Abstract
The central complement inhibitor factor I (FI) degrades activated complement factors C4b and C3b in the presence of cofactors such as C4b-binding protein, factor H, complement receptor 1, and membrane cofactor protein. FI is a serine protease composed of two chains. The light chain comprises the serine protease domain, whereas the heavy chain contains several domains; that is, the FI and membrane attack complex domain (FIMAC), CD5, low density lipoprotein receptor 1 (LDLr1) and LDLr2 domains. To understand better how FI acts as a complement inhibitor, we used homology-based models of FI domains to predict potential binding sites. Specific amino acids were then mutated to yield 16 well expressed mutants, which were then purified from media of eukaryotic cells for functional analyses. The Michaelis constant (Km) of all FI mutants toward a small substrate was not altered, whereas some mutants showed increased maximum initial velocity (Vmax). All the mutations in the FIMAC domain affected the ability of FI to degrade C4b and C3b irrespective of the cofactor used, whereas only some mutations in the CD5 and LDLr1/2 domains had a similar effect. These same mutants also showed impaired binding to C3met. In conclusion, the FIMAC domain appears to harbor the main binding sites important for the ability of FI to degrade C4b and C3b.
Keywords: Complement, Inflammation, Protease, Protein Domains, Protein-Protein Interactions, Complement Inhibitors, Factor I
Introduction
The complement system is a key component of the innate immune system. The main functions of complement are defense against infections and clearance of immune complexes and apoptotic cells as well as alerting the adaptive immune system. Complement can be activated through three different pathways; that is, the classical pathway, which is initiated by binding of C1q to antigen-antibody complexes, the lectin pathway triggered by binding of mannose-binding lectin or ficolins to carbohydrate structures, and the alternative pathway, which is activated by the spontaneous activation of C3 in the absence of inhibition via factor H (FH).2 All three pathways converge at the cleavage of C3 into C3a and C3b. In the next step, C5-convertases are formed that cleave C5 into C5a and C5b (1). To regulate complement on self-tissue and to prevent spontaneous activation and systemic depletion, complement is controlled by both fluid-phase and membrane-bound inhibitors (2). Several complement inhibitors accelerate decay of the C3 and C5 convertases, but their actions are strongly enhanced by the fluid-phase inhibitor factor I (FI). FI is a key serine protease that inactivates all complement pathways by degrading activated complement factors C4b and C3b. FI does not cleave intact C4 or C3, and it degrades C4b and C3b only in the presence of specific cofactors such as FH (3), C4b-binding protein (C4BP) (4), membrane-cofactor protein (MCP) (5), and complement receptor 1 (CR1) (6). The physiological importance of FI is shown by the fact that patients lacking FI have increased susceptibility to recurrent infection with encapsulated microorganisms, glomerulonephritis, and rheumatologic diseases (7–10). Heterozygous mutations have been also identified in FI in patients with atypical hemolytic uremic syndrome (11–14).
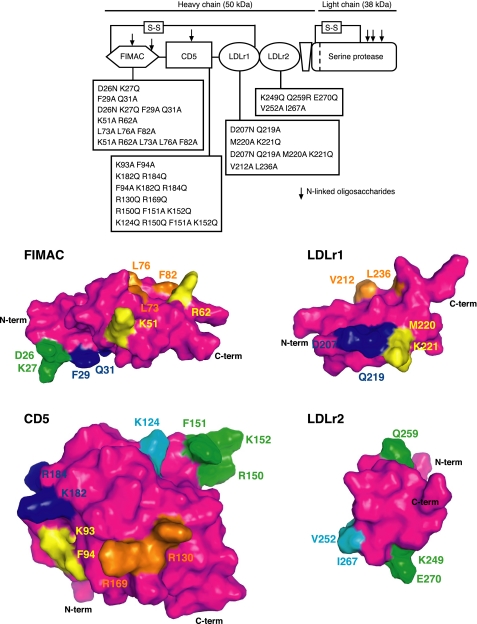
FI is a multidomain protein (88 kDa) produced as a single polypeptide chain and translationally modified by the addition of six N-linked glycans (15). Furthermore, FI is proteolytically processed, which results in removal of the signal peptide and excision of four basic amino acids. The latter cleavage generates the two FI chains, the heavy (50 kDa) and the light chain (38 kDa), which remain covalently linked by a disulfide bond (16). The non-catalytic heavy chain is composed of several domains that show structural homology to other proteins; that is, the FI and membrane attack domain (FIMAC), the CD5 domain, the low density lipoprotein receptor domains 1 and 2 (LDLr1 and -2), and a small region of unknown homology (17) (Fig. 1). The light chain comprises the serine protease (SP) domain, which contains the catalytic triad His-362, Asp-411, and Ser-507 (18).
FIGURE 1.
Predicted three-dimensional structure of the individual domains of FI. FI is composed of the following domains: FIMAC, a CD5-like domain, LDLr1 and -2, a small region of no known homology, and an SP domain. The positions of N-linked glycosylations are marked with arrows, and two important interdomain disulfide bridges are indicated. The mutations are numbered without the signal peptide. The putative models of FIMAC, CD5, and LDLr1 and -2, domains were constructed using homology modeling. The constructed mutants containing several simultaneous amino acid changes are listed for each domain. The figure was prepared with PyMol. The scale and orientation of the different domains were modified to facilitate the overall location of the mutations. Mutants D26N/K27Q and K249Q/Q259R/E270Q that were not secreted by cells, possibly due to folding/destabilization problems, are also shown for consistency.
Both x-ray, neutron scattering (19), and electron microscopy (20) have been used to elucidate the low resolution three-dimensional structure of FI, but so far there is no high resolution structure of FI. FI is a unique serine protease as it circulates in its active form in blood and does not have any known circulating inhibitor. This in addition to the fact that FI cannot cleave its substrates without a cofactor suggests that the active site of FI is obscured or conformationally altered by the heavy chain and only becomes exposed when a substrate/cofactor binds to FI (20).
Because there is no three-dimensional structure of FI available yet, our group has previously developed homology-based models of the individual domains of FI (9, 14). These models were used in the current study to rationally design point mutations to study relationships between the structure and function of FI. To elucidate which parts of the heavy chain are involved in the function of FI, putative binding patches on the surface of the individual domains were mutated. In each recombinant mutant protein, 2–5 amino acids were changed from charged amino acids to polar ones (most often to glutamine), whereas the hydrophobic amino acids were altered to alanine residues. Functional analyses of purified FI variants showed that particularly the FIMAC domain is important for the ability of FI to degrade C4b and C3b in the fluid phase as well as C3b deposited on the surface. These data are important for understanding the structural requirements for the function of this important complement inhibitor of which alterations are associated with severe diseases.
