | ++ | ++ | ND | ND | |
| T288K | ++ | ++ | ND | ND | |
| G291K | − | − | 68,2 | 92 ± 5,6 | |
| G293R | − | − | 134 | 96 ± 4,8 | |
| D301K | ++ | ++ | ND | ND | |
| V309K | + | + | 3,2 | ND | |
| E310K | ++ | + | ND | ND | |
| T311K | ++ | + | ND | ND | |
| S329K | ++ | ++ | ND | ND | |
| Y332K | ++ | ++ | ND | ND | |
| L334K | + | ++ | 54,5 | ND | |
| Mutants not expressed at the cell surface | G307K | − | − | 0 | ND |
| L317K | − | − | 0 | ND |
a Response of TLR2 mutants to PAM3CSK4. −, no response; +, intermediate response compared with wild type; ++, no significant difference between mutant and wild type.
b The expression of TLR2 mutants at the surface of transiently transfected cells was measured by flow cytometry with an antibody that recognizes the FLAG epitope expressed at the N terminus of the protein. The results are expressed as the percentages of cells with fluorescence intensity above vector control compared with wild type.
c Binding of fluorescent A488-Pam3CSK4 to the cell surface was measured as described under “Experimental Procedures” and expressed as a percentage of wild type.
d ND, not done.
Binding of Pam3CSK4 by TLR2 Mutants
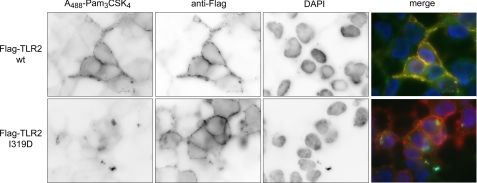
We have previously shown that binding of a fluorescent and active derivative of the Pam3CSK4 lipopeptide to TLR2 at the surface of cells could be observed when the lipopeptide was presented as a complex with sCD14 (19). Using wild type TLR2 as a reference, we measured the ability of the nine mutants defective for responses to Pam3CSK4 to bind this lipopeptide (Table 1). The I319D mutant completely lost the ability to bind Pam3CSK4 (Fig. 3). In contrast, the other eight mutants were still able to significantly bind the lipopeptide, albeit less efficiently than wild type for some. For example, the ability of L328K and Y323K mutants to bind Pam3CSK4 is reduced by ∼60% as compared with wild type.
FIGURE 3.
Pam3CSK4 binds the surface of cells expressing wild type TLR2 but does not bind cells expressing TLR2 I319D. The 293 cells transiently transfected with human TLR2 or TLR2 I319D mutant were incubated for 15 min at 37 °C with A488-Pam3CSK4-sCD14 (80 ng/ml A488-Pam3CSK4, 1 μg/ml sCD14) complexes and then stained for surface expressed FLAG-tagged TLR2 proteins. In addition, the cells were stained for the nucleus by the addition of 4′,6′-diamino-2-phenylindole (DAPI) in the mounting medium. CCD images for the fluorescence of A488-Pam3CSK4, TLR2 (wild type (wt) or I319D mutant), and the nucleus are shown for representative cells as well as the merged images.
Mapping of Mutations on TLR2 Crystal Structures
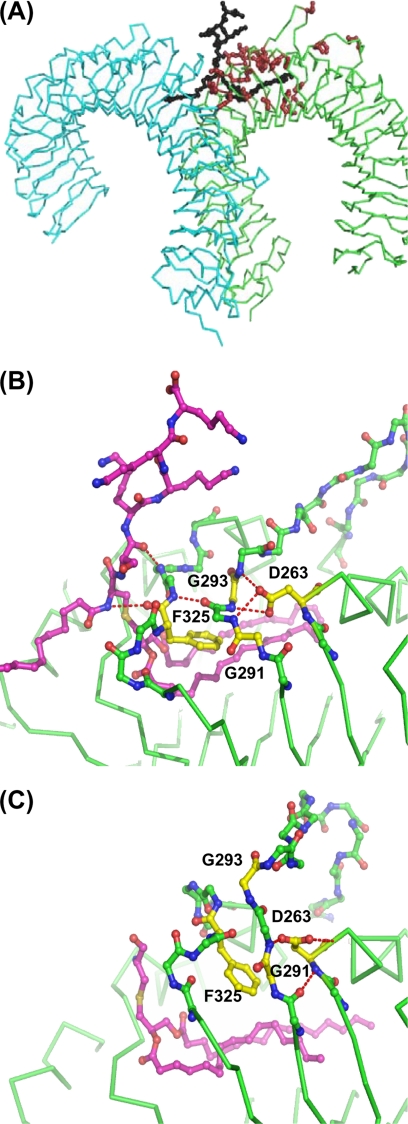
The recent crystal structures of a human TLR2-TLR1-Pam3CSK4, mouse TLR2-Pam3CSK4, and mouse TLR2-Pam2CSK4 complexes (18) validated our strategy of the mutant design and confirmed, at least in part, our prediction of the lipopeptide-binding pocket location. Indeed, the replaced residues of our TLR2 mutants are clustered at the area of LRR8–11, where, in accordance with the crystal structures, key functional interactions between TLR2, TLR1, and the Pam3CSK4 lipopeptide occur (Fig. 4A).