EXPERIMENTAL PROCEDURES
Homology-based Model of FI and Structural Analysis of the Mutations
The three-dimensional models of the CD5, LDLr1, LDLr2, and SP domains of human FI were described in Nilsson et al. (9), whereas the model of the FIMAC domain was presented in Nilsson et al. (14). The numbering of the amino acid residues in FI is based on the mature protein without signal sequence. Hot spots and regions putatively involved in macromolecular interactions were investigated using the online electrostatic computation server PCE (21) and the ODA (Optimal Docking Areas) online application that predicts protein-protein binding sites (22). These data together with interactive structural analysis of the models guided us to select a limited number of residues to assess experimentally. The residues that were targets for the mutagenesis were solvent-exposed and not involved in any clear stabilizing interactions with the remaining parts of each domain (e.g. salt bridges), and the amino acid substitutions were, therefore, expected to be well tolerated. However, as the various domains were modeled separately and because it is not known how they are arranged in the intact FI, it is possible that some of the mutated sites are involved in interdomain interactions rather than in the binding of substrates and cofactors. In each case several amino acids were mutated simultaneously, and these were in close structural proximity, forming patches that were either hydrophobic or charged (Fig. 1).
Proteins
Human C4BP (23) and FH (24) were purified as described previously. C1, C4, C2, C3, C3b, C4b, factor B (FB), factor D (FD), and properdin were purchased from Complement Technology (Tyler, TX). C3b and C4b were labeled with 125I using the chloramine T method as described before (25). C3 was treated with 100 mm methylamine, pH 8.0, at 37 °C for 1 h to hydrolyze the internal thioester bond, thus changing the conformation to C3met. C3met was then dialyzed overnight in 50 mm Tris-HCl, 150 mm NaCl, pH 8.0, at 4 °C. C3met still retains the anaphylatoxin domain but resembles C3b in its overall conformation and properties.
CFI cDNA Clones for Recombinant Proteins
Full-length cDNA encoding the human CFI gene was cloned into the eukaryotic expression vector pcDNA3 (Invitrogen) with addition of an N-terminal His tag as described previously (13). The mutations were introduced in the CFI gene using primers listed in Table 1 and the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA). The mutations were confirmed by automated DNA sequencing using the Big dye terminator kit (Applied Biosystems, Foster City, CA).
TABLE 1.
Primers used for site-directed mutagenesis
The mutations are marked in bold and underlined.
| Mutationsa | Sequence (5′ to 3′) |
|---|---|
| D26N/K27Q | CT CAC CTC TCC TGC AAT CAA GTC TTC TGC CAG C |
| F29A/Q31A | CC TGC GAT AAA GTC GCC TGC GCG CCA TGG CAG AGA TG |
| D26N/K27Q/F29A/Q31A | TC TCC TGC AAT CAA GTC GCC TGC GCG CCA TGG CAG AG |
| K51A | CCG TAT CAG TGC CCA GCG AAT GGC ACT GCA GTG |
| R62A | G TGT GCA ACT AAC AGG GCA AGC TTC CCA ACA TAC |
| L73A/L76A | C TGT CAA CAA AAG AGT GCG GAA TGT GCT CAT CCA GGG ACA AAG GC |
| F82A | CTT CAT CCA GGG ACA AAG GCT TTA AAT AAC GGA ACA TG |
| K93Q/F94A | C ACA GCC GAA GGA CAG GCT AGT GTT TCC TTG AAG |
| F94A | C ACA GCC GAA GGA AAG GCT AGT GTT TCC TTG AAG |
| K182Q/R184Q | GAA TGT ACT TTT ACT CAG AGA CAA ACT ATG GGT TAC CAG G |
| R130Q | GC AGC TGG AGC ATG CAG GAA GCC AAC GTG G |
| R169Q | TA CAT GTG CAT TGC CAA GGA TTA GAG ACC AG |
| R150Q/F151A/K152Q | GCT GAT ACT CAA AGA CAG GCT CAG TTG TCT GAT CTC TC |
| K124Q | G ACA ATG TTC ATA TGC CAA AGC AGC TGG AGC AT |
| D207N | CA GAT TCT CCA ATG GAT AAC TTC TTT CAG TGT GTG |
| Q219A | AT GGG AAA TAC ATT TCT GCG ATG AAA GCC TGT GAT G |
| M220A/K221Q | G AAT GGG AAA TAC ATT TCT CAG GCG CAA GCC TGT GAT GG |
| Q219A/M220A/K221Q | G AAT GGG AAA TAC ATT TCT GCG GCG CAA GCC TGT GAT GG |
| V212A | GAC TTC TTT CAG TGT GCG AAT GGG AAA TAC ATT |
| L236A | A GAC CAA AGT GAT GAA GCG TGT TGT AAA GCA TGC |
| K249Q | AA GGC TTC CAT TGC CAA TCG GGT GTT TGC |
| Q259R | T CCA AGC CAG TAT CGA TGC AAT GGT GAG G |
| E270Q | AC TGC ATT ACA GGG CAA GAT GAA GTT GGC |
| V252A | TGC AAA TCG GGT GCT TGC ATT CCA AGC |
| I267A | GT GAG GTG GAC TGC GCT ACA GGG GAA GAT G |
a Numbering without the signal peptide.
Purification of Recombinant FI
FI wt and the mutants were expressed in human embryonic kidney (HEK 293) cells and purified by affinity chromatography using nickel-nitrilotriacetic acid Superflow resin (Qiagen, Hilden, Germany) as described (9). The purified recombinant FI mutants were visualized using goat antibodies (Quidel, San Diego, CA) on Western blot as described before (9). All proteins were stored at −80 °C. Protein concentrations were determined by measuring the absorbance at 280 nm and subtracting the absorbance at 320 nm. The concentrations were then verified by SDS/PAGE electrophoresis followed by Coomassie staining of proteins.
Amidolytic Assay
The amidolytic assay was essentially performed as described (26). Briefly, wt FI or the individual mutants (all at a final concentration of 25 μg/ml) were combined with different concentrations (200, 100, 25, 6.25 μm final concentration) of DPR-AMC substrate (t-butoxycarbonyl-Asp(benzyl)-Pro-Arg-amidomethylcoumarin·HCl; Bachem, Bubendorf, Switzerland) in Bicine buffer (25 mm Bicine, 0.5 mm EDTA, 146 mm NaCl, H2O, pH 8.25). These were then transferred to black optical plates (Nunc, Roskilde, Denmark), and the activity of each enzyme was subsequently determined using a microplate reader (Tecan infinite 200, Tecan, Männedorf, Switzerland) with excitation at 360 nm and an emission readout at 465 nm at 37 °C for 1 h. Using Graphpad Prism 5.0 (Graphpad Software, La Jolla, CA), both maximum initial velocity (Vmax) and the Michaelis constant (Km) of all enzymes were determined using nonlinear regression analysis. The amount of substrate converted in the reaction was calculated from the fluorescence units using a conversion factor, which was determined from complete substrate turnover using thrombin as a converting enzyme.
C4b and C3b Degradation Assay
To assay degradation of C4b, recombinant FI wt or mutant proteins were diluted to a final concentration of 1, 2.5, 5, or 10 μg/ml and mixed with 100 μg/ml C4BP, 50 μg/ml C4b, and trace amounts of 125I-labeled C4b. The positive control contained 100 μg/ml C4BP and 20 μg/ml wt FI, whereas in the negative control FI was omitted. The C3b degradation assay was similar except that 20 μg/ml FH, 150 μg/ml C3b, 1, 2.5, 5, or 10 μg/ml of FI, and trace amounts of 125I-labeled C3b were mixed together. To determine FI activity in the presence of CR1 as a cofactor, 18 μl of human erythrocyte ghosts were prepared as described previously (27) and added as a source of CR1. Two concentrations of FI were tested: 0.5 and 10 μg/ml. We further used lung cancer cell line H2087 as a source of the co-factor MCP, a cell line that we have previously shown to express MCP but no CR1 (28). The H2087 cells were harvested with versene (Invitrogen) and solubilized at 8 × 107 cells/ml in phosphate-buffered saline containing 1% Nonidet P-40 and 2 mm phenylmethylsulfonyl fluoride. After centrifugation (25,000 rpm, 30 min, 4 °C), 12 μl of clear cell extract was added to 1 or 10 μg/ml of FI together with 150 μg/ml C3b and trace amounts of 125I-labeled C3b. Irrespective of the cofactor, in all assays where degradation of C3b was tested, 20 μg/ml FI and 20 μg/ml FH were used as a positive control, whereas in the negative controls FI was omitted. The samples were incubated at 37 °C for 90 min, and the reactions were stopped by adding reduced sample buffer and boiling for 3 min. The proteins were separated on a 10–15% gradient SDS-PAGE gel and visualized using a Fluorescent image analyzer (Fujifilm, Tokyo, Japan). The intensity of the cleavage products (C4d and 46/43 kDa) of C4b and C3b were analyzed using ImageGauge software (Fujifilm). Each experiment was repeated three times.
Kinetics of Degradation of Fluid-phase C4b and C3b
To determine the kinetics of the mutants that were still functional but less efficient than wt FI, samples were taken from a reaction mixture containing 5 μg/ml wt or mutant FI, 100 μg/ml C4BP and 50 μg/ml C4b or 20 μg/ml FH and 150 μg/ml C3b, and trace amounts of 125I-labeled C4b or C3b. Incubations were done at 37 °C, and samples were withdrawn at 5, 15, 45, and 90 min. Each experiment was conducted in triplicate.
FI Degradation of Surface-bound C3b
C3b was deposited on sheep erythrocytes by incubation with C3, FB, and FD as described previously (13). This was followed by incubation with 20 μg/ml FH and 0.1, 0.5, and 1 μg/ml recombinant wt or mutant FI at 37 °C for 60 min. After washing, iC3b and C3d were detected using murine monoclonal anti-human iC3b and anti-human C3d antibodies, respectively (both at 5 μg/ml, Quidel) followed by goat anti-mouse antibodies conjugated to fluorescein isothiocyanate (diluted 1:100, Dako) and analyzed by flow cytometry (Partec, Germany, Münster). The experiment was repeated three times, and the results were analyzed using FlowJo software (Three Star, Ashland, OR).
Binding of FI to C3met
C3met was coated overnight at 4 °C onto microtiter plates (Maxisorp, Nunc) at a concentration of 5 μg/ml in 75 mm sodium carbonate buffer, pH 9.6. The wells were subsequently blocked with 1% bovine serum albumin in phosphate-buffered saline for 2 h at room temperature. wt and mutant FI (100 μg/ml) were diluted in 0.5× phosphate-buffered saline containing 1% bovine serum albumin and 0.05% Tween 20 and added to the wells for 1 h at 37 °C. The same buffer was used for washing at each step. The binding of FI to C3met was detected using goat-anti-FI antibodies (Quidel) followed by horseradish peroxidase-labeled rabbit-anti goat antibodies (Dako) and the OPD development kit (Dako). The absorbance was measured at 490 nm. The absorbance values obtained for each mutant without C3met coating (1% bovine serum albumin) and those with only FI omitted were subtracted as background. Each experiment was repeated three times in duplicate.
Statistical Methods
One-way ANOVA with Tukey's multiple comparison test was performed using GraphPad Prism to calculate the p values.