FIGURE 4.
Location of mutations in the TLR2 structure. A, a general view of the crystal structure of TLR1-TLR2-Pam3CSK4 complex (Protein Data Bank code 2Z7X) and locations of TLR2 residues (in brown) that were mutated in this study. TLR2, TLR1 and Pam3CSK4 are in green, blue, and black, respectively. B and C, H-bond networks on the surfaces of LRRs 9–11 stabilize two conformations of TLR2 in the region adjacent to the ligand-binding pocket. B and C, human TLR1-TLR2-lipopeptide complex (B; Protein Data Bank code 2Z7X; conformation 1) and mouse TLR2-lipopeptide complex (C; Protein Data Bank code 2Z81, conformation 2). H-bonds are shown by red dotted lines. The regions of LRR10 and 11 that are in different conformations in the two structures are outlined by a ball-and-stick backbone. The remaining parts of the chains are shown by green traces of Cα atoms. Side chains of mutated residues are shown in yellow.
The mutated residues can be subdivided into three groups according to their location within the structure. The first group consists of six apolar residues that are located inside the binding pocket of TLR2 and interact with the two ester-bound lipid chains of Pam3CSK4 (Table 1). In this group, replacement of the residues located near the opening of the pocket (Ile319, Phe325, and Leu328) by a charged residue rendered TLR2 unable to mediate cellular response to Pam3CSK4. The I319D mutant also lost the ability to bind Pam3CSK4, whereas the F325K and L328K mutants showed a modest decrease in their ability to bind the lipopeptide at a concentration that allows strong NF-κ activation. Thus, changes at the opening of the pocket can affect the ability of TLR2 to bind Pam3CSK4. The modest decrease in binding efficiency of F325K and L328K cannot, however, account for the complete loss of response of these two mutants. The most likely explanation, given the location of these mutations in the region of contact with TLR1, is that they also interfere indirectly with the ability of TLR2 to interact functionally with TLR1 and induce signaling. In contrast, replacement of residues located near the bottom of the pocket (Phe284, Phe295, and Leu331) by a lysine did not alter the ability of TLR2 to bind or to induce a response to Pam3CSK4. Analysis of the three-dimensional structure of TLR2 shows that the charged amino group of lysine in positions 295 and 331 may find a way out of the structure without large rearrangement of the polypeptide backbone. However, the ϵ-amino group of Lys in position 284 cannot be exposed to the solvent and be outside of the pocket without serious structural rearrangement. One possibility is that this Lys forms H-bonds with the peptide groups of the backbone or (and) water molecules inside the pocket. Indeed, the pocket with the two ester-bound lipid chains of Pam3CSK4 still has ∼10% of the solvent-accessible volume (18). Altogether these mutations show that the anterior part of the pocket is more sensitive to changes than sites located near the pocket bottom. This observation is in agreement with the fact that TLR2 can recognize lipids with fatty acyl chains longer than Pam3CSK4 (25), because this implies that the TLR2 pocket can have some free space at its bottom to accommodate longer acyl chains.
The second group comprises residues Phe322, Tyr323, Leu324, and Asp327, which are located on the surface near the entrance into the lipopeptide-binding pocket (Table 1). In addition, the Phe322, Tyr323, and Leu324 residues are a part of the interface between TLR2 and TLR1. The F322K mutant was still able to mediate the response to Pam3CSK4. The L324K mutant did it less efficiently than the wild type protein, and the Y323K mutant lost completely the ability to mediate responses to the lipopeptide while retaining significant binding capacities. These results suggest that Tyr323 plays an important role in the interaction of TLR2 with TLR1 and that some other residues at the TLR1-TLR2 interface can be replaced without significant functional effect. The fourth mutant, D327K, failed to respond to Pam3CSK4 while preserving its ability to bind the lipopeptide. The side chain of Asp327 is not in contact with TLR1 but is at the proximity of the positively charged Lys residues of the lipopeptide. So, the replacement of negatively charged Asp by positively charged Lys may affect orientation of the peptide head group of Pam3CSK4 because of the electrostatic repulsion.