RESULTS
Expression and Characterization of Recombinant FI Proteins
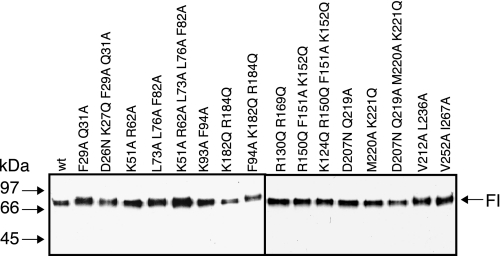
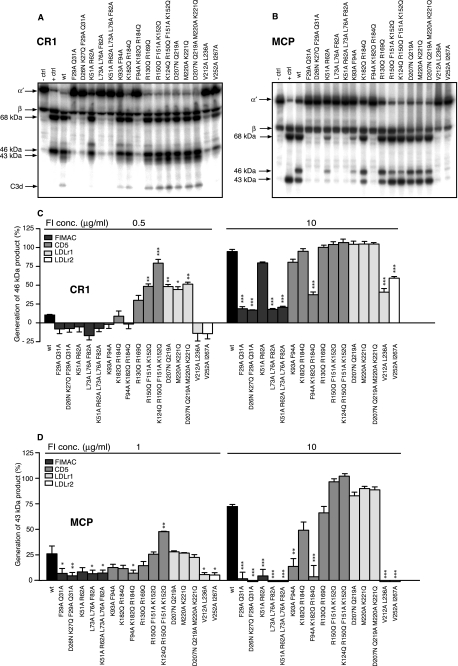
The amino acids that were mutated (highlighted in Fig. 1) were selected based on the well established observation that protein-protein interaction sites often contain solvent-exposed hydrophobic clusters and in some cases charged patches (29). We combined several point mutations to cover the different faces of the domains in the heavy chain and to better address the possible molecular functions of these patches with a minimal number of recombinant mutants. To study how the mutations affect the functional activity of FI, the wt and mutant FI constructs were transfected in a stable manner in HEK 293 cells. 16 of 18 mutants were successfully expressed and purified. The two mutants (D26N/K27Q, K249Q/Q259R/E270Q) that did not express, most likely due to folding problems, were located in the FIMAC and LDLr2 domains, respectively. The 16 purified mutants are visualized using Western blotting in Fig. 2. All mutants had the same molecular mass (88 kDa) and migrated similarly to wt FI both under non-reducing (Fig. 2) and reducing conditions (not shown).
FIGURE 2.
Characterization of purified recombinant wt and structural mutants of FI. The purified proteins (0.1 μg/well) were separated by SDS/PAGE under non-reducing conditions and transferred to polyvinylidene difluoride membranes. The FI protein was detected using polyclonal goat anti-human FI and polyclonal rabbit anti-goat antibody conjugated to horseradish peroxidase.
Using the same amounts of protein, the enzymatic activity of the FI mutants toward a small substrate was compared with the wt control in an amidolytic assay. None of the mutants showed statistically significant lower Vmax than the wt control, indicating that all the mutants preserved the enzymatic activity toward the small substrate despite the introduced mutations (Table 2). However, the mutants F29A/Q31A and D26N/K27Q/F29A/Q31A (both FIMAC), F94A/K182Q/R184Q (CD5), V212A/L236A (LDLr1), and V252A/I267A (LDLr2) all showed a significant increase (p < 0.001) of the Vmax of degradation of the small DPR substrate compared with wt FI, indicating enhanced enzymatic activity toward the DPR substrate. No significant differences could be observed in the Km of all the mutants compared with the wt FI control.
TABLE 2.
Kinetic parameters for DPR-AMC substrate cleavage by wt FI and the mutants
Vmax is expressed as pmol of amidomethylcoumarin released/min/μg of enzyme.
| Mutations | n |
Vmax |
km |
||
|---|---|---|---|---|---|
| Mean | S.D. | Mean | S.D. | ||
| μm | |||||
| wt | 6 | 0.090 | 0.013 | 149.6 | 30.03 |
| F29A/Q31A | 3 | 0.854a | 0.210 | 199.1 | 94.76 |
| D26N/K27Q/F29A/Q31A | 3 | 0.834a | 0.177 | 140.1 | 57.17 |
| K51A/R62A | 3 | 0.373 | 0.121 | 177 | 96.92 |
| L73A/L76A/F82A | 3 | 0.053 | 0.012 | 83 | 42.79 |
| K51A/R62A/L73A/L76A/F82A | 3 | 0.085 | 0.016 | 54.32 | 36.42 |
| K93Q/F94A | 3 | 0.147 | 0.016 | 74.1 | 31.27 |
| K182Q/R184Q | 3 | 0.058 | 0.080 | 41.37 | 18.21 |
| F94A/K182Q/R184Q | 3 | 0.672a | 0.160 | 168.9 | 76.82 |
| R130Q/R169Q | 3 | 0.160 | 0.030 | 103.6 | 42.22 |
| R150Q/F151A/K152Q | 3 | 0.087 | 0.008 | 122.2 | 22.18 |
| K124Q/R150Q/F151A/K152Q | 3 | 0.100 | 0.007 | 112.1 | 39.45 |
| D207N/Q219A | 3 | 0.044 | 0.007 | 114.9 | 43.1 |
| M220A/K221Q | 3 | 0.010 | 0.002 | 50.84 | 25.99 |
| D207N/Q219A/M220A/K221Q | 3 | 0.032 | 0.010 | 195.1 | 116.1 |
| V212A/L236A | 3 | 2.126a | 0.585 | 203.8 | 105.9 |
| V252A/I267A | 3 | 1.284a | 0.319 | 121.6 | 72.38 |
a p < 0.001 by ANOVA and Dunnett's post-hoc test.