The third group includes 16 mutants with residues that are not in direct contact with either TLR1 or the lipopeptide (Table 1). All of them are accessible to the solvent. Most of these mutants, as expected from their location within the three-dimensional structure, do not affect the ability of TLR2 to bind lipopeptides and to mediate the responses. Surprising exceptions are three mutants, D263K, G291K, and G293K, that were still able to bind the lipopeptide but lost the ability to respond to the ligand. The mutated residues are located too far from the dimer interface or the entrance of the hydrophobic pocket to interfere directly with TLR2-TLR1 heterodimerization or TLR2 binding of Pam3CSK4 (Fig. 4). Possible roles for these residues in TLR2 functionality are described in the next section.
Details of Molecular Mechanism of Bacterial Lipid Recognition by TLR2-TLR1 Heterodimers Suggested by Mutagenesis
Superposition of the crystal structures of the human TLR2-TLR1-Pam3CSK4 and mouse TLR2-Pam3CSK4 revealed that TLR2 ectodomains have very similar structures except in the two loop regions of LRR10 (amino acids 291–305) and LRR11 (amino acids 322–329) on the convex surface of the solenoid. As a consequence, the entrance of the lipopeptide-binding pocket of TLR2, which is located between LRR11 and LRR12, is narrower in the human TLR2/TLR1-Pam3CSK4 structure (conformation 1) than in the mouse TLR2-Pam3CSK4 structure (conformation 2). In conformation 1, the LRR11 loop also provides additional contacts with TLR1. Given the high degree of sequence conservation between human and mouse TLR2, Jin et al. (18) suggested that the change in conformation of LRR10 and 11 of TLR2 is induced by TLR1 binding. These two regions of TLR2 were intensively mutated in the present study. Analysis of the mutant phenotypes showed that replacement of a majority of residues in the LRR11 loop that is in contact with TLR1 renders TLR2 unable to mediate responses.
As stated above, the most surprising observation in our mutational analysis was the absence of response of the D263K, G291K, and G293R mutants. Close examination of the crystal structures (18) reveals that these residues can participate in specific networks of H-bonds on the convex surface of TLR2 (Fig. 4, B and C). In conformation 1, the network starts with Asp263 located on LRR9. The side chain of Asp263 forms H-bonds with the main chain NH groups of Val292, Gly293, and Asn294 of LRR10. This network continues via backbone H-bonds between the carbonyl oxygen of Val292 and the NH group of Tyr326 of LRR11 and ends by H-bonds of the backbone oxygen of Phe325 and the NH group of Asp327 of LRR11 with the NH group of the cysteine and the carbonyl oxygen of the serine of Pam3CSK4 (Fig. 4B). The H-bonds between LRR11 and the lipopeptide stabilize the amide-bound lipid chain of Pam3CSK4 in an orientation that is proper for its insertion in the lipid-binding pocket of TLR1. In addition, in conformation 1, the LRR11 loop of TLR2 contacts TLR1 via side chains of the Phe322, Tyr323, and Leu324 residues. In contrast, in the complexes without TLR1, Asp263 forms a H-bond with the backbone NH group of Leu292 that stabilizes conformation 2, in which TLR2 does not orient the third tail of the lipopeptide toward TLR1, and the LRR11 loop moves away from the dimerization site. Thus, Asp263 serves as a pivot of both H-bond networks. The crucial role of Asp263 is supported by its strict conservation among TLR2 molecules. In the D263K mutant, the lysine side chain cannot form H-bonds with the main chain NH groups of LRR10. As a result, this mutation should destroy H-bonds networks via a “domino effect” mechanism. To be a part of the network in conformation 1, Gly291 and Gly293 must have conformations that are sterically disallowed for any other residues. Similarly, in conformation 2, the presence of Gly in position 291 may also be important because more bulky residues should create steric hindrance. Thus, the D263K, G291K, and G293K mutations should destabilize both conformations observed in the crystal structures (18) and allow free transition between them. Our experiments show that mutations abolishing this H-bond network render TLR2 unable to signal in response to Pam3CSK4. Because these mutants still bind the lipopeptide very efficiently, the only likely explanation is that they lose the ability to interact with TLR1. Indeed, disruption of the H-bond network of conformation 1 is likely to affect the ability of TLR2 to properly orient the amide-bound lipid chain toward the TLR1 pocket. In addition, the Phe322, Tyr323, and Leu324 loop may be in a loose conformation.
The loss of response of the mutants thus underlines the importance of the surface H-bond network, which involves several residues (Asp263, Gly291, Gly293, Val292, Phe325, and Tyr326) and uncovers a possible mechanism for the switching of TLR2 between conformations 1 and 2. Our finding that these mutants still efficiently bind their ligand suggests that stabilization of these TLR2 conformations is not necessary for ligand binding. However, the ability of TLR2 to stabilize the ligand in the orientation that is proper for binding to TLR1 and therefore favorable for heteromerization seems to be of paramount importance.
Ability of TLR2 Mutants to Mediate Responses to Di-acylated Lipopeptide MALP-2 and Mycobacterial Lipomannan
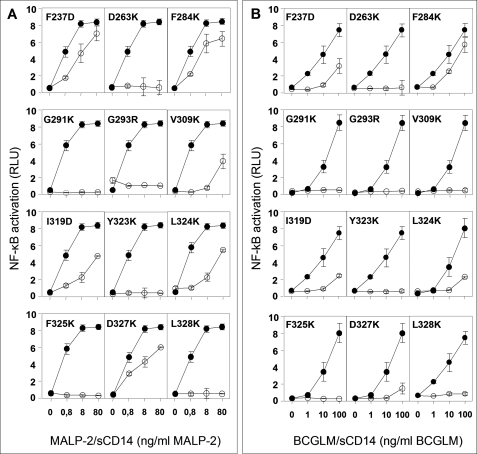
To better understand the structure-function relationship of TLR2 with its agonists, we measured the ability of TLR2 mutants to mediate response to the di-acylated synthetic mycoplasma-derived lipopeptide MALP-2 and to BCGLM. Di-acylated MALP-2 is structurally close to PAM3CSK4 with a conserved PAM2CS-peptide moiety and activates TLR2-TLR6 heterodimers (10). In contrast, BCGLM, a lipoglycan consisting in a conserved mannosylphosphatidyl myo-inositol anchor that is mainly tri-acylated and polymannosylated, activates TLR2-TLR1 heterodimers (26). All of the mutants defective for response to PAM3CSK4 were also defective for responses to MALP-2 and BCGLM (Fig. 5). In addition, no new defects were found by screening the mutants with these agonists (supplemental Fig. S1). These observations suggest that residues of TLR2 involved in interacting with TLR1 and TLR6 upon binding microbial lipids are mostly identical whether the polar head is a polypeptidic or a glycosidic chain and the co-receptor is TLR1 or TLR6.
FIGURE 5.
Activation of NF-κB by TLR2 mutants in response to a synthetic di-acylated lipopeptide MALP-2 and BCGLM. Mutants defective for responses to Pam3CSK4-sCD14 were tested for responses to MALP-2-sCD14 complexes (A) and BCGLM-sCD14 complexes (B) as described for Fig. 2. On each panel, responses mediated by the indicated mutant (○) are compared with those of wild type TLR2 (●).
Conclusions
In this work, we designed and produced a series of TLR2 mutants and assessed their ability to bind and to induce response to the Pam3CSK4 lipopeptide. Our mutational analysis provides evidence that the recently published crystal structure of the TLR2-TLR1-Pam3CSK4 complex (18) represents a functional signal-inducing complex. Indeed, in agreement with this structure, we found that several mutations in the anterior part of the hydrophobic lipopeptide-binding pocket or at the TLR1-TLR2 interface diminished or blocked signaling.