Mutations in FI, Especially in the FIMAC Domain, Cause Reduced Cleavage of Fluid-phase C4b and C3b
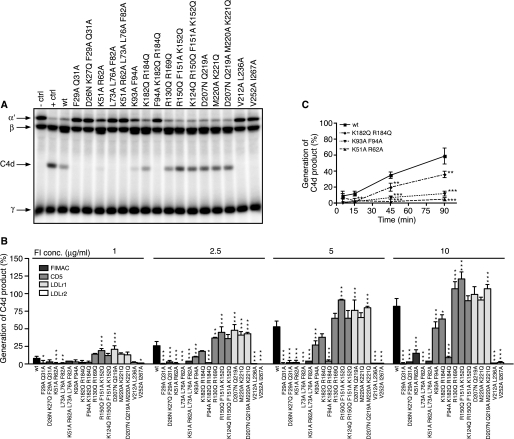
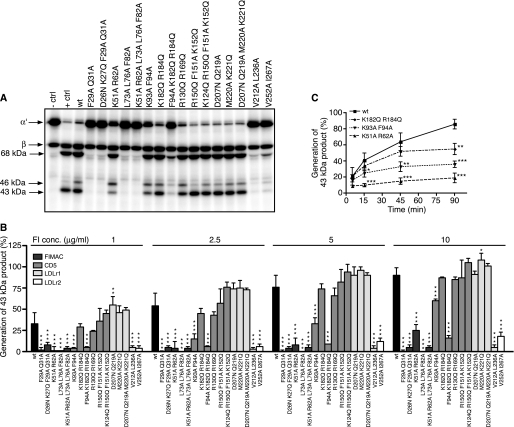
To elucidate if the different mutations affect the function of FI, fluid-phase degradation assays of C4b and C3b were performed. Different concentrations of the wt or mutant FI were mixed with fluid phase cofactors (C4BP or FH) and 125I-labeled C4b/C3b, incubated at 37 °C, and then analyzed by SDS-PAGE. In the representative gel picture of C4b, where 5 μg/ml FI was used, the negative control shows intact bands for the α′, β, and γ chains, and the positive control shows that the α′ chain is degraded, whereas the C4d product is formed in the presence of C4BP and FI (Fig. 3A). Intensities of bands corresponding to the C4d product were calculated from three different gels, and the obtained mean values related to the positive control present on each gel are presented in Fig. 3B. The kinetics of degradation by wt and three mutants are shown in Fig. 3C. In the gel picture of C3b (5 μg/ml FI), the negative control shows intact bands corresponding to the α′ and β chains, and the positive control shows degradation of the α′ chain and generation of 68-, 46-, and 43-kDa products (Fig. 4A). Intensities of bands corresponding to the 43-kDa product were calculated from three different gels, and the obtained mean values related to the positive control are presented in Fig. 4B. The kinetics of degradation by wt and three mutants are shown in Fig. 4C. We found that all of the analyzed mutations in the FIMAC domain resulted in strongly impaired or near complete loss of activity of FI in degradation of both C4b and C3b in the presence of C4BP and FH. The K51A/R62A mutant was the only mutant in the FIMAC domain that had some activity, but it was strongly impaired compared with wt. Also some of the mutations in the CD5, LDLr1, and LDLr2 domains affected the activity of FI. The F94A/K182Q/R184Q mutant in CD5, V212A/L236A in LDLr1, and V525A/I267A in LDLr2 were severely impaired in activity, whereas the K93A/F94A mutant (CD5 domain) showed significantly decreased but not abolished activity. The kinetics of degradation of C4b and C3b by the K51A/R62A, K93A/F94A, and K128Q/R184Q mutants were similar to wt FI even though they showed impaired functional activity.
FIGURE 3.
Degradation of C4b by wt and mutant FI in the fluid phase. A, recombinant wt or mutant FI (5 μg/ml) was incubated with C4b, C4BP, and trace amounts of 125I-C4b for 90 min at 37 °C and then separated on a 10–15% gradient SDS-PAGE. FI was omitted in the negative control (− ctrl) and 20 μg/ml FI was used as a positive control (+ ctrl). B, different concentrations (1, 2.5, 5, and 10 μg/ml) of FI were analyzed, and the generation of the C4d product was quantified by densitometry. C, the kinetics of C4b degradation was analyzed for wt and three chosen mutants, and the generation of the C4d product was quantified for samples that were withdrawn at different time points (5, 15, 45, and 90 min). The experiments in A–C were repeated three times, and the results are presented as % of the positive control present on each gel as means ± S.D. One-way ANOVA with Tukey's multiple comparison tests were preformed to evaluate the statistical significance of differences observed between wt and mutant FI. *, p < 0.05; **, p < 0.01; ***, p < 0.001.
FIGURE 4.
Degradation of C3b by wt and mutant FI in the fluid phase. A, recombinant wt or mutant FI (5 μg/ml) was incubated with C3b, FH, and trace amounts of 125I-C3b for 90 min at 37 °C and then separated on a 10–15% gradient SDS-PAGE. FI was omitted in the negative control (− ctrl), and 20 μg/ml FI was used as a positive control (+ ctrl). B, different concentrations (1, 2.5, 5, and 10 μg/ml) of FI were analyzed, and the generation of the 43-kDa product was quantified by densitometry. C, the kinetics of C3b degradation was analyzed for wt and three chosen mutants, and the generation of the 43-kDa product was quantified for samples that were withdrawn at different time points (5, 15, 45, and 90 min). The experiments in A–C were repeated three times, and the results are presented as the % of the positive control present on each gel as the means ± S.D. One-way ANOVA with Tukey's multiple comparison tests were preformed to evaluate the statistical significance of differences observed between wt and mutant FI. *, p < 0.05; **, p < 0.01; ***, p < 0.001.
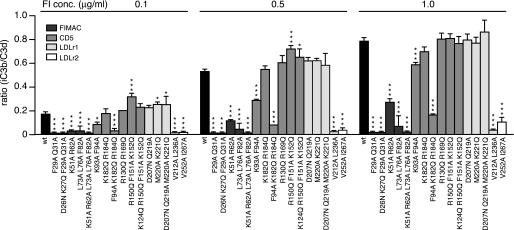
FI Degradation of Surface-bound C3b
The degradation of C3b deposited on sheep erythrocytes was analyzed using flow cytometry to analyze if the mutations had a different effect when FI was degrading a substrate on a surface as opposed to the fluid phase. Sheep erythrocytes opsonized with C3b were incubated with FI and FH, and the cleavage products, iC3b and C3d, were detected with specific antibodies. A high ratio of iC3b:C3d indicates degradation by FI. Also in this experiment, the mutants located in the FIMAC domain showed strongly impaired or near complete loss of activity (Fig. 5). The K51A/R62A mutant was the only mutant in the FIMAC domain that had activity, but it was strongly impaired. In the CD5 domain, the K182Q/R184Q mutant showed normal activity, and the K93Q/F94A mutant showed impaired activity, whereas the combined F94A/K182Q/R184Q mutant had greatly impaired function. Only one mutant (V212A/L236A) in the LDLr1 domain showed no activity, whereas the other amino acids that were mutated did not alter the function of FI. The V252A/I267A mutant in LDLr2 was not able to degrade C3b deposited on surface of erythrocytes. These results show that similar binding sites were important for activity of FI toward both soluble and deposited C3b.
FIGURE 5.
Cleavage of C3b on the surface. C3b was deposited on the surface of sheep erythrocytes by incubation of erythrocytes with C3, FB, and FD. The C3b was degraded by adding 0.1, 0.5, and 1.0 μg/ml FI and 20 μg/ml FH and incubating for 60 min at 37 °C. The amounts of deposited C3b and formed iC3b were detected using specific monoclonal antibodies, and the results are presented as a ratio of iC3b:C3d. The experiments were repeated three times, and the means ± S.D. are shown. One-way ANOVA with Tukey's multiple comparison tests were preformed to evaluate the statistical significance of differences observed between wt and mutant FI. *, p < 0.05; ***, p < 0.001.