The most surprising finding was that some mutations that were located on the surface of the TLR2 structure and 10–15 Å away from the site of contact with the lipopeptide or TLR1 induced a complete block of signaling. Close examination of the crystal structures (18) revealed that the mutated residues participate in two specific H-bond networks on the convex surface of TLR2. The networks stabilize LRR10 and LRR11 in conformations 1 and 2 in the presence and absence of TLR1 correspondingly. The first H-bond network extends from pivot residue Asp263 of LRR9 via LRR10 and terminates by H-bonding of LRR11 with lipopeptide. The H-bonding with LRR11 stabilizes the amide-bound lipid chain of Pam3CSK4 in an orientation that is suitable for interaction with TLR1. This conformation also provides additional contacts for interaction of the LRR11 with TLR1. In conformation 2, the same residues of LRR9–11 form a different H-bond network, which makes the entrance of the hydrophobic pocket slightly wider and does not orient the amide-bound lipid chain of the lipopeptide toward TLR1. In accordance with the results of our mutation analysis, the integrity of these H-bond networks is not necessary for binding the lipopeptide but is critical for signaling. These data allow us to suggest that binding of ligands possessing fatty acyl moieties by TLR2 have two principal steps: first, a nonspecific binding of fatty acids with the TLR2 hydrophobic pocket and second, a specific H-bonding of the ligand polar head with the anterior part of the pocket, which is critical for the signal induction. Our data also allowed us to suggest a dynamic model of interaction, in which LRR10 and LRR11 of TLR2 fluctuate between two conformations stabilized through the above mentioned H-bond networks. One of them with the wider pocket entrance is more suitable for the nonspecific step of the ligand binding, whereas the other, via a specific orientation of the ligand, favors formation of a signal-inducing ternary complex.
We found that residues of TLR2 that play an important role in TLR2-TLR1-mediated response to Pam3CSK4 appear to be also critical for the TLR2-TLR1-dependent response to tri-acylated BCGLM and for the TLR2-TLR6-dependent response to di-acylated MALP-2. These observations are in agreement with structural data that showed a common binding site for a di- and tri-acylated agonists (18). In addition, we have recently shown that despite structural differences, tri-acylated lipoglycan BCGLM can compete with Pam3CSK4 for binding to TLR2 (26). The network of hydrogen bonds that controls positioning of Pam3CSK4 is thus likely to play a similar role for other ligands. This suggests that positioning of ligand is also important for TLR2-TLR6-mediated cell signaling.
In conclusion, our study not only validates the TLR2-TLR1-Pam3CSK4 complex seen in the crystal structure as a signal-inducing complex but also allowed us to gain insight into the dynamics of the recognition process. Future studies will be required to understand how TLR2 agonists such as bacterial peptidoglycan and viral glycoproteins that do not possess fatty acids are recognized.
Supplementary Material
Acknowledgments
We thank Drs. Edouard Bertrand and Naomi Taylor for invaluable help with performing this work and Dr. Jérôme Nigou for critical reading of this manuscript.
This work was supported in part by the Agence Nationale de Recherche sur le Sida et les Hépatites Virales (to T. V.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
- TLR
- Toll-like receptor
- TIR
- Toll/interleukin-1 receptor
- LRR
- leucine-rich repeats
- MAMP
- microbial-associated molecular patterns
- sCD14
- soluble CD14
- A488
- Alexa Fluor 488-labeled
- BCGLM
- lipomannan from M. bovis
- PBS
- phosphate-buffered saline
- HSA
- human serum albumin
- EGFP
- enhanced green fluorescent protein.
REFERENCES
- 1.Akira S., Uematsu S., Takeuchi O. (2006) Cell 124, 783–801 [DOI] [PubMed] [Google Scholar]
- 2.O'Neill L. A., Bowie A. G. (2007) Nat. Rev. Immunol. 7, 353–364 [DOI] [PubMed] [Google Scholar]