FI Activity in the Presence of CR1 and MCP as Cofactors
To establish if the functional effects of different mutations in FI depend on the cofactor used, we also performed the C3b degradation assay using CR1 and MCP as cofactors. Wild type or mutant FI in different concentrations were incubated with CR1 or MCP and 125I-C3b and then analyzed by SDS-PAGE. Fig. 6 shows representative gels obtained with 10 μg/ml CR1 (A) and MCP (B) used as cofactors. As described previously for FH, intensities of bands corresponding to the 46- or 43-kDa product were calculated from three independent experiments, and the mean values of the positive control in percent (%) are presented in Fig. 6C (CR1) and Fig. 6D (MCP). In the case of CR1, as additional degradation product C3d was observed as expected. Similar to the results obtained with C4BP and FH as cofactors, we found that all of mutations occurring in the FIMAC domain resulted in a significant loss of activity of FI. Furthermore, The F94A/K182Q/R184Q mutant in CD5, V212A/L236A mutant in LDLr1, and V525A/I267A mutant in LDLr2 were severely impaired in activity. We conclude that the currently investigated mutations affect the activity of FI in the same manner irrespective of the cofactor used.
FIGURE 6.
Degradation of C3b by wt and mutant FI in the presence of CR1 and MCP as cofactors. Recombinant wt or mutant FI (10 μg/ml) was incubated with C3b, CR1 (A), or MCP (B) and trace amounts of 125I-C3b for 90 min at 37 °C and then separated on a 10–15% gradient SDS-PAGE. FI was omitted in the negative control (− ctrl), and 20 μg/ml FI with 20 μg/ml FH was used as a positive control (+ ctrl). The indicated concentrations (0.5 and 10 μg/ml for CR1 and 1 and 10 μg/ml for MCP) of FI were analyzed, and the generation of the 46-kDa (CR1, C) or 43-kDa (MCP, D) products were quantified by densitometry. The experiments were repeated three times, and the results are presented as the % of the positive control present on each gel as the means ± S.D. One-way ANOVA with Tukey's multiple comparison tests were preformed to evaluate the statistical significance of differences observed between wt and mutant FI. *, p < 0.05; **, p < 0.01; ***, p < 0.001.
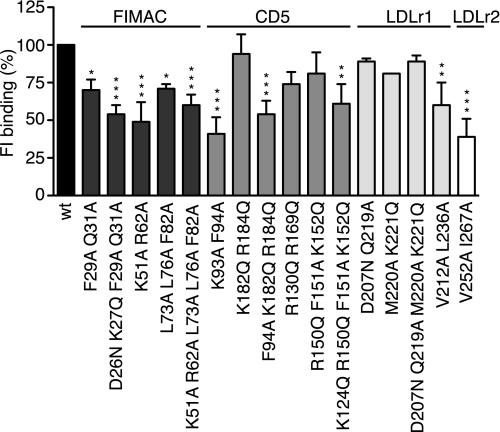
Mutations in Several Domains of the Heavy Chain Have Impaired Binding to C3met
From functional assays we could see that mutations in several domains, especially the FIMAC domain, had impaired activity when cleaving C3b and C4b. To elucidate if the reduced function could be caused by impaired binding to C3b, a direct binding assay was performed. wt and mutant FI were allowed to bind to C3met-coated microtiter plates, and the binding was detected with antibodies against FI (Fig. 7). The negative controls for the bovine serum albumin coating for each single mutant as well as values obtained for samples without FI were subtracted, and the results were normalized against wt FI. This was necessary because the interaction between C3b and FI is very difficult to study because it has low affinity. We found that all mutants in the FIMAC domain showed reduced binding to C3met, whereas only some mutations in the CD5 domain and single mutants in the LDLr1 and the LDLr2 domains had reduced binding.
FIGURE 7.
Binding of wt and mutant FI to C3met. C3met was coated on microtiter plates and incubated with wt and mutant FI, and bound FI was detected with specific antibodies. Background signals originating from antibodies alone incubated with C3met on plates as well as FI and antibodies incubated with empty well were subtracted from the original values. Thus, the obtained values were normalized to the value obtained for the wt FI in each experiment. The original absorbance values for wt FI were in the range of 0.3–0.4 units. The experiments were repeated three times in duplicate, and means ± S.D. are shown. One-way ANOVA with Tukey's multiple comparison tests were preformed to evaluate the statistical significance of differences observed between wt and mutant FI. *, p < 0.05; **, p < 0.01; ***, p < 0.001.
DISCUSSION
Complement inhibitors act on key components of the converging pathways of complement to prevent pathological consequences of unwanted complement activation. The inhibitors bind C4b and C3b, and these interactions have two consequences: decay acceleration of C3-convertases that contain C4b/C3b and degradation of these factors by the protease FI. In recent years significant efforts have been made to elucidate the binding sites for C3b and C4b on various cofactor proteins such as C4BP (24, 30), CR1 (31), MCP (32), and FH (33). However, the mechanism by which the cofactors assist in the cleavage of FI is far from clear.
In the present study we have evaluated several potential binding sites on the surface of the FI heavy chain that are important for its ability to degrade C3b and C4b. It is known that the SP domain of the FI light chain contains the catalytically active site; however, in vivo FI is strictly dependent on its cofactors to be able to function as a protease. The molecular events leading to the cleavage of C3b/C4b by FI in the presence of cofactors are not well defined at present. The substrates themselves may induce a conformational change in the FI protein that would make the active site accessible and would allow degradation of the substrate. In support of this hypothesis, it has been shown that diisopropylfluorophosphate (DFP) can inhibit the activity of FI in complex with C3b; however, no inhibition could be observed when FI was incubated with DFP in the absence of C3b (34). This indicates that FI is able to weakly interact with C3b even in the absence of cofactors and that this binding induces a conformational change in FI. Direct, albeit low affinity (0.7 mm at physiological ionic strength) binding between FI and C3b was demonstrated to be ionic strength-dependent (20). In the presence of FH, the affinity between C3b and FI increased manyfold. Furthermore, it has been suggested that C3b undergoes a conformational change upon binding to FH that results in the accessibility of peptide bonds in C3b that can be cleaved by FI (35), and this has now been confirmed using x-ray crystallography of the complex between C3b and complement control protein (CCP) domains 1–4 of FH (33). Intact SP domain was generated by proteolysis, and it was shown to have the same activity toward small substrates as intact FI (18). However, in contrast to intact FI that was absolutely dependent on the presence of a cofactor, the SP domain alone cleaved C3b slowly in the absence of cofactors at more than the two usual sites. This suggests that the heavy chain is perhaps involved in directly blocking the active site of FI or in keeping the SP domain in an inactive conformation. Furthermore, the SP domain may be important together with the cofactor in orienting the natural substrates to restrict cleavage of C3b to the two specific sites. It is also possible that a cofactor contributes to another interaction site for FI in the three-molecule complex.
To gain new insight into the relationships between structure and function of FI, we have analyzed the potential binding sites predicted after analysis of the three-dimensional -structure of the different FI domains predicted by molecular modeling. Sixteen recombinant variants of FI were expressed in eukaryotic cells and subjected to functional analyses. One inherent problem of such an experimental approach is that it is difficult to distinguish if an observed functional defect related to a mutation is caused by a direct alteration of the binding site or, rather, by a change in overall or local folding in turn affecting the binding site. We can at least exclude that mutations analyzed in this study caused a gross misfolding of FI as they all were active in degradation of a small substrate. Furthermore, a functional defect caused by a mutation in FI resulting in impaired degradation of C3b can be related to impairment of the binding site for C3b or cofactor as well as a structural alteration precluding conformational change upon formation of the trimolecular complex. In this study we analyzed binding of the FI mutants to C3b, but we were not able to set a reliable assay testing interaction of FI with cofactors or C3b in complex with cofactors. However, even keeping these caveats in mind it is possible to draw a number of conclusions regarding the relationship between structure and function of FI from our results.
We found that the majority of mutations located in the FIMAC domain resulted in nearly abolished activity of FI in all functional assays. The loss of function could be partially explained by the reduced binding that these mutants to C3met. K51A/R62A was the only mutant in the FIMAC domain that still showed detectable but decreased activity. It seems that the FIMAC domain, particularly its N terminus, is involved in the complement inhibitory function of the FI protein. Interestingly, it was observed that the FIMAC domains of C6/C7 bind the C345C domain of C5 (36), which is highly homologous to C3, so it could be that the FI FIMAC binds the C345C of C3b/C4b. It is tempting to hypothesize that the FIMAC domain partially blocks the active site located in the SP domain or that it packs against the rest of the protein to maintain the SP in an inactive conformation. Binding of the FIMAC domain to substrates then induces a conformational change in FI that allows proteolytic activity toward C3b/C4b presented in a proper conformation by the cofactors.
The mutations K93Q and F94A close to the N terminus of the CD5 domain caused impaired activity compared with wt FI, whereas the K182Q/R184Q mutant had normal function. Interestingly, the combination of these, F94A/K182Q/R184Q, showed strongly reduced function, especially in the fluid-phase degradation of C4b and C3b. R150Q/F151A/K152Q, K124Q/R150Q/F151A/K152Q, and R130Q/R169Q are three mutants localized on other faces of the predicted structure of the CD5 domain (Fig. 1). These substitutions did not affect the function of FI, but the K124Q/R150Q/F151A/K152Q mutant showed reduced binding to C3met. All of the above suggests that the N-terminal part of the CD5 domain and maybe the Lys-124 residue have interactions with substrates/cofactors and are important for the functional activity of FI, whereas the other regions of CD5 investigated here most likely are less important.
In the LDLr1 domain the analyzed mutations are located on different faces of the modeled three-dimensional structure. The FI mutant involving amino acids Asp-207, Gln-219, Met-220, and Lys-221 does not have an altered function. This suggests that these amino acids are all solvent-exposed in the complete three-dimensional structure of FI and that they do not participate in any interaction crucial for degradation of C4b/C3b. The other mutant, involving amino acids Val-212 and Leu-236, expressed at a somewhat lower level than wt and was not active in most of the assays used in this study. This part of the LDLr1 domain may be involved in interactions with other domains, or it might be important for the stability of the domain.
The LDLr2 domain seems to be closely involved in interactions with the other domains as one mutant (K249Q/Q259R/E270Q) was not secreted at all, whereas the V252A/I267A mutant did, but at lower levels than wt, and had nearly a complete loss of function. It is possible that these mutations mainly affect the folding of the FI protein.
Our results show that for each mutant the changes in ability to degrade C3b are mirrored by the effects on cleavage of C4b. We have observed a similar phenomenon when analyzing FI mutations found in patients suffering from an inherited factor I deficiency (9). In the current study we found that the enzymatic activity of the mutants generally followed the results of their binding capacity to C3met, i.e. reduced binding led to reduced function. Therefore, we hypothesize that both substrates, C4b and C3b, interact with similar parts of the FI molecule. The same could be true for all the cofactors, as we did not observe differences in the activity of mutants when four different cofactors were used. Interestingly, a previously analyzed mutant A222G, initially identified in an atypical hemolytic uremic syndrome patient, caused normal degradation of fluid-phase C4b and C3b, but the degradation of surface-bound C3b was impaired (14). This result implies that there could be discrete differences in FI activity against C3b in solution as opposed to C3b deposited on the surface. In conclusion, in this study we show that the FIMAC domain is particularly important for the ability of FI to degrade C4b and C3b in the presence of several cofactors.
Acknowledgment
We thank Dr. Pietro Roversi (University of Oxford) for critical reading of this manuscript and advice.
This work was supported by the United States Immunodeficiency Network, the Swedish Research Council, the Söderberg Foundation, the Swedish Foundation for Strategic Research, and the Foundations of Österlund, Greta and Johan Kock, Knut and Alice Wallenberg, and Inga-Britt and Arne Lundberg.
- FH
- factor H
- FI
- factor I
- FB
- factor B
- FD
- factor D
- C4BP
- C4b-binding protein
- CR1
- complement receptor 1
- FIMAC
- factor I membrane attack complex (domain)
- LDLr
- low density lipoprotein receptor
- MCP
- membrane cofactor protein
- SP
- serine protease (domain)
- DPR-AMC
- (Boc-Asp(benzyl)-Pro-Arg-amidomethylcoumarin·HCl
- Bicine
- N,N-bis(2hydroxyethyl)glycine)
- wt
- wild type
- ANOVA
- analysis of variance.