- 3.Trinchieri G., Sher A. (2007) Nat. Rev. Immunol. 7, 179–190 [DOI] [PubMed] [Google Scholar]
- 4.Bell J. K., Mullen G. E., Leifer C. A., Mazzoni A., Davies D. R., Segal D. M. (2003) Trends Immunol. 24, 528–533 [DOI] [PubMed] [Google Scholar]
- 5.Kajava A. V. (1998) J. Mol. Biol. 277, 519–527 [DOI] [PubMed] [Google Scholar]
- 6.Kobe B., Kajava A. V. (2001) Curr. Opin. Struct. Biol. 11, 725–732 [DOI] [PubMed] [Google Scholar]
- 7.Kobe B., Kajava A. V. (2000) Trends Biochem. Sci. 25, 509–515 [DOI] [PubMed] [Google Scholar]
- 8.Ozinsky A., Underhill D. M., Fontenot J. D., Hajjar A. M., Smith K. D., Wilson C. B., Schroeder L., Aderem A. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 13766–13771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alexopoulou L., Thomas V., Schnare M., Lobet Y., Anguita J., Schoen R. T., Medzhitov R., Fikrig E., Flavell R. A. (2002) Nat. Med. 8, 878–884 [DOI] [PubMed] [Google Scholar]
- 10.Takeuchi O., Kawai T., Mühlradt P. F., Morr M., Radolf J. D., Zychlinsky A., Takeda K., Akira S. (2001) Int. Immunol. 13, 933–940 [DOI] [PubMed] [Google Scholar]
- 11.Takeuchi O., Sato S., Horiuchi T., Hoshino K., Takeda K., Dong Z., Modlin R. L., Akira S. (2002) J. Immunol. 169, 10–14 [DOI] [PubMed] [Google Scholar]
- 12.Henneke P., Morath S., Uematsu S., Weichert S., Pfitzenmaier M., Takeuchi O., Müller A., Poyart C., Akira S., Berner R., Teti G., Geyer A., Hartung T., Trieu-Cuot P., Kasper D. L., Golenbock D. T. (2005) J. Immunol. 174, 6449–6455 [DOI] [PubMed] [Google Scholar]
- 13.Krishnegowda G., Hajjar A. M., Zhu J., Douglass E. J., Uematsu S., Akira S., Woods A. S., Gowda D. C. (2005) J. Biol. Chem. 280, 8606–8616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sandor F., Latz E., Re F., Mandell L., Repik G., Golenbock D. T., Espevik T., Kurt-Jones E. A., Finberg R. W. (2003) J. Cell Biol. 162, 1099–1110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tapping R. I., Tobias P. S. (2003) J. Endotoxin Res. 9, 264–268 [DOI] [PubMed] [Google Scholar]
- 16.Gilleron M., Nigou J., Nicolle D., Quesniaux V., Puzo G. (2006) Chem. Biol. 13, 39–47 [DOI] [PubMed] [Google Scholar]
- 17.Buwitt-Beckmann U., Heine H., Wiesmüller K. H., Jung G., Brock R., Akira S., Ulmer A. J. (2006) J. Biol. Chem. 281, 9049–9057 [DOI] [PubMed] [Google Scholar]
- 18.Jin M. S., Kim S. E., Heo J. Y., Lee M. E., Kim H. M., Paik S. G., Lee H., Lee J. O. (2007) Cell 130, 1071–1082 [DOI] [PubMed] [Google Scholar]
- 19.Vasselon T., Detmers P. A., Charron D., Haziot A. (2004) J. Immunol. 173, 7401–7405 [DOI] [PubMed] [Google Scholar]
- 20.Kim J. I., Lee C. J., Jin M. S., Lee C. H., Paik S. G., Lee H., Lee J. O. (2005) J. Biol. Chem. 280, 11347–11351 [DOI] [PubMed] [Google Scholar]
- 21.Aliprantis A. O., Yang R. B., Mark M. R., Suggett S., Devaux B., Radolf J. D., Klimpel G. R., Godowski P., Zychlinsky A. (1999) Science 285, 736–739 [DOI] [PubMed] [Google Scholar]
- 22.Brightbill H. D., Libraty D. H., Krutzik S. R., Yang R. B., Belisle J. T., Bleharski J. R., Maitland M., Norgard M. V., Plevy S. E., Smale S. T., Brennan P. J., Bloom B. R., Godowski P. J., Modlin R. L. (1999) Science 285, 732–736 [DOI] [PubMed] [Google Scholar]
- 23.Kirschning C. J., Wesche H., Merrill Ayres T., Rothe M. (1998) J. Exp. Med. 188, 2091–2097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vasselon T., Hanlon W. A., Wright S. D., Detmers P. A. (2002) J. Leukocyte Biol. 71, 503–510 [PubMed] [Google Scholar]
- 25.Buwitt-Beckmann U., Heine H., Wiesmüller K. H., Jung G., Brock R., Ulmer A. J. (2005) FEBS J. 272, 6354–6364 [DOI] [PubMed] [Google Scholar]
- 26.Nigou J., Vasselon T., Ray A., Constant P., Gilleron M., Besra G. S., Sutcliffe I., Tiraby G., Puzo G. (2008) J. Immunol. 180, 6696– 6702 [DOI] [PubMed] [Google Scholar]
- 27.Higgins D. G., Thompson J. D., Gibson T. J. (1996) Methods Enzymol. 266, 383–402 [DOI] [PubMed] [Google Scholar]
- 28.Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. (1990) J. Mol. Biol. 215, 403–410 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.