REFERENCES
- 1.Walport M. J. (2001) N. Engl. J. Med. 344, 1058–1066 [DOI] [PubMed] [Google Scholar]
- 2.Sjöberg A. P., Trouw L. A., Blom A. M. (2009) Trends Immunol. 30, 83–90 [DOI] [PubMed] [Google Scholar]
- 3.Weiler J. M., Daha M. R., Austen K. F., Fearon D. T. (1976) Proc. Natl. Acad. Sci. U.S.A. 73, 3268–3272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gigli I., Fujita T., Nussenzweig V. (1979) Proc. Natl. Acad. Sci. U.S.A. 76, 6596–6600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Seya T., Turner J. R., Atkinson J. P. (1986) J. Exp. Med. 163, 837–855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Medof M. E., Nussenzweig V. (1984) J. Exp. Med. 159, 1669–1685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vyse T. J., Morley B. J., Bartok I., Theodoridis E. L., Davies K. A., Webster A. D., Walport M. J. (1996) J. Clin. Invest. 97, 925–933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Amadei N., Baracho G. V., Nudelman V., Bastos W., Florido M. P., Isaac L. (2001) Scand. J. Immunol. 53, 615–621 [DOI] [PubMed] [Google Scholar]
- 9.Nilsson S. C., Trouw L. A., Renault N., Miteva M. A., Genel F., Zelazko M., Marquart H., Muller K., Sjöholm A. G., Truedsson L., Villoutreix B. O., Blom A. M. (2009) Eur. J. Immunol. 39, 310–323 [DOI] [PubMed] [Google Scholar]
- 10.Sadallah S., Gudat F., Laissue J. A., Spath P. J., Schifferli J. A. (1999) Am. J. Kidney Dis. 33, 1153–1157 [DOI] [PubMed] [Google Scholar]
- 11.Fremeaux-Bacchi V., Dragon-Durey M. A., Blouin J., Vigneau C., Kuypers D., Boudailliez B., Loirat C., Rondeau E., Fridman W. H. (2004) J. Med. Genet. 41, e84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kavanagh D., Richards A., Noris M., Hauhart R., Liszewski M. K., Karpman D., Goodship J. A., Fremeaux-Bacchi V., Remuzzi G., Goodship T. H., Atkinson J. P. (2008) Mol. Immunol. 45, 95–105 [DOI] [PubMed] [Google Scholar]
- 13.Nilsson S. C., Karpman D., Vaziri-Sani F., Kristoffersson A. C., Salomon R., Provot F., Fremeaux-Bacchi V., Trouw L. A., Blom A. M. (2007) Mol. Immunol. 44, 1835–1844 [DOI] [PubMed] [Google Scholar]
- 14.Nilsson S. C., Kalchishkova N., Trouw L. A., Fremeaux-Bacchi V., Villoutreix B. O., Blom A. M. (2009) Eur. J. Immunol. 40, 172–185 [DOI] [PubMed] [Google Scholar]
- 15.Tsiftsoglou S. A., Arnold J. N., Roversi P., Crispin M. D., Radcliffe C., Lea S. M., Dwek R. A., Rudd P. M., Sim R. B. (2006) Biochim. Biophys. Acta 1764, 1757–1766 [DOI] [PubMed] [Google Scholar]
- 16.Fearon D. T. (1979) Proc. Natl. Acad. Sci. U.S.A. 76, 5867–5871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chamberlain D., Ullman C. G., Perkins S. J. (1998) Biochemistry 37, 13918–13929 [DOI] [PubMed] [Google Scholar]
- 18.Tsiftsoglou S. A., Willis A. C., Li P., Chen X., Mitchell D. A., Rao Z., Sim R. B. (2005) Biochemistry 44, 6239–6249 [DOI] [PubMed] [Google Scholar]
- 19.Perkins S. J., Smith K. F., Sim R. B. (1993) Biochem. J. 295, 101–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.DiScipio R. G. (1992) J. Immunol. 149, 2592–2599 [PubMed] [Google Scholar]
- 21.Miteva M. A., Tufféry P., Villoutreix B. O. (2005) Nucleic Acids Res. 33, W372–W375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fernandez-Recio J., Totrov M., Skorodumov C., Abagyan R. (2005) Proteins 58, 134–143 [DOI] [PubMed] [Google Scholar]
- 23.Dahlbäck B. (1983) Biochem. J. 209, 847–856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Blom A. M., Kask L., Dahlbäck B. (2003) Mol. Immunol. 39, 547–556 [DOI] [PubMed] [Google Scholar]
- 25.Greenwood F. C., Hunter W. M., Glover J. S. (1963) Biochem. J. 89, 114–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tsiftsoglou S. A., Sim R. B. (2004) J. Immunol. 173, 367–375 [DOI] [PubMed] [Google Scholar]
- 27.Spiller O. B., Blackbourn D. J., Mark L., Proctor D. G., Blom A. M. (2003) J. Biol. Chem. 278, 9283–9289 [DOI] [PubMed] [Google Scholar]
- 28.Okroj M., Hsu Y. F., Ajona D., Pio R., Blom A. M. (2008) Mol. Immunol. 45, 169–179 [DOI] [PubMed] [Google Scholar]
- 29.Bahadur R. P., Zacharias M. (2008) Cell Mol. Life Sci. 65, 1059–1072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Blom A. M., Kask L., Dahlbäck B. (2001) J. Biol. Chem. 276, 27136–27144 [DOI] [PubMed] [Google Scholar]
- 31.Krych M., Hauhart R., Atkinson J. P. (1998) J. Biol. Chem. 273, 8623–8629 [DOI] [PubMed] [Google Scholar]
- 32.Adams E. M., Brown M. C., Nunge M., Krych M., Atkinson J. P. (1991) J. Immunol. 147, 3005–3011 [PubMed] [Google Scholar]
- 33.Wu J., Wu Y. Q., Ricklin D., Janssen B. J., Lambris J. D., Gros P. (2009) Nat. Immunol. 10, 728–733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ekdahl K. N., Nilsson U. R., Nilsson B. (1990) J. Immunol. 144, 4269–4274 [PubMed] [Google Scholar]
- 35.Isenman D. E. (1983) J. Biol. Chem. 258, 4238–4244 [PubMed] [Google Scholar]
- 36.Thai C. T., Ogata R. T. (2005) J. Immunol. 174, 6227–6232 [DOI] [PubMed] [Google Scholar